Abstract
We describe a method for the simulation of the growth of elongated plant organs, such as seedling roots. By combining a midline representation of the organ on a tissue scale and a vertex-based representation on the cell scale, we obtain a multiscale method, which is able to both simulate organ growth and incorporate cell-scale processes. Equations for the evolution of the midline are obtained, which depend on the cell-wall properties of individual cells through appropriate averages over the vertex-based representation. The evolution of the organ midline is used to deform the cellular-scale representation. This permits the investigation of the regulation of organ growth through the cell-scale transport of the plant hormone auxin. The utility of this method is demonstrated in simulating the early stages of the response of a root to gravity, using a vertex-based template acquired from confocal imaging. Asymmetries in the concentrations of auxin between the upper and lower sides of the root lead to bending of the root midline, reflecting a gravitropic response.
Keywords: multicellular, multiscale, biomechanics, auxin, growth, gravitropism
1. Introduction
Growth is one of the main responses plants possess with which they adapt to their environment. To extract resources from their surroundings efficiently, plants change their growth patterns in response to external stimuli such as light, touch and the direction of gravity. As a result, the simulation of many processes in plant development requires an integrative model for organ growth. Plant organs usually have a multicellular structure; for example, the primary root of Arabidopsis thaliana contains multiple concentric cylindrical layers of cells. As adjacent cells are tightly bound to each other, the overall growth of the organ must be continuous (symplastic). However, different tissue layers have different responses to plant hormones such as auxin and giberellic acid; this facilitates integrated responses to simultaneous environmental and intrinsic stimuli [1].
The rate at which the plant hormone auxin accumulates in a cell is regulated by auxin efflux carriers, such as those of the PIN-FORMED (PIN) auxin efflux carrier family; these have an asymmetric, polar distribution on the plasma membrane of the cell that leads to directed auxin transport on a tissue scale. Simulations of auxin transport in the tip of the primary root [2–5], particularly in the complex region containing the columella cells and lateral root cap, require a geometrical representation that accurately identifies individual cells and their spatial relationship with other cells. Individual cell geometries must be represented with a sufficient level of detail to (at least) permit the calculation of the volumes (areas in two dimensions) of cells and the surface areas (lengths) of the interfaces between neighbouring cells. Early models for auxin transport in the root tip [4,6] used idealized templates to represent the geometries of individual cells. With recent advances in imaging and image analysis, it has become possible to generate templates for simulations from actual cellular geometries, both in two [2] and three dimensions [7,8].
Current mechanical models for plant organs generally fall into one of two categories: those that treat the organ as a continuum, and those that consider individual cells. Treating the organ as a continuum has computational and conceptual advantages, but fails to capture the precise multicellular structure. A range of models that consider individual cells have been developed for plant organs. These include, but are not limited to, cellular Potts, vertex [9,10], vertex-element [11] or finite-element [7,12–14] formulations. Those models which explicitly represent individual cell walls (i.e. vertex-element and finite-element models) can be sensitive to the precise positioning of cell walls [11], which is difficult to obtain with sufficient accuracy for a whole plant organ from microscopy images.
Here, we present a hybrid model, which resolves the geometries of individual cells through a vertex-based representation, but uses a coarser-scale midline representation to track organ growth. The evolution of the organ in its midline representation depends upon the properties of individual cells through averaged constitutive relationships, thereby allowing cell-scale changes in cell-wall mechanical properties, e.g. caused by changes in the concentration of the plant hormone auxin in a particular tissue layer, to affect growth on the whole organ scale. In turn, growth and deformation of the organ midline affects the cell-scale geometry, changing the lengths of cell walls and volumes of cells, thereby modifying hormone transport. It also changes the distance of cells from the organ tip, which in the model used here modifies the mechanical properties of cell walls. This serves as a simple, but concrete, example of a multiresolution method for multicellular organs. The effectiveness of this approach is illustrated by its application to the early stages of the bending of a growing root in response to gravity.
The model approach is in many ways similar to those adopted in implementing multiscale homogenization methods, such as classical techniques [15] and methods such as representative volume elements [16]. Such methods typically make the assumption that the microscale problem is spatially periodic, or can be represented as a realization of a random process, and do not explicitly track all the changes to the microscale structure. While plant tissue is often highly organized, the specific geometries that we wish to use do not satisfy these assumptions. Numerical methods based upon representative volume elements, in which mechanical properties of the material are obtained from simulations of small regions around the quadrature points of a coarse finite-element discretization, have been applied to plant organs [17,18]. However, this technique requires numerical simulation of the microscale model.
Moreover, unlike classical homogenization problems in continuum mechanics, the microscale problem here is not necessarily well characterized; we have an incomplete understanding of the material properties of cell walls, and it is challenging to measure accurately the geometries of individual cells in a multicellular tissue. As a result, the principal aim is not to solve the microscale problem accurately (as may be the case for homogenization or multigrid methods, for example), but instead to generate effective macroscale equations which capture the qualitative and, to an appropriate degree, the quantitative behaviour of the organ. Our approach exploits the fact that, for symplastic growth, strain fields typically vary smoothly over the tissue scale. By contrast, stress fields can share the heterogeneity of the microstructure [19], supporting integration of stress up to the tissue level.
The method used here is loosely based upon the Cauchy–Born rule for the mechanical properties of periodic structures: while the cell-scale structure here is not periodic, we assume that the deformation on the cell-scale is a smooth interpolation of the deformation on the tissue scale. The Cauchy–Born rule has been shown to have deficiencies when applied to models for animal tissues [18,20,21], largely because of the potential for lattice-scale deformations. However, plant tissues are highly ordered and grow in a symplastic fashion, which we expect to mitigate such deficiencies. Related methods have been developed in computer graphics, where the simulation of the mechanical behaviour of objects with highly complex (and sometimes topologically defective) geometries is of interest. One approach, e.g. used in [22], is to generate a simpler, coarse geometry; the mechanical behaviour of this coarse geometry is simulated and, through suitable interpolation, used to deform the detailed geometry.
The paper is organized as follows. Section 2 briefly describes the cellular-scale (vertex-based) representation and details the midline approach and computational implementation. Section 3 describes illustrative numerical results, and we summarize the advantages and limitations of the results in §4. Details of the auxin transport model, the coordinate transformations used for the midline model and the precise form of the growth profile used are described in the appendices.
2. Material and methods
Here, in order to describe the method in a simple context, we consider a two-dimensional model of a plant organ, where all points under consideration lie in a plane; therefore, individual cells occupy two-dimensional regions, and the organ midline is a curve lying in this two-dimensional plane.
2.1. Vertex-based organ representation
We use a vertex-based representation of the cellular-scale geometry of the organ (figure 1), as has previously been employed in a large number of other simulations (e.g. [23]). Cells occupy polygonal regions, described by the positions of their vertices; neighbouring cells have common vertices, which enforce the symplastic nature of organ growth. In the simulations of this paper, the implementation of a vertex-based tissue from the OpenAlea software platform [24] was used. Using such a representation, it is straightforward to calculate geometrical quantities such as cell areas and the lengths of the interfaces between neighbouring cells, which are needed for models of auxin transport and other cell-scale processes.
Figure 1.
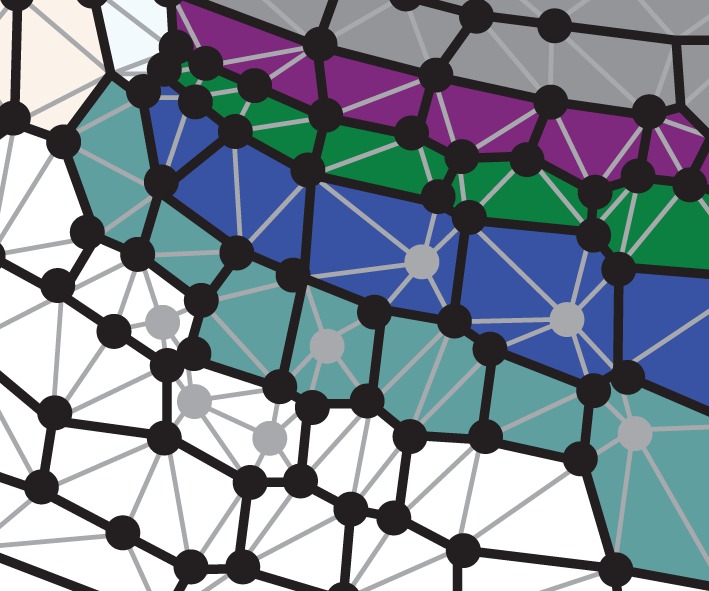
Vertex-based organ representation. Circles indicate vertices. Grey lines show triangulation of the polygonal regions. Grey circles are vertices inserted within cells for the purposes of triangulation. Coloured shading indicates cell type within the organ.
While the method described here can be used without triangulating the polygonal regions occupied by each cell, for the purposes of the simulations described below it proves very useful to do so. One reason for this is that the growth of organs varies over length scales that are comparable with the size of cells [25], which suggests that it is likely that the mechanical properties of cell walls are not uniform on each cell. Another reason is that it simplifies the calculation of the integrals appearing in the discretized versions of the midline equations. Triangularization of the vertex-based tissue representation is perfomed using gmsh [26], which uses a Delaunay algorithm. Additional vertices are inserted within cell walls and in the interior of cells to enforce a maximum triangle area.
2.2. Midline organ representation
Here, we outline the mathematical details of our midline description. A more general kinematic framework for three-dimensional deformations of elongating rods can be found in [27], for example. Compared with that study, we consider the midline of a two-dimensional object (rather than a three-dimensional rod) restricted to deform in a plane, and as we consider a viscous (rather than elastic) constitutive model we will be concerned with the strain rates (rather than strains) experienced by the material.
