The title compounds C17H14BrNO2 (I), and C17H15NO3 (II), were obtained from the reaction of 6-methoxy-3,4-dihydro-2H-naphthalen-1-one and 2-bromonicotinaldehyde in ethanol. Compound (I) was the expected product and compound (II) was the oxidation product from air exposure.
Keywords: crystal structure; chalcone; 3-[(E)-(6-Methoxy-3,4-dihydronaphth-2-oylidene)methyl]-1H-pyridin-2-one and (E)-2-[(2-bromo-3-pyridyl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1-one
Abstract
The title compounds C17H14BrNO2, (I), and C17H15NO3, (II), were obtained from the reaction of 6-methoxy-3,4-dihydro-2H-naphthalen-1-one and 2-bromonicotinaldehyde in ethanol. Compound (I) was the expected product and compound (II) was the oxidation product from air exposure. In the crystal structure of compound (I), there are no short contacts or hydrogen bonds. The structure does display π–π interactions between adjacent benzene rings and adjacent pyridyl rings. Compound (II) contains two independent molecules, A and B, in the asymmetric unit; both are non-planar, the dihedral angles between the methoxybenzene and 1H-pyridin-2-one mean planes being 35.07 (9)° in A and 35.28 (9)°in B. In each molecule, the 1H-pyridin-2-one unit participates in intermolecular N—H⋯O hydrogen bonding to another molecule of the same type (A to A or B to B). The structure also displays π–π interactions between the pyridyl and the benzene rings of non-equivalent molecules (viz., A to B and B to A).
Chemical context
In order to address the need for new therapeutic agents, medicinal chemists have often looked to nature for inspiration. Our research strategy to synthesize novel compounds considered analogs of the natural product chalcone, which contains two aromatic rings and an α-β-unsaturated ketone. Chalcones, bioactive defense molecules found in plants and used in traditional Chinese medicine, have demonstrated anticancer, antibacterial, antifungal, and anti-inflammatory properties (Nowakowska, 2007 ▸; Katsori et al., 2011 ▸). Chalcones that contain methoxy groups (Shenvi et al., 2013 ▸; Bandgar et al., 2010 ▸) and/or pyridine groups (Prasad et al., 2008 ▸; Yee et al., 2005 ▸) have demonstrated activity against a variety of cancer cell lines and antibiotic-resistant bacteria. Thus, we set out to create a library of chalcones that combine those two functional groups. During the synthesis of the title compound (I) by the Claisen–Schmidt condensation of 6-methoxy-3,4-dihydro-2H-naphthalen-1-one and 2-bromonicotinaldehyde, two different types of crystals were obtained – those of the desired chalcone (I) and those of the oxidized product (II). The title compound (I) is a chalcone analog of one currently being studied for its potential anticancer and antibacterial activity [unpublished results].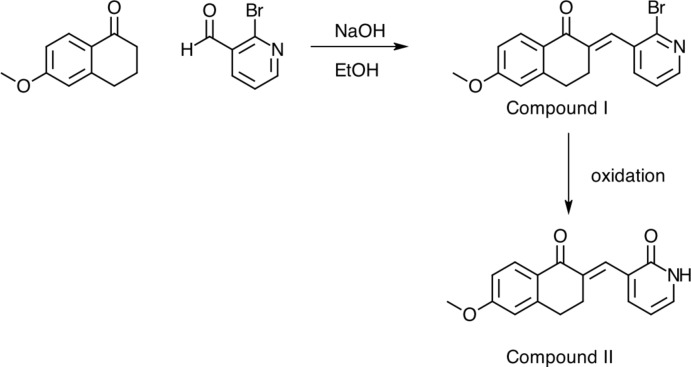
Structural commentary
Compound (I) is non-planar (Fig. 1 ▸) with the pyridyl and the benzene ring being rotated by 73.61 (11)°. The C1—Br1 bond distance is 1.916 (4) Å. In compound (II), which presents two independent molecules in the asymmetric unit (A and B, Fig. 2 ▸), the Br atom is replaced by an oxygen atom, with C—O distances O1A—C1A = 1.258 (3) and O1B—C1B = 1.257 (3) Å. The molecules are also non-planar, the benzene–pyridyl angle being 36.18 (10)° in A and 35.91 (10)° in B.
Figure 1.
A view of the molecular structure of compound (I), showing the atom and ring labeling. Displacement ellipsoids are drawn at the 50% probability level.
Figure 2.
A view of the molecular structure of compound (II), showing the atom and ring labeling. Displacement ellipsoids are drawn at the 50% probability level.
Supramolecular features
In the crystal structure of (I), molecules are linked by Br⋯π and π–π interactions. The Br1⋯Cg1i distance is 3.635 (3) Å [symmetry code: (i) −1 + x, y, z; Cg1 is the centroid of the benzene ring] and has a ‘face-on’ geometry. There are two π–π interactions in the crystal between adjacent benzene rings, Cg1⋯Cg1ii = 3.944 (4) Å [symmetry code: (ii) 1 − x, 1 − y, 1 − z] and between adjacent pyridyl rings, Cg2⋯Cg2iii = 3.639 (4) Å [symmetry code: (iii) −x, 1 − y, −z]. The π–π interactions form ribbons in the ( 01) plane (Fig. 3 ▸), which are held together by the Br⋯π interactions (Fig. 4 ▸).
01) plane (Fig. 3 ▸), which are held together by the Br⋯π interactions (Fig. 4 ▸).
Figure 3.
A view of hydrogen-bonded dimers formed in compound (II). Only molecule A is shown, for simplicity. Hydrogen bonds (see Table1) are drawn with dashed lines.
Figure 4.
N—H⋯O hydrogen bonding in (II) between 1H-pyridin-2-one unit between molecule of the same type
In each one of the independent molecules in (II), the 1H-pyridin-2-one unit participates in intermolecular N—H⋯O hydrogen bonding, with a classical  (8) synthon, to another molecule of the same type (A to A or B to B), see Fig. 5 ▸ and Table 1 ▸ for details. These hydrogen-bonding interactions form dimers that are reminiscent of those frequently observed between carboxylic acids. The hydrogen-bonded units are linked by π–π stacking interactions between the benzene and pyridyl rings in adjacent molecules of different type (A–B or B–A interactions) (Fig. 6 ▸); Cg3⋯Cg4i = 3.875 (4) and Cg5⋯Cg6ii = 3.857 (4) Å [symmetry codes: (i) 3 − x, 1 − y, −z; (ii) 1 − x, 1 − y, 1 − z; Cg3 and Cg4 are the centroids of the pyridyl and benzene rings of molecule A, Cg5 and Cg6 are the corresponding centroids in molecule B].
(8) synthon, to another molecule of the same type (A to A or B to B), see Fig. 5 ▸ and Table 1 ▸ for details. These hydrogen-bonding interactions form dimers that are reminiscent of those frequently observed between carboxylic acids. The hydrogen-bonded units are linked by π–π stacking interactions between the benzene and pyridyl rings in adjacent molecules of different type (A–B or B–A interactions) (Fig. 6 ▸); Cg3⋯Cg4i = 3.875 (4) and Cg5⋯Cg6ii = 3.857 (4) Å [symmetry codes: (i) 3 − x, 1 − y, −z; (ii) 1 − x, 1 − y, 1 − z; Cg3 and Cg4 are the centroids of the pyridyl and benzene rings of molecule A, Cg5 and Cg6 are the corresponding centroids in molecule B].
Figure 5.
Dimers formed by hydrogen-bonding interactions in (II).
Table 1. Hydrogen-bond geometry (Å, °) for (II) .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1A—H1A⋯O1A i | 0.96 (3) | 1.82 (3) | 2.778 (3) | 178 (3) |
| N1B—H1B⋯O1B ii | 0.98 (3) | 1.80 (3) | 2.778 (3) | 176 (3) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 6.
The hydrogen-bonded units in (II) are linked by π–π stacking interactions between the phenyl and pyridyl rings in adjacent molecules of different type.
Database survey
A search of the Cambridge Structural Database (Version 5.37 with four updates, Groom et al., 2016 ▸) for structures containing the combined tetralone and pyridine backbone returned no hits. The search was broadened by changing the nitrogen to carbon, which returned 43 hits. The carbon–containing version of (I) has been reported (Dimmock et al., 2002 ▸; Yee et al., 2005 ▸). Many of these similar chalcones also demonstrated biological activities (Dimmock et al., 2002 ▸).
