The title p-hydroxy Schiff base, is the product of a condensation reaction between benzocaine and vanillin. The benzylidine and benzoate rings are inclined to one another by 24.58 (8)°, and the conformation about the C=N bond is E.
Keywords: crystal structure, p-hydroxy Schiff base, hydrogen bonding
Abstract
The title p-hydroxy Schiff base, C17H17NO4, was synthesized via the condensation reaction of benzocaine with vanillin. The benzylidine and benzoate rings are inclined to one another by 24.58 (8)°, and the conformation about the C=N bond is E. In the crystal, molecules are linked by O—H⋯N hydrogen bonds, forming zigzag chains propagating along [010]. Adjacent chains are linked by C—H⋯π and weak offset π–π interactions [intercentroid distance = 3.819 (1) Å], forming sheets parallel to (10-2).
Chemical context
The pharmaceutical industry generally seeks to formulate crystalline forms of their active ingredient by their inherent stability (Yadav et al., 2009 ▸; Paul et al., 2005 ▸). Increasing attention is now being paid to crystal engineering for improving crystal properties (Byrn et al., 1999 ▸). One such strategy is co-crystallization due to its potential for enhancing the physicochemical properties of an API, such as solubility, bioavailability, dissolution, and chemical and physical stability (Shan & Zaworotko, 2008 ▸; Good & Rodríguez-Hornedo, 2009 ▸). The term co-crystal does not have a clear and consistent definition in the literature (Desiraju, 2003 ▸; Bond, 2007 ▸; Shan & Zaworotko, 2008 ▸). Generally, a co-crystal is defined as a homogeneous crystalline phase consisting of two or more discrete chemical entities bound together in the crystal lattice through non-covalent, non-ionic molecular interactions.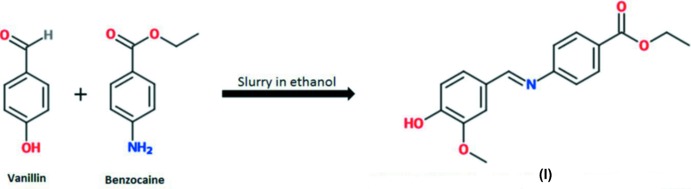
Benzocaine, the ethyl ester of p-aminobenzoic acid, is a local anaesthetic which is used to subside pain perception. It relieves pain by inhibiting the voltage-dependent sodium channels on the nerve membrane, which results in stopping the propagation of the action potential. (Neumcke et al., 1981 ▸). In this study, we intended to formulate co-crystals of benzocaine and determine the impact on its physicochemical properties. Vanillin was selected as a potential co-former as it is FDA approved and has the potential to form a strong hydrogen bond between the amine and hydroxy groups of benzocaine and vanillin, respectively. However, during crystallization a chemical reaction between the two was observed, the product of which is a novel p-hydroxy Schiff base. Schiff bases are an important class of organic compounds with significant biological and chemical importance. In general, they are synthesized by the condensation reaction of an aliphatic or aromatic amine with a carbonyl containing compound, such as an aldehyde, via nucleophilic addition. Herein, we report on the crystal structure of the title compound, a new p-hydroxy Schiff base, synthesized from benzocaine and vanillin by slurry crystallization.
Structural commentary
The title Schiff base, (I), is the product of the reaction of benzocaine with vanillin (Scheme). In the title compound, Fig. 1 ▸, the conformation of the C10=N1 imine bond is E. The molecule is non-planar, with a dihedral angle between the aryl rings of 24.58 (8)°. The m-methoxy group (O1/C13/C16) is slightly out of the plane of the benzene ring (C11–C14/C20/C21) to which it is attached by 5.37 (18)°, while the mean plane of the ethylacetate group (O3/O17/C1/C2/C4) is inclined to the benzene ring (C5–C8/C18/C19) to which it is attached by 10.23 (11)°. This non-linearity is consistent for Schiff bases.