The geometry is described on a larger scale by a curved line through the midline of the organ, as can be seen in figure 2. For the purposes of simulation, the midline of the organ is parametrized using a Lagrangian representation, i.e.
| 2.1 |
where 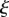 is constant following a material point;
is constant following a material point; 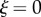 corresponds to a fixed point on the organ (the base) and
corresponds to a fixed point on the organ (the base) and 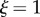 corresponds to the tip or apex of the organ. Using such a representation, common measures of organ geometry can be easily calculated; distance along the midline is given by the integral of the stretch
corresponds to the tip or apex of the organ. Using such a representation, common measures of organ geometry can be easily calculated; distance along the midline is given by the integral of the stretch
| 2.2 |
so the total length of the organ is given by
| 2.3 |
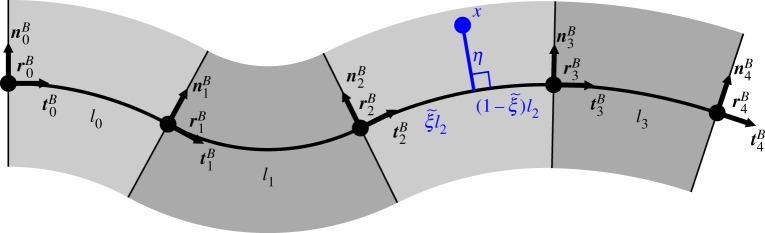
Figure 2.
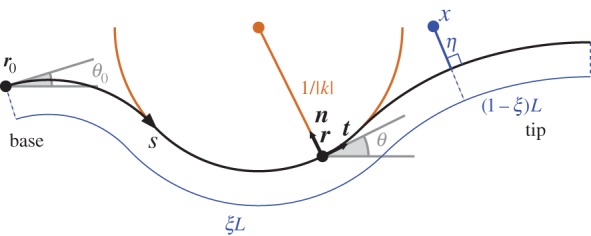
Midline organ representation in two spatial dimensions. The variable s describes distance along the midline (thick black line), which is of total length L. At a point r on the midline of the tissue, t and n are the tangent and normal vectors, respectively. The angle made between the positive x-direction and the tangent is denoted by 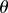 . The curvature
. The curvature  is the rate of change of
is the rate of change of 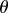 with respect to s, or equivalently the reciprocal of the radius of the osculating circle meeting the surface at a point (brown). The variable
with respect to s, or equivalently the reciprocal of the radius of the osculating circle meeting the surface at a point (brown). The variable 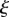 is the proportion of the distance of a point along the midline in the initial configuration; a point at constant
is the proportion of the distance of a point along the midline in the initial configuration; a point at constant 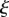 corresponds to a material point. This coordinate system is extended to points away from the midline through the variable
corresponds to a material point. This coordinate system is extended to points away from the midline through the variable 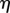 , which is the (signed) distance from a point x to the midline along a line normal to the midline.
, which is the (signed) distance from a point x to the midline along a line normal to the midline.
Note also that 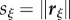 . Some plant organs, such as roots, grow mostly in a region near the tip, so for the purposes of specifying growth profiles we also introduce the distance from the tip
. Some plant organs, such as roots, grow mostly in a region near the tip, so for the purposes of specifying growth profiles we also introduce the distance from the tip 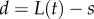 . The local unit tangent, t, and normal, n, to the midline are given by
. The local unit tangent, t, and normal, n, to the midline are given by
| 2.4 |
where k is a unit vector in the positive z-direction, normal to the plane of interest. The angle 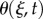 , between the positive x-axis and the tangent to the midline, is chosen such that
, between the positive x-axis and the tangent to the midline, is chosen such that
| 2.5 |
One key measurement of the local shape of the organ is its curvature, defined by
| 2.6 |
In the simulations, we will fix the position and angle at the end of the midline where 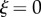 , so
, so 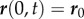 and
and 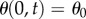 for all t. The midline path is then fully specified, and can be found from integrating (2.4)–(2.6) with respect to
for all t. The midline path is then fully specified, and can be found from integrating (2.4)–(2.6) with respect to 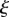 .
.
A number of measures have been proposed to describe the local kinematics of growth. The (Lagrangian) velocities of material points on the midline (relative to 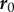 ) are given by
) are given by
| 2.7 |
The speed of a point along the midline is similarly given by
| 2.8 |
We also define the spin (angular velocity) of a point on the midline to be
| 2.9 |
The most common measure for the rate of growth along the organ axis is the relative elemental growth rate or relative elongation rate (RER), given by
| 2.10 |
Here, we choose to describe bending of the root midline through the rate of change of curvature (following a material point), 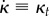 .
.
2.2.1. Relating midline and Cartesian coordinates
The midline representation describes the locations and trajectories of material points lying on the midline of the organ. By using curvilinear coordinates defined relative to the midline, we may extend this description to the whole of the organ in the form
| 2.11 |
For each point x in the organ, we find the nearest point r on the midline; 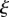 is then the midline coordinate of r, and
is then the midline coordinate of r, and 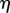 is the signed distance from the point x to the midline at r. Assuming that the tissue does not undergo shear deformation, these coordinates are orthogonal, i.e.
is the signed distance from the point x to the midline at r. Assuming that the tissue does not undergo shear deformation, these coordinates are orthogonal, i.e. 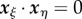 everywhere, and uniquely defined in a local neighbourhood of the midline (provided
everywhere, and uniquely defined in a local neighbourhood of the midline (provided 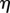 is less than the maximum value of
is less than the maximum value of 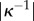 ), assuming that the midline does not turn through more than 180° (i.e. does not double back upon itself). The vertices in the vertex-based representation will be assumed to remain at fixed positions in the midline coordinate system.
), assuming that the midline does not turn through more than 180° (i.e. does not double back upon itself). The vertices in the vertex-based representation will be assumed to remain at fixed positions in the midline coordinate system.
As described in more detail in appendix A, the rate of strain tensor is given by
| 2.12 |
i.e. it is diagonal in these curvilinear coordinates, with zero strain rate normal to the midline, and strain rate  parallel to the midline, where this is given by
parallel to the midline, where this is given by
| 2.13 |
2.3. Viscous and elastic growth models
Here, we will consider plant organs over sufficiently long time scales and sufficiently short length scales that the effects of inertia are negligible. As a consequence, each material element in the tissue is in mechanical quasi-equilibrium. This will require a model for the constitutive law, which relates the deformation in the tissue to material stresses.
Multicellular models, which explicitly represent individual cell walls, have tended to use one of two main classes of constitutive laws: elastic growth models and viscous growth models.
Elastic growth models treat the cell wall on short time scales as an elastic material. The strain generated by cell-wall deformation relaxes over longer time scales through growth. Various different models for strain relaxation have been proposed [12,13,28].
Viscous growth models treat the cell wall as a viscous material, with cell-wall stresses depending on the strain rate in the wall rather than the strain [29,30]. Such a model is appropriate to capture growth processes, which occur over time scales longer than those on which elastic stresses relax, and has the advantages of being computationally and conceptually simpler than elastic growth models, as the viscous constitutive law in some respects (see below) combines the elastic constitutive law and the growth law. However, viscous models do have the weaknesses that they are not appropriate to study deformations occurring over short time scales (such as indentation experiments).
2.3.1. Relationship between viscous and elastic growth models
A number of different elastic growth models are possible. The simplest possible one-dimensional model consists of a linearly elastic spring, with undeformed length 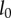 that grows at a rate proportional to the strain (or stress) in the spring, for which
that grows at a rate proportional to the strain (or stress) in the spring, for which
| 2.14 |
where l is the length of the spring and 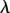 the spring constant. We assume that l varies over time scales much longer than the relaxation time scale
the spring constant. We assume that l varies over time scales much longer than the relaxation time scale 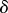 , which we take to be small.
, which we take to be small.
The second part of (2.14) gives us that, to leading order in 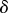 ,
, 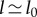 . To next order in
. To next order in 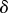 , this equation becomes
, this equation becomes 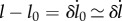 . Substituting these approximations into the first part of equation (2.14) gives
. Substituting these approximations into the first part of equation (2.14) gives
| 2.15 |
so the forces in the spring can be seen to be approximated by a viscous element (or dashpot) with viscosity 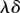 , provided that the time scale of deformation is much longer than the relaxation time for viscous stresses. In this sense, viscoelastic growth models of the form (2.14) can be approximated by viscous growth models over long time scales. In modelling plant tissues, it is common for viscous relationships such as (2.15) to be supplemented with a threshold stress (or strain) above which the material yields irreversibly [31], but we do not incorporate this feature here.
, provided that the time scale of deformation is much longer than the relaxation time for viscous stresses. In this sense, viscoelastic growth models of the form (2.14) can be approximated by viscous growth models over long time scales. In modelling plant tissues, it is common for viscous relationships such as (2.15) to be supplemented with a threshold stress (or strain) above which the material yields irreversibly [31], but we do not incorporate this feature here.
2.4. Midline evolution equations
The midline evolution equations that will be obtained are similar to those in [29]. However, in that work the equations were derived from consideration of the forces acting on a cross-section normal to the axis of the root, which requires that both the cross-sectional cellular geometry and the mechanical properties of cells vary slowly along the organ axis. Here, we wish to derive the midline equations in a variational form, as this makes it simpler to deduce the discrete forms of the midline equations that we use for simulation. This is made more complicated by the fact that variational principles for dissipative systems (away from equilibrium) may not be as well grounded as those for elastic solids, for example. Below, we sketch out a plausible approach to the derivation of the governing system that we then implement computationally; given our focus on the practical implementation of the hybrid model, we here leave more rigorous considerations on one side.
