Abstract
A biomarker can be a substance or structure measured in body parts, fluids or products that can affect or predict disease incidence. As age-related macular degeneration (AMD) is the leading cause of blindness in the developed world, much research and effort has been invested in the identification of different biomarkers to predict disease incidence, identify at risk individuals, elucidate causative pathophysiological etiologies, guide screening, monitoring and treatment parameters, and predict disease outcomes. To date, a host of genetic, environmental, proteomic, and cellular targets have been identified as both risk factors and potential biomarkers for AMD. Despite this, their use has been confined to research settings and has not yet crossed into the clinical arena. A greater understanding of these factors and their use as potential biomarkers for AMD can guide future research and clinical practice. This article will discuss known risk factors and novel, potential biomarkers of AMD in addition to their application in both academic and clinical settings.
1. Introduction to AMD
Age-related macular degeneration (AMD) was first described in the medical literature as a “symmetrical central choroido-retinal disease occurring in senile persons” in 1874. Later, the terms “age-related maculopathy”, “age-related macular degeneration” (de Jong, 2006), and “age-related macular disease” (Bird, 1996) were acknowledged as descriptions of age-related central visual impairment. AMD is characterized by central visual loss due to degenerative and neovascular alteration in the macular region of the retina (Gehrs et al., 2006).
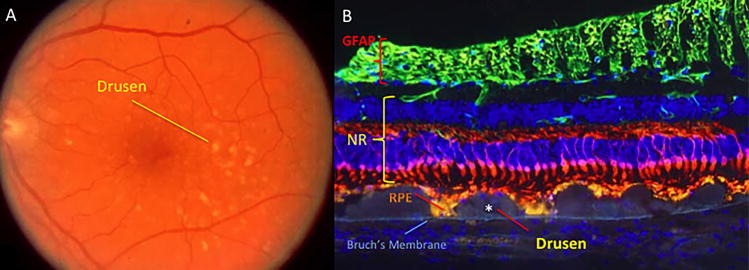
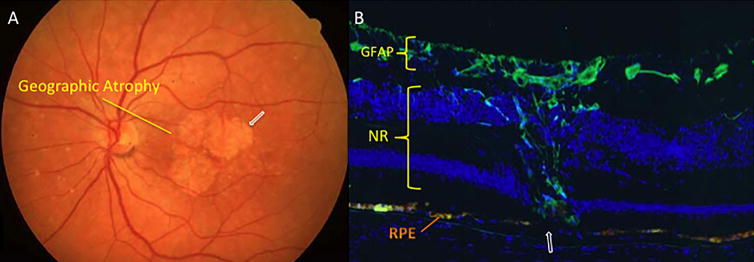
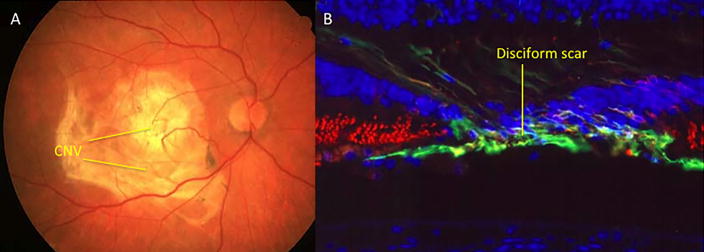
AMD is the predominant cause of blindness in developed countries (Evans et al., 2004). It is becoming similarly important in the developing world in association with increasing longevity and Westernisation of diet and lifestyle (Krishnan et al., 2010). AMD may be classified as dry or wet, with geographic atrophy commonly seen with the former and choroidal neovascularization (CNV) commonly seen in the latter. Wet or neovascular AMD (nvAMD) is a result of CNV, or the growth of new blood vessels from the choroid into the sub-retinal space and sub-RPE, eventually leading to vision loss (Kokotas et al., 2011). AMD may also be classified into early, intermediate, or advanced disease (Bourla and Young, 2006). The Age-Related Eye Disease Study (AREDS) divided AMD into 5 different categories based on amount, size, and nature of drusen, location and area of retinal pigmented epithelium (RPE) atrophy, and neovascularization (Age-Related Eye Disease Study Research, 2001). Early AMD initially manifests as pigmentary irregularities of the retina and deposits of extracellular material called drusen (Figure 1.2) that collect at the RPE-choroidal interface. Drusen can be categorized as small (63mm or less), intermediate (>63mm but <125mm), or large (>125mm) (Age-Related Eye Disease Study Research, 2000). Intermediate AMD often involves more confluent collections of intermediate and large size drusen and poses a greater risk for the development of late or advanced AMD (Age-Related Eye Disease Study Research, 2001). Approximately 1 in 2 persons with extensive macular drusen will progress within 5 years to vision threatening geographic atrophy (GA) (Figure 1.3) and/or neovascularisation (Figure 1.4) (Davis et al., 2005), which are late stage manifestations of the condition (Sunness et al., 1999; Wong et al., 2008). GA is currently untreatable and although nvAMD may now be controlled with antiangiogenic agents, the majority of patients so treated have residual visual disability due to varying degrees of retinal tissue disruption, scarring and/or atrophy. Furthermore, antiangiogenic treatments are invasive and costly (Raftery et al., 2007), requiring monthly intravitreal injections and long-term follow-up, and involve increased risk of intra-ocular infection. In the US, it is estimated that visual impairment due to AMD will double by the year 2050 and that the use of antiangiogenic agents will only reduce that by 17% (Rein et al., 2009). Healthcare costs in the UK for nvAMD patients have been shown to be seven-fold that of age-matched controls when factors such as falls, depression and help with daily tasks are accounted for (Raftery et al., 2007). These facts make a compelling case for identification and development of robust biomarkers to enable earlier, more accurate diagnosis and better prediction of likely prognosis.
2. Biomarkers: Definition and Conceptual Framework
A biomarker can be a substance, structure, or biochemical or molecular alteration measured in human body parts, fluids, or products that can affect or predict disease incidence. It has been formally defined as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention” (Biomarkers Definitions Working, 2001). This formal definition was developed by the Biomarkers and Surrogate Endpoint Working Group (under the direction of the Office of the Director, National Institutes of Health) to provide a conceptual framework and classification system within a rapidly burgeoning field. The formal classification system further divided biomarkers into three main groups:
Type 0: those used to estimate the emergence or development of a disease. These may be used in combination to produce a panel of biomarkers that reflect a specific disease state and are used in the prediction, early onset, progression, regression, treatment efficacy and diagnosis of disease.
Type 1: those that predict the responses to therapeutic interventions. These are most commonly associated with the pharmaceutical and biotechnology industries.
Type 2: those that in principle could be used as surrogate clinical endpoints in the course of clinical trials. In this situation, usually one principal biomarker is chosen which has been comprehensively characterized and validated.
In practice, biomarkers can provide the means to further understand the prediction, cause, diagnosis, progression or regression of disease as well as outcome of treatment (Mayeux, 2004). The substantial interest in biomarker identification and assay development stems from their potential to address significant scientific problems including: more efficient and specific end points in drug discovery and development, more direct measurement of exposures in epidemiological and clinical studies which are free from recall bias, as well as providing information on absorption and metabolism. Biomarkers serve this purpose for other major disease processes, such as in cancer (Perera and Weinstein, 2000).
Biomarker development for AMD is focused on identifying disease risk factors, both developing assays for screening and diagnosis, and using this information for prognostic application and guiding treatment decision-making. Biomarkers are vital to identifying susceptible patient populations and exposures that may lead to disease prior to AMD development. Sensitive, validated biomarkers are vital for early detection of AMD and influencing late outcomes of the disease process with appropriate treatment.
3. Biomarkers of Susceptibility
3.1 Age
Age is the strongest demographic risk factor associated with age-related macular degeneration (Age-Related Eye Disease Study Research, 2000; Blumenkranz et al., 1986; Buch et al., 2005; Choudhury et al., 2011; Erke et al., 2012; Klein et al., 1997; Klein et al., 2007; Klein et al., 2002; Miyazaki et al., 2005; Owen et al., 2012; Rein et al., 2009; Tomany et al., 2003; Wang et al., 2007; Woo et al., 2015). Table 1 combines data from four different studies estimating the prevalence of age-related macular degeneration by age group (Friedman et al., 2004; Klein et al., 1992; Smith et al., 2001a; Vingerling et al., 1995b). These data show the overwhelming increase in risk of AMD with age.
Table 1. Prevalence of AMD by Age.
Pooled data on the relationship between age and prevalence of age-related macular degeneration. The single bold number represents the % of the population, in the given age range, with AMD; while the range of numbers within parentheses describes the ranges reported by the various studies.
| Prevalence of AMD by Age (% of age-specific population) | |||
|---|---|---|---|
| Age | Geographic atrophy | Neovascular AMD | Any Late AMD |
| <55 | 0.08 (0–0.15) | 0.14 (0–0.28) | 0.2 (0–0.39) |
| 55–64 | 0.25 (0.04–0.47) | 0.37 (0.09–0.65) | 0.25 (0.13–0.38) |
| 65–74 | 1.37 (0.17–2.57) | 0.68 (0.3–1.05) | 1.62 (0.67–2.57) |
| 75–84 | 2.25 (1.26–3.24) | 2.52 (1.7–3.33) | 4.93 (3.19–6.67) |
| 85+ | 7.54 (3.31–11.77) | 8.49 (5.41–11.57) | 14.47 (11.57–17.36) |
| All Ages | 0.63 (0.44–0.81) | 0.96 (0.72–1.2) | 1.64 (1.47–1.81) |
Due to the time-dependent mechanisms of AMD, specifically the formation of drusen and choroidal neovascularization, age will likely remain the leading risk factor for the development of age-related macular degeneration.
3.2 Gender
Current evidence concerning gender and the associated risk of developing AMD is conflicting. Many studies have concluded that there is no significant increase risk for developing AMD based on gender alone (Age-Related Eye Disease Study Research, 2000; Erke et al., 2012; Frank et al., 2000; Owen et al., 2012).
One study found that early AMD incidence is slightly, but not significantly, higher in men than women, and that the incidence of late AMD is significantly higher in men than women with an odds ratio of 2.62 (Miyazaki et al., 2005). Another study, a meta-analysis of AMD in Europe, found some evidence to suggest a higher risk of late-stage wet AMD in women compared to men (Owen et al., 2012). In addition, the Age Related Eye Disease Research Group (AREDS) found that intermediate drusen, extensive small drusen, or the pigment abnormalities associated with AMD were found more often in females (Age-Related Eye Disease Study Research, 2000). In the UK, Owen et al. (2012) found that the prevalence of AMD in women was 60% higher than that of men (314,000 cases in women, 192,000 men). In addition, women had an incidence of late-AMD (both dry and wet AMD) of 4.1 per 1000 compared to 2.3 per thousand for men. However, this study admitted that the difference in gender prevalence could have been be due to the fact that there are a greater number of older women in the UK than in other parts of the world (Owen et al., 2012).
Certain AMD risk SNPs may also be gender specific (Grassmann et al., 2015). These results suggest that women may be at a higher risk of developing the disease, but more epidemiological studies are needed to draw definitive conclusions on the association between gender and AMD.
3.3 Race
Data support the conclusion that race is a significant risk factor in the development of AMD. In the year 2000, AMD was the leading cause of blindness among Caucasian Americans, accounting for 54% of cases (Congdon et al., 2004). Many studies support the conclusion that whites are significantly more likely to develop AMD (especially wet AMD) than blacks (Attebo et al., 1996; Bressler et al., 2008; Clemons et al., 2005; Congdon et al., 2004; Frank et al., 2000; Friedman et al., 1999; Rein et al., 2009). The Baltimore Eye Study reported a nearly four-fold higher risk of developing any form of AMD for whites compared to blacks and late AMD was significantly more prevalent in whites (Friedman et al., 1999).
The AREDS Group found higher rates of choroidal neovascularization (CNV) in whites compared to blacks (Clemons et al., 2005). Another study also suggested that choroidal neovascularization (CNV) was more associated with whites, but that late-stage dry AMD (with geographic atrophy) was more associated with blacks (Bressler et al., 2008). In contrast, for dry AMD, one research group suggested that whites were more likely to progress from medium to large foveal drusen, but this study did not find any significant data concerning the progression to GA or CNV in blacks versus whites (Chang et al., 2008). Notably, correlations of race with AMD prevalence might suggest that the degree of ocular pigmentation could be a significant protective factor, with white Caucasians showing the highest rates (5.4%), followed by Chinese (4.6%), Hispanics (4.2%) and blacks (2.4%) (Klein et al., 2006; Priya et al., 2012).
Although early stages of AMD seem to be race-independent, evidence suggests that those of Caucasian descent are significantly more likely to develop the later, more symptomatic stages of AMD that result in a loss of central vision. In addition, race appears to impact the role of various genetic influences on AMD (Restrepo et al., 2014). The effect of different genetic variants on AMD among different racial or ethnic groups will be discussed later.
3.4 Iris Color
Iris color has also been shown to be associated with AMD risk. Patients with lighter colored irides have been shown to have a two-fold higher incidence of AMD (Frank et al., 2000; Nicolas et al., 2003) than those with darker irides. One study showed that those with brown eyes, compared to those with blue eyes, were more likely to develop early stage AMD, but those with brown eyes were significantly less likely to have RPE depigmentation, a distinct feature of late stage AMD (Tomany et al., 2003). Another study supported this conclusion with evidence that Caucasians with blue irides are significantly more likely to develop AMD than Caucasians with brown irides (Frank et al., 2000). A meta-analysis showed that there seemed to be a protective effect of brown irides compared to blue, however this result was not significant (Chakravarthy et al., 2010).
More recently, Klein et al. found a modest association of iris color with incidence of early AMD, however there was no association between AMD development and gene markers of iris pigmentation variability (Klein et al., 2014). Another recent study by Schick et al (2015) found no association between iride color and early or late AMD (Schick et al., 2015). Iris color may be associated with AMD risk, however more definitive research is necessary.
3.5 Obesity
Obesity may be associated with the risk of developing AMD, although a causal relationship has not been established. Seddon et al. (2003) found that the relative risk for AMD was 2.35 for those with a BMI over 30, and 2.32 for those with a BMI between 25 and 29 (P=0.007) (Seddon et al., 2003a). They reported a 2-fold increase in progression of AMD when comparing patients in the highest and lowest tertiles for waist circumference (P=0.02). Comparing these two groups’ waist to-hip-ratio yielded a relative risk of 1.84 (P=0.02). In addition, those that exercised vigorously three times per week had a 25% risk reduction in AMD progression when compared to those that did not exercise (Seddon et al., 2003a). In addition, Adams et al. found that a 0.1 waist/hip ratio increase was correlated with a 13% chance increase of developing early AMD (P=0.03), and a 75% chance increase of developing late AMD (P=0.02). However, an inverse relationship was seen between waist/hip ratio and risk of AMD with women (Adams et al., 2011). Schaumberg et al. found that increased body mass index was correlated with dry AMD in men. However, lean individuals were also found at risk, hinting that the data was controversial (Schaumberg et al., 2001). Overall it appears that obesity is associated with AMD, but it is unclear whether there is any causative relationship between the two. In addition, it appears that there is a stronger correlation between AMD and obesity in men than in women.
3.6 Hypertension
Whether elevated blood pressure increases the risk of developing AMD is a matter of controversy. Some studies have shown that elevated blood pressure contributes to the pathophysiology of AMD (Churchill et al., 2006; Klein et al., 2003). One retrospective study used blood pressure record data from 1,828 subjects and found a “small and consistent significant association between age-related maculopathy and systemic hypertension” (Sperduto and Hiller, 1986). In comparison to normotensive individuals, another study found that patients with both treated and uncontrolled blood pressure have a three-fold increase in the development of exudative macular degeneration (Klein et al., 2003). An additional study added to the complexity of the matter, concluding that even patients with controlled hypertension were at an increased risk of AMD (Hyman et al., 2000).
Drusen size has also been correlated with hypertension. The AREDS group reported that the presence of large drusen in the macula, or extensive intermediate size drusen, is associated with hypertension and the use of hypertensive medication such as hydrochlorothiazide (Age-Related Eye Disease Study Research, 2000). On the other hand, another group noted that the presence of cardiovascular disease, including hypertension, did not increase the risk of late AMD (Delcourt et al., 2001). An additional study concluded that hypertension does not contribute to CNV (Blumenkranz et al., 1986).
Hypertension may be associated with an increased risk of AMD, however more definitive research is necessary.
4. Biomarkers in Heritability and Genetics
Studies have shown that there is a strong correlation between a family history of AMD and the subsequent development of both early and late forms of the disease, as judged by higher concordance rates among monozygotic twins when compared with dizygotic, and segregation analyses comparing first degree relatives of affected individuals as compared with the general population (Priya et al., 2012; Smith and Mitchell, 1998). One study showed that the risk of developing late AMD was increased nearly 4-fold for those with a family history of AMD, particularly in cases of nvAMD (Smith and Mitchell, 1998). This risk is amplified when immediate family members have the disease, with one study estimating a 27.8 times increase in risk with an affected parent and 12 times increase in risk for those with an affected sibling (Shahid et al., 2012).
Despite the strong genetic influence on the development of AMD, there is much dispute between associated versus causative pathological genes. For monogenic disorders, where one mutation is necessary and sufficient to cause symptoms of disease, there are accepted criteria whereby a mutation is judged to be causative of pathogenesis. First, the putative mutation should correspond with symptoms of disease in a family with a number of affected individuals. Second, it should be absent from a large number of ethnically-matched control individuals. Third, similar mutations in the same gene should cause similar disorders in different kindreds. Fourth and finally, the mutation should have a predicted impact on protein structure and function (Gorin, 2012). The gold standard proof-positive of causality is that, when engineered into animal models, such mutations should produce similar symptoms of disease to those seen in affected patients, and therefore be amenable to appropriate gene therapies.
For complex disorders, these standards of proving causality are much more difficult to obtain. Given that most cases of AMD occur later in life, large family cohorts are difficult to obtain and most AMD studies have focused on groups of smaller families and sibling pair or genome-wide association studies (GWAS) using larger numbers of less closely related individuals (Priya et al., 2012). While statistical analysis of large datasets can be incredibly powerful, linkage disequilibrium (close proximity of variants on the same chromosome, meaning that alleles are largely segregated together without recombination when passed from one generation to another) can make it difficult to identify which variant is, in fact, responsible for disease risk. This is even more problematic when the variants in question do not have an obvious predicted effect on protein structure or function (Gorin, 2012). A factor that is merely contributory to a complex, multi-factorial disorder is likely to have a much more subtle effect on disease progression than a mutation which is necessary and sufficient, on its own, to cause symptoms of disease. This makes positive identification more difficult. Furthermore, the logistics associated with engineering these variants into animal models, and then crossbreeding them such that a number of additive variants are sufficient to cause symptoms are also exponentially more challenging.
Despite these complexities, progress has been made in identifying approximately 40 genes that may be associated with the development of AMD. Some of these genes fall into broad groups based on their function such as genes with retinal-specific function, immune function, neovascularization, or lipoprotein-related function. These gene groups will be discussed below. Additionally, seven new loci with an association to AMD were recently identified by the AMD consortium (Fritsche et al., 2013). Genes found in these loci will be introduced although their specific function and role in AMD development and progression is currently not well understood.
4.1 Genes with Retinal-Specific Function
Unlike retinitis pigmentosa (RP) and other early onset retinal degenerations, retinal specific genes have minimal impact on AMD susceptibility. Only three monogenic macular dystrophy-related genes (ABCA4, ApoE, and TIMP-3) show any association with AMD risk. Of these genes, only ABCA4 expression is restricted to the retina while ApoE and TIMP-3 have additional, systemic functions. ABCA4 will be discussed presently while TIMP-3 and ApoE will be discussed later under sections related to their systemic function.
4.1.1 ABCA4
ABCA4 is an ATP-binding cassette (ABC) reporter protein belonging to the ABC1 subfamily (Allikmets et al., 1997b). It is a specific marker for photoreceptor cells and found in the outer segment disks of this retinal layer. Its function involves the transportation of N-retinylidene phophatidylethanolamine (PE) across membranes in photoreceptor cells (Beharry et al., 2004). In the absence of functional ABCA4, a N-retinylidene PE complex accumulates, leading to a buildup of lipofuscin fluorophore A2E (Sparrow et al., 1999), and eventual aberrant cholesterol metabolism in the RPE cells (Lakkaraju et al., 2007). Retinal degeneration in those with compromised ABCA4 may result from loss of the ability to transport N-retinylidene PE out of photoreceptor cells, or RPE toxicity associated with A2E buildup (Beharry et al., 2004).
Mutations in ABCA4 are associated with a host of retinal diseases including Stargardt disease (Allikmets et al., 1997b), autosomal recessive Retinitis pigmentosis 19 (Martinez-Mir et al., 1998), cone-rod dystrophy3 (Maugeri et al., 2000), fundus flavimaculatus (Shroyer et al., 1999), severe retinal dystrophy (Singh et al., 2006) and age-related macular degeneration (Allikmets et al., 1997a; Allikmets et al., 1997b). ABCA4 does not represent a common risk factor for AMD. It is likely associated with a specific AMD subtype that manifests a fine granular pattern with peripheral punctate spots and which shares some features with Stargardt disease (Fritsche et al., 2012). Notably, carriers of Stargardt disease are thought to be at higher risk for the development of AMD (Allikmets, 2000; Allikmets et al., 1997a; Shroyer et al., 1999). Of note, the phenotypic similarity between AMD and Stargardt disease (Westeneng-van Haaften et al., 2012) may lead to misdiagnosis in primary care settings and overrepresentation of ABCA4 mutations in large cohorts of AMD patients where phenotyping was not highly rigorous.
In summary, ABCA4 mutations cause a range of retinopathies that vary in severity related to the degree of residual retinal function. Despite this, they are not a common cause of AMD (Fritsche et al., 2012; Guymer et al., 2001) and more research is needed to clarify its actual role in AMD development.
4.1.2 ApoE
To be discussed under Lipoprotein-related genes (section 4.4.2).
4.1.3 TIMP-3
To be discussed in Neovascularization-related genes (section 4.3.2).
4.2 Immune System-Related Genes
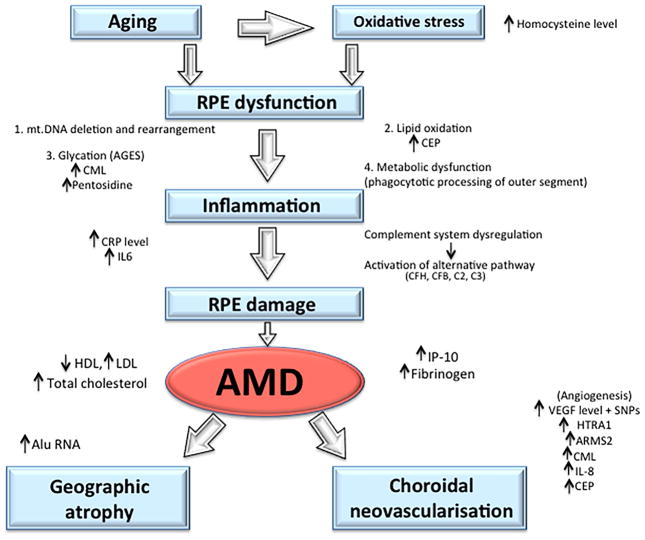
An increasing body of evidence implicates aberrant inflammatory processes as culprits in a long list of diseases associated with aging, including AMD. The immune system primarily exists to mount innate and adaptive defences against pathogens. However, it can also be activated by particulate matter accumulation and altered host material such as drusen, a hallmark of early AMD. This response is typically chronic and represents a “sterile” form of inflammation that is unconnected with infection (Campbell and Doyle, 2013; Doyle et al., 2012).
Immunological responses have long been implicated in AMD progression, but the pathways involved have remained unclear. Many polymorphisms of various genes coding for components of the immune system have been implicated in AMD. In this section we will discuss the function of genes coding for various components of the complement cascade in addition to other immune-related genes including: ARMS2/HTRA1, EFEMP1, and Hemicentin1.
4.2.1 Complement
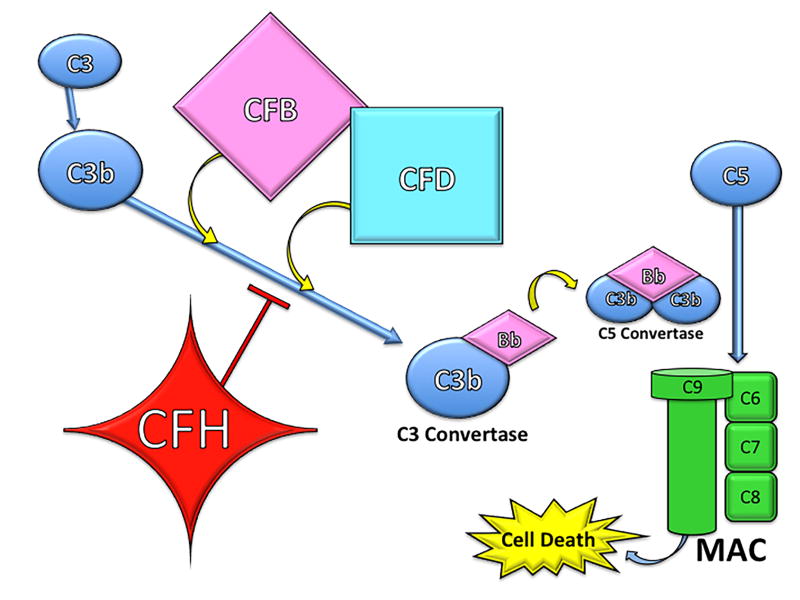
Many of the immune-related genetic polymorphisms associated with AMD code for various components of the complement cascade. Complement consists of a series of soluble proteins (over 25 in total), circulated via the hematopoietic system to almost all parts of the body. The complement cascade can be activated by a number of triggers, usually related to infection, and aids in the destruction of foreign pathogens. There are three complement pathways: classical pathway, alternative pathway, and mannose-binding lectin pathway. Although different in their initiating events, all three pathways result in complement activation and eventual formation of transmembrane pores, called the membrane attack complex (MAC), leading to rapid influx of extracellular fluid and subsequent cell death (Figure 2). Additionally, various complement components (i.e. C3b and C5a) act as opsonins (marking pathogen for ingestion by phagocytes) and also serve as chemoattractants to recruit appropriate inflammatory cells to the site of infection (Manthey et al., 2009; Thompson et al., 2007).
Different components of the complement pathways have been associated with AMD, namely factor H, factor H-related complement, factor B, factor D, and factors 2, 3, and 5. The functions of these proteins will be discussed below. In general, mutations leading to dysregulation of the complement system result in increased inflammation, aberrant activation of the immune system, and subsequent increased risk of AMD development; contrastingly, mutations in complement activators lead to decreased complement activity and are protective against developing AMD.
4.2.1.1 Complement Factor H
In 2005, Complement Factor H (CFH) was first identified as a regulator of innate immunity in the human retina, with individuals with a variant form of this factor having an increased risk of age-related macular degeneration (Edwards et al., 2005; Haines et al., 2005; Klein et al., 2005). CFH is a regulatory control of the complement system and gene polymorphisms leading to its dysfunction have some of the strongest genetic ties to the development of AMD (Babanejad et al., 2015; Garcia et al., 2015; Horie-Inoue and Inoue, 2014; Klein et al., 2013; Shen et al., 2015; Wong et al., 2015).
CFH plays two critical roles in the innate immune system. First, it regulates the conversion of C3 to C3a and C3b. Secondly, CFH inactivates C3b to iC3b, preventing the formation of C3 convertase and subsequently opsonizes pathogens (Rodriguez de Cordoba et al., 2004). Through inactivation of C3b to iC3b, CFH prevents formation of the downstream membrane attack complex (MAC). Hageman et al. suggested that at-risk alleles decreasing the function of CFH may lead to greater levels of MAC complex at the RPE-choroid interface, compromising the integrity of Bruch’s membrane, a common pathology seen in nvAMD (Hageman et al., 2005). By inactivating C3b, CFH acts as a potent negative regulator of the alternative complement pathway. Dysregulation in this inflammatory pathway caused by CFH polymorphisms can increase the risk of developing AMD, while proper CFH function serves as a predictive protecting factor.
It is estimated that nearly 50% of the risk of developing AMD can be attributed to CFH variants, resulting in decreased function of CFH (Edwards et al., 2005; Klein et al., 2013; Shen et al., 2015). CFH variants have been consistently found as a component of drusen in those with a history of AMD (Hageman et al., 2005). Several studies have found positive associations between sequence polymorphisms in the CFH gene and early stage AMD (Chen et al., 2010; Habibi et al., 2013; Holliday et al., 2013; Quan et al., 2012; Rodriguez de Cordoba et al., 2004; Yuan et al., 2013), although it appears that CFH variants play a more apparent role in late stage geographic atrophy and neovascular AMD (Hageman et al., 2005; Klein et al., 2013). High levels of variant CFH DNA transcript can be found in RPE and choroid layers of the eye (Hageman et al., 2005).
In addition, the presence of certain variant CFH alleles has shown to increase levels of lipid metabolism products responsible for oxidative stress such as malondialdehyde (MDA) (Weismann et al., 2011). Increased levels of MDA provide one probable mechanism responsible for RPE cell death (Klein et al., 2013; Weismann et al., 2011). Many of the CFH SNPs encode regions of the functional CFH protein that contain binding sites for C3b, heparin, sialic acid, and C-reactive protein, alluding to the possibility that these SNPs may influence binding affinities of different molecules to CFH (Hageman et al., 2005). Although not entirely understood, CFH variants likely lead to increased risk of AMD development through their negative impact on proper CFH function and subsequent dysregulation of the inflammatory complement pathway.
Much of the risk conferred by CFH mutations comes from the Y402H variant (rs1061170). In this high-risk variant, a tyrosine residue is replaced by a histidine at the 402 amino acid position, limiting its regulatory function (Ormsby et al., 2008). Edwards et al. estimated that this specific allele variant may account for up to 50% of the attributable risk of AMD (Edwards et al., 2005). Those that possess at least one high-risk C allele in the Y402H variant are two to three times more likely to develop AMD compared to homozygote individuals with T alleles (Quan et al., 2012). CFH variants are associated with both early and late stage AMD. Gangnon et al. found that the CC genotype of the CFH Y402H SNP was associated with a statistically significant increase in incidence of early AMD (Hazard Ratio=1.98) and progression of early AMD to late AMD (Hazard Ratio = 1.72) when compared to the TT genotype (Gangnon et al., 2012). A genome-wide association study (GWAS) found that the CFH SNP rs10737680 was strongly associated with early stage AMD (OR=3.1, P < 1.6 × 10−75) (Chen et al., 2010). In another recent GWAS, CFH SNP rs1329424 showed an odds ratio of 1.41 (P=1.5x10−31) for the early stage AMD (Holliday et al., 2013). These results affirm that there are many CFH polymorphisms associated with early stage AMD. Although CFH variants are linked with early AMD, they are more strongly and frequently associated with late-stage AMD (Edwards et al., 2005; Gangnon et al., 2012; Haines et al., 2005; Klein et al., 2011; Klein et al., 2013; Klein et al., 2005; Magnusson et al., 2006). Again, the Y402H polymorphism appears to be the most prominent genetic risk factor. Haines et al. reported odds ratios for nvAMD of 3.45 and 5.57 for those that were heterozygotes and homozygotes for the C allele, respectively (Haines et al., 2005). Magnusson et al. reported odds ratios of 2.39 for advanced AMD in patients possessing at least one risk-allele compared to controls (Magnusson et al., 2006). Seddon et al. found that patients homozygous for the C allele were 3.5 times more likely to have AMD when compared to controls (Seddon et al., 2009).
In 2013, Klein et al. published data from a 20-year follow-up in the cohort Beaver Dam Eye Study. This data showed strong genetic correlations between the combined effects of CFH SNP rs1061170 (Y402H) and ARMS2 SNP rs10490924 and AMD. In this study, patients age 45 who had zero or one allele from either of the CFH or ARMS2 SNPs had a cumulative incidence of AMD of 1.4% by age 80. Contrastingly, those age 45 possessing three to four alleles from the CFH or ARMS2 SNPs had a cumulative incidence of late AMD of 15.3%. These two SNPs have shown consistent correlation with late-stage AMD than with earlier stages. When at least one CFH risk allele was present, the population-attributable risk for late AMD was 53 percent (Klein et al., 2013). The strong association with risk alleles of CFH rs1061170 has also been validated in recent studies (Babanejad et al., 2015; Fauser and Lambrou, 2015; Hautamaki et al., 2015).
These data support complement factor H as being one of the strongest genetic risk factors for the development of AMD, accounting for 50% of the attributable risk for the disease. Some groups have begun to make recombinant forms of CFH in hopes of simulating its inhibitory role on the complement pathway and decreasing the risk of AMD development (Rohrer et al., 2010).
4.2.1.2 Complement Factor H-related 1 and 3
Complement Factor H-related 1 and 3 (CFHR1 and CFHR3) are secreted complement regulatory proteins that work in conjunction with complement factor H (Gene ID: 3078, 10878, OMIM: 134371, OMIM: 605336). Each of these proteins are secreted plasma proteins synthesized by hepatocytes and contain domains known as complement control factor modules. CFHR1 is a complement regulator that acts downstream of CFH through inhibition of C5 convertase and subsequent assembly and membrane insertion of the membrane attack complex (MAC). Although a complete understanding of its function is not clear, it is believed that CFHR3 acts as cofactor for factor I in inactivating C3b, thus limiting C3 convertase activity (Fritsche et al., 2010). This blocks downstream generation of C5a as well as its associated inflammatory effects like subsequent neutrophil recruitment. CFHR1 and CFHR3 share similar binding locations on C3b with CFH (Fritsche et al., 2010). Although these proteins have a similar role to CFH in complement regulation, due to their similar binding sites, they actually compete with CFH. As such, deficiency of CFHR1 and CFHR3 implies a theoretical loss of their role in complement regulation but also enhanced local regulation by CFH (Fritsche et al., 2010). Interestingly, an 84kb deletion in CFHR1 and CFHR3 genes, leading to absence of their associated proteins in serum, actually demonstrated a decreased risk of AMD development (Hughes et al., 2006). Of note, this deletion has also been shown to carry an increased risk of atypical hemolytic uremic syndrome (Martinez-Barricarte et al., 2008).
The protective effect attributed to mutations in CFHR1 and CFHR3 against AMD progression was initially thought to be tied to the protective effects of normal CFH function. However, recent evidence suggests that CNP ΔCFHR3/CFHR1 on chromosome 1q32 is protective against AMD independent of common CFH genotypes (Fritsche et al., 2010).
In conclusion, deletion mutations in CFHR1 and CFHR3, and subsequent absence of associated plasma proteins, confer a protective effect on AMD development. This is thought to be due to enhanced regulation of complement activity by functional CFH at the C3 convertase level (Fritsche et al., 2010), thus decreasing overall inflammation and lower the risk of AMD development. However, there may be various CFHR1 allotypes that carry increased risk of AMD development. CFHR1*A is associated with increased risk of AMD, and its effects are thought to be linked and additive to high-risk Y402H variant of CFH (Martinez-Barricarte et al., 2012).
4.2.1.3 Complement Factor B
Complement factor B (CFB) polymorphisms have shown to be protective against the development of AMD (Gold et al., 2006b; Mantel et al., 2014; Spencer et al., 2007; Sun et al., 2012; Thakkinstian et al., 2012). In the first steps of the alternative pathway, CFB is bound to C3b. When the alternative pathway is initiated, C3b-bound factor B is cleaved, resulting in the formation of the C3 convertase. Because CFB is one of the main activators of the alternative complement pathway, mutations limiting CFB function may result in decreased risk of complement-induced drusen formation in the retina (Gold et al., 2006b). The rare alleles in CFB SNPs rs9332739, rs547154, rs4151667 (L9H variant), and rs641153 (R32Q) have been shown to exhibit protective effects against AMD (Spencer et al., 2007; Sun et al., 2012). The risk of developing AMD was decreased in half for patients carrying at least one of the rare alleles for any of the previously mentioned CFB SNPs (Sun et al., 2012). In another study, CFB SNP rs641153 (R32Q) was seen only in patients with drusen ≤250 μm and appeared to be protective against the formation of larger drusen >250 μm (Mantel et al., 2014). A meta-analysis found that patients with CFB SNPs rs4151667 and rs614153 carrying the minor A allele had a significantly smaller likelihood of having AMD (OR=0.54 and 0.41 respectively) (Thakkinstian et al., 2012). Overall it appears that rare alleles in certain CFB SNPs are responsible for decreasing activation of the alternative pathway and may be protective against the development of AMD.
