Abstract
We combined nucleic acid-based molecular methods, biogeochemical measurements, and physicochemical characteristics to investigate microbial sedimentary ecosystems of Laguna Tebenquiche, Atacama Desert, Chile. Molecular diversity, and biogeochemistry of hypersaline microbial mats, rhizome-associated concretions, and an endoevaporite were compared with: The V4 hypervariable region of the 16S rRNA gene was amplified by pyrosequencing to analyze the total microbial diversity (i.e., bacteria and archaea) in bulk samples, and in addition, in detail on a millimeter scale in one microbial mat and in one evaporite. Archaea were more abundant than bacteria. Euryarchaeota was one of the most abundant phyla in all samples, and particularly dominant (97% of total diversity) in the most lithified ecosystem, the evaporite. Most of the euryarchaeal OTUs could be assigned to the class Halobacteria or anaerobic and methanogenic archaea. Planctomycetes potentially also play a key role in mats and rhizome-associated concretions, notably the aerobic organoheterotroph members of the class Phycisphaerae. In addition to cyanobacteria, members of Chromatiales and possibly the candidate family Chlorotrichaceae contributed to photosynthetic carbon fixation. Other abundant uncultured taxa such as the candidate division MSBL1, the uncultured MBGB, and the phylum Acetothermia potentially play an important metabolic role in these ecosystems. Lithifying microbial mats contained calcium carbonate precipitates, whereas endoevoporites consisted of gypsum, and halite. Biogeochemical measurements revealed that based on depth profiles of O2 and sulfide, metabolic activities were much higher in the non-lithifying mat (peaking in the least lithified systems) than in lithifying mats with the lowest activity in endoevaporites. This trend in decreasing microbial activity reflects the increase in salinity, which may play an important role in the biodiversity.
Keywords: hypersaline lakes, microbial mats, endoevaporites, concretions, Atacama, pyrosequencing
Introduction
The Salar de Atacama, located in the Chilean Central Andes, is the largest Quaternary halite deposit in the world (3064 km2 and >900 m thick; Warren, 2010). This Salar is comprised of a porous halide (90%), the interstices of which are permeated with a sodium chloride brine rich in lithium, potassium, magnesium, and boron (Bevacqua, 1992; Risacher and Alonso, 1996). Laguna Tebenquiche, located in the northern part close to the core zone of the Salar (Risacher et al., 2003), is one of the largest water bodies in this system (Demergasso et al., 2008). The lake is fed by groundwater of Tertiary and Quaternary volcanic origin (Risacher and Alonso, 1996). Bacterial and archaeal microorganisms inhabiting this lake are subject to extreme environmental conditions, such as high solar radiation (incl. UV), extreme diel temperature fluctuations, extreme changes in salinity due to net evaporation, and high lithium, boron, and arsenic concentrations (Lara et al., 2012; Farías et al., 2014). “Extreme” environmental conditions are generally conducive of microbial mat development (Rothschild and Mancinelli, 2001; Dupraz and Visscher, 2005) and a variety of photosynthetic organosedimentary ecosystems were found in Laguna Tebenquiche (Farías et al., 2014). These ecosystems included non-lithifying laminated photosynthetic microbial mats characterized by copious amounts of exopolymeric substances similar to other hypersaline mats (Dupraz et al., 2004, 2009) and also endoevaporitic domes made up of gypsum harboring endolithic phototrophs such as those described by Canfield et al. (2004) and Oren et al. (1995).
Previous studies based on culture-dependent methods recovered a large number of bacterial and archaeal strains from Laguna Tebenquiche's water column and sediments. Isolates included moderately halophilic Gram-negative rods (Prado et al., 1991), moderately halophilic Gram-positive cocci (Valderrama et al., 1991), heterotrophic halophilic microorganisms (Prado et al., 1993), and extremely halophilic archaea (Lizama et al., 2001, 2002). Using morphological and physiological properties, the bacterial isolates were assigned to the genera Vibrio, Halomonas, Acinetobacter, Alteromonas, Psychrobacter, and Marinococcus, all of which grouped within the class Gammaproteobacteria. Archaeal strains were assigned to the genera Halorubrum, Haloarcula, Halobacterium, and Haloferax (phylum Euryarchaeota).
Demergasso et al. (2008) analyzed the bacterial community of the water column by DGGE fingerprinting at several locations of Laguna Tebenquiche during the winter and summer season. Their study revealed a heterogeneous community composition of which changed along a salinity gradient in the water column of the lake. The dominant phylum was Bacteroidetes, which in the most saline part of the lake comprised a cluster related to Salinibacter relatives and at intermediate salinities consisted of clusters distantly related to Psychroflexus spp. Within the Gammaproteobacteria, which encompassed the most abundant class, a cluster related to uncultured bacteria from Mono Lake (USA) dominated. In addition, a few clones could be assigned to the candidate division OP1 (currently reclassified within the phylum Parcubacteria), to uncultivated clones CS_B020 and BD1-5 from marine sediments and to sequences of the KB1 group found in sediments from hypersaline brines.
Recently, we reported on the bacterial diversity, mineral composition, and key metabolic activities in two lakes in the Salar de Atacama (Farías et al., 2014), Laguna La Brava, and Laguna Tebenquiche. In both lakes, discrete bacterial communities, and mineral compositions developed along the salinity gradient of the overlying water. In Laguna Tebenquiche, the most abundant bacterial 16S rRNA amplicons in mats and endoevaporites resembled Bacteroidetes, and the second-most abundant amplicons could be assigned to Proteobacteria (Alphaproteobacteria and Deltaproteobacteria). Surprisingly low cyanobacterial diversity was found, which was corroborated by a low abundance of chlorophyll a (Chla). Another recent study, focusing on phototrophic bacteria (Thiel et al., 2010) was motivated by the low Chla concentration and low abundance of cyanobacteria in Laguna Tebenquiche. That investigation found evidence for a new gammaproteobacterial lineage based on pufLM gene analyses and furthermore found that green sulfur bacteria could not be detected with molecular techniques but could be revealed by culture-dependent methods.
Our previous study focused on the bacterial 16S rRNA sequences in two lakes and included a non-lithifying microbial mat and a gypsum endoevaporite in Laguna Tebenquiche (Farías et al., 2014). In a consecutive field campaign documented here, and with the purpose of covering all microbial ecosystems associated to mineral precipitation, we increased the number different benthic microbial ecosystems to include one endoevaporitic domal mat, two microbial mats with different degrees of lithification, and two rhizome-associated concretions. These concretions were included in our study because they present lithified structures similar to microbialites, but in contrast form in association with plants. Also, in the present investigation we deployed primers, which amplified the V4 hypervariable region of both archaeal and bacterial 16S rDNA genes, in order to cover the total microbial diversity in each system with the purpose to investigate the low cyanobacterial presence determined in a previous study, as well as the vertical distribution of key functional groups, we determined the microbial diversity in discrete depth horizons in a non-lithifying microbial mat and an endoevaporitic mat. Our investigation enables a correlation of taxonomic diversity with geochemical gradients (e.g., oxygen and sulfide profiles) on a small (vertical) scale and with physicochemical and geochemical characteristics (e.g., salinity, water depth) on a large (horizontal) scale. The results of this study increase our knowledge of the genetic and metabolic diversity of the benthic microbial ecosystems in Laguna Tebenquiche and provide novel insights into the microbial processes in extreme ecosystems at high altitude.
Materials and methods
Sample collection
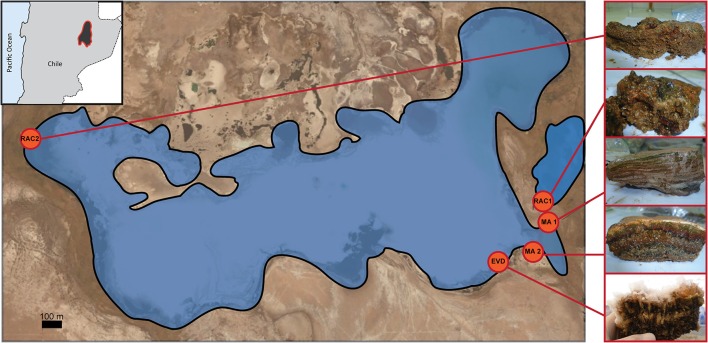
Samples were obtained from Laguna Tebenquiche in November 2013 and selected based on a preliminary inspection of sedimentary structures present along a salinity gradient (Figure 1). Five different locations were sampled: one endoevaporitic dome, EVD (23°08′24.6″ S, 68°15′0.2″ W); two microbial mats, MA1 (23°08′18.5″S, 68°14′49.9″W) and MA2 (23°08′23.44″S, 68°14′53.89″W); and two rhizome-associated concretions, RAC1 (23°08′15.42″S, 68°14′49.89″W) and RAC2 (23°7′47.50″S, 68°16′22.8″W) from opposite sides of the lake. Water samples (1 L) were collected immediately (ca. 1 cm above) over the sampling sites of the corresponding sediment systems. The endoevaporitic gypsum dome was recovered from ca. 15 cm water depth. The organic-rich microbial mats MA1 and MA2 were submersed in ca. 15 and 5 cm of water, respectively. MA1 was very gelatinous and contained only a few trapped minerals, contrary to MA2, which was leathery, and comprised a semi-lithified subsurface layer. The rhizome-associated concretions forming around the root system of the grass Distichlis spicata are commonly found near the shoreline of the lake. Samples for a detailed depth analysis of the microbial diversity were collected from the MA1 and EVD sites, respectively, which represent the end members of lithification (non-lithified and fully lithified, respectively).
Figure 1.
Location of microbial mats (MA1 and MA2), rhizome-associated lithified concretions (RAC1 and RAC2), and evaporite (EVD) in Laguna Tebenquiche.
Water column characteristics
The temperature and pH of the water column were determined in situ. Samples were stored in acid-cleaned bottles on ice in the dark until analyses in the laboratory within 48 h. Dissolved oxygen, salinity, conductivity, total P, , , dissolved Si, Ca2+, Mg2+, K+, , and Na+, according to the methodology described by Eaton et al. (2005). , orthophosphates, and Total Organic Nitrogen (TON) were analyzed using a Merck Nova 60 Spectro Photometer by following standard methods, as described by American Public Health Association (1998).
Sediment characteristics
Bulk samples of all microbial sediments were taken for mineral analyses and kept at 4°C in the dark prior to analysis. The mineral composition was determined by X-ray diffraction (XRD) analysis of finely ground (<20 μm) samples of dried mats and endoevaporites with a PANalyticalX'Pert PRO diffractometer, with Cu lamp (kα = 1.5403 Å) operated at 40 mÅ, and 40 kV at Centro de Investigaciones Geológicas (La Plata, Argentina).
Microelectrode measurements
Depth profiles of the oxygen and sulfide concentration were measured in situ (Taillefert and Rozan, 2002; Visscher et al., 2002) during the peak of photosynthesis when the intensity of photosynthetically active radiation (PAR) was 1850–2550 μmol quanta.m−2.s−1. Oxygen was determined with a Clark-type probe and sulfide using an amperometric sensor (Unisense, Arhus, Denmark). Both O2 and H2S needle probes had internal reference, guard, and measuring electrodes and were connected to a modified portable picoammeter (Unisense PA 2000, Arhus, Denmark). The electrodes were calibrated in the laboratory before and after field measurements and in between measurements checked by a two-point calibration. Electrodes could not be deployed in rhizome-associated lithified concretions due to the hardness of these samples. Three to five replicate profiles covering the upper 10–15 mm of each sample were determined. The oxygen and sulfide concentrations were corrected for altitude according to Sherwood et al. (1991).