2.4.1. Energy dissipation for viscous models
In analogy with Stokes' law for viscous flow, we will use an energy dissipation principle to derive the evolution equations of the organ midline. Introducing a wall viscosity 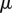 , the energy dissipation rate per unit area is
, the energy dissipation rate per unit area is 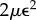 , so the overall rate of energy dissipation is given by
, so the overall rate of energy dissipation is given by
| 2.16 |
where this integral is over the region in two dimensions R occupied by the organ. We expect that the evolution of the midline will be an extremum of D, subject to the constraint that the work done by the internal pressure forces (turgor) be equal to the viscous dissipation, i.e. that
| 2.17 |
Thus, using the method of Lagrange multipliers, we find that the midline evolution must be a stationary point of
| 2.18 |
2.4.2. Midline equations from variational principle
The evolution of the organ midline is a stationary point of (2.18) at each time t, with respect to variations in the RER 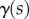 and curvature generation rate
and curvature generation rate 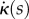 . Using (2.13) gives
. Using (2.13) gives
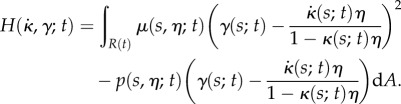 |
2.19 |
Here, for clarity we show the explicit dependence of all quantities on s, 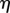 and t; in the remainder of this section we will omit the dependence on t for conciseness. We desire that the midline evolution be an extremum of H. Writing
and t; in the remainder of this section we will omit the dependence on t for conciseness. We desire that the midline evolution be an extremum of H. Writing 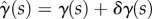 ,
, 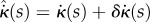 , we have
, we have
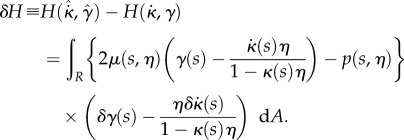 |
2.20 |
Splitting this into two gives
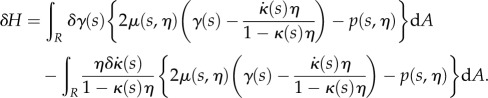 |
2.21 |
To be a stationary point, these two integrals must independently be zero for all variations 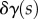 and
and 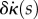 . When we change the integration variables to midline coordinates
. When we change the integration variables to midline coordinates 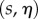 , for which
, for which 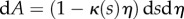 , this condition becomes
, this condition becomes
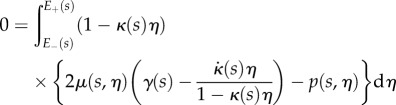 |
2.22a |
and
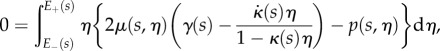 |
2.22b |
for all 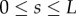 , where these integrals at each s are over the portion of the line normal to the midline that lies within the region occupied by the organ,
, where these integrals at each s are over the portion of the line normal to the midline that lies within the region occupied by the organ, 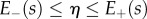 (i.e. these amount to integrated versions of the Euler–Lagrange equations arising from (2.19)). Identifying
(i.e. these amount to integrated versions of the Euler–Lagrange equations arising from (2.19)). Identifying
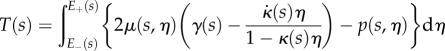 |
2.23 |
and
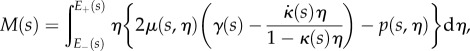 |
2.24 |
as the net tension and moment acting on each cross-section normal to the root midline, these equations are
| 2.25 |
for all 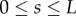 . Using the expressions for
. Using the expressions for 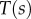 and
and 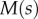 , we can find
, we can find 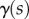 and
and 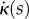 at each point s as the solution of the two-dimensional linear system
at each point s as the solution of the two-dimensional linear system
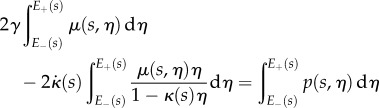 |
2.26 |
and
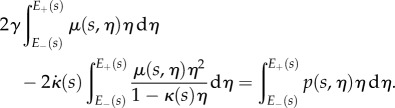 |
2.27 |
When the organ curvature is small relative to its thickness, i.e. 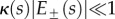 , we have
, we have
| 2.28 |
if the midline is chosen such that 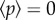 , where here
, where here
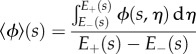 |
2.29 |
denotes the average of a quantity 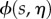 over the line normal to the midline at each s. These correspond to the two-dimensional version of the midline equations from [29].
over the line normal to the midline at each s. These correspond to the two-dimensional version of the midline equations from [29].
2.5. Midline discretization
Simulation of the midline equations for complex geometries generally requires discretization of the midline geometry. The simplest approach is to treat the midline as a piecewise circular curve, as shown in figure 3; such a choice is natural as it leads to a piecewise constant curvature  , although other discretizations with sufficient smoothness such as piecewise polynomial splines would be feasible. This consists of N sections with length
, although other discretizations with sufficient smoothness such as piecewise polynomial splines would be feasible. This consists of N sections with length 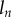 and curvature
and curvature 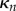 , for
, for  . We then replace the midline coordinates for each vertex,
. We then replace the midline coordinates for each vertex, 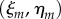 by
by 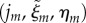 , where j is the index of the piecewise circular section containing the vertex and
, where j is the index of the piecewise circular section containing the vertex and 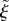 is the proportion of the distance along the circular section that the nearest point on the midline lies.
is the proportion of the distance along the circular section that the nearest point on the midline lies.
Figure 3.
Piecewise circular discretization of midline coordinate system. The midline curve is divided into N circular arcs, with lengths  and curvatures
and curvatures  . The location of a point x in this coordinate system is described by the index j of the circular section, the normal distance
. The location of a point x in this coordinate system is described by the index j of the circular section, the normal distance 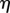 from the midline, and
from the midline, and 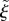 , the proportion of the way of the along the midline that the foot of the normal lies.
, the proportion of the way of the along the midline that the foot of the normal lies.
Substituting this discretization into (2.20) gives
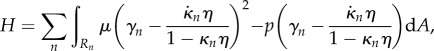 |
2.30 |
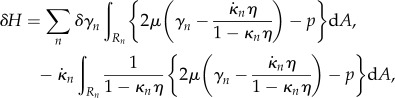 |
2.31 |
where 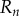 is the region
is the region 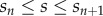 ,
, 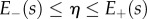 occupied by the organ within the nth midline section, where
occupied by the organ within the nth midline section, where
| 2.32 |
On transforming the integration to midline coordinates, this can be written as
| 2.33 |
where
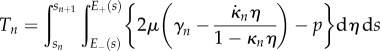 |
2.34 |
and
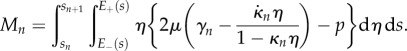 |
2.35 |
Thus, we find that the net tension and moment vanish over each section. This gives equations for the evolution of 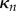 and
and 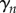 , namely that for each
, namely that for each  ,
, 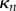 and
and 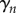 are the solutions of the two-dimensional linear system
are the solutions of the two-dimensional linear system
| 2.36a |
and
| 2.36b |
Here
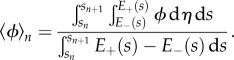 |
2.37 |
For the purposes of simulation, it is easier to compute all integrals in terms of spatial coordinates, in particular this is because edges between vertices will be treated as straight lines in Cartesian coordinates, rather than in the midline coordinate system. Transforming back into Cartesian coordinates for the integration variables, and for simplicity subtracting 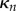 times (2.36b) from (2.36a), the equations for each section become
times (2.36b) from (2.36a), the equations for each section become
| 2.38a |
and
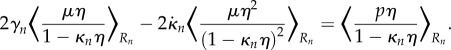 |
2.38b |
Here,
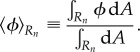 |
2.39 |
These integrals are approximated by numerical quadrature using the triangularization of the vertex-based tissue representation. Integrands are evaluated at the three vertices of each triangle; each vertex contributes 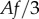 to the integral corresponding to the midline section that contains it, where A is the area of the triangle and f the value of the integrand at the vertex.
to the integral corresponding to the midline section that contains it, where A is the area of the triangle and f the value of the integrand at the vertex.
2.6. Regulation of cell-wall mechanical properties
One key feature of this computational approach is that it can incorporate regulation of cell-wall mechanical properties through cell-scale processes. The regulation of growth in the primary root tip is likely to be the consequence of the combination of many different signals, some of which are typically diffusible hormones. Modelling the complex interactions between multiple hormones is beyond the scope of this study. We instead choose a simple model for the regulation of cell-wall properties. The hormone auxin is thought to be the key signal generating growth differences between the upper and lower sides of the root during gravitropic bending; evidence indicates that the epidermal (outermost) cell tissue plays a primary role in responding to this auxin asymmetry and generating bending [6]. All cells are taken to have uniform constant turgor pressure P, and the viscosities of the cell walls (apart from those in the epidermis) are chosen such that the RER of the root adopts the Peters-Baskin step-stool function 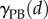 ([25], and here described in appendix D)
([25], and here described in appendix D)
| 2.40 |
where 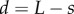 is the distance from the root tip. However, as noted above, we wish to let the cell-wall properties of epidermal cells outside the meristem be regulated by auxin. As a simple method, we choose epidermal cell viscosities to be
is the distance from the root tip. However, as noted above, we wish to let the cell-wall properties of epidermal cells outside the meristem be regulated by auxin. As a simple method, we choose epidermal cell viscosities to be
| 2.41 |
where a is the cellular auxin concentration (in arb. units, scaled such that the concentration in the central vasculature at the base of the organ is 2), 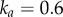 is the half-saturation constant,
is the half-saturation constant, 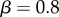 controls the amplitude of the response and
controls the amplitude of the response and 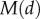 is chosen to be a smooth function which is approximately zero within the meristem and one outside it, namely
is chosen to be a smooth function which is approximately zero within the meristem and one outside it, namely
| 2.42 |
where 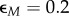 ,
, 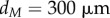 , and erfc denotes the complementary error function. Cellular auxin concentrations are computed using a model described in appendix C.