4.2.1.4 Complement Factor D
In addition to CFB, Complement factor D (CFD) is another alternative pathway protein with variant mutations implicated in the development of AMD (Gold et al., 2006a; Jakobsdottir et al., 2008). Its role in the innate immune system is to cleave and subsequently activate CFB. There is evidence that CFD is the rate-limiting protein in the activation of the alternative pathway (Stanton et al., 2011).
Increase in CFD activity may lead to a greater likelihood of developing AMD. Stanton et al. (2011) found that AMD patients had an average of an 11% increase in CFD compared to the control group (P=0.00025) (Stanton et al., 2011). Another study aimed to determine the association between six CFD SNPs (rs1683564, rs35186399, rs1683563, rs3826945, rs34337649, and rs1651896) and advanced AMD in a Caucasian population. Researchers did not find any significant correlation between any of the CFD SNPs and wet AMD (Zeng et al., 2010).
Although CFD is an established regulator of the complement pathway, more research is needed to elucidate its role and relationship in the development of AMD. Despite this, several groups have begun to develop CFD-inhibitors, geared to decrease alternative complement pathway activity and treat AMD (Abdel-Magid, 2012; Katschke et al., 2012).
4.2.1.5 Complement Components C2, C3 and C5
Drusen are made up of many different constituent proteins, including C2, C3 and C5 complement (Hageman et al., 2001; Mullins et al., 2000). C2, C3 and C5 are a part of the classical complement pathway and are pro-inflammatory proteins. Activated C1 cleaves C2 into C2a and C2b. C2a binds C4b to form the C3 or C5 convertase, meaning that C2 is instrumental in raising levels of activated C3 and C5 (Gene ID: 717).
Numerous studies have indicated the involvement of C2 variants such as rs9332739 and rs547154 in AMD risk (Gold et al., 2006a; Jakobsdottir et al., 2008; Thakkinstian et al., 2012; Wu et al., 2013). Early subretinal deposits of C3 and C5 have been found in AMD mouse models, indicating that these inflammatory complement proteins may play a role in the pathogenesis of AMD. This role was further clarified by evidence indicating that C3a and C3b induce vascular endothelial growth factor (VEGF) expression, recruit leukocytes, and promote CNV (Nozaki et al., 2006). Genetic C3 SNPs R102G and L314P have been shown to contribute to an increased risk of AMD development (Spencer et al., 2008). Additionally, a meta-analysis also found association between AMD and C3 SNPs rs11569536, rs2230199, and rs1047286, while a negative association was found between rs2250656 (Qian-Qian et al., 2015). One study showed that high-risk alleles in C3 SNP Arg80Gly (rs2230199) were 2.6 times more likely to be found in patients with AMD compared to controls (Yates et al., 2007). A Boston study also found that this SNP was significantly associated with AMD (P<10−12)(Maller et al., 2007). It has been estimated that C3 SNPs may contribute as much as 22% of the population’s attributable risk of AMD (Yates et al., 2007). Overall C2, C3, and C5 variants lead to increased risk of AMD.
4.2.1.6 Complement Summary
Genes related to proper function of the complement pathway hold some of the greatest influence on and association with AMD. CFH is responsible for regulation of the complement system, and mutations leading to decrease in its inhibitory function can account for nearly 50% of the attributable risk of AMD. Other SNPs leading to increase in activity of CFD and CFB result in increased complement activity and an associated increase in AMD risk. Finally variants of C2, C3, and C5 may affect pro-inflammatory cell recruitment, VEGF levels, and regulation of complement pathway and have a significant impact on attributable AMD risk.
4.2.2 ARMS2/HTRA1
Early genetic studies identified a susceptibility locus on chromosome 10q26 for the development of AMD (Yang et al., 2006). Further genomic studies of this chromosome demonstrated two AMD susceptible loci. One is known as rs10490924, which lies within the gene LOC387715/ARMS2, now known as Age-Related Maculopathy Susceptibility 2 (ARMS2) (Hautamaki et al., 2015; Kanda et al., 2007; Shen et al., 2015). The other, rs11200638, lies within the promoter region of the gene known as High Temperature Requirement Factor A1 (HTRA1) gene (Dewan et al., 2006; Francis and Klein, 2011; Yang et al., 2006). These two SNPs in ARMS2 and HTRA1, rs10490924 and rs11200638, have been found to be in strong linkage disequilibrium (LD) that confer virtually identical risks for AMD, making it difficult to differentiate the two AMD risk alleles in these genes and their effects (Wang et al., 2009).
ARMS2 is a 12kDa protein that contains nine phosphorylation sites and localizes to the outer mitochondrial membrane when expressed in mammalian cells (Yang et al., 2006). Its function is unknown, but the gene is a highly conserved ortholog in chimpanzees. ARMS2 is present in mammalian placental and retinal tissues, specifically in the mitochondria of photoreceptor cells. Specific mitochondrial changes including accumulation of cytochrome-c oxidase cones, deletion of mitochondrial DNA (mt.DNA), decreases in the size and number of mitochondria, and destruction of mitochondrial cristae are commonly seen with AMD pathology. The retina has the highest energy demand of any tissue in the body and is symptomatically affected in 50% of primary mitochondrial disorders (those caused by mutations in the mitochondrial genome) (Farrar et al., 2013; Yu-Wai-Man et al., 2011). It is understood that oxidative damage plays a significant role in these changes. Photoreceptors are exposed to high levels of light and oxygen, and as such, suffer frequent damage via oxidative injury. Such oxidative stress can damage the mitochondrial genome, resulting in mutations in both from mutations in both mitochondria-encoded genes and those with mitochondrial functions that are encoded by the nuclear genome (Farrar et al., 2013; Yu-Wai-Man et al., 2011).
The cellular localization of ARMS2 is currently unclear. While ARMS2 is thought to be a mitochondria-encoded gene found in photoreceptor cells, where it may play a role in the amount of oxidative stress in the retina (Smailhodzic et al., 2012), ARMS2 has also been reported to be mainly distributed in the cytosol, not necessarily in the mitochondrial outer membrane, which would suggest that ARMS2 may confer risk to AMD through an alternative non-mitochondrial pathway (Wang et al., 2009). ARMS2 localization needs further clarification and investigation.
In the 20-year follow-up report of the Beaver Dam Study, Klein et al found the population-attributable risk for late AMD was 53% when at least one risk-allele for ARMS2 was present (Klein et al., 2013). It has also been reported that ARMS2 variants may play a role in the activation of the complement cascade (Smailhodzic et al., 2012). ARMS2 variants combined with those in the CFH encoding gene on loci 1q31 account for over 50% of the attributable genetic risk associated with AMD (Holliday et al., 2013; Marmorstein et al., 2007; Stanton et al., 2011; Wyatt et al., 2013). ARMS2 variants combined with those in CFH and C3 SNPs account for 76% of the attributable genetic risk for AMD (Spencer et al., 2008). Although the entire function and mechanism of ARMS2 action remains unclear, it is a critical player in the development of AMD.
HTRA1 has been shown to be associated with an increased risk of wet AMD in certain populations (Dewan et al., 2006; Yang et al., 2006). The HTRA1 gene encodes a serine protease expressed in RPE and drusen. The protein appears to regulate the degradation of extracellular matrix proteoglycans and works in conjunction with other extracellular matrix degradation enzymes (i.e. collagenases and metalloproteinases). HTRA1 also binds to and inhibits transforming growth factor-beta (TGF-β), a factor known to play a crucial role in extracellular matrix deposition and angiogenesis (Yang et al., 2006; Zhang and Marmorstein, 2010). Therefore, it is possible that HTRA1 may play a role in the regulation of the Bruch’s membrane and growth of vessels into the RPE. One study concluded that HTRA1 SNP rs11200638 is the most likely causal variant of AMD at the 10q26 locus among the Han Chinese population, and estimated the combined population attributable risk for CFH and HTRA1 alleles to be 75% (Yang et al., 2006).
Recently, two rs2284665 SNP in ARMS2/HTRA1 was also recently identified as affecting the growth of CNV in AMD (Akagi-Kurashige et al., 2015). Overall, variants of ARMS2/HTRA1 genes confer a major risk for the development of AMD.
4.2.3 EFEMP1 (Fibulin 3)
Epidermal growth factor (EGF) containing fibulin-like extracellular matrix protein (ECM) 1 (EFEMP1) is a member of the fibulin family of matrix glycoproteins, and it is also known as fibulin 3. Fibulins contain a number of EGF-like repeats, followed by a C-terminus fibulin-like domain. EFEMP1 can be found in several locations including RPE cells and endothelial basement membrane of choroidal vessels (Zhang and Marmorstein, 2010). EFEMP1 stimulates expression of TIMP-1 and TIMP-3 (discussed later) but also inhibits various matrix metalloproteinases (MMPs). Of note, EFEMP1 is also an inhibitor of angiogenesis (Zhang and Marmorstein, 2010). EFEMP1 has been identified as an innate immune responsive gene that is upregulated after optic crush injury (Templeton et al., 2013). EFEMP1 is also upregulated in gliomas and is mutated in Doyne honeycomb dystrophy (DHD, otherwise known as malattia Leventinese, an autosomal dominant maculopathy) (OMIM: 601548), and this pathology has shed light onto its possible effect on progression of AMD. A single mutation, R345W, appears to be responsible for DHD symptoms in five families studied (Stone et al., 1999), and causes similar problems in EFEMP1 R345W-knock-in mice (Fu et al., 2007; Marmorstein et al., 2007). In these knock in mice, the EFEMP1 mutation resulted in misfolding, inefficient secretion, and retention of the translated protein in cells (Marmorstein et al., 2002). Additionally subretinal deposits found in these mice also contained EFEMP1 and TIMP-3. In mutant mice, EFEMP1 is deposited between the RPE and drusen. Copy number variants involving EFEMP1 have been associated with AMD (Meyer et al., 2011). In AMD eyes, EFEMP1 is deposited beneath the RPE immediately overlying drusen although it was not a major component of the drusen itself in either case. However, accumulation of misfolded EFEMP1 may lead to drusen formation and to subsequent cellular degeneration (Marmorstein et al., 2007; Marmorstein et al., 2002).
Additional studies using a yeast-2-hybrid approach have shown that EFEMP1 interacts with and binds to CFH, and has an even higher affinity for the AMD related CFH 402H variant (Wyatt et al., 2013). Wyatt et al., demonstrated that Fibulin 3 and CFH proteins co-localize in cholesterol rich regions of soft drusen in patients homozygous for the CFH 402H variant (Wyatt et al., 2013). This may support why defects in EFEMP1 have been shown to lead to extracellular matrix alterations and depositions within the retina. These alterations are key to the associated AMD risk of EFEMP1 as they result in excessive complement activation (Fu et al., 2007). Although the actual attributable risk of EFEMP1 on AMD development is not known, its role in ECM alteration and retinal deposits make it a likely player in AMD pathogenesis. Further studies are needed to more precisely define this risk.
4.2.4 Hemicentin-1
Hemicentin-1 (HMCN1) is a large, extracellular member of the immunoglobulin (Ig) superfamily. Like EFEMP1, HMNC1 contains EGF-like domains, as well as 48 tandem Ig modules. Its function is likely structural as a C. elegans homolog has been noted to be involved in attaching neurons to substrates, anchoring of the germ line syncytium and epidermal hemidesmosome organization (Gene ID: 83872). HMCN1 is one the few genes which is thought to cause AMD in a Mendelian fashion, a mutation segregated exclusively with the disease phenotype in a large multigenerational family with AMD (Schultz et al., 2003) (OMIM: 608548). HMCN1 also interacts directly with ARMS2 (Kortvely et al., 2010). While the familial AMD mutation is rare, and is therefore not a major contributor to AMD risk (Fisher et al., 2007), other polymorphisms may influence the longitudinal rate of AMD development (Thompson et al., 2007).
4.3 Genes Related to Neovascularization
An additional category of AMD risk factors includes genes producing proteins (VEGF, TIMP-3, Fibulin 5) that directly and indirectly influence the growth of new blood vessels. Of these, VEGF is the most important. VEGF is the target of current anti-AMD therapies ranibizumab (Lucentis) and bevacizumab (Avastin). Genes that directly or indirectly influence the levels of VEGF are likely to have an influence on AMD development.
4.3.1 Vascular Endothelial Growth Factor
Vascular endothelial cell growth factor (VEGF) is a member of the platelet-derived growth factors. It acts in numerous ways to promote pathological neovascularization. First, it functions as an endothelial cell mitogen to initiate angiogenesis and inhibits apoptosis in order to sustain angiogenesis (Ng and Adamis, 2005). As a chemo-attractant, VEGF increases endothelial cell migration, proliferation, and vessel formation. VEGF also acts as a pro-inflammatory molecule, increasing leukocyte migration, which in turn increases VEGF expression (Storkebaum and Carmeliet, 2004). In opposition to its negative pathological effects, VEGF also has a neuroprotective function relating to its ability to increase neuronal survival and Schwann cell proliferation (Storkebaum and Carmeliet, 2004).
VEGF plays an important role in maintaining a healthy retina. Five different retinal cell types produce VEGF including: vascular endothelium, retinal pigmented epithelium, Muller cells, ganglion cells, and astrocytes (Kinnunen et al., 2012; Ng and Adamis, 2005). In addition, VEGFR-2 expression has been localized to the inner nuclear layer (Muller cells and amacrine cells), ganglion cells, and retinal vasculature (Penn et al., 2008). There are seven biologically active isoforms in the VEGF family, but VEGF-A is thought to be most critical in the angiogenesis process. VEGF-A is also referred to as VEGF-165 to specify the number of amino acids in the protein. It interacts with specific tyrosine-kinase receptors VEGFR-2, producing a subsequent downstream cascade. Vascular endothelial growth factor receptor 2 (VEGFR2) activation results in increased release of nitric oxide and prostacyclin I2, which alter vascular permeability and endothelial cell proliferation (Galan et al., 2010; Penn et al., 2008).
VEGF-A has been linked to many ocular neovascular diseases (Ambati and Fowler, 2012) and previous studies have demonstrated increased intraocular levels of VEGF in patients with wet AMD, diabetic retinopathy, retinopathy of prematurity, retinal vein occlusion and neovascular glaucoma (Miller et al., 2013). All of these ocular diseases share a similar pathological process: vascular permeability and neovascularization. VEGF is released by the retina and retinal pigment epithelium in response to tissue hypoxia. Increased VEGF production results in subsequent upregulation of endothelial nitric oxide synthase, metalloproteinases and decreased tissue inhibitors of metalloproteinase expression, which together enhance choroidal neovascularization.
Single nucleotide polymorphisms at the genetic level are presumed to contribute to altered VEGF and VEGFR-2 expression, leading to the pathogenesis of AMD and even diabetic retinopathy. These SNPs and others associated with AMD are summarized in Table 2.
Table 2.
Genetic Biomarkers
| Biomarker | Type of Biomarker | SNPs ID | P. Value | Odds ratio | References |
|---|---|---|---|---|---|
|
| |||||
| CFH | DNA | rs1410996 | 1.4 × 10−16 | 2.9 | (Babanejad et al., 2015; Chen et al., 2011; Fritsche et al., 2010; Hautamaki et al., 2015; Woo et al., 2015; Yang et al., 2010) |
| rs1061170 (Y402H) | 2.4 × 10−14 | 2.5 | |||
| rs800292 (V62I) | 0.003/0.02 | 0.57 (0.40–0.83) | |||
| rs2274700:A | NA | 0.36 | |||
|
| |||||
| CFB | DNA | rs641153 | 0.0001 | 0.40 | (Bergeron-Sawitzke et al., 2009; McKay et al., 2009) |
| rs4151657 | 0.01 | 1.43 | |||
| rs4151672 | 0.04 | 0.52 | |||
| rs4151667 | 0.0020 | 0.36 | |||
|
| |||||
| C3 | DNA | R102G (rs2230199) | 0.02–0.009 | 1.4 | (Bergeron-Sawitzke et al., 2009; Park et al., 2009; Zerbib et al., 2010) |
| rs1047286 | 9.2E-05 | NA | |||
| rs3745565 | 0.009 | NA | |||
| rs11569536 | 0.014 | NA | |||
| rs171094 | 0.00030 | 1.3 | |||
|
| |||||
| C2 | DNA | rs1042663 | 0.001 | 0.47 | (Bergeron-Sawitzke et al., 2009; McKay et al., 2009) |
| rs3020644 | 0.01 | 1.43 | |||
| rs2072632 | 0.01 | 1.43 | |||
| rs9332739 | 0.04 | 0.52 | |||
| rs547154 | 0.0016 | 0.49 | |||
|
| |||||
| ARMS2 | DNA | rs10490924 | 2.8 × 10−29 | 2.86 | (Bergeron-Sawitzke et al., 2009; Fritsche et al., 2008; Hautamaki et al., 2015; Shen et al., 2015; Woo et al., 2015) |
| rs3750848 | 2.8 × 10−29 | 2.86 | |||
| del443ins54 | 4.1 × 10−29 | 2.85 | |||
| rs10490923 | 0.040 | 0.70 | |||
|
| |||||
| HTRA1 | DNA | rs11200638 | 6.9 × 10−29 | 2.85 | (Fritsche et al., 2008; Yang et al., 2010) |
| rs932275 | 6.1 × 10−28 | 2.83 | |||
|
| |||||
| VEGF | DNA | rs699947 | 0.025 | 1.02–1.81 | (Fang et al., 2009; Ross et al., 2007) |
| rs1413711 | 0.042 | 0.62-0.59 | |||
| rs2010963 | 0.02 CNV | NA | |||
|
| |||||
| CD36 | DNA | rs3173798 | 9.96 × 10−4 | 0.55 | (Kondo et al., 2009a) |
| rs3211883 | 2.09 × 10−4 | 0.50 | |||
| rs10499862 | 0.00895 | 0.51 | |||
| rs3173800 | 0.00427 | 1.67 | |||
| rs17154232 | 0.0250 | 0.54 | |||
4.3.2 Tissue Inhibitor of Matrix Metalloproteinase-3
Tissue Inhibitor of Matrix Metalloproteinase (TIMP) refers to a group of proteins that inhibit matrix metalloproteinases (MMPs), a group of proteins involved in extracellular matrix (ECM) degradation (Gene ID: 7078). There are four variations of TIMP include: TIMP-1, TIMP-2, TIMP-3 and TIMP-4. Polymorphisms in TIMP-3, specifically, have been linked to varying AMD susceptibility in several studies (Ardeljan and Chan, 2013; Chen et al., 2010; Fritsche et al., 2016). Localized to RPE cells of Bruch’s membrane (Della et al., 1996; Kamei and Hollyfield, 1999), TIMP-3 contains a netrin (SanGiovanni et al., 2015) responsive domain and is induced in response to mitogens. TIMP-3 is unique from its other TIMP family members in that it binds directly to components of the extracellular matrix ECM (Leco et al., 1994).
Macular Bruch’s membrane concentrations of TIMP-3 appear to be age dependent (Kamei and Hollyfield, 1999). Additionally, patients with AMD often have supranormal levels of TIMP-3 both in macular Bruch’s membrane and in macular drusen. As TIMP-3 inhibits MMPs, excess TIMP-3 may retard Bruch’s membrane renewal and result in the thickening of Bruch’s membrane. By reducing Bruch’s membrane permeability, the trafficking of metabolites and nutrients between the choroid and RPE is also reduced, ultimately resulting in RPE and photoreceptor atrophy(Kamei and Hollyfield, 1999).
Most notably, TIMP-3 is a potent inhibitor of angiogenesis and is mutated in Sorsby’s fundus dystrophy, which includes features similar to AMD such as submacular CNV (Weber et al., 1994), but typically presents before age 40 (Chen et al., 2010) (OMIM: 188826). This matrix-bound inhibitor has various functions, including regulating the turnover of Bruch’s membrane, and perhaps most importantly, acting as a local inhibitor of VEGF thus limiting CNV (Qi et al., 2003). TIMP-3 blocks angiogenesis by inhibiting the binding of VEGF to the VEGF receptor (Qi et al., 2003). Mice lacking TIMP-3 have increased choroidal vasculature (Rodriguez de Cordoba et al., 2004). Mutations leading to decreased activity in TIMP-3 result in increased VEGF levels and a subsequent increase in the growth of pathological blood vessels in the eye.
Although many hypotheses have been proposed regarding the direct mechanism and causative relationship between TIMP-3 and AMD development, the direct mechanistic relationship remains unknown. While TIMP-3 is known to inhibit CNV, elevated levels have also been associated with thickened macular Bruch’s membrane and subsequent RPE atrophy (Kamei and Hollyfield, 1999). A recent GWAS study by Fritsche et al. (2016) demonstrated that a very rare coding variant of TIMP-3 may have a causal role for AMD development (Fritsche et al., 2016). Despite this, more research is needed to address the mechanistic relationship between these associative findings.
4.3.3 Fibulin 5
The fibulin 5 (Fbln5) gene is a member of the same family as EFEMP1/Fbln3. Fibulin 5 is a secreted ECM protein that promotes cell adhesion via interactions between integrins and its tripeptide Arg-Gly-Asp (RGD) motif (Nakamura et al., 1999). Fibulin 5 contains similar EGF-like domains to fibulin 3, and normally localizes to Bruch membrane and the intercapillary pillars of the choriocapillaris. In AMD, fibulin 5 localizes to pathological sub-RPE deposits as well as to small drusen (Kucukevcilioglu et al., 2015; Mullins et al., 2007). This gene is expressed in developing arteries and is upregulated in adult vessels in response to injury or atherosclerosis, implying a role in vascular development and remodeling (Gene ID: 10516). Mutations in fibulin 5 are also associated with cutis laxa (Markova et al., 2003) and AMD3 (Stone et al., 2004). There is evidence to suggest that fibulin 5 may downregulate VEGF and other promoters of angiogenesis. Meanwhile, overexpression of fibulin 5 in choroidal endothelial cells has been shown to alter their proliferation and migration (Li et al., 2012). As properly functioning fibulin 5 strengthens cell adhesions, down-regulates VEGF, and controls choroidal endothelial cell proliferation, mutations leading to misfolding and subsequent dysfunction of this protein confer an increased risk of AMD development (Schneider et al., 2010).
4.4 Lipoprotein-Related Genes
A number of AMD-predisposing variants are in components of lipoprotein and circulating cholesterol metabolism. Some of these variants may also alter risk with respect to atherosclerosis and other disorders associated with aging.
4.4.1 Hepatic Lipase
Hepatic Lipase (LIPC) is responsible for lipoprotein production. Both in a large GWAS and subsequent meta-analysis, the presence of an HDL-elevating allele of the LIPC gene was found to be associated with decreased risk of AMD (Neale et al., 2010; Reynolds et al., 2010; Wang et al., 2015b). Reynolds et al. found that the LIPC gene may be responsible for increasing HDL numbers in the blood, as well as a decreased risk of developing AMD. While high HDL levels are protective against AMD, individuals with high LDL counts have a significantly higher risk of developing AMD (Reynolds et al., 2010). The exact mechanism for the positive effects of HDL is unknown. Genetic analysis of the LIPC gene revealed that there are several different genotypes. In particular, the TT genotype has been linked to a significantly decreased risk of developing both wet and late-stage dry AMD. The discovery of LIPC, a genetic variant in the HDL pathway, may serve as a potential marker to be used in laboratory testing and individual risk analysis for the development of AMD (Reynolds et al., 2010).
4.4.2 Apolipoprotein E
In 1995, Klaver et al identified the Apolipoprotein E (ApoE) gene polymorphism as a strong risk factor for age-related macular degeneration, among various other neurodegenerative diseases. ApoE is the major apolipoprotein of the CNS and an important regulator of cholesterol and lipid transport (Klaver et al., 1998). Since then, the ApoE gene, found on chromosome 19q13.2 (Adams et al., 2011), has been consistently shown to play a significant role in the development of AMD (Baird et al., 2004; Levy et al., 2015; Paun et al., 2015). The ApoE polymorphism rs2075650 has been strongly associated with early AMD (P=1.1x10−6) (Holliday et al., 2013).
ApoE is responsible for the movement and transportation of lipids and cholesterol throughout cells in the body. Specifically, ApoE facilitates the binding of lipoproteins to low-density lipoprotein (LDL) receptors. This is crucial to fulfil cell requirements for lipoprotein cholesterol (Baird et al., 2004; Mahley, 1988). ApoE also has non-lipid functions such as immunoregulation and modulation of cell growth and differentiation. It is produced in high amounts in the liver, brain, and eye. High levels of ApoE mRNA can be found in the retina as well as the RPE, Bruch’s membrane, and choroid. Interestingly, ApoE is also a ubiquitous component of drusen (Mahley, 1988).
There are three common allelic variants frequently associated with the development of AMD (ε2, ε3, and ε4) (Adams et al., 2012). The ε4 allele has shown to be protective against the development of AMD while the ε2 allele has been implicated in moderately increasing the risk of AMD development (Anderson et al., 2001; Baird et al., 2004; Shen et al., 2015). The ε3 allele is often used as a baseline control in studies to determine risk. In a study by Baird et al., patients possessing allelic variants ε3ε4 were nearly 42% less likely to have AMD when compared to patients with allelic variants ε3ε3 (OR=0.58). The ε3ε4 genotype was most protective against late-stage dry AMD, lowering the risk of geographic atrophy by 65% (OR=0.35 compared to ε3ε3 patients with geographic atrophy). In contrast, the ε2 allele appears to increase the incidence of AMD. Those with the ε2ε3 genotype were diagnosed with AMD nearly four years earlier than those with the ε3ε3 genotype (P=0.15) (Baird et al., 2004). In a later study by Baird et al. (2006), patients who possessed the ε2 allele had nearly a 4.8-fold increased relative risk of developing AMD compared to patients containing the ε4 allele (Baird et al., 2006). Adams et al. found that patients with ε2ε3 genotype had a 33% increased risk of early AMD (the presence of drusen ≥125 μm with or without the presence of pigmentary abnormalities) when compared to those with the control ε3ε3 genotype (P=0.004) (Adams et al., 2012). In addition, patients with the ε2ε2 genotype were 83% more likely to have late AMD than patients with the ε3ε3 genotype (OR=1.83, P=0.04) (McKay et al., 2011). Wickremasinghe found that one copy of the ε2 allele was associated with intraretinal fluid at baseline (Wickremasinghe et al., 2014). In 2015, Shen et al. found that one copy of the ε2 T-allele (rs7412) had an increased risk of overall AMD (OR= 1.17, P= 5.7×10−3) and nonexudative AMD (OR= 1.32, P= 0.077), but not CNV (OR= 1.03, P= 0.153). Two copies of the ε2 increased the risk of developing all forms of AMD, while the ε4 allele was protective against all forms of AMD, especially CNV (Shen et al., 2015). In a pooled analysis of 15 studies (n=21,160), McKay et al., also showed that the ε4 allele was protective against late AMD (OR= 0.72, P= 4.41x10−11) (McKay et al., 2011).
Interestingly, the ApoE ε4 allele, while protective in AMD, is associated with increased risk for Alzheimer’s disease (AD), while the converse is true for ApoE2, which is protective against AD (Kovacs et al., 2007). To complicate matters further, there appears to be an interplay between the ε4 allele and the CFH-Y402H variant which may predispose to co-morbidity for both AD and AMD (Zetterberg et al., 2008). Toops et al (2016) suggests that the reason the allelic risk factors are opposite in AD and AMD, is that that ApoE serves a fundamentally different purpose in regulating cholesterol homeostasis in RPE versus neurons (Toops et al., 2016).
In sum, the ApoE gene is consistently associated with AMD and can have either protective or susceptible affects based on the presence of certain alleles.
4.4.3 Cholesteryl Ester Transfer Protein
A number of studies have linked cholesteryl ester transfer protein (CETP) to increased AMD or polypoidal choroidal vasculopathy (PCV) risk (Chen et al., 2010; Paun et al., 2015; Peter and Seddon, 2010; Wang et al., 2015b; Yu et al., 2011; Zhang et al., 2013). CETP is responsible for transferring cholesteryl esters between lipoproteins. Cholesterol is then transferred away from peripheral tissues via high-density lipoproteins (HDL), eventually resulting in uptake of cholesterol by the liver (Gene ID: 1071, OMIM: 118470). CETP mutations may influence susceptibility to atherosclerosis and coronary heart disease (Zhong et al., 1996).
In the retina, CETP is localized mainly to the inter-photoreceptor matrix, suggesting that the retina has an intra-retinal mechanism for processing and maturation of HDL-like lipoproteins (Tserentsoodol et al., 2006). This mechanism may be important, as photoreceptor outer segments are engulfed by the RPE at a significant daily rate and are lipid rich, indicating a high rate of lipid membrane renewal. In one population, the A allele of CETP polymorphism rs3764261 was associated with slightly lower rates of advanced AMD (OR= 0.49, P= 0.011) in addition to unilateral (OR= 0.52, P= 0.043) and bilateral AMD (OR= 0.45, P= 0.026) (Wang et al., 2015a). Unfortunately, many of the mentioned studies focused primarily on a particular ethnic population, and the results may not widely apply. Although CETP may play a contributory protective role against development of AMD in certain populations, studies involving patients with more diverse ethnic backgrounds are necessary.
4.4.4 CD36
CD36 is a major surface component of platelets and serves as a thrombospondin receptor in platelets and other cell lines (Gene ID: 948). Only limited evidence has been shown demonstrating its link to AMD. Thrombospondins are a diverse family, involved in mainly adhesive processes, implicating CD36 as a cell adhesion molecule. CD36 also binds oxidized LDL, collagen and phospholipids and may act in transport of fatty acids and/or as a regulator of fatty acid transport. Mutations in CD36 can lead to platelet glycolipid deficiency. Additionally, certain CD36 polymorphisms result in varied susceptibility to malaria, as CD36 acts as the major receptor for Plasmodium falciparum infected erythrocytes. CD36 deficiency is correlated with various insulin resistance syndromes in humans and rats (OMIM: 173510). CD36 variants have also been associated with nvAMD (Kondo et al., 2009b) and differ in frequency between nvAMD patients and those with polypoidal choroidal vasculopathy (PCV) (Bessho et al., 2012).
CD36 is expressed in the RPE and may participate in phagocytosis of photoreceptor outer segments. Deficiency of CD36 has been shown to lead to significant photoreceptor degeneration in mice (Houssier et al., 2008; Picard et al., 2010). One of the initial pathogenic changes in AMD involves drusen formation and the laying down of laminar deposits along Bruch’s membrane. These deposits have been shown to contain oxidized LDL, to which CD36 binds. Increases in oxidized plasma LDL are observed with age and high cholesterol diet. CD36 serves as the primary receptor in RPE cells for oxidized plasma LDL. Interestingly, mice with CD36 deficiency showed Bruch’s membrane thickening with deposition of oxidized LDL, despite consuming a regular diet. Conversely, treating high cholesterol model ApoE null mice with a CD36 agonist resulted in reduced thickening and preservation of photoreceptor function, despite a high fat diet (Picard et al., 2010).
CD36 deficiency also leads to decreased COX-2 production in the RPE. COX-2 is pro-angiogenic and can stimulate VEGF production. Mice with a COX-2 null mutation develop choroidal degeneration similar to that seen in geographic atrophy (Houssier et al., 2008). Although many indirect methods have linked CD36 to AMD, more rigorous experimentation is required to further solidify this association.
4.5 Potential AMD-Related Loci
The AMD consortium identified seven new loci that are associated with AMD risk. These include: ADAMTS9, Col8A1-FILIP1L, IER3-DDR1, SLC16A8, TGFBR1, RAD51B, and B3GALTL (Fritsche et al., 2013). Each will be discussed separately below. Additionally several other recently identified polymorphisms of various genes that confer increased risk of AMD development will be addressed.
4.5.1 ADAMTS9
ADAMTS9 is a member of the disintegrin and metalloproteinase with thrombospondin motifs family, members of which have been implicated in proteoglycan cleavage, control of organ shape during development and inhibition of angiogenesis (Gene ID: 56999). It maps to a region of chromosome 3 often lost in various tumours (OMIM: 605421) and may be associated with AMD development (Fritsche et al., 2013). Deletion of this gene in HEK293 cells inhibits endoplasmic reticulum to Golgi apparatus transport of some proteins (Yoshina et al., 2012). A C. elegans homologue, Gon1, is involved in gonad development and extracellular matrix degradation; the two proteins are functionally interchangeable (Yoshina et al., 2012). No obvious retinal function has so far been noted for this gene and further studies are required to confirm its association with AMD.
4.5.2 Col8A1-FILIP1L
Collagen Type 8, alpha 1 (Col8A1) is a short chain collagen and a major component of the basement membrane of the corneal endothelium (Gene ID: 1295, OMIM: 120251) and may play a role in development of AMD (Yu et al., 2011). Filamin interacting protein 1 (FILIP1) (Gene ID: 27145) is involved in extracellular remodelling and linking glycoproteins to actin filaments (FLNA, Gene ID 2136). FILIP1L (OMIM: 612993; Gene ID: 11259) is a relatively uncharacterised protein homologous to FILIP1 may have a role in inhibition of angiogenesis and cell proliferation (Kwon et al., 2008). Col8A1/FILIP1L comprises a genetic interval on chromosome 3 (human) identified by the AMD consortium as containing an AMD susceptibility locus (Fritsche et al., 2013). It is as yet unclear which of these transcripts (or regulatory regions within this area) harbours the predisposing mutation, however, FILIP1L may be an angiogenesis inhibitor and Col8a1 is a constituent of blood vessel cell walls, making both plausible potential candidates as players in AMD pathogenesis.
4.5.3 IER3-DDR1
The IER3-DDR1 locus comprises a genetic interval on chromosome 6p21 (human) identified by the AMD consortium as containing an AMD susceptibility locus (Fritsche et al., 2013). This interval contains a number of transcribed loci, including immediate early response 3 (IER3), a transcription factor of the NF-kappa beta family, which is thought to function as an anti-apoptotic factor (Gene ID: 8870). IER3 is also highly expressed in monocytes and macrophages and has a role in the immune response. Knockout mice lacking this gene showed increased susceptibility to Leishmania infection and an aggravated inflammatory response, thought to result from decreased TNF production by both macrophages and T cells (Akilov et al., 2009).
Discoidin domain receptor tyrosine kinase 1 (DDR1) is also present in this interval. DDR1, is a receptor tyrosine kinase that is overexpressed in various tumour types and belongs to a subfamily that interacts with collagens (Gene ID: 780). DDR1 may be involved in neurite extension and has a functional role in arterial vessel walls (OMIM: 600408). The functions of LINC00243 (long intergenic non-protein coding RNA 243) (Gene ID: 401247) and the microRNA MIR4640 (Gene ID: 100616237), also transcribed from this interval, are not known. The immunological role of IER3 and the presence of DDR1 in the vasculature would make both feasible candidates for AMD susceptibility. However, it is currently unclear which of these transcripts (or regulatory regions within this area) harbours an AMD-predisposing mutation.
