Abstract
RNA interference and CRISPR/Cas9 technologies now enable systematic discovery of genes that regulate key pathways in the complex interaction between immune cells and tumor cells. Discovery screens are feasible in an in vivo setting, allowing identification of genes that limit the effectiveness of anti-tumor immunity. In vivo discovery screens can be informed by single-cell RNA-seq experiments that define the differentially expressed genes between functionally distinct immune cell subpopulations, both in humans and relevant animal models. Novel targets for cancer immunotherapy are being defined by the in depth functional annotation of immunosuppressive pathways in the tumor microenvironment.
I. Introduction
Recent clinical trials have demonstrated that targeting of inhibitory pathways in the immune system can enhance endogenous anti-tumor immunity and result in durable clinical benefit even in patients with advanced cancer. Most impressive is the fact that immunotherapy can enable long-term survival of patients with metastatic cancer, as first shown with the CTLA-4 blocking antibody Ipilimumab [1-3]. Antibodies that target the inhibitory PD-1 receptor on T cells or its PD-L1 ligand induce clinical responses in many human cancers, including melanoma, lung cancer, renal cancer, bladder cancer and Hodgkin's lymphoma [4-6]. These exciting advances are based on careful mechanistic studies that defined these immunological pathways and their role in autoimmunity, tumor immunity, and infectious diseases [7].
The discovery of such immuno-regulatory pathways has in the past been a very slow process in which each step, such as the cloning of the relevant genes and the generation of knockout mice, required years of effort. However, a series of recent technological advances are now greatly accelerating the pace of discovery and enable systematic identification of key regulators of immune responses. Importantly, such discovery efforts can now be performed in vivo in the relevant tissue microenvironment and thereby enable identification of regulatory mechanisms that control immune processes in particular pathologic states. This review will provide a synopsis of these new technological and conceptual approaches and the exciting discoveries that have been made in this process.
The tremendous complexity of biological systems has frequently necessitated the development of highly reductionist systems. However, it is this complexity that makes the study of biology so intriguing. Genome-scale discovery tools enable the delineation of regulatory circuits in a systematic manner and thereby help investigators to unravel the complexity of their biological system. Our growing understanding of complex immuno-regulatory mechanisms will create opportunities for specific intervention in human diseases for which no therapeutic opportunities are currently available.
II. Key advances in the development of genome-scale discovery approaches
Development of pooled shRNA screens
The discovery of the RNA (RNAi) interference pathway enabled the development of systematic genetic screens that could be widely implemented. Short-hairpin RNAs (shRNA) take advantage of the microRNA pathway: precursor microRNAs are first cleaved in the nucleus by the enzyme Drosha and further processed in the cytosol by Dicer, resulting in the formation of a miRNA duplex. This duplex is unwound, and one of the strands (the guide strand) is packaged into the RNA-induced silencing complex (RISC) formed by Argonaute and other proteins. The microRNA then guides the RISC to the 3’ UTR of the relevant mRNA, leading to decreased mRNA stability and/or inhibition of translation [8]. ShRNAs have a short, synthetic hairpin between the guide and the passenger strands which is cleaved by Dicer into an active duplex for loading into RISC. ShRNAs can be readily synthesized by annealing of two long oligonucleotides, which has enabled the generation of large libraries in lentiviral vectors.
Early shRNA screens were performed in an arrayed format in which the effect of each shRNA was tested individually in 96 or 384 well plates - a time-consuming and expensive process [9,10]. A breakthrough in this field was the development of pooled screening formats in which large numbers of shRNAs could be examined simultaneously in populations of cells [11]. The lentiviral vector used to introduce a shRNA library into cells of interest integrates into the genome, enabling tracking of shRNA representation before and after a selection step that is designed to interrogate a particular biological pathway. Early approaches introduced separate barcodes into such libraries which were quantified by hybridization to custom microarrays. Later, it was shown that the shRNAs themselves could be utilized for quantification [12,13]. A second breakthrough in the development of pooled screens was the use of deep sequencing for accurate quantification of shRNA representation [14]. Sequencing provides two important advantages: the signal is linear over a wide concentration range, and the background is substantially lower compared to the microarray-based readouts. These two aspects have greatly improved the sensitivity of shRNA screens, which is particularly important for depletion screens in which shRNAs of interest are lost as the result of the selection process.