, and erfc denotes the complementary error function. Cellular auxin concentrations are computed using a model described in appendix C.
2.7. Effect of midline evolution on cell-level vertex-based representation
The equations (2.38) govern the evolution in time of the discrete midline; here, we describe how this is used to update the vertex-based representation of the organ.
At the start of each time step, we calculate the discrete midline coordinates 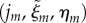 for each of the vertices in the cell-scale representation using the method described in appendix Ba. Note that, while usually constant between time steps, these coordinates may change owing to subdivision of the discrete midline, and new vertices may be introduced following cell division. Cell-level properties (such as auxin concentrations) are updated (e.g. using the auxin transport model described in appendix C) to permit the calculation of cell-wall viscosities.
for each of the vertices in the cell-scale representation using the method described in appendix Ba. Note that, while usually constant between time steps, these coordinates may change owing to subdivision of the discrete midline, and new vertices may be introduced following cell division. Cell-level properties (such as auxin concentrations) are updated (e.g. using the auxin transport model described in appendix C) to permit the calculation of cell-wall viscosities.
Then, using the discretized midline equations (2.38), the evolution of the organ midline over one time step is calculated. The new positions of the vertices in Cartesian coordinates are calculated using the discrete midline coordinates and the relationship (B 1). As a result of this step, cell areas and cell-wall lengths may change, and these are recomputed. Concentrations of substances contained by cells (such as hormones) are updated to reflect the effect of dilution. Vertex velocites can also be calculated using the derivatives of (B 1) with respect to the midline variables; these are used for the specification of the axis of cell division.
From this, we see that the evolution of the macroscopic midline affects the cell-level properties of the simulation in a number of different ways. Changes in the areas of cells and the lengths of the walls between neighbouring cells modify the transport of auxin between cells, thereby affecting the auxin levels experienced by cells. These changes in cell area also change hormone concentrations through dilution, although for auxin this effect is small compared with transport. The evolution of the macroscopic midline also changes the distance of cells from the apex of the tissue, and, therefore, cell-wall mechanical properties.
2.8. Simulation overview
At each time step, the following operations are performed in order.
— Processes on the vertex-based mesh, such as auxin transport, are evolved for one time step.
— The viscosity
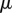 for each (triangle of the) cell wall is calculated.
for each (triangle of the) cell wall is calculated.— The mean viscosity
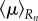 , the corresponding moments
, the corresponding moments  ,
, 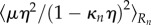 , and the mean
, and the mean 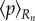 and moment
and moment 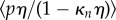 of the pressure are calculated for each section of the midline.
of the pressure are calculated for each section of the midline.— The lengths and curvatures of each of the midline sections are updated from (2.38) using a simple first-order forward Euler method.
— The positions of each of the vertices in Cartesian coordinates is updated using the coordinate transformation (B 1).
— Cell-level quantities, such as auxin concentration, are updated based on the change in cell size over the growth step.
— Cell division is performed.
— Midline sections, which have exceeded a specified threshold in length, are divided into two, and midline coordinates for the vertices in this section updated to reflect this change.
We note here that the numerical approximations made in this simulation are numerical integration the system of the ordinary differential equations for auxin transport [32], quadrature over the triangles to approximate the integrals occurring in (2.36), the piecewise constant approximation of the midline, and the forward Euler method used for the evolution of the lengths and curvatures of the midline sections.
3. Results
3.1. Organ bending
We first investigate the behaviour of this method when applied to the growth of an organ with a relatively simple, artificial geometry. The initial shape of the organ was taken to be a rectangle of height 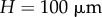 and length
and length 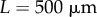 . This was subdivided in the y-direction into
. This was subdivided in the y-direction into 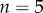 files of cells (of height
files of cells (of height 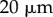 ). Each file of cells was further subdivided in the x-direction into cells, starting at the left-hand end of the tissue (smallest x-coordinate), with each cell length being drawn from a uniform distribution on
). Each file of cells was further subdivided in the x-direction into cells, starting at the left-hand end of the tissue (smallest x-coordinate), with each cell length being drawn from a uniform distribution on 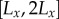 , where
, where 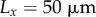 . Cell-wall viscosities were chosen to be
. Cell-wall viscosities were chosen to be 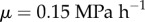 , except for the uppermost layer of cells, which we took to be easier to extend, with
, except for the uppermost layer of cells, which we took to be easier to extend, with 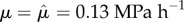 . Cellular turgor pressures were taken to be
. Cellular turgor pressures were taken to be 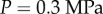 . (Note that similar simulations have been carried out in the context of a vertex-element method; see [11, fig. 6].) The midline was initially discretized into
. (Note that similar simulations have been carried out in the context of a vertex-element method; see [11, fig. 6].) The midline was initially discretized into 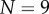 sections of equal length.
sections of equal length.
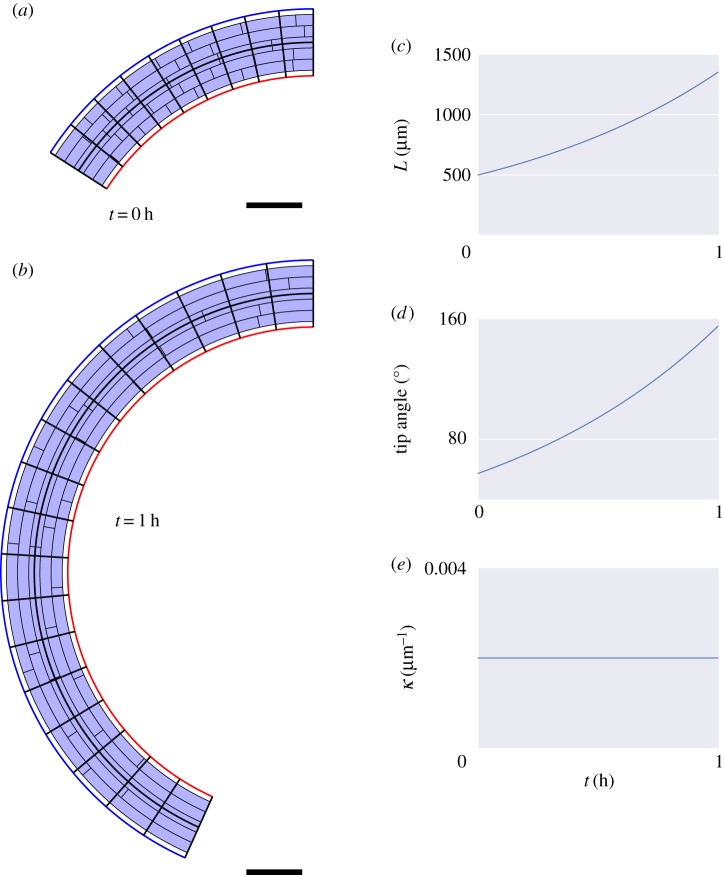
Figure 6.
Simulations with axial variation in cell-wall properties. (a) Elongation rate 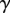 as a function of the distance
as a function of the distance 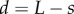 from the organ tip. (b) Initial artificial geometry (as in figure 4). The organ is anchored (
from the organ tip. (b) Initial artificial geometry (as in figure 4). The organ is anchored (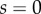 ,
, 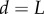 ) at its right-hand end, and the tip of the organ (
) at its right-hand end, and the tip of the organ (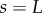 ,
, 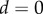 ) is at the left. (c) Simulation at
) is at the left. (c) Simulation at 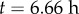 . (d) Simulation with cell division (within the pink region of (a)). Colour bars indicate cell areas (
. (d) Simulation with cell division (within the pink region of (a)). Colour bars indicate cell areas (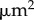 ). Scale bar,
). Scale bar, 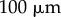 .
.
Results of these simulations are shown in figure 4. From figure 4d, we see that the organ extends, and from figure 4e that the tip angle of the organ increases with time. The number of midline sections, N, increases adaptively with time, through midline sections being divided into two when they exceed 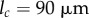 . Simulations with increased number of initial midline sections (
. Simulations with increased number of initial midline sections (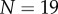 ) and smaller threshold for division (
) and smaller threshold for division (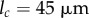 ) gave results which were in close numerical agreement; the relative difference in both midline length and tip angle after
) gave results which were in close numerical agreement; the relative difference in both midline length and tip angle after 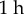 were less than
were less than 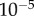 .
.
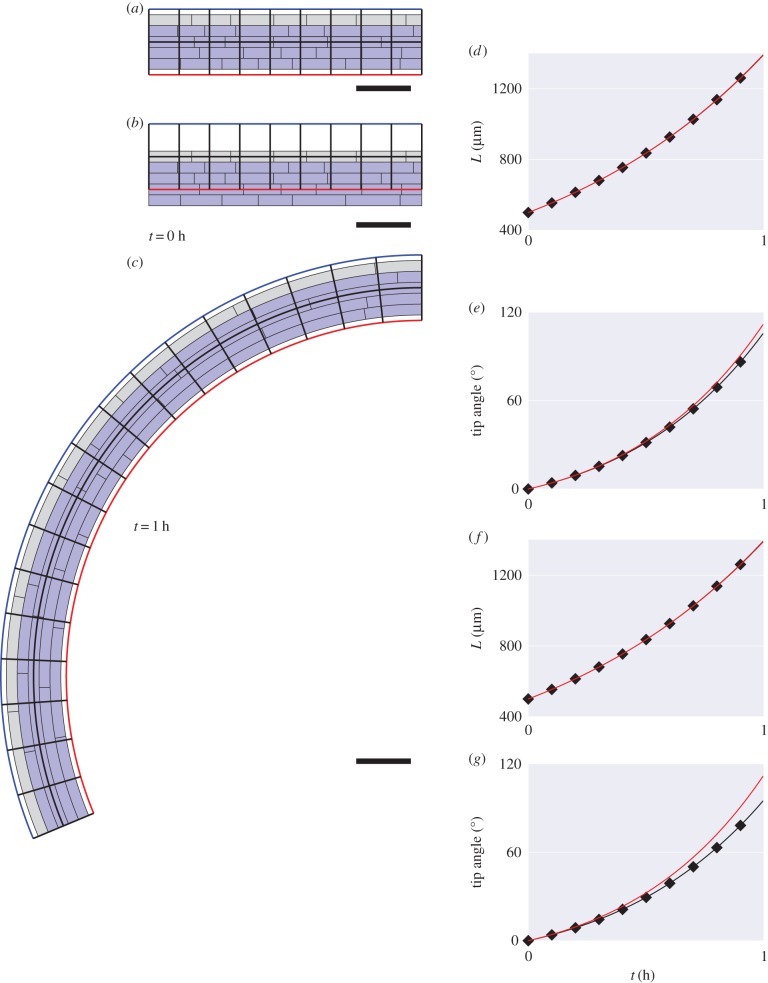
Figure 4.