4.5.4 SLC16A8
Solute carrier family 16 member 8 (SLC16A8) is a member of a family of proton-coupled monocarboxylate transporters involved in transporting lactate across cell membranes (Gene ID: 23539; OMIM: 610409). This gene was cloned from an RPE-derived library and may be specifically expressed in the RPE. Knockout mice showed a four-fold increase in lactate in parts of the retina and a reduced amplitude ERG, despite showing histologically normal retinas up to 36 weeks. Physiological changes may possibly relate to a decrease in the pH of the subretinal space, causing functional problems (Daniele et al., 2008). Recent evidence in a Nature Genetics study by Fritsche et al (2016) demonstrated that a splice variant in SLC16A8 may be strongly linked to AMD (Fritsche et al., 2016). In any case, the restricted expression of SLC16A8 in the RPE makes it a plausible candidate for AMD susceptibility and a potential focus of future investigation.
4.5.5 RAD51, RAD51B
Rad51 has been associated with AMD in a Chinese population (Zhou et al., 2013), while Rad51B was recently found to be associated with AMD risk by the AMD consortium (Fritsche et al., 2013). Rad51 family members interact with each other to carry out DNA repair via homologous recombination. Rad51 binds to tumour suppressors BRCA1 and 2 as part of a possible cellular response to DNA damage (Gene ID: 5888), while overexpression of Rad51B causes cell cycle delay and apoptosis, suggesting a role in DNA damage detection (Gene ID: 5890). A SNP in Rad51B has been associated with increased risk of male breast cancer (Orr et al., 2012). Rad51B is associated with both non-exudative AMD and CNV (Shen et al., 2015). No retinal specific functions have been noted to date, however, variants in genes that have apoptotic and/or cell cycle related functions would have obvious implications in retinopathies where RPE and/or photoreceptor cells die by toxicity related apoptosis.
4.5.6 FRK/Col10A1
An AMD susceptibility polymorphism, rs1999930 on 6q21-q22.3 near FRK/COL10A1, has been recently identified near the FRK and Col10A1 genes (Yu et al., 2011). Collagen 10A1 is a short-chain minor component of cartilage, expressed by hypertrophic chondrocytes during endochondral ossification. Mutations of Col10A1 result in Schmid-type metaphyseal chondrodysplasia (SMCD) and Japanese-type spondylometaphyseal dysplasia (SMD) in humans. Mimicked mutations in Col10A1 knockout mice result in variable skeletohematopoietic defects. In mice, these mutations have been shown to be dominant negative mutations and result in phenotypes similar to those seen in humans (Gress and Jacenko, 2000). The relevance of Col10A1 to AMD pathogenesis remains unclear. While the C-terminal region of the gene has been implicated in angiogenesis inhibition, a number of other collagens (Col8a1, Col15a1) have also been implicated in AMD pathogenesis, suggesting a possible retinal function of the Col10A1 gene (Yu et al., 2011).
FRK is a nuclear tyrosine kinase that has been implicated in suppression of the cell cycle during G1 and S phase (Gene ID: 2444). It has been shown to induce nerve growth factor (NGF) and enhance neurite outgrowth in a cell line that models neuronal differentiation. Knockout mice show subtle changes in thyroid hormone regulation with age, but are otherwise normal and fertile (OMIM: 606573). Indirect negative effects on retinal microvasculature and repression of the VEGF pathway via AKT signalling have been postulated to be of possible relevance in AMD pathogenesis (Yu et al., 2011), although further confirmatory investigation is requisite.
4.5.7 CACNG3
Calcium channel, voltage-dependent, gamma subunit 3 (CACNG3) is a type I transmembrane AMPA receptor regulatory protein (TARP), a class of proteins which regulate trafficking and channel gating of AMPA receptors. This gene is associated with susceptibility to familial infantile convulsive disorder (Gene ID: 10368, OMIM606403). One study reported a potential association between AMD and CACNG3 variants rs2283550, and rs4787924, although more studies are needed to confirm this association and clarify the role of the gene in AMD pathogenesis (Spencer et al., 2011)
4.5.8 MYRIP
A novel protective variant in myosin VIIA and Rab interacting protein (MYRIP), rs2679798, was identified as part of a scan involving families with an increased cohort of AMD affected members and confirmed as part of a case control cohort (Kopplin et al., 2010). MYRIP interacts with myosin Va and VIIA (OMIM: 611790); mutations in myosin VIIa cause deafness and Usher syndrome 1B, of which the primary symptoms include deafness and retinitis pigmentosa (OMIM: 276903).
MYRIP is expressed in the RPE and also interacts with Rab27a and Rab27b in a GTP dependent manner. As all three proteins are associated with melanosomes, it has been proposed that they form a complex involved in melanosome trafficking (El-Amraoui et al., 2002), in which MYRIP acts as a myosin receptor (Kuroda and Fukuda, 2005). Notably, increased pigmentation may also have a protective effect on AMD, as previously discussed. MYRIP also functions as a protein kinase A anchoring protein in zebrafish and can interact with the Sec6 and Sec8 components of the exocyst complex, which controls protein trafficking and exocytosis. In this instance, MYRIP was hypothesized to link PKA to the exocytosis machinery (Goehring et al., 2007).
One of the major roles of the retinal pigment epithelium (RPE) is the degradation of phagosomes, which are derived from the ingestion of photoreceptor outer segment (POS) disk membranes. Jiang et al. found that POS phagosomes associate with myosin-VIIa and then kinesin-1 light chain 1 (KLC1), as they moved from the apical region of the RPE, and lack of KLC1 impairs phagosome degradation and RPE pathogenesis that was comparable to aspects of AMD, with an excessive accumulation of RPE and sub-RPE deposits, as well as oxidative and inflammatory stress responses. This research group demonstrated that defective microtubule motor transport in the RPE, such as transport of POS phagosomes in relation to degradation, leads to phenotypes associated with AMD (Jiang et al., 2015).
4.5.9 Skiv2L
A novel protective variant, an intronic polymorphism rs429608 within the Skiv2L gene, was identified as part of a scan involving families with an increased cohort of AMD affected members and confirmed as part of a case control cohort (Kopplin et al., 2010). Superkiller viralicidic activity 2-like (Skiv2L) gene codes for an RNA helicase. Skiv2L is expressed at higher levels in T and B lymphocytes and dendritic cells (Fernando et al., 2007; Liu et al., 2013; Liu et al., 2011), hypothesizing that it may be protective in development of AMD through immune response modulation (Kondo et al., 2009a; Liu et al., 2013). Interestingly, while the minority A allele is thought to be protective in an American study (Kopplin et al., 2010), more recent studies in Han Chinese have implicated it as being pathogenic in AMD (Liu et al., 2011; Lu et al., 2013). As differences in genetic background can account for wider phenotypic variation with regard to the effect of the same mutation in different mouse strains, it is possible that the same applies to humans. Skiv2L contains a DEAD box and may therefore function as an RNA helicase. It is a human homolog of Ski2, suggesting an antiviral function, and may block translation of polyA deficient RNAs (Gene ID: 6499) Ski2 is one of a group of 6 yeast proteins that acts to constrain the copy number of RNA viruses. Human Skiv2L is associated with the exosome, which is known to mediate 3′–5′ RNA degradation in yeast. However, the yeast Ski2 is not an exosomal protein (OMIM: 600478). Mutations cause trichohepatoenteric syndrome, which results in growth retardation, hair abnormalities, facial dysmorphism, immunodeficiency and liver disease (OMIM: 614602). Consistent with an immunological role, the Skiv2L gene maps to the MHCIII region and is in linkage disequilibrium with other loci implicated in AMD (C2, CFB), but is also postulated to exert an independent effect (Kondo et al., 2009b). Skiv2L is expressed in the retina and RPE in both humans and mice (Liu et al., 2011), however, given that the polymorphism associated with AMD is intronic, it is difficult to speculate with regard to its impact on protein structure and function. A different Skiv2L polymorphism, rs2075702, conferred a reduced risk of PCV in a Japanese cohort (Kondo et al., 2009b). Another Skiv2L polymorphism, rs429608, exerted a protective effect on nvAMD in a Han Chinese cohort of 490 nvAMD patients (Ye et al., 2016). Further research is required to better understand the association of Skiv2L with the risk of AMD development.
4.5.10 IGFR1
Cha et al. found that the pattern of cytokine expression in the aqueous humor of exudative AMD patients varies from that of normal control subjects, where exudative AMD eyes were found to have increased levels of insulin-like growth factor binding protein 2 (IGFBP-2) and insulin-like growth factor-1 (IGF-1), indicating that the altered expression of IGF-related molecules may be involved in the disease pathogenesis for exudative AMD (Cha et al., 2013).
The rs2872060 SNP in insulin-like growth factor-1 receptor (IGFR1) has been significantly associated with AMD (Chiu et al., 2011). This gene has tyrosine kinase activity and acts as a receptor for IGF1, which it binds with high affinity. It acts as an anti-apoptotic agent in enhancing cell survival and is therefore overexpressed in a wide range of tumors (Gene ID: 3480). Some IGFR1 polymorphisms are positively associated with longevity, while mutations in this gene have resulted in growth retardation. Knockout mice die at birth from respiratory failure and are severely reduced in size; however, heterozygotes show an increase in lifespan (OMIM: 147370). Specific knockout in vascular endothelial cells resulted in a 34% decrease in retinal vascularization following hypoxia (Kondo et al., 2003). IGF1 and IGFR1 are found in the retina, RPE, and in some retinocapillary and choriocapillary endothelial cells. Their co-localization would point towards an autocrine function in the retina and associated ocular tissues (Lambooij et al., 2003). AMD pathology almost certainly results from the participation of IGF1 in neovascularization as inhibition of the IGF signaling pathway results in a reduction in VEGF levels (Sall et al., 2004), and IGF inhibitors have been shown to decrease VEGF levels in mice with laser induced CNV (Economou et al., 2008). IGFR1 SNP rs2872060 may serve as a fruitful target of further investigation evaluating risk factors for AMD.
4.5.11 REST-C4/F14-PolR2B-IGFBP7
An association has been identified with AMD risk between TNFRSF10A-LOC389641 rs1713985 and the multi-gene locus REST-C4/F14-POLR2B-IGFBP7 on chromosome 4q12 (Arakawa et al., 2011). This region contains the REST, NOA1, Pol2RB and IGFBP7 genes. This association was initially identified in a Japanese population and has not since been verified elsewhere (Nakata et al., 2012; Zhou et al., 2013). REST is a transcriptional repressor of neural specific genes is expressed in undifferentiated progenitors and may be a master negative regulator of neurogenesis (Gene ID: 5978). NOA1 is a large protein peripherally associated with the mitochondrial inner membrane, which may play a role in apoptosis and in mitochondrial respiratory processes (OMIM: 614919). Pol2RB is the second largest subunit of RNA polymerase II, which synthesizes mRNA in eukaryotes (Gene ID: 5431). IGFBP7 is a low affinity IGF binding protein that stimulates prostacyclin production and cell adhesion (OMIM: 602867).
Interestingly, a splice variant mutation in this gene causes retinal arterial macroaneurysm, probably caused by vascular endothelium malfunction, where IGFBP7 is normally highly expressed (Abu-Safieh et al., 2011). Given the role of IGF1 in stimulating VEGF signaling, which is in turn responsible for neovascularization, IGFBP7 is perhaps of most interest in this locus with regard to AMD.
4.5.12 SCARB1
Polymorphisms in the scanger receptor class B, member 1 (SCARB1) gene, such as rs9919713 (Meyers et al., 2014) and rs5888 have been associated with AMD (Zerbib et al., 2009). SCARB1 is a membrane receptor for HDL and is thought to mediate cholesterol transfer to and from HDL (Gene ID: 949). SCARB1 is closely related to CD36 and genetic variants affect susceptibility to atherosclerosis and insulin resistance, while knockout mice are infertile (OMIM: 601040). SCARB1 knockouts fed with a high cholesterol diet develop retinal changes closely related to AMD (Provost et al., 2009). SCARB1 binds lutein in the retina which, together with zeaxanthin, forms macular pigment. A shortage of lutein in the diet can result in degenerative signs in the retina. Notably, lutein is transported via HDL in the blood, while SCARB1 is an HDL receptor (Kijlstra et al., 2012). Lutein is also thought to diminish ocular, and possibly systemic inflammation levels, making it a very relevant component of retinal and macular function (Kijlstra et al., 2012). As such the intimate dynamics between SCARB1 and cholesterol and lutein transport contribute to its postulated effects on AMD development.
4.5.13 SerpinG1
In 2008, Ennis et al found that genetic variation in SerpinG1 significantly alters susceptibility to AMD (Ennis et al., 2008). Polymorphisms within the SerpinG1 gene have been associated with AMD in Caucasians (Gibson et al., 2010; Lee et al., 2010), with less influence in other ethnicities (Dong et al., 2015; Liu et al., 2015). Serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (SerpinG1) is localized to photoreceptor cells, inner nuclear layer neurons, choriocapillaris, and choroidal extracellular matrix within the retina, where it plays a crucial role in the regulation of complement activation (Mullins et al., 2009). SerpinG1 regulates the initiating steps of the complement cascade through its inhibition of activated C1r and C1s (Gene ID: 710). Mutations in SerpinG1 cause hereditary angioedema, as seen in other conditions poor C1 regulation (Carugati et al., 2001) (OMIM: 606860). It is also a component of innate immunity activated by optic crush injury (Templeton et al., 2013). Liu et al. conducted a meta-analysis suggesting that SerpinG1 is not a major component of AMD in East Asians, but is a genetic risk factor for AMD in Caucasians, providing evidence for an ethnic diversity in the genetic etiology of AMD (Liu et al., 2015). SerpinG1 may exert its influence on AMD development through regulation of the classical pathway of complement activation and other immune system functions in this disease process.
4.5.14 TNFRSF10A
A polymorphism which alters tumor necrosis factor receptor superfamily, member 10a (TNFRSF10A) transcription levels, rs13278062 at the locus TNFRSF10A-LOC389641 has been identified as a risk factor for AMD in a Japanese population (Arakawa et al., 2011). This finding has since been confirmed in other Asian populations (Nakata et al., 2012). Another group found that SNPs at AMD loci TNFRSF10A showed a trend toward association with typical chronic central serous chorioretinopathy (de Jong et al., 2015). TNFRSF10A is a member of the TNF receptor superfamily and is activated by tumor necrosis factor-related apoptosis inducing ligand (TNFSF10/TRAIL). It utilizes FADD (Fas-Associated protein with Death Domain) as an adapter protein in the pro-apoptotic pathway. As such, it plays critical role in apoptosis regulation.
4.5.15 CCL2
SNPs in chemokine (C-C motif) ligand 2 (CCL2), rs4586 and its receptor CCR2 rs1799865, combine to increase risk of AMD in individuals carrying both variants (Anand et al., 2012). Anand et al. (2012) noted that CCL2 and CCR2 levels were elevated in peripheral blood mononuclear cells (PBMCs) in individuals with AMD compared to controls (Anand et al., 2012).
CCL2 is a cytokine which displays chemotactic activity for monocytes and basophils and has been implicated in a number of diseases (atherosclerosis, psoriasis and rheumatoid arthritis) which are characterized by monocytic infiltrates (Gene ID: 6347). Mutations are associated with resistance to HIV. Knockout mice have shown some degree of immunodeficiency (related to issues with monocyte recruitment and an inability to mount a type II helper cell response) but were also 83% more resistant to atherosclerosis when placed on a high cholesterol diet (OMIM: 158105). Knockout mice also developed signs of AMD (drusen, lipofuscin accumulation, choroidal neovascularization and photoreceptor degeneration), possibly implicating macrophage dysfunction in the pathogenesis of AMD (Ambati et al., 2003).
Crossbreeding of the CCL2 knockout mouse with a knockout of Cx3CR1 (see below) resulted in mice with symptoms of AMD by 6 weeks of age (Tuo et al., 2007). However, these symptoms may have been exacerbated by the presence of an rd8 mutation in the genetic background of the mice studied, without which, a mild phenotype indicative of some retinal stress was observed (Vessey et al., 2012). In the absences of the rd8 allele, deficiency in CCR2 in mice was found to lead to a mild form of retinal degeneration which is associated with the recruitment of macrophages, particularly to the subretinal space; though this model enabled the research group to assess consequences of perturbed chemokine signaling, it did not recapitulate cardinal AMD features (Hagbi-Levi et al., 2016). CCL2 expression by retinal Mueller cells promotes macrophage infiltration of the retina and accelerates retinal damage and photoreceptor death, following injury (Rutar et al., 2012). CCL2 activated monocytes have also been shown to accelerate apoptosis in RPE cells (Yang et al., 2011). In summary, patients that have both CCL2 (rs4586) and CCR2 (rs1799865) SNPs may have an increased risk of AMD, though the evidence is not definitive.
4.5.16 Cx3CR1
Various studies have shown association of SNPs within chemokine (C-x3-C motif) receptor 1 (Cx3CR1) with AMD susceptibility in Chinese and Greek populations (Anastasopoulos et al., 2012; Ma et al., 2015; Yang et al., 2010). This gene is a receptor for fractalkine, a chemokine which is involved in the migration and adhesion of leukocytes. Variants are associated with increased susceptibility to HIV (Gene ID: 1524). Cx3CR1 is expressed by all retinal microglia and accumulate at retinal lesions in patients with AMD (Combadiere et al., 2007). Knockout mice showed accumulation of microglial cells at sites of retinal degeneration and choroidal neovascularization (OMIM: 601470). Its potential relation to AMD pathogenesis may be related to its effect on immune system modulation on a chemokine signaling level.
4.5.17 ERCC6
Polymorphism ERCC6 C-6530>G has been associated with AMD susceptibility (Tuo et al., 2006). Copy number variations involving this gene may also influence AMD susceptibility (Liu et al., 2011). Excision repair cross-complementation group 6 (ERCC6) is involved in transcription coupled excision repair of new DNA mutations, interacting with a complex of functionally similar proteins at DNA repair sites (Gene ID: 2074). Mutations in this gene cause UV sensitivity and cerebrooculofacioskeletal syndrome 1, in addition to AMD susceptibility (OMIM: 609413). ERCC6 functions in universal transcription as a component of RNA polymerase 1 transcription complex, and may therefore interact with another susceptibility allele in the CFH intron, by epistasis (Tuo et al., 2006). Initial data support ERCC6’s perceived effect on AMD pathogenesis as homozygous inheritance of both CFH and ERCC6 polymorphisms confers a disease odds ratio of 23 (Tuo et al., 2006). Polymorphisms in the ERCC6 and ERCC2 DNA repair genes reported to be associated with AMD confirms the importance of the cellular reaction to DNA damage, influenced by variability in DNA repair genes, in AMD pathogenesis (Blasiak et al., 2012). Further investigation is necessary to determine the magnitude of DNA repair genes’ importance in AMD.
4.5.18 FSCN2
Fascins have a role in crosslinking actin into filamentous bundles during dynamic cell extension. Fascin 2 (FSCN2) is specific to photoreceptor cells and may play a role in photoreceptor outer segment disk morphogenesis. A 208delG mutation was thought to result in autosomal dominant retinitis pigmentosa (adRP) or macular degeneration in a Japanese population (Wada et al., 2003), although the mutation in question was later found in Chinese normal controls (Zhang et al., 2007) (Gene ID: 25794; OMIM: 607643). Janik-Papis et al. (2009) also reported that the gene products of FSCN2 may play a role in the pathogenesis of AMD in the Polish population (Janik-Papis et al., 2009). FSCN2 knockout mice have been shown to exhibit progressive retinal degeneration (Yokokura et al., 2005), hinting at the crucial role FSCN2 plays in retinal and photoreceptor maintenance. Because of conflicting data, further investigation is necessary to understand whether there is a significant association between FSCN2 and AMD.
4.5.19 AMD-Related Loci Summary
The goal of a GWAS is to use genetic risk factors to make predictions about who is at risk and to identify the biological underpinnings of disease susceptibility for developing new prevention and treatment strategies (Bush and Moore, 2012). Application of this method was instrumental in first identifying CFH as a key associated factor in AMD (Hageman et al., 2005; Haines et al., 2005; Klein et al., 2005). A majority of the previously discussed factors have also been identified and associated with AMD based on this method of investigation (Fritsche et al., 2013; Fritsche et al., 2016). Although more focused research is required to confirm the identified loci and their reported association with AMD, it is vital to recognize their emerging importance. Additionally, several genes mentioned have only demonstrated association to AMD in particular ethnic populations, preventing universal conclusions from being applied. Although many of the mentioned genes are not currently confirmed as AMD risk factors, they may serve as attractive and fruitful targets for future research and investigation.
4.6 Effects of Genetic Polymorphism on Anti-VEGF Therapy
Not only can a person’s genetic makeup predispose them to the development and progression of AMD, but it can also determine the degree of response to AMD treatments. Current AMD therapies include intravitreal anti-VEGF antibody treatment such as bevacizumab and ranibizumab. As VEGF is responsible for vascular permeability and neovascularization, anti-VEGF therapies counteract the negative effects of VEGF leading to stabilization and quiescence of the choroidal vasculature. As with many other therapies, patients experience a broad degree of response to this therapy. The genetic makeup of each patient plays an influential role in the degree of positive or negative response to anti-VEGF therapy. By understanding the response of anti-VEGF treatment on people with different phenotypes, physicians can provide a more targeted and cost-effective therapy to their patients
4.6.1 VEGF and VEGFR polymorphisms
VEGF polymorphisms are also associated with the response to an anti-VEGF treatment. VEGF SNPs rs3025000 and rs699946 have been associated with improved anti-VEGF treatments (Abedi et al., 2013; Fauser and Lambrou, 2015).
For the VEGF SNP rs302500 (with alleles T & C), at least one T allele appears to be advantageous in anti-VEGF treatment (P<1 × 10−4). Abedi et al. observed that patients with either a TT or TC genotypes showed significantly better visual acuity improvement (7+ letters) at 3, 6, and 12 months than did patients with the CC genotype (Abedi et al., 2013). In addition, patients with the T allele for rs3025000 needed less anti-VEGF injections to attain comparable visual acuity outcomes compared to other groups (Abedi et al., 2013).
Regarding VEGF SNP rs699946, Agosta et al. found that patients with the G allele responded better to anti-VEGF therapy compared to patients carrying the A allele (Agosta et al., 2012). VEGF-A SNPs rs833069 and rs833068 have been associated with the number of injections received for an anti-VEGF treatment. For VEGF-A SNP rs833069, genotypes of GG received an average of 2 injections compared to AG and AA genotypes, which received an average of 6.57 and 6.40 injections respectively (Smith and Mitchell, 1998). For VEGF-A SNP rs833068, genotypes of AA received an average of 2.67 injections compared to AG and GG genotypes, which received an average of 6.57 each (Francis and Klein, 2011). Park et al. found improvement of visual acuity by ≥15 letters in patients with TT genotype of VEGF-A rs3025039 (Park et al., 2014). However, one study found no association with other VEGF polymorphisms rs1413711, rs3025039, rs2010963, rs833061, rs699947, rs3024997, rs833069 and rs1005230 and visual acuity (VA) response after anti-VEGF treatment (Smailhodzic et al., 2012). Overall, it appears that the VEGF SNPs rs3025000, rs833068, rs844069, and rs699946 have exhibited the strongest response to anti-VEGF treatment.
In addition to the effects of VEGF SNPs on patient response to anti-VEGF treatment, recent evidence supports positive treatment effects due to various vascular endothelial growth factor receptor (VEGFR) SNPs. Hermann et al. identified 2 SNPs (rs4576072 and rs6828477) in the VEGFR2 gene that were independently associated with a significantly improved visual acuity response over control at 3 and 12 months of treatment with ranibizumab. After 12 months, the mean increase in VA for both VEGFR2 SNPs combined on the logMAR scale was 0.26 (P=0.006), 0.08 (P=0.012), and 0.02 (P=0.097) for patients with 3, 2, or 1 minor alleles, respectively. Patients with no contributing minor alleles had an average VA decrease of 0.03 (Hermann et al., 2014).
Two large randomized clinical trials (Comparison of AMD Treatments Trials (CATT) and Alternative Treatment to Inhibitor VEGF in Patients with Age-Related Macular Degeneration (IVAN)) were performed to investigate the effects of various VEGF polymorphisms and their association with change in visual acuity in response to anti-VEGF treatment (CATT Research Group et al., 2011; IVAN Study Investigators et al., 2012). Recently Hagstrom et al. reemphasized earlier results that data from the CATT and IVAN trials do not support pharmacogenetic associations between VEGFR2 SNPs, rs4576072 and rs6828477, and change in visual acuity after anti-VEGF treatment (Hagstrom et al., 2015).
4.6.2 Complement Factor H polymorphisms
SNP alleles for Complement Factor H (CFH) have shown the strongest pharmacogenetic association to anti-VEGF treatment. The CFH Y402H genotype (with alleles, T and C) may influence response to anti-VEGF therapy. Homozygosity for the C allele in CFH gene has been associated with poor response to anti-VEGF treatments (Brantley et al., 2007; Kloeckener-Gruissem et al., 2011; Nischler et al., 2011; Teper et al., 2010). Patients with at least one T allele appear to respond better to anti-VEGF treatment than do CFH CC homozygote patients (Agosta et al., 2012). Patients with the CC Y402H genotype for CFH show significantly less improvements in visual acuity after anti-VEGF treatment than compared to CFH genotypes of TC and TT (Brantley et al., 2007; Kloeckener-Gruissem et al., 2011; Nischler et al., 2011; Teper et al., 2010). One study reported that patients with the CFH TT genotype experienced an average visual acuity improvement from 20/248 to 20/166; patients with the CFH TC genotype experienced an average visual acuity improvement from 20/206 to 20/170; patients with the CFH CC genotype experienced an average visual acuity decline from 20/206 to 20/341 (P=0.016). A total of 53.7% of patients with CFH TT and TC genotypes gained visual acuity with treatment, while only 10.5% of patients with the CFH CC genotype gained visual acuity with treatment (P=0.004) (Brantley et al., 2007). A Chinese meta-analysis found that six out of ten studies showed the C-allele to be a good predictor of poor response in patients receiving anti-VEGF treatment (Chen et al., 2012). Thus, in addition to the CC Y402H genotype being strongly associated with the development of AMD, it is also indicative a poor response to anti-VEGF treatment. An additional study cited that patients having the minor allele (A) in CFH SNP rs1065489 (CFH) had a significantly worse visual outcome compared to control. However, one study found no statistically significant positive or negative response differences to anti-VEGF therapy for various CFH genotypes (Orlin et al., 2012). In the recent Comparison of AMD Treatments Trial (CATT), there was no statistically significant difference between different allelic genotypes in patients with CFH SNP rs1061170 (Hagstrom et al., 2013) – a CFH SNP strongly associated with AMD development (Klein et al., 2013). In sum, certain polymorphisms of the gene encoding complement factor H, especially those with the CC homozygote genotype, appear to respond poorly to VEGF treatment.
4.6.3 Complement Factor 3 polymorphisms
SNPs that encode complement factor 3 (C3) have been implicated in the development of AMD. Those who are homozygous for the high-risk allele (GG) in SNP rs2230199 have a greater chance of developing AMD than those without the risk alleles. In addition, this genotype may indicate a more positive (although weak) response to anti-VEGF treatment. In March 2013, Hagstrom et al. found that patients homozygous for the high risk alleles had better visual acuity compared to those with one or zero risk alleles (P=0.03). However, these results did not reach the study’s statistical significance parameters of P< 0.01. In addition, other indicators of an improved response to treatment (retinal thickness, >15 letter increase, leakage, etc.) did not reach statistical significance (Hagstrom et al., 2013).
4.6.4 ARMS2/HTRA1 polymorphisms
The ARMS2 gene has been strongly implicated in the development of AMD (Kanda et al., 2007; Klein et al., 2013; Shen et al., 2015). Further, the ARMS2 SNP rs10490924 has been shown to be a strong predictor for the development of late AMD (Klein et al., 2013; Shen et al., 2015). Although this polymorphism may play a role in the development and progression of AMD, its presence does not predict how a patient will respond to anti-VEGF treatment. A recent study by Hagstrom et al. found that there was no statistically significant change in retinal thickness, visual acuity, mean number of injections, foveal thickness, vascular leakage, or lesion size following anti-VEGF treatment for those with rs10490924 (Hagstrom et al., 2013). Although this allele is strongly associated with late AMD, it does not a play a major influential role in a patient’s response to treatment.
Certain High Temperature Requirement Factor (HTRA1) genotypes may also provide short-term benefit in response to anti-VEGF treatment. HTRA1 genotypes GG, GA, and AA, showed mean letter score changes of +2.2, +7.5, and +2.9 respectively (McKibbin et al., 2012). However, several studies showed no association between positive or negative responders to anti-VEGF treatment (Orlin et al., 2012; Spencer et al., 2007). Although HTRA1 is also associated with a greater risk of AMD development, it may be associated with improved short-term response to current therapies for wet AMD. The high risk T allele of HTRA1 LOC387715 corresponded with a lower average number of bevacizumab injections (GG, 2.143; GT, 2.000; TT, 1.575; P= 0.064) in a Korean population. It was also associated with greater improvement in visual acuity 6 months after treatment (logarithm of the minimum angle of resolution; TT, 0.346; GT, 0.264; GG, 0.188; P= 0.037) (Kang et al., 2012). A recent study published by Hagstrom et al. did not find a significant correlation between genotypes of the HRTA1 SNP rs11200638 and improved response to anti-VEGF treatment (Hagstrom et al., 2013), even though this allele has been thought to influence the development of AMD.
4.6.5 PLAG12A polymorphisms
One study determined the effects of 21 different SNPs in six genes that had previously been associated with AMD (PLAG12A, IL23R, STAT3, VEGF-A, KDR, and HIF1A). Researchers found that only the SNP rs2285714 from the phospholipase A2 group XII A (PLA2G12A) gene showed an association (although weak) with poor response to anti-VEGF treatment. Initially, patients with at least one T allele of rs2285714 were nearly three times more likely to be poor responders than good responders to anti-VEGF treatment. However, after Bonferroni corrections were performed, these results no longer maintained statistical significance (Wang et al., 2012b).
4.6.6 Apolipoprotein E polymorphisms
Different alleles of the gene encoding ApoE showed different responses in preserved visual acuity to anti-VEGF treatment. Patients with the APOE ε4 allele showed stronger visual acuity after anti-VEGF treatment than did those with the APOE ε2 allele. In addition, patients with the APOE ε4 allele had an odds ratio for a 2-line gain in vision from baseline that was 1.5 times higher than patients with the ε2 allele (OR=4.04 compared to OR=2.54). This data suggest a significantly better visual response to anti-VEGF treatment for patients with the APOE ε4 allele (Wickremasinghe et al., 2011).
4.6.7 Effects of Genetic Polymorphisms on Anti-VEGF Therapy Summary
The data presented here suggest that each person’s response to AMD treatments may be influenced by individual genetics, and as such, understanding each patient’s individual genetic makeup holds prognostic benefit. Potentially, a doctor could predict a patient’s likelihood and degree of response to anti-VEGF therapy based on their genotype for certain alleles. This information may also decrease cost and discomfort to patients by preventing ineffective and unnecessary treatments for their genetic profile.
Many of the studies previously mentioned were retrospective and should not serve as standards to influence current clinical practice. Some studies even suggest that although certain alleles may predict development of AMD, they do not predict response to anti-VEGF therapy (Hagstrom et al., 2013). Additionally, there have multi-center clinical trials evaluating responses to current anti-VEGF therapies that have led to current best practice guidelines in treating AMD (CATT Research Group et al., 2011). Despite this, these data may serve as grounds to conduct future prospective, randomized clinical trials examining the effect of individual genetics on a patient’s response to anti-VEGF therapy. In addition to prognostic value, such studies could provide novel recommendations in terms of quantity and course of anti-VEGF treatment (or recommendation against treatment). In addition, such information would provide added incentive for gene therapy as a treatment for AMD in the future.
4.7 Biomarkers in Heritability and Genetics Summary
Overall, it appears that genetic polymorphisms of the CFH and ARMS2 genes show the strongest correlation with the development of AMD. Screening for high-risk alleles may provide at-risk patients with motivating information to make adequate lifestyle changes to eliminate environmental AMD risk factors. However, it is debatable whether early genetic screening of younger individuals without AMD is beneficial. In a 20-year follow-up of the Beaver Dam Eye Study, Klein et al., found that 80% of people with high-risk CFH and ARMS2 alleles never progressed to late AMD if they lived to 80 years old (Klein et al., 2013). Although high-risk genotypes make a significant contribution to the development of AMD, early genotyping of patients may not provide any extra benefit to prevention or care over traditional phenotypic staging of the disease.
5. Biomarkers of Exposure
It is believed that behavioural and nutritional factors are included in the etiology of AMD, in addition to genetic susceptibility (Seddon et al., 2011). As AMD is a multi-factorial disease, both genetic and environmental aspects determine development and progression of AMD. Environmental exposure may influence expression of genetic determinants of AMD and genetic susceptibility can worsen effects of environmental exposure (Klein et al., 2014). Both of these factors may also be influenced by epigenetic changes as well (Hutchinson et al., 2014). This section will discuss known environmental exposures – specifically smoking, sunlight, and dietary factors – and their effects on AMD development and progression.
5.1 Smoking
Cigarette smoking is the strongest environmental risk factor for wet AMD (Pons and Marin-Castano, 2011), and is responsible for a two- to three- fold risk increase of developing AMD (Evans et al., 2005; Thornton et al., 2005). Smoking is associated with both dry and wet forms of AMD, but there is a stronger correlation between smoking and the development of nvAMD (Clemons et al., 2005; Delcourt et al., 1998; Smith et al., 1996; Thornton et al., 2005; Vingerling et al., 1995a; Vingerling et al., 1995b; Woo et al., 2015). In addition, the number of packs and pack years smoked is directly correlated to the degree of AMD risk (Chang et al., 2008; Khan et al., 2006; Thornton et al., 2005).
The cessation of smoking is greatly beneficial to health in general, but whether ex-smokers have an increased risk of developing AMD is inconclusive. Some studies have significant findings to suggest that former smokers are still at an increased risk (Smith et al., 1996; Thornton et al., 2005; Vingerling et al., 1995a; Vingerling et al., 1995b), while other studies did not find statistically significant evidence of any increase in risk of AMD for ex-smokers (Klein et al., 1998; Klein et al., 2002).
There is also minimal evidence concerning the effect of second hand or passive smoking and risk of AMD (Smith et al., 1996; Thornton et al., 2005; Vingerling et al., 1995a; Vingerling et al., 1995b), although there is some evidence to support an increased risk of AMD development from exposure to secondhand smoke (Pons and Marin-Castano, 2011).
The exact mechanism of how and why smoking affects the pathophysiology of AMD is not fully understood. Solberg et al. theorized that “smoking may also reduce choroidal blood flow in the eye, and promote ischemia, hypoxia, and micro-infarctions, all of which could increase the susceptibility of the macula to degenerative changes” (Solberg et al., 1998). This is supported by evidence that smoking decreases choriocapillaris blood flow associated with AMD (Ciulla et al., 2001; Lutty et al., 1999). In addition, Kew et al. found that smoking may work through a complement mechanism, activating C3 in the alternative pathway (Kew et al., 1985). Decreased plasma levels of complement factor H have been observed in smokers (Esparza-Gordillo et al., 2004). In mouse models, nicotine administration increases the severity of CNV (Suner et al., 2004).
Although more research is needed to fully elucidate the specific mechanisms of AMD development, smoking remains as the strongest environmental exposure risk factor associated with the development and aggravation of AMD.
5.2 Sunlight
It has been suggested that sunlight exposure could be a risk factor for the development of AMD (Mitchell et al., 1998; Schick et al., 2015; Taylor et al., 1992). Excessive exposure to light has been reported to cause retinal damage (Coleman et al., 2008), and it has been suggested that wearing sunglasses may be considerably protective against this risk factor (Cruickshanks et al., 1993; Schick et al., 2015).
Other studies have shown little or no association between sunlight exposure and the risk of AMD (Age-Related Eye Disease Study Research, 2000; Fletcher et al., 2008; The Eye Disease Case-Control Study Group, 1992).