Figure 1.
The molecular structure of compound (I), with atom labeling. Displacement ellipsoids are drawn at the 50% probability level.
Supramolecular features
In the crystal, molecules are linked by O—H⋯N hydrogen bonds, forming zigzag chains propagating along [010]; see Table 1 ▸ and Fig. 2 ▸. Adjacent chains are linked by C—H⋯π interactions (Table 1 ▸, Fig. 2 ▸), and weak offset π-π- interactions, forming sheets parallel to (10 ) [Cg1⋯Cg1i = 3.819 (1) Å, interplanar distance = 3.672 (2) Å, slippage = 1.05 Å, Cg1 is the centroid of ring C5–C8/C18/C19; symmetry code: (i) −x + 2, −y + 1, −z + 1],
) [Cg1⋯Cg1i = 3.819 (1) Å, interplanar distance = 3.672 (2) Å, slippage = 1.05 Å, Cg1 is the centroid of ring C5–C8/C18/C19; symmetry code: (i) −x + 2, −y + 1, −z + 1],
Table 1. Hydrogen-bond geometry (Å, °).
Cg2 is the centroid of the C11–C14/C20/C21 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O15—H15⋯N1i | 0.88 (2) | 2.00 (2) | 2.828 (2) | 156 (2) |
| C2—H2B⋯Cg2ii | 0.97 | 2.87 | 3.766 (2) | 154 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 2.
A view along the c axis of the crystal packing of compound (I), with hydrogen bonds shown as dashed lines and C—H⋯π interactions as blue arrows (see Table 1 ▸).
The crystal structure analysis of compound (I), has shown that, due to the aromatic hydroxy group being located in the para rather than the ortho position, this Schiff base cannot form the intramolecular C=N⋯O—H hydrogen bond responsible for keto–enol tautomerism. However, the close proximity of the C=N and O—H groups gives rise to the possibility that external stimulation of the material by heat or light may lead to the zwitterionic form. The potential for compound (I) to form a zwitterionic state, coupled with the non-linear conformation of the molecule in the solid state, suggest that this Schiff base may exhibit interesting physical properties, that we are currently in the process of evaluating.
Database survey
In the Cambridge Structural Database (CSD, V53.7; Groom et al., 2016 ▸), there are three known Schiff bases synthesized from benzocaine (CSD ref codes: VABSUO; Shakir et al., 2010 ▸, and ZOZROV and ZOZRUB; Kurogoshi & Hori, 1996 ▸), and one derived from vanillin (CSD ref code: LEFVID; Fejfarová et al., 2012 ▸). The dihedral angles between the aryl rings in VABSUO, ZOZROV, ZOZRUB and LEFVID were found to be 24.85 (9), 59.7 (2), 53.94 (9), and 37.87 (10)°, respectively. The N1=C10 and C8—N1 bond lengths of the imine group of the title compound are 1.274 (2) and 1.415 (2) Å, respectively. They are comparable to the imine bond lengths observed for VABSUO, ZOZROV, ZOZRUB and LEFVID, which vary between 1.262 (4)–1.283 (3) Å and 1.414 (7)–1.428 (3) Å, respectively.