Bending through differences in cell-wall viscosity. (a) Initial artificial geometry with five files of cells. The uppermost file has (grey) lower viscosity (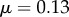 ) relative to the other cells (blue,
) relative to the other cells (blue, 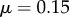 ). The discretized midline is overlaid with black lines between red and blue boundaries indicating cross-sectional orientations. (b) Same geometry as (a) but with midline offset from centre of the organ. (c) Simulation at
). The discretized midline is overlaid with black lines between red and blue boundaries indicating cross-sectional orientations. (b) Same geometry as (a) but with midline offset from centre of the organ. (c) Simulation at 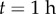 . Length (d) and tip angle (e), with midline at the organ centre (red) or offset (black with diamonds). (f,g) Comparison of midline (red) and vertex-element (black with diamonds) simulations (based on [11]). Scale bar,
. Length (d) and tip angle (e), with midline at the organ centre (red) or offset (black with diamonds). (f,g) Comparison of midline (red) and vertex-element (black with diamonds) simulations (based on [11]). Scale bar, 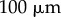 .
.
We also explored the effect of the choice of the organ midline. The midline of an extended body is not well defined; one candidate for the organ midline, namely the medial axis [33], tends to branch towards the tip of the organ. Using a midline offset by a distance 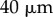 from the centre of the organ figure 4b, it can be seen that the length figure 4d and tip angle (figure 4e) of the resulting simulation are very close to those obtained using the centreline of the organ. Note that, in the simulations of figs 4–7, the small curvature correction terms (1 − κη) were omitted from the calculations of the averaged quantities in (2.38); including them makes the agreement even better.
from the centre of the organ figure 4b, it can be seen that the length figure 4d and tip angle (figure 4e) of the resulting simulation are very close to those obtained using the centreline of the organ. Note that, in the simulations of figs 4–7, the small curvature correction terms (1 − κη) were omitted from the calculations of the averaged quantities in (2.38); including them makes the agreement even better.
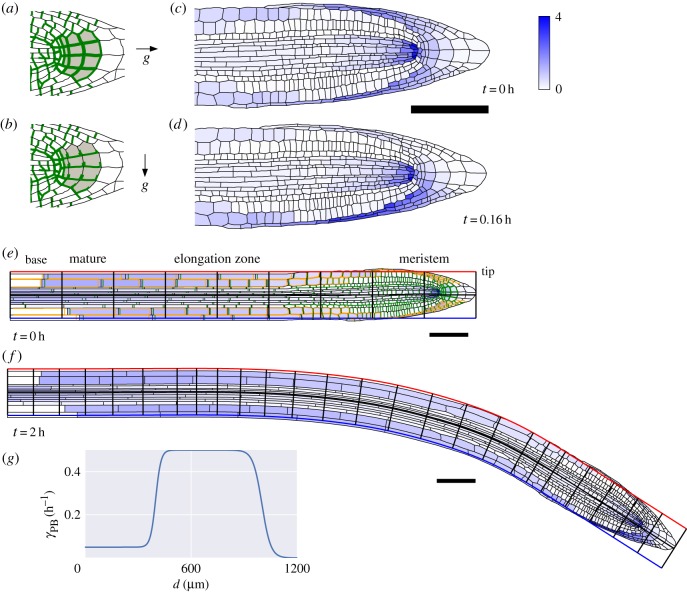
Figure 7.
Simulations of gravitropic bending. (a) PIN auxin efflux carriers (green) have apolar localization around the boundary of columella cells (grey) when the root is growing vertically. (b) PINs localize to lower sides of columella cells following change in the relative gravity vector g (i.e. through reorientation of the root). (c) Initial auxin concentrations (blue shading) found from the steady state of the auxin transport equations (appendix C) with collumella PINs as in (a). (d) Auxin concentrations at 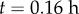 following reorientation of root and change in columella PIN localization. (e) Initial root geometry. Auxin influx and efflux carriers indicated by orange and green lines. Discretized root midline (black) overlaid. (f) Root geometry at
following reorientation of root and change in columella PIN localization. (e) Initial root geometry. Auxin influx and efflux carriers indicated by orange and green lines. Discretized root midline (black) overlaid. (f) Root geometry at 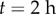 , with bending of the root evident. (g) Step-stool elongation rate function
, with bending of the root evident. (g) Step-stool elongation rate function 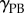 as a function of distance
as a function of distance 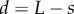 from root tip. All scale bars
from root tip. All scale bars 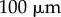 . Same colour scale used for auxin concentrations in all panels.
. Same colour scale used for auxin concentrations in all panels.
The results were also compared with those generated by the vertex-element model previously described in [11]. This model uses triangular elements with anisotropic viscous mechanical properties to describe cell walls in the plane of the simulation, and viscoelastic elements to describe cell walls perpendicular to the plane of plane of the simulation. Choosing the parameters of the vertex-element model such that axial elongation was resisted only by the isotropic viscosity of cell walls in the plane of the simulation, we found in figure 4f that the length of the organ was in close agreement between the two approaches, but that the tip angle figure 4g increased more slowly in the vertex-element simulations than in midline simulations.
A second simulation was performed in which the initial organ geometry was curved. This initial geometry was generated using the rectangular geometry used before, with the midline through the centre of the object. The positions of the vertices were found in the discrete midline coordinates. A curvature of 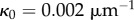 was then applied to the midline, and the new positions of the vertices in Cartesian components calculated. Unlike the earlier simulations, we took all cell walls to have the same viscosity
was then applied to the midline, and the new positions of the vertices in Cartesian components calculated. Unlike the earlier simulations, we took all cell walls to have the same viscosity 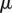 . From figure 5c–e, it can be seen that while the organ extends, and the tip angle increases, the average curvature of the root midline remains almost constant over the course of the simulations. Such behaviour is consistent with (2.28). Again, the number of midline sections N increases as the organ extends (
. From figure 5c–e, it can be seen that while the organ extends, and the tip angle increases, the average curvature of the root midline remains almost constant over the course of the simulations. Such behaviour is consistent with (2.28). Again, the number of midline sections N increases as the organ extends (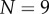 initially, and
initially, and 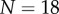 at
at 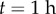 ).
).
Figure 5.
Simulations with curved initial geometry. (a) Initial synthetic curved geometry. (b) Simulation at 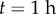 . Evolution of length (c), tip angle (d) and average curvature (e). Scale bar,
. Evolution of length (c), tip angle (d) and average curvature (e). Scale bar, 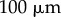 .
.
3.2. Axial variation of cell-wall properties
To illustrate that the method is applicable to situations in which cell-wall properties vary with time, we consider a simulation in which the properties of cell walls were function of their distance from the tip of the organ. We take the dependence of cell-wall viscosity on distance to be a function of the distance from the tip 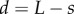 , namely
, namely
| 3.1 |
where 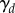 is as shown in figure 6a. (Note that we use a different elongation rate function as the organ considered here is smaller than the full-root geometry used later.) We see the results of this simulation in figure 6c. The non-uniformity of the elongation rate profile leads to a non-uniform distribution of cell lengths along the organ axis, with the largest cells at the end of the simulation being found roughly
is as shown in figure 6a. (Note that we use a different elongation rate function as the organ considered here is smaller than the full-root geometry used later.) We see the results of this simulation in figure 6c. The non-uniformity of the elongation rate profile leads to a non-uniform distribution of cell lengths along the organ axis, with the largest cells at the end of the simulation being found roughly 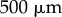 from the tip (left-hand end of the organ). This is to be expected, as these cells have spent most of the simulation in the region of rapid expansion (i.e. in the elongation zone, which here lies between
from the tip (left-hand end of the organ). This is to be expected, as these cells have spent most of the simulation in the region of rapid expansion (i.e. in the elongation zone, which here lies between 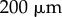 and
and 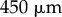 from the tip). Although the model has no variation in cell-wall properties normal to the organ midline, slight bending of the midline can be seen in the simulations. The polygons occupied by each cell are divided into triangles, and cell-wall properties are assigned depending upon the positions of these triangles; this gives a discrete approximation to the smooth growth rate shown in figure 6a. Differences in cellular geometries lead to differences in the triangulation, and therefore cell-wall properties are not exactly uniform over each line normal to the root midline. In addition, the simulation currently uses a crude method to handle triangles which overlap the ends of each piecewise circular section, and differences in the degree of overlap across the root also contribute towards bending of the root midline. Such effects become more significant as the axial gradient in cell-wall properties increases, and indicate the importance of both the discretization of the cells and the midline being sufficiently fine in regions where cell-wall properties change rapidly along the root axis.
from the tip). Although the model has no variation in cell-wall properties normal to the organ midline, slight bending of the midline can be seen in the simulations. The polygons occupied by each cell are divided into triangles, and cell-wall properties are assigned depending upon the positions of these triangles; this gives a discrete approximation to the smooth growth rate shown in figure 6a. Differences in cellular geometries lead to differences in the triangulation, and therefore cell-wall properties are not exactly uniform over each line normal to the root midline. In addition, the simulation currently uses a crude method to handle triangles which overlap the ends of each piecewise circular section, and differences in the degree of overlap across the root also contribute towards bending of the root midline. Such effects become more significant as the axial gradient in cell-wall properties increases, and indicate the importance of both the discretization of the cells and the midline being sufficiently fine in regions where cell-wall properties change rapidly along the root axis.