Recently however, in a retrospective study by Schick et al (2015), there appeared to be no association between current sun exposure and the development of AMD, but a significant association between previous sun exposure (outside >8hours daily) and early AMD (OR=5.54, P=0.02) and late AMD (OR=2.77, P=0.01) (Schick et al., 2015).
Owing to the complexity in quantifying sun exposure differences among people living in the same location, Smith et al., criticised many studies which reported an association between sunlight exposure and AMD (Smith et al., 2001b).
5.3 Diet
Both increased dietary fat intake and obesity are positively associated with increased risk of AMD incidence and progression. Additionally, a protective effect from antioxidants, nuts, fish, and omega-3 polysaccharide unsaturated fatty acids has been described (Biomarkers Definitions Working, 2001; Seddon et al., 2003b).
The Age Related Eye Disease Study (AREDS) demonstrated the efficacy of zinc-antioxidant supplements for preventing or delaying progression of early AMD to late AMD in patients who are at high risk (Biomarkers Definitions Working, 2001). A population-based cohort study in Australia showed that high dietary lutein and zeaxanthin intake reduced the risk of the long-term incident AMD and that high beta-carotene intake was associated with an increased risk of AMD (Tan et al., 2008). However, In the U.S., the third National Health and Nutrition Examination Survey (NHANES) illustrated that age-related maculopathy is not significantly associated with dietary fat in a large cross-sectional survey (Heuberger et al., 2001). The clinical evidence suggested that the omega-3 fatty acid may be protect against AMD, although, it is not evidently supported by the current literature (Hodge et al., 2006). Awh et al. suggested that treatment of AREDS supplements should be tailored according to patient’s specific genotypes (Awh et al., 2015; Awh et al., 2013); however this was later challenged (Chew et al., 2015). It is believed that behavioural and nutritional factors are included in the aetiology of AMD, in addition to genetic susceptibility (Seddon et al., 2011). AREDS and AREDS2 supplements, the combination of antioxidants (lutein and zeaxanthin) and zinc, are currently the only evidenced based dietary supplement to decrease risk of progression to late-stage AMD (Age-Related Eye Disease Study 2 Research et al., 2013; Biomarkers Definitions Working, 2001; Chew et al., 2015).
6. Biomarkers of Disease & Progression
Although, several candidate genes for AMD have already been identified, many individuals carrying these AMD genes never go on to develop AMD. As such, there are other factors at work. Other limitations to genetic biomarkers of AMD include the necessity to develop a simple and inexpensive methodology to identify people who carry SNPs of the relevant AMD genes. Although genetic biomarkers can be valuable predictors of AMD susceptibility, given these limitations, they are currently inappropriate for screening and guiding treatment until further efficiency and cost-effectiveness for tailored testing is developed.
Developing analytical procedures and immunoassays to detect biomarkers of early and late AMD, both for screening and monitoring therapeutic progress of patients with disease, would contribute significantly to reducing blindness from this debilitating disease.
An interesting use of proteomics is using specific protein biomarkers to diagnose disease, and serum biomarkers have been used extensively in screening and monitoring other disease processes (Table 3). Metabolomics is another relatively new area of research that may also shed new insights on potential biomarkers for AMD screening and progression. MicroRNA (miRNA) profiling has also become a vital technology to help identify potential disease biomarkers (Zhao et al., 2010), elucidate the pathogenesis of many diseases, and reveal valuable diagnostic and prognostic information, as well as promising treatment targets (Bentwich et al., 2005).
Table 3.
Proteomic and Serum Biomarkers
| Biomarker | Type | AMD Assessment | Level (% higher than control plasma) | Odds Ratio | P. Value | Sensitivity, Specificity (%) | References |
|---|---|---|---|---|---|---|---|
| CRP | Systemic inflammatory marker | Prognosis Higher risk CNV | Elevated | 2.6 | .046–.06 | NA | (Kikuchi et al., 2007; Seddon et al., 2010a; Seddon et al., 2005) |
| CEP, CEP autoantibody titers | End product of lipid oxidation | Diagnosis CNV | Elevated in plasma 60%, 30% | 3.17 | <0.001 | 73,65 | (Ebrahem et al., 2006; Ni et al., 2009a; Ni et al., 2009b) |
| CML | Advanced glycation end products | Diagnosis | 54% | 6.3 | <0.0001 | 84,72 | (Ni et al., 2009b) |
| Pentosidine | Advanced glycation end products | Diagnosis | 64% | 10.6 | <0.0001 | 84,88 | (Ni et al., 2009b) |
| VEGF | Protein stimulates the growth of new blood vessels | Prognosis (neovascular form) | Elevated | NA | 0.019 | NA | (Lip et al., 2001; Tsai et al., 2006) |
| IL-6 | Inflammatory marker | Prognosis | Elevated (AMD progression) | NA | 0.02 –0.03 | NA | (Seddon et al., 2005) |
| IP-10 | Chemokine | Diagnosis | Elevated intermediate dry AMD | NA | NA | NA | (Zhang and Marmorstein, 2010) |
| Fibrinogen level | Plasma glycoprotein | Prognosis | Elevated | 6.7 | 0.0001 | NA | (Lip et al., 2001; Smith et al., 1998) |
| Homocysteine | Amino acid in the blood | Diagnosis Prognosis | Elevated | 1.6 | .001 | NA | (Seddon et al., 2006) |
These biomarkers are discussed below.
6.1 Proteomic biomarkers
Proteomic biomarker discovery from human plasma has had a significant impact on clinical applications as well as early detection of disease. Below, we discuss carboxyethylpyrrole, a promising proteomic biomarker of early AMD, as well as homocysteine.
6.1.1 Carboxyethylpyrrole
The research interest of early identification of AMD-susceptible individuals has found carboxyethylpyrrole (CEP), a protein generated from free radical induced oxidation of docosahexaenoate (DHA)-containing lipids levels and CEP autoantibody titers to be possible biomarkers for AMD (Gu et al., 2010; Hollyfield et al., 2010; Ni et al., 2009a). Gu et al. suggested that there is a significant elevation in both serum CEP oxidative protein modifications and serum CEP autoantibodies (vicarious markers of CEP) in AMD by 60 and 30%, respectively. Additionally, prospective assessment CEP markers can be utilized to distinguish between AMD plasma and normal individual’s plasma with 76% accuracy (Gu et al., 2010; Ni et al., 2009a), while the accuracy is increased to 92% if CEP and pentosidine (a marker of protein damage and advanced glycation end products) (Sell et al., 1991) have been measured (Ni et al., 2009b). The Clinical Genomic and Proteomic AMD Study Group (2009) suggested that the combined measurement may be more accurate to differentiate between patients and control individuals (Gu et al., 2009). Additionally, plasma protein Nε-carboxymethyllysine (CML) and pentosidine had been quantified in order to use them as AMD biomarkers; where the level of CML and pentosidine was higher in AMD (Ni et al., 2009b).
6.1.2 Homocysteine
It is believed that an elevated homocysteine level is associated with advanced AMD (Axer-Siegel et al., 2004; Ghosh et al., 2013; Seddon et al., 2006). Homocysteine has been suggested as a biomarker of cardiovascular disease (Boushey et al., 1995). Additionally, studies not only found that homocysteine was significantly elevated in AMD patients (Kamburoglu et al., 2006; Rochtchina et al., 2007; Vine et al., 2005) but that the levels were higher in nvAMD vs. the dry form of AMD (Coral et al., 2006; Ghosh et al., 2013; Nowak et al., 2005). Although these data may support using serum homocysteine levels as a surrogate marker and screening for AMD, the studies had small numbers of cohorts yielding low statistical power.
In 2015, Christen et al. conducted an average 10-year prospective study to examine the association of plasma homocysteine levels and AMD in over 27.000 females over the age of 40. Comparing the highest and lowest quartiles of plasma homocysteine levels in women that developed AMD, there was a modest, yet non-significant difference (HR 1.24, P=0.07) (Christen et al., 2015). Overall, the data on the relationship between plasma homocysteine levels and AMD remains controversial (Huang et al., 2015; Keles et al., 2014; Pinna et al., 2016) and future well-designed trials are needed to establish homocysteine as a viable biomarker of AMD.
6.1.3 Proteomic Biomarkers Summary
While the relationship between plasma homocysteine levels and AMD is controversial (Huang et al., 2015; Keles et al., 2014; Pinna et al., 2016), proteomic research shows promise with respect to developing biomarkers for AMD, such as CEP, particularly when used in combination with pentosidine (Gu et al., 2009). Additionally, plasma protein Nε-carboxymethyllysine (CML) and pentosidine had been quantified in order to use them as AMD biomarkers, where the level of CML and pentosidine was higher in patients with AMD (Ni et al., 2009b). Additional studies must be undertaken to validate proteomic biomarkers of AMD and develop assays for clinical application.
6.2 Serum biomarkers
Commonly used for other disease processes, the use of serum biomarkers may be a useful tool to one day improve screening and distinguish AMD patients from healthy individuals, as well as for monitoring disease progression and response to treatment. Below, we discuss possible serum biomarkers for early detection of AMD, such as specific circulating autoantibodies, interferon γ-inducible protein 10 (IP-10), eotaxin, and soluble FMS-like tyrosine kinase-1 (sFlt-1), for which we present promising recent findings from our lab. We also discuss possible serum biomarkers for monitoring AMD disease progression, such as C-reactive protein (CRP), cholesterol, and VEGF.
6.2.1 C-Reactive Protein
C-reactive protein (CRP) is an acute-phase reactant that is predominantly synthesized in liver hepatocytes and adipocyte cells. When elevated, it acts as a non-specific serum marker for subclinical inflammation (Bhutto et al., 2011). As an inflammatory agent, CRP functions to activate complement factors, macrophages and platelets via increased expression of pro-inflammatory factors. Elevated CRP is a risk factor for heart disease, type 2 diabetes and AMD (Seddon et al., 2005).
CRP interacts with various proteins in the complement pathway, including one of the alternative complement’s main regulators, complement factor H (CFH). Given that CRP plays pro-inflammatory and pro-thrombotic roles in the pathogenesis of atherosclerosis, it is thought that CRP may play a similar role in the progression of AMD (Seddon et al., 2004).As previously discussed, CFH has been strongly associated with AMD and its interaction with CRP may be responsible for one mechanism of AMD development. With elevated levels of CRP, there is an increased interaction between CRP and CFH (Bhutto et al., 2011). CRP interferes with the regulatory function of CFH, and thereby can indirectly activate the inflammatory response to foreign body-like material, such as drusen. This process increases susceptibility to AMD. It has been shown that CRP levels were significantly higher in individuals with intermediate or advanced stages of AMD (Bhutto et al., 2011). High levels of CRP, along with low levels of CFH at the retina-choroid interface may lead to uncontrolled complement activation with associated cell and tissue damage (Bhutto et al., 2011).
Several studies have found higher levels of plasma CRP levels in AMD patients compared to healthy controls (Colak et al., 2011; Hong et al., 2011; Robman et al., 2010; Seddon et al., 2004; Yasuma et al., 2010). Seddon et al. found that CRP was significantly associated with the presence of both intermediate and advanced stages of AMD (OR=1.65, P=0.02) (Seddon et al., 2004). A meta-analysis by Hong et al. found that patients with serum levels of CRP >3 mg/L had a two-fold increase in likelihood of late onset AMD than did patients with serum CRP levels <1mg/L (Hong et al., 2011). Additionally, increased levels of CRP levels have been associated with ARMS2/HTRA1 high-risk alleles (Robman et al., 2010; Seddon et al., 2010b; Yasuma et al., 2010).
Chronic inflammation plays a critical role in the development of age-related macular degeneration. Some complement factors that have been implicated in the pathogenesis of AMD include CRP and CFH. Immunohistochemical analysis has demonstrated that the sub-retinal pigment epithelial and choroidal spaces contain drusen and complement components, including CRP and CFH (Mullins et al., 2000). Interestingly, while drusen show high levels of both CFH and CRP, levels of CRP are reduced in areas affected by geographic atrophy (Bhutto et al., 2011). CRP is an important activator of the classical complement pathway and the pathogenesis of AMD is thought to involve the activation of the complement pathway (Edwards et al., 2005; Hageman et al., 2005; Haines et al., 2005; Klein et al., 2005). It has been shown that CRP may enhance NF-kB nuclear translocation (Wang et al., 2010) and induce secretion of pro-inflammatory cytokine interleukin 8 (IL-8) (Wang et al., 2010; Xie et al., 2005).
Overall, CRP plasma levels appear to be significantly correlated with AMD. Because CRP interferes with the regulatory function of CFH on the complement pathway, it can induce inflammation; thus high plasma CRP levels are a risk factor for AMD. Despite this, further studies are necessary to inquire into the role, efficacy, and accuracy of CRP as a screening and prognostic tool in patients with AMD.
6.2.2 Circulating Autoantibodies
Heckenlively et al has described anti-retinal antibodies in pigmentary retinopathy and other retinal degenerations (Heckenlively et al., 1996). Serum autoantibodies may also be useful biomarkers that play a mechanistic role in AMD (Adamus et al., 2014; Donoso et al., 2006; Iannaccone et al., 2012; Nussenblatt and Ferris, 2007; Penfold et al., 2001). Iannaccone et al. (2015) identified autoantibodies in AMD sera, even in early-stage disease, including Annexin A5 (ANXA5), Protein S100-A9 (also known as calgranulin B that, when complexed with S100A8, forms calprotectin), and heat shock proteins, HSPA8, HSPA9, and HSPB4 (also known as alpha-crystallin A chain), which may play a role in AMD via autophagy compromise and downstream activation of the inflammasome (Iannaccone et al., 2012).
Serum antibodies against cyclic nucleotide phosphodiesterase phosphatidylserine (PS) have been strongly associated with various stages of AMD (Morohoshi et al., 2012). Elevated antibody ratios have been found in patients with AMD when compared to control groups. In one study, serum from AMD patients was found to contain immunoglobins G and M that were reactive to self-antigen PS (Morohoshi et al., 2012). IgG/IgM ratios can act as indicators of drusen formation in AMD. The molecular components of drusen found in Bruch’s membrane contain possible autoantigens that may be responsible for the pathogenesis of AMD. It has been observed that IgG/IgM ratio levels increase with increasing staging in autoimmune diseases. This study found that serum IgG/IgM ratios were elevated in both dry and wet AMD. In addition, the anti-PS levels were significantly higher in wet AMD than dry AMD. This study found that IgG/IgM ratios over 7 showed up to a 44-fold increased risk for advanced choroidal neovascular AMD. In addition, it was found that the levels of IgG/IgM elevation were directly correlated to the stage of AMD (Morohoshi et al., 2012). Careful monitoring of serum antibody ratios may provide an accurate and efficient way to monitor progression of AMD.
6.2.3 Cholesterol
Serum cholesterol levels may be associated with increased risk of AMD. One study reported that low-density lipoprotein (LDL) levels were increased and high density lipoprotein (HDL) levels were decreased in patients with both late-stage dry and wet AMD compared to controls. In addition, there was a significant increase in the risk of developing AMD in patients with higher total cholesterol levels (Reynolds et al., 2010). Recently, Paun et al (2015) demonstrated a correlation between serum apolipoprotein, cholesterol, and triglyceride and patients with certain high-risk AMD genotypes (CETP, ApoE, and FADS1). Patients with these high-risk genotypes had higher serum levels of Apo-A1 and HDLC with lower levels of triglycerides compared to controls (Paun et al., 2015). More prospective, randomized trials are needed to establish serum lipid levels as a clinically suitable biomarker for patients at risk for AMD.
6.2.4 Interferon γ-Inducible Protein 10 and Eotaxin
Serum Interferon γ-Inducible Protein 10 (IP-10) and eotaxin may serve as serum biomarkers for early detection of AMD. IP-10 is a chemoattractant for activated T lymphocytes and is thought to play an important role effect T cell generation and trafficking to sites of tissue inflammation (Dufour et al., 2002). Eotaxin serves as a potent chemoattractant in eosinophil and other inflammatory cell recruitment (Menzies-Gow et al., 2002). As immunomodulators, IP-10 and eotaxin may serve as possible biomarkers for AMD development.
Mo et al. found that those with early stages of AMD had significantly higher levels of IP-10 and eotaxin compared to controls. These cytokines were elevated in subjects with all early forms of AMD. Specifically, eotaxin levels were significantly higher in AREDS stage 1 AMD patients compared to control (Mo et al., 2010). In addition, IP-10 levels were significantly elevated above control levels in all stages of early AMD, reaching their peak at AREDS Stage 3. However, patients with wet AMD actually showed lower levels of IP-10 and eotaxin compared to patients with AREDS stage 3 and geographic atrophy AMD (Mo et al., 2010).
Eotaxin-1 (CCL11) and Eotaxin-2 (CCL24) are associated with the inflammatory processes seen in AMD. CCL24 may be of particular importance as a biomarker of wet neovascular AMD. One study noted that eotaxin-2 levels were significantly higher in wet AMD patients than in patients with dry AMD or healthy controls. This study also compared eotaxin-2 levels before and after anti-VEGF (bevacizumab) treatment. Of particular interest is the finding that eotaxin-2 levels were significantly increased in AMD patients compared to controls even after anti-VEGF treatment. This suggests that eotaxin-2 may play a role in CNV together with, yet independent of VEGF. Future research should investigate the efficacy of anti-eotaxin-CCL24 in conjunction with current anti-VEGF treatment (Sharma et al., 2012). IP-10 and eotaxin may serve as early biomarkers of AMD and may aid in detection during early stages of AMD (Mo et al., 2010).
6.2.5 Vascular Endothelial Growth Factor
Vascular Endothelial Growth Factor (VEGF) levels are often elevated in patients with AMD (Lip et al., 2001; Tsai et al., 2006). VEGF is found at elevated levels in RPE cells of AMD patients (Lip et al., 2001; Tsai et al., 2006). Lip et al. found that plasma levels of VEGF were significantly increased from baseline in 78 AMD patients compared to 25 healthy controls (P=0.0196) (Lip et al., 2001). Another study showed elevated plasma VEGF levels in 77 AMD patients compared to 42 healthy controls (P<0.001) and noted that patients with wet AMD had significantly higher VEGF levels relative to patients with dry AMD (P<0.05), suggesting that high serum VEGF levels may be indicative of the progression of the disease (Tsai et al., 2006). Apart from the VEGF genetic SNPs that may confer increased AMD risk, elevated serum VEGF levels are one of the key culprits of AMD pathogenesis.
6.2.6 Soluble FMS-like Tyrosine Kinase-1
Our lab has recently determined that soluble FMS-like tyrosine kinase-1 (sFlt-1), also known as soluble vascular endothelial growth factor receptor-1 (sVEGFR-1), may be a promising, novel biomarker for AMD. sFlt-1 has previously been linked to inhibition of choroidal neovascularization (Gehlbach et al., 2003). sFlt-1 is an alternatively spliced, secreted isoform of the cell-surface receptor membrane-bound flt-1 (mbflt-1). sFlt-1 acts as a ‘manacle’ for VEGF-A, sequestering the proangiogenic growth factor, essentially neutralizing its effect to stimulate its receptor and thus preventing downstream, angiogenic signalling (Ambati et al., 2006; Kendall and Thomas, 1993). In other words, sFlt-1 acts as a trap for secreted VEGF-A extracellularly (Ambati et al., 2006).
We first determined in 2006, that sFlt-1 (also known as sVEGFR-1) is responsible for the avascular nature of the cornea, leading to corneal clarity and optimal vision (Ambati et al., 2006). In 2013, we demonstrated that sFlt-1 is also responsible for photoreceptor avascular privilege, protecting the outer retina from invasion by vessels from either the choroid or inner retinal vasculature (Luo et al., 2013). Because of this earlier work elucidating the ability of sFlt-1 to bind and neutralize VEGF-A, we sought to identify whether sFlt-1 played a protective role against CNV and AMD.
Working with the CARMA clinical trial dataset in Northern Ireland in collaboration with Dr. Ruth Hogg’s group, we compared serum sFlt-1 levels in patients without AMD, and in patients with early AMD, and nvAMD. sFlt-1 levels were significantly lower in patients with nvAMD compared to those with early AMD and those without AMD (Uehara et al., 2015). Additionally, we noted that with each 10-point increase (pg/mL) in serum sFlt-1 concentrations, the odds of having nvAMD decreased by 27.8% (OR = 0.722, P = 0.002) and 27% (OR = 0.730, P= 0.003) compared to patients without AMD and those with early AMD, respectively (Uehara et al., 2015). In patients over 73 years of age, sFlt-1 levels <80 pg/ml were associated with a 6.7 fold increased risk of nvAMD. To our knowledge, this is the strongest association of a serum biomarker with nvAMD. Although unclear whether reduced sFlt-1 levels are a by-product of, or were directly causative of neovascularization in AMD patients, these data indicate that monitoring for sFlt-1 levels could potentially serve as a promising biomarker for potentially identifying patients at risk for progression to nvAMD.
6.2.7 Serum Biomarkers Summary
Research on serum biomarkers for AMD to date has been promising, particularly with respect to circulating autoantibodies, IP-10, eotaxin, and as we presented our lab’s findings, sFlt-1. These serum biomarkers and others presented, such as cholesterol, CRP, and VEGF, may shed light on AMD pathogenesis and risk. As reliable serum biomarkers are discovered, a screening tool combining multiple biomarkers may one day provide accurate predictive results for AMD development in clinical settings.
6.3 Metabolomics
Individual blood-based biomarkers can be helpful in understanding the pathophysiology of AMD and for detecting and monitoring disease progression. Yet, each of these individual markers is also significantly impacted by multiple genetic, systemic, environmental and demographic factors. Attempts to assess “the collective set of metabolites produced or present in a biosystem” known as the metabolome, have emerged since the late 1990’s (Smolinska et al., 2012). The rapidly developing field of metabolomics (the study of the quantitative complement of metabolites in a biological system and changes in metabolite concentration or fluxes related to genetic or environmental perturbations) aims to take a more holistic approach to the investigation of pathology and has already been applied to a number of conditions including Alzheimer’s disease (Graham et al., 2013), diabetes (Maher et al., 2009) and multiple sclerosis (Moussallieh et al., 2014).
Nuclear Magnetic Resonance (NMR) spectroscopy and Mass Spectroscopy (MS) are the main methodologies used in metabolomics. Advances in NMR and MS have been accompanied by the development of analytical tools to handle the complex high-dimensional multivariate data generated by these platforms. Studies commonly investigate urine, plasma and serum, however other biofluids such as cerebrospinal fluid (Lista et al., 2014), amniotic fluid (Amorini et al., 2012) and vitreous humor (Barba et al., 2010) are made possible by metabolomics (Alonso et al., 2015; Mamas et al., 2011; Smolinska et al., 2012).
Several studies have already begun to incorporate this methodology for AMD. One group identified neuroprotectin D1 (NPD1), a DHA-derived bioactive lipid, as associated with AMD (Bazan et al., 2011), and another group performed a metabolome-wide association study (MWAS) to identify a map of diverse metabolites, ranging from peptides, bile acids, and vitamin D to broader pathways like that of tyrosine metabolism, that may be unique to patients with AMD (Osborn et al., 2013).
As MWAS is still a fairly new methodology for identifying potential biomarkers in AMD, metabolomic research may continue to reveal new insights for AMD screening and monitoring in the future.
6.4 MicroRNAs as Biomarkers
Another new area of research for AMD biomarker identification includes the study of microRNAs (miRNAs). During retinal development, a specific interaction occurs between different molecules to develop the appropriate structure that guarantee the integrity of cellular functions (Maiorano and Hindges, 2012). miRNAs play an important role for both retinal development and maintenance (Arora et al., 2007).
miRNAs are endogenous, non-coding, single stranded RNAs, which are approximately 22 nucleotides long (Bartel, 2004) and believed to be essential role regulation of gene expression (Bartel, 2004) (Bartel, 2009; O’Connell et al., 2008) (Lim et al., 2005; Valencia-Sanchez et al., 2006), thereby influencing cellular functions and altering biological processes such as cell growth and proliferation, death (apoptosis), development, and differentiation (Jovanovic and Hengartner, 2006) in numerous species, including humans (Lee et al., 1993). Figure 3 demonstrates the series of intracellular events that generate miRNAs.
It is believed that a single miRNA can regulate expression levels of hundreds of genes (Shivdasani, 2006). Regulation and deregulation of cellular functions by miRNAs may result in development of normal cells into diseased cells (Gregory and Shiekhattar, 2005; McManus, 2003) leading to the development and progression of human diseases (Urbich et al., 2008). miRNAs play an important role in immune response (O’Connell et al., 2008), cell-cycle control (Croce and Calin, 2005), metabolism (Krützfeldt and Stoffel, 2006), and cell development (Cheng et al., 2005). It is likely that miRNA expression or function is significantly changed in many disease states, including cancer, inflammatory diseases, and metabolic diseases (Keller et al., 2009)(Krützfeldt and Stoffel, 2006).
In addition, miRNAs play an essential role in normal and abnormal development of the eye, like miR204 involvement in microphthalmia (Conte et al., 2010), cornea (Ryan et al., 2006), and retina (Li et al., 2009) (Vecino et al., 2004) (Walker and Harland, 2009) (Hackler et al., 2010) (Yan et al., 2010). In the retina, miRNAs are involved in regulation of retinal cell differentiation (Arora et al., 2010; Decembrini et al., 2009), including neuronal and retinal ganglion cell (RGC) development (Baudet et al., 2012). While it has been proposed that deregulation of miRNAs may induce retinal degeneration, manipulation of miRNAs expression levels may serve as a therapeutic target for degenerative retinal diseases, in addition to various eye diseases (Sundermeier and Palczewski, 2012). Table 4 outlines the various miRNA-profiling studies with the predominant miRNA expression associated with specific retinal disorders. Below, we discuss the potential of circulating miRNAs as biomarkers for AMD screening and monitoring.
Table 4.
miRNA Profiling Studies
| Reference | Tissue source | Disease Model | Predominant miRNA Expression |
|---|---|---|---|
| (Szemraj et al., 2015) | Human | AMD | miR-661, miR-3121, miR-4258, miR-889 |
| (Lukiw et al., 2012) | Human | AMD | miR-9, miR-125b, miR-146a, miR-155 |
| (Lin et al., 2011) | Human | macular RPE cells from AMD eyes | miR-23a |
| (Kutty et al., 2010b) | Human RPE cell (line ARPE-19) | 4HPR-induced apoptosis | miR-9, miR-16, miR-26b, miR-23a, and miR-15b |
| (Kutty et al., 2010a) | HRPE cells | HRPE cells exposed to inflammatory cytokine | miR-155 |
| (Zhu et al., 2011) | Mice | light-induced retinal degeneration | miR-96, miR-182, miR-183 |
| (Bai et al., 2011) | Mice | ischemia-induced retinal neovascularization | miR-126 |
| (Shen et al., 2008) | Mice | ischemia-induced retinal neovascularization (NV) | miR-106a, miR-146, miR-181, miR-199a, miR-214, miR-424, miR-451, miR-31, miR-150, miR-184 |
| (Zhou et al., 2011) | Mice | laser-induced choroidal neovascularization | miR-23, miR-27 |
| (Ishida et al., 2011) | Mice | experimental autoimmune uveoretinitis (EAU) | miR-142-5p, miR-21, miR-182 |
| (Loscher et al., 2007) | Mice | mouse model of RP | miR-1, miR-133, miR-96, miR-183 |
| (Kovacs et al., 2011) | Rats | (STZ)-induced diabetic retinopathy | miR-146 |
| (Wu et al., 2012) | Rats | (STZ)-induced diabetic retinopathy | miR-182, miR-96, miR-183, miR-211, miR-204, and miR-124miR-10b, miR-10a, miR-219-2-3p, miR-144, miR-338, miR-199a-3p |
HRPE =Human retinal pigment epithelial cells, STZ =Streptozotocin, 4HPR =N-(4-hydroxyphenyl)-retinamide, RP= retinitis pigmentosa.
6.4.1 miRNAs in AMD
While the research on circulating miRNAs associated with AMD is still in its infancy, a few individual miRNAs have been shown to play a functional role in AMD development and progression. miRNAs play important roles in several pathophysiological processes, including immune and inflammatory response, pathological angiogenesis and the response to oxidative stress, all of which have been suggested to be associated with AMD pathogenesis and progression (Wang et al., 2012a). Figure 4 depicts possible effects of miRNAs on AMD development.
Urbich, et al. has shown that miRNAs are involved in vascular biology (Urbich et al., 2008). Zhou et al. (2011) demonstrated that endothelial cells and vascularized tissues are highly enriched with miRNAs encoded by the miR-23, 27, and 24 gene clusters (Zhou et al., 2011). Inhibition of miR-23 and miR-27 may serve to repress both angiogenesis in vitro and postnatal retinal vascular development in vivo. It has been speculated that manipulation of miR-23/27 levels could provide a novel therapeutic target for patients with vascular disorders, such as neovascular AMD (Nakamura et al., 1999). Lin et al. demonstrated that miR-23a expression was significantly downregulated in macular RPE cells from AMD eyes. RPE cells are also protected against oxidative damage by miR-23a, which may also provide a potential therapeutic target in retinal degenerative diseases (Lin et al., 2011). Others have demonstrated that intraocular injection of pre-miR-31, −150, and −184 in mice considerably reduces ischemia-induced retinal neovascularisation (NV), and injection of pre-miR-31 and −150 reduced levels of choroidal NV (Shen et al., 2008). These data propose that changing of miRNA levels contributes to both retinal and choroidal neovascularization. Alteration of various gene product levels in the angiogenesis cascade and their function may be controlled by miRNAs, which have the ability to self-regulate and control their own expression. As such manipulation of miRNAs may serve as an effective therapeutic approach for future AMD treatment.
Polisena et al (2006) revealed that in-vitro angiogenesis may be regulated by miRNAs and their receptors which target angiogenic factors such as VEGF, bFGF, HGF, and SCF (Poliseno et al., 2006). Another study showed that miR-23b, miR-23a, miR-let-7a, miR-let-7b, miR-125b, miR-221, miR-222, miR-125a, miR-24, miR-106a, miR-20, miR-16, miR-let-7c, miR-103, and miR-133a could possibly target some of these angiogenic factors. Whereas, miR-221 and miR-222 regulate specifically the angiogenic activity of stem cells, it has been demonstrated that vascular integrity and angiogenesis of endothelial cells is also regulated by miRNAs (Fish et al., 2008). miR-126 directly represses negative regulators of VEGF pathway, supporting its role to regulate vascular integrity and angiogenesis. miR-126 knockdown in zebrafish leads to loss of blood vessels integrity and bleeding during embryonic development (Fish et al., 2008). These results suggest miR-126 may be an additional, new therapeutic target.
The final stage of dry AMD, geographic atrophy (GA), is a result of RPE degradation. A key enzyme in microRNA processing, DICER, is depleted in GA; furthermore, conditional knockdown of DICER in mice has been shown to cause RPE degeneration. The mechanism of degeneration is linked to small retrotransposons known as Alu elements. Alu elements are short interspersed repeats, or SINES, which make up 10% of the human genome, mostly shared with other primates, indicating a relatively distant evolutionary origin. Retrotransposons are mobile “junk” or parasitic DNA elements that are transcribed as RNA, and in limited circumstances when reverse-transcribed into DNA again, can re-insert in random genomic locations, sometimes resulting in deleterious mutations. Their transcription is maintained under strict cellular controls, probably mediated via DICER. Alu RNA is increased in the RPE of patients with GA; meanwhile, treatment of DICER ablated mice with Alu antisense RNA prevents RPE degeneration (Kaneko et al., 2011). DICER ablation, or Alu RNA exposure activates the NLRP3 inflammasome, while inhibition of inflammasome components or downstream signalling inhibits RPE degradation caused by DICER depletion or Alu exposure (Dridi et al., 2012). Alu exposure or DICER depletion also results in increased ERK1/2 phosphorylation, which is integral to pathogenesis; inhibition of this signalling pathway arrested RPE degradation. ERK1/2 phosphorylation occurs secondary to IL18 signalling, which results from activation of IL18 by the NLRP3 inflammasome (Dridi et al., 2012). Elucidation of this pathogenic mechanism is of academic importance in assigning DICER a role in cellular RNA policing, above and beyond its function in miRNA activation (Kaneko et al., 2011). This is also of importance to the treatment of AMD as a variety of pharmacological targets can now be investigated (Dridi et al., 2012).
6.4.2 Summary of miRNAs in AMD
miRNA may serve as valuable target for future investigation both in AMD pathogenesis, monitoring, and even treatment avenues, although current evidence supporting association and role of various miRNAs with AMD is limited and in preliminary stages. Additional research is necessary to not only confirm these preliminary findings, but also elucidate the exact role miRNAs may serve as potential biomarkers in a clinical setting.
6.5 Summary of Biomarkers of Disease & Progression
In summary, recent advancements in proteomics, metabolomics, miRNA profiling, in addition to serum studies, point toward multiple potential biomarkers for AMD, which may one day be used in concert for powerful disease screening and surveillance. Technical obstacles (assay development, developing of statistical parameters, etc.) discussed in section 7 must be overcome before applying these promising biomarkers for the whole population or at least for individuals at risk for AMD.
7. Process of Biomarker Development and Validation
Although a multitude of potential biomarkers have been identified, no biomarkers are currently being used to screen, guide clinical management, or predict disease outcomes or response to treatment in patients with AMD. This vast disparity of identified potential biomarkers and their actual clinical use is not specific to AMD as a disease, nor ophthalmology as a field of medicine. In oncologic medicine, biomarkers have been consistently used to screen for early malignancies, develop a differential diagnosis, guide treatment decisions, measure response to therapy, as well as monitor for recurrent disease (Duffy et al., 2015). As new oncologic biomarkers are being consistently and aggressively pursued, several theoretical frameworks for biomarker development, validation, and clinical use have been articulated (Duffy et al., 2015; Moons et al., 2009; Pavlou et al., 2013; Taylor et al., 2008). Building upon previously established frameworks, we have described our own process of biomarker validation and development in a six-step process (Figure 5):
Identification of potential biomarker
Development of assay to accurately test or measure biomarker
Intrinsic validation of the biomarker
Determination of statistical parameters of biomarker in clinical setting
Determination of tests effect on patient outcomes
Approval of test through governing regulatory agency
7.1 Identification of Potential Biomarker
This review has addressed numerous biomarkers that have been identified and associated with disease risk-modulation, progression of disease, and response to treatment, in patients with AMD. Environmental, genetic, proteomic, and other cellular biomarkers have been identified to date. Identification of these biomarkers in laboratory settings is the first step in the process. Ensuring identified biomarkers are sensitive and valid for AMD is also important, and this may often require additional investigation, particularly where the quality of evidence on potential biomarkers is weak or contradictory.
7.2 Development of Assay to Accurately Test or Measure Biomarker
Once a sensitive and valid biomarker has been identified, a corresponding assay must be developed to both accurately and reliably measure the marker. Assays must be able to accurately and reliably measure biomarkers to allow for their routine use and distribution into practice at clinical laboratories (Pavlou et al., 2013).
7.3 Intrinsic Validation of the Biomarker
Once an assay has been developed to measure a particular biomarker, the test must undergo strict analytical validation. This involves determining the various intrinsic statistical properties and parameters of the test including: accuracy, precision, repeatability, reproducibility, analytical specificity and sensitivity, limit of detection, linearity, and robustness (Duffy et al., 2015). Assays must undergo validation by comparison to their associated gold standard measurement tools (i.e. ELISA for protein, DNA sequencing for genes, Transcription-PCR or microarray for mRNA, etc.) (Pavlou et al., 2013). This step is vital to ensure that the biomarker test has the necessary statistical properties to accurately influence clinical decision-making.
7.4 Determination of Statistical Parameters of Biomarker in Clinical Setting
After an assay has been intrinsically validated, the test must be applied in a clinical setting. Clinical validation involves the process of describing both the clinical accuracy and efficacy of a given test (Duffy et al., 2015). During this process, statistical measurements of the test’s performance are developed, including: sensitivity, specificity, positive predictive and negative predictive values, likelihood ratio, and receiver operating characteristic (ROC) analysis (Duffy et al., 2015; Moons et al., 2009).