Synthesis and crystallization
Compound (I) was prepared by slurrying an equimolar mixture of benzocaine (1.16 g, 7 mmol) and vanillin (1.07 g, 7 mmol) in 2 ml of anhydrous ethanol (see Scheme). The slurry was stirred continuously for 18 h at room temperature (296 K). The product was then filtered and air dried before being analysed by powder X-ray diffraction to determine the presence of a new crystalline phase. Single crystals were then prepared by dissolving an equimolar mixture of benzocaine (0.83 g, 5 mmol) and vanillin (0.77 g, 5 mmol) in 10 ml of ethanol. The solution was allowed to evaporate under ambient conditions and yellow block-like crystals were obtained after four days.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. Two H atoms, H15 and H10, were located in a difference Fourier map and freely refined. The remaining H atoms were placed in geometrically calculated positions and included in the refinement process using a riding model: C—H = 0.93–0.97 Å with U iso(H) = 1.5U eq(C-methyl) and 1.2U eq(C) for other H atoms.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C17H17NO4 |
| M r | 299.31 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 296 |
| a, b, c (Å) | 12.4229 (5), 9.6392 (5), 13.2384 (6) |
| β (°) | 102.457 (3) |
| V (Å3) | 1547.94 (12) |
| Z | 4 |
| Radiation type | Cu Kα |
| μ (mm−1) | 0.76 |
| Crystal size (mm) | 0.26 × 0.11 × 0.04 |
| Data collection | |
| Diffractometer | Bruker SMART APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Bruker, 2013 ▸) |
| T min, T max | 0.599, 0.753 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18263, 2895, 2277 |
| R int | 0.037 |
| (sin θ/λ)max (Å−1) | 0.614 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.040, 0.121, 1.05 |
| No. of reflections | 2895 |
| No. of parameters | 210 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.23, −0.14 |
Supplementary Material
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S2056989016008999/su5304sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016008999/su5304Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016008999/su5304sup3.pdf
Supporting information file. DOI: 10.1107/S2056989016008999/su5304Isup4.cml
CCDC reference: 1483394
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C17H17NO4 | F(000) = 632 |
| Mr = 299.31 | Dx = 1.284 Mg m−3 |
| Monoclinic, P21/c | Cu Kα radiation, λ = 1.54178 Å |
| a = 12.4229 (5) Å | Cell parameters from 3549 reflections |
| b = 9.6392 (5) Å | θ = 11.5–68.2° |
| c = 13.2384 (6) Å | µ = 0.76 mm−1 |
| β = 102.457 (3)° | T = 296 K |
| V = 1547.94 (12) Å3 | Block, colorless |
| Z = 4 | 0.26 × 0.11 × 0.04 mm |