In vivo screens for discovery of tumor suppressor genes
The development of such pooled screening approaches enabled in vivo discovery of key regulators in tumor and immune cells. Early in vivo shRNA screens focused on the discovery of tumor suppressor genes because loss of function enhances tumor growth in a cell autonomous manner. The strong enrichment of shRNAs targeting tumor suppressor genes is the key factor for the success of such screens. The first of these studies focused on the analysis of putative tumor suppressors located in focal deletions in human liver cancer [15]. Libraries with 48 shRNAs per pool were introduced into premalignant liver cell progenitors (overexpression of Myc and deletion of p53). Control shRNA pools did not induce tumors in immunodeficient mice, but shRNA pools targeting potential tumor suppressor genes resulted in tumor growth in most mice. These experiments led to the identification of 13 tumor suppressor genes of which only one had previously been linked to cancer. This approach has since been utilized to discover a substantial number of tumor suppressors in other types of cancer [16,17].
Several key aspects need to be considered in the design of informative in vivo screens. First, it is critical to ensure a sufficient representation of shRNAs (or gRNAs for CRISPR screens) in the cells or interest. At least 100 cells should carry each of the individual shRNAs to prevent bias in screening by stochastic events. Second, it is very important to consider the bottlenecks in the in vivo survival/migration/function of the interrogated cells. For example, even though large numbers of tumor cells are injected into a mouse, only relatively few survive and form a tumor. It is the number of surviving cells that determine how many shRNAs can be interrogated in one pool. Third, it is important to consider that shRNAs can have off-target effects, making it essential to identify multiple shRNAs with the same phenotype and to rigorously demonstrate that the observed phenotype is indeed on-target.
III. In vivo discovery of regulators of T cell function in tumors and infectious diseases
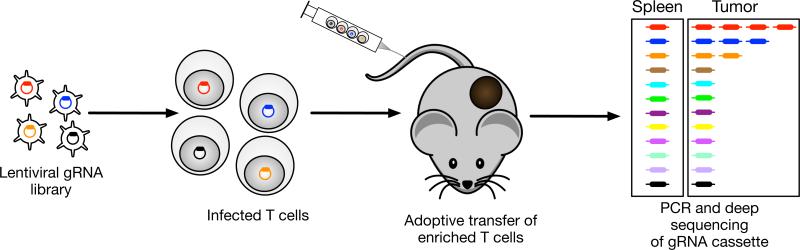
The proliferation and effector function of cytotoxic T cells is strongly inhibited by multiple immunosuppressive cell populations in the tumor microenvironment. A pooled shRNA screening approach was used to discover key regulatory pathways in CD8 T cells that inhibit their response following tumor antigen recognition (Figure 1) [18]. Pools of ~1,000 shRNAs were introduced into CD8 T cells (from OT-I transgenic mice) that recognized an ovalbumin peptide presented by B16-Ova melanoma cells. ShRNAs that targeted negative regulators became strongly enriched in tumors but not control organs by improving the ability of T cells to proliferate in response to tumor antigen recognition. The screen included genes upregulated in dysfunctional T cells (T cell anergy and exhaustion) as well as kinases and phosphatases. A total of 43 genes were identified for which at least three shRNAs enhanced T cell accumulation in tumors relative to a control organ. Several known negative regulators of TCR signaling were identified (Cbl-b, Dgka), validating this approach. The most strongly enriched shRNAs targeted Ppp2r2d which encodes a regulatory subunit of the PP2A family of phosphatases. Silencing of Ppp2r2d enhanced T cell proliferation, increased cytokine production and protected T cells from apoptosis within tumors. A shRNA targeting Ppp2r2d was also shown to enhance the anti-tumor activity of transferred T cells in the B16-Ova melanoma model [18]. The discovered genes could therefore be targeted to improve the efficacy of adoptive T cell therapy. An shRNA expression cassette could be readily included in the lentiviral vector used to introduce a chimeric antigen receptor (CAR) or TCR into peripheral blood T cells of cancer patients.