DNA extraction and sequencing
For DNA analyses, triplicate cores (2 cm2 each) were taken to a depth of 3 cm and pooled prior to homogenizing in order to obtain a representative sample. For the depth distribution of diversity, samples of MA1, and EVD were dissected following visible layers with depth: MA1 layers were taken from 0 to 1.5 mm (layer 1), 1.5 to 4 mm (layer 2), 4 to 6.5 mm (layer 3), 6.5 to 9.5 mm (layer 4), and 9.5 to 11.5 mm (layer 5). EVD samples were taken from 0 to 10 mm (layer 1; predominantly the halite crust), 10 to 20 mm (layer 2; the gypsum crust); 20 to 30 mm (layer 3; top of the endolithic mat), 30 to 37 mm (layer 4; bottom of the endolithic mat), 37 to 57 mm (layer 5; sediment underlying the mat). Homogenates used for DNA extraction were stored at −20°C in the dark and processed within a week.
Total DNA was isolated from 0.2 g material following the protocol supplied in the Power Biofilm DNA Isolation Kit (MO BIO Laboratories, Inc.).
The V4 hypervariable region of the bacterial and archaeal 16S rRNA gene was amplified using the RK primers (F515 and R806) that contain adaptors A and B required for 454 FLX pyrosequencing (Roche Applied Science) and a 10 nucleotide “multiple identifier” (MID; Bates et al., 2010). Bates et al. (2010) designed this primer set to be universal for a broad range of archaeal and bacterial taxa with few biases or excluded groups. Five independent PCRs were performed to reduce bias. The PCR mixture (25 μl final volume) contained 2.5 μl FastStart High Fidelity 10X Reaction Buffer (Roche Applied Science, Mannheim, Germany), 20 ng of template DNA, 0.4 μM of each primer, and 1.25 U FastStart High Fidelity Enzyme Blend (Roche Applied Science), and 0.2 mM dNTPs. The PCR conditions were 95°C for 5 min for initial denaturalization, followed by 95°C for 45 s, 57°C for 45 s, 72°C for 60 s in 30 cycles, and a final elongation step at 72°C for 4 min. Two negative control reactions containing all components except the template were performed. The five reactions products were pooled and purified using AMPure beads XP. Quantification of the purified PCR product was performed using the Quant-IT Pico Green dsDNA Kit (Invitrogen Molecular Probes Inc, Oregon, USA). Purified PCR product was pyrosequenced on a Roche 454 GS-FLX system, Titanium chemistry. Sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under the accession number: SRP066553.
16S rRNA amplicons processing
All analyses of the V4 hypervariable region of the microbial 16S rRNA amplicons were conducted within the QIIME software package (Caporaso et al., 2010). Raw 454 reads were demultiplexed and quality filtered by removing low quality or ambiguous reads. Sequences shorter than 150 bp were discarded and Roche adapters, linkers, primers, and sample barcodes were removed. The 454 reads were denoised to reduce possible sequencing errors and clustered at 97% identity in operational taxonomic unit (OTU) using uclust (Edgar, 2010). One representative sequence of each cluster was aligned to the Greengenes database with PyNAST 1.1 (DeSantis et al., 2006). Chimeric sequences were detected using the ChimeraSlayer algorithm and subsequently removed (Haas et al., 2011). OTUs observed in only one sample or represented by only one sequence were discarded. Finally, the number of sequences assigned to each OTU was summarized in a table generated by QIIME.
In order to analyze the diversity, the OTU table was subsampled using 10 replicates for each sampling effort at increasing intervals of 100 sequences. Alpha diversity indexes were calculated on each subsample and on the OTU table. Alpha diversity metrics calculated included Observed OTUs, Chao1 (estimates the species richness), Shannon (the entropic information of the abundances of observed OTUs, accounting for both richness and evenness), Equitability (Shannon index corrected for the number of species, “pure” evenness), Dominance (calculated as the sum of the squares of the frequencies of each OTU), and Simpson (1-Dominance) indexes.
Canonical correspondence analysis
A constrained ordination was carried out by a Canonical Correspondence Analysis (CCA) to correlate environmental variables with microbial phyla and samples. A Monte Carlo test with 499 permutations was carried out to ensure the significance of canonical axes. CANOCO 4.5 software package (Microcomputer Power, Ithaca, NY, USA) was used to perform the CCA and the tool CANODRAW for triplot visualization (ter Braak and Smilauer, 2002).
Results
Environmental characteristics
Water column
Physicochemical analyses of the water column overlying the sediment samples (Table 1; Table S1; no data were obtained for RAC2) showed an increase of the salinity (i.e., conductivity) from the RAC1 site to the MA2 site, with intermediate values for sites MA1 and EVD. This observation was supported by trends in total alkalinity as well as major cation (e.g., sodium, magnesium) and anion (e.g., chloride, sulfate) concentrations. The calcium concentration was the highest at the RAC1 site, intermediate at the MA1, and EVD sites and the lowest at the MA2 site, which pattern followed the amount of calcium incorporation in minerals present in these samples (i.e., no calcium was incorporated in MA2). The phosphate concentration was the highest in RAC1, intermediate in MA1 and EVD, and the lowest in MA2 water samples. Nitrate concentrations followed the opposite trend, with the highest values in water overlying MA2 and the lowest values present at RAC1. Metals and metalloids such as lithium, and boron where in lower amount in RAC1 where conductivity is lower, arsenic is almost constant in all the samples sites while silica is lower only in MA2 (Table S1).
Table 1.
Physico-chemical parameters for the overlying water from the different samples studied.
| Physico-chemical parameter | Unit | MA1 | MA2 | RAC1 | EVD |
|---|---|---|---|---|---|
| Biochemical oxygen demand (BOD) | mg/L | 3.9 | 1.9 | 4.0 | 4.2 |
| Chemical oxygen demand (COD) | mg/L | 269 | 191 | 169 | 224 |
| Chlorophyll a | μg/L | <0.1 | <0.1 | <0.1 | 2 |
| Conductivity | mS/cm | 161 | 228 | 94 | 177 |
| Hardness | mg/L | 14,909 | 31,785 | 7640 | 16,662 |
| pH | – | 7.8 | 7.6 | 7.4 | 7.8 |
| Total Alkalinity | mg CaCO3/L | 487 | 697 | 380 | 525 |
| Temperature | °C | 23.3 | 27.0 | 27.6 | 31.0 |
| Turbidity | NTU | 15.17 | 4.40 | 4.52 | 19.11 |
| Salinity | g/L | 106 | 150 | 62 | 117 |
| Organic matter | mg/L | 17 | 14 | 7 | 12 |
Sedimentary structures
In the microbial sedimentary structures, the total mineral content per volume (corresponding to the degree of lithification) was the highest in EVD and RAC1, intermediate in MA2 and the lowest in the non-lithifying MA1 samples. The XRD analysis of mineralogy revealed that RAC1 and RAC2 comprised halite and to a lesser extent gypsum and calcite (Figure 2). MA2 contained gypsum and halite with traces of aragonite and calcite, MA1 mainly halite with lower amounts of calcite, and aragonite. EVD consisted of predominantly of gypsum, with halite being less abundant. It should be noted that a fraction of minerals, especially halite, observed in MA1 and MA2 could be due to drying artifacts. Hand samples, inspected under a dissection microscope showed calcite grains, as demonstrated by dissolution upon addition of 2N HCl, but with the exception of EVD did not reveal halite as a major mineral component. We postulate that the hypersaline porewater upon drying of the samples, cause halite to precipitate.
Figure 2.
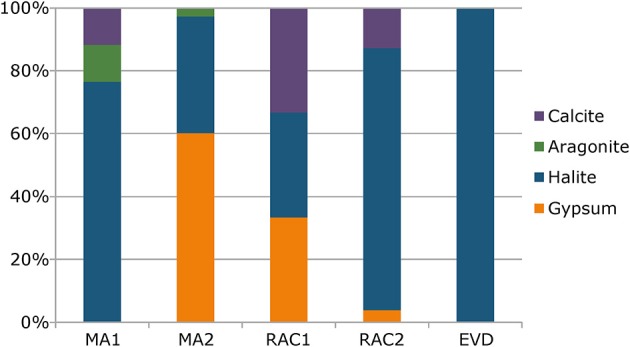
Mineral composition of microbial mats (MA1 and MA2), rhizome-associated lithified concretions (RAC1 and RAC2), and evaporite (EVD) obtained by X-ray diffraction (XRD) analyses.
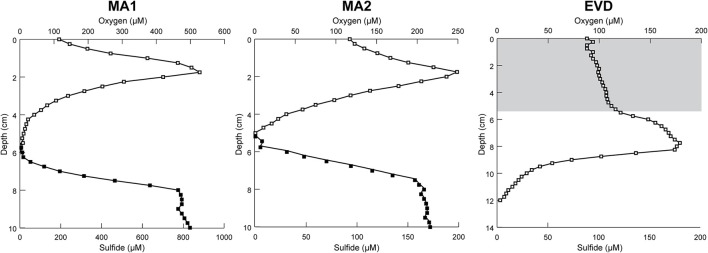
Oxygen and sulfide concentrations were measured in MA1, MA2, and EVD (Figure 3). The oxygen concentration in both mats peaked at 1.75–2 mm depth (527 and 249 μM in MA1 and MA2, respectively) and decreased to zero at 5.25–6 mm. The higher O2 peak in MA1 compared to MA2 was indicative of a higher rate of photosynthesis, and the steepness of the O2 depth profile suggested high respiration and/or sulfide oxidation rates. Much higher sulfide concentrations were observed in MA1 compared to MA2 (830 and 173 μM, respectively), supporting the notion of higher metabolic activities in MA1. A 4–8 mm thick salt crust (gray shaded area) covered the surface of EVD, and a metabolically-active endolithic mat was present underneath this halite layer. The oxygen concentration increased to 179 μM at 7.75 mm, after which it slowly decrease to zero at ca. 12 mm. No sulfide could be detected in the upper 15 mm of EVD.
Figure 3.
Representative depth profiles of the concentration of O2 (squares) and sulfide (triangles) measured in situ with microelectrodes during the light period. The gray shaded area depicts the mineral crust in RAC1.
Composition of bacterial and archaeal communities
A total of 19,320 raw reads were obtained from all five samples by using 454 pyrosequencing. After quality filtering, denoising, and removing chimeras, a total of 15,438 rRNA sequences representing 926 OTUs clustered at 97% sequence similarity remained. The rarefaction curves of observed OTUs vs. sequence number per sample are shown in Figure S1. Rarefaction curves revealed that only the EVD sample approached saturation, indicating that the sequencing depths for the other samples were insufficient to cover the microbial diversity. Using the rarefied or random subsampled sequences (i.e., 1070 per sample to remove sampling depth heterogeneity), the highest and the lowest number of OTUs were observed in RAC1 and EVD, respectively (Table S2). The diversity of microbial communities in the samples was further evaluated using Chao1, Shannon, Equitability, Dominance, and Simpson indexes. The Shannon, Equitability, and Simpson indexes ranged from 3.00 to 6.55, 0.814 to 0.456 and 0.963 to 0.595, respectively and showed the highest values in RAC1 and the lowest values in EVD. However, of all samples studied the Chao index was the highest in MA1 and RAC1. This implies that the samples RAC1 and MA1 have the highest microbial diversity with a larger equitability of OTUs and the sample EVD has the lowest microbial diversity with a few dominant OTUs.
Most of the 16S rRNA sequences in EVD were classified as archaea (97%). Furthermore, EVD was the only sample in which all sequences could be assigned to a domain level. All samples were composed of several phyla or groups, however, only a few were dominant (Figure 4). The Euryarchaeota phylum was ubiquitous in all samples, notably EVD where it comprised 97% of 16S rRNA sequences. This high proportion of euryarchaeal sequences was also reflected in a low diversity and in a large dominance of a few OTUs in EVD. Both mat samples MA1 and MA2 showed a relatively similar phylum composition, differing only in the respective proportions, with Euryarchaeota being the most abundant (33 and 62% of 16S rRNA sequences in MA1 and MA2, respectively). Other phyla present in these samples include Crenarchaeota, Planctomycetes, Firmicutes, Acetothermia, and Chloroflexi. The rhizome-associated concretions RAC1 and RAC2 exhibited disparate 16S rRNA profiles. The most abundant phylum in RAC1 was Chloroflexi (21% of 16S rRNA sequences) and in the two dominant phyla in RAC2 were Acetothermia and Firmicutes (accounting for 25 and 22% of 16S rRNA sequences, respectively). Euryarchaeota, Crenarchaeota, and Planctomycetes were present in both rhizome-associated concretions but comprised lower percentages.