Data collection
| Bruker SMART APEXII CCD diffractometer | 2277 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.037 |
| ω scans | θmax = 71.1°, θmin = 3.6° |
| Absorption correction: multi-scan (SADABS; Bruker, 2013) | h = −15→15 |
| Tmin = 0.599, Tmax = 0.753 | k = −11→11 |
| 18263 measured reflections | l = −15→16 |
| 2895 independent reflections |
Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.040 | w = 1/[σ2(Fo2) + (0.0681P)2 + 0.1332P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.121 | (Δ/σ)max = 0.001 |
| S = 1.05 | Δρmax = 0.23 e Å−3 |
| 2895 reflections | Δρmin = −0.14 e Å−3 |
| 210 parameters | Extinction correction: SHELXL2013 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.0018 (4) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.40310 (9) | 0.79734 (13) | 0.24104 (8) | 0.0566 (3) | |
| C1 | 1.3086 (2) | −0.0296 (3) | 0.5552 (2) | 0.1030 (8) | |
| H1A | 1.2688 | −0.0661 | 0.4904 | 0.155* | |
| H1B | 1.3627 | −0.0959 | 0.5878 | 0.155* | |
| H1C | 1.3447 | 0.0551 | 0.5433 | 0.155* | |
| C2 | 1.23138 (15) | −0.00168 (18) | 0.62283 (15) | 0.0675 (5) | |
| H2A | 1.1963 | −0.0870 | 0.6375 | 0.081* | |
| H2B | 1.2702 | 0.0384 | 0.6877 | 0.081* | |
| O3 | 1.14975 (10) | 0.09453 (12) | 0.56841 (10) | 0.0647 (3) | |
| C4 | 1.07204 (13) | 0.13648 (19) | 0.61646 (13) | 0.0577 (4) | |
| C5 | 0.99507 (12) | 0.23783 (16) | 0.55419 (12) | 0.0512 (4) | |
| C6 | 0.92059 (14) | 0.3074 (2) | 0.60045 (14) | 0.0636 (5) | |
| H6 | 0.9217 | 0.2925 | 0.6701 | 0.076* | |
| C7 | 0.84532 (14) | 0.3980 (2) | 0.54458 (14) | 0.0609 (4) | |
| H7 | 0.7960 | 0.4438 | 0.5767 | 0.073* | |
| C8 | 0.84222 (11) | 0.42174 (15) | 0.44046 (12) | 0.0461 (3) | |
| N1 | 0.75716 (9) | 0.50849 (13) | 0.38633 (9) | 0.0464 (3) | |
| C10 | 0.77038 (12) | 0.57994 (15) | 0.30896 (12) | 0.0472 (3) | |
| C11 | 0.68431 (12) | 0.66679 (15) | 0.24840 (11) | 0.0442 (3) | |
| C12 | 0.58421 (12) | 0.68797 (15) | 0.27877 (11) | 0.0449 (3) | |
| H12 | 0.5723 | 0.6466 | 0.3389 | 0.054* | |
| C13 | 0.50380 (11) | 0.76950 (14) | 0.22013 (11) | 0.0416 (3) | |
| C14 | 0.52011 (11) | 0.83080 (14) | 0.12814 (10) | 0.0419 (3) | |
| O15 | 0.44049 (9) | 0.90702 (12) | 0.06788 (8) | 0.0514 (3) | |
| C16 | 0.38242 (16) | 0.7484 (3) | 0.33611 (15) | 0.0777 (6) | |
| H16A | 0.3899 | 0.6493 | 0.3391 | 0.117* | |
| H16B | 0.3089 | 0.7735 | 0.3410 | 0.117* | |
| H16C | 0.4344 | 0.7893 | 0.3925 | 0.117* | |
| O17 | 1.06660 (13) | 0.09730 (18) | 0.70144 (12) | 0.0924 (5) | |
| C18 | 0.99445 (13) | 0.26473 (18) | 0.45153 (13) | 0.0565 (4) | |
| H18 | 1.0455 | 0.2211 | 0.4202 | 0.068* | |
| C19 | 0.91880 (13) | 0.35567 (18) | 0.39485 (12) | 0.0545 (4) | |