Figure 1. Design of in vivo CRISPR screen for discovery of negative regulators of immune function in the tumor microenvironment.
T cells specific for a tumor antigen are transduced with a lentiviral vector encoding a pooled gRNA library. T cells are isolated from Cas9 transgenic mice because the large size of the Cas9 cDNA greatly diminishes lentiviral titers. T cells in which a gRNA inactivates a negative regulator proliferate in response to tumor antigen recognition; such proliferation results in accumulation of the corresponding gRNAs in the tumor. The representation of all gRNAs is quantified by deep sequencing following isolation of T cells from tumors and control tissues. A similar strategy was previously used for an in vivo shRNA screen that identified negative regulators of T cell function in the tumor microenvironment [18].
In the course of an infection, T cells differentiate into short-lived effector and long-lived memory populations. A focused in vivo shRNA screen was performed in T cells to discover regulators that drive these competing cellular fates [19]. CD4 or CD8 T cells specific for LCMV were infected in vitro with focused pools of shRNAs in a retroviral vector (~110 shRNAs), and mice were infected with LCMV clone 13 following T cell transfer. T cells were sorted on day 7 into short-lived effector and memory precursor cells based on well-defined surface markers. Two components of the positive transcription elongation factor (P-TEFb) were discovered as novel genes required for efficient CD4 and CD8 effector T cell differentiation. Silencing of Cyclin T1 or its catalytic partner Cdk9 impaired development of short-term CD8 effector cells and CD4 cells with a Th1 profile [19]. The function of this complex is to stimulate the transition of paused RNA polymerase II complexes into productive elongation. This study shows how the use of focused shRNA pools enables the functional interrogation of genes that are differentially expressed in cells with alternative fates.
Both of these projects required introduction of shRNAs into primary T cells by viral transduction. Only proliferating primary T cells can be infected by a lentiviral/retroviral vector, requiring either exposure to homeostatic cytokines (IL-7 or IL-15) or crosslinking of TCR-CD3 and CD28 receptors. It was recently shown that the differentiation of T cells from naïve to effector states can be studied by introducing a lentiviral vector into hematopoietic stem cells. Use of the Lac operon system allowed induction of shRNA expression by administration of IPTG. The system was used to examine the role of the BATF transcription factor in the transition of naïve to effector T cells. The authors found that knockdown of BATF induced T cell apoptosis within the first 72 hours following LCMV infection [20]. This general approach could be useful for focused in vivo shRNA or CRISPR screens, in particular for immune cell types that do not appropriately home to tumors following adoptive transfer (Figure 2), and multiple distinct readouts could be used to interrogate immune cell function (Figure 3).
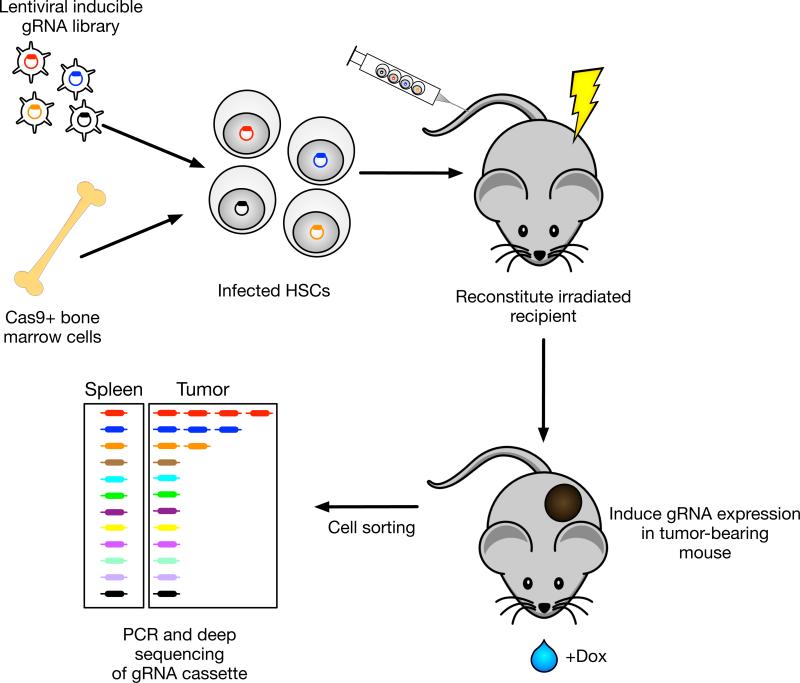
Figure 2. In vivo induction of gRNA expression in defined immune cell types for focused CRISPR screens.