Cell division can readily be incorporated in this framework. At each time step (here with length 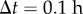 ) cells divide with probability
) cells divide with probability 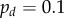 , provided that their area is greater than
, provided that their area is greater than 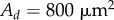 and that the distance from their centroid to the tip of the organ (measured along the centreline) is less than some threshold
and that the distance from their centroid to the tip of the organ (measured along the centreline) is less than some threshold 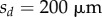 . The resulting distribution of cell areas is shown in figure 6d. Again, the largest cells can be found just beyond the region of rapid expansion. However, there is now a region of small cells near the tip owing to cell division events. This indicates the possibility of applying models of these types to explore the effects of changes in the spatial distribution of growth and division rates upon the resulting distribution of cell lengths in a multicellular tissue.
. The resulting distribution of cell areas is shown in figure 6d. Again, the largest cells can be found just beyond the region of rapid expansion. However, there is now a region of small cells near the tip owing to cell division events. This indicates the possibility of applying models of these types to explore the effects of changes in the spatial distribution of growth and division rates upon the resulting distribution of cell lengths in a multicellular tissue.
3.3. Whole root geometry with auxin transport
We now apply this method to a more complex scenario, in which changes in the cellular levels of the hormone auxin regulate the mechanical properties of individual cells.
As a cell-scale geometry, we use a template based upon a longitudinal cross-section of an A. thaliana root (from [2]). Owing to limitations of the field of view of the microscope, only part of the growing region of the root was acquired. The geometry was therefore extended by stretching the cells furthest from the tip, and then subdividing these cells using a length distribution that depended upon the cell file and the distance from the tip.
Cell-wall viscosity of epidermal cells (shootwards of the meristem at the root tip) was regulated by auxin in a manner described in §2.6. During gravitropism, it has been observed that the PIN efflux proteins (which transport auxin out of cells) change their localization in certain columella cells within the tip of the root [34]. When a root is growing vertically, PIN efflux proteins were found to be equally distributed on all sides of the columella cells (as illustrated in figure 7a). However, Kleine-Vehn et al. [34] observed that PIN proteins become localized to the lower sides of columella cells soon after gravistimulation (i.e. when a vertical root is rotated by a quarter-turn such that it is oriented in a horizontal direction).
In our simulation, we first find the steady state of the auxin transport equations on the static initial geometry with equal abundance of PIN proteins on all sides of the columella cells (figure 7c). These auxin concentrations were used as the initial conditions for the simulation. We then changed the localization of PINs on columella cells such that, after gravistimulation, they were only found on the lower faces of the columella cells (figure 7b). Soon after the start of the simulation, a difference in auxin concentrations between the upper and lower sides of the root tip can be observed (figure 7d). These differences are most pronounced in the cells bordering the columella cells (known as lateral root cap cells), and are smaller in magnitude in the epidermal cells, reflecting observations made using auxin sensors [35,36].
As the simulation proceeds, the increase in cell-wall viscosity for epidermal cells on the lower side of the root and the decrease on the upper-side of the root leads to the generation of curvature (figure 7e), and thereby, reorients the root tip. We terminate the simulations after the first 2 h; at later times during the gravitropic response, the asymmetry in auxin concentrations between the upper and lower sides of the root is thought to be switched off [35].
4. Discussion
Here, we have described a multiscale computational method that can be used for the simulation of multicellular plant organs undergoing axial elongation. This method captures the multicellular geometry of the plant tissue throughout the simulation, which permits simulation of cell-scale processes such as auxin transport. The macroscale equations for the evolution of the organ midline incorporate appropriately weighted averages of cell-level mechanical properties, which allows the simulation to capture the effects of changes in the mechanical properties of individual cells. This framework provides a convenient tool with which to investigate the intricate internal control systems that regulate growth patterns. However, the model is still vulnerable to the effect of geometric imperfections, whereby internal structural asymmetries can induce bending. This raises the possibility of internal regulatory mechanisms correcting such biases, analogous to the proprioception proposed in other models of gravitropic bending [37], which are naturally implemented in terms of the evolving centreline of the organ.
The method has promise for the investigation of a range of biological problems, in particular responses to gravity, and also responses to other stimuli such as water and light. The midline representation is well suited to long slender organs but it presents a restriction on the deformations that the organ can undergo. The model is unable to explain the complex pattern of growth and cell division that occurs near the root tip [38], or during the development of a new lateral root primordium [39]. Such deficiencies require either a different organ-scale description of growth in these regions [40,41], or coupling the method to the solution of a (finite-element) cell-scale simulation [12,13] in a local region. However, the theoretical details and practical implementation of such a coupled method have not yet been studied. The model is also unable to describe situations in which the organ can widen, for example the increases in root diameter that occur when a root is impeded by soil [42]. Nevertheless, the type of hybrid framework that we have described would seem to have promise for the description of a wide variety of multiscale and multicellular processes. It would also be of interest to extend this approach to study branching root structures, in which the root midline is represented by a (graph-theoretic) tree of circular sections rather than a one-dimensional list, in a similar manner to root architecture simulations (see the review of [43]). Additional consideration of the appropriate behaviour at branching points would be necessary in this case.
We note here that the model for gravitropism is deliberately simplistic, and here to illustrate how cell-scale models can be effectively integrated into this framework. There are still many details of the gravitropic response that are yet to be explained [44]; the sensing of auxin by cells is complex [45], and it is also thought that other mechanisms such as ion channels may play key roles [46].
Appendix A. Derivation of rate of strain tensor in midline coordinates
From the definitions (2.4) of t and n,
| A 1 |
and also
| A 2 |
The first part of (A 2) can be written as
| A 3 |
Using (2.10) and (A 3) gives
| A 4 |
From the chain rule, temporal derivatives following material points and at fixed distances along the midline are related by
| A 5 |
and similarly
| A 6 |
Here, the subscript 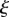 indicates that the partial derivative with respect to t is to be taken with
indicates that the partial derivative with respect to t is to be taken with 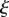 held constant (also known as a material or Lagrangian derivative), while the subscript s denotes a derivative with respect to t at constant distance s along the midline. Using these gives the compatibility conditions
held constant (also known as a material or Lagrangian derivative), while the subscript s denotes a derivative with respect to t at constant distance s along the midline. Using these gives the compatibility conditions
| A 7 |
Elsewhere in this paper, all temporal derivatives are following a material point, and we omit this notation.
The velocity in Cartesian coordinates of a point at fixed midline coordinates 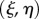 is given by
is given by
| A 8 |
Using (A 4), we have
| A 9 |
which can be combined with (A 7) to give
| A 10 |
where we define
| A 11 |
(As we will see below,  is the strain rate component parallel to the organ midline.) Also, from (A 8)
is the strain rate component parallel to the organ midline.) Also, from (A 8)
| A 12 |
Differentiating (2.11), we have that
| A 13 |
and from (2.4) and (A 1) this becomes
| A 14 |
Using these, we find that
| A 15 |
and
| A 16 |
so
| A 17 |
The rate of strain tensor is therefore given by
| A 18 |
Appendix B. Discrete coordinate transformations
Here, we list for reference the explicit form of the transformations between the discrete piecewise circular midline coordinates and Cartesian coordinates.
(a) Midline to Cartesian coordinates
There is a relatively simple transformation between discrete midline coordinates 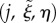 and Cartesian coordinates x, given by
and Cartesian coordinates x, given by
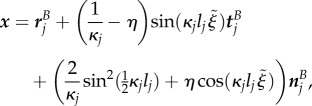 |
B 1a |
where 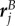 ,
, 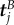 and
and 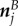 denote the position, tangent and normal to the start of the jth circular section (figure 3), given by
denote the position, tangent and normal to the start of the jth circular section (figure 3), given by
| B 1b |
with
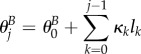 |
B 1c |
and
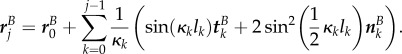 |
B 1d |
Note that, for 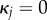 , (B 1a) becomes
, (B 1a) becomes
| B 1e |
(b) Cartesian to midline coordinates
The inverse transformation from Cartesian coordinates x to midline coordinates 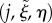 first involves identifying a circular section for which x lies in the region between the normals to the circle at either end of the section (figure 8), i.e. j such that
first involves identifying a circular section for which x lies in the region between the normals to the circle at either end of the section (figure 8), i.e. j such that
| B 2 |
If multiple suitable sections exist, the one whose midpoint is closest to the point x is chosen. The values of 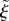 and
and 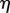 are then given by
are then given by
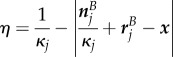 |
B 3a |
and
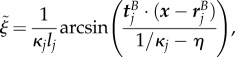 |
B 3b |
if 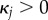 ,
,
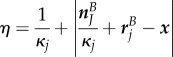 |
B 3c |
and
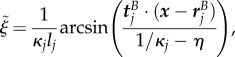 |
B 3d |
if 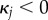 , and
, and
| B 3e |
and
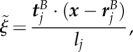 |
B 3f |
if 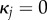 . Note that the expressions here suffer from round-off errors for small
. Note that the expressions here suffer from round-off errors for small  , in which case they are instead calculated in the form
, in which case they are instead calculated in the form
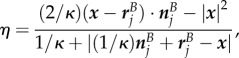 |
B 3g |
for 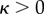 and
and
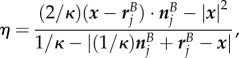 |
B 3h |
for 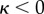 .