| H19 | 0.9193 | 0.3726 | 0.3258 | 0.065* | |
| C20 | 0.70063 (12) | 0.73037 (17) | 0.15876 (12) | 0.0504 (4) | |
| H20 | 0.7673 | 0.7185 | 0.1387 | 0.060* | |
| C21 | 0.61918 (12) | 0.81096 (16) | 0.09903 (11) | 0.0492 (4) | |
| H21 | 0.6312 | 0.8521 | 0.0389 | 0.059* | |
| H15 | 0.3862 (18) | 0.925 (2) | 0.0989 (16) | 0.077 (6)* | |
| H10 | 0.8383 (16) | 0.5785 (18) | 0.2855 (14) | 0.060 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0488 (6) | 0.0743 (7) | 0.0522 (6) | 0.0172 (5) | 0.0227 (5) | 0.0181 (5) |
| C1 | 0.1014 (17) | 0.1064 (18) | 0.1052 (18) | 0.0560 (15) | 0.0308 (14) | 0.0244 (15) |
| C2 | 0.0615 (10) | 0.0546 (9) | 0.0794 (12) | 0.0128 (8) | −0.0002 (9) | 0.0111 (8) |
| O3 | 0.0589 (7) | 0.0652 (7) | 0.0676 (7) | 0.0188 (6) | 0.0084 (6) | 0.0122 (6) |
| C4 | 0.0489 (8) | 0.0607 (10) | 0.0612 (10) | 0.0016 (7) | 0.0069 (7) | 0.0083 (7) |
| C5 | 0.0413 (7) | 0.0544 (8) | 0.0556 (9) | −0.0004 (6) | 0.0057 (7) | 0.0063 (7) |
| C6 | 0.0572 (9) | 0.0845 (12) | 0.0516 (9) | 0.0134 (9) | 0.0171 (8) | 0.0164 (8) |
| C7 | 0.0524 (9) | 0.0764 (11) | 0.0585 (9) | 0.0149 (8) | 0.0217 (8) | 0.0141 (8) |
| C8 | 0.0360 (7) | 0.0497 (8) | 0.0512 (8) | −0.0022 (6) | 0.0067 (6) | 0.0026 (6) |
| N1 | 0.0388 (6) | 0.0501 (7) | 0.0490 (7) | 0.0018 (5) | 0.0061 (5) | 0.0021 (5) |
| C10 | 0.0381 (7) | 0.0510 (8) | 0.0520 (8) | −0.0007 (6) | 0.0086 (6) | 0.0004 (6) |
| C11 | 0.0416 (7) | 0.0456 (7) | 0.0451 (7) | −0.0002 (6) | 0.0085 (6) | 0.0000 (6) |
| C12 | 0.0472 (8) | 0.0480 (8) | 0.0405 (7) | 0.0020 (6) | 0.0114 (6) | 0.0049 (6) |
| C13 | 0.0409 (7) | 0.0450 (7) | 0.0405 (7) | 0.0005 (6) | 0.0125 (6) | −0.0003 (6) |
| C14 | 0.0439 (7) | 0.0435 (7) | 0.0376 (7) | −0.0011 (6) | 0.0071 (6) | 0.0002 (5) |
| O15 | 0.0483 (6) | 0.0630 (7) | 0.0439 (6) | 0.0092 (5) | 0.0122 (5) | 0.0125 (5) |
| C16 | 0.0667 (11) | 0.1140 (17) | 0.0626 (11) | 0.0195 (11) | 0.0365 (9) | 0.0240 (11) |
| O17 | 0.0848 (10) | 0.1206 (13) | 0.0759 (9) | 0.0343 (9) | 0.0268 (8) | 0.0444 (9) |
| C18 | 0.0503 (8) | 0.0636 (10) | 0.0564 (9) | 0.0106 (7) | 0.0131 (7) | 0.0017 (7) |
| C19 | 0.0544 (9) | 0.0625 (9) | 0.0462 (8) | 0.0100 (7) | 0.0102 (7) | 0.0034 (7) |
| C20 | 0.0415 (7) | 0.0605 (9) | 0.0523 (8) | 0.0005 (7) | 0.0170 (6) | 0.0022 (7) |
| C21 | 0.0487 (8) | 0.0596 (9) | 0.0420 (7) | −0.0011 (7) | 0.0152 (6) | 0.0068 (6) |
Geometric parameters (Å, º)
| O1—C13 | 1.3650 (17) | N1—C10 | 1.2739 (19) |
| O1—C16 | 1.418 (2) | C10—C11 | 1.455 (2) |
| C1—C2 | 1.471 (3) | C10—H10 | 0.960 (19) |
| C1—H1A | 0.9600 | C11—C20 | 1.389 (2) |
| C1—H1B | 0.9600 | C11—C12 | 1.402 (2) |