Synthesis and crystallization
6-Methoxy-3,4-dihydro-2H-naphthalen-1-one (1 mmol) and 2-bromonicotinaldehyde (1 mmol) were dissolved in ethanol (5 mL). An NaOH solution (5 M, 1 mL) was added and the reaction was stirred until a precipitate formed. The reaction mixture was cooled in an ice bath for 20 minutes. The solids were filtered off and recrystallized from MeOH/H2O. Slow evaporation of a methanolic solution gave dark purple/brown crystals, which proved to be 3-[(E)-(6-methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-idene)methyl]pyridin-2(1H)-one, (II), and lighter purple crystals which proved to be (E)-2-[(2-bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one, (I).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were positioned geometrically and refined as riding with C—H = 0.95 or 0.98 Å and U iso(H) = 1.2U eq(C).
Table 2. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C17H14BrNO2 | C17H15NO3 |
| M r | 344.21 | 281.31 |
| Crystal system, space group | Monoclinic, P21/c | Triclinic, P

|
| Temperature (K) | 173 | 173 |
| a, b, c (Å) | 8.885 (8), 14.253 (13), 11.583 (11) | 8.079 (8), 12.296 (12), 14.009 (13) |
| α, β, γ (°) | 90, 92.760 (9), 90 | 88.85 (3), 76.969 (16), 89.43 (3) |
| V (Å3) | 1465 (3) | 1356 (3) |
| Z | 4 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 2.82 | 0.10 |
| Crystal size (mm) | 0.45 × 0.30 × 0.10 | 0.50 × 0.20 × 0.20 |
| Data collection | ||
| Diffractometer | Rigaku XtaLAB mini | Rigaku XtaLAB mini |
| Absorption correction | Multi-scan (REQAB; Rigaku, 1998 ▸) | Multi-scan (REQAB; Rigaku, 1998 ▸) |
| T min, T max | 0.587, 0.754 | 0.808, 0.981 |
| No. of measured, independent and observed [F 2 > 2.0σ(F 2)] reflections | 15395, 3363, 2583 | 14459, 6203, 3987 |
| R int | 0.056 | 0.049 |
| (sin θ/λ)max (Å−1) | 0.650 | 0.649 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.041, 0.088, 0.99 | 0.058, 0.157, 1.03 |
| No. of reflections | 3363 | 6203 |
| No. of parameters | 190 | 387 |
| H-atom treatment | H-atom parameters constrained | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.26, −0.46 | 0.21, −0.23 |
Supplementary Material
Crystal structure: contains datablock(s) General, I, II. DOI: 10.1107/S2056989016009300/bg2587sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016009300/bg2587Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016009300/bg2587IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
MEM and ADG are grateful for the financial support of an ASU Undergraduate Research Grant and SKZ for an ASU Research and Scholarship Grant.
supplementary crystallographic information
(I) (E)-2-[(2-Bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one . Crystal data
| C17H14BrNO2 | F(000) = 696.00 |
| Mr = 344.21 | Dx = 1.560 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71075 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3472 reflections |
| a = 8.885 (8) Å | θ = 2.3–27.5° |
| b = 14.253 (13) Å | µ = 2.82 mm−1 |
| c = 11.583 (11) Å | T = 173 K |
| β = 92.760 (9)° | Prism, dark-purple/brown |
| V = 1465 (3) Å3 | 0.45 × 0.30 × 0.10 mm |
| Z = 4 |
(I) (E)-2-[(2-Bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one . Data collection
| Rigaku XtaLAB mini diffractometer | 2583 reflections with F2 > 2.0σ(F2) |
| Detector resolution: 6.827 pixels mm-1 | Rint = 0.056 |
| ω scans | θmax = 27.5° |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | h = −11→11 |
| Tmin = 0.587, Tmax = 0.754 | k = −18→18 |
| 15395 measured reflections | l = −15→15 |
| 3363 independent reflections |
(I) (E)-2-[(2-Bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.041 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.088 | H-atom parameters constrained |
| S = 0.99 | w = 1/[σ2(Fo2) + (0.0211P)2 + 1.2385P] where P = (Fo2 + 2Fc2)/3 |
| 3363 reflections | (Δ/σ)max = 0.001 |
| 190 parameters | Δρmax = 0.26 e Å−3 |
| 0 restraints | Δρmin = −0.46 e Å−3 |
| Primary atom site location: structure-invariant direct methods |
(I) (E)-2-[(2-Bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one . Special details
| Refinement. Refinement was performed using all reflections. The weighted R-factor (wR) and goodness of fit (S) are based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 sigma(F2) is used only for calculating R-factor (gt). |
(I) (E)-2-[(2-Bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Br1 | −0.13688 (4) | 0.66365 (2) | 0.18318 (3) | 0.05196 (12) | |
| O1 | 0.4068 (3) | 0.77510 (13) | 0.2922 (2) | 0.0510 (6) | |
| O2 | 0.8298 (3) | 0.48452 (15) | 0.56282 (19) | 0.0521 (6) | |
| N1 | −0.1337 (3) | 0.6115 (2) | −0.0441 (3) | 0.0519 (7) | |
| C1 | −0.0406 (4) | 0.63829 (19) | 0.0420 (3) | 0.0399 (7) | |
| C2 | −0.0701 (5) | 0.5917 (3) | −0.1437 (3) | 0.0569 (9) | |
| C3 | 0.0819 (4) | 0.5971 (3) | −0.1590 (3) | 0.0511 (8) | |
| C4 | 0.1750 (4) | 0.6256 (2) | −0.0669 (3) | 0.0441 (8) | |
| C5 | 0.1155 (4) | 0.64673 (18) | 0.0395 (3) | 0.0366 (7) | |
| C6 | 0.2119 (4) | 0.67894 (19) | 0.1378 (3) | 0.0382 (7) | |
| C7 | 0.3472 (3) | 0.64406 (18) | 0.1728 (3) | 0.0355 (7) | |
| C8 | 0.4221 (4) | 0.55788 (19) | 0.1273 (3) | 0.0397 (7) | |