Figure 4.
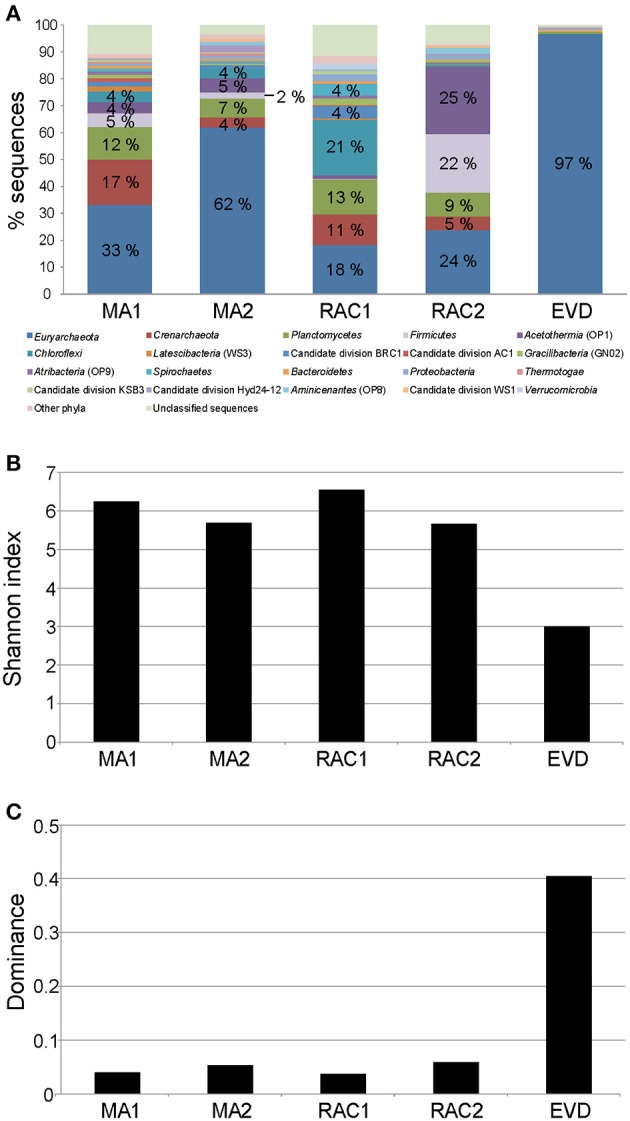
Comparison of the microbial diversity in microbial mats (MA1 and MA2), rhizome-associated lithified concretions (RAC1 and RAC2) and endoevaporite (EVD). (A) Stacked column graph representing the relative distribution of the dominant phyla in the different samples. Sequences were assigned taxonomically using Greengenes database with a minimum percentage similarity of 97% and a minimum e-value of 10−5. (B) Shannon index of the estimated richness of OTUs. (C) Dominance index of OTUs.
OTUs related to the phylum Cyanobacteria, well known for their important role in mats (Visscher et al., 1991, 1992; van Gemerden, 1993; Baumgartner et al., 2009b), were detected in low number in all samples.
The most abundant OTUs (higher than 1% 16S rRNA sequences) were assigned to taxonomic levels (Table 2). In MA1, the most abundant phylum, Euryarchaeota, was made up by five OTUs, three of which belonging to the class Thermoplasmata, within which one OTU classified as DHVEG-1 containing 23% of the 16S rRNA sequences, and two OTUs to the class Methanobacteria. Within Crenarchaeota, one OTU was assigned to the marine benthic group B (MBGB) with 16% of the 16S rRNA sequences. Two of three abundant OTUs from the phylum Planctomycetes were related to the class Phycisphaerae, and two of the Firmicutes OTUs belonged to the family Halobacteroidaceae, with one of these specifically classified to the genus Halanaerobacter (1% 16S rRNA sequences). The taxa Acetothermia, Chloroflexi (class Anaerolineae), Latescibacteria, candidate division AC1, Gracilibacteria, and Atribacteria were represented in MA1 by one OTU with more than 1% of the total number of 16S rRNA sequences.
Table 2.
Abundant microbial OTUs per sample classified at the lowest possible taxonomic level.
| MA1 | MA2 | RAC1 | RAC2 | EVD | |||||
|---|---|---|---|---|---|---|---|---|---|
| DHVEG-1 (Phyl. Euryarchaeota/ Class Thermoplasmata) |
23.1 | DHVEG-1 (Phyl. Euryarchaeota/ Class Thermoplasmata) |
19.8 | Cand. fam. Chlorothrixaceae (Phyl. Chloroflexi/ Class Chloroflexia) |
17.1 | KB1 (Phyl. Acetothermia) |
25.0 | Gen. Halonotius (Phyl. Euryarchaeota/ Class Halobacteria) |
62.7 |
| MBGB (Phyl. Crenarchaeota) |
16.4 | Gen. Halonotius (Phyl. Euryarchaeota/ Class Halobacteria) |
14.8 | DHVEG-1 (Phyl. Euryarchaeota/ Class Thermoplasmata) |
12.6 | Gen. Halanaerobium (Phyl. Firmicutes/ Class Clostridia) |
16.2 | Gen. Halorhabdus (Phyl. Euryarchaeota/ Class Halobacteria) |
6.4 |
| AKAU3564 (Phyl. Planctomycetes/ Class Phycisphaerae) |
6.9 | Fam. Halobacteriaceae (Phyl. Euryarchaeota/ Class Halobacteria) |
6.5 | MBGB (Phyl. Crenarchaeota) |
10.0 | AKAU3564 (Phyl. Planctomycetes/ Class Phycisphaerae) |
6.5 | Fam. Halobacteriaceae (Phyl. Euryarchaeota/ Class Halobacteria) |
6.2 |
| KB1 (Phyl. Acetothermia) |
4.1 | KB1 (Phyl. Acetothermia) |
5.3 | AKAU3564 (Phyl. Planctomycetes/ Class Phycisphaerae) |
4.5 | MBGB (Phyl. Crenarchaeota) |
5.2 | Gen. Halorubrum (Phyl. Euryarchaeota/ Class Halobacteria) |
5.4 |
| Fam. Halanaerobiaceae (Phyl. Firmicutes/ Class Clostridia) |
3.2 | Gen. Halorhabdus (Phyl. Euryarchaeota/ Class Halobacteria) |
4.3 | Uncultured soil bacterium PRR-11 (Cand. div. BRC1) | 2.1 | Fam. Halanaerobiaceae (Phyl. Firmicutes/ Class Clostridia) |
3.8 | XKL75 (Phyl. Euryarchaeota/ Class Halobacteria) |
5.2 |
| 20c–4 (Phyl. Euryarchaeota/ Class Thermoplasmata) |
2.8 | Gen. Halorubrum (Phyl. Euryarchaeota/ Class Halobacteria) |
4.2 | NPL-UPA2 (Cand. div. BRC1) | 2.0 | Fam. Halobacteriaceae (Phyl. Euryarchaeota/ Class Halobacteria) |
3.7 | Gen. Haloarcula (Phyl. Euryarchaeota/ Class Halobacteria) |
4.0 |
| OPB11 (Phyl. Chloroflexi/ Class Anaerolineae) |
2.6 | XKL75 (Phyl. Euryarchaeota/ Class Halobacteria) |
4.1 | Fam. Methanomassiliicoccaceae (Phyl. Euryarchaeota/ Class Thermoplasmata) |
1.9 | Cand. div. MSBL1 (Phyl. Euryarchaeota) |
3.6 | MSP41 (Phyl. Euryarchaeota/ Ord. Halobacteriales) |
2.4 |
| MSBL9 (Phyl. Planctomycetes/ Class Phycisphaerae) |
2.2 | MBGB (Phyl. Crenarchaeota) |
3.7 | 3BR-5F (Phyl. Gracilibacteria) |
1.8 | Fam. Methanomassiliicoccaceae (Phyl. Euryarchaeota/ Class Thermoplasmata) |
3.5 | Gen. Haloplanus (Phyl. Euryarchaeota/ Class Halobacteria) |
1.3 |
| Ord. Methanobacteriales (Phyl. Euryarchaeota/ Class Methanobacteria) |
1.8 | Cand. fam. Chlorothrixaceae (Phyl. Chloroflexi/ Class Chloroflexia) |
2.9 | MSBL9 (Phyl. Planctomycetes/ Class Phycisphaerae) |
1.6 | Gen. Halonotius (Phyl. Euryarchaeota/ Class Halobacteria) |
2.5 | ||
| Uncultured soil bacterium PRR-12 (Phyl. Latescibacteria) |
1.7 | AKAU3564 (Phyl. Planctomycetes/ Class Phycisphaerae) |
2.3 | Fam Pirellulaceae (Phyl. Planctomycetes/ Class Planctomycetia) |
1.5 | Gen. Halorhabdus (Phyl. Euryarchaeota/ Class Halobacteria) |
2.4 | ||
| Fam. Methanomassiliicoccaceae (Phyl. Euryarchaeota/ Class Thermoplasmata) |
1.5 | WM88 (Cand. div. Hyd24-12) | 2.3 | SBYZ_6080 (Phyl. Spirochaetes) |
1.5 | ArcA07 (Phyl. Euryarchaeota) |
2.3 | ||
| B04R032 (Cand. div. AC1) | 1.5 | Fam. Halobacteriaceae (Phyl. Euryarchaeota/ Class Halobacteria) |
2.0 | Ord. Phycisphaerales (Phyl. Planctomycetes/ Class Phycisphaerae) |
1.3 | HMMVPog-54 (Phyl. Aminicenantes) |
2.0 | ||
| Gen. Halanaerobacter (Phyl. Firmicutes/ Class Clostridia) |
1.4 | MSP41 (Phyl. Euryarchaeota/ Ord. Halobacteriales) |
1.5 | Ord. Methanobacteriales (Phyl. Euryarchaeota/ Class Methanobacteria) |
1.2 | Gen. Halanaerobacter (Phyl. Firmicutes/ Class Clostridia) |
1.3 | ||
| 3BR–5F (Phyl. Gracilibacteria) |
1.2 | HMMVPog-54 (Phyl. Aminicenantes) |
1.4 | BA021 (Atribacteria) | 1.1 | XKL75 (Phyl. Euryarchaeota/ Fam. Halobacteriaceae) |
1.1 | ||
| BA021 (Phyl. Atribacteria) |
1.2 | Gen. Haloarcula (Phyl. Euryarchaeota/ Class Halobacteria) |
1.4 | Sediment-4 (Phyl. Spirochaetes/ Ord. Leptospirales) |
1.1 | ||||
| Phyl. Planctomycetes | 1.1 | Cand. div. WS1 | 1.2 | Uncultured crenarchaeote MCG (Phyl. Crenarchaeota) |
1.1 | ||||
| Cand. div. MSBL1 (Phyl. Euryarchaeota) |
1.1 | Phyl. Planctomycetes | 1.1 | MSBL6 (Phyl. Acetothermia) |
1.0 | ||||
| SC103 (Phyl. Thermotogae/ Fam. Thermotogaceae) |
1.0 | ODP1230B3009 (Phyl. Planctomycetes/ Class Phycisphaerae) |
1.0 | ||||||
Each OTU contains at less 1% 16S rRNA sequences.