| C1—H1C | 0.9600 | C12—C13 | 1.372 (2) |
| C2—O3 | 1.446 (2) | C12—H12 | 0.9300 |
| C2—H2A | 0.9700 | C13—C14 | 1.4070 (19) |
| C2—H2B | 0.9700 | C14—O15 | 1.3469 (17) |
| O3—C4 | 1.329 (2) | C14—C21 | 1.380 (2) |
| C4—O17 | 1.202 (2) | O15—H15 | 0.88 (2) |
| C4—C5 | 1.486 (2) | C16—H16A | 0.9600 |
| C5—C18 | 1.382 (2) | C16—H16B | 0.9600 |
| C5—C6 | 1.388 (2) | C16—H16C | 0.9600 |
| C6—C7 | 1.373 (2) | C18—C19 | 1.382 (2) |
| C6—H6 | 0.9300 | C18—H18 | 0.9300 |
| C7—C8 | 1.390 (2) | C19—H19 | 0.9300 |
| C7—H7 | 0.9300 | C20—C21 | 1.381 (2) |
| C8—C19 | 1.387 (2) | C20—H20 | 0.9300 |
| C8—N1 | 1.4152 (18) | C21—H21 | 0.9300 |
| C13—O1—C16 | 117.64 (12) | C11—C10—H10 | 115.0 (11) |
| C2—C1—H1A | 109.5 | C20—C11—C12 | 118.87 (13) |
| C2—C1—H1B | 109.5 | C20—C11—C10 | 119.95 (13) |
| H1A—C1—H1B | 109.5 | C12—C11—C10 | 121.18 (13) |
| C2—C1—H1C | 109.5 | C13—C12—C11 | 120.24 (13) |
| H1A—C1—H1C | 109.5 | C13—C12—H12 | 119.9 |
| H1B—C1—H1C | 109.5 | C11—C12—H12 | 119.9 |
| O3—C2—C1 | 107.05 (16) | O1—C13—C12 | 125.80 (13) |
| O3—C2—H2A | 110.3 | O1—C13—C14 | 113.73 (12) |
| C1—C2—H2A | 110.3 | C12—C13—C14 | 120.46 (13) |
| O3—C2—H2B | 110.3 | O15—C14—C21 | 119.69 (12) |
| C1—C2—H2B | 110.3 | O15—C14—C13 | 121.15 (12) |
| H2A—C2—H2B | 108.6 | C21—C14—C13 | 119.15 (13) |
| C4—O3—C2 | 117.42 (14) | C14—O15—H15 | 111.7 (14) |
| O17—C4—O3 | 123.07 (16) | O1—C16—H16A | 109.5 |
| O17—C4—C5 | 124.49 (16) | O1—C16—H16B | 109.5 |
| O3—C4—C5 | 112.43 (14) | H16A—C16—H16B | 109.5 |
| C18—C5—C6 | 118.72 (15) | O1—C16—H16C | 109.5 |
| C18—C5—C4 | 122.37 (15) | H16A—C16—H16C | 109.5 |
| C6—C5—C4 | 118.91 (15) | H16B—C16—H16C | 109.5 |
| C7—C6—C5 | 120.74 (15) | C19—C18—C5 | 120.78 (15) |
| C7—C6—H6 | 119.6 | C19—C18—H18 | 119.6 |
| C5—C6—H6 | 119.6 | C5—C18—H18 | 119.6 |
| C6—C7—C8 | 120.60 (15) | C18—C19—C8 | 120.32 (15) |
| C6—C7—H7 | 119.7 | C18—C19—H19 | 119.8 |
| C8—C7—H7 | 119.7 | C8—C19—H19 | 119.8 |
| C19—C8—C7 | 118.78 (14) | C21—C20—C11 | 120.90 (13) |
| C19—C8—N1 | 123.94 (14) | C21—C20—H20 | 119.6 |
| C7—C8—N1 | 117.23 (13) | C11—C20—H20 | 119.6 |
| C10—N1—C8 | 120.99 (12) | C14—C21—C20 | 120.36 (13) |
| N1—C10—C11 | 123.17 (13) | C14—C21—H21 | 119.8 |
| N1—C10—H10 | 121.8 (11) | C20—C21—H21 | 119.8 |
| C1—C2—O3—C4 | −178.81 (19) | C16—O1—C13—C12 | 5.9 (2) |
| C2—O3—C4—O17 | −0.7 (3) | C16—O1—C13—C14 | −175.42 (16) |
| C2—O3—C4—C5 | 178.40 (14) | C11—C12—C13—O1 | 179.61 (14) |
| O17—C4—C5—C18 | −170.41 (19) | C11—C12—C13—C14 | 1.1 (2) |
| O3—C4—C5—C18 | 10.6 (2) | O1—C13—C14—O15 | −1.10 (19) |
| O17—C4—C5—C6 | 9.2 (3) | C12—C13—C14—O15 | 177.62 (13) |
| O3—C4—C5—C6 | −169.87 (16) | O1—C13—C14—C21 | 179.55 (13) |