To continue past this stage of development, novel biomarkers are compared to current tests to determine the added value to current measurement techniques. Put simply by Duffy et al., a novel biomarker must address one of the following questions:
Does it address a currently unmet need in disease detection or management? (Duffy et al., 2015)
Does it provide a “clinical advantage” over existing biomarkers in terms of cost, simplicity, accuracy, or rapidity? (Duffy et al., 2015)
Determining the added value of a biomarker compared to current measurement techniques helps determine whether there is clinical utility for its development (Duffy et al., 2015).
7.5 Determination of Test’s Effect on Patient Outcomes
However, determining and validating the intrinsic and clinical statistical measures of a biomarker assay is insufficient in justifying its immediate transition into the clinical arena. After intrinsic validity and clinical utility have been established, the test must now demonstrate the ability to impact and improve outcomes in clinical practice (Duffy et al., 2015). This stage of the development and validation is the most financially burdensome and time intensive stage of the process (Pavlou et al., 2013). Controlled, randomized clinical trials addressing specific end-points are required to accurately articulate the effect of a particular biomarker on patient outcomes (i.e. mortality, morbidity, etc.). This process can take years and decades before accuracy, efficacy, and safety have been established through well-designed trials.
7.6 Approval of Test By Governing Regulatory Agency
After demonstrating that a biomarker assay has a favorable impact on patient outcomes, approval by federal regulatory agencies is the last and final step in the development process. These governing bodies include the Food Drug Administration (FDA) in the U.S. and meeting certain manufacturing, marketing, safety, and legal criteria in Europe, qualifying for the Conformité Européenne (CE) mark. (Duffy et al., 2015). Once approval has been granted, use of the biomarker assay can commence in clinical practice.
7.7 Biomarker Development and Validation Summary
The process of biomarker development and validation is both financially burdensome and time-intensive. However, this tedious process is necessary to ensure that only accurate and efficacious biomarkers are permitted in clinical settings. Developers of biomarker assays for AMD should bear in mind that such assays are useful only to the extent that they can influence clinical management (Cammann et al., 2011; Pepe et al., 2004; Sutcliffe et al., 2009). Currently, no biomarkers are being used for detection or management of AMD. However, a deepened understanding of the theoretical framework for biomarker development and validation can help guide researchers and clinicians into a future of biomarker application in clinical medicine. A summary of current AMD biomarkers and list factors and where each is along the pathway to clinical implementation is found in Figure 6.
8. Future Prospects and Challenges
Further identification of causative AMD genotypes with prospective randomized clinical trials will help refine genomic profiles and potentially lay the foundation for development of targeted genetic therapies targeting high-risk genes in the RPE, choroid, and outer retina. Although it might seem otherwise, routine genotyping certainly carries significant risks.
There are definite risks associated with routine genetic testing of patients with AMD. Dr. Edwin Stone in a recent JAMA article categorized these as decision-making risks, psychological risks, and economic risks (Stone, 2015). Decision-making risks involve patients making unwise decisions based upon results of genetic studies. For example, a patient with strong family history AMD may defer close ophthalmologic monitoring because of testing that shows he has a low-risk genotype (Stone, 2015). Additionally, as positive genetic tests do not guarantee development of a disease, patients with unfavorable genetic testing results may have increased levels of unnecessary anxiety and make life-changing decisions based upon these results. Finally, genetic testing can be financially burdensome and any increase in allocation of resources towards genetic testing siphons funds away from other vital services, such as regular visits to a clinical ophthalmologist (Stone, 2015). Currently, the American Academy of Ophthalmology has made the following recommendation: “Avoid routine genetic testing for genetically complex disorders like age-related macular degeneration…until specific treatment or surveillance strategies have been shown in one or more published clinical trials to be of benefit to individuals with specific disease-associated genotypes. In the meantime, ophthalmologists should confine the genotyping of such patients to research studies” (Stone et al., 2012). Despite this, products such as RetnaGene and others continue to be marketed to ophthalmic practice with little, if any clinical utility.
With greater and more efficient abilities to identify the genetic underpinnings of disease and treatment response, a greater focus on personalized medicine must be emphasized. A greater understanding of how an individual’s genetic makeup will influence their response to current AMD treatments can help determine the outcome and cost-effectiveness of such treatments. As research improves treatment for AMD, biomarkers will allow for earlier detection of AMD cases and at-risk AMD patients, allowing for better prognoses and a decrease in vision loss for age-related macular degeneration patients in the future. At this current time, biomarkers remain as a research tool but are on the precipice of emergence into clinical practice. Genetic testing and gene therapies may be the future for screening and treatment of AMD. Current technological advancements have made genetic testing a valuable screening tool for a variety of diseases and a similar approach may be applied to AMD in the near future. One current drawback to genetic testing is the expensive nature of the tests make them inaccessible for many patients, especially the uninsured and underinsured. Development of non-genetic biomarkers as a screening tool could help to identify at-risk individuals who would most likely benefit from costly genetic testing. The possibilities and benefits of using biomarkers as surveillance for AMD are endless, and hopefully will one day be a part of a screening regimen that will help identify those at risk of developing this devastating disease and/or a monitoring regimen for patients undergoing treatment. While current genetic biomarker testing may not be recommended for patients at this time, knowing the susceptibilities, environmental exposures, demographic risk factors, and possibilities for future biomarkers related to the development of AMD is well within the reach of both patients and practicing clinicians. Patient education on risk factors and protective behaviors associated with AMD can lead to prevention and identification of earlier stages of the disease and better outcomes for patients.
9. Conclusion
The risk factors for AMD (both genetic and environmental) lead to inflammation and anatomical changes and dysfunction, which can eventually lead to choroidal neovascularization and geographic atrophy with eventual loss of vision. These changes and the role of the different proposed biomarkers and risk factors are outlined in Figure 7. More research is required to validate previous studies suggesting clinical efficacy for many of the biomarkers listed. We hope that our summary and discussion of several potential biomarkers of AMD may serve as a framework for future studies. We anticipate that many of these biomarkers will be used to diagnose, monitor, and prognosticate AMD in the future.
Highlights.
Biomarkers can be used to predict disease incidence, identify at risk individuals, elucidate causative pathophysiological etiologies, guide screening, monitoring and treatment parameters, and predict disease outcome.
The process of biomarker development has been outlined, from biomarker identification to clinical implementation.
Soluble FMS-like tyrosine kinase-1 (sFlt-1), also known as soluble vascular endothelial growth factor receptor-1 (sVEGFR-1), has been identified as a novel, and perhaps the most clinically significant, serum biomarker for identifying patients at risk for neovascular AMD
Summary of current risk factors and potential biomarkers as well as identifying which stage in the biomarker development process each is currently situated.
Acknowledgments
Supported in part by an Unrestricted Grant from Research to Prevent Blindness, a Diabetes Metabolism Consortium Training Grant, and a National Institutes of Health Diabetes T32 Training Grant. Financial support was also provided by the Irish Research Council for Science, Engineering & Technology/EMBARK Initiative, Mater Vision Institute, Libyan Ministry of Higher Education and Scientific Research, Medical Charities Research Group (MRCG), Health Research Board (HRB), Science Foundation Ireland (SFI), Fighting Blindness Ireland, the Macular Society, and an Irish Research Council (IRD) studentship. An additional generous thanks to Dr. Ann Milam, Scheie Eye Institute, University of Pennsylvania for contribution of clinical images (Figure 1).
Appendix
Figure 1. Forms of AMD, (see appendix).
Figure 1.1: Normal Human Retina
A) Posterior pole view of a normal human retina. The fovea is arrowed. B) Immunofluorescent image of macular cones (labelled red), cell nuclei are blue (DAPI) and RPE (orange) due to autofluorescence. NR: neuroretina, RPE: retinal pigment epithelium (Courtesy of Dr Ann Milam, Scheie Eye Institute, University of Pennsylvania Philadelphia, USA)
Figure 1.2: Drusen in AMD
A) Fundus view of a retina with drusen (white deposits). B) Section of retina showing confluent drusen (*) near optic nerve head. Astrocytes, positive for glial fibrillary acidic protein (GFAP), define the nerve fibre layer (NFL). The cones (red) in the photoreceptor layer (PRL) are shortened and decreased in number over the drusen. The RPE are autofluorescent (orange) due to lipofuscin build up. Cell nuclei are labelled blue. There is loss of cone cells (red) adjacent to drusen. The RPE (orange) is thinned and abnormal over the drusen. (Courtesy of Dr Ann Milam, Scheie Eye Institute, University of Pennsylvania, Philadelphia, USA)
Figure 1.3: Geographic Atrophy
A) Fundal view of a patient with geographic atrophy, note the pale areas of retinal atrophy (arrow). B) Section of retina showing RPE cell loss (arrowed) with overlying loss of photoreceptors (PRL) at the macula. This area is replaced by intra-retinal glial tissue (anti-GFAP: green) (Courtesy of Dr Ann Milam, Scheie Eye Institute, University of Pennsylvania, Philadelphia, USA)
Figure 1.4: Advanced Neovascular Age-related Macular Degeneration
A) Fundal view of patient with advanced age related macular degeneration. A disciform scar is located at the macula. B) A immunofluorescence image of a macula with AMD, rods (red) surround an area of RPE and photoreceptor cell loss, replaced by a sub-retinal and intra-retinal glial (anti-GFAP: green) scar. (Courtesy of Dr Ann Milam, Scheie Eye Institute, University of Pennsylvania, Philadelphia, USA)
Figure 2. Complement Cascade (see appendix).
Simplified summary of the alternative complement cascade. Diagram shows the stimulatory role of complement factors B (CFB) and D (CFD), in opposition to the inhibitory role of complement factor H (CFH). The end result of this cascade is the formulation of the membrane attack complex (MAC), which leads to cell lysis and death.
Figure 3. miRNA Processing Pathway (see appendix).
miRNA-processing pathway. miRNAs are generated through a series of cleavage events, which take place in the nucleus and the cytoplasm, forming pri-miRNA and pre-miRNA before forming the mature structure. Hairpin miRNA originates from precursor miRNA within the nucleus as part of a longer primary transcript. At this stage, the pri-miRNA includes the characteristic 5′ 7-methyl guanosine cap and a 3′ poly-A tail, believed to originate from RNA polymerase II activity (Zeng et al., 2006). Pre-miRNA formation also occurs within the nucleus. The microprocessor complex containing the enzymes Drosha, RNase III-like enzyme, and its co-factor DGCR-8, (double-stranded RNA binding protein) all contribute to miRNAs processing. Drosha is comprised of two RNase II domains. Of these, one is believed to cleave the 3′ end of the pre-miRNA while the 5′ end is cleaved by the other domain (Jha et al., 2011). Hairpin release occurs in the nucleus, and then the miRNA is exported to cytoplasm. Dicer enzyme processing occurs, followed by strand selection by RISC (Winter et al., 2009). (RISC= RNA-induced silencing complex, pre-miRNA= Precursor microRNA, pri-miRNA = Primary microRNA)
Figure 4. Effects of microRNA on AMD Development (see appendix).
Simplified diagram depicting the role of microRNA on AMD development. microRNAs regulate different growth and angiogenic factors as well as various elements influencing inflammation, leading to many of the processes responsible for age-related macular degeneration.
Figure 5. Biomarker Development (see appendix).
Process of biomarker development, from inception to clinical implementation.
Figure 6. Biomarker Progress to Clinical Use (see appendix).
Table of biomarkers and their progression in stages of developmental process. The most promising genetic biomarkers include Complement Factor H (CFH) and Age-Related Maculopathy Susceptibility 2 (ARMS2). Patients possessing certain CFH or ARMS2 genetic SNPs have shown positive outcomes after taking AREDs supplements compared to controls. Several complement associated genes, ApoE genes, and genes relating to HTRA1, have been used in clinical predictive models of AMD (Klein et al., 2011). Most of the risk factors and potential biomarkers listed have recently been identified, and future research should be focused on confirming their association with AMD and elucidating their role as a viable marker for AMD. To date, there have been no official randomized clinical trials showing any significant correlation between a specific biomarker and patient outcomes.
Figure 7. Diagnostic and Prognostic Biomarkers for AMD (see appendix).
Summary of development and progression of AMD and depiction of potential biomarkers to predict susceptibility, progression, and prognosis. As individuals age, they experience increased oxidative stress, which can result in elevated homocysteine levels in plasma. As RPE function worsens, various events occur such as mitochondrial DNA (mt.DNA) deletion and rearrangement (such as in ARMS2), increase in lipid oxidation and its end products (i.e. carboxyethypyrrole (CEP)). Proteins begin to break down and are glycated, resulting in elevated levels of advanced glycation end products, such as carboxymethyllysine and pentosidine. Retinal metabolic dysfunction occurs leading to phagocytosis of photoreceptor outer segments. These events lead to inflammation, which can be noted by inflammatory cytokines such as CRP and IL-6. Various mutations in key regulatory complement cascade proteins (CFH, CFB, C2, C3) lead to activation and dysregulation of the complement system, further increasing inflammation and leading to damage of the RPE. Damage to macular RPE and photoreceptor cells results in AMD, either through geographic atrophy or choroidal neovascularisation. Serum IP-10 elevation is seen in early dry-AMD. Changes in serum lipoprotein levels and cholesterol have been associated with geographic atrophy and CNV, while increased levels of Alu RNA in RPE cells is seen mainly in geographic atrophy. Elevated fibrinogen levels can be seen in late-AMD. Increased angiogenesis and worsening CNV can be seen with elevated levels of various factors (VEGF, CML, IL-8, CEP) and expression of certain high-risk genotypes (CFH, HTRA1, ARMS2).
Footnotes
The authors have no proprietary interests or conflicts
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Nathan G. Lambert, Email: Nathan.lambert@hsc.utah.edu.
Malkit K. Singh, Email: mona.singh@hsc.utah.edu.
Hanan ElShelmani, Email: elshelh@tcd.ie.
Fiona C. Mansergh, Email: mansergf@tcd.ie.
Michael A. Wride, Email: wridem@tcd.ie.
Maximilian Padilla, Email: maximilianrpadilla413@gmail.com.
David Keegan, Email: djkeegan71@hotmail.com.
Ruth E. Hogg, Email: r.e.hogg@qub.ac.uk.
Balamurali K. Ambati, Email: bala.ambati@utah.edu.
Bibliography
- Abdel-Magid AF. Factor D Inhibitors for the Treatment of AMD: Patent Highlight. ACS Med Chem Lett. 2012;3:781–782. doi: 10.1021/ml300233n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedi F, Wickremasinghe S, Richardson AJ, Makalic E, Schmidt DF, Sandhu SS, Baird PN, Guymer RH. Variants in the VEGFA gene and treatment outcome after anti-VEGF treatment for neovascular age-related macular degeneration. Ophthalmology. 2013;120:115–121. doi: 10.1016/j.ophtha.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Abu-Safieh L, Abboud EB, Alkuraya H, Shamseldin H, Al-Enzi S, Al-Abdi L, Hashem M, Colak D, Jarallah A, Ahmad H, Bobis S, Nemer G, Bitar F, Alkuraya FS. Mutation of IGFBP7 causes upregulation of BRAF/MEK/ERK pathway and familial retinal arterial macroaneurysms. American journal of human genetics. 2011;89:313–319. doi: 10.1016/j.ajhg.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams MK, Simpson JA, Aung KZ, Makeyeva GA, Giles GG, English DR, Hopper J, Guymer RH, Baird PN, Robman LD. Abdominal obesity and age-related macular degeneration. American journal of epidemiology. 2011;173:1246–1255. doi: 10.1093/aje/kwr005. [DOI] [PubMed] [Google Scholar]
- Adams MK, Simpson JA, Richardson AJ, English DR, Aung KZ, Makeyeva GA, Guymer RH, Giles GG, Hopper J, Robman LD, Baird PN. Apolipoprotein E gene associations in age-related macular degeneration: the Melbourne Collaborative Cohort Study. American journal of epidemiology. 2012;175:511–518. doi: 10.1093/aje/kwr329. [DOI] [PubMed] [Google Scholar]
- Adamus G, Chew EY, Ferris FL, Klein ML. Prevalence of anti-retinal autoantibodies in different stages of Age-related macular degeneration. BMC ophthalmology. 2014;14:154. doi: 10.1186/1471-2415-14-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study 2 Research G. Chew EY, SanGiovanni JP, Ferris FL, Wong WT, Agron E, Clemons TE, Sperduto R, Danis R, Chandra SR, Blodi BA, Domalpally A, Elman MJ, Antoszyk AN, Ruby AJ, Orth D, Bressler SB, Fish GE, Hubbard GB, Klein ML, Friberg TR, Rosenfeld PJ, Toth CA, Bernstein P. Lutein/zeaxanthin for the treatment of age-related cataract: AREDS2 randomized trial report no. 4. JAMA Ophthalmol. 2013;131:843–850. doi: 10.1001/jamaophthalmol.2013.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study Research G. Risk factors associated with age-related macular degeneration. A case-control study in the age-related eye disease study: Age-Related Eye Disease Study Report Number 3. Ophthalmology. 2000;107:2224–2232. doi: 10.1016/s0161-6420(00)00409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Age-Related Eye Disease Study Research G. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agosta E, Lazzeri S, Orlandi P, Figus M, Fioravanti A, Di Desidero T, Sartini MS, Nardi M, Danesi R, Bocci G. Pharmacogenetics of antiangiogenic and antineovascular therapies of age-related macular degeneration. Pharmacogenomics. 2012;13:1037–1053. doi: 10.2217/pgs.12.77. [DOI] [PubMed] [Google Scholar]
- Akagi-Kurashige Y, Yamashiro K, Gotoh N, Miyake M, Morooka S, Yoshikawa M, Nakata I, Kumagai K, Tsujikawa A, Yamada R, Matsuda F, Saito M, Iida T, Sugahara M, Kurimoto Y, Cheng CY, Khor CC, Wong TY, Yoshimura N Nagahama Cohort Research G. MMP20 and ARMS2/HTRA1 Are Associated with Neovascular Lesion Size in Age-Related Macular Degeneration. Ophthalmology. 2015;122:2295–2302. e2292. doi: 10.1016/j.ophtha.2015.07.032. [DOI] [PubMed] [Google Scholar]
- Akilov OE, Ustyugova IV, Zhi L, Hasan T, Wu MX. Enhanced susceptibility to Leishmania infection in resistant mice in the absence of immediate early response gene X-1. J Immunol. 2009;183:7994–8003. doi: 10.4049/jimmunol.0900866. [DOI] [PubMed] [Google Scholar]
- Allikmets R. Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. American journal of human genetics. 2000;67:487–491. doi: 10.1086/303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A, Dean M, Lupski JR, Leppert M. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science. 1997a;277:1805–1807. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- Allikmets R, Singh N, Sun H, Shroyer NF, Hutchinson A, Chidambaram A, Gerrard B, Baird L, Stauffer D, Peiffer A, Rattner A, Smallwood P, Li Y, Anderson KL, Lewis RA, Nathans J, Leppert M, Dean M, Lupski JR. A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat Genet. 1997b;15:236–246. doi: 10.1038/ng0397-236. [DOI] [PubMed] [Google Scholar]
- Alonso A, Marsal S, Julia A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol. 2015;3:23. doi: 10.3389/fbioe.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati BK, Nozaki M, Singh N, Takeda A, Jani PD, Suthar T, Albuquerque RJ, Richter E, Sakurai E, Newcomb MT, Kleinman ME, Caldwell RB, Lin Q, Ogura Y, Orecchia A, Samuelson DA, Agnew DW, St Leger J, Green WR, Mahasreshti PJ, Curiel DT, Kwan D, Marsh H, Ikeda S, Leiper LJ, Collinson JM, Bogdanovich S, Khurana TS, Shibuya M, Baldwin ME, Ferrara N, Gerber HP, De Falco S, Witta J, Baffi JZ, Raisler BJ, Ambati J. Corneal avascularity is due to soluble VEGF receptor-1. Nature. 2006;443:993–997. doi: 10.1038/nature05249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati J, Anand A, Fernandez S, Sakurai E, Lynn BC, Kuziel WA, Rollins BJ, Ambati BK. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nature medicine. 2003;9:1390–1397. doi: 10.1038/nm950. [DOI] [PubMed] [Google Scholar]
- Ambati J, Fowler BJ. Mechanisms of age-related macular degeneration. Neuron. 2012;75:26–39. doi: 10.1016/j.neuron.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorini AM, Giorlandino C, Longo S, D’Urso S, Mesoraca A, Santoro ML, Picardi M, Gullotta S, Cignini P, Lazzarino D, Lazzarino G, Tavazzi B. Metabolic profile of amniotic fluid as a biochemical tool to screen for inborn errors of metabolism and fetal anomalies. Mol Cell Biochem. 2012;359:205–216. doi: 10.1007/s11010-011-1015-y. [DOI] [PubMed] [Google Scholar]
- Anand A, Sharma NK, Gupta A, Prabhakar S, Sharma SK, Singh R, Gupta PK. Single nucleotide polymorphisms in MCP-1 and its receptor are associated with the risk of age related macular degeneration. PloS one. 2012;7:e49905. doi: 10.1371/journal.pone.0049905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasopoulos E, Kakoulidou A, Coleman AL, Sinsheimer JS, Wilson MR, Yu F, Salonikiou A, Koskosas A, Pappas T, Founti P, Lambropoulos A, Topouzis F. Association of sequence variation in the CX3CR1 gene with geographic atrophy age-related macular degeneration in a Greek population. Curr Eye Res. 2012;37:1148–1155. doi: 10.3109/02713683.2012.705413. [DOI] [PubMed] [Google Scholar]
- Anderson DH, Ozaki S, Nealon M, Neitz J, Mullins RF, Hageman GS, Johnson LV. Local cellular sources of apolipoprotein E in the human retina and retinal pigmented epithelium: implications for the process of drusen formation. Am J Ophthalmol. 2001;131:767–781. doi: 10.1016/s0002-9394(00)00961-2. [DOI] [PubMed] [Google Scholar]
- Arakawa S, Takahashi A, Ashikawa K, Hosono N, Aoi T, Yasuda M, Oshima Y, Yoshida S, Enaida H, Tsuchihashi T, Mori K, Honda S, Negi A, Arakawa A, Kadonosono K, Kiyohara Y, Kamatani N, Nakamura Y, Ishibashi T, Kubo M. Genome-wide association study identifies two susceptibility loci for exudative age-related macular degeneration in the Japanese population. Nat Genet. 2011;43:1001–1004. doi: 10.1038/ng.938. [DOI] [PubMed] [Google Scholar]
- Ardeljan D, Chan CC. Aging is not a disease: distinguishing age-related macular degeneration from aging. Prog Retin Eye Res. 2013;37:68–89. doi: 10.1016/j.preteyeres.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora A, Guduric-Fuchs J, Harwood L, Dellett M, Cogliati T, Simpson D. Prediction of microRNAs affecting mRNA expression during retinal development. BMC Developmental Biology. 2010;10:1. doi: 10.1186/1471-213X-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora A, McKay GJ, Simpson DA. Prediction and verification of miRNA expression in human and rat retinas. Invest Ophthalmol Vis Sci. 2007;48:3962–3967. doi: 10.1167/iovs.06-1221. [DOI] [PubMed] [Google Scholar]
- Attebo K, Mitchell P, Smith W. Visual acuity and the causes of visual loss in Australia. The Blue Mountains Eye Study. Ophthalmology. 1996;103:357–364. doi: 10.1016/s0161-6420(96)30684-2. [DOI] [PubMed] [Google Scholar]
- Awh CC, Hawken S, Zanke BW. Treatment response to antioxidants and zinc based on CFH and ARMS2 genetic risk allele number in the Age-Related Eye Disease Study. Ophthalmology. 2015;122:162–169. doi: 10.1016/j.ophtha.2014.07.049. [DOI] [PubMed] [Google Scholar]
- Awh CC, Lane AM, Hawken S, Zanke B, Kim IK. CFH and ARMS2 genetic polymorphisms predict response to antioxidants and zinc in patients with age-related macular degeneration. Ophthalmology. 2013;120:2317–2323. doi: 10.1016/j.ophtha.2013.07.039. [DOI] [PubMed] [Google Scholar]
- Axer-Siegel R, Bourla D, Ehrlich R, Dotan G, Benjamini Y, Gavendo S, Weinberger D, Sela BA. Association of neovascular age-related macular degeneration and hyperhomocysteinemia. American Journal of Ophthalmology. 2004;137:84–89. doi: 10.1016/s0002-9394(03)00864-x. [DOI] [PubMed] [Google Scholar]
- Babanejad M, Moein H, Akbari MR, Badiei A, Yaseri M, Soheilian M, Najmabadi H. Investigating the CFH Gene Polymorphisms as a Risk Factor for Age-related Macular Degeneration in an Iranian Population. Ophthalmic genetics. 2015;21:1–6. doi: 10.3109/13816810.2014.955585. [DOI] [PubMed] [Google Scholar]
- Bai Y, Bai X, Wang Z, Zhang X, Ruan C, Miao J. MicroRNA-126 inhibits ischemia-induced retinal neovascularization via regulating angiogenic growth factors. Exp Mol Pathol. 2011;91:471–477. doi: 10.1016/j.yexmp.2011.04.016. [DOI] [PubMed] [Google Scholar]
- Baird PN, Guida E, Chu DT, Vu HT, Guymer RH. The epsilon2 and epsilon4 alleles of the apolipoprotein gene are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2004;45:1311–1315. doi: 10.1167/iovs.03-1121. [DOI] [PubMed] [Google Scholar]
- Baird PN, Richardson AJ, Robman LD, Dimitrov PN, Tikellis G, McCarty CA, Guymer RH. Apolipoprotein (APOE) gene is associated with progression of age-related macular degeneration (AMD) Hum Mutat. 2006;27:337–342. doi: 10.1002/humu.20288. [DOI] [PubMed] [Google Scholar]
- Barba I, Garcia-Ramirez M, Hernandez C, Alonso MA, Masmiquel L, Garcia-Dorado D, Simo R. Metabolic fingerprints of proliferative diabetic retinopathy: an 1H-NMR-based metabonomic approach using vitreous humor. Invest Ophthalmol Vis Sci. 2010;51:4416–4421. doi: 10.1167/iovs.10-5348. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudet ML, Zivraj KH, Abreu-Goodger C, Muldal A, Armisen J, Blenkiron C, Goldstein LD, Miska EA, Holt CE. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat Neurosci. 2012;15:29–38. doi: 10.1038/nn.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan NG, Molina MF, Gordon WC. Docosahexaenoic acid signalolipidomics in nutrition: significance in aging, neuroinflammation, macular degeneration, Alzheimer’s, and other neurodegenerative diseases. Annual review of nutrition. 2011;31:321–351. doi: 10.1146/annurev.nutr.012809.104635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beharry S, Zhong M, Molday RS. N-retinylidene-phosphatidylethanolamine is the preferred retinoid substrate for the photoreceptor-specific ABC transporter ABCA4 (ABCR) J Biol Chem. 2004;279:53972–53979. doi: 10.1074/jbc.M405216200. [DOI] [PubMed] [Google Scholar]
- Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- Bergeron-Sawitzke J, Gold B, Olsh A, Schlotterbeck S, Lemon K, Visvanathan K, Allikmets R, Dean M. Multilocus analysis of age-related macular degeneration. Eur J Hum Genet. 2009;17:1190–1199. doi: 10.1038/ejhg.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho H, Honda S, Kondo N, Kusuhara S, Tsukahara Y, Negi A. The association of CD36 variants with polypoidal choroidal vasculopathy compared to typical neovascular age-related macular degeneration. Mol Vis. 2012;18:121–127. [PMC free article] [PubMed] [Google Scholar]
- Bhutto IA, Baba T, Merges C, Juriasinghani V, McLeod DS, Lutty GA. C-reactive protein and complement factor H in aged human eyes and eyes with age-related macular degeneration. Br J Ophthalmol. 2011;95:1323–1330. doi: 10.1136/bjo.2010.199216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biomarkers Definitions Working G. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- Bird A. Age-related macular disease. Br J Ophthalmol. 1996;80:2–3. doi: 10.1136/bjo.80.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasiak J, Synowiec E, Salminen A, Kaarniranta K. Genetic variability in DNA repair proteins in age-related macular degeneration. Int J Mol Sci. 2012;13:13378–13397. doi: 10.3390/ijms131013378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenkranz MS, Russell SR, Robey MG, Kott-Blumenkranz R, Penneys N. Risk factors in age-related maculopathy complicated by choroidal neovascularization. Ophthalmology. 1986;93:552–558. doi: 10.1016/s0161-6420(86)33702-3. [DOI] [PubMed] [Google Scholar]
- Bourla DH, Young TA. Age-related macular degeneration: a practical approach to a challenging disease. J Am Geriatr Soc. 2006;54:1130–1135. doi: 10.1111/j.1532-5415.2006.00771.x. [DOI] [PubMed] [Google Scholar]
- Boushey CJ, Beresford SA, Omenn GS, Motulsky AG. A quantitative assessment of plasma homocysteine as a risk factor for vascular disease. Probable benefits of increasing folic acid intakes. JAMA. 1995;274:1049–1057. doi: 10.1001/jama.1995.03530130055028. [DOI] [PubMed] [Google Scholar]
- Brantley MA, Jr, Fang AM, King JM, Tewari A, Kymes SM, Shiels A. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to intravitreal bevacizumab. Ophthalmology. 2007;114:2168–2173. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Bressler SB, Munoz B, Solomon SD, West SK. Racial differences in the prevalence of age-related macular degeneration: the Salisbury Eye Evaluation (SEE) Project. Arch Ophthalmol. 2008;126:241–245. doi: 10.1001/archophthalmol.2007.53. [DOI] [PubMed] [Google Scholar]
- Buch H, Vinding T, la Cour M, Jensen GB, Prause JU, Nielsen NV. Risk factors for age-related maculopathy in a 14-year follow-up study: the Copenhagen City Eye Study. Acta Ophthalmol Scand. 2005;83:409–418. doi: 10.1111/j.1600-0420.2005.00492.x. [DOI] [PubMed] [Google Scholar]
- Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammann H, Jung K, Meyer HA, Stephan C. Avoiding pitfalls in applying prediction models, as illustrated by the example of prostate cancer diagnosis. Clin Chem. 2011;57:1490–1498. doi: 10.1373/clinchem.2011.166959. [DOI] [PubMed] [Google Scholar]
- Campbell M, Doyle SL. An eye on the future of inflammasomes and drug development in AMD. J Mol Med (Berl) 2013;91:1059–1070. doi: 10.1007/s00109-013-1050-0. [DOI] [PubMed] [Google Scholar]
- Carugati A, Pappalardo E, Zingale LC, Cicardi M. C1-inhibitor deficiency and angioedema. Mol Immunol. 2001;38:161–173. doi: 10.1016/s0161-5890(01)00040-2. [DOI] [PubMed] [Google Scholar]
- Cha DM, Woo SJ, Kim HJ, Lee C, Park KH. Comparative analysis of aqueous humor cytokine levels between patients with exudative age-related macular degeneration and normal controls. Invest Ophthalmol Vis Sci. 2013;54:7038–7044. doi: 10.1167/iovs.13-12730. [DOI] [PubMed] [Google Scholar]
- CATT Research Group. Martin DF, Maguire MG, Ying GS, Grunwald JE, Fine SL, Jaffe GJ. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N Engl J Med. 2011;364:1897–1908. doi: 10.1056/NEJMoa1102673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy U, Wong TY, Fletcher A, Piault E, Evans C, Zlateva G, Buggage R, Pleil A, Mitchell P. Clinical risk factors for age-related macular degeneration: a systematic review and meta-analysis. BMC ophthalmology. 2010;10:31. doi: 10.1186/1471-2415-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MA, Bressler SB, Munoz B, West SK. Racial differences and other risk factors for incidence and progression of age-related macular degeneration: Salisbury Eye Evaluation (SEE) Project. Invest Ophthalmol Vis Sci. 2008;49:2395–2402. doi: 10.1167/iovs.07-1584. [DOI] [PubMed] [Google Scholar]