.
Figure 8.
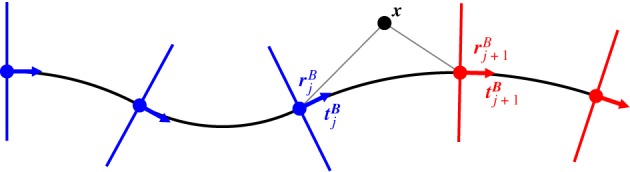
Criterion for determining the index of the section that a point lies in for the piecewise circular coordinate system. Lines normal to the breakpoints (ends of each of the circular sections) are coloured red and blue depending on the side of the normal, relative to the direction of the curve, that the point x lies.
Appendix C. Auxin transport
Auxin transport was simulated using the model of [2]; we summarize this briefly here. All parameter values for auxin transport are as listed there.
Auxin is considered to be uniformly distributed within each of the N cells, with concentration 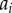 , where
, where 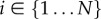 denotes the index of the cell (figure 9). Apoplastic compartments are associated with the common edges between each pair of adjacent polygons, and have auxin concentrations
denotes the index of the cell (figure 9). Apoplastic compartments are associated with the common edges between each pair of adjacent polygons, and have auxin concentrations  , where i and j are the indices of the two neighbouring cells (we identify
, where i and j are the indices of the two neighbouring cells (we identify 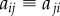 ). The volume of each cell is taken to be proportional to its area
). The volume of each cell is taken to be proportional to its area 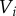 , while the volume of each apoplastic compartment is taken to be proportional to its length
, while the volume of each apoplastic compartment is taken to be proportional to its length 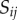 times its thickness
times its thickness 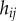 (
(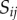 can be calculated from the polygonal network;
can be calculated from the polygonal network; 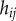 is a property associated with each apoplastic compartment).
is a property associated with each apoplastic compartment).
Figure 9.
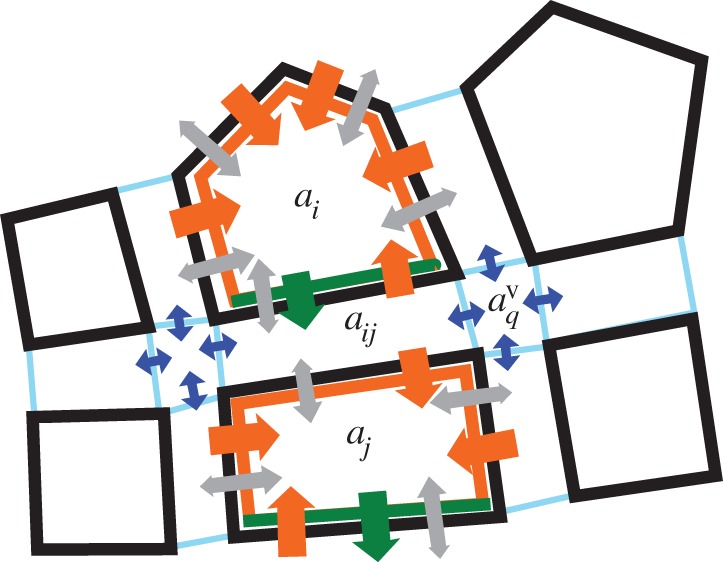
Auxin transport on the vertex-based geometrical representation. Solid black lines denote cell boundaries; spaces between cells are expanded to show to apoplastic and vertex compartments. Auxin concentrations within each cell 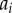 ,
, 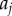 are taken to be uniform. Auxin is imported by AUX/LAX influx carriers (orange), undergoes directional efflux by PIN efflux carriers (green) and is able to diffuse (grey). This leads to a flux between a cell and the neighbouring apoplastic compartment, the latter with auxin concentration
are taken to be uniform. Auxin is imported by AUX/LAX influx carriers (orange), undergoes directional efflux by PIN efflux carriers (green) and is able to diffuse (grey). This leads to a flux between a cell and the neighbouring apoplastic compartment, the latter with auxin concentration 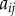 . Diffusive auxin fluxes (blue) permit apoplastic auxin diffusion via vertex compartments (
. Diffusive auxin fluxes (blue) permit apoplastic auxin diffusion via vertex compartments (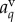 ).
).
Auxin influx and efflux carriers facilitate the movement of auxin ions through the cell plasma membrane. Auxin fluxes from apoplast compartments (with label ij) into adjacent cells (with label i), 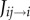 , are given by
, are given by
| C 4 |
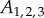 ,
, 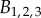 and
and 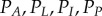 are constants,
are constants, 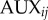 ,
, 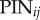 and LAXij are 1 if AUX/PIN/LAX proteins are present on the plasma membrane domain of cell i facing cell j, and 0 otherwise. For each cell i we have
and LAXij are 1 if AUX/PIN/LAX proteins are present on the plasma membrane domain of cell i facing cell j, and 0 otherwise. For each cell i we have
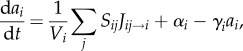 |
C 5 |
where 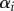 is the auxin production rate in each cell,
is the auxin production rate in each cell, 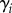 is the auxin degradation rate and the sum runs over all cells j that neighbour cell i.
is the auxin degradation rate and the sum runs over all cells j that neighbour cell i.
Auxin transport between adjacent apoplastic compartments (i.e. compartments that meet a common vertex) occurs by diffusion. A (small) ‘vertex compartment’ is associated with each vertex, and the diffusive flux between a vertex compartment and the neighbouring wall compartment (per unit wall thickness) is taken to be
| C 6 |
where 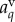 is the auxin concentration in the vertex compartment with index q. As the vertex compartment is small, the net flux into the compartment must be approximately zero. This provides equations specifying the vertex compartment concentrations
is the auxin concentration in the vertex compartment with index q. As the vertex compartment is small, the net flux into the compartment must be approximately zero. This provides equations specifying the vertex compartment concentrations
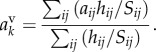 |
C 7 |
Substituting this into (B 6), and again considering conservation of auxin, equations for the wall compartment auxin concentrations are given by
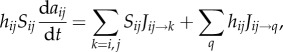 |
C 8 |
where the first sum (k) runs over the pair of cells (i,j) adjacent to the wall compartment, while the second sum (q) is over the two vertices at either end of the wall compartment. Auxin production and PIN efflux carrier locations were specified on the root template using the same rules as in [2], except that during our simulation of the response to gravity we modify the PIN efflux carrier locations on specific (columella) cells located within the tip of the root. Boundary conditions were applied at the base of the root; cells in the central files were set to have constant auxin concentration 1, while cells in the outermost three cell files (from the outside in, these are known as the epidermis, cortex and endodermis) were set to have constant zero auxin concentration.
Appendix D. Peters–Baskin step-stool function
For reference, here we describe the function 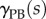 that we use to specify growth rates as a function of the distance from the organ tip. Using the definitions given in [25], we have
that we use to specify growth rates as a function of the distance from the organ tip. Using the definitions given in [25], we have
| D 1a |
| D 1b |
| D 1c |
| D 1d |
and
| D 9e |
for some constants 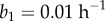 (RER in meristem),
(RER in meristem), 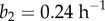 (RER in elongation zone),
(RER in elongation zone), 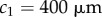 (length of meristem),
(length of meristem), 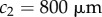 (distance of end of elongation zone from tip),
(distance of end of elongation zone from tip), 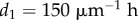 (controls width of meristem-elongation zone transition),
(controls width of meristem-elongation zone transition), 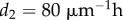 (controls width of elongation zone to mature transition).
(controls width of elongation zone to mature transition).
Competing interests
We declare we have no competing interests.
Funding
We would like to thank the Biological and Biotechnology Science Research Council (BBSRC) for responsive mode (BB/J009717/1) and CISB awards to the Centre for Plant Integrative Biology. J.R.K. would like to thank the Isaac Newton Institute for Mathematical Sciences for its hospitality during the programme Coupling Geometric PDEs with Physics for Cell Morphology, Motility and Pattern Formation supported by EPSRC grant no. EP/K032208/1 and the Simons Foundation for a Fellowship during his participation in the programme.