| C9 | 0.4650 (4) | 0.49170 (19) | 0.2271 (3) | 0.0431 (7) | |
| C10 | 0.5546 (3) | 0.53914 (19) | 0.3242 (3) | 0.0349 (7) | |
| C11 | 0.5366 (3) | 0.63552 (18) | 0.3440 (3) | 0.0352 (7) | |
| C12 | 0.4297 (3) | 0.69188 (19) | 0.2712 (3) | 0.0380 (7) | |
| C13 | 0.6523 (3) | 0.48629 (19) | 0.3960 (3) | 0.0370 (7) | |
| C14 | 0.7322 (4) | 0.5294 (2) | 0.4867 (3) | 0.0411 (7) | |
| C15 | 0.7157 (4) | 0.6258 (3) | 0.5061 (3) | 0.0477 (8) | |
| C16 | 0.6194 (4) | 0.6772 (2) | 0.4356 (3) | 0.0447 (8) | |
| C17 | 0.8613 (4) | 0.3880 (3) | 0.5410 (3) | 0.0539 (9) | |
| H2 | −0.1340 | 0.5728 | −0.2077 | 0.0683* | |
| H3 | 0.1219 | 0.5817 | −0.2311 | 0.0613* | |
| H4 | 0.2804 | 0.6309 | −0.0758 | 0.0530* | |
| H6 | 0.1750 | 0.7297 | 0.1814 | 0.0459* | |
| H8A | 0.5135 | 0.5761 | 0.0871 | 0.0476* | |
| H8B | 0.3525 | 0.5256 | 0.0710 | 0.0476* | |
| H9A | 0.3720 | 0.4651 | 0.2576 | 0.0517* | |
| H9B | 0.5248 | 0.4391 | 0.1975 | 0.0517* | |
| H13 | 0.6639 | 0.4210 | 0.3828 | 0.0445* | |
| H15 | 0.7712 | 0.6553 | 0.5682 | 0.0572* | |
| H16 | 0.6088 | 0.7425 | 0.4493 | 0.0536* | |
| H17A | 0.9360 | 0.3650 | 0.5993 | 0.0646* | |
| H17B | 0.9011 | 0.3816 | 0.4640 | 0.0646* | |
| H17C | 0.7684 | 0.3513 | 0.5450 | 0.0646* |
(I) (E)-2-[(2-Bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Br1 | 0.04010 (19) | 0.0613 (3) | 0.0558 (3) | 0.01067 (15) | 0.01591 (15) | 0.01474 (16) |
| O1 | 0.0488 (14) | 0.0315 (12) | 0.0735 (16) | 0.0008 (9) | 0.0093 (12) | −0.0123 (11) |
| O2 | 0.0533 (14) | 0.0551 (14) | 0.0471 (13) | 0.0012 (11) | −0.0050 (11) | −0.0050 (11) |
| N1 | 0.0415 (16) | 0.0614 (18) | 0.0525 (17) | −0.0020 (13) | −0.0003 (14) | 0.0138 (14) |
| C1 | 0.0361 (16) | 0.0365 (16) | 0.0479 (18) | 0.0030 (12) | 0.0094 (14) | 0.0138 (13) |
| C2 | 0.062 (3) | 0.063 (3) | 0.045 (2) | −0.0061 (18) | −0.0079 (17) | 0.0126 (17) |
| C3 | 0.061 (3) | 0.051 (2) | 0.0415 (18) | 0.0011 (16) | 0.0087 (16) | 0.0097 (15) |
| C4 | 0.0470 (19) | 0.0387 (17) | 0.0478 (19) | −0.0008 (14) | 0.0126 (15) | 0.0097 (14) |
| C5 | 0.0355 (16) | 0.0307 (15) | 0.0443 (17) | 0.0021 (11) | 0.0084 (13) | 0.0097 (12) |
| C6 | 0.0387 (17) | 0.0308 (15) | 0.0462 (17) | −0.0009 (12) | 0.0131 (14) | 0.0029 (12) |
| C7 | 0.0348 (16) | 0.0294 (14) | 0.0434 (17) | −0.0018 (11) | 0.0130 (13) | −0.0027 (12) |
| C8 | 0.0348 (16) | 0.0411 (17) | 0.0436 (17) | 0.0037 (12) | 0.0052 (13) | −0.0104 (13) |
| C9 | 0.0448 (18) | 0.0315 (15) | 0.0525 (19) | 0.0045 (13) | −0.0029 (15) | −0.0117 (14) |
| C10 | 0.0304 (15) | 0.0348 (15) | 0.0406 (16) | −0.0043 (11) | 0.0111 (12) | −0.0073 (12) |
| C11 | 0.0301 (15) | 0.0323 (14) | 0.0439 (17) | −0.0047 (11) | 0.0103 (13) | −0.0085 (12) |
| C12 | 0.0347 (16) | 0.0307 (15) | 0.0499 (18) | −0.0049 (12) | 0.0162 (13) | −0.0085 (13) |
| C13 | 0.0341 (16) | 0.0346 (15) | 0.0432 (16) | −0.0048 (12) | 0.0103 (13) | −0.0059 (13) |
| C14 | 0.0366 (16) | 0.0471 (18) | 0.0404 (17) | −0.0030 (13) | 0.0098 (13) | −0.0049 (14) |
| C15 | 0.0425 (19) | 0.0508 (19) | 0.0498 (19) | −0.0093 (15) | 0.0016 (15) | −0.0161 (15) |
| C16 | 0.0418 (18) | 0.0391 (17) | 0.0543 (19) | −0.0063 (13) | 0.0124 (15) | −0.0162 (14) |
| C17 | 0.054 (3) | 0.058 (3) | 0.049 (2) | 0.0080 (17) | 0.0013 (16) | 0.0003 (16) |
(I) (E)-2-[(2-Bromopyridin-3-yl)methylidene]-6-methoxy-3,4-dihydronaphthalen-1(2H)-one . Geometric parameters (Å, º)
| Br1—C1 | 1.916 (4) | C11—C16 | 1.395 (5) |
| O1—C12 | 1.230 (4) | C13—C14 | 1.383 (5) |
| O2—C14 | 1.366 (4) | C14—C15 | 1.400 (5) |
| O2—C17 | 1.428 (4) | C15—C16 | 1.367 (5) |
| N1—C1 | 1.322 (4) | C2—H2 | 0.950 |
| N1—C2 | 1.339 (5) | C3—H3 | 0.950 |
| C1—C5 | 1.394 (5) | C4—H4 | 0.950 |
| C2—C3 | 1.373 (6) | C6—H6 | 0.950 |
| C3—C4 | 1.379 (5) | C8—H8A | 0.990 |
| C4—C5 | 1.396 (5) | C8—H8B | 0.990 |
| C5—C6 | 1.466 (4) | C9—H9A | 0.990 |
| C6—C7 | 1.345 (4) | C9—H9B | 0.990 |
| C7—C8 | 1.504 (4) | C13—H13 | 0.950 |
| C7—C12 | 1.490 (4) | C15—H15 | 0.950 |
| C8—C9 | 1.526 (5) | C16—H16 | 0.950 |
| C9—C10 | 1.507 (4) | C17—H17A | 0.980 |
| C10—C11 | 1.403 (4) | C17—H17B | 0.980 |
| C10—C13 | 1.394 (4) | C17—H17C | 0.980 |
| C11—C12 | 1.477 (4) | ||
| Br1···C6 | 3.175 (5) | C7···H17Cvii | 3.4744 |
| O1···C6 | 2.788 (4) | C8···H2ix | 3.3343 |
| O1···C16 | 2.824 (4) | C8···H8Axii | 3.2064 |
| O2···C16 | 3.597 (5) | C8···H8Bxii | 3.3383 |
| N1···C4 | 2.776 (5) | C9···H2ix | 3.0789 |
| C1···C3 | 2.682 (5) | C9···H4xii | 3.4093 |
| C2···C5 | 2.739 (5) | C10···H3xii | 3.5613 |
| C4···C7 | 3.115 (5) | C11···H17Cvii | 3.0608 |
| C4···C8 | 3.215 (5) | C12···H4ii | 3.3921 |
| C5···C8 | 3.129 (5) | C12···H9Biii | 3.5629 |
| C7···C10 | 2.898 (5) | C12···H17Cvii | 2.8936 |
| C8···C11 | 2.883 (5) | C13···H3xii | 2.9961 |
| C9···C12 | 2.918 (5) | C14···H9Avii | 3.1465 |
| C10···C15 | 2.779 (5) | C14···H17Aiv | 3.4966 |
| C11···C14 | 2.786 (5) | C14···H17Biv | 3.5174 |
| C13···C16 | 2.777 (5) | C15···H2xi | 3.5940 |
| C13···C17 | 2.816 (5) | C15···H9Avii | 3.1588 |
| Br1···C11i | 3.545 (4) | C15···H17Aiv | 3.3838 |
| O1···C3ii | 3.482 (5) | C15···H17Biv | 3.4081 |
| O1···C4ii | 3.039 (5) | C16···H17Cvii | 3.4878 |
| O1···C9iii | 3.302 (5) | C17···H6vi | 3.4285 |