In MA2, nine OTUs could be assigned to Euryarchaeota, one of which with 19% 16S rRNA sequences to the class Thermoplasmata. The remaining OTUs were assigned to the orders Halobacteriales and Haloferacales with four specifically to the genera Halorhabdus and Haloarcula and Halonotius and Halorubrum, respectively (4 and 1 and 15 and 4% 16S rRNA sequences, respectively). One OTU in MA2 was related to the crenarchaeotal MBGB (4% 16S rRNA sequences), two OTUs were related to Planctomycetes, one of which to the class Phycisphaerae (2% 16S rRNA sequences), and Acetothermia, Chloroflexi (candidate family Chlorothrixaceae), candidate division Hyd24-12, Aminicenantes, candidate division WS1, and Thermotogae were represented by one OTU.
In RAC1, the dominant phylum Chloroflexi was represented by one OTU, which accounted for 17% of the 16S rRNA sequences. Three abundant euryarchaeal OTUs were found in this sample, two OTUs were assigned to Thermoplasmata and one OTU to the class Methanobacteria. Four of five OTUs classified into Planctomycetes were assigned to the class Phycisphaerae and the remaining OTU to the class Planctomycetia (family Pirellulaceae). Other OTUs were assigned to candidate division BRC1 (two OTUs), Gracilibacteria (one OTU), Spirochaetes (two OTUs), and Acetothermia (one OTU). RAC2 comprised one Acetothermia OTU (with 25% 16S rRNA sequences), seven euryarchaeotal OTUs related to the classes Halobacteria (two of these OTUs were assigned to the genera Halorhabdus and Halonotius), Methanobacteria and Thermoplasmata; and three Firmicutes OTUs classified to the family Halanaerobiaceae (two of which to the genera Halanaerobium and Halanaerobacter). One OTU each with more than 1% of the total 16S rRNA sequences was assigned to the phyla Planctomycetes, Crenarchaeota and Aminicenantes, respectively.
In the EVD sample, all dominant OTUs belonged to the orders Halobacteriales and Haloferacales, some of which could be classified to the genera Halorhabdus and Haloarcula and Halonotius, Halorubrum, and Haloplanus, respectively (6 and 4 and 63, 5, and 1% 16S rRNA sequences, respectively).
The OTUs related to the taxa MBGB (Crenarchaeota), AKAU3564 (phylum Planctomycetes, class Phycisphaerae) and KB1 or MSBL6 (Acetothermia) were present in mat and rhizome-associated concretion samples. A dominance of DHVEG-1 (phylum Euryarchaeota, class Thermoplasmata) was observed in the mat samples and RAC1.
Microbial structure and their relation to water physicochemical parameters
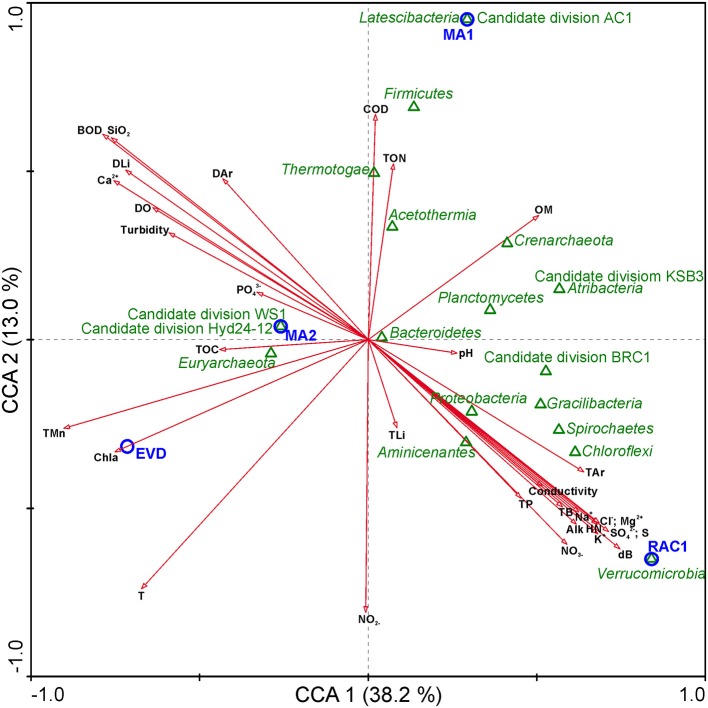
A canonical correspondence analysis (CCA) was carried out to investigate the relationships between several physicochemical and geochemical characteristics and the 10 most abundant phyla present in each different sample (Figure 5). In this analysis, CCA1, and CCA2 ordination axes could explain 51.2% of the total variance data. Conductivity is positively correlated to geochemical properties such as sodium, chloride, potassium, magnesium, sulfate, and nitrate concentrations and the hardness of the water appeared stronger correlated to magnesium than to calcium. Such physicochemical characteristics were negatively correlated with RAC1 and with representatives of the phylum Verrucomicrobia in this sample. The relative proportion of sequences assigned to Euryarchaeota, in each sample, which peaked in samples EVD and MA2, could likely be contributed to an increase in conductivity. Biodiversity in MA2 showed a negative correlation with TON but a positive correlation with the nitrite and nitrate concentration. Bacteroidetes was a minor phylum in all samples and in the CCA triplot this phylum appeared in the center, suggesting that this phylum prevailed under a wide range of physicochemical conditions.
Figure 5.
Canonical correspondence analysis (CCA) of microbial community, samples, and environmental parameters. Arrows indicate the direction and magnitude of environmental parameters associated with phyla (open green triangles) and samples studied (open blue circles). BOD, Biochemical oxygen demand; COD, Chemical oxygen demand; Chla, Chlorophyll a; HN, Hardness; Alk, Total alkalinity; T, Temperature; OM, Organic matter; TOC, Total organic Carbon; DO, Dissolved oxygen; , Nitrate; , Nitrite; TON, Total organic nitrogen; TP, Total phosphorus; OP, Orthophosphate; , Sulfate; S, Sulfur; S2−, Total sulfide; Na+, Sodium; Cl−, Chloride; K+, Potassium; Mg2+, Magnesium; Ca2+, Calcium; DB, Dissolved boron; TB, Total boron; DLi, Dissolved lithium; TLi, Total lithium; SiO2, Silica; Dar, Dissolved Arsenic; Tar, Total Arsenic.
Phylogenetic distribution with depth
The mat sample MA1 and the endoevaporite sample EVD were selected for a more detailed study as representatives of ecosystems in opposite conditions lithification. The samples were dissected according to the uppermost five visible layers. We obtained a total of 35,383 raw reads from the two samples combined (i.e., 10 layers). A total of 27,479 rRNA sequences that passed sequence processing (i.e., quality control criteria, denoising, and removing chimeras) were clustered in 1018 OTUs with a minimum of 97% sequence similarity. The rarefaction curves for MA1 and EVD samples with depth revealed that the surface layer and layer second to the surface of both samples reached an asymptote. In contrast, the bottom three layers in both samples did not reach an asymptote, suggesting that the sequencing depths in these samples were insufficient (Figure S2). The number of sequences was normalized to 1900 sequences per layer in order to allow comparison of the different depth horizons in MA1 and EVD (Table 3). In both samples, the lowest number of OTUs was detected in the surface layer (layer 1). The largest number of OTUs was found in the middle (third) layer of MA1 and in the bottom layer (layer 5) of EVD. The Shannon diversity index and Chao1 estimator confirmed that in MA1 the lowest microbial diversity with a high dominance was located in the top layer. The highest diversity was found in the third layer. The oxic-anoxic interface during the daytime fell within this layer and likely facultative and thus obligate anaerobic and microaerophilic microorganisms proliferated here. In EVD, Shannon, Chao1, and Simpson indices all increased with depth. The surface layer in both samples exhibited the lowest diversity with a few strongly dominant OTUs, likely resulting from the most extreme physicochemical conditions to which this layer was exposed.
Table 3.
Observed microbial richness and diversity estimates based on 97% OTU clusters by layers in MA1 and EVD.
| Sample | Layer | Number of total reads | Seqs/Sample | Observed OTUs | Chao1 | Shannon | Equitability | Dominance | Simpson |
|---|---|---|---|---|---|---|---|---|---|
| MA1 | 1 | 2340 | 1900 | 121 | 154 | 4.437 | 0.641 | 0.154 | 0.846 |
| 2 | 2685 | 1900 | 159 | 183 | 5.284 | 0.723 | 0.065 | 0.935 | |
| 3 | 2348 | 1900 | 316 | 443 | 6.721 | 0.809 | 0.025 | 0.975 | |
| 4 | 3668 | 1900 | 177 | 280 | 5.176 | 0.693 | 0.067 | 0.933 | |
| 5 | 2091 | 1900 | 283 | 408 | 6.217 | 0.763 | 0.042 | 0.958 | |
| EVD | 1 | 3259 | 1900 | 77 | 102 | 2.927 | 0.467 | 0.317 | 0.683 |
| 2 | 2625 | 1900 | 106 | 138 | 3.294 | 0.490 | 0.331 | 0.669 | |
| 3 | 3196 | 1900 | 197 | 239 | 5.624 | 0.738 | 0.071 | 0.929 | |
| 4 | 1903 | 1900 | 193 | 246 | 5.861 | 0.772 | 0.049 | 0.951 | |
| 5 | 3364 | 1900 | 241 | 301 | 5.606 | 0.709 | 0.082 | 0.918 |
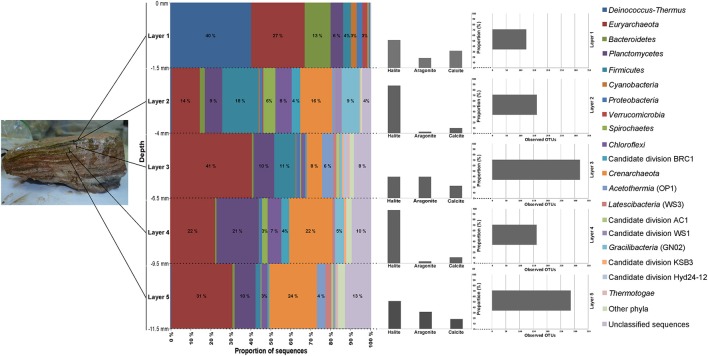
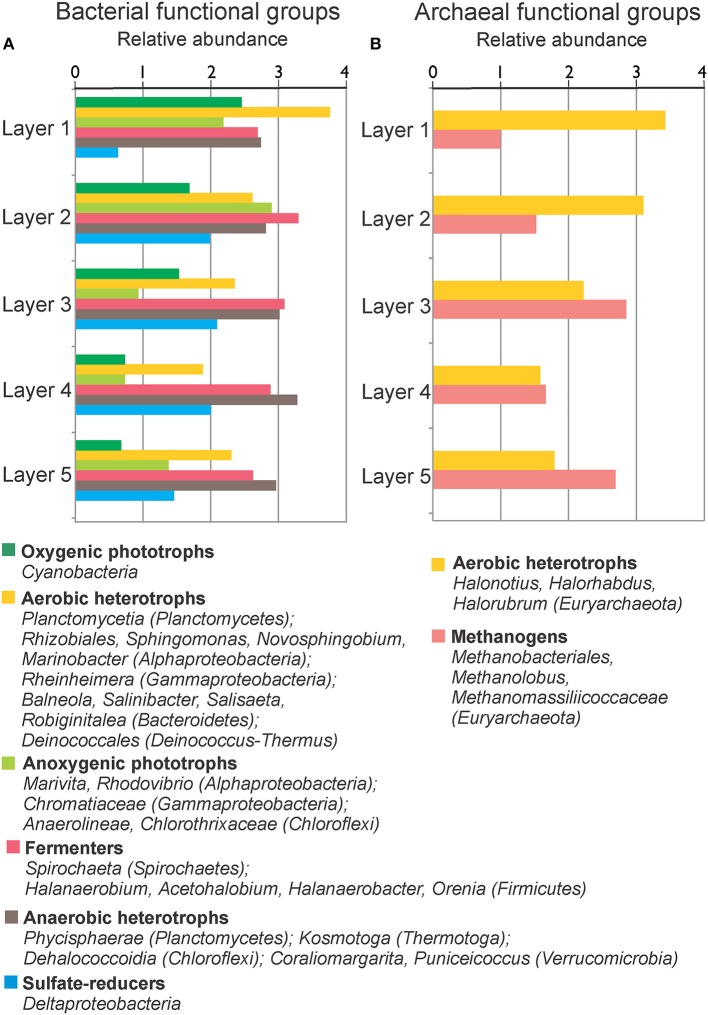
The OTUs in the different layers of MA1 could be classified to the phylum level (Figure 6). Commonly shared phyla in the five mat layers included Deinococcus-Thermus, Euryarchaeota, Bacteroidetes, Planctomycetes, Firmicutes, Proteobacteria, Spirochaetes, Chloroflexi, and candidate division BRC1. Euryarchaeota was the most abundant phylum followed by Planctomycetes, especially in the three deepest layers. In the top two layers the euryarchaeotal OTUs were mainly assigned to the class Halobacteria. In contrast, in the two bottom layers the euryarchaeotal OTUs were principally designated to the classes Methanobacteria and Thermoplasmata, distantly followed by Methanomicrobia (Methanolobus) (Table S3; Figure 7B). In the uppermost two layers, some OTUs related to Halobacteria with more than 1% 16S rRNA sequences could be classified at genus level as Halonotius, Halorhabdus, and Halorubrum. The Planctomycetes OTUs were mainly associated to classes as Phycisphaerae and Planctomycetia. Crenarchaeota and to a lesser extent, Acetothermia were two most abundant phyla (Figure 4). OTUs for these phyla were detected in all except in the surface layer (Figure 6). Firmicutes were very abundant in the second and third layers, with OTUs classified within the family Halanaerobiaceae (Class Clostridia) and the genera Halanaerobium and Halanaerobacter (Table S3; Figure 7A). Chloroflexi were found in the highest amounts in the second and fourth layers with OTUs belonging to the classes Anaerolineae and Chloroflexia (Table S3). Deinococcus-Thermus (with OTUs related to family Trueperaceae) were abundant in the surface layer of MA1. The presence of Cyanobacteria (OTUs related to family Cyanobacteraceae) decreased sharply from the surface to the second layer, and was undetectable in the deeper layers (Figures 6, 7A). A high proportion of OTUs in the top layer could be classified within Bacteroidetes, especially to the class Rhodothermi and the genus Salisaeta. Similarly, Verrucomicrobia OTUs assigned to the class Opitutae were present in relatively high numbers in the surface layer as well (Table S3; Figure 7A). Compared to other layers, a larger number of OTUs attributed to Gracilibacteria, Spirochaetes and candidate division BCR1 were present in the second and fourth layers. The number of OTUs that could not be assigned to any taxon increased with depth.