- Chen H, Yu KD, Xu GZ. Association between variant Y402H in age-related macular degeneration (AMD) susceptibility gene CFH and treatment response of AMD: a meta-analysis. PloS one. 2012;7:e42464. doi: 10.1371/journal.pone.0042464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, Tosakulwong N, Pericak-Vance MA, Campochiaro PA, Klein ML, Tan PL, Conley YP, Kanda A, Kopplin L, Li Y, Augustaitis KJ, Karoukis AJ, Scott WK, Agarwal A, Kovach JL, Schwartz SG, Postel EA, Brooks M, Baratz KH, Brown WL, Brucker AJ, Orlin A, Brown G, Ho A, Regillo C, Donoso L, Tian L, Kaderli B, Hadley D, Hagstrom SA, Peachey NS, Klein R, Klein BE, Gotoh N, Yamashiro K, Ferris F, III, Fagerness JA, Reynolds R, Farrer LA, Kim IK, Miller JW, Corton M, Carracedo A, Sanchez-Salorio M, Pugh EW, Doheny KF, Brion M, Deangelis MM, Weeks DE, Zack DJ, Chew EY, Heckenlively JR, Yoshimura N, Iyengar SK, Francis PJ, Katsanis N, Seddon JM, Haines JL, Gorin MB, Abecasis GR, Swaroop A Complications of Age-Related Macular Degeneration Prevention Trial Research G. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zeng J, Zhao C, Wang K, Trood E, Buehler J, Weed M, Kasuga D, Bernstein PS, Hughes G, Fu V, Chin J, Lee C, Crocker M, Bedell M, Salasar F, Yang Z, Goldbaum M, Ferreyra H, Freeman WR, Kozak I, Zhang K. Assessing susceptibility to age-related macular degeneration with genetic markers and environmental factors. Arch Ophthalmol. 2011;129:344–351. doi: 10.1001/archophthalmol.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Research. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew EY, Klein ML, Clemons TE, Agron E, Abecasis GR. Genetic testing in persons with age-related macular degeneration and the use of the AREDS supplements: to test or not to test? Ophthalmology. 2015;122:212–215. doi: 10.1016/j.ophtha.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CJ, Conley YP, Gorin MB, Gensler G, Lai CQ, Shang F, Taylor A. Associations between genetic polymorphisms of insulin-like growth factor axis genes and risk for age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:9099–9107. doi: 10.1167/iovs.11-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury F, Varma R, McKean-Cowdin R, Klein R, Azen SP Los Angeles Latino Eye Study G. Risk factors for four-year incidence and progression of age-related macular degeneration: the los angeles latino eye study. Am J Ophthalmol. 2011;152:385–395. doi: 10.1016/j.ajo.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen WG, Cook NR, Ridker PM, Buring JE. Prospective study of plasma homocysteine level and risk of age-related macular degeneration in women. Ophthalmic epidemiology. 2015;22:85–93. doi: 10.3109/09286586.2015.1012272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill AJ, Carter JG, Lovell HC, Ramsden C, Turner SJ, Yeung A, Escardo J, Atan D. VEGF polymorphisms are associated with neovascular age-related macular degeneration. Human molecular genetics. 2006;15:2955–2961. doi: 10.1093/hmg/ddl238. [DOI] [PubMed] [Google Scholar]
- Ciulla TA, Harris A, Martin BJ. Ocular perfusion and age-related macular degeneration. Acta Ophthalmol Scand. 2001;79:108–115. doi: 10.1034/j.1600-0420.2001.079002108.x. [DOI] [PubMed] [Google Scholar]
- Clemons TE, Milton RC, Klein R, Seddon JM, Ferris FL., 3rd Risk factors for the incidence of Advanced Age-Related Macular Degeneration in the Age-Related Eye Disease Study (AREDS) AREDS report no. 19. Ophthalmology. 2005;112:533–539. doi: 10.1016/j.ophtha.2004.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colak E, Kosanovic-Jakovic N, Zoric L, Radosavljevic A, Stankovic S, Majkic-Singh N. The association of lipoprotein parameters and C-reactive protein in patients with age-related macular degeneration. Ophthalmic Res. 2011;46:125–132. doi: 10.1159/000323815. [DOI] [PubMed] [Google Scholar]
- Coleman HR, Chan CC, Ferris FL, III, Chew EY. Age-related macular degeneration. The Lancet. 2008;372:1835–1845. doi: 10.1016/S0140-6736(08)61759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combadiere C, Feumi C, Raoul W, Keller N, Rodero M, Pezard A, Lavalette S, Houssier M, Jonet L, Picard E, Debre P, Sirinyan M, Deterre P, Ferroukhi T, Cohen SY, Chauvaud D, Jeanny JC, Chemtob S, Behar-Cohen F, Sennlaub F. CX3CR1-dependent subretinal microglia cell accumulation is associated with cardinal features of age-related macular degeneration. J Clin Invest. 2007;117:2920–2928. doi: 10.1172/JCI31692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congdon N, O’Colmain B, Klaver CC, Klein R, Munoz B, Friedman DS, Kempen J, Taylor HR, Mitchell P Eye Diseases Prevalence Research G. Causes and prevalence of visual impairment among adults in the United States. Arch Ophthalmol. 2004;122:477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P, Banfi S. miR-204 is required for lens and retinal development via Meis2 targeting. Proceedings of the National Academy of Sciences. 2010;107:15491–15496. doi: 10.1073/pnas.0914785107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coral K, Raman R, Rathi S, Rajesh M, Sulochana KN, Angayarkanni N, Paul PG, Ramakrishnan S. Plasma homocysteine and total thiol content in patients with exudative age-related macular degeneration. Eye (Lond) 2006;20:203–207. doi: 10.1038/sj.eye.6701853. [DOI] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, Cancer, and Stem Cell Division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Cruickshanks KJ, Klein R, Klein BEK. Sunlight and Age-Related Macular Degeneration: The Beaver Dam Eye Study. Arch Ophthalmol. 1993;111:514–518. doi: 10.1001/archopht.1993.01090040106042. [DOI] [PubMed] [Google Scholar]
- Daniele LL, Sauer B, Gallagher SM, Pugh EN, Jr, Philp NJ. Altered visual function in monocarboxylate transporter 3 (Slc16a8) knockout mice. Am J Physiol Cell Physiol. 2008;295:C451–457. doi: 10.1152/ajpcell.00124.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MD, Gangnon RE, Lee LY, Hubbard LD, Klein BE, Klein R, Ferris FL, Bressler SB, Milton RC. The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS Report No. 17. Arch Ophthalmol. 2005;123:1484–1498. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong EK, Breukink MB, Schellevis RL, Bakker B, Mohr JK, Fauser S, Keunen JE, Hoyng CB, den Hollander AI, Boon CJ. Chronic central serous chorioretinopathy is associated with genetic variants implicated in age-related macular degeneration. Ophthalmology. 2015;122:562–570. doi: 10.1016/j.ophtha.2014.09.026. [DOI] [PubMed] [Google Scholar]
- de Jong PTVM. Age-Related Macular Degeneration. New England Journal of Medicine. 2006;355:1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, Wang X, Evangelista M, Barsacchi G, Cremisi F. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci U S A. 2009;106:21179–21184. doi: 10.1073/pnas.0909167106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcourt C, Diaz JL, Ponton-Sanchez A, Papoz L. Smoking and age-related macular degeneration. The POLA Study. Pathologies Oculaires Liees a l’Age. Arch Ophthalmol. 1998;116:1031–1035. doi: 10.1001/archopht.116.8.1031. [DOI] [PubMed] [Google Scholar]
- Delcourt C, Michel F, Colvez A, Lacroux A, Delage M, Vernet MH, Group PS. Associations of cardiovascular disease and its risk factors with age-related macular degeneration: the POLA study. Ophthalmic epidemiology. 2001;8:237–249. doi: 10.1076/opep.8.4.237.1613. [DOI] [PubMed] [Google Scholar]
- Della NG, Campochiaro PA, Zack DJ. Localization of TIMP-3 mRNA expression to the retinal pigment epithelium. Invest Ophthalmol Vis Sci. 1996;37:1921–1924. [PubMed] [Google Scholar]
- Dewan A, Liu M, Hartman S, Zhang SS, Liu DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, Barnstable C, Pang CP, Hoh J. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- Dong Y, Li ZD, Fang XY, Shi XF, Chen S, Tang X. Association between SERPING1 rs2511989 polymorphism and age-related macular degeneration: Meta-analysis. Int J Ophthalmol. 2015;8:385–394. doi: 10.3980/j.issn.2222-3959.2015.02.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoso LA, Kim D, Frost A, Callahan A, Hageman G. The role of inflammation in the pathogenesis of age-related macular degeneration. Survey of ophthalmology. 2006;51:137–152. doi: 10.1016/j.survophthal.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SL, Campbell M, Ozaki E, Salomon RG, Mori A, Kenna PF, Farrar GJ, Kiang AS, Humphries MM, Lavelle EC, O’Neill LA, Hollyfield JG, Humphries P. NLRP3 has a protective role in age-related macular degeneration through the induction of IL-18 by drusen components. Nature medicine. 2012;18:791–798. doi: 10.1038/nm.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi S, Hirano Y, Tarallo V, Kim Y, Fowler BJ, Ambati BK, Bogdanovich S, Chiodo VA, Hauswirth WW, Kugel JF, Goodrich JA, Ponicsan SL, Hinton DR, Kleinman ME, Baffi JZ, Gelfand BD, Ambati J. ERK1/2 activation is a therapeutic target in age-related macular degeneration. Proc Natl Acad Sci U S A. 2012;109:13781–13786. doi: 10.1073/pnas.1206494109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy MJ, Sturgeon CM, Soletormos G, Barak V, Molina R, Hayes DF, Diamandis EP, Bossuyt PM. Validation of New Cancer Biomarkers: A Position Statement from the European Group on Tumor Markers. Clin Chem. 2015;61:809–820. doi: 10.1373/clinchem.2015.239863. [DOI] [PubMed] [Google Scholar]
- Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168:3195–3204. doi: 10.4049/jimmunol.168.7.3195. [DOI] [PubMed] [Google Scholar]
- Ebrahem Q, Renganathan K, Sears J, Vasanji A, Gu X, Lu L, Salomon RG, Crabb JW, Anand-Apte B. Carboxyethylpyrrole oxidative protein modifications stimulate neovascularization: Implications for age-related macular degeneration. Proc Natl Acad Sci U S A. 2006;103:13480–13484. doi: 10.1073/pnas.0601552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou MA, Wu J, Vasilcanu D, Rosengren L, All-Ericsson C, van der Ploeg I, Menu E, Girnita L, Axelson M, Larsson O, Seregard S, Kvanta A. Inhibition of VEGF secretion and experimental choroidal neovascularization by picropodophyllin (PPP), an inhibitor of the insulin-like growth factor-1 receptor. Invest Ophthalmol Vis Sci. 2008;49:2620–2626. doi: 10.1167/iovs.07-0742. [DOI] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- El-Amraoui A, Schonn JS, Kussel-Andermann P, Blanchard S, Desnos C, Henry JP, Wolfrum U, Darchen F, Petit C. MyRIP, a novel Rab effector, enables myosin VIIa recruitment to retinal melanosomes. EMBO Rep. 2002;3:463–470. doi: 10.1093/embo-reports/kvf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis S, Jomary C, Mullins R, Cree A, Chen X, Macleod A, Jones S, Collins A, Stone E, Lotery A. Association between the SERPING1 gene and age-related macular degeneration: a two-stage case-control study. Lancet. 2008;372:1828–1834. doi: 10.1016/S0140-6736(08)61348-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erke MG, Bertelsen G, Peto T, Sjolie AK, Lindekleiv H, Njolstad I. Prevalence of age-related macular degeneration in elderly Caucasians: the Tromso Eye Study. Ophthalmology. 2012;119:1737–1743. doi: 10.1016/j.ophtha.2012.03.016. [DOI] [PubMed] [Google Scholar]
- Esparza-Gordillo J, Soria JM, Buil A, Almasy L, Blangero J, Fontcuberta J, Rodriguez de Cordoba S. Genetic and environmental factors influencing the human factor H plasma levels. Immunogenetics. 2004;56:77–82. doi: 10.1007/s00251-004-0660-7. [DOI] [PubMed] [Google Scholar]
- Evans JR, Fletcher AE, Wormald RP. Age-related macular degeneration causing visual impairment in people 75 years or older in Britain: an add-on study to the Medical Research Council Trial of Assessment and Management of Older People in the Community. Ophthalmology. 2004;111:513–517. doi: 10.1016/j.ophtha.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Evans JR, Fletcher AE, Wormald RP. 28,000 Cases of age related macular degeneration causing visual loss in people aged 75 years and above in the United Kingdom may be attributable to smoking. Br J Ophthalmol. 2005;89:550–553. doi: 10.1136/bjo.2004.049726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang AM, Lee AY, Kulkarni M, Osborn MP, Brantley MA., Jr Polymorphisms in the VEGFA and VEGFR-2 genes and neovascular age-related macular degeneration. Mol Vis. 2009;15:2710–2719. [PMC free article] [PubMed] [Google Scholar]
- Farrar GJ, Chadderton N, Kenna PF, Millington-Ward S. Mitochondrial disorders: aetiologies, models systems, and candidate therapies. Trends Genet. 2013;29:488–497. doi: 10.1016/j.tig.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Fauser S, Lambrou GN. Genetic predictive biomarkers of anti-VEGF treatment response in patients with neovascular age-related macular degeneration. Survey of ophthalmology. 2015;60:138–152. doi: 10.1016/j.survophthal.2014.11.002. [DOI] [PubMed] [Google Scholar]
- Fernando MM, Stevens CR, Sabeti PC, Walsh EC, McWhinnie AJ, Shah A, Green T, Rioux JD, Vyse TJ. Identification of two independent risk factors for lupus within the MHC in United Kingdom families. PLoS genetics. 2007;3:e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SA, Rivera A, Fritsche LG, Keilhauer CN, Lichtner P, Meitinger T, Rudolph G, Weber BH. Case-control genetic association study of fibulin-6 (FBLN6 or HMCN1) variants in age-related macular degeneration (AMD) Hum Mutat. 2007;28:406–413. doi: 10.1002/humu.20464. [DOI] [PubMed] [Google Scholar]
- Fletcher AE, Bentham GC, Agnew M, Young IS, Augood C, Chakravarthy U, de Jong PTVM, Rahu M, Seland J, Soubrane G, Tomazzoli L, Topouzis F, Vingerling JR, Vioque J. Sunlight Exposure, Antioxidants, and Age-Related Macular Degeneration. Arch Ophthalmol. 2008;126:1396–1403. doi: 10.1001/archopht.126.10.1396. [DOI] [PubMed] [Google Scholar]
- Francis PJ, Klein ML. Update on the role of genetics in the onset of age-related macular degeneration. Clin Ophthalmol. 2011;5:1127–1133. doi: 10.2147/OPTH.S11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank RN, Puklin JE, Stock C, Canter LA. Race, iris color, and age-related macular degeneration. Trans Am Ophthalmol Soc. 2000;98:109–115. discussion 115–107. [PMC free article] [PubMed] [Google Scholar]
- Friedman DS, Katz J, Bressler NM, Rahmani B, Tielsch JM. Racial differences in the prevalence of age-related macular degeneration: the Baltimore Eye Survey. Ophthalmology. 1999;106:1049–1055. doi: 10.1016/S0161-6420(99)90267-1. [DOI] [PubMed] [Google Scholar]
- Friedman DS, O’Colmain BJ, Munoz B, Tomany SC, McCarty C, de Jong PT, Nemesure B, Mitchell P, Kempen J. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, Igo RP, Jr, Buitendijk GH, Sim X, Weeks DE, Guymer RH, Merriam JE, Francis PJ, Hannum G, Agarwal A, Armbrecht AM, Audo I, Aung T, Barile GR, Benchaboune M, Bird AC, Bishop PN, Branham KE, Brooks M, Brucker AJ, Cade WH, Cain MS, Campochiaro PA, Chan CC, Cheng CY, Chew EY, Chin KA, Chowers I, Clayton DG, Cojocaru R, Conley YP, Cornes BK, Daly MJ, Dhillon B, Edwards AO, Evangelou E, Fagerness J, Ferreyra HA, Friedman JS, Geirsdottir A, George RJ, Gieger C, Gupta N, Hagstrom SA, Harding SP, Haritoglou C, Heckenlively JR, Holz FG, Hughes G, Ioannidis JP, Ishibashi T, Joseph P, Jun G, Kamatani Y, Katsanis N, CNK, Khan JC, Kim IK, Kiyohara Y, Klein BE, Klein R, Kovach JL, Kozak I, Lee CJ, Lee KE, Lichtner P, Lotery AJ, Meitinger T, Mitchell P, Mohand-Said S, Moore AT, Morgan DJ, Morrison MA, Myers CE, Naj AC, Nakamura Y, Okada Y, Orlin A, Ortube MC, Othman MI, Pappas C, Park KH, Pauer GJ, Peachey NS, Poch O, Priya RR, Reynolds R, Richardson AJ, Ripp R, Rudolph G, Ryu E, Sahel JA, Schaumberg DA, Scholl HP, Schwartz SG, Scott WK, Shahid H, Sigurdsson H, Silvestri G, Sivakumaran TA, Smith RT, Sobrin L, Souied EH, Stambolian DE, Stefansson H, Sturgill-Short GM, Takahashi A, Tosakulwong N, Truitt BJ, Tsironi EE, Uitterlinden AG, van Duijn CM, Vijaya L, Vingerling JR, Vithana EN, Webster AR, Wichmann HE, Winkler TW, Wong TY, Wright AF, Zelenika D, Zhang M, Zhao L, Zhang K, Klein ML, Hageman GS, Lathrop GM, Stefansson K, Allikmets R, Baird PN, Gorin MB, Wang JJ, Klaver CC, Seddon JM, Pericak-Vance MA, Iyengar SK, Yates JR, Swaroop A, Weber BH, Kubo M, Deangelis MM, Leveillard T, Thorsteinsdottir U, Haines JL, Farrer LA, Heid IM, Abecasis GR Consortium AMDG. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–439. 439e431–432. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Fleckenstein M, Fiebig BS, Schmitz-Valckenberg S, Bindewald-Wittich A, Keilhauer CN, Renner AB, Mackensen F, Mossner A, Pauleikhoff D, Adrion C, Mansmann U, Scholl HP, Holz FG, Weber BH. A subgroup of age-related macular degeneration is associated with mono-allelic sequence variants in the ABCA4 gene. Invest Ophthalmol Vis Sci. 2012;53:2112–2118. doi: 10.1167/iovs.11-8785. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, Burdon KP, Hebbring SJ, Wen C, Gorski M, Kim IK, Cho D, Zack D, Souied E, Scholl HP, Bala E, Lee KE, Hunter DJ, Sardell RJ, Mitchell P, Merriam JE, Cipriani V, Hoffman JD, Schick T, Lechanteur YT, Guymer RH, Johnson MP, Jiang Y, Stanton CM, Buitendijk GH, Zhan X, Kwong AM, Boleda A, Brooks M, Gieser L, Ratnapriya R, Branham KE, Foerster JR, Heckenlively JR, Othman MI, Vote BJ, Liang HH, Souzeau E, McAllister IL, Isaacs T, Hall J, Lake S, Mackey DA, Constable IJ, Craig JE, Kitchner TE, Yang Z, Su Z, Luo H, Chen D, Ouyang H, Flagg K, Lin D, Mao G, Ferreyra H, Stark K, von Strachwitz CN, Wolf A, Brandl C, Rudolph G, Olden M, Morrison MA, Morgan DJ, Schu M, Ahn J, Silvestri G, Tsironi EE, Park KH, Farrer LA, Orlin A, Brucker A, Li M, Curcio CA, Mohand-Said S, Sahel JA, Audo I, Benchaboune M, Cree AJ, Rennie CA, Goverdhan SV, Grunin M, Hagbi-Levi S, Campochiaro P, Katsanis N, Holz FG, Blond F, Blanche H, Deleuze JF, Igo RP, Jr, Truitt B, Peachey NS, Meuer SM, Myers CE, Moore EL, Klein R, Hauser MA, Postel EA, Courtenay MD, Schwartz SG, Kovach JL, Scott WK, Liew G, Tan AG, Gopinath B, Merriam JC, Smith RT, Khan JC, Shahid H, Moore AT, McGrath JA, Laux R, Brantley MA, Jr, Agarwal A, Ersoy L, Caramoy A, Langmann T, Saksens NT, de Jong EK, Hoyng CB, Cain MS, Richardson AJ, Martin TM, Blangero J, Weeks DE, Dhillon B, van Duijn CM, Doheny KF, Romm J, Klaver CC, Hayward C, Gorin MB, Klein ML, Baird PN, den Hollander AI, Fauser S, Yates JR, Allikmets R, Wang JJ, Schaumberg DA, Klein BE, Hagstrom SA, Chowers I, Lotery AJ, Leveillard T, Zhang K, Brilliant MH, Hewitt AW, Swaroop A, Chew EY, Pericak-Vance MA, DeAngelis M, Stambolian D, Haines JL, Iyengar SK, Weber BH, Abecasis GR, Heid IM. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, Pandey MK, Kohl J, Zipfel PF, Weber BH, Skerka C. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD) Human molecular genetics. 2010;19:4694–4704. doi: 10.1093/hmg/ddq399. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BHF. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- Fu L, Garland D, Yang Z, Shukla D, Rajendran A, Pearson E, Stone EM, Zhang K, Pierce EA. The R345W mutation in EFEMP1 is pathogenic and causes AMD-like deposits in mice. Human molecular genetics. 2007;16:2411–2422. doi: 10.1093/hmg/ddm198. [DOI] [PubMed] [Google Scholar]
- Galan A, Ferlin A, Caretti L, Buson G, Sato G, Frigo AC, Foresta C. Association of age-related macular degeneration with polymorphisms in vascular endothelial growth factor and its receptor. Ophthalmology. 2010;117:1769–1774. doi: 10.1016/j.ophtha.2010.01.030. [DOI] [PubMed] [Google Scholar]
- Gangnon RE, Lee KE, Klein BE, Iyengar SK, Sivakumaran TA, Klein R. Effect of the Y402H variant in the complement factor H gene on the incidence and progression of age-related macular degeneration: results from multistate models applied to the Beaver Dam Eye Study. Arch Ophthalmol. 2012;130:1169–1176. doi: 10.1001/archophthalmol.2012.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Alvarez L, Nogacka AM, Gonzalez-Iglesias H, Escribano J, Fernandez-Vega B, Fernandez-Vega A, Fernandez-Vega L, Coca-Prados M. CFH polymorphisms in a Northern Spanish population with neovascular and dry forms of age-related macular degeneration. Acta ophthalmologica. 2015;93:e658–e666. doi: 10.1111/aos.12790. [DOI] [PubMed] [Google Scholar]
- Gehlbach P, Demetriades AM, Yamamoto S, Deering T, Xiao WH, Duh EJ, Yang HS, Lai H, Kovesdi I, Carrion M, Wei L, Campochiaro PA. Periocular gene transfer of sFlt-1 suppresses ocular neovascularization and vascular endothelial growth factor-induced breakdown of the blood-retinal barrier. Hum Gene Ther. 2003;14:129–141. doi: 10.1089/104303403321070829. [DOI] [PubMed] [Google Scholar]
- Gehrs KM, Anderson DH, Johnson LV, Hageman GS. Age-related macular degeneration--emerging pathogenetic and therapeutic concepts. Ann Med. 2006;38:450–471. doi: 10.1080/07853890600946724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Saha M, Das D. A study on plasma homocysteine level in age-related macular degeneration. Nepal J Ophthalmol. 2013;5:195–200. doi: 10.3126/nepjoph.v5i2.8728. [DOI] [PubMed] [Google Scholar]
- Gibson J, Cree A, Collins A, Lotery A, Ennis S. Determination of a gene and environment risk model for age-related macular degeneration. Br J Ophthalmol. 2010;94:1382–1387. doi: 10.1136/bjo.2010.182568. [DOI] [PubMed] [Google Scholar]
- Goehring AS, Pedroja BS, Hinke SA, Langeberg LK, Scott JD. MyRIP anchors protein kinase A to the exocyst complex. J Biol Chem. 2007;282:33155–33167. doi: 10.1074/jbc.M705167200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, Group AMDGCS, Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006a;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006b;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin MB. Genetic insights into age-related macular degeneration: controversies addressing risk, causality, and therapeutics. Mol Aspects Med. 2012;33:467–486. doi: 10.1016/j.mam.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SF, Chevallier OP, Roberts D, Holscher C, Elliott CT, Green BD. Investigation of the human brain metabolome to identify potential markers for early diagnosis and therapeutic targets of Alzheimer’s disease. Anal Chem. 2013;85:1803–1811. doi: 10.1021/ac303163f. [DOI] [PubMed] [Google Scholar]
- Grassmann F, Friedrich U, Fauser S, Schick T, Milenkovic A, Schulz HL, von Strachwitz CN, Bettecken T, Lichtner P, Meitinger T, Arend N, Wolf A, Haritoglou C, Rudolph G, Chakravarthy U, Silvestri G, McKay GJ, Freitag-Wolf S, Krawczak M, Smith RT, Merriam JC, Merriam JE, Allikmets R, Heid IM, Weber BH. A Candidate Gene Association Study Identifies DAPL1 as a Female-Specific Susceptibility Locus for Age-Related Macular Degeneration (AMD) Neuromolecular medicine. 2015;17:111–120. doi: 10.1007/s12017-015-8342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Shiekhattar R. MicroRNA Biogenesis and Cancer. Cancer research. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- Gress CJ, Jacenko O. Growth plate compressions and altered hematopoiesis in collagen X null mice. J Cell Biol. 2000;149:983–993. doi: 10.1083/jcb.149.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Pauer GJ, Yue X, Narendra U, Sturgill GM, Bena J, Gu X, Peachey NS, Salomon RG, Hagstrom SA, Crabb JW. Proteomic and Genomic Biomarkers for Age-Related Macular Degeneration. Adv Exp Med Biol. 2010;664:411–417. doi: 10.1007/978-1-4419-1399-9_47. [DOI] [PubMed] [Google Scholar]
- Gu J, Pauer GJ, Yue X, Narendra U, Sturgill GM, Bena J, Gu X, Peachey NS, Salomon RG, Hagstrom SA, Crabb JW, Clinical G, Proteomic AMDSG. Assessing susceptibility to age-related macular degeneration with proteomic and genomic biomarkers. Mol Cell Proteomics. 2009;8:1338–1349. doi: 10.1074/mcp.M800453-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guymer RH, Heon E, Lotery AJ, Munier FL, Schorderet DF, Baird PN, McNeil RJ, Haines H, Sheffield VC, Stone EM. Variation of codons 1961 and 2177 of the Stargardt disease gene is not associated with age-related macular degeneration. Arch Ophthalmol. 2001;119:745–751. doi: 10.1001/archopht.119.5.745. [DOI] [PubMed] [Google Scholar]
- Habibi I, Sfar I, Kort F, Aounallah-Skhiri H, Chebil A, Chouchene I, Bouraoui R, Limaiem R, Largheche L, Jendoubi-Ayed S, Makhlouf M, Ben Abdallah T, Ayed K, El Matri L, Gorgi Y. Y402H polymorphism in complement factor H and age-related macular degeneration in the Tunisian population. Ophthalmic Res. 2013;49:177–184. doi: 10.1159/000345068. [DOI] [PubMed] [Google Scholar]
- Hackler L, Jr, Wan J, Swaroop A, Qian J, Zack DJ. MicroRNA profile of the developing mouse retina. Invest Ophthalmol Vis Sci. 2010;51:1823–1831. doi: 10.1167/iovs.09-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagbi-Levi S, Grunin M, Elbaz-Hayoun S, Tal D, Obolensky A, Hanhart J, Banin E, Burstyn-Cohen T, Chowers I. Retinal Phenotype following Combined Deletion of the Chemokine Receptor CCR2 and the Chemokine CX3CL1 in Mice. Ophthalmic Res. 2016;55:126–134. doi: 10.1159/000441794. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJ, Silvestri G, Russell SR, Klaver CC, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20:705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- Hagstrom SA, Ying GS, Maguire MG, Martin DF, Gibson J, Lotery A, Chakravarthy U CATT Research Group, IVAN Study Investigators. VEGFR2 Gene Polymorphisms and Response to Anti-Vascular Endothelial Growth Factor Therapy in Age-Related Macular Degeneration. Ophthalmology. 2015;122:1563–1568. doi: 10.1016/j.ophtha.2015.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom SA, Ying GS, Pauer GJ, Sturgill-Short GM, Huang J, Callanan DG, Kim IK, Klein ML, Maguire MG, Martin DF Comparison of AMDTTRG. Pharmacogenetics for genes associated with age-related macular degeneration in the Comparison of AMD Treatments Trials (CATT) Ophthalmology. 2013;120:593–599. doi: 10.1016/j.ophtha.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proceedings of the National Academy of Sciences. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautamaki A, Seitsonen S, Holopainen JM, Moilanen JA, Kivioja J, Onkamo P, Jarvela I, Immonen I. The genetic variant rs4073 A-->T of the Interleukin-8 promoter region is associated with the earlier onset of exudative age-related macular degeneration. Acta ophthalmologica. 2015;93:726–733. doi: 10.1111/aos.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckenlively JR, Aptsiauri N, Nusinowitz S, Peng C, Hargrave PA. Investigations of antiretinal antibodies in pigmentary retinopathy and other retinal degenerations. Trans Am Ophthalmol Soc. 1996;94:179–200. [PMC free article] [PubMed] [Google Scholar]
- Hermann MM, van Asten F, Muether PS, Smailhodzic D, Lichtner P, Hoyng CB, Kirchhof B, Grefkes C, den Hollander AI, Fauser S. Polymorphisms in vascular endothelial growth factor receptor 2 are associated with better response rates to ranibizumab treatment in age-related macular degeneration. Ophthalmology. 2014;121:905–910. doi: 10.1016/j.ophtha.2013.10.047. [DOI] [PubMed] [Google Scholar]
- Heuberger RA, Mares-Perlman JA, Klein R, Klein BEK, Millen AE, Palta M. Relationship of dietary fat to age-related maculopathy in the Third National Health and Nutrition Examination Survey. Arch Ophthalmol. 2001;119:1833–1838. doi: 10.1001/archopht.119.12.1833. [DOI] [PubMed] [Google Scholar]
- Hodge WG, Schachter HM, Barnes D, Pan Y, Lowcock EC, Zhang L, Sampson M, Morrison A, Tran K, Miguelez M, Lewin G. Efficacy of [omega]-3 Fatty Acids in Preventing Age-Related Macular Degeneration: A Systematic Review. Ophthalmology. 2006;113:1165–1173. doi: 10.1016/j.ophtha.2006.02.043. [DOI] [PubMed] [Google Scholar]
- Holliday EG, Smith AV, Cornes BK, Buitendijk GH, Jensen RA, Sim X, Aspelund T, Aung T, Baird PN, Boerwinkle E, Cheng CY, van Duijn CM, Eiriksdottir G, Gudnason V, Harris T, Hewitt AW, Inouye M, Jonasson F, Klein BE, Launer L, Li X, Liew G, Lumley T, McElduff P, McKnight B, Mitchell P, Psaty BM, Rochtchina E, Rotter JI, Scott RJ, Tay W, Taylor K, Teo YY, Uitterlinden AG, Viswanathan A, Xie S, Vingerling JR, Klaver CC, Tai ES, Siscovick D, Klein R, Cotch MF, Wong TY, Attia J, Wang JJ Wellcome Trust Case Control C. Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis. PloS one. 2013;8:e53830. doi: 10.1371/journal.pone.0053830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollyfield JG, Perez VL, Salomon RG. A hapten generated from an oxidation fragment of docosahexaenoic acid is sufficient to initiate age-related macular degeneration. Mol Neurobiol. 2010;41:290–298. doi: 10.1007/s12035-010-8110-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong T, Tan AG, Mitchell P, Wang JJ. A review and meta-analysis of the association between C-reactive protein and age-related macular degeneration. Survey of ophthalmology. 2011;56:184–194. doi: 10.1016/j.survophthal.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Horie-Inoue K, Inoue S. Genomic aspects of age-related macular degeneration. Biochemical and biophysical research communications. 2014;452:263–275. doi: 10.1016/j.bbrc.2014.08.013. [DOI] [PubMed] [Google Scholar]
- Houssier M, Raoul W, Lavalette S, Keller N, Guillonneau X, Baragatti B, Jonet L, Jeanny JC, Behar-Cohen F, Coceani F, Scherman D, Lachapelle P, Ong H, Chemtob S, Sennlaub F. CD36 deficiency leads to choroidal involution via COX2 down-regulation in rodents. PLoS Med. 2008;5:e39. doi: 10.1371/journal.pmed.0050039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Wang F, Sah BK, Jiang J, Ni Z, Wang J, Sun X. Homocysteine and the risk of age-related macular degeneration: a systematic review and meta-analysis. Sci Rep. 2015;5:10585. doi: 10.1038/srep10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- Hutchinson JN, Fagerness J, Kirby A, Reynolds R, Zak A, Gimelbrant A, Plenge R, Daly M, Chess A, Seddon JM. (Epi)Genetic analyses of age-related macular degeneration: case-control and discordant twin studies. Human heredity. 2014;78:59–72. doi: 10.1159/000362814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L, Schachat AP, He Q, Leske MC. Hypertension, cardiovascular disease, and age-related macular degeneration. Age-Related Macular Degeneration Risk Factors Study Group. Arch Ophthalmol. 2000;118:351–358. doi: 10.1001/archopht.118.3.351. [DOI] [PubMed] [Google Scholar]
- Iannaccone A, Neeli I, Krishnamurthy P, Lenchik NI, Wan H, Gerling IC, Desiderio DM, Radic MZ. Autoimmune biomarkers in age-related macular degeneration: a possible role player in disease development and progression. Adv Exp Med Biol. 2012;723:11–16. doi: 10.1007/978-1-4614-0631-0_2. [DOI] [PubMed] [Google Scholar]
- Ishida W, Fukuda K, Higuchi T, Kajisako M, Sakamoto S, Fukushima A. Dynamic changes of microRNAs in the eye during the development of experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 2011;52:611–617. doi: 10.1167/iovs.10-6115. [DOI] [PubMed] [Google Scholar]
- IVAN Study Investigators. Chakravarthy U, Harding SP, Rogers CA, Downes SM, Lotery AJ, Wordsworth S, Reeves BC. Ranibizumab versus bevacizumab to treat neovascular age-related macular degeneration: one-year findings from the IVAN randomized trial. Ophthalmology. 2012;119:1399–1411. doi: 10.1016/j.ophtha.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J, Conley YP, Weeks DE, Ferrell RE, Gorin MB. C2 and CFB genes in age-related maculopathy and joint action with CFH and LOC387715 genes. PloS one. 2008;3:e2199. doi: 10.1371/journal.pone.0002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik-Papis K, Zaras M, Sklodowska A, Ulinska M, Borucka AI, Blasiak J. Genetic aspects of age-related macular degeneration. Klin Oczna. 2009;111:178–182. [PubMed] [Google Scholar]
- Jha A, Mehra M, Shankar R. The regulatory epicenter of miRNAs. J Biosci. 2011;36:621–638. doi: 10.1007/s12038-011-9109-y. [DOI] [PubMed] [Google Scholar]
- Jiang M, Esteve-Rudd J, Lopes VS, Diemer T, Lillo C, Rump A, Williams DS. Microtubule motors transport phagosomes in the RPE, and lack of KLC1 leads to AMD-like pathogenesis. J Cell Biol. 2015;210:595–611. doi: 10.1083/jcb.201410112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene. 2006;25:6176–6187. doi: 10.1038/sj.onc.1209912. [DOI] [PubMed] [Google Scholar]
- Kamburoglu G, Gumus K, Kadayifcilar S, Eldem B. Plasma homocysteine, vitamin B12 and folate levels in age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol. 2006;244:565–569. doi: 10.1007/s00417-005-0108-2. [DOI] [PubMed] [Google Scholar]
- Kamei M, Hollyfield JG. TIMP-3 in Bruch’s membrane: changes during aging and in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1999;40:2367–2375. [PubMed] [Google Scholar]
- Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, Zhang Q, Grossniklaus HE, Provis JM, Madigan MC, Milam AH, Justice NL, Albuquerque RJ, Blandford AD, Bogdanovich S, Hirano Y, Witta J, Fuchs E, Littman DR, Ambati BK, Rudin CM, Chong MM, Provost P, Kugel JF, Goodrich JA, Dunaief JL, Baffi JZ, Ambati J. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HK, Yoon MH, Lee DH, Chin HS. Pharmacogenetic influence of LOC387715/HTRA1 on the efficacy of bevacizumab treatment for age-related macular degeneration in a Korean population. Korean journal of ophthalmology. 2012;26:414–422. doi: 10.3341/kjo.2012.26.6.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali M, Peluso I, Marigo V, Banfi S. Identification and Characterization of MicroRNAs Expressed in the Mouse Eye. Investigative Ophthalmology & Visual Science. 2007;48:509–515. doi: 10.1167/iovs.06-0866. [DOI] [PubMed] [Google Scholar]