| O2···O2iv | 3.447 (5) | H2···O2xiii | 2.9448 |
| O2···C17iv | 3.549 (5) | H2···C8ix | 3.3343 |
| C3···O1v | 3.482 (5) | H2···C9ix | 3.0789 |
| C4···O1v | 3.039 (5) | H2···C15xiii | 3.5940 |
| C5···C17iii | 3.572 (6) | H2···H8Bix | 2.9228 |
| C9···O1vi | 3.302 (5) | H2···H9Aix | 2.2314 |
| C10···C14vii | 3.576 (5) | H2···H9Bix | 3.4838 |
| C11···Br1viii | 3.545 (4) | H2···H15xiii | 2.9364 |
| C14···C10vii | 3.576 (5) | H3···Br1ix | 3.5417 |
| C17···O2iv | 3.549 (5) | H3···O1v | 3.2529 |
| C17···C5vi | 3.572 (6) | H3···C10xii | 3.5613 |
| Br1···H6 | 2.9275 | H3···C13xii | 2.9961 |
| O1···H6 | 2.4600 | H3···H6v | 2.9190 |
| O1···H16 | 2.5365 | H3···H9Bxii | 3.1583 |
| O2···H13 | 2.6545 | H3···H13xii | 2.6509 |
| O2···H15 | 2.4904 | H3···H17Bxii | 2.7453 |
| N1···H3 | 3.2414 | H4···O1v | 2.3578 |
| C1···H2 | 3.1134 | H4···C9xii | 3.4093 |
| C1···H4 | 3.2223 | H4···C12v | 3.3921 |
| C1···H6 | 2.7708 | H4···H6v | 3.5320 |
| C2···H4 | 3.2239 | H4···H8Axii | 3.4784 |
| C4···H2 | 3.2145 | H4···H9Bxii | 2.4924 |
| C4···H6 | 3.2363 | H4···H16v | 3.4309 |
| C4···H8A | 3.4942 | H4···H17Ciii | 3.1940 |
| C4···H8B | 2.6124 | H6···C3ii | 3.2152 |
| C5···H3 | 3.2719 | H6···C4ii | 3.5709 |
| C5···H8B | 2.7337 | H6···C17iii | 3.4285 |
| C6···H4 | 2.6663 | H6···H3ii | 2.9190 |
| C6···H8A | 3.1347 | H6···H4ii | 3.5320 |
| C6···H8B | 2.6513 | H6···H13iii | 3.1833 |
| C7···H4 | 2.9181 | H6···H17Avii | 3.0803 |
| C7···H9A | 2.7383 | H6···H17Biii | 2.8049 |
| C7···H9B | 3.3265 | H6···H17Ciii | 3.2031 |
| C8···H4 | 2.8141 | H6···H17Cvii | 3.3881 |
| C8···H6 | 3.3683 | H8A···Br1viii | 3.4801 |
| C9···H13 | 2.6622 | H8A···N1viii | 3.5846 |
| C10···H8A | 2.8022 | H8A···C8xii | 3.2064 |
| C10···H8B | 3.3699 | H8A···H4xii | 3.4784 |
| C10···H16 | 3.2663 | H8A···H8Axii | 2.9651 |
| C11···H8A | 3.0904 | H8A···H8Bxii | 2.6613 |
| C11···H9A | 2.9829 | H8A···H9Bxii | 3.3050 |
| C11···H9B | 3.2731 | H8A···H16v | 3.1746 |
| C11···H13 | 3.2840 | H8B···N1ix | 2.7636 |
| C11···H15 | 3.2626 | H8B···C2ix | 3.1634 |
| C12···H6 | 2.5034 | H8B···C8xii | 3.3383 |
| C12···H8A | 2.8241 | H8B···H2ix | 2.9228 |
| C12···H8B | 3.3631 | H8B···H8Axii | 2.6613 |
| C12···H9A | 3.2750 | H8B···H8Bxii | 3.2432 |
| C12···H16 | 2.6445 | H8B···H9Bxii | 3.3817 |
| C13···H9A | 2.9123 | H9A···O1vi | 3.4120 |
| C13···H9B | 2.6028 | H9A···O2vii | 2.9006 |
| C13···H15 | 3.2694 | H9A···N1ix | 3.3590 |
| C13···H17B | 2.7501 | H9A···C2ix | 3.0408 |
| C13···H17C | 2.7511 | H9A···C14vii | 3.1465 |
| C14···H16 | 3.2512 | H9A···C15vii | 3.1588 |
| C14···H17A | 3.2009 | H9A···H2ix | 2.2314 |
| C14···H17B | 2.6071 | H9A···H15vii | 2.9801 |
| C14···H17C | 2.6424 | H9B···O1vi | 2.4164 |
| C15···H13 | 3.2725 | H9B···C3xii | 3.5819 |
| C17···H13 | 2.5192 | H9B···C4xii | 3.2624 |
| H2···H3 | 2.3066 | H9B···C12vi | 3.5629 |
| H3···H4 | 2.3381 | H9B···H2ix | 3.4838 |
| H4···H6 | 3.4655 | H9B···H3xii | 3.1583 |
| H4···H8A | 2.8438 | H9B···H4xii | 2.4924 |
| H4···H8B | 2.3341 | H9B···H8Axii | 3.3050 |
| H6···H8B | 3.5753 | H9B···H8Bxii | 3.3817 |
| H8A···H9A | 2.8675 | H9B···H16vi | 3.4571 |
| H8A···H9B | 2.3336 | H13···O1vi | 2.9497 |
| H8B···H9A | 2.3265 | H13···C3xii | 3.5284 |
| H8B···H9B | 2.4075 | H13···H3xii | 2.6509 |
| H9A···H13 | 2.9774 | H13···H6vi | 3.1833 |
| H9B···H13 | 2.4383 | H15···Br1xiv | 2.9998 |
| H13···H17A | 3.4907 | H15···C1xiv | 3.4050 |
| H13···H17B | 2.3340 | H15···H2xi | 2.9364 |
| H13···H17C | 2.2821 | H15···H9Avii | 2.9801 |
| H15···H16 | 2.3091 | H15···H17Aiv | 3.3316 |
| Br1···H3ix | 3.5417 | H15···H17Biv | 3.0003 |
| Br1···H8Ai | 3.4801 | H16···N1xiv | 3.0906 |
| Br1···H15x | 2.9998 | H16···H4ii | 3.4309 |
| Br1···H17Avii | 3.0445 | H16···H8Aii | 3.1746 |
| O1···H3ii | 3.2529 | H16···H9Biii | 3.4571 |
| O1···H4ii | 2.3578 | H17A···Br1vii | 3.0445 |
| O1···H9Aiii | 3.4120 | H17A···O2iv | 3.5822 |
| O1···H9Biii | 2.4164 | H17A···C4vi | 3.5660 |
| O1···H13iii | 2.9497 | H17A···C5vi | 3.5209 |
| O1···H17Cvii | 3.0841 | H17A···C6vii | 3.4317 |
| O2···H2xi | 2.9448 | H17A···C14iv | 3.4966 |
| O2···H9Avii | 2.9006 | H17A···C15iv | 3.3838 |
| O2···H17Aiv | 3.5822 | H17A···H6vii | 3.0803 |
| O2···H17Biv | 3.0868 | H17A···H15iv | 3.3316 |
| N1···H8Ai | 3.5846 | H17B···O2iv | 3.0868 |
| N1···H8Bix | 2.7636 | H17B···C3xii | 3.5563 |
| N1···H9Aix | 3.3590 | H17B···C5vi | 3.3507 |
| N1···H16x | 3.0906 | H17B···C6vi | 3.2597 |
| C1···H15x | 3.4050 | H17B···C14iv | 3.5174 |
| C2···H8Bix | 3.1634 | H17B···C15iv | 3.4081 |
| C2···H9Aix | 3.0408 | H17B···H3xii | 2.7453 |
| C3···H6v | 3.2152 | H17B···H6vi | 2.8049 |
| C3···H9Bxii | 3.5819 | H17B···H15iv | 3.0003 |
| C3···H13xii | 3.5284 | H17C···O1vii | 3.0841 |
| C3···H17Bxii | 3.5563 | H17C···C4vi | 3.2643 |
| C4···H6v | 3.5709 | H17C···C5vi | 3.2590 |
| C4···H9Bxii | 3.2624 | H17C···C6vi | 3.2537 |
| C4···H17Aiii | 3.5660 | H17C···C7vii | 3.4744 |
| C4···H17Ciii | 3.2643 | H17C···C11vii | 3.0608 |
| C5···H17Aiii | 3.5209 | H17C···C12vii | 2.8936 |
| C5···H17Biii | 3.3507 | H17C···C16vii | 3.4878 |
| C5···H17Ciii | 3.2590 | H17C···H4vi | 3.1940 |
| C6···H17Avii | 3.4317 | H17C···H6vi | 3.2031 |
| C6···H17Biii | 3.2597 | H17C···H6vii | 3.3881 |