A lentiviral gRNA library is introduced into hematopoietic stem cells (HSC) from mice that are transgenic for Cas9 and Cre The promoter chosen for Cre expression determines the cell type in which Cas9 is expressed (Cre-mediated excision loxP-stop-loxP cassette). The expression of gRNAs is induced by doxycycline when tumors are established. This approach enables a focused in vivo screen without ex vivo manipulation of differentiated immune cells.
Figure 3. Multi-dimensional CRISPR screen for regulators of immune function.
Many different readouts can be used to select for phenotypes of interest. In this particular example, T cells are transduced with a lentiviral gRNA library and co-cultured with dendritic cells and tumor cells. Multiple readouts can be used to examine the impact of gene inactivation on T cell function, including proliferation (CFSElow cells), cytokine production (IFNγproducing cells), activation (CD69 expression) or exhaustion (PD-1 expression). Such multi-dimensional readouts can help to functionally annotate genes of interest.
IV. CRISPR/Cas9 discovery technologies
Development of genome-scale CRISPR libraries
The utility of RNAi is limited by the incompleteness of protein depletion and confounding off-target effects. Recent publications have reported the generation of genome-scale CRISPR libraries which significantly improve complex screens [21-23]. Cas9 can induce double strand breaks at specific genomic loci depending on the sequence of the bound guide RNA (gRNA), and the resulting frame shift deletion/insertion mutations result in loss of function alleles. Genome-scale libraries can be readily generated by synthesis of oligonucleotides on chips. One of these recent reports demonstrated the robustness of such CRISPR-based screens using a positive selection screen for gene knockouts that confer resistance to the BRAF kinase inhibitor vemurafenib in a human melanoma cell line [21]. The authors confirmed previously characterized resistance genes (NF1 and MED12) and discovered a series of novel hits including neurofibromin (NF2), Cullin E3 ligase (CUL3), and components of the STAGA histone acetyltransferase complex (TADA1 and TADA2B). Comparison to a previous shRNA screen performed in the same cell line documented the robustness of the CRISPR-based approach. For the top 10 hits, 78% of gRNAs targeting each gene were among the top 5% enriched gRNAs, while a much smaller fraction (20%) of shRNAs for each gene ranked among the top 5% enriched shRNAs[21]. The major strengths of CRISPR-based screens are therefore the consistency with which distinct gRNAs inactivate the same gene as well as the ability to study the phenotype of complete knockout alleles with pooled libraries. These improvements will be particularly relevant for screens that rely on complex biological readouts.
A genome-scale CRISPR library was recently used to identify inhibitors of metastasis in a mouse model of lung cancer [24]. The parental tumor cell lines (oncogenic Kras, homozygous p53 deletion) did not form metastases, but 89% of mice had lung metastases when tumor cells expressing a gRNA library and Cas9 were injected. The gRNAs that were most strongly enriched in metastases targeted Nf2, Pten, Cdkn2a, which are well-known tumor suppressor genes. Most lung metastases were dominated by one or a few gRNAs demonstrating that rare events can be detected in a genome-scale in vivo CRISPR screen if a powerful positive selection mechanism is employed (such as formation of metastases by a non-metastatic tumor).