Figure 6.
Taxonomic composition by layers in MA1. Sequences were assigned taxonomically using Greengenes database with a minimum percentage similarity of 97% and a minimum e-value of 10−5.
Figure 7.
Relative abundance (log 10 base) of bacterial (A) and archaeal (B) functional groups by layers in MA1. Functional groups are formed with the available metabolic information from the microorganisms present in the sample.
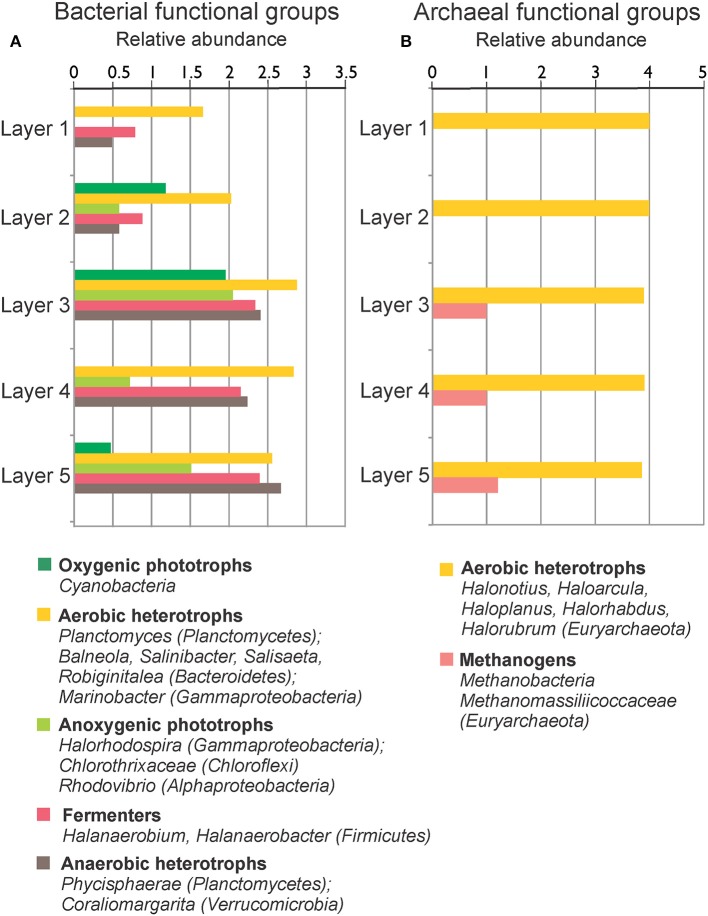
EVD displayed a limited diversity and dominance at phylum level (Figure 8). The phylum Euryarchaeota was ubiquitous in all layers, particularly in the first and second layers. This phylum was almost exclusively composed of the class Halobacteria (Figure 9B). In all layers of EVD, a large proportion of the total 16S rRNA sequences was assigned to the genus Halonotius (58–24%), followed by minor genera such as Halorhabdus, Halorubrum, or Haloarcula (Table S4). A minor proportion of OTUs classified within other phyla: low proportions of 16S rRNA sequences belonging to Bacteroidetes, Proteobacteria, and Firmicutes were detected in all layers. The highest representation of Bacteroidetes was found in the third layer, although in the second and the third layers some OTUs classified within the genus Salinibacter (Table S4), and Proteobacteria (OTUs classified within the class Gammaproteobacteria) and Firmicutes (OTUs belonging to the class Clostridia and orders Thermoanaerobacterales and Halanaerobiales) were more abundant in the bottom layers [from middle (third) layer to the bottom (fifth) layer; Table S4; Figure 9A]. Planctomycetes was also observed in the three deepest layers, with OTUs associated to the orders Phycisphaerales, Pirellulales, and Planctomycetales (the genus Planctomyces is assigned to this latter order), and Acetothermia in the two deepest layers. Similar to MA1 as noted above, the proportion of unclassified OTUs increased with depth in EVD.
Figure 8.
Taxonomic composition by layers in EVD. Sequences were assigned taxonomically using Greengenes database with a minimum percentage similarity of 97% and a minimum e-value of 10−5.
Figure 9.
Relative abundance (log 10 base) of bacterial (A) and archaeal (B) functional groups by layers in EVD. Functional groups are formed with the available metabolic information from the microorganisms present in the sample.
Discussion
Contemporary microbial mats, including microbialites, typically thrive in extreme environments such as High Altitude Andean Lakes (HAAL). Several of the HAAL ecosystems have been described (Thiel et al., 2010; Farías et al., 2013; Gomez et al., 2014; Rasuk et al., 2016), but relatively little is known about the microbial diversity of the community in relation to biogeochemistry, especially that of the benthic ecosystems. The current investigation is the first in which the total microbial diversity was evaluated and interpreted in relation to the geochemical and physicochemical characteristics. The comparison of different types of benthic microbial ecosystems (e.g., with varying degrees of lithification) in Laguna Tebenquiche, and also an in-depth analysis of separate depth horizons in two contrasting laminated systems unambiguously revealed that archaea comprise the bulk of the microbial diversity. Previous diversity studies in benthic ecosystems of HAAL using high-throughput genome sequencing focused on the bacterial composition only (Farías et al., 2014). Furthermore, the archaeal and bacterial diversity in the water column of several HAAL including Laguna Tebenquiche was determined using DGGE in combination with specific primers for each Domain (Demergasso et al., 2004). A high-throughput genomic sequencing approach also deploying different archaeal and bacterial primers was used in a lithifying microbial mat from a hypersaline lake in Kiribati (i.e., not a HAAL) and revealed that the bacterial diversity was approximately three times higher than archaeal (Schneider et al., 2013). Thus, to our knowledge, an accurate assessment of the relative contribution of archaea to the total community composition in any hypersaline microbial sedimentary system was lacking until the current study.
A variety of benthic microbial ecosystems (e.g., mats, rhizome-associated concretions, and endoevaporites) showing an increased degree of lithification developed along a salinity gradient in Laguna Tebenquiche. Archaea were dominant in all five sedimentary structures, with Euryarchaeota the single largest contributing phylum and the presence of Crenarchaeota reported for first time in HAAL ecosystems. Euryarchaeal OTUs comprised 62 and 97% of the total diversity in MA2 and EVD, respectively, which represent the sites at with the highest conductivity. The class Halobacteria, which organisms require Na+ for growth and therefore can be expected to thrive in hypersaline Laguna Tebenquiche (Grant, 2001; Oren, 2010), was a major contributor to this phylum. The water column of Laguna Tebenquiche contains a high arsenic concentration (0.07 mM; Table S1), which can promote halobacterial growth. A potential role for As cycling was documented in biofilms of another hypersaline, alkaline HAAL, Laguna Diamante, where halobacterial abundance reached 94% of total OTUs (Rascovan et al., 2016). Analysis of the Diamante biofilm metagenome revealed that genes for As(III) oxidation and As(V) reduction were ubiquitous, indicating that As cycling supported energy conservation (Rascovan et al., 2016). Similar to Laguna Diamante, we postulate that Halobacteria inhabiting Laguna Tebenquiche could use arsenic for bioenergetics purposes.
MA1, RAC1, and RAC2 contained euryarchaeal OTUs with gene sequences similar to those of methanogenic archaea from the class Methanobacteria, the family Methanomassiliicoccaceae (class Thermoplasmata) and the candidate division MSBL1. 16S rRNA sequences and fosmids of the uncultured candidate division MSBL1 have been retrieved from hypersaline anoxic environments such as the deep-sea hypersaline anoxic brines in the Mediterranean Sea (van der Wielen et al., 2005; Daffonchio et al., 2006; Borin et al., 2009; Yakimov et al., 2013) and a hypersaline microbial mat of a solar saltern (López-López et al., 2013). The majority of archaeal 16S rRNA sequences recovered from these ecosystems belonged to the uncultured candidate division MSBL1 and combined methane production that was observed suggests that this archaeal lineage is involved in methanogenesis at extreme salinities. Methane production has been observed in MA1 samples (Visscher, unpublished), validating a potential biogeochemical role for MSBL1-like organisms in Laguna Tebenquiche.
The most abundant euryarchaeal OTUs in RAC1, MA1, and MA2 were classified into the deep-sea hydrothermal vent Euryarchaeotic group 1 (DHVEG-1). The uncultured DHVEG-1 members were found in the water column and sediment of anoxic deep-sea hydrothermal vents no additional information is available (Takai and Horikoshi, 1999).
The crenarchaeal OTUs make up 4–17% of the sequences in mats and rhizome-associated concretions. Many of these OTUs could be classified as MBGB, the biogeochemical function of which is highly speculative, but, based on observations in hypersaline microbial mats, could involve sulfate reduction (Robertson et al., 2009).