| C6···H17Ciii | 3.2537 | ||
| C14—O2—C17 | 117.4 (3) | C11—C16—C15 | 121.2 (3) |
| C1—N1—C2 | 115.9 (3) | N1—C2—H2 | 118.055 |
| Br1—C1—N1 | 114.2 (3) | C3—C2—H2 | 118.055 |
| Br1—C1—C5 | 119.2 (3) | C2—C3—H3 | 120.838 |
| N1—C1—C5 | 126.6 (3) | C4—C3—H3 | 120.843 |
| N1—C2—C3 | 123.9 (4) | C3—C4—H4 | 119.759 |
| C2—C3—C4 | 118.3 (4) | C5—C4—H4 | 119.753 |
| C3—C4—C5 | 120.5 (3) | C5—C6—H6 | 116.619 |
| C1—C5—C4 | 114.8 (3) | C7—C6—H6 | 116.628 |
| C1—C5—C6 | 123.7 (3) | C7—C8—H8A | 109.712 |
| C4—C5—C6 | 121.4 (3) | C7—C8—H8B | 109.712 |
| C5—C6—C7 | 126.8 (3) | C9—C8—H8A | 109.713 |
| C6—C7—C8 | 126.9 (3) | C9—C8—H8B | 109.719 |
| C6—C7—C12 | 117.4 (3) | H8A—C8—H8B | 108.195 |
| C8—C7—C12 | 115.6 (3) | C8—C9—H9A | 108.968 |
| C7—C8—C9 | 109.8 (3) | C8—C9—H9B | 108.968 |
| C8—C9—C10 | 113.1 (3) | C10—C9—H9A | 108.972 |
| C9—C10—C11 | 120.1 (3) | C10—C9—H9B | 108.971 |
| C9—C10—C13 | 119.6 (3) | H9A—C9—H9B | 107.765 |
| C11—C10—C13 | 120.3 (3) | C10—C13—H13 | 120.158 |
| C10—C11—C12 | 121.0 (3) | C14—C13—H13 | 120.170 |
| C10—C11—C16 | 118.7 (3) | C14—C15—H15 | 120.081 |
| C12—C11—C16 | 120.3 (3) | C16—C15—H15 | 120.079 |
| O1—C12—C7 | 120.8 (3) | C11—C16—H16 | 119.393 |
| O1—C12—C11 | 121.3 (3) | C15—C16—H16 | 119.404 |
| C7—C12—C11 | 117.8 (3) | O2—C17—H17A | 109.466 |
| C10—C13—C14 | 119.7 (3) | O2—C17—H17B | 109.477 |
| O2—C14—C13 | 124.7 (3) | O2—C17—H17C | 109.473 |
| O2—C14—C15 | 115.1 (3) | H17A—C17—H17B | 109.467 |
| C13—C14—C15 | 120.3 (3) | H17A—C17—H17C | 109.469 |
| C14—C15—C16 | 119.8 (3) | H17B—C17—H17C | 109.476 |
| C17—O2—C14—C13 | 5.8 (4) | C12—C7—C8—C9 | 50.7 (3) |
| C17—O2—C14—C15 | −174.6 (3) | C7—C8—C9—C10 | −52.0 (3) |
| C1—N1—C2—C3 | 0.6 (5) | C8—C9—C10—C11 | 27.9 (4) |
| C2—N1—C1—Br1 | −179.0 (3) | C8—C9—C10—C13 | −153.9 (3) |
| C2—N1—C1—C5 | −0.8 (5) | C9—C10—C11—C12 | 0.4 (4) |
| Br1—C1—C5—C4 | 179.22 (15) | C9—C10—C11—C16 | 179.0 (3) |
| Br1—C1—C5—C6 | −3.3 (4) | C9—C10—C13—C14 | −178.6 (3) |
| N1—C1—C5—C4 | 1.1 (4) | C11—C10—C13—C14 | −0.4 (4) |
| N1—C1—C5—C6 | 178.6 (3) | C13—C10—C11—C12 | −177.9 (3) |
| N1—C2—C3—C4 | −0.7 (5) | C13—C10—C11—C16 | 0.8 (4) |
| C2—C3—C4—C5 | 1.1 (5) | C10—C11—C12—O1 | 175.7 (3) |
| C3—C4—C5—C1 | −1.2 (4) | C10—C11—C12—C7 | −2.8 (4) |
| C3—C4—C5—C6 | −178.7 (3) | C10—C11—C16—C15 | −0.6 (5) |
| C1—C5—C6—C7 | 139.3 (3) | C12—C11—C16—C15 | 178.1 (3) |
| C4—C5—C6—C7 | −43.4 (4) | C16—C11—C12—O1 | −2.9 (5) |
| C5—C6—C7—C8 | −7.3 (5) | C16—C11—C12—C7 | 178.5 (3) |
| C5—C6—C7—C12 | 176.3 (3) | C10—C13—C14—O2 | 179.4 (3) |
| C6—C7—C8—C9 | −125.8 (3) | C10—C13—C14—C15 | −0.2 (5) |
| C6—C7—C12—O1 | −25.6 (4) | O2—C14—C15—C16 | −179.3 (3) |
| C6—C7—C12—C11 | 152.9 (3) | C13—C14—C15—C16 | 0.4 (5) |
| C8—C7—C12—O1 | 157.5 (3) | C14—C15—C16—C11 | 0.0 (5) |
| C8—C7—C12—C11 | −23.9 (4) |
Symmetry codes: (i) x−1, y, z; (ii) x, −y+3/2, z+1/2; (iii) −x+1, y+1/2, −z+1/2; (iv) −x+2, −y+1, −z+1; (v) x, −y+3/2, z−1/2; (vi) −x+1, y−1/2, −z+1/2; (vii) −x+1, −y+1, −z+1; (viii) x+1, y, z; (ix) −x, −y+1, −z; (x) x−1, −y+3/2, z−1/2; (xi) x+1, y, z+1; (xii) −x+1, −y+1, −z; (xiii) x−1, y, z−1; (xiv) x+1, −y+3/2, z+1/2.
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Crystal data
| C17H15NO3 | Z = 4 |
| Mr = 281.31 | F(000) = 592.00 |
| Triclinic, P1 | Dx = 1.378 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71075 Å |
| a = 8.079 (8) Å | Cell parameters from 3099 reflections |
| b = 12.296 (12) Å | θ = 1.7–27.5° |
| c = 14.009 (13) Å | µ = 0.10 mm−1 |
| α = 88.85 (3)° | T = 173 K |
| β = 76.969 (16)° | Prism, purple |
| γ = 89.43 (3)° | 0.50 × 0.20 × 0.20 mm |
| V = 1356 (3) Å3 |
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Data collection
| Rigaku XtaLAB mini diffractometer | 3987 reflections with F2 > 2.0σ(F2) |
| Detector resolution: 6.827 pixels mm-1 | Rint = 0.049 |
| ω scans | θmax = 27.5° |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | h = −10→10 |
| Tmin = 0.808, Tmax = 0.981 | k = −15→15 |
| 14459 measured reflections | l = −18→18 |
| 6203 independent reflections |
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.058 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.157 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0585P)2 + 0.2529P] where P = (Fo2 + 2Fc2)/3 |
| 6203 reflections | (Δ/σ)max < 0.001 |
| 387 parameters | Δρmax = 0.21 e Å−3 |
| 0 restraints | Δρmin = −0.23 e Å−3 |
| Primary atom site location: structure-invariant direct methods |
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Special details
| Refinement. Refinement was performed using all reflections. The weighted R-factor (wR) and goodness of fit (S) are based on F2. R-factor (gt) are based on F. The threshold expression of F2 > 2.0 sigma(F2) is used only for calculating R-factor (gt). |
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1A | 1.35958 (19) | 0.48754 (12) | 0.11653 (11) | 0.0326 (4) | |
| O1B | 0.63793 (19) | 0.98881 (12) | 0.38219 (11) | 0.0332 (4) | |
| O2A | 0.9197 (3) | 0.46536 (12) | 0.39203 (12) | 0.0402 (4) | |
| O2B | 1.0769 (2) | 0.96666 (12) | 0.10487 (11) | 0.0387 (4) | |
| O3A | 0.6064 (3) | 0.15815 (13) | 0.75027 (11) | 0.0442 (5) | |