- Katschke KJ, Jr, Wu P, Ganesan R, Kelley RF, Mathieu MA, Hass PE, Murray J, Kirchhofer D, Wiesmann C, van Lookeren Campagne M. Inhibiting alternative pathway complement activation by targeting the factor D exosite. J Biol Chem. 2012;287:12886–12892. doi: 10.1074/jbc.M112.345082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keles S, Ates O, Kartal B, Alp HH, Ekinci M, Ceylan E, Ondas O, Arpali E, Dogan S, Yildirim K, Keles MS. Evaluation of cardiovascular biomarkers in patients with age-related wet macular degeneration. Clin Ophthalmol. 2014;8:1573–1578. doi: 10.2147/OPTH.S66160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, Lenhof HP, Ruprecht K, Meese E. Multiple Sclerosis: MicroRNA Expression Profiles Accurately Differentiate Patients with Relapsing-Remitting Disease from Healthy Controls. PloS one. 2009;4:e7440. doi: 10.1371/journal.pone.0007440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall RL, Thomas KA. Inhibition of vascular endothelial cell growth factor activity by an endogenously encoded soluble receptor. Proc Natl Acad Sci U S A. 1993;90:10705–10709. doi: 10.1073/pnas.90.22.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew RR, Ghebrehiwet B, Janoff A. Cigarette smoke can activate the alternative pathway of complement in vitro by modifying the third component of complement. J Clin Invest. 1985;75:1000–1007. doi: 10.1172/JCI111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan JC, Thurlby DA, Shahid H, Clayton DG, Yates JR, Bradley M, Moore AT, Bird AC Genetic Factors in AMDS. Smoking and age related macular degeneration: the number of pack years of cigarette smoking is a major determinant of risk for both geographic atrophy and choroidal neovascularisation. Br J Ophthalmol. 2006;90:75–80. doi: 10.1136/bjo.2005.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijlstra A, Tian Y, Kelly ER, Berendschot TT. Lutein: more than just a filter for blue light. Prog Retin Eye Res. 2012;31:303–315. doi: 10.1016/j.preteyeres.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Kikuchi M, Nakamura M, Ishikawa K, Suzuki T, Nishihara H, Yamakoshi T, Nishio K, Taki K, Niwa T, Hamajima N, Terasaki H. Elevated C-reactive protein levels in patients with polypoidal choroidal vasculopathy and patients with neovascular age-related macular degeneration. Ophthalmology. 2007;114:1722–1727. doi: 10.1016/j.ophtha.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Kinnunen K, Petrovski G, Moe MC, Berta A, Kaarniranta K. Molecular mechanisms of retinal pigment epithelium damage and development of age-related macular degeneration. Acta ophthalmologica. 2012;90:299–309. doi: 10.1111/j.1755-3768.2011.02179.x. [DOI] [PubMed] [Google Scholar]
- Klaver CC, Kliffen M, van Duijn CM, Hofman A, Cruts M, Grobbee DE, van Broeckhoven C, de Jong PT. Genetic association of apolipoprotein E with age-related macular degeneration. American journal of human genetics. 1998;63:200–206. doi: 10.1086/301901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BE, Howard KP, Iyengar SK, Sivakumaran TA, Meyers KJ, Cruickshanks KJ, Klein R. Sunlight exposure, pigmentation, and incident age-related macular degeneration. Invest Ophthalmol Vis Sci. 2014;55:5855–5861. doi: 10.1167/iovs.14-14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein ML, Francis PJ, Ferris FL, 3rd, Hamon SC, Clemons TE. Risk assessment model for development of advanced age-related macular degeneration. Arch Ophthalmol. 2011;129:1543–1550. doi: 10.1001/archophthalmol.2011.216. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Jensen SC, Meuer SM. The five-year incidence and progression of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology. 1997;104:7–21. doi: 10.1016/s0161-6420(97)30368-6. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Knudtson MD, Meuer SM, Swift M, Gangnon RE. Fifteen-year cumulative incidence of age-related macular degeneration: the Beaver Dam Eye Study. Ophthalmology. 2007;114:253–262. doi: 10.1016/j.ophtha.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Knudtson MD, Wong TY, Cotch MF, Liu K, Burke G, Saad MF, Jacobs DR., Jr Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the multi-ethnic study of atherosclerosis. Ophthalmology. 2006;113:373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Linton KL. Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology. 1992;99:933–943. doi: 10.1016/s0161-6420(92)31871-8. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Moss SE. Relation of smoking to the incidence of age-related maculopathy. The Beaver Dam Eye Study. American journal of epidemiology. 1998;147:103–110. doi: 10.1093/oxfordjournals.aje.a009421. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Tomany SC, Cruickshanks KJ. The association of cardiovascular disease with the long-term incidence of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology. 2003;110:1273–1280. doi: 10.1016/S0161-6420(03)00599-2. [DOI] [PubMed] [Google Scholar]
- Klein R, Klein BE, Tomany SC, Meuer SM, Huang GH. Ten-year incidence and progression of age-related maculopathy: The Beaver Dam eye study. Ophthalmology. 2002;109:1767–1779. doi: 10.1016/s0161-6420(02)01146-6. [DOI] [PubMed] [Google Scholar]
- Klein R, Myers CE, Meuer SM, Gangnon RE, Sivakumaran TA, Iyengar SK, Lee KE, Klein BE. Risk alleles in CFH and ARMS2 and the long-term natural history of age-related macular degeneration: the Beaver Dam Eye Study. JAMA Ophthalmol. 2013;131:383–392. doi: 10.1001/jamaophthalmol.2013.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B, Barthelmes D, Labs S, Schindler C, Kurz-Levin M, Michels S, Fleischhauer J, Berger W, Sutter F, Menghini M. Genetic association with response to intravitreal ranibizumab in patients with neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52:4694–4702. doi: 10.1167/iovs.10-6080. [DOI] [PubMed] [Google Scholar]
- Kokotas H, Grigoriadou M, Petersen MB. Age-related macular degeneration: genetic and clinical findings. Clin Chem Lab Med. 2011;49:601–616. doi: 10.1515/CCLM.2011.091. [DOI] [PubMed] [Google Scholar]
- Kondo N, Honda S, Kuno S, Negi A. Positive association of common variants in CD36 with neovascular age-related macular degeneration. Aging (Albany NY) 2009a;1:266–274. doi: 10.18632/aging.100006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Honda S, Kuno S, Negi A. Role of RDBP and SKIV2L variants in the major histocompatibility complex class III region in polypoidal choroidal vasculopathy etiology. Ophthalmology. 2009b;116:1502–1509. doi: 10.1016/j.ophtha.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Kondo T, Vicent D, Suzuma K, Yanagisawa M, King GL, Holzenberger M, Kahn CR. Knockout of insulin and IGF-1 receptors on vascular endothelial cells protects against retinal neovascularization. J Clin Invest. 2003;111:1835–1842. doi: 10.1172/JCI17455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopplin LJ, Igo RP, Jr, Wang Y, Sivakumaran TA, Hagstrom SA, Peachey NS, Francis PJ, Klein ML, SanGiovanni JP, Chew EY, Pauer GJ, Sturgill GM, Joshi T, Tian L, Xi Q, Henning AK, Lee KE, Klein R, Klein BE, Iyengar SK. Genome-wide association identifies SKIV2L and MYRIP as protective factors for age-related macular degeneration. Genes Immun. 2010;11:609–621. doi: 10.1038/gene.2010.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortvely E, Hauck SM, Duetsch G, Gloeckner CJ, Kremmer E, Alge-Priglinger CS, Deeg CA, Ueffing M. ARMS2 is a constituent of the extracellular matrix providing a link between familial and sporadic age-related macular degenerations. Invest Ophthalmol Vis Sci. 2010;51:79–88. doi: 10.1167/iovs.09-3850. [DOI] [PubMed] [Google Scholar]
- Kovacs B, Lumayag S, Cowan C, Xu S. microRNAs in Early Diabetic Retinopathy in Streptozotocin-Induced Diabetic Rats. Investigative Ophthalmology & Visual Science. 2011;52:4402–4409. doi: 10.1167/iovs.10-6879. [DOI] [PubMed] [Google Scholar]
- Kovacs KA, Pamer Z, Kovacs A, Fekete S, Miseta A, Kovacs B, Kovacs GL. Association of apolipoprotein E polymorphism with age-related macular degeneration and Alzheimer’s disease in south-western Hungary. Ideggyogy Sz. 2007;60:169–172. [PubMed] [Google Scholar]
- Krishnan T, Ravindran RD, Murthy GV, Vashist P, Fitzpatrick KE, Thulasiraj RD, John N, Maraini G, Camparini M, Chakravarthy U, Fletcher AE. Prevalence of early and late age-related macular degeneration in India: the INDEYE study. Invest Ophthalmol Vis Sci. 2010;51:701–707. doi: 10.1167/iovs.09-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krützfeldt J, Stoffel M. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metabolism. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Kucukevcilioglu M, Patel CB, Stone EM, Russell SR. Clinically detectable drusen domains in fibulin-5-associated age-related macular degeneration (AMD): Drusen subdomains in fibulin-5 AMD. Int Ophthalmol. 2015 doi: 10.1007/s10792-015-0164-5. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Kuroda TS, Fukuda M. Functional analysis of Slac2-c/MyRIP as a linker protein between melanosomes and myosin VIIa. J Biol Chem. 2005;280:28015–28022. doi: 10.1074/jbc.M501465200. [DOI] [PubMed] [Google Scholar]
- Kutty RK, Nagineni CN, Samuel W, Vijayasarathy C, Hooks JJ, Redmond TM. Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway. Biochemical and biophysical research communications. 2010a;402:390–395. doi: 10.1016/j.bbrc.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutty RK, Samuel W, Jaworski C, Duncan T, Nagineni CN, Raghavachari N, Wiggert B, Redmond TM. MicroRNA expression in human retinal pigment epithelial (ARPE-19) cells: increased expression of microRNA-9 by N-(4-hydroxyphenyl)retinamide. Mol Vis. 2010b;16:1475–1486. [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Hanna E, Lorang D, He M, Quick JS, Adem A, Stevenson C, Chung JY, Hewitt SM, Zudaire E, Esposito D, Cuttitta F, Libutti SK. Functional characterization of filamin a interacting protein 1-like, a novel candidate for antivascular cancer therapy. Cancer Research. 2008;68:7332–7341. doi: 10.1158/0008-5472.CAN-08-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkaraju A, Finnemann SC, Rodriguez-Boulan E. The lipofuscin fluorophore A2E perturbs cholesterol metabolism in retinal pigment epithelial cells. Proc Natl Acad Sci U S A. 2007;104:11026–11031. doi: 10.1073/pnas.0702504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambooij AC, van Wely KH, Lindenbergh-Kortleve DJ, Kuijpers RW, Kliffen M, Mooy CM. Insulin-like growth factor-I and its receptor in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2003;44:2192–2198. doi: 10.1167/iovs.02-0410. [DOI] [PubMed] [Google Scholar]
- Larsson J, Goumans MJ, Sjostrand LJ, van Rooijen MA, Ward D, Leveen P, Xu X, ten Dijke P, Mummery CL, Karlsson S. Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J. 2001;20:1663–1673. doi: 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leco KJ, Khokha R, Pavloff N, Hawkes SP, Edwards DR. Tissue inhibitor of metalloproteinases-3 (TIMP-3) is an extracellular matrix-associated protein with a distinctive pattern of expression in mouse cells and tissues. J Biol Chem. 1994;269:9352–9360. [PubMed] [Google Scholar]
- Lee AY, Kulkarni M, Fang AM, Edelstein S, Osborn MP, Brantley MA. The effect of genetic variants in SERPING1 on the risk of neovascular age-related macular degeneration. Br J Ophthalmol. 2010;94:915–917. doi: 10.1136/bjo.2009.172007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lesnik Oberstein SA, Kriek M, White SJ, Kalf ME, Szuhai K, den Dunnen JT, Breuning MH, Hennekam RC. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. American journal of human genetics. 2006;79:562–566. doi: 10.1086/507567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy O, Lavalette S, Hu SJ, Housset M, Raoul W, Eandi C, Sahel JA, Sullivan PM, Guillonneau X, Sennlaub F. APOE Isoforms Control Pathogenic Subretinal Inflammation in Age-Related Macular Degeneration. J Neurosci. 2015;35:13568–13576. doi: 10.1523/JNEUROSCI.2468-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Xu H, Zeng Y, Yin ZQ. Overexpression of fibulin-5 in retinal pigment epithelial cells inhibits cell proliferation and migration and downregulates VEGF, CXCR4, and TGFB1 expression in cocultured choroidal endothelial cells. Curr Eye Res. 2012;37:540–548. doi: 10.3109/02713683.2012.665561. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin H, Qian J, Castillo AC, Long B, Keyes KT, Chen G, Ye Y. Effect of miR-23 on oxidant-induced injury in human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2011;52:6308–6314. doi: 10.1167/iovs.10-6632. [DOI] [PubMed] [Google Scholar]
- Lip PL, Blann AD, Hope-Ross M, Gibson JM, Lip GYH. Age-related macular degeneration is associated with increased vascular endothelial growth factor, hemorheology and endothelial dysfunction. Ophthalmology. 2001;108:705–710. doi: 10.1016/s0161-6420(00)00663-1. [DOI] [PubMed] [Google Scholar]
- Lista S, Zetterberg H, Dubois B, Blennow K, Hampel H. Cerebrospinal fluid analysis in Alzheimer’s disease: technical issues and future developments. J Neurol. 2014;261:1234–1243. doi: 10.1007/s00415-014-7366-z. [DOI] [PubMed] [Google Scholar]
- Liu K, Chen LJ, Tam PO, Shi Y, Lai TY, Liu DT, Chiang SW, Yang M, Yang Z, Pang CP. Associations of the C2-CFB-RDBP-SKIV2L locus with age-related macular degeneration and polypoidal choroidal vasculopathy. Ophthalmology. 2013;120:837–843. doi: 10.1016/j.ophtha.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Liu K, Lai TY, Ma L, Lai FH, Young AL, Brelen ME, Tam PO, Pang CP, Chen LJ. Ethnic differences in the association of SERPING1 with age-related macular degeneration and polypoidal choroidal vasculopathy. Sci Rep. 2015;5:9424. doi: 10.1038/srep09424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MM, Agron E, Chew E, Meyerle C, Ferris FL, 3rd, Chan CC, Tuo J. Copy number variations in candidate genes in neovascular age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:3129–3135. doi: 10.1167/iovs.10-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, Farrar GJ. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8:R248. doi: 10.1186/gb-2007-8-11-r248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Shi Y, Qu C, Zhao P, Liu X, Gong B, Ma S, Zhou Y, Zhang Q, Fei P, Xu Y, Hu J, Fan Y, Lin Y, Zhu X, Yang Z. A genetic variant in the SKIV2L gene is significantly associated with age-related macular degeneration in a Han Chinese population. Invest Ophthalmol Vis Sci. 2013;54:2911–2917. doi: 10.1167/iovs.12-11381. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Surjyadipta B, Dua P, Alexandrov PN. Common micro RNAs (miRNAs) target complement factor H (CFH) regulation in Alzheimer’s disease (AD) and in age-related macular degeneration (AMD) Int J Biochem Mol Biol. 2012;3:105–116. [PMC free article] [PubMed] [Google Scholar]
- Luo L, Uehara H, Zhang X, Das SK, Olsen T, Holt D, Simonis JM, Jackman K, Singh N, Miya TR, Huang W, Ahmed F, Bastos-Carvalho A, Le YZ, Mamalis C, Chiodo VA, Hauswirth WW, Baffi J, Lacal PM, Orecchia A, Ferrara N, Gao G, Young-Hee K, Fu Y, Owen L, Albuquerque R, Baehr W, Thomas K, Li DY, Chalam KV, Shibuya M, Grisanti S, Wilson DJ, Ambati J, Ambati BK. Photoreceptor avascular privilege is shielded by soluble VEGF receptor-1. Elife. 2013;2:e00324. doi: 10.7554/eLife.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutty G, Grunwald J, Majji AB, Uyama M, Yoneya S. Changes in choriocapillaris and retinal pigment epithelium in age-related macular degeneration. Mol Vis. 1999;5:35. [PubMed] [Google Scholar]
- Ma B, Dang G, Yang S, Duan L, Zhang Y. CX3CR1 polymorphisms and the risk of age-related macular degeneration. Int J Clin Exp Pathol. 2015;8:9592–9596. [PMC free article] [PubMed] [Google Scholar]
- Magnusson KP, Duan S, Sigurdsson H, Petursson H, Yang Z, Zhao Y, Bernstein PS, Ge J, Jonasson F, Stefansson E, Helgadottir G, Zabriskie NA, Jonsson T, Bjornsson A, Thorlacius T, Jonsson PV, Thorleifsson G, Kong A, Stefansson H, Zhang K, Stefansson K, Gulcher JR. CFH Y402H confers similar risk of soft drusen and both forms of advanced AMD. PLoS Med. 2006;3:e5. doi: 10.137/journal.pmed.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher AD, Lindon JC, Nicholson JK. (1)H NMR-based metabonomics for investigating diabetes. Future Med Chem. 2009;1:737–747. doi: 10.4155/fmc.09.54. [DOI] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Maiorano NA, Hindges R. Non-coding RNAs in retinal development. Int J Mol Sci. 2012;13:558–578. doi: 10.3390/ijms13010558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- Mamas M, Dunn WB, Neyses L, Goodacre R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch Toxicol. 2011;85:5–17. doi: 10.1007/s00204-010-0609-6. [DOI] [PubMed] [Google Scholar]
- Mantel I, Ambresin A, Moetteli L, Droz I, Roduit R, Munier FL, Schorderet DF. Complement factor B polymorphism and the phenotype of early age-related macular degeneration. Ophthalmic genetics. 2014;35:12–17. doi: 10.3109/13816810.2013.766217. [DOI] [PubMed] [Google Scholar]
- Manthey HD, Woodruff TM, Taylor SM, Monk PN. Complement component 5a (C5a) Int J Biochem Cell Biol. 2009;41:2114–2117. doi: 10.1016/j.biocel.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Markova D, Zou Y, Ringpfeil F, Sasaki T, Kostka G, Timpl R, Uitto J, Chu ML. Genetic heterogeneity of cutis laxa: a heterozygous tandem duplication within the fibulin-5 (FBLN5) gene. American journal of human genetics. 2003;72:998–1004. doi: 10.1086/373940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein LY, McLaughlin PJ, Peachey NS, Sasaki T, Marmorstein AD. Formation and progression of sub-retinal pigment epithelium deposits in Efemp1 mutation knock-in mice: a model for the early pathogenic course of macular degeneration. Human molecular genetics. 2007;16:2423–2432. doi: 10.1093/hmg/ddm199. [DOI] [PubMed] [Google Scholar]
- Marmorstein LY, Munier FL, Arsenijevic Y, Schorderet DF, McLaughlin PJ, Chung D, Traboulsi E, Marmorstein AD. Aberrant accumulation of EFEMP1 underlies drusen formation in Malattia Leventinese and age-related macular degeneration. Proc Natl Acad Sci U S A. 2002;99:13067–13072. doi: 10.1073/pnas.202491599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Barricarte R, Pianetti G, Gautard R, Misselwitz J, Strain L, Fremeaux-Bacchi V, Skerka C, Zipfel PF, Goodship T, Noris M, Remuzzi G, de Cordoba SR European Working Party on the Genetics of HUS. The complement factor H R1210C mutation is associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol. 2008;19:639–646. doi: 10.1681/ASN.2007080923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Barricarte R, Recalde S, Fernandez-Robredo P, Millan I, Olavarrieta L, Vinuela A, Perez-Perez J, Garcia-Layana A, Rodriguez de Cordoba S Spanish Multicenter Group on AMD. Relevance of complement factor H-related 1 (CFHR1) genotypes in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2012;53:1087–1094. doi: 10.1167/iovs.11-8709. [DOI] [PubMed] [Google Scholar]
- Martinez-Mir A, Paloma E, Allikmets R, Ayuso C, del Rio T, Dean M, Vilageliu L, Gonzalez-Duarte R, Balcells S. Retinitis pigmentosa caused by a homozygous mutation in the Stargardt disease gene ABCR. Nat Genet. 1998;18:11–12. doi: 10.1038/ng0198-11. [DOI] [PubMed] [Google Scholar]
- Maugeri A, Klevering BJ, Rohrschneider K, Blankenagel A, Brunner HG, Deutman AF, Hoyng CB, Cremers FP. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. American journal of human genetics. 2000;67:960–966. doi: 10.1086/303079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeux R. Biomarkers: potential uses and limitations. NeuroRx : the journal of the American Society for Experimental NeuroTherapeutics. 2004;1:182–188. doi: 10.1602/neurorx.1.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay GJ, Patterson CC, Chakravarthy U, Dasari S, Klaver CC, Vingerling JR, Ho L, de Jong PT, Fletcher AE, Young IS, Seland JH, Rahu M, Soubrane G, Tomazzoli L, Topouzis F, Vioque J, Hingorani AD, Sofat R, Dean M, Sawitzke J, Seddon JM, Peter I, Webster AR, Moore AT, Yates JR, Cipriani V, Fritsche LG, Weber BH, Keilhauer CN, Lotery AJ, Ennis S, Klein ML, Francis PJ, Stambolian D, Orlin A, Gorin MB, Weeks DE, Kuo CL, Swaroop A, Othman M, Kanda A, Chen W, Abecasis GR, Wright AF, Hayward C, Baird PN, Guymer RH, Attia J, Thakkinstian A, Silvestri G. Evidence of association of APOE with age-related macular degeneration: a pooled analysis of 15 studies. Hum Mutat. 2011;32:1407–1416. doi: 10.1002/humu.21577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay GJ, Silvestri G, Patterson CC, Hogg RE, Chakravarthy U, Hughes AE. Further assessment of the complement component 2 and factor B region associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:533–539. doi: 10.1167/iovs.08-2275. [DOI] [PubMed] [Google Scholar]
- McKibbin M, Ali M, Bansal S, Baxter PD, West K, Williams G, Cassidy F, Inglehearn CF. CFH, VEGF and HTRA1 promoter genotype may influence the response to intravitreal ranibizumab therapy for neovascular age-related macular degeneration. Br J Ophthalmol. 2012;96:208–212. doi: 10.1136/bjo.2010.193680. [DOI] [PubMed] [Google Scholar]
- McManus MT. MicroRNAs and cancer. Seminars in Cancer Biology. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- Menzies-Gow A, Ying S, Sabroe I, Stubbs VL, Soler D, Williams TJ, Kay AB. Eotaxin (CCL11) and eotaxin-2 (CCL24) induce recruitment of eosinophils, basophils, neutrophils, and macrophages as well as features of early- and late-phase allergic reactions following cutaneous injection in human atopic and nonatopic volunteers. J Immunol. 2002;169:2712–2718. doi: 10.4049/jimmunol.169.5.2712. [DOI] [PubMed] [Google Scholar]
- Meyer KJ, Davis LK, Schindler EI, Beck JS, Rudd DS, Grundstad AJ, Scheetz TE, Braun TA, Fingert JH, Alward WL, Kwon YH, Folk JC, Russell SR, Wassink TH, Stone EM, Sheffield VC. Genome-wide analysis of copy number variants in age-related macular degeneration. Hum Genet. 2011;129:91–100. doi: 10.1007/s00439-010-0904-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers KJ, Mares JA, Igo RP, Jr, Truitt B, Liu Z, Millen AE, Klein M, Johnson EJ, Engelman CD, Karki CK, Blodi B, Gehrs K, Tinker L, Wallace R, Robinson J, LeBlanc ES, Sarto G, Bernstein PS, SanGiovanni JP, Iyengar SK. Genetic evidence for role of carotenoids in age-related macular degeneration in the Carotenoids in Age-Related Eye Disease Study (CAREDS) Invest Ophthalmol Vis Sci. 2014;55:587–599. doi: 10.1167/iovs.13-13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JW, Le Couter J, Strauss EC, Ferrara N. Vascular endothelial growth factor a in intraocular vascular disease. Ophthalmology. 2013;120:106–114. doi: 10.1016/j.ophtha.2012.07.038. [DOI] [PubMed] [Google Scholar]
- Mitchell P, Smith W, Wang JJ. Iris color, skin sun sensitivity, and age-related maculopathy - The Blue Mountains Eye Study. Ophthalmology. 1998;105:1359–1363. doi: 10.1016/S0161-6420(98)98013-7. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Kiyohara Y, Yoshida A, Iida M, Nose Y, Ishibashi T. The 5-year incidence and risk factors for age-related maculopathy in a general Japanese population: the Hisayama study. Invest Ophthalmol Vis Sci. 2005;46:1907–1910. doi: 10.1167/iovs.04-0923. [DOI] [PubMed] [Google Scholar]
- Mo FM, Proia AD, Johnson WH, Cyr D, Lashkari K. Interferon gamma-inducible protein-10 (IP-10) and eotaxin as biomarkers in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51:4226–4236. doi: 10.1167/iovs.09-3910. [DOI] [PubMed] [Google Scholar]
- Moons KG, Royston P, Vergouwe Y, Grobbee DE, Altman DG. Prognosis and prognostic research: what, why, and how? BMJ. 2009;338:b375. doi: 10.1136/bmj.b375. [DOI] [PubMed] [Google Scholar]
- Morohoshi K, Patel N, Ohbayashi M, Chong V, Grossniklaus HE, Bird AC, Ono SJ. Serum autoantibody biomarkers for age-related macular degeneration and possible regulators of neovascularization. Exp Mol Pathol. 2012;92:64–73. doi: 10.1016/j.yexmp.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Moussallieh FM, Elbayed K, Chanson JB, Rudolf G, Piotto M, De Seze J, Namer IJ. Serum analysis by 1H nuclear magnetic resonance spectroscopy: a new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult Scler. 2014;20:558–565. doi: 10.1177/1352458513504638. [DOI] [PubMed] [Google Scholar]
- Mullins RF, Faidley EA, Daggett HT, Jomary C, Lotery AJ, Stone EM. Localization of complement 1 inhibitor (C1INH/SERPING1) in human eyes with age-related macular degeneration. Experimental eye research. 2009;89:767–773. doi: 10.1016/j.exer.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Olvera MA, Clark AF, Stone EM. Fibulin-5 distribution in human eyes: relevance to age-related macular degeneration. Experimental eye research. 2007;84:378–380. doi: 10.1016/j.exer.2006.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- Nakamura T, Ruiz-Lozano P, Lindner V, Yabe D, Taniwaki M, Furukawa Y, Kobuke K, Tashiro K, Lu Z, Andon NL, Schaub R, Matsumori A, Sasayama S, Chien KR, Honjo T. DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J Biol Chem. 1999;274:22476–22483. doi: 10.1074/jbc.274.32.22476. [DOI] [PubMed] [Google Scholar]
- Nakata I, Yamashiro K, Akagi-Kurashige Y, Miyake M, Kumagai K, Tsujikawa A, Liu K, Chen LJ, Liu DT, Lai TY, Sakurada Y, Yoneyama S, Cheng CY, Cackett P, Yeo IY, Tay WT, Cornes BK, Vithana EN, Aung T, Matsuo K, Matsuda F, Wong TY, Iijima H, Pang CP, Yoshimura N. Association of genetic variants on 8p21 and 4q12 with age-related macular degeneration in Asian populations. Invest Ophthalmol Vis Sci. 2012;53:6576–6581. doi: 10.1167/iovs.12-10219. [DOI] [PubMed] [Google Scholar]
- Neale BM, Fagerness J, Reynolds R, Sobrin L, Parker M, Raychaudhuri S, Tan PL, Oh EC, Merriam JE, Souied E, Bernstein PS, Li B, Frederick JM, Zhang K, Brantley MA, Jr, Lee AY, Zack DJ, Campochiaro B, Campochiaro P, Ripke S, Smith RT, Barile GR, Katsanis N, Allikmets R, Daly MJ, Seddon JM. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc Natl Acad Sci U S A. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng EW, Adamis AP. Targeting angiogenesis, the underlying disorder in neovascular age-related macular degeneration. Can J Ophthalmol. 2005;40:352–368. doi: 10.1016/S0008-4182(05)80078-X. [DOI] [PubMed] [Google Scholar]
- Ni J, Yuan X, Gu J, Yue X, Gu X, Nagaraj RH, Crabb JW. Plasma protein pentosidine and carboxymethyllysine, biomarkers for age-related macular degeneration. Mol Cell Proteomics. 2009a;8:1921–1933. doi: 10.1074/mcp.M900127-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Yuan X, Gu J, Yue X, Gu X, Nagaraj RH, Crabb JW, Genomic TC, Group PAS. Plasma Protein Pentosidine and Carboxymethyllysine, Biomarkers for Age-related Macular Degeneration. Molecular & Cellular Proteomics. 2009b;8:1921–1933. doi: 10.1074/mcp.M900127-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas CM, Robman LD, Tikellis G, Dimitrov PN, Dowrick A, Guymer RH, McCarty CA. Iris colour, ethnic origin and progression of age-related macular degeneration. Clin Experiment Ophthalmol. 2003;31:465–469. doi: 10.1046/j.1442-9071.2003.00711.x. [DOI] [PubMed] [Google Scholar]
- Nischler C, Oberkofler H, Ortner C, Paikl D, Riha W, Lang N, Patsch W, Egger SF. Complement factor H Y402H gene polymorphism and response to intravitreal bevacizumab in exudative age-related macular degeneration. Acta ophthalmologica. 2011;89:e344–349. doi: 10.1111/j.1755-3768.2010.02080. [DOI] [PubMed] [Google Scholar]
- Nowak M, Swietochowska E, Wielkoszynski T, Marek B, Kos-Kudla B, Szapska B, Kajdaniuk D, Glogowska-Szelag J, Sieminska L, Ostrowska Z, Koziol H, Klimek J. Homocysteine, vitamin B12, and folic acid in age-related macular degeneration. Eur J Ophthalmol. 2005;15:764–767. [PubMed] [Google Scholar]
- Nozaki M, Raisler BJ, Sakurai E, Sarma JV, Barnum SR, Lambris JD, Chen Y, Zhang K, Ambati BK, Baffi JZ, Ambati J. Drusen complement components C3a and C5a promote choroidal neovascularization. Proc Natl Acad Sci U S A. 2006;103:2328–2333. doi: 10.1073/pnas.0408835103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenblatt RB, Ferris F., 3rd Age-related macular degeneration and the immune response: implications for therapy. Am J Ophthalmol. 2007;144:618–626. doi: 10.1016/j.ajo.2007.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. The Journal of Experimental Medicine. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlin A, Hadley D, Chang W, Ho AC, Brown G, Kaiser RS, Regillo CD, Godshalk AN, Lier A, Kaderli B, Stambolian D. Association between high-risk disease loci and response to anti-vascular endothelial growth factor treatment for wet age-related macular degeneration. Retina. 2012;32:4–9. doi: 10.1097/IAE.0b013e31822a2c7c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormsby RJ, Ranganathan S, Tong JC, Griggs KM, Dimasi DP, Hewitt AW, Burdon KP, Craig JE, Hoh J, Gordon DL. Functional and structural implications of the complement factor H Y402H polymorphism associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:1763–1770. doi: 10.1167/iovs.07-1297. [DOI] [PubMed] [Google Scholar]
- Orr N, Lemnrau A, Cooke R, Fletcher O, Tomczyk K, Jones M, Johnson N, Lord CJ, Mitsopoulos C, Zvelebil M, McDade SS, Buck G, Blancher C, Trainer AH, James PA, Bojesen SE, Bokmand S, Nevanlinna H, Mattson J, Friedman E, Laitman Y, Palli D, Masala G, Zanna I, Ottini L, Giannini G, Hollestelle A, Ouweland AM, Novakovic S, Krajc M, Gago-Dominguez M, Castelao JE, Olsson H, Hedenfalk I, Easton DF, Pharoah PD, Dunning AM, Bishop DT, Neuhausen SL, Steele L, Houlston RS, Garcia-Closas M, Ashworth A, Swerdlow AJ Consortium KC. Genome-wide association study identifies a common variant in RAD51B associated with male breast cancer risk. Nat Genet. 2012;44:1182–1184. doi: 10.1038/ng.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MP, Park Y, Parks MB, Burgess LG, Uppal K, Lee K, Jones DP, Brantley MA., Jr Metabolome-wide association study of neovascular age-related macular degeneration. PloS one. 2013;8:e72737. doi: 10.1371/journal.pone.0072737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen CG, Jarrar Z, Wormald R, Cook DG, Fletcher AE, Rudnicka AR. The estimated prevalence and incidence of late stage age related macular degeneration in the UK. Br J Ophthalmol. 2012;96:752–756. doi: 10.1136/bjophthalmol-2011-301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KH, Fridley BL, Ryu E, Tosakulwong N, Edwards AO. Complement Component 3 (C3) Haplotypes and Risk of Advanced Age-Related Macular Degeneration. Investigative Ophthalmology & Visual Science. 2009;50:3386–3393. doi: 10.1167/iovs.08-3231. [DOI] [PubMed] [Google Scholar]
- Park UC, Shin JY, McCarthy LC, Kim SJ, Park JH, Chung H, Yu HG. Pharmacogenetic associations with long-term response to anti-vascular endothelial growth factor treatment in neovascular AMD patients. Mol Vis. 2014;20:1680–1694. [PMC free article] [PubMed] [Google Scholar]
- Paun CC, Ersoy L, Schick T, Groenewoud JM, Lechanteur YT, Fauser S, Hoyng CB, de Jong EK, den Hollander AI. Genetic Variants and Systemic Complement Activation Levels Are Associated With Serum Lipoprotein Levels in Age-Related Macular Degeneration. Invest Ophthalmol Vis Sci. 2015;56:7766–7773. doi: 10.1167/iovs.15-17035. [DOI] [PubMed] [Google Scholar]
- Pavlou MP, Diamandis EP, Blasutig IM. The long journey of cancer biomarkers from the bench to the clinic. Clin Chem. 2013;59:147–157. doi: 10.1373/clinchem.2012.184614. [DOI] [PubMed] [Google Scholar]
- Penfold PL, Madigan MC, Gillies MC, Provis JM. Immunological and aetiological aspects of macular degeneration. Prog Retin Eye Res. 2001;20:385–414. doi: 10.1016/s1350-9462(00)00025-2. [DOI] [PubMed] [Google Scholar]
- Penn JS, Madan A, Caldwell RB, Bartoli M, Caldwell RW, Hartnett ME. Vascular endothelial growth factor in eye disease. Prog Retin Eye Res. 2008;27:331–371. doi: 10.1016/j.preteyeres.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. American journal of epidemiology. 2004;159:882–890. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- Perera FP, Weinstein IB. Molecular epidemiology: recent advances and future directions. Carcinogenesis. 2000;21:517–524. doi: 10.1093/carcin/21.3.517. [DOI] [PubMed] [Google Scholar]
- Peter I, Seddon JM. Genetic epidemiology: successes and challenges of genome-wide association studies using the example of age-related macular degeneration. Am J Ophthalmol. 2010;150:450–452. doi: 10.1016/j.ajo.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard E, Houssier M, Bujold K, Sapieha P, Lubell W, Dorfman A, Racine J, Hardy P, Febbraio M, Lachapelle P, Ong H, Sennlaub F, Chemtob S. CD36 plays an important role in the clearance of oxLDL and associated age-dependent sub-retinal deposits. Aging (Albany NY) 2010;2:981–989. doi: 10.18632/aging.100218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinna A, Zinellu A, Tendas D, Blasetti F, Carru C, Castiglia P. Plasma Homocysteine and Asymmetrical Dimethyl-l-Arginine (ADMA) and Whole Blood DNA Methylation in Early and Neovascular Age-Related Macular Degeneration: A Pilot Study. Curr Eye Res. 2016;41:88–96. doi: 10.3109/02713683.2014.1002044. [DOI] [PubMed] [Google Scholar]
- Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108:3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- Pons M, Marin-Castano ME. Nicotine increases the VEGF/PEDF ratio in retinal pigment epithelium: a possible mechanism for CNV in passive smokers with AMD. Invest Ophthalmol Vis Sci. 2011;52:3842–3853. doi: 10.1167/iovs.10-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya RR, Chew EY, Swaroop A. Genetic studies of age-related macular degeneration: lessons, challenges, and opportunities for disease management. Ophthalmology. 2012;119:2526–2536. doi: 10.1016/j.ophtha.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost AC, Vede L, Bigot K, Keller N, Tailleux A, Jais JP, Savoldelli M, Ameqrane I, Lacassagne E, Legeais JM, Staels B, Menasche M, Mallat Z, Behar-Cohen F, Abitbol M. Morphologic and electroretinographic phenotype of SR-BI knockout mice after a long-term atherogenic diet. Invest Ophthalmol Vis Sci. 2009;50:3931–3942. doi: 10.1167/iovs.08-2527. [DOI] [PubMed] [Google Scholar]
- Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, Baker A, Anand-Apte B. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nature medicine. 2003;9:407–415. doi: 10.1038/nm846. [DOI] [PubMed] [Google Scholar]
- Qian-Qian Y, Yong Y, Jing Z, Xin B, Tian-Hua X, Chao S, Jia C. Nonsynonymous single nucleotide polymorphisms in the complement component 3 gene are associated with risk of age-related macular degeneration: a meta-analysis. Gene. 2015;561:249–255. doi: 10.1016/j.gene.2015.02.039. [DOI] [PubMed] [Google Scholar]