In contrast to earlier work (Demergasso et al., 2008; Farías et al., 2014) that reported predominance of ubiquitous Bacteroidetes and Proteobacteria in Laguna Tebenquiche, the current study found that these phyla were not dominant. Instead, Plantomycetes, Firmicutes, and Acetotermia were the main contributors to bacterial diversity. This finding could result from primer bias: earlier studies (5,7) focused on the bacterial community, whereas the current investigation analyzed the total microbial community (bacteria and archaea). OTUs belonging to Planctomycetes, which was the most abundant phylum within the bacteria in MA1 and MA2 and the second and third-most abundant phylum in RAC1 and RAC2, respectively, were mainly associated with the class Phycisphaerae. This class was recently described by one cultured and several uncultured representatives retrieved from marine environments and soils (Fukunaga et al., 2009). Diverse organoheterotrophic capabilities allows organisms belonging to this class to colonize of a wide variety of ecosystems, ranging from aquatic to terrestrial habitats including several extreme environments. Phycisphaerae were previously found in microbial mats of hypersaline lakes in the Bahamas (Baumgartner et al., 2009a) and Kiribati (Schneider et al., 2013).
The OTUs assigned to the phylum Firmicutes, which were prevalent in RAC2 (Figure 4), comprised mainly of fermenter halophilic anaerobic members of the order Halanaerobiales. OTUs assigned to the phylum Acetothermia, especially the taxon KB1, were ubiquitous in RAC2 (25% of total sequences) and less in both mats (4–5% of total sequences; Figure 4). Members of this phylum are uncultivated thermophiles (Hugenholtz et al., 1998; Costa et al., 2009; Kim et al., 2012; Németh et al., 2014) and the partial reconstructed genome indicates that the reductive acetyl-CoA pathway is used for CO2 fixation. It has been suggested that members of this phylum contribute significantly to primary production under anoxic oligocarbophylic conditions (Takami et al., 2012). It should be noted that mats are organic rich, but the bulk of this organic matter is made up by the complex structure of expolymeric substances (EPS) (Decho, 2000; Decho et al., 2005). The majority of this EPS is recalcitrant, notably in deeper layers, resisting microbial degradation (Braissant et al., 2009). As a result, especially the anoxic parts of EPS-rich mats and microbialites could be deprived of readily available organic substrates for respiration, thereby increasing the importance of anaerobic CO2 fixation and methanogenesis in these systems.
Typically, cyanobacteria are the principle autotrophs in microbial mats (Visscher and van Gemerden, 1993; Visscher and Stolz, 2005). However, given the limited presence of cyanobacterial as well as proteobacterial OTUs in our samples, it is plausible that, in addition to the Halobacteria discussed above, other organisms such as Acetothermia contribute to community CO2 fixation in Tebenquiche. The scarcity of cyanobacteria in the current study corroborates earlier observations of a low cyanobacterial presence in mats and evaporites from the Salar de Atacama (Farías et al., 2013, 2014; Rasuk et al., 2014, 2016). However, we cannot rule out that the cyanobacterial community comprises a few species with high abundance, and high metabolic activity. Similar observations of low cyanobacterial diversity were made previously by other investigators in a variety of mats and microbialites (McKay et al., 2003; Ley et al., 2006; Baumgartner et al., 2009a,b; Lynch et al., 2012). Alphaproteobacteria and Gammaproteobacteria, prevalent in MA1 and EVD, are typical inhabitants of microbial mats and microbialites (Dupraz et al., 2011) and may significantly contribute to primary production (van Gemerden, 1993). The depth profiles of sulfide (Figure 3) support the notion that sulfide-oxidizing bacteria, including both chemolithoautotrophs and photolithoautrophs, could also contribute to the organic carbon pool.
OTUs belonging to Chlorothixaceae and Anaerolinaeae (Chloroflexi), Rhodovibrio (Alphaproteobacteria), and purple sulfur bacteria classified into the family Chromatiaceae and Ectothiorhodospiraceae (Gammaproteobacteria) were found and most likely carrying out the anoxygenic photosynthesis in Laguna Tebenquiche.
Members of Bacteroidetes were present but scarce in all samples. Bacteroidetes are believed to be among the best-adapted organisms to growth under the wide range of physicochemical conditions found in Atacama Desert (Demergasso et al., 2004). In addition, this phylum was observed in a variety of other hypersaline systems including microbial mats (Sørensen et al., 2005; Ley et al., 2006), water and sediment samples (Demergasso et al., 2004, 2008, 2010; Dorador, 2007), and evaporites (Stivaletta et al., 2011; Farías et al., 2014; Rasuk et al., 2014).
The presence of the phylum Verrucomicrobia was strongly correlated to RAC1. In similar microbialites found under less saline conditions [e.g., stromatolites from Laguna Socompa (Argentina) and a microbialites in Laguna La Brava in Atacama Desert (Chile)], with a conductivity of less than conductivity 115 mS/cm (Farías et al., 2013, 2014), OTUs related to this phylum were also found. It is plausible that members of this phylum are ubiquitous in microbialites found in high altitude whenever the salinity is not excessively high.
To further investigate the impact of the limited cyanobacterial diversity discussed above and unusually high abundance of archaea, we determined the microbial diversity in discrete depth horizons.
Deinococcus sp. has been a model organism for investigating UV resistance (Arrage et al., 1993), and is frequently found in a variety of microbial mats (Skirnisdottir et al., 2000; Pagaling et al., 2012; Abed et al., 2014; Tytgat et al., 2014), including stromatolites of Shark Bay, Australia (Goh et al., 2009). In the non-lithifying mat MA1, a large proportion of 16S rRNA sequences in the surface layer were associated with the phylum Deinococcus-Thermus. Farías et al. (2013) suggested that elevated levels of UV radiation associated with high altitude ecosystems limit microbial abundance and diversity, e.g., in stromatolites found in Laguna Socompa (Argentina). Therefore, the high dominance of the phylum Deinococcus-Thermus in the surface layer was likely due to their UV resistance mechanism. We also found that crenarchaeal and acetothermial OTUs that classified as MBGB and KB1, respectively, both prevalent in deeper layers, were absent in the surface layer of MA1 where the oxygen concentrations peaked (Table S3).
Some metabolic quandaries could be addressed through analysis in individual layers of MA1 and EVD. As outlined above, the cyanobacterial contribution to primary production appeared limited. Anoxygenic phototrophic Alphaproteobacteria and Gammaproteobacteria and Chloroflexi were present in the surface and second layers of MA1 and in the second and third layers of EVD, immediately underneath the surface halite layer. This distribution was further substantiated by microelectrode measurements of oxygen distribution (Figure 3), showing a maximum concentration at 1.5–1.75 mm of depth in MA1 (corresponding to surface and second layer) and between 6 and 8 mm of depth in EVD (coinciding with the second and third layers). Some gammaproteobacterial OTUs in both MA1 and EVD were related to the order Chromatiales (Gammaproteobacteria), which has representatives containing bacteriochlorophyll a and b. Some of these Chromatiales grow under both oxic and anoxic conditions and are capable of photolithoautotrophic and chemolithoautotrophic metabolism (van Gemerden, 1993).
Both aerobic and anaerobic heterotrophic microorganisms were detected in MA1 and EVD. The main heterotrophs in all layers of EVD were aerobic haloarchaea. Euryarchaeal OTUs related to anaerobic and methanogenic archaea were found in MA1 and EVD, with increasing proportions of 16S rRNA gene sequences with increasing depth, where permanently anoxic conditions prevail. A plethora of anaerobic fermenters resided in all layers of MA1 and EVD, however mainly represented by Firmicutes. Sulfate reduction was likely carried out by Deltaproteobacteria in MA1, including in the surface and second layers, both of which are oxic. Studies performed in several microbial mats, including hypersaline mats in Guerrero Negro, Solar Lake, Kiritimati Atoll, Shark Bay, Texel, and the Bahamas revealed high sulfate reduction rates in oxic layers as well (Canfield and Des Marais, 1991; Fründ and Cohen, 1992; Visscher et al., 1992, 1998; Bühring et al., 2009; Pages et al., 2014; Wong et al., 2015).
Most diversity studies in microbial mats and microbialites to date focused only on bacteria and showed important contributions of Cyanobateria, Proteobacteria, and Bacteroidetes to the total diversity in these system. Although primer bias, mentioned above, clearly impact the outcome of molecular investigations, the primacy of bacteria over archaea has been reported in numerous benthic microbial ecosystems, including hypersaline mats of Kribati (Schneider et al., 2013), open marine stromatolites (Baumgartner et al., 2009a), and hypersaline microbialites in the Bahamas (Baumgartner et al., 2009b) and Cuatro Cienegas, Guerrero Negro, and Shark Bay (Burns et al., 2004; Ley et al., 2006; Valeria et al., 2007; Demergasso et al., 2008). The high relative abundance of archaea in the samples from Laguna Tebenquiche warrants a reassessment of the role of archaea in these benthic microbial ecosystems and necessitates a similar approach when investigating other microbial mats.
Conclusion
In conclusion, in Laguna Tebenquiche, Euryarchaeota was one of the most abundant phyla in all samples studied, notably in EVD, in which it represented 97% of the 16S rRNA sequences. Most of these euryarchaeal OTUs were classified within the class Halobacteria or anaerobic and methanogenic archaea. Halobacteria mainly grow by aerobic oxidation of amino acids, carbohydrates or alcohol (i.e., glycerol), but can also photophosphorylate under anoxic conditions (Hartmann et al., 1980). This suggests an important role for (an)aerobic heterotrophy and potentially methanogenesis carried out by Euryarchaeota in these benthic ecosystems. Planctomycetes played also a key role in mats and rhizome-associated concretion samples, notably the class Phycisphaerae, which are aerobic organoheterotrophs. In addition to cyanobacterial primary production, anoxygenic photosynthesis by Chromatiales, possibly Chloroflexi and the candidate family Chlorotrichaceae (in RAC1 and mat samples) could contribute to CO2 fixation in the mat. Other taxa present that could play a metabolic role include the uncultured candidate division MSBL1, possibly be involved in methanogenesis, and the uncultured MBGB. Some members of Proteobacteria could contribute to sulfate reduction and the uncultured phylum Acetothermia could perform CO2 fixation through the reductive acetyl-CoA pathway. The importance of understanding the taxonomic and metabolic diversity in Laguna Tebenquiche resides in the extreme conditions of this environment, which may provide new insights into microbial processes in the Earth's early history and potentially on habitable exoplanets.
Author contributions
AF performed data analysis, interpreted data and wrote paper. MR contributed in sampling and data analysis. PV contributed in work proposal, sampling, performed oxygen and sulfide profile analysis and wrote paper. MC, FN obtained funding for the original project idea and performed physicochemical analysis. DP performed mineral analysis. MP contributed in oxygen and sulfide profile analysis. AV contributed in data analysis. MF obtained funding for the original project idea, contributed in work proposal, sampling and wrote paper. All authors read and approved this manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by Sociedad Química y Minera de Chile and Centro de Ecología Aplicada. AF, MR are recipients of a CONICET fellowship. We also want to thank Lic. Cecilia Genazzini and Mr. Pablo García of CONICET for their assistance in the XRD laboratory. PV acknowledges support of NSF grant # OCE1052974.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01284
Rarefaction curves plotting the number of observed OTUs as a function of the number of sequences in microbial mats (MA1 and MA2), rhizome-associated lithified concretions (RAC1 and RAC2) and evaporite (EVD).
Rarefaction curves plotting the number of observed OTUs as a function of the number of sequences. (A) Rarefaction curves by layers in MA1. (B) Rarefaction curves by layers in EVD.
Physico-chemical parameters for the overlying water from the different samples studied.
Observed microbial richness and diversity estimates based on 97% OTU clusters in microbial mats (MA1 and MA2), rhizome-associated lithified mats (RAC1 and RAC2) and evaporite (EVD).
Abundant microbial OTUs classified at the lowest possible taxonomic level in MA1. Each OTU contains at less 1% 16S rRNA sequences.
Abundant microbial OTUs classified at the lowest possible taxonomic level in EVD. Each OTU contains at less 1% 16S rRNA sequences.