Cas9 transgenic mice
The large size of the Cas9 cDNA is a major issue for screens involving primary cells because it greatly reduces lentiviral titers. Recently reported Cas9 transgenic mice provide an excellent solution for this important technical issue [25]. In this mouse strain, Cas9 expression is induced by Cre-mediated excision of a loxP-stop-loxP cassette, enabling either cell type specific or ubiquitous expression of Cas9 expression, depending on the promoter chosen for Cre expression. The transgene also drives EGFP expression for visualization of Cas9-expressing cells. These mice were used to develop a multigenic mouse model of lung cancer. A single adeno-associated viral (AAV) vector was built that contained three tandem gRNA expression cassettes for simultaneous targeting of multiple genes, a homology repair template for introduction of the KrasG12D mutation as well as Cre for induction of Cas9 expression. Intra-tracheal injection of this AAV resulted in lung tumors in all mice [25]. Cas9 transgenic mice represent a versatile tool for genetic screens, either for focused in vivo screens or as a source of primary Cas9-expressing cells. For example, inducible cell type specific gene knockout screens could be performed by introducing a gRNA library into hematopoietic stem cells under the control of an inducible promoter and by restricting Cas9 expression to an immune cell population of interest based on the promoter used to drive Cre expression.
V. Moving from individual genes to biological pathways
Our tools have now improved to the point where it is feasible to discover many components of a biological pathway. An excellent example is a recent CRISPR screen in primary dendritic cells that interrogated the response to LPS [26]. Bone-marrow derived dendritic cells were generated from the Cas9-transgenic mice described above, infected with a lentiviral gRNA library, stimulated with LPS and then sorted based on the level of intracellular TNFα expression. Most of the known positive regulators of LPS signaling were identified among the top 100 ranked genes, demonstrating how an unbiased genome-scale screen can result in identification of many components of a biological pathway. Five of six components of the PAF complex were identified, which is a regulator of transcription elongation and 3’ mRNA processing. A gRNA targeting PAFc, a core component of the complex, significantly reduced sustained inflammatory signatures in dendritic cells.
An exciting complementary approach for the discovery of biological circuits is the definition of the transcriptome of single cells (single-cell RNA-seq). Co-variation of gene expression across cells enables identification of genes associated with distinct functional states [27]. This approach was recently applied to discover key regulators of the pathogenicity of Th17 cells in the EAE model of multiple sclerosis [28]. Single-cell RNA-seq was performed on T cells isolated from the CNS of mice with EAE as well as from T cells differentiated in vitro into Th17 cells under conditions that yielded pathogenic cells (IL-1β, IL-6 and IL-23) or non-pathogenic cells (IL-6 and TGFβ). Analysis of co-variation of gene transcripts across cells identified two key transcript modules: one that co-varied with known pro-inflammatory Th17 cytokines (including IL-17A) and another that co-varied with regulatory cytokines (such as IL-10). This approach led to the discovery of novel genes required for the pathogenicity of Th17 cells, including the glycosphingolipid receptor Gpr65. This approach could of course be combined with a focused CRISPR screen in which genes belonging to such a module are functionally interrogated.
Outlook
Immuno-oncology is now one of the most exciting frontiers in cancer research. The new technologies discussed here open the possibility for a much deeper understanding of the complex interactions between immune cells and tumor cells. Highly informative genetic screens can now be performed in vivo to discover key regulators of these interactions, and it is likely that novel therapeutic strategies can be formulated based on such scientific insights.
Highlights.