- Quan YL, Zhou AY, Feng ZH. Association between complementary factor H Y402H polymorphisms and age-related macular degeneration in Chinese: Systematic review and meta-analysis. Int J Ophthalmol. 2012;5:242–246. doi: 10.3980/j.issn.2222-3959.2012.02.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery J, Clegg A, Jones J, Tan SC, Lotery A. Ranibizumab (Lucentis) versus bevacizumab (Avastin): modelling cost effectiveness. Br J Ophthalmol. 2007;91:1244–1246. doi: 10.1136/bjo.2007.116616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein DB, Wittenborn JS, Zhang X, Honeycutt AA, Lesesne SB, Saaddine J. Forecasting age-related macular degeneration through the year 2050: the potential impact of new treatments. Arch Ophthalmol. 2009;127:533–540. doi: 10.1001/archophthalmol.2009.58. [DOI] [PubMed] [Google Scholar]
- Restrepo NA, Spencer KL, Goodloe R, Garrett TA, Heiss G, Buzkova P, Jorgensen N, Jensen RA, Matise TC, Hindorff LA, Klein BE, Klein R, Wong TY, Cheng CY, Cornes BK, Tai ES, Ritchie MD, Haines JL, Crawford DC. Genetic determinants of age-related macular degeneration in diverse populations from the PAGE study. Invest Ophthalmol Vis Sci. 2014;55:6839–6850. doi: 10.1167/iovs.14-14246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R, Rosner B, Seddon JM. Serum lipid biomarkers and hepatic lipase gene associations with age-related macular degeneration. Ophthalmology. 2010;117:1989–1995. doi: 10.1016/j.ophtha.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robman L, Baird PN, Dimitrov PN, Richardson AJ, Guymer RH. C-reactive protein levels and complement factor H polymorphism interaction in age-related macular degeneration and its progression. Ophthalmology. 2010;117:1982–1988. doi: 10.1016/j.ophtha.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Rochtchina E, Wang JJ, Flood VM, Mitchell P. Elevated serum homocysteine, low serum vitamin B12, folate, and age-related macular degeneration: the Blue Mountains Eye Study. Am J Ophthalmol. 2007;143:344–346. doi: 10.1016/j.ajo.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Rodriguez de Cordoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. The human complement factor H: functional roles, genetic variations and disease associations. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Rohrer B, Long Q, Coughlin B, Renner B, Huang Y, Kunchithapautham K, Ferreira VP, Pangburn MK, Gilkeson GS, Thurman JM, Tomlinson S, Holers VM. A targeted inhibitor of the complement alternative pathway reduces RPE injury and angiogenesis in models of age-related macular degeneration. Adv Exp Med Biol. 2010;703:137–149. doi: 10.1007/978-1-4419-5635-4_10. [DOI] [PubMed] [Google Scholar]
- Ross RJ, Verma V, Rosenberg KI, Chan CC, Tuo J. Genetic markers and biomarkers for age-related macular degeneration. Expert Rev Ophthalmol. 2007;2:443–457. doi: 10.1586/17469899.2.3.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutar M, Natoli R, Provis JM. Small interfering RNA-mediated suppression of Ccl2 in Muller cells attenuates microglial recruitment and photoreceptor death following retinal degeneration. J Neuroinflammation. 2012;9:221. doi: 10.1186/1742-2094-9-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184. [PubMed] [Google Scholar]
- Sall JW, Klisovic DD, O’Dorisio MS, Katz SE. Somatostatin inhibits IGF-1 mediated induction of VEGF in human retinal pigment epithelial cells. Experimental eye research. 2004;79:465–476. doi: 10.1016/j.exer.2004.06.007. [DOI] [PubMed] [Google Scholar]
- SanGiovanni JP, Chen J, Gupta AS, Smith LE, Sapieha P, Lee PH. Netrin-1 - DCC Signaling Systems and Age-Related Macular Degeneration. PloS one. 2015;10:e0125548. doi: 10.1371/journal.pone.0125548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaumberg DA, Christen WG, Hankinson SE, Glynn RJ. Body mass index and the incidence of visually significant age-related maculopathy in men. Arch Ophthalmol. 2001;119:1259–1265. doi: 10.1001/archopht.119.9.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick T, Ersoy L, Lechanteur YT, Saksens NT, Hoyng CB, den Hollander AI, Kirchhof B, Fauser S. History of Sunlight Exposure Is a Risk Factor for Age-Related Macular Degeneration. Retina Retina. 2016;36:787–90. doi: 10.1097/IAE.0000000000000756. [DOI] [PubMed] [Google Scholar]
- Schneider R, Jensen SA, Whiteman P, McCullagh JS, Redfield C, Handford PA. Biophysical characterisation of fibulin-5 proteins associated with disease. J Mol Biol. 2010;401:605–617. doi: 10.1016/j.jmb.2010.06.039. [DOI] [PubMed] [Google Scholar]
- Schultz DW, Klein ML, Humpert AJ, Luzier CW, Persun V, Schain M, Mahan A, Runckel C, Cassera M, Vittal V, Doyle TM, Martin TM, Weleber RG, Francis PJ, Acott TS. Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Human molecular genetics. 2003;12:3315–3323. doi: 10.1093/hmg/ddg348. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Cote J, Davis N, Rosner B. Progression of age-related macular degeneration: association with body mass index, waist circumference, and waist-hip ratio. Arch Ophthalmol. 2003a;121:785–792. doi: 10.1001/archopht.121.6.785. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Cote J, Rosner B. Progression of age-related macular degeneration: association with dietary fat, transunsaturated fat, nuts, and fish intake. Arch Ophthalmol. 2003b;121:1728–1737. doi: 10.1001/archopht.121.12.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Gensler G, Klein ML, Milton RC. Evaluation of Plasma Homocysteine and Risk of Age-Related Macular Degeneration. American Journal of Ophthalmology. 2006;141:201–203. doi: 10.1016/j.ajo.2005.07.059. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Gensler G, Milton RC, Klein ML, Rifai N. Association between C-reactive protein and age-related macular degeneration. JAMA. 2004;291:704–710. doi: 10.1001/jama.291.6.704. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Gensler G, Rosner B. C-reactive protein and CFH, ARMS2/HTRA1 gene variants are independently associated with risk of macular degeneration. Ophthalmology. 2010a;117:1560–1566. doi: 10.1016/j.ophtha.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, George S, Rosner B, Rifai N. Progression of age-related macular degeneration: prospective assessment of C-reactive protein, interleukin 6, and other cardiovascular biomarkers. Arch Ophthalmol. 2005;123:774–782. doi: 10.1001/archopht.123.6.774. [DOI] [PubMed] [Google Scholar]
- Seddon JM, Reynolds R, Rosner B. Peripheral retinal drusen and reticular pigment: association with CFHY402H and CFHrs1410996 genotypes in family and twin studies. Invest Ophthalmol Vis Sci. 2009;50:586–591. doi: 10.1167/iovs.08-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Reynolds R, Rosner B. Associations of smoking, body mass index, dietary lutein, and the LIPC gene variant rs10468017 with advanced age-related macular degeneration. Mol Vis. 2010b;16:2412–2424. [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Reynolds R, Shah HR, Rosner B. Smoking, dietary betaine, methionine, and vitamin D in monozygotic twins with discordant macular degeneration: epigenetic implications. Ophthalmology. 2011;118:1386–1394. doi: 10.1016/j.ophtha.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sell DR, Nagaraj RH, Grandhee SK, Odetti P, Lapolla A, Fogarty J, Monnier VM. Pentosidine: a molecular marker for the cumulative damage to proteins in diabetes, aging, and uremia. Diabetes/metabolism reviews. 1991;7:239–251. doi: 10.1002/dmr.5610070404. [DOI] [PubMed] [Google Scholar]
- Shahid H, Khan JC, Cipriani V, Sepp T, Matharu BK, Bunce C, Harding SP, Clayton DG, Moore AT, Yates JR Genetic Factors in AMDSG. Age-related macular degeneration: the importance of family history as a risk factor. Br J Ophthalmol. 2012;96:427–431. doi: 10.1136/bjophthalmol-2011-300193. [DOI] [PubMed] [Google Scholar]
- Sharma NK, Prabhakar S, Gupta A, Singh R, Gupta PK, Gupta PK, Anand A. New biomarker for neovascular age-related macular degeneration: eotaxin-2. DNA Cell Biol. 2012;31:1618–1627. doi: 10.1089/dna.2012.1786. [DOI] [PubMed] [Google Scholar]
- Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, Campochiaro PA. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16:1208–1216. doi: 10.1038/mt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Hoffmann TJ, Melles RB, Sakoda LC, Kvale MN, Banda Y, Schaefer C, Risch N, Jorgenson E. Differences in the Genetic Susceptibility to Age-Related Macular Degeneration Clinical Subtypes. Invest Ophthalmol Vis Sci. 2015;56:4290–4299. doi: 10.1167/iovs.15-16533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroyer NF, Lewis RA, Allikmets R, Singh N, Dean M, Leppert M, Lupski JR. The rod photoreceptor ATP-binding cassette transporter gene, ABCR, and retinal disease: from monogenic to multifactorial. Vision research. 1999;39:2537–2544. doi: 10.1016/s0042-6989(99)00037-1. [DOI] [PubMed] [Google Scholar]
- Singh HP, Jalali S, Hejtmancik JF, Kannabiran C. Homozygous null mutations in the ABCA4 gene in two families with autosomal recessive retinal dystrophy. Am J Ophthalmol. 2006;141:906–913. doi: 10.1016/j.ajo.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Smailhodzic D, Klaver CC, Klevering BJ, Boon CJ, Groenewoud JM, Kirchhof B, Daha MR, den Hollander AI, Hoyng CB. Risk alleles in CFH and ARMS2 are independently associated with systemic complement activation in age-related macular degeneration. Ophthalmology. 2012;119:339–346. doi: 10.1016/j.ophtha.2011.07.056. [DOI] [PubMed] [Google Scholar]
- Smith W, Assink J, Klein R, Mitchell P, Klaver CC, Klein BE, Hofman A, Jensen S, Wang JJ, de Jong PT. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology. 2001a;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Smith W, Assink J, Klein R, Mitchell P, Klaver CCW, Klein BEK, Hofman A, Jensen S, Wang JJ, de Jong PTVM. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology. 2001b;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- Smith W, Mitchell P. Family history and age-related maculopathy: the Blue Mountains Eye Study. Aust N Z J Ophthalmol. 1998;26:203–206. doi: 10.1111/j.1442-9071.1998.tb01311.x. [DOI] [PubMed] [Google Scholar]
- Smith W, Mitchell P, Leeder SR. Smoking and age-related maculopathy. The Blue Mountains Eye Study. Arch Ophthalmol. 1996;114:1518–1523. doi: 10.1001/archopht.1996.01100140716016. [DOI] [PubMed] [Google Scholar]
- Smith W, Mitchell P, Leeder SR, Wang JJ. Plasma fibrinogen levels, other cardiovascular risk factors, and age-related maculopathy: the Blue Mountains Eye Study. Arch Ophthalmol. 1998;116:583–587. doi: 10.1001/archopht.116.5.583. [DOI] [PubMed] [Google Scholar]
- Smolinska A, Blanchet L, Buydens LM, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Anal Chim Acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- Solberg Y, Rosner M, Belkin M. The association between cigarette smoking and ocular diseases. Survey of ophthalmology. 1998;42:535–547. doi: 10.1016/s0039-6257(98)00002-2. [DOI] [PubMed] [Google Scholar]
- Sparrow JR, Parish CA, Hashimoto M, Nakanishi K. A2E, a lipofuscin fluorophore, in human retinal pigmented epithelial cells in culture. Invest Ophthalmol Vis Sci. 1999;40:2988–2995. [PubMed] [Google Scholar]
- Spencer KL, Hauser MA, Olson LM, Schmidt S, Scott WK, Gallins P, Agarwal A, Postel EA, Pericak-Vance MA, Haines JL. Protective effect of complement factor B and complement component 2 variants in age-related macular degeneration. Human molecular genetics. 2007;16:1986–1992. doi: 10.1093/hmg/ddm146. [DOI] [PubMed] [Google Scholar]
- Spencer KL, Olson LM, Anderson BM, Schnetz-Boutaud N, Scott WK, Gallins P, Agarwal A, Postel EA, Pericak-Vance MA, Haines JL. C3 R102G polymorphism increases risk of age-related macular degeneration. Human molecular genetics. 2008;17:1821–1824. doi: 10.1093/hmg/ddn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer KL, Olson LM, Schnetz-Boutaud N, Gallins P, Wang G, Scott WK, Agarwal A, Jakobsdottir J, Conley Y, Weeks DE, Gorin MB, Pericak-Vance MA, Haines JL. Dissection of chromosome 16p12 linkage peak suggests a possible role for CACNG3 variants in age-related macular degeneration susceptibility. Invest Ophthalmol Vis Sci. 2011;52:1748–1754. doi: 10.1167/iovs.09-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperduto RD, Hiller R. Systemic hypertension and age-related maculopathy in the Framingham Study. Arch Ophthalmol. 1986;104:216–219. doi: 10.1001/archopht.1986.01050140070022. [DOI] [PubMed] [Google Scholar]
- Stanton CM, Yates JR, den Hollander AI, Seddon JM, Swaroop A, Stambolian D, Fauser S, Hoyng C, Yu Y, Atsuhiro K, Branham K, Othman M, Chen W, Kortvely E, Chalmers K, Hayward C, Moore AT, Dhillon B, Ueffing M, Wright AF. Complement factor D in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:8828–8834. doi: 10.1167/iovs.11-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EM. Genetic testing for age-related macular degeneration: not indicated now. JAMA Ophthalmol. 2015;133:598–600. doi: 10.1001/jamaophthalmol.2015.0369. [DOI] [PubMed] [Google Scholar]
- Stone EM, Aldave AJ, Drack AV, Maccumber MW, Sheffield VC, Traboulsi E, Weleber RG. Recommendations for genetic testing of inherited eye diseases: report of the American Academy of Ophthalmology task force on genetic testing. Ophthalmology. 2012;119:2408–2410. doi: 10.1016/j.ophtha.2012.05.047. [DOI] [PubMed] [Google Scholar]
- Stone EM, Braun TA, Russell SR, Kuehn MH, Lotery AJ, Moore PA, Eastman CG, Casavant TL, Sheffield VC. Missense variations in the fibulin 5 gene and age-related macular degeneration. N Engl J Med. 2004;351:346–353. doi: 10.1056/NEJMoa040833. [DOI] [PubMed] [Google Scholar]
- Stone EM, Lotery AJ, Munier FL, Heon E, Piguet B, Guymer RH, Vandenburgh K, Cousin P, Nishimura D, Swiderski RE, Silvestri G, Mackey DA, Hageman GS, Bird AC, Sheffield VC, Schorderet DF. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- Storkebaum E, Carmeliet P. VEGF: a critical player in neurodegeneration. J Clin Invest. 2004;113:14–18. doi: 10.1172/JCI200420682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Zhao M, Li X. CFB/C2 gene polymorphisms and risk of age-related macular degeneration: a systematic review and meta-analysis. Curr Eye Res. 2012;37:259–271. doi: 10.3109/02713683.2011.635401. [DOI] [PubMed] [Google Scholar]
- Sundermeier TR, Palczewski K. The physiological impact of microRNA gene regulation in the retina. Cell Mol Life Sci. 2012;69:2739–2750. doi: 10.1007/s00018-012-0976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suner IJ, Espinosa-Heidmann DG, Marin-Castano ME, Hernandez EP, Pereira-Simon S, Cousins SW. Nicotine increases size and severity of experimental choroidal neovascularization. Invest Ophthalmol Vis Sci. 2004;45:311–317. doi: 10.1167/iovs.03-0733. [DOI] [PubMed] [Google Scholar]
- Sunness JS, Gonzalez-Baron J, Applegate CA, Bressler NM, Tian Y, Hawkins B, Barron Y, Bergman A. Enlargement of atrophy and visual acuity loss in the geographic atrophy form of age-related macular degeneration. Ophthalmology. 1999;106:1768–1779. doi: 10.1016/S0161-6420(99)90340-8. [DOI] [PubMed] [Google Scholar]
- Sutcliffe P, Hummel S, Simpson E, Young T, Rees A, Wilkinson A, Hamdy F, Clarke N, Staffurth J. Use of classical and novel biomarkers as prognostic risk factors for localised prostate cancer: a systematic review. Health Technol Assess. 2009;13:iii, xi–xiii, 1–219. doi: 10.3310/hta13050. [DOI] [PubMed] [Google Scholar]
- Szemraj M, Bielecka-Kowalska A, Oszajca K, Krajewska M, Gos R, Jurowski P, Kowalski M, Szemraj J. Serum MicroRNAs as Potential Biomarkers of AMD. Med Sci Monit. 2015;21:2734–2742. doi: 10.12659/MSM.893697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JSL, Wang JJ, Flood V, Rochtchina E, Smith W, Mitchell P. Dietary Antioxidants and the Long-term Incidence of Age-Related Macular Degeneration: The Blue Mountains Eye Study. Ophthalmology. 2008;115:334–341. doi: 10.1016/j.ophtha.2007.03.083. [DOI] [PubMed] [Google Scholar]
- Taylor HR, West S, Munoz B, Rosenthal FS, Bressler SB, Bressler NM. The Long-Term Effects of Visible-Light On The Eye. Arch Ophthalmol. 1992;110:99–104. doi: 10.1001/archopht.1992.01080130101035. [DOI] [PubMed] [Google Scholar]
- Taylor JM, Ankerst DP, Andridge RR. Validation of biomarker-based risk prediction models. Clin Cancer Res. 2008;14:5977–5983. doi: 10.1158/1078-0432.CCR-07-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton JP, Freeman NE, Nickerson JM, Jablonski MM, Rex TS, Williams RW, Geisert EE. Innate immune network in the retina activated by optic nerve crush. Invest Ophthalmol Vis Sci. 2013;54:2599–2606. doi: 10.1167/iovs.12-11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper SJ, Nowinska A, Pilat J, Palucha A, Wylegala E. Involvement of genetic factors in the response to a variable-dosing ranibizumab treatment regimen for age-related macular degeneration. Mol Vis. 2010;16:2598–2604. [PMC free article] [PubMed] [Google Scholar]
- Thakkinstian A, McEvoy M, Chakravarthy U, Chakrabarti S, McKay GJ, Ryu E, Silvestri G, Kaur I, Francis P, Iwata T, Akahori M, Arning A, Edwards AO, Seddon JM, Attia J. The association between complement component 2/complement factor B polymorphisms and age-related macular degeneration: a HuGE review and meta-analysis. American journal of epidemiology. 2012;176:361–372. doi: 10.1093/aje/kws031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Eye Disease Case-Control Study Group. Risk Factors for Neovascular Age-Related Macular Degeneration. Arch Ophthalmol. 1992;110:1701–1708. doi: 10.1001/archopht.1992.01080240041025. [DOI] [PubMed] [Google Scholar]
- Thompson CL, Klein BE, Klein R, Xu Z, Capriotti J, Joshi T, Leontiev D, Lee KE, Elston RC, Iyengar SK. Complement factor H and hemicentin-1 in age-related macular degeneration and renal phenotypes. Human molecular genetics. 2007;16:2135–2148. doi: 10.1093/hmg/ddm164. [DOI] [PubMed] [Google Scholar]
- Thornton J, Edwards R, Mitchell P, Harrison RA, Buchan I, Kelly SP. Smoking and age-related macular degeneration: a review of association. Eye (Lond) 2005;19:935–944. doi: 10.1038/sj.eye.6701978. [DOI] [PubMed] [Google Scholar]
- Tomany SC, Klein R, Klein BE. The relationship between iris color, hair color, and skin sun sensitivity and the 10-year incidence of age-related maculopathy: the Beaver Dam Eye Study. Ophthalmology. 2003;110:1526–1533. doi: 10.1016/s0161-6420(03)00539-6. [DOI] [PubMed] [Google Scholar]
- Toops KA, Tan LX, Lakkaraju A. Apolipoprotein E Isoforms and AMD. Adv Exp Med Biol. 2016;854:3–9. doi: 10.1007/978-3-319-17121-0_1. [DOI] [PubMed] [Google Scholar]
- Tsai DC, Charng MJ, Lee FL, Hsu WM, Chen SJ. Different plasma levels of vascular endothelial growth factor and nitric oxide between patients with choroidal and retinal neovascularization. Ophthalmologica. 2006;220:246–251. doi: 10.1159/000093079. [DOI] [PubMed] [Google Scholar]
- Tserentsoodol N, Gordiyenko NV, Pascual I, Lee JW, Fliesler SJ, Rodriguez IR. Intraretinal lipid transport is dependent on high density lipoprotein-like particles and class B scavenger receptors. Mol Vis. 2006;12:1319–1333. [PubMed] [Google Scholar]
- Tuo J, Bojanowski CM, Zhou M, Shen D, Ross RJ, Rosenberg KI, Cameron DJ, Yin C, Kowalak JA, Zhuang Z, Zhang K, Chan CC. Murine ccl2/cx3cr1 deficiency results in retinal lesions mimicking human age-related macular degeneration. Invest Ophthalmol Vis Sci. 2007;48:3827–3836. doi: 10.1167/iovs.07-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuo J, Ning B, Bojanowski CM, Lin ZN, Ross RJ, Reed GF, Shen D, Jiao X, Zhou M, Chew EY, Kadlubar FF, Chan CC. Synergic effect of polymorphisms in ERCC6 5′ flanking region and complement factor H on age-related macular degeneration predisposition. Proc Natl Acad Sci U S A. 2006;103:9256–9261. doi: 10.1073/pnas.0603485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara H, Mamalis C, McFadden M, Taggart M, Stagg B, Passi S, Earle P, Chakravarthy U, Hogg RE, Ambati BK. The reduction of serum soluble Flt-1 in patients with neovascular age-related macular degeneration. Am J Ophthalmol. 2015;159:92–100. e101–102. doi: 10.1016/j.ajo.2014.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79:581–588. doi: 10.1093/cvr/cvn156. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vecino E, Hernandez M, Garcia M. Cell death in the developing vertebrate retina. Int J Dev Biol. 2004;48:965–974. doi: 10.1387/ijdb.041891ev. [DOI] [PubMed] [Google Scholar]
- Vessey KA, Greferath U, Jobling AI, Phipps JA, Ho T, Waugh M, Fletcher EL. Ccl2/Cx3cr1 knockout mice have inner retinal dysfunction but are not an accelerated model of AMD. Invest Ophthalmol Vis Sci. 2012;53:7833–7846. doi: 10.1167/iovs.12-10650. [DOI] [PubMed] [Google Scholar]
- Vine AK, Stader J, Branham K, Musch DC, Swaroop A. Biomarkers of Cardiovascular Disease as Risk Factors for Age-Related Macular Degeneration. Ophthalmology. 2005;112:2076–2080. doi: 10.1016/j.ophtha.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Vingerling JR, Dielemans I, Hofman A, Grobbee DE, Hijmering M, Kramer CF, de Jong PT. The prevalence of age-related maculopathy in the Rotterdam Study. Ophthalmology. 1995a;102:205–210. doi: 10.1016/s0161-6420(95)31034-2. [DOI] [PubMed] [Google Scholar]
- Vingerling JR, Klaver CC, Hofman A, de Jong PT. Epidemiology of age-related maculopathy. Epidemiol Rev. 1995b;17:347–360. doi: 10.1093/oxfordjournals.epirev.a036198. [DOI] [PubMed] [Google Scholar]
- Wada Y, Abe T, Itabashi T, Sato H, Kawamura M, Tamai M. Autosomal dominant macular degeneration associated with 208delG mutation in the FSCN2 gene. Arch Ophthalmol. 2003;121:1613–1620. doi: 10.1001/archopht.121.11.1613. [DOI] [PubMed] [Google Scholar]
- Walker JC, Harland RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev. 2009;23:1046–1051. doi: 10.1101/gad.1777709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhou J, Hou X, Nguyen DH, Cao G, Li G, Qiu G, Zhang K, Zhang M, Su Z. CETP Gene may be Associated with Advanced Age-related Macular Degeneration in the Chinese Population. Ophthalmic genetics. 2015a;21:1–6. doi: 10.3109/13816810.2014.881506. [DOI] [PubMed] [Google Scholar]
- Wang G, Spencer KL, Court BL, Olson LM, Scott WK, Haines JL, Pericak-Vance MA. Localization of age-related macular degeneration-associated ARMS2 in cytosol, not mitochondria. Invest Ophthalmol Vis Sci. 2009;50:3084–3090. doi: 10.1167/iovs.08-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JJ, Rochtchina E, Lee AJ, Chia EM, Smith W, Cumming RG, Mitchell P. Ten-year incidence and progression of age-related maculopathy: the Blue Mountains Eye Study. Ophthalmology. 2007;114:92–98. doi: 10.1016/j.ophtha.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Wang S, Koster KM, He Y, Zhou Q. miRNAs as potential therapeutic targets for age-related macular degeneration. Future Med Chem. 2012a;4:277–287. doi: 10.4155/fmc.11.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VM, Rosen RB, Meyerle CB, Kurup SK, Ardeljan D, Agron E, Tai K, Pomykala M, Chew EY, Chan CC, Tuo J. Suggestive association between PLA2G12A single nucleotide polymorphism rs2285714 and response to anti-vascular endothelial growth factor therapy in patients with exudative age-related macular degeneration. Mol Vis. 2012b;18:2578–2585. [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bian ZM, Yu WZ, Yan Z, Chen WC, Li XX. Induction of interleukin-8 gene expression and protein secretion by C-reactive protein in ARPE-19 cells. Experimental eye research. 2010;91:135–142. doi: 10.1016/j.exer.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Wang YF, Han Y, Zhang R, Qin L, Wang MX, Ma L. CETP/LPL/LIPC gene polymorphisms and susceptibility to age-related macular degeneration. Sci Rep. 2015b;5:15711. doi: 10.1038/srep15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BH, Vogt G, Pruett RC, Stohr H, Felbor U. Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby’s fundus dystrophy. Nat Genet. 1994;8:352–356. doi: 10.1038/ng1294-352. [DOI] [PubMed] [Google Scholar]
- Weismann D, Hartvigsen K, Lauer N, Bennett KL, Scholl HP, Charbel Issa P, Cano M, Brandstatter H, Tsimikas S, Skerka C, Superti-Furga G, Handa JT, Zipfel PF, Witztum JL, Binder CJ. Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature. 2011;478:76–81. doi: 10.1038/nature10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westeneng-van Haaften SC, Boon CJ, Cremers FP, Hoefsloot LH, den Hollander AI, Hoyng CB. Clinical and genetic characteristics of late-onset Stargardt’s disease. Ophthalmology. 2012;119:1199–1210. doi: 10.1016/j.ophtha.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Wickremasinghe SS, Sandhu SS, Amirul-Islam FM, Abedi F, Richardson AJ, Baird PN, Guymer RH. Polymorphisms in the APOE gene and the location of retinal fluid in eyes with neovascular age-related macular degeneration. Retina. 2014;34:2367–2375. doi: 10.1097/IAE.0000000000000258. [DOI] [PubMed] [Google Scholar]
- Wickremasinghe SS, Xie J, Lim J, Chauhan DS, Robman L, Richardson AJ, Hageman G, Baird PN, Guymer R. Variants in the APOE gene are associated with improved outcome after anti-VEGF treatment for neovascular AMD. Invest Ophthalmol Vis Sci. 2011;52:4072–4079. doi: 10.1167/iovs.10-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Wong CW, Liao J, Cheung GC, Khor CC, Vithana EN, Wang JJ, Mitchell P, Aung T, Wong TY, Cheng CY. Lens status influences the association between CFH polymorphisms and age-related macular degeneration: findings from two population-based studies in Singapore. PloS one. 2015;10:e0119570. doi: 10.1371/journal.pone.0119570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TY, Chakravarthy U, Klein R, Mitchell P, Zlateva G, Buggage R, Fahrbach K, Probst C, Sledge I. The natural history and prognosis of neovascular age-related macular degeneration: a systematic review of the literature and meta-analysis. Ophthalmology. 2008;115:116–126. doi: 10.1016/j.ophtha.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Woo SJ, Ahn J, Morrison MA, Ahn SY, Lee J, Kim KW, DeAngelis MM, Park KH. Analysis of Genetic and Environmental Risk Factors and Their Interactions in Korean Patients with Age-Related Macular Degeneration. PloS one. 2015;10:e0132771. doi: 10.1371/journal.pone.0132771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Gao Y, Ren AJ, Zhao SH, Zhong M, Peng YJ, Shen W, Jing M, Liu L. Altered microRNA expression profiles in retinas with diabetic retinopathy. Ophthalmic Res. 2012;47:195–201. doi: 10.1159/000331992. [DOI] [PubMed] [Google Scholar]
- Wu L, Tao Q, Chen W, Wang Z, Song Y, Sheng S, Li P, Zhou J. Association between polymorphisms of complement pathway genes and age-related macular degeneration in a Chinese population. Invest Ophthalmol Vis Sci. 2013;54:170–174. doi: 10.1167/iovs.12-10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt MK, Tsai JY, Mishra S, Campos M, Jaworski C, Fariss RN, Bernstein SL, Wistow G. Interaction of complement factor h and fibulin3 in age-related macular degeneration. PloS one. 2013;8:e68088. doi: 10.1371/journal.pone.0068088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Chang L, Guan Y, Wang X. C-reactive protein augments interleukin-8 secretion in human peripheral blood monocytes. J Cardiovasc Pharmacol. 2005;46:690–696. doi: 10.1097/01.fjc.0000183568.48389.a1. [DOI] [PubMed] [Google Scholar]
- Yan N, Ma K, Ma J, Chen W, Wang Y, Cao G, Man-Kit Lam D, Liu X. Profiling microRNAs differentially expressed in rabbit retina. Adv Exp Med Biol. 2010;664:203–209. doi: 10.1007/978-1-4419-1399-9_23. [DOI] [PubMed] [Google Scholar]
- Yang D, Elner SG, Chen X, Field MG, Petty HR, Elner VM. MCP-1-activated monocytes induce apoptosis in human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2011;52:6026–6034. doi: 10.1167/iovs.10-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Hu J, Zhang J, Guan H. Polymorphisms in CFH, HTRA1 and CX3CR1 confer risk to exudative age-related macular degeneration in Han Chinese. Br J Ophthalmol. 2010;94:1211–1214. doi: 10.1136/bjo.2009.165811. [DOI] [PubMed] [Google Scholar]
- Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar V, Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- Yasuma TR, Nakamura M, Nishiguchi KM, Kikuchi M, Kaneko H, Niwa T, Hamajima N, Terasaki H. Elevated C-reactive protein levels and ARMS2/HTRA1 gene variants in subjects without age-related macular degeneration. Mol Vis. 2010;16:2923–2930. [PMC free article] [PubMed] [Google Scholar]
- Yates JR, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- Ye Z, Shuai P, Zhai Y, Li F, Jiang L, Lu F, Wen F, Huang L, Zhang D, Liu X, Lin Y, Luo H, Zhang H, Zhu X, Wu Z, Yang Z, Gong B, Shi Y. Associations of 6p21.3 Region with Age-related Macular Degeneration and Polypoidal Choroidal Vasculopathy. Sci Rep. 2016;6:20914. doi: 10.1038/srep20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokura S, Wada Y, Nakai S, Sato H, Yao R, Yamanaka H, Ito S, Sagara Y, Takahashi M, Nakamura Y, Tamai M, Noda T. Targeted disruption of FSCN2 gene induces retinopathy in mice. Invest Ophthalmol Vis Sci. 2005;46:2905–2915. doi: 10.1167/iovs.04-0856. [DOI] [PubMed] [Google Scholar]
- Yoshina S, Sakaki K, Yonezumi-Hayashi A, Gengyo-Ando K, Inoue H, Iino Y, Mitani S. Identification of a novel ADAMTS9/GON-1 function for protein transport from the ER to the Golgi. Mol Biol Cell. 2012;23:1728–1741. doi: 10.1091/mbc.E11-10-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Wai-Man P, Griffiths PG, Chinnery PF. Mitochondrial optic neuropathies - disease mechanisms and therapeutic strategies. Prog Retin Eye Res. 2011;30:81–114. doi: 10.1016/j.preteyeres.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Bhangale TR, Fagerness J, Ripke S, Thorleifsson G, Tan PL, Souied EH, Richardson AJ, Merriam JE, Buitendijk GH, Reynolds R, Raychaudhuri S, Chin KA, Sobrin L, Evangelou E, Lee PH, Lee AY, Leveziel N, Zack DJ, Campochiaro B, Campochiaro P, Smith RT, Barile GR, Guymer RH, Hogg R, Chakravarthy U, Robman LD, Gustafsson O, Sigurdsson H, Ortmann W, Behrens TW, Stefansson K, Uitterlinden AG, van Duijn CM, Vingerling JR, Klaver CC, Allikmets R, Brantley MA, Jr, Baird PN, Katsanis N, Thorsteinsdottir U, Ioannidis JP, Daly MJ, Graham RR, Seddon JM. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Human molecular genetics. 2011;20:3699–3709. doi: 10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan D, Yang Q, Liu X, Yuan D, Yuan S, Xie P, Liu Q. Complement factor H Val62Ile variant and risk of age-related macular degeneration: a meta-analysis. Mol Vis. 2013;19:374–383. [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Chen Y, Tong Z, Zhou X, Zhao C, Wang K, Hughes G, Kasuga D, Bedell M, Lee C, Ferreyra H, Kozak I, Haw W, Guan J, Shaw R, Stevenson W, Weishaar PD, Nelson MH, Tang L, Zhang K. Lack of association of CFD polymorphisms with advanced age-related macular degeneration. Mol Vis. 2010;16:2273–2278. [PMC free article] [PubMed] [Google Scholar]
- Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- Zerbib J, Richard F, Puche N, Leveziel N, Cohen SY, Korobelnik JF, Sahel J, Munnich A, Kaplan J, Rozet JM, Souied EH. R102G polymorphism of the C3 gene associated with exudative age-related macular degeneration in a French population. Mol Vis. 2010;16:1324–1330. [PMC free article] [PubMed] [Google Scholar]
- Zerbib J, Seddon JM, Richard F, Reynolds R, Leveziel N, Benlian P, Borel P, Feingold J, Munnich A, Soubrane G, Kaplan J, Rozet JM, Souied EH. rs5888 variant of SCARB1 gene is a possible susceptibility factor for age-related macular degeneration. PloS one. 2009;4:e7341. doi: 10.1371/journal.pone.0007341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterberg M, Landgren S, Andersson ME, Palmer MS, Gustafson DR, Skoog I, Minthon L, Thelle DS, Wallin A, Bogdanovic N, Andreasen N, Blennow K, Zetterberg H. Association of complement factor H Y402H gene polymorphism with Alzheimer’s disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:720–726. doi: 10.1002/ajmg.b.30668. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Li S, Xiao X, Jia X, Guo X. The 208delG mutation in FSCN2 does not associate with retinal degeneration in Chinese individuals. Invest Ophthalmol Vis Sci. 2007;48:530–533. doi: 10.1167/iovs.06-0669. [DOI] [PubMed] [Google Scholar]
- Zhang X, Li M, Wen F, Zuo C, Chen H, Wu K, Zeng R. Different impact of high-density lipoprotein-related genetic variants on polypoidal choroidal vasculopathy and neovascular age-related macular degeneration in a Chinese Han population. Experimental eye research. 2013;108:16–22. doi: 10.1016/j.exer.2012.12.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Marmorstein LY. Focus on molecules: fibulin-3 (EFEMP1) Experimental eye research. 2010;90:374–375. doi: 10.1016/j.exer.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Shen J, Medico L, Wang D, Ambrosone CB, Liu S. A Pilot Study of Circulating miRNAs as Potential Biomarkers of Early Stage Breast Cancer. PloS one. 2010;5:e13735. doi: 10.1371/journal.pone.0013735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Sharp DS, Grove JS, Bruce C, Yano K, Curb JD, Tall AR. Increased coronary heart disease in Japanese-American men with mutation in the cholesteryl ester transfer protein gene despite increased HDL levels. J Clin Invest. 1996;97:2917–2923. doi: 10.1172/JCI118751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wang D, Zhang J, Zhang M, Lu F, Qiu G, Zhao L, Nguyen DH, Luo H, Cao G, Zhang W, Jiang W, Li G, Zhang K, Zhang M, Su Z. RAD51 gene is associated with advanced age-related macular degeneration in Chinese population. Clin Biochem. 2013;46:1689–1693. doi: 10.1016/j.clinbiochem.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci U S A. 2011;108:8287–8292. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Sun W, Okano K, Chen Y, Zhang N, Maeda T, Palczewski K. Sponge transgenic mouse model reveals important roles for the microRNA-183 (miR-183)/96/182 cluster in postmitotic photoreceptors of the retina. J Biol Chem. 2011;286:31749–31760. doi: 10.1074/jbc.M111.259028. [DOI] [PMC free article] [PubMed] [Google Scholar]