| O3B | 1.3821 (2) | 0.65941 (13) | −0.24660 (11) | 0.0412 (5) | |
| N1A | 1.4242 (3) | 0.36168 (15) | −0.00284 (13) | 0.0298 (4) | |
| N1B | 0.5793 (3) | 0.86238 (15) | 0.50526 (13) | 0.0316 (5) | |
| C1A | 1.3324 (3) | 0.39499 (17) | 0.08683 (15) | 0.0280 (5) | |
| C1B | 0.6684 (3) | 0.89632 (17) | 0.41435 (15) | 0.0278 (5) | |
| C2A | 1.4049 (3) | 0.26451 (18) | −0.04361 (17) | 0.0354 (6) | |
| C2B | 0.6040 (3) | 0.76448 (18) | 0.54789 (16) | 0.0350 (6) | |
| C3A | 1.2856 (3) | 0.19377 (18) | 0.00387 (16) | 0.0347 (6) | |
| C3B | 0.7238 (3) | 0.69400 (18) | 0.50207 (16) | 0.0342 (6) | |
| C4A | 1.1847 (3) | 0.22236 (17) | 0.09530 (16) | 0.0307 (5) | |
| C4B | 0.8213 (3) | 0.72330 (18) | 0.40892 (16) | 0.0321 (5) | |
| C5A | 1.2066 (3) | 0.31888 (17) | 0.13941 (15) | 0.0272 (5) | |
| C5B | 0.7950 (3) | 0.81991 (17) | 0.36243 (15) | 0.0280 (5) | |
| C6A | 1.1081 (3) | 0.35595 (17) | 0.23403 (15) | 0.0274 (5) | |
| C6B | 0.8909 (3) | 0.85753 (17) | 0.26640 (15) | 0.0270 (5) | |
| C7A | 1.0120 (3) | 0.29973 (17) | 0.30996 (15) | 0.0277 (5) | |
| C7B | 0.9845 (3) | 0.80123 (17) | 0.19149 (15) | 0.0266 (5) | |
| C8A | 0.9851 (3) | 0.17786 (17) | 0.31644 (15) | 0.0326 (5) | |
| C8B | 1.0132 (3) | 0.67944 (17) | 0.18803 (16) | 0.0333 (5) | |
| C9A | 0.9762 (3) | 0.13030 (17) | 0.41899 (15) | 0.0287 (5) | |
| C9B | 1.0185 (3) | 0.63272 (17) | 0.08736 (15) | 0.0283 (5) | |
| C10A | 0.8586 (3) | 0.19351 (17) | 0.49738 (15) | 0.0261 (5) | |
| C10B | 1.1337 (3) | 0.69533 (16) | 0.00597 (15) | 0.0252 (5) | |
| C11A | 0.8353 (3) | 0.30523 (17) | 0.48528 (15) | 0.0288 (5) | |
| C11B | 1.1553 (3) | 0.80721 (17) | 0.01437 (15) | 0.0280 (5) | |
| C12A | 0.9227 (3) | 0.36508 (17) | 0.39546 (15) | 0.0281 (5) | |
| C12B | 1.0721 (3) | 0.86650 (17) | 0.10352 (15) | 0.0279 (5) | |
| C13A | 0.7811 (3) | 0.14120 (17) | 0.58544 (15) | 0.0285 (5) | |
| C13B | 1.2104 (3) | 0.64271 (17) | −0.08067 (15) | 0.0278 (5) | |
| C14A | 0.6831 (3) | 0.20038 (18) | 0.66097 (16) | 0.0334 (5) | |
| C14B | 1.3057 (3) | 0.70229 (18) | −0.15880 (16) | 0.0316 (5) | |
| C15A | 0.6584 (4) | 0.3122 (2) | 0.64862 (18) | 0.0454 (7) | |
| C15B | 1.3265 (4) | 0.8140 (2) | −0.15060 (17) | 0.0421 (7) | |
| C16A | 0.7334 (4) | 0.36361 (19) | 0.56189 (17) | 0.0417 (6) | |
| C16B | 1.2532 (3) | 0.86551 (19) | −0.06559 (16) | 0.0374 (6) | |
| C17A | 0.6263 (4) | 0.04374 (19) | 0.76739 (17) | 0.0396 (6) | |
| C17B | 1.3661 (4) | 0.54477 (19) | −0.25898 (17) | 0.0399 (6) | |
| H1A | 1.501 (4) | 0.414 (3) | −0.041 (2) | 0.054 (8)* | |
| H1B | 0.499 (4) | 0.914 (3) | 0.543 (2) | 0.058 (9)* | |
| H2A | 1.4750 | 0.2463 | −0.1053 | 0.0424* | |
| H2B | 0.5360 | 0.7457 | 0.6106 | 0.0419* | |
| H3A | 1.2704 | 0.1259 | −0.0242 | 0.0417* | |
| H3B | 0.7417 | 0.6263 | 0.5321 | 0.0410* | |
| H4A | 1.0986 | 0.1738 | 0.1278 | 0.0368* | |
| H4B | 0.9080 | 0.6752 | 0.3770 | 0.0385* | |
| H6A | 1.1124 | 0.4321 | 0.2436 | 0.0329* | |
| H6B | 0.8865 | 0.9338 | 0.2550 | 0.0324* | |
| H8A1 | 1.0793 | 0.1423 | 0.2697 | 0.0391* | |
| H8A2 | 0.8781 | 0.1612 | 0.2966 | 0.0391* | |
| H8B1 | 0.9209 | 0.6437 | 0.2368 | 0.0400* | |
| H8B2 | 1.1217 | 0.6624 | 0.2068 | 0.0400* | |
| H9A1 | 0.9368 | 0.0541 | 0.4216 | 0.0344* | |
| H9A2 | 1.0915 | 0.1297 | 0.4322 | 0.0344* | |
| H9B1 | 1.0579 | 0.5562 | 0.0866 | 0.0340* | |
| H9B2 | 0.9022 | 0.6332 | 0.0756 | 0.0340* | |
| H13A | 0.7958 | 0.0651 | 0.5935 | 0.0341* | |
| H13B | 1.1973 | 0.5665 | −0.0860 | 0.0333* | |
| H15A | 0.5898 | 0.3524 | 0.7002 | 0.0545* | |
| H15B | 1.3918 | 0.8545 | −0.2042 | 0.0505* | |
| H16A | 0.7160 | 0.4394 | 0.5538 | 0.0501* | |
| H16B | 1.2687 | 0.9415 | −0.0606 | 0.0449* | |
| H17A | 0.7475 | 0.0252 | 0.7539 | 0.0475* | |
| H17B | 0.5710 | 0.0026 | 0.7242 | 0.0475* | |
| H17C | 0.5738 | 0.0254 | 0.8359 | 0.0475* | |
| H17D | 1.2455 | 0.5259 | −0.2472 | 0.0479* | |
| H17E | 1.4186 | 0.5047 | −0.2122 | 0.0479* | |
| H17F | 1.4230 | 0.5254 | −0.3259 | 0.0479* |
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1A | 0.0333 (9) | 0.0271 (8) | 0.0320 (8) | −0.0061 (7) | 0.0041 (7) | −0.0016 (7) |
| O1B | 0.0326 (9) | 0.0290 (9) | 0.0324 (9) | 0.0070 (7) | 0.0039 (7) | 0.0002 (7) |
| O2A | 0.0543 (11) | 0.0227 (8) | 0.0368 (9) | 0.0025 (7) | 0.0041 (8) | 0.0012 (7) |
| O2B | 0.0513 (11) | 0.0219 (8) | 0.0369 (9) | −0.0033 (7) | 0.0032 (8) | −0.0018 (7) |
| O3A | 0.0536 (11) | 0.0373 (10) | 0.0307 (9) | 0.0040 (8) | 0.0132 (8) | 0.0055 (8) |
| O3B | 0.0462 (11) | 0.0365 (10) | 0.0317 (9) | −0.0032 (8) | 0.0113 (8) | −0.0075 (7) |
| N1A | 0.0258 (10) | 0.0283 (10) | 0.0302 (10) | −0.0019 (8) | 0.0045 (8) | −0.0008 (8) |
| N1B | 0.0299 (11) | 0.0310 (10) | 0.0289 (10) | 0.0008 (8) | 0.0041 (8) | −0.0011 (8) |
| C1A | 0.0240 (11) | 0.0277 (11) | 0.0298 (11) | 0.0010 (9) | −0.0006 (9) | 0.0012 (9) |
| C1B | 0.0265 (11) | 0.0276 (11) | 0.0268 (11) | −0.0036 (9) | −0.0009 (9) | 0.0001 (9) |
| C2A | 0.0347 (13) | 0.0337 (13) | 0.0334 (12) | 0.0006 (10) | 0.0018 (10) | −0.0062 (10) |
| C2B | 0.0348 (13) | 0.0345 (13) | 0.0300 (12) | −0.0063 (10) | 0.0042 (10) | 0.0039 (10) |