Powerful genetic tools are now available to study immune pathways in tumors
Pooled screens enable simultaneous investigation of thousands of genes
CRISPR/Cas9 screens enable functional interrogation of genetic circuits
Cas9 transgenic mice greatly facilitate CRISPR screens in primary cells
Acknowledgements
This work was supported by grants from the Melanoma Research Alliance, the Bridge Project of MIT/Koch Institute and Dana-Farber/Harvard Cancer Center, and the NIH (1R01CA173750).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolchok JD, Hodi FS, Weber JS, Allison JP, Urba WJ, Robert C, O'Day SJ, Hoos A, Humphrey R, Berman DM, et al. Development of ipilimumab: a novel immunotherapeutic approach for the treatment of advanced melanoma. Ann N Y Acad Sci. 2013;1291:1–13. doi: 10.1111/nyas.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 4.Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ, Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, Schuster SJ, Millenson MM, Cattry D, Freeman GJ, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baumeister SH, Freeman GJ, Dranoff G, Sharpe AH. Coinhibitory Pathways in Immunotherapy for Cancer. Annu Rev Immunol. 2016 doi: 10.1146/annurev-immunol-032414-112049. [DOI] [PubMed] [Google Scholar]
- 8.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 9.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 10.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 11.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 12.Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva JM, Marran K, Parker JS, Silva J, Golding M, Schlabach MR, Elledge SJ, Hannon GJ, Chang K. Profiling essential genes in human mammary cells by multiplex RNAi screening. Science. 2008;319:617–620. doi: 10.1126/science.1149185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassik MC, Lebbink RJ, Churchman LS, Ingolia NT, Patena W, LeProust EM, Schuldiner M, Weissman JS, McManus MT. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods. 2009;6:443–445. doi: 10.1038/nmeth.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zender L, Xue W, Zuber J, Semighini CP, Krasnitz A, Ma B, Zender P, Kubicka S, Luk JM, Schirmacher P, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bric A, Miething C, Bialucha CU, Scuoppo C, Zender L, Krasnitz A, Xuan Z, Zuber J, Wigler M, Hicks J, et al. Functional identification of tumor-suppressor genes through an in vivo RNA interference screen in a mouse lymphoma model. Cancer Cell. 2009;16:324–335. doi: 10.1016/j.ccr.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Zhou P, Shaffer DR, Alvarez Arias DA, Nakazaki Y, Pos W, Torres AJ, Cremasco V, Dougan SK, Cowley GS, Elpek K, et al. In vivo discovery of immunotherapy targets in the tumour microenvironment. Nature. 2014;506:52–57. doi: 10.1038/nature12988. [An in vivo pooled shRNA screening approach was used to discover negative regulators of T cell function in the tumor microenvironment. The study demonstrated feasibility of systematic discovery of immunoregulatory processes in the relevant tissue.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Chen R, Belanger S, Frederick MA, Li B, Johnston RJ, Xiao N, Liu YC, Sharma S, Peters B, Rao A, et al. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity. 2014;41:325–338. doi: 10.1016/j.immuni.2014.08.002. [The authors combined an in vivo shRNA screening approach with flow cytometric selection to discover novel regulators of T cell differentiation into effector or memory cells.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godec J, Cowley GS, Barnitz RA, Alkan O, Root DE, Sharpe AH, Haining WN. Inducible RNAi in vivo reveals that the transcription factor BATF is required to initiate but not maintain CD8+ T-cell effector differentiation. Proc Natl Acad Sci U S A. 2015;112:512–517. doi: 10.1073/pnas.1413291112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [The authors showed that genome-scale CRISPR-Cas9 screens generate robust datasets and enable discovery of novel genes even in systems that have been extensively studied previously, such as resistance of human melanoma cells to a BRAF inhibitor.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 24**.Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [The authors performed the first genome-scale in vivo CRISPR-Cas9 screen and demonstrated that even rare gRNAs which inactivate important tumor suppressors can be identified in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159:440–455. doi: 10.1016/j.cell.2014.09.014. [Cas9 knock-in mice have become a valuable tool for loss of function screens in primary cells. The authors demonstrate that gene inactivation can be induced locally by injection of an AAV vector encoding a gRNA cassette.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Parnas O, Jovanovic M, Eisenhaure TM, Herbst RH, Dixit A, Ye CJ, Przybylski D, Platt RJ, Tirosh I, Sanjana NE, et al. A Genome-wide CRISPR Screen in Primary Immune Cells to Dissect Regulatory Networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [The authors demonstrate that single-cell RNA-seq data enable definition of gene modules responsible for alternative functional states of T cells. This approach can be used to select candidate genes for in vivo CRISPR screens.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat Rev Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 28.Gaublomme JT, Yosef N, Lee Y, Gertner RS, Yang LV, Wu C, Pandolfi PP, Mak T, Satija R, Shalek AK, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163:1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]