References
- Abed R. M. M., Al-Kharusi S., Prigent S., Headley T. (2014). Diversity, distribution and hydrocarbon biodegradation capabilities of microbial communities in oil-contaminated cyanobacterial mats from a constructed wetland. PLoS ONE 9:e114570. 10.1371/journal.pone.0114570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Public Health Association (1998). Standard Methods for the Examination of Water and Wastewater, 20th Edn. Baltimore: Port City Press. [Google Scholar]
- Arrage A. A., Phelps T. J., Benoit R. E., White D. C. (1993). Survival of subsurface microorganisms exposed to UV radiation and hydrogen peroxide. Appl. Environ. Microbiol. 59, 3545–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S. T., Berg-Lyons D., Caporaso J. G., Walters W. A., Knight R., Fierer N. (2010). Examining the global distribution of dominant archaeal populations in soil. ISME J. 5, 908–917. 10.1038/ismej.2010.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner L. K., Dupraz C., Buckley D. H., Spear J. R., Pace N. R., Visscher P. T. (2009a). Microbial species richness and metabolic activities in hypersaline microbial mats: insight into biosignature formation through lithification. Astrobiology 9, 861–874. 10.1089/ast.2008.0329 [DOI] [PubMed] [Google Scholar]
- Baumgartner L. K., Spear J. R., Buckley D. H., Pace N. R., Reid R. P., Dupraz C., et al. (2009b). Microbial diversity in modern marine stromatolites, Highborne Cay, Bahamas. Environ. Microbiol. 11, 2710–2719. 10.1111/j.1462-2920.2009.01998.x [DOI] [PubMed] [Google Scholar]
- Bevacqua P. (1992). Geomorfología del Salar de Atacama y Estratigrafía de su Núcleo y Delta, Segunda Región de Antofagasta. Thesis, Universidad Católica del Norte, Antofagasta. [Google Scholar]
- Borin S., Brusetti L., Daffonchio D., Delaney E., Baldi F. (2009). Biodiversity of prokaryotic communities in sediments of different sub-basins of the Venice lagoon. Res. Microbiol. 160, 307–314. 10.1016/j.resmic.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Braissant O., Decho A. W., Przekop K. M., Gallagher K. L., Glunk C., Dupraz C., et al. (2009). Characteristics and turnover of exopolymeric substances in a hypersaline microbial mat. FEMS Microbiol. Ecol. 67, 293–307. 10.1111/j.1574-6941.2008.00614.x [DOI] [PubMed] [Google Scholar]
- Bühring S. I., Smittenberg R. H., Sachse D., Lipp J. S., Golubic S., Sachs J. P., et al. (2009). A hypersaline microbial mat from the Pacific Atoll Kiritimati: insights into composition and carbon fixation using biomarker analyses and a 13C-labeling approach. Geobiology 7, 308–323. 10.1111/j.1472-4669.2009.00198.x [DOI] [PubMed] [Google Scholar]
- Burns B. P., Goh F., Allen M., Neilan B. A. (2004). Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ. Microbiol. 6, 1096–1101. 10.1111/j.1462-2920.2004.00651.x [DOI] [PubMed] [Google Scholar]
- Canfield D. E., Des Marais D. (1991). Aerobic sulfate reduction in microbial mats. Science 251, 1471–1473. 10.1126/science.11538266 [DOI] [PubMed] [Google Scholar]
- Canfield D. E., Sørensen K. B., Oren A. (2004). Biogeochemistry of a gypsum-encrusted microbial ecosystem. Geobiology 2, 133–150. 10.1111/j.1472-4677.2004.00029.x [DOI] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa K. C., Navarro J. B., Shock E. L., Zhang C. L., Soukup D., Hedlund B. P. (2009). Microbiology and geochemistry of great boiling and mud hot springs in the United States Great Basin. Extremophiles 13, 447–459. 10.1007/s00792-009-0230-x [DOI] [PubMed] [Google Scholar]
- Daffonchio D., Borin S., Brusa T., Brusetti L., van der Wielen P. W. J. J., Bolhuis H., et al. (2006). Stratified prokaryote network in the oxic-anoxic transition of a deep-sea halocline. Nature 440, 203–207. 10.1038/nature04418 [DOI] [PubMed] [Google Scholar]
- Decho A. W. (2000). Microbial biofilms in intertidal systems: an overview. Cont. Shelf Res. 20, 1257–1273. 10.1016/S0278-4343(00)00022-4 [DOI] [Google Scholar]
- Decho A. W., Visscher P. T., Reid R. P. (2005). Production and cycling of natural microbial exopolymers (EPS) within a marine stromatolite. Palaeogeogr. Palaeoclimatol. Palaeoecol. 219, 71–86. 10.1016/j.palaeo.2004.10.015 [DOI] [Google Scholar]
- Demergasso C., Casamayor E. O., Chong G., Galleguillos P., Escudero L., Pedrós-Alió C. (2004). Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, Northern Chile. FEMS Microbiol. Ecol. 48, 57–69. 10.1016/j.femsec.2003.12.013 [DOI] [PubMed] [Google Scholar]
- Demergasso C., Dorador C., Meneses D., Blamey J., Cabrol N., Escudero L., et al. (2010). Prokaryotic diversity pattern in high-altitude ecosystems of the Chilean Altiplano. J. Geophys. Res. 115, G00D09 10.1029/2008JG000836 [DOI] [Google Scholar]
- Demergasso C., Escudero L., Casamayor E. O., Chong G., Balagué V., Pedrós-Alió C. (2008). Novelty and spatio-temporal heterogeneity in the bacterial diversity of hypersaline Lake Tebenquiche (Salar de Atacama). Extremophiles 12, 491–504. 10.1007/s00792-008-0153-y [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorador C. (2007). Microbial Diversity in High Altitude Wetlands of the Chilean Altiplano: Phylogeny, Diversity, and Function. Thesis, University of Kiel, Kiel.
- Dupraz C., Reid R. P., Braissant O., Decho A. W., Norman R. S., Visscher P. T. (2009). Processes of carbonate precipitation in modern microbial mats. Earth Sci. Rev. 96, 141–162. 10.1016/j.earscirev.2008.10.005 [DOI] [Google Scholar]
- Dupraz C., Reid R. P., Visscher P. T. (2011). Microbialite, Modern, in Encyclopedia of Geobiology, eds Reitner J., Thiel V. (New York, NY: Springer-Verlag; ), 617–635. 10.1007/978-1-4020-9212-1 [DOI] [Google Scholar]
- Dupraz C., Visscher P. T. (2005). Microbial lithification in marine stromatolites and hypersaline mats. Trends Microbiol. 13, 429–438. 10.1016/j.tim.2005.07.008 [DOI] [PubMed] [Google Scholar]
- Dupraz C., Visscher P. T., Baumgartner L. K., Reid R. P. (2004). Microbe-mineral interactions: early carbonate precipitation in a hypersaline lake (Eleuthera Island, Bahamas). Sedimentology 51, 745–765. 10.1111/j.1365-3091.2004.00649.x [DOI] [Google Scholar]
- Eaton A., Clesceri L. S., Rice L., Greenberg A., Franson M. (2005). Standard Methods for the Examination of Water and Wastewater, 21st Edn. Washington, DC: American Public Health Association. [Google Scholar]
- Edgar R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Farías M. E., Contreras M., Rasuk M. C., Kurth D., Flores M. R., Poiré D. G., et al. (2014). Characterization of bacterial diversity associated with microbial mats, gypsum evaporites and carbonate microbialites in thalassic wetlands: tebenquiche and La Brava, Salar de Atacama, Chile. Extremophiles 18, 311–329. 10.1007/s00792-013-0617-6 [DOI] [PubMed] [Google Scholar]
- Farías M. E., Rascovan N., Toneatti D. M., Albarracín V. H., Flores M. R., Poiré D. G., et al. (2013). The discovery of stromatolites developing at 3570 m above sea level in a high-altitude volcanic lake Socompa, Argentinean Andes. PLoS ONE 8:e53497. 10.1371/journal.pone.0053497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fründ C., Cohen Y. (1992). Diurnal cycles of sulfate reduction under oxic conditions in cyanobacterial mats. Appl. Envir. Microbiol. 58, 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga Y., Kurahashi M., Sakiyama Y., Ohuchi M., Yokota A., Harayama S. (2009). Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J. Gen. Appl. Microbiol. 55, 267–275. 10.2323/jgam.55.267 [DOI] [PubMed] [Google Scholar]
- Goh F., Allen M. A., Leuko S., Kawaguchi T., Decho A. W., Burns B. P., et al. (2009). Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J. 3, 383–396. 10.1038/ismej.2008.114 [DOI] [PubMed] [Google Scholar]
- Gomez F. J., Kah L. C., Bartley J. K., Astini R. A. (2014). Microbialites in a high-altitude andean lake: multiple controls on carbonate precipitation and lamina accretion. Palaios 29, 233–249. 10.2110/palo.2013.049 [DOI] [Google Scholar]
- Grant W. D. (2001). Genus I. halobacterium elazari-volcani 1957, 207, AL emend. larsen and grant 1989, 2222, in Bergey's Manual of Systematic Bacteriology: Vol. 1, The Archaea and the Deeply Branching and Phototrophic Bacteria, eds Boone D. R., Castenholz W. (New York, NY: Springer-Verlag; ), 301–305. [Google Scholar]
- Haas B. J., Gevers D., Earl A. M., Feldgarden M., Ward D. V., Giannoukos G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. 10.1101/gr.112730.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann R., Sickinger H. D., Oesterhelt D. (1980). Anaerobic growth of halobacteria. Proc. Natl. Acad. Sci. U.S.A. 77, 3821–3825. 10.1073/pnas.77.7.3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P., Pitulle C., Hershberger K. L., Pace N. R. (1998). Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180, 366–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-S., Makama M., Petito J., Park N.-H., Cohan F. M., Dungan R. S. (2012). Diversity of Bacteria and Archaea in hypersaline sediment from Death Valley National Park, California. Microbiologyopen 1, 135–148. 10.1002/mbo3.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara J., Escudero González L., Ferrero M., Chong Díaz G., Pedrós-Alió C., Demergasso C. (2012). Enrichment of arsenic transforming and resistant heterotrophic bacteria from sediments of two salt lakes in Northern Chile. Extremophiles 16, 523–538. 10.1007/s00792-012-0452-1 [DOI] [PubMed] [Google Scholar]
- Ley R. E., Harris J. K., Wilcox J., Spear J. R., Miller S. R., Bebout B. M., et al. (2006). Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72, 3685–3695. 10.1128/AEM.72.5.3685-3695.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizama C., Monteoliva-Sánchez M., Prado B., Ramos-Cormenzana A., Weckesser J., Campos V. (2001). Taxonomic study of extreme halophilic archaea isolated from the “Salar de Atacama,” Chile. Syst. Appl. Microbiol. 24, 464–474. 10.1078/0723-2020-00053 [DOI] [PubMed] [Google Scholar]
- Lizama C., Monteoliva-Sánchez M., Suárez-García A., Roselló-Mora R., Aguilera M., Campos V., et al. (2002). Halorubrum tebenquichense sp. nov., a novel halophilic archaeon isolated from the Atacama Saltern. Chile. Int. J. Syst. Evol. Microbiol. 52, 149–155. 10.1099/00207713-52-1-149 [DOI] [PubMed] [Google Scholar]
- López-López A., Richter M., Peña A., Tamames J., Rosselló-Móra R. (2013). New insights into the archaeal diversity of a hypersaline microbial mat obtained by a metagenomic approach. Syst. Appl. Microbiol. 36, 205–214. 10.1016/j.syapm.2012.11.008 [DOI] [PubMed] [Google Scholar]
- Lynch R. C., King A. J., Farías M. E., Sowell P., Vitry C., Schmidt S. K. (2012). The potential for microbial life in the highest-elevation (>6000 m.a.s.l.) mineral soils of the Atacama region. J. Geophys. Res. 117:G02028 10.1029/2012JG001961 [DOI] [Google Scholar]