| C3A | 0.0417 (14) | 0.0251 (12) | 0.0331 (12) | −0.0036 (10) | 0.0007 (10) | −0.0022 (10) |
| C3B | 0.0383 (13) | 0.0278 (12) | 0.0328 (12) | 0.0007 (10) | −0.0004 (10) | 0.0027 (10) |
| C4A | 0.0304 (12) | 0.0276 (12) | 0.0314 (12) | −0.0044 (9) | −0.0015 (9) | 0.0035 (9) |
| C4B | 0.0321 (12) | 0.0276 (12) | 0.0334 (12) | 0.0009 (9) | −0.0001 (10) | −0.0067 (10) |
| C5A | 0.0263 (11) | 0.0263 (11) | 0.0267 (11) | −0.0002 (9) | −0.0014 (9) | 0.0031 (9) |
| C5B | 0.0261 (11) | 0.0270 (11) | 0.0283 (11) | −0.0001 (9) | −0.0005 (9) | −0.0047 (9) |
| C6A | 0.0295 (12) | 0.0222 (11) | 0.0285 (11) | −0.0012 (9) | −0.0027 (9) | 0.0006 (9) |
| C6B | 0.0277 (11) | 0.0224 (10) | 0.0283 (11) | 0.0004 (8) | −0.0008 (9) | −0.0002 (9) |
| C7A | 0.0261 (11) | 0.0255 (11) | 0.0286 (11) | 0.0026 (9) | −0.0003 (9) | 0.0003 (9) |
| C7B | 0.0260 (11) | 0.0243 (11) | 0.0272 (11) | −0.0015 (8) | −0.0009 (9) | −0.0006 (9) |
| C8A | 0.0371 (13) | 0.0258 (12) | 0.0283 (12) | 0.0003 (9) | 0.0067 (10) | −0.0010 (9) |
| C8B | 0.0406 (13) | 0.0247 (11) | 0.0289 (12) | −0.0018 (10) | 0.0044 (10) | −0.0016 (9) |
| C9A | 0.0301 (12) | 0.0221 (11) | 0.0300 (11) | 0.0006 (9) | 0.0015 (9) | 0.0003 (9) |
| C9B | 0.0304 (12) | 0.0218 (11) | 0.0300 (11) | −0.0024 (9) | −0.0010 (9) | −0.0005 (9) |
| C10A | 0.0235 (11) | 0.0248 (11) | 0.0276 (11) | 0.0009 (8) | −0.0006 (9) | −0.0010 (9) |
| C10B | 0.0214 (10) | 0.0242 (11) | 0.0274 (11) | −0.0009 (8) | −0.0004 (9) | 0.0001 (9) |
| C11A | 0.0311 (12) | 0.0233 (11) | 0.0285 (11) | 0.0039 (9) | 0.0000 (9) | 0.0009 (9) |
| C11B | 0.0287 (12) | 0.0230 (11) | 0.0292 (11) | −0.0044 (9) | 0.0002 (9) | −0.0028 (9) |
| C12A | 0.0279 (11) | 0.0244 (11) | 0.0293 (11) | 0.0023 (9) | −0.0010 (9) | 0.0017 (9) |
| C12B | 0.0281 (11) | 0.0236 (11) | 0.0300 (11) | −0.0019 (9) | −0.0022 (9) | −0.0024 (9) |
| C13A | 0.0283 (11) | 0.0224 (11) | 0.0316 (11) | 0.0012 (9) | −0.0004 (9) | 0.0008 (9) |
| C13B | 0.0274 (11) | 0.0245 (11) | 0.0290 (11) | −0.0022 (9) | −0.0010 (9) | −0.0027 (9) |
| C14A | 0.0365 (13) | 0.0299 (12) | 0.0279 (11) | 0.0020 (10) | 0.0048 (10) | 0.0029 (10) |
| C14B | 0.0289 (12) | 0.0334 (12) | 0.0273 (11) | −0.0015 (9) | 0.0051 (9) | −0.0054 (10) |
| C15A | 0.0554 (17) | 0.0346 (14) | 0.0347 (13) | 0.0151 (12) | 0.0137 (12) | −0.0008 (11) |
| C15B | 0.0507 (16) | 0.0337 (13) | 0.0319 (13) | −0.0136 (11) | 0.0122 (11) | −0.0006 (10) |
| C16A | 0.0537 (16) | 0.0247 (12) | 0.0368 (13) | 0.0109 (11) | 0.0099 (12) | 0.0027 (10) |
| C16B | 0.0450 (15) | 0.0276 (12) | 0.0333 (12) | −0.0084 (10) | 0.0054 (11) | −0.0029 (10) |
| C17A | 0.0469 (15) | 0.0341 (13) | 0.0335 (13) | −0.0079 (11) | −0.0006 (11) | 0.0078 (10) |
| C17B | 0.0470 (15) | 0.0342 (13) | 0.0355 (13) | 0.0066 (11) | −0.0023 (11) | −0.0078 (11) |
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Geometric parameters (Å, º)
| O1A—C1A | 1.258 (3) | C11B—C12B | 1.480 (3) |
| O1B—C1B | 1.257 (3) | C11B—C16B | 1.405 (3) |
| O2A—C12A | 1.233 (3) | C13A—C14A | 1.385 (3) |
| O2B—C12B | 1.233 (3) | C13B—C14B | 1.388 (3) |
| O3A—C14A | 1.360 (3) | C14A—C15A | 1.400 (4) |
| O3A—C17A | 1.436 (4) | C14B—C15B | 1.395 (4) |
| O3B—C14B | 1.360 (3) | C15A—C16A | 1.374 (4) |
| O3B—C17B | 1.434 (4) | C15B—C16B | 1.369 (4) |
| N1A—C1A | 1.375 (3) | N1A—H1A | 0.96 (3) |
| N1A—C2A | 1.360 (4) | N1B—H1B | 0.98 (3) |
| N1B—C1B | 1.373 (3) | C2A—H2A | 0.950 |
| N1B—C2B | 1.366 (4) | C2B—H2B | 0.950 |
| C1A—C5A | 1.451 (3) | C3A—H3A | 0.950 |
| C1B—C5B | 1.459 (3) | C3B—H3B | 0.950 |
| C2A—C3A | 1.354 (4) | C4A—H4A | 0.950 |
| C2B—C3B | 1.351 (4) | C4B—H4B | 0.950 |
| C3A—C4A | 1.404 (3) | C6A—H6A | 0.950 |
| C3B—C4B | 1.406 (4) | C6B—H6B | 0.950 |
| C4A—C5A | 1.380 (4) | C8A—H8A1 | 0.990 |
| C4B—C5B | 1.381 (4) | C8A—H8A2 | 0.990 |
| C5A—C6A | 1.463 (3) | C8B—H8B1 | 0.990 |
| C5B—C6B | 1.462 (3) | C8B—H8B2 | 0.990 |
| C6A—C7A | 1.349 (3) | C9A—H9A1 | 0.990 |
| C6B—C7B | 1.347 (3) | C9A—H9A2 | 0.990 |
| C7A—C8A | 1.514 (4) | C9B—H9B1 | 0.990 |
| C7A—C12A | 1.496 (3) | C9B—H9B2 | 0.990 |
| C7B—C8B | 1.513 (4) | C13A—H13A | 0.950 |
| C7B—C12B | 1.497 (3) | C13B—H13B | 0.950 |
| C8A—C9A | 1.527 (4) | C15A—H15A | 0.950 |
| C8B—C9B | 1.525 (4) | C15B—H15B | 0.950 |
| C9A—C10A | 1.504 (3) | C16A—H16A | 0.950 |
| C9B—C10B | 1.503 (3) | C16B—H16B | 0.950 |
| C10A—C11A | 1.397 (4) | C17A—H17A | 0.980 |
| C10A—C13A | 1.398 (3) | C17A—H17B | 0.980 |
| C10B—C11B | 1.399 (4) | C17A—H17C | 0.980 |
| C10B—C13B | 1.400 (3) | C17B—H17D | 0.980 |
| C11A—C12A | 1.482 (3) | C17B—H17E | 0.980 |
| C11A—C16A | 1.402 (4) | C17B—H17F | 0.980 |
| O1A···C2A | 3.542 (4) | C11A···H3Bvii | 3.4817 |
| O1A···C6A | 2.812 (4) | C11A···H4Bvii | 3.1490 |
| O1B···C2B | 3.542 (4) | C11B···H2Ai | 3.5524 |
| O1B···C6B | 2.821 (4) | C11B···H3Av | 3.5010 |
| O2A···C6A | 2.754 (4) | C11B···H4Av | 3.1694 |
| O2A···C16A | 2.792 (4) | C12A···H3Bvii | 3.1055 |
| O2B···C6B | 2.750 (4) | C12A···H17Dv | 3.0137 |
| O2B···C16B | 2.798 (4) | C12A···H17Fv | 3.4151 |
| O3B···C16B | 3.599 (4) | C12B···H3Av | 3.2054 |
| N1A···C4A | 2.710 (4) | C12B···H17Avii | 3.0581 |
| N1B···C4B | 2.721 (4) | C12B···H17Cvii | 3.4482 |
| C1A···C3A | 2.819 (4) | C13A···H1Biii | 3.28 (4) |