| C18—C5—C6—C7 | 2.0 (3) | C12—C13—C14—C21 | −1.7 (2) |
| C4—C5—C6—C7 | −177.54 (17) | C6—C5—C18—C19 | −2.1 (3) |
| C5—C6—C7—C8 | 0.0 (3) | C4—C5—C18—C19 | 177.46 (16) |
| C6—C7—C8—C19 | −2.0 (3) | C5—C18—C19—C8 | 0.1 (3) |
| C6—C7—C8—N1 | 175.45 (16) | C7—C8—C19—C18 | 1.9 (3) |
| C19—C8—N1—C10 | −31.1 (2) | N1—C8—C19—C18 | −175.34 (15) |
| C7—C8—N1—C10 | 151.56 (15) | C12—C11—C20—C21 | −1.3 (2) |
| C8—N1—C10—C11 | 177.71 (13) | C10—C11—C20—C21 | 178.69 (14) |
| N1—C10—C11—C20 | −173.29 (14) | O15—C14—C21—C20 | −178.47 (14) |
| N1—C10—C11—C12 | 6.7 (2) | C13—C14—C21—C20 | 0.9 (2) |
| C20—C11—C12—C13 | 0.5 (2) | C11—C20—C21—C14 | 0.6 (2) |
| C10—C11—C12—C13 | −179.54 (13) |
Hydrogen-bond geometry (Å, º)
Cg2 is the centroid of the C11–C14/C20/C21 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O15—H15···N1i | 0.88 (2) | 2.00 (2) | 2.828 (2) | 156 (2) |
| C2—H2B···Cg2ii | 0.97 | 2.87 | 3.766 (2) | 154 |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2; (ii) −x+2, −y+1, −z+1.
References
- Bond, A. D. (2007). CrystEngComm, 9, 833–834.
- Bruker (2013). APEX2, SAINT, XPREP and SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Byrn, S. R., Pfeiffer, R. R. & Stowell, J. G. (1999). West Lafayette: SSCI.
- Desiraju, G. R. (2003). CrystEngComm, 5, 466–467.
- Fejfarová, K., Dušek, M., Maghsodlou Rad, S. & Khalaji, A. D. (2012). Acta Cryst. E68, o2466. [DOI] [PMC free article] [PubMed]
- Good, D. J. & Rodríguez-Hornedo, N. (2009). Cryst. Growth Des. 9, 2252–2264.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Kurogoshi, S. & Hori, K. (1996). Acta Cryst. C52, 660–663.
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Neumcke, B., Schwarz, W. & Stampfli, R. (1981). Pflugers Arch. 390, 230–236. [DOI] [PubMed]
- Paul, E. L., Tung, H. H. & Midler, M. (2005). Powder Technol. 150, 133–143.
- Shakir, R. M., Ariffin, A. & Ng, S. W. (2010). Acta Cryst. E66, o2915. [DOI] [PMC free article] [PubMed]
- Shan, N. & Zaworotko, M. J. (2008). Drug Discovery Today, 13, 440–446. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Yadav, A. V., Shete, A. S., Dabke, A. P., Kulkarni, P. V. & Sakhare, S. S. (2009). Indian J. Pharm. Sci. 71, 359–370. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, Global. DOI: 10.1107/S2056989016008999/su5304sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016008999/su5304Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989016008999/su5304sup3.pdf
Supporting information file. DOI: 10.1107/S2056989016008999/su5304Isup4.cml
CCDC reference: 1483394
Additional supporting information: crystallographic information; 3D view; checkCIF report