- McKay C. P., Friedmann E. I., Gómez-Silva B., Cáceres-Villanueva L., Andersen D. T., Landheim R. (2003). Temperature and moisture conditions for life in the extreme arid region of the Atacama desert: four years of observations including the El Niño of 1997-1998. Astrobiology 3, 393–406. 10.1089/153110703769016460 [DOI] [PubMed] [Google Scholar]
- Németh A., Szirányi B., Krett G., Janurik E., Kosáros T., Pekár F., et al. (2014). Prokaryotic phylogenetic diversity of Hungarian deep subsurface geothermal well waters. Acta Microbiol. Immunol. Hung. 61, 363–377. 10.1556/AMicr.61.2014.3.9 [DOI] [PubMed] [Google Scholar]
- Oren A. (2010). Industrial and environmental applications of halophilic microorganisms. Environ. Technol. 31, 825–834. 10.1080/09593330903370026 [DOI] [PubMed] [Google Scholar]
- Oren A., Kühl M., Karsten U. (1995). An endoevaporitic microbial mat within a gypsum crust: zonation of phototrophs, photopigments, and light penetration. Mar. Ecol. Prog. Ser. 128, 151–159. 10.3354/meps128151 [DOI] [Google Scholar]
- Pagaling E., Grant W. D., Cowan D. A., Jones B. E., Ma Y., Ventosa A., et al. (2012). Bacterial and archaeal diversity in two hot spring microbial mats from the geothermal region of Tengchong, China. Extremophiles 16, 607–618. 10.1007/s00792-012-0460-1 [DOI] [PubMed] [Google Scholar]
- Pages A., Welsh D. T., Teasdale P. R., Grice K., Vacher M., Bennett W. W., et al. (2014). Diel fluctuations in solute distributions and biogeochemical cycling in a hypersaline microbial mat from Shark Bay, WA. Mar. Chem. 167, 102–112. 10.1016/j.marchem.2014.05.003 [DOI] [Google Scholar]
- Prado B., del Moral A., Campos V. (1993). Distribution and types of Heterotrophyc halophilic flora from Salar de Atacama, Chile. Toxicol. Environ. Chem. 38, 163–166. 10.1080/02772249309357887 [DOI] [Google Scholar]
- Prado B., Del Moral A., Quesada E., Ríos R., Monteoliva-Sanchez M., Campos V., et al. (1991). Numerical taxonomy of moderately halophilic Gram-negative rods isolated from the Salar de Atacama, Chile. Syst. Appl. Microbiol. 14, 275–281. 10.1016/S0723-2020(11)80381-4 [DOI] [Google Scholar]
- Rascovan N., Maldonado J., Vazquez M. P., Eugenia Farías M. (2016). Metagenomic study of red biofilms from Diamante Lake reveals ancient arsenic bioenergetics in haloarchaea. ISME J. 10, 299–309. 10.1038/ismej.2015.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasuk M. C., Fernández A. B., Kurth D., Contreras M., Novoa F., Poiré D., et al. (2016). Bacterial diversity in microbial mats and sediments from the Atacama Desert. Microb. Ecol. 71, 44–56. 10.1007/s00248-015-0649-9 [DOI] [PubMed] [Google Scholar]
- Rasuk M. C., Kurth D., Flores M. R., Contreras M., Novoa F., Poire D., et al. (2014). Microbial characterization of microbial ecosystems associated to evaporites domes of gypsum in Salar de Llamara in Atacama desert. Microb. Ecol. 68, 483–494. 10.1007/s00248-014-0431-4 [DOI] [PubMed] [Google Scholar]
- Risacher F., Alonso H. (1996). Geoquimica del Salar de Atacama, parte 2: evolucion de las aguas. Andean Geol. 23, 123–134. 10.5027/andgeoV23n2-a02 [DOI] [Google Scholar]
- Risacher F., Alonso H., Salazar C. (2003). The origin of brines and salts in Chilean salars: a hydrochemical review. Earth Sci. Rev. 63, 249–293. 10.1016/S0012-8252(03)00037-0 [DOI] [Google Scholar]
- Robertson C. E., Spear J. R., Harris J. K., Pace N. R. (2009). Diversity and stratification of archaea in a hypersaline microbial mat. Appl. Environ. Microbiol. 75, 1801–1810. 10.1128/AEM.01811-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild L. J., Mancinelli R. L. (2001). Life in extreme environments. Nature 409, 1092–1101. 10.1038/35059215 [DOI] [PubMed] [Google Scholar]
- Schneider D., Arp G., Reimer A., Reitner J., Daniel R. (2013). Phylogenetic analysis of a microbialite-forming microbial mat from a hypersaline lake of the Kiritimati atoll, Central Pacific. PLoS ONE 8:e66662. 10.1371/journal.pone.0066662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood J. E., Stagnitti F., Kokkinn M. J., Williams W. D. (1991). Dissolved oxygen concentrations in hypersaline waters. Limnol. Oceanogr. 36, 235–250. 10.4319/lo.1991.36.2.0235 [DOI] [Google Scholar]
- Skirnisdottir S., Hreggvidsson G. O., Hjörleifsdottir S., Marteinsson V. T., Petursdottir S. K., Holst O., et al. (2000). Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66, 2835–2841. 10.1128/AEM.66.7.2835-2841.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen K. B., Canfield D. E., Teske A. P., Oren A. (2005). Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 71, 7352–7365. 10.1128/AEM.71.11.7352-7365.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivaletta N., Barbieri R., Cevenini F., López-García P. (2011). Physicochemical conditions and microbial diversity associated with the evaporite deposits in the Laguna de la Piedra (Salar de Atacama, Chile). Geomicrobiol. J. 28, 83–95. 10.1080/01490451003653102 [DOI] [Google Scholar]
- Taillefert M., Rozan T. F. (2002). Electrochemical methods for the environmental analyses of trace element biogeochemistry, in Environmental Electrochemistry (Washington, DC: American Chemical Society; ). [Google Scholar]
- Takai K., Horikoshi K. (1999). Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics 152, 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami H., Noguchi H., Takaki Y., Uchiyama I., Toyoda A., Nishi S., et al. (2012). A deeply branching thermophilic bacterium with an ancient acetyl-CoA pathway dominates a subsurface ecosystem. PLoS ONE 7:e30559. 10.1371/journal.pone.0030559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak C. J. F., Smilauer P. (2002). CANOCO Reference Manual and CanoDraw for Windows User's Guide: Software for Canonical Community Ordination (version 4.5). Ithaca, NY: Microcomputer Power. [Google Scholar]
- Thiel V., Tank M., Neulinger S. C., Gehrmann L., Dorador C., Imhoff J. F. (2010). Unique communities of anoxygenic phototrophic bacteria in saline lakes of Salar de Atacama (Chile): evidence for a new phylogenetic lineage of phototrophic Gammaproteobacteria from pufLM gene analyses. FEMS Microbiol. Ecol. 74, 510–522. 10.1111/j.1574-6941.2010.00966.x [DOI] [PubMed] [Google Scholar]
- Tytgat B., Verleyen E., Obbels D., Peeters K., De Wever A., D'hondt S., et al. (2014). Bacterial diversity assessment in Antarctic terrestrial and aquatic microbial mats: a comparison between bidirectional pyrosequencing and cultivation. PLoS ONE 9:e97564. 10.1371/journal.pone.0097564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valderrama M. J., Prado B., del Moral A., Ríos R., Ramos-Cormenzana A., Campos V. (1991). Numerical taxonomy of moderately halophilic Gram-positive cocci isolated from the Salar de Atacama (Chile). Microbiologia 7, 35–41. [PubMed] [Google Scholar]
- Valeria S., Falcon L., Elser J., Eguiarte L. (2007). Protecting a Window into the Ancient Earth: Towards a Precambrian Park at Cuatro Cienegas, Mexico. The Citizen's Page, Evolutionary Ecology Research. Available online at: http://www.evolutionary-ecology.com/citizen/citizen.html
- van der Wielen P. W. J. J., Bolhuis H., Borin S., Daffonchio D., Corselli C., Giuliano L., et al. (2005). The enigma of prokaryotic life in deep hypersaline anoxic basins. Science 307, 121–123. 10.1126/science.1103569 [DOI] [PubMed] [Google Scholar]
- van Gemerden H. (1993). Microbial mats: a joint venture. Mar. Geol. 113, 3–25. [Google Scholar]
- Visscher P. T., Beukema J., van Gemerden H. (1991). In situ characterization of sediments: measurements of oxygen and sulfide profiles with a novel combined needle electrode. Limnol. Oceanogr. 36, 1476–1480. 10.4319/lo.1991.36.7.1476 [DOI] [Google Scholar]
- Visscher P. T., Hoeft S. E., Surgeon T. M. L., Rogers D. R., Bebout B. M., Thompson J. S. J., et al. (2002). Microelectrode measurements in stromatolites: Unraveling the Earth's past?, in Environmental Electrochemistry: Analyses of Trace Element Biogeochemistry, eds Taillefert M., Rozan T. (Washington, DC: Oxford University Press; ), 265–282. [Google Scholar]
- Visscher P. T., Prins R. A., Gemerden H. (1992). Rates of sulfate reduction and thiosulfate consumption in a marine microbial mat. FEMS Microbiol. Lett. 86, 283–294. 10.1111/j.1574-6968.1992.tb04820.x [DOI] [Google Scholar]
- Visscher P. T., Reid R. P., Bebout B. M., Hoeft S. E., Macintyre I. G., Thompson J. A. (1998). Formation of lithified micritic laminae in modern marine stromatolites (Bahamas); the role of sulfur cycling. Am. Mineral. 83, 1482–1493. [Google Scholar]
- Visscher P. T., Stolz J. F. (2005). Microbial mats as bioreactors: populations, processes, and products. Palaeogeogr. Palaeoclimatol. Palaeoecol. 219, 87–100. 10.1016/j.palaeo.2004.10.016 [DOI] [Google Scholar]
- Visscher P. T., van Gemerden H. (1993). Sulfur cycling in laminated marine microbial ecosystems, in Biogeochemistry of Global Change, ed Oremland R. S. (Boston, MA: Springer US; ), 672–690. [Google Scholar]
- Warren J. K. (2010). Evaporites through time: tectonic, climatic and eustatic controls in marine and nonmarine deposits. Earth Sci.Rev. 98, 217–268. 10.1016/j.earscirev.2009.11.004 [DOI] [Google Scholar]
- Wong H. L., Smith D.-L., Visscher P. T., Burns B. P. (2015). Niche differentiation of bacterial communities at a millimeter scale in Shark Bay microbial mats. Sci. Rep. 5:15607. 10.1038/srep15607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakimov M. M., La Cono V., Slepak V. Z., La Spada G., Arcadi E., Messina E., et al. (2013). Microbial life in the Lake Medee, the largest deep-sea salt-saturated formation. Sci. Rep. 3:3554. 10.1038/srep03554 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Rarefaction curves plotting the number of observed OTUs as a function of the number of sequences in microbial mats (MA1 and MA2), rhizome-associated lithified concretions (RAC1 and RAC2) and evaporite (EVD).
Rarefaction curves plotting the number of observed OTUs as a function of the number of sequences. (A) Rarefaction curves by layers in MA1. (B) Rarefaction curves by layers in EVD.
Physico-chemical parameters for the overlying water from the different samples studied.
Observed microbial richness and diversity estimates based on 97% OTU clusters in microbial mats (MA1 and MA2), rhizome-associated lithified mats (RAC1 and RAC2) and evaporite (EVD).
Abundant microbial OTUs classified at the lowest possible taxonomic level in MA1. Each OTU contains at less 1% 16S rRNA sequences.
Abundant microbial OTUs classified at the lowest possible taxonomic level in EVD. Each OTU contains at less 1% 16S rRNA sequences.