References
- 1.Ubeda-Tomas S, Beemster GT, Bennett MJ. 2012. Hormonal regulation of root growth: integrating local activities into global behaviour. Trends Plant Sci. 17, 326–331. ( 10.1016/j.tplants.2012.02.002) [DOI] [PubMed] [Google Scholar]
- 2.Band LR, et al. 2014. Systems analysis of auxin transport in the Arabidopsis root apex. Plant Cell 26, 862–875. ( 10.1105/tpc.113.119495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cruz-Ramirez A, et al. 2012. A bistable circuit involving SCARECROW-RETINOBLASTOMA integrates cues to inform asymmetric stem cell division. Cell 150, 1002–1015. ( 10.1016/j.cell.2012.07.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B. 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013. ( 10.1038/nature06215) [DOI] [PubMed] [Google Scholar]
- 5.Santuari L, Scacchi E, Rodriguez-Villalon A, Salinas P, Dohmann EM, Brunoud G, Vernoux T, Smith RS, Hardtke CS. 2011. Positional information by differential endocytosis splits auxin response to drive Arabidopsis root meristem growth. Curr. Biol. 21, 1918–1923. ( 10.1016/j.cub.2011.10.002) [DOI] [PubMed] [Google Scholar]
- 6.Swarup R, Kramer EM, Perry P, Knox K, Leyser HM, Haseloff J, Beemster GT, Bhalerao R, Bennett MJ. 2005. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nat. Cell Biol. 7, 1057–1065. ( 10.1038/ncb1316) [DOI] [PubMed] [Google Scholar]
- 7.Bassel GW, Stamm P, Mosca G, Barbier de Reuille P, Gibbs DJ, Winter R, Janka A, Holdsworth MJ, Smith RS. 2014. Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc. Natl Acad. Sci. USA 111, 8685–8690. ( 10.1073/pnas.14046161.11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peret B, et al. 2013. Sequential induction of auxin efflux and influx carriers regulates lateral root emergence. Mol. Syst. Biol. 9, 699 ( 10.1038/msb.2013.43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merks RM, Guravage M, Inze D, Beemster GT. 2011. VirtualLeaf: an open-source framework for cell-based modeling of plant tissue growth and development. Plant Physiol. 155, 656–666. ( 10.1104/pp.110.167619) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uyttewaal M, et al. 2012. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell 149, 439–451. ( 10.1016/j.cell.2012.02.048) [DOI] [PubMed] [Google Scholar]
- 11.Fozard JA, Lucas M, King JR, Jensen OE. 2013. Vertex-element models for anisotropic growth of elongated plant organs. Front. Plant Sci. 4, 233 ( 10.3389/fpls.2013.00233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boudon F, Chopard J, Ali O, Gilles B, Hamant O, Boudaoud A, Traas J, Godin C. 2015. A computational framework for 3D mechanical modeling of plant morphogenesis with cellular resolution. PLoS Comput. Biol. 11, e1003950 ( 10.1371/journal.pcbi.1003950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bozorg B, Krupinski P, Jonsson H. 2014. Stress and strain provide positional and directional cues in development. PLoS Comput. Biol. 10, e1003410 ( 10.1371/journal.pcbi.1003410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jonsson H, Meyerowitz EM. 2014. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. Elife 3, e01967 ( 10.7554/eLife.01967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bensoussan A, Papanicolau G, Lions J-L. 1978. Asymptotic analysis for periodic structures. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 16.Hill R. 1963. Elastic properties of reinforced solids: some theoretical principles. J. Mech. Phys. Solids 11, 357–372. ( 10.1016/0022-5096(63)90036-X) [DOI] [Google Scholar]
- 17.Ghysels P, Samaey G, Tijskens B, Van Liedekerke P, Ramon H, Roose D. 2009. Multi-scale simulation of plant tissue deformation using a model for individual cell mechanics. Phys. Biol. 6, 016009 ( 10.1088/1478-3975/6/1/016009) [DOI] [PubMed] [Google Scholar]
- 18.Ghysels P, Samaey G, Van Liedekerke P, Tijskens E, Ramon H, Roose D. 2010. Multiscale modeling of viscoelastic plant tissue. Int. J. Multiscale Comp. Eng. 8, 379–396. ( 10.1615/IntJMultCompEng.v8.i4.30) [DOI] [Google Scholar]
- 19.Baskin TI, Jensen OE. 2013. On the role of stress anisotropy in the growth of stems. J. Exp. Bot. 64, 4697–4707. ( 10.1093/jxb/ert176) [DOI] [PubMed] [Google Scholar]
- 20.Davit Y, Osborne JM, Byrne HM, Gavaghan D, Pitt-Francis J. 2013. Validity of the cauchy-born rule applied to discrete cellular-scale models of biological tissues. Phys. Rev. E 87, 042724 ( 10.1103/PhysRevE.87.042724) [DOI] [PubMed] [Google Scholar]
- 21.Murisic N, Hakim V, Kevrekidis IG, Shvartsman SY, Audoly B. 2015. From discrete to continuum models of three-dimensional deformations in epithelial sheets. Biophys. J. 109, 154–163. ( 10.1016/j.bpj.2015.05.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller M, Teschner M, Gross M. 2004. Physically-based simulation of objects represented by surface meshes. In Proc. of the Computer Graphics International, 2004, pp. 26–33. New York, NY: IEEE.
- 23.Nagai T, Honda H. 2001. A dynamic cell model for the formation of epithelial tissues. Philos. Mag. B 81, 699–719. ( 10.1080/13642810108205772) [DOI] [Google Scholar]
- 24.Pradal C, Dufour-Kowalski S, Boudon F, Fournier C, Godin C. 2008. Openalea: a visual programming and component-based software platform for plant modelling. Funct. Plant Biol. 35, 751–760. ( 10.1071/FP08084) [DOI] [PubMed] [Google Scholar]
- 25.Peters WS, Baskin TI. 2006. Tailor-made composite functions as tools in model choice: the case of sigmoidal vs bi-linear growth profiles. Plant Methods 2, 11 ( 10.1186/1746-4811-2-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geuzaine C, Remacle J-F. 2009. Gmsh: A 3-D finite element mesh generator with built-in pre-and post-processing facilities. Int. J. Num. Meth. Eng. 79, 1309–1331. ( 10.1002/nme.2579) [DOI] [Google Scholar]
- 27.Moulton DE, Lessinnes TH, Goriely A. 2013. Morphoelastic rods. Part I: a single growing elastic rod. J. Mech. Phys. Solids 61, 398–427. ( 10.1016/j.jmps.2012.09.017) [DOI] [Google Scholar]
- 28.Kennaway R, Coen E, Green A, Bangham A. 2011. Generation of diverse biological forms through combinatorial interactions between tissue polarity and growth. PLoS Comput. Biol. 7, e1002071 ( 10.1371/journal.pcbi.1002071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyson RJ, et al. 2014. Mechanical modelling quantifies the functional importance of outer tissue layers during root elongation and bending. New Phytol. 202, 1212–1222. ( 10.1111/nph.12764) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyson RJ, Jensen OE. 2010. A fibre-reinforced fluid model of anisotropic plant cell growth. J. Fluid Mech. 655, 472–503. ( 10.1017/S002211201000100X) [DOI] [Google Scholar]
- 31.Lockhart JA. 1965. An analysis of irreversible plant cell elongation. J. Theor. Biol. 8, 264–275. ( 10.1016/0022-5193(65)90077-9) [DOI] [PubMed] [Google Scholar]
- 32.Hindmarsh AC. 1983. Odepack, a systematized collection of ode solvers. In Scientific computing (ed. Stepleman RS.), pp. 55–64. Amsterdam, The Netherlands: North-Holland. [Google Scholar]
- 33.Blum H. 1967. A transformation for extracting new descriptors of shape. In Models for the perception of speech and visual form. (ed Dunn W.), pp. 362–380. Cambridge, MA: MIT Press. [Google Scholar]
- 34.Kleine-Vehn J, Ding Z, Jones AR, Tasaka M, Morita MT, Friml J. 2010. Gravity-induced PIN transcytosis for polarization of auxin fluxes in gravity-sensing root cells. Proc. Natl Acad. Sci. USA 107, 22 344–22 349. ( 10.1073/pnas.1013145107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Band LR, et al. 2012. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl Acad. Sci. USA 109, 4668–4673. ( 10.1073/pnas.1201498109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brunoud G, et al. 2012. Anovel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 482, 103–106. ( 10.1038/nature10791) [DOI] [PubMed] [Google Scholar]
- 37.Bastien R, Bohr T, Moulia B, Douady S. 2013. Unifying model of shoot gravitropism reveals proprioception as a central feature of posture control in plants. Proc. Natl Acad. Sci. USA 110, 755–760. ( 10.1073/pnas.1214301109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390, 287–289. ( 10.1038/36856) [DOI] [PubMed] [Google Scholar]
- 39.von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EH, Maizel A. 2016. Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr. Biol. 26, 439–449. ( 10.1016/j.cub.2015.12.047) [DOI] [PubMed] [Google Scholar]
- 40.Nakielski J, Lipowczan M. 2013. Spatial and directional variation of growth rates in Arabidopsis root apex: a modelling study. PLoS ONE 8, e84337 ( 10.1371/journal.pone.0084337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szymanowska-Pulka J, Potocka I, Karczewski J, Jiang K, Nakielski J, Feldman LJ. 2012. Principal growth directions in development of the lateral root in Arabidopsis thaliana. Annu. Bot. 110, 491–501. ( 10.1093/aob/mcs129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin K, Shen J, Ashton RW, Dodd IC, Parry MA, Whalley WR. 2013. How do roots elongate in a structured soil? J. Exp. Bot. 64, 4761–4777. ( 10.1093/jxb/ert286) [DOI] [PubMed] [Google Scholar]
- 43.Dunbabin VM, et al. 2013. Modelling root–soil interactions using three–dimensional models of root growth, architecture and function. Plant and Soil 372, 93–124. ( 10.1007/s11104-013-1769-y) [DOI] [Google Scholar]
- 44.Baldwin KL, Strohm AK, Masson PH. 2013. Gravity sensing and signal transduction in vascular plant primary roots. Am. J. Bot. 100, 126–142. ( 10.3732/ajb.1200318) [DOI] [PubMed] [Google Scholar]
- 45.Weijers D, Wagner D. 2016. Transcriptional responses to the auxin hormone. Annu. Rev. Plant Biol. 67, 539–574. ( 10.1146/annurev-arplant-043015-112122) [DOI] [PubMed] [Google Scholar]
- 46.Shih HW, DePew CL, Miller ND, Monshausen GB. 2015. The cyclic nucleotide-gated channel CNGC14 regulates root gravitropism in Arabidopsis thaliana. Curr. Biol. 25, 3119–3125. ( 10.1016/j.cub.2015.10.025) [DOI] [PubMed] [Google Scholar]