| C1B···C3B | 2.829 (4) | C13A···H4Bvii | 3.5271 |
| C2A···C5A | 2.790 (4) | C13A···H9A1xi | 3.2812 |
| C2B···C5B | 2.782 (4) | C13A···H9A2xi | 3.4733 |
| C4A···C7A | 3.177 (4) | C13B···H1Ai | 3.25 (3) |
| C4A···C8A | 3.193 (4) | C13B···H4Av | 3.5120 |
| C4B···C7B | 3.167 (4) | C13B···H9B1v | 3.2984 |
| C4B···C8B | 3.187 (5) | C13B···H9B2v | 3.5161 |
| C5A···C8A | 3.201 (4) | C14A···H2Aviii | 3.3823 |
| C5B···C8B | 3.198 (4) | C14A···H4Bvii | 3.5794 |
| C7A···C10A | 2.926 (4) | C14A···H8B2vii | 3.2033 |
| C7B···C10B | 2.932 (4) | C14B···H2Bix | 3.3802 |
| C8A···C11A | 2.887 (4) | C14B···H4Av | 3.5312 |
| C8B···C11B | 2.886 (4) | C14B···H8A2v | 3.1403 |
| C9A···C12A | 2.938 (5) | C15A···H2Aviii | 3.5133 |
| C9B···C12B | 2.932 (5) | C15A···H4Bvii | 3.4446 |
| C10A···C15A | 2.786 (4) | C15A···H8B2vii | 3.0027 |
| C10B···C15B | 2.782 (4) | C15A···H17Eviii | 3.3979 |
| C11A···C14A | 2.788 (4) | C15A···H17Fviii | 3.2020 |
| C11B···C14B | 2.791 (4) | C15B···H2Bix | 3.5065 |
| C13A···C16A | 2.781 (5) | C15B···H4Av | 3.3787 |
| C13A···C17A | 2.823 (4) | C15B···H8A2v | 2.9149 |
| C13B···C16B | 2.783 (5) | C15B···H17Bxii | 3.2749 |
| C13B···C17B | 2.819 (4) | C15B···H17Cxii | 3.2706 |
| O1A···O1Ai | 3.540 (4) | C16A···H3B | 3.2475 |
| O1A···O3Bi | 3.528 (4) | C16A···H4Bvii | 3.2293 |
| O1A···N1Ai | 2.778 (3) | C16A···H17Fviii | 3.3113 |
| O1A···C9B | 3.366 (4) | C16B···H3Aiv | 3.2754 |
| O1A···C17Bi | 3.316 (5) | C16B···H4Av | 3.1988 |
| O1B···O1Bii | 3.556 (4) | C16B···H17Cxii | 3.2954 |
| O1B···O3Aiii | 3.530 (4) | C17A···H2Aviii | 3.1643 |
| O1B···N1Bii | 2.778 (3) | C17A···H8A1xi | 3.2466 |
| O1B···C9Aiv | 3.395 (4) | C17A···H15Bxiii | 2.9788 |
| O1B···C17Aiii | 3.343 (5) | C17A···H16Bxiii | 3.5415 |
| O2A···C3B | 3.431 (4) | C17B···H2Bix | 3.1756 |
| O2A···C4B | 3.262 (5) | C17B···H8B1v | 3.2548 |
| O2A···C17Bv | 3.282 (5) | C17B···H15Aix | 2.9481 |
| O2B···O2Bvi | 3.519 (4) | H1A···O1Ai | 1.82 (3) |
| O2B···C3Aiv | 3.391 (4) | H1A···N1Ai | 2.93 (3) |
| O2B···C4Aiv | 3.263 (5) | H1A···C1Ai | 2.71 (3) |
| O2B···C16Bvi | 3.490 (5) | H1A···C10Bi | 3.36 (3) |
| O2B···C17Avii | 3.300 (5) | H1A···C13Bi | 3.25 (3) |
| O3A···O1Biii | 3.530 (4) | H1A···H1Ai | 2.42 (4) |
| O3A···C2Aviii | 3.269 (4) | H1A···H8B2i | 3.5253 |
| O3B···O1Ai | 3.528 (4) | H1A···H9B1i | 3.5001 |
| O3B···C2Bix | 3.277 (4) | H1A···H9B2v | 3.4543 |
| O3B···C7Av | 3.522 (5) | H1A···H13B | 3.2363 |
| O3B···C12Av | 3.586 (4) | H1A···H13Bi | 3.3392 |
| N1A···O1Ai | 2.778 (3) | H1A···H17E | 2.8349 |
| N1A···C13Bi | 3.411 (5) | H1B···O1Bii | 1.80 (3) |
| N1A···C14Bi | 3.554 (5) | H1B···N1Bii | 2.92 (3) |
| N1B···O1Bii | 2.778 (3) | H1B···C1Bii | 2.70 (3) |
| N1B···C13Aiii | 3.430 (5) | H1B···C10Aiii | 3.36 (3) |
| N1B···C14Aiii | 3.584 (5) | H1B···C13Aiii | 3.28 (4) |
| C1A···C14Bi | 3.494 (5) | H1B···H1Bii | 2.42 (4) |
| C1B···C14Aiii | 3.473 (5) | H1B···H8A2iii | 3.4722 |
| C2A···O3Aix | 3.269 (4) | H1B···H9A1iii | 3.4644 |
| C2B···O3Bviii | 3.277 (4) | H1B···H9A2vii | 3.4365 |
| C3A···O2Bx | 3.391 (4) | H1B···H13Aiv | 3.2562 |
| C3B···O2A | 3.431 (4) | H1B···H13Aiii | 3.3828 |
| C3B···C12Avii | 3.537 (5) | H1B···H15Bviii | 3.5086 |
| C4A···O2Bx | 3.263 (5) | H1B···H17Biv | 2.9558 |
| C4A···C10Bv | 3.345 (5) | H2A···O1Ai | 3.5344 |
| C4A···C11Bv | 3.461 (5) | H2A···O3Aix | 2.3436 |
| C4B···O2A | 3.262 (5) | H2A···C11Bi | 3.5524 |
| C4B···C10Avii | 3.331 (5) | H2A···C14Aix | 3.3823 |
| C4B···C11Avii | 3.440 (5) | H2A···C15Aix | 3.5133 |
| C7A···O3Bv | 3.522 (5) | H2A···C17Aix | 3.1643 |
| C9A···O1Bx | 3.395 (4) | H2A···H8B2i | 3.4372 |
| C9B···O1A | 3.366 (4) | H2A···H9B2v | 3.3194 |
| C10A···C4Bvii | 3.331 (5) | H2A···H15Aix | 2.9548 |
| C10B···C4Av | 3.345 (5) | H2A···H17Cix | 2.9064 |
| C11A···C4Bvii | 3.440 (5) | H2A···H17E | 3.5542 |
| C11B···C4Av | 3.461 (5) | H2B···O1Bii | 3.5356 |
| C12A···O3Bv | 3.586 (4) | H2B···O3Bviii | 2.3441 |
| C12A···C3Bvii | 3.537 (5) | H2B···C14Bviii | 3.3802 |
| C12A···C17Bv | 3.493 (5) | H2B···C15Bviii | 3.5065 |
| C12B···C17Avii | 3.543 (5) | H2B···C17Bviii | 3.1756 |
| C13A···N1Biii | 3.430 (5) | H2B···H8A2iii | 3.4956 |
| C13B···N1Ai | 3.411 (5) | H2B···H9A2vii | 3.3172 |
| C14A···N1Biii | 3.584 (5) | H2B···H15Bviii | 2.9363 |
| C14A···C1Biii | 3.473 (5) | H2B···H17Fviii | 2.9233 |
| C14B···N1Ai | 3.554 (5) | H3A···O2Bx | 2.8622 |
| C14B···C1Ai | 3.494 (5) | H3A···O2Bv | 3.4629 |
| C16B···O2Bvi | 3.490 (5) | H3A···C7Bv | 3.5482 |
| C17A···O1Biii | 3.343 (5) | H3A···C11Bv | 3.5010 |
| C17A···O2Bvii | 3.300 (5) | H3A···C12Bv | 3.2054 |
| C17A···C12Bvii | 3.543 (5) | H3A···C16Bx | 3.2754 |
| C17B···O1Ai | 3.316 (5) | H3A···H9B2v | 3.3943 |
| C17B···O2Av | 3.282 (5) | H3A···H16Bx | 2.3352 |
| C17B···C12Av | 3.493 (5) | H3A···H17Cix | 3.0415 |
| O1A···H1A | 2.43 (3) | H3B···O2A | 2.9423 |
| O1A···H6A | 2.4489 | H3B···O2Avii | 3.3296 |
| O1B···H1B | 2.45 (3) | H3B···C7Avii | 3.4330 |
| O1B···H6B | 2.4642 | H3B···C11Avii | 3.4817 |
| O2A···H6A | 2.3405 | H3B···C12Avii | 3.1055 |
| O2A···H16A | 2.4985 | H3B···C16A | 3.2475 |
| O2B···H6B | 2.3395 | H3B···H9A2vii | 3.3936 |
| O2B···H16B | 2.5041 | H3B···H16A | 2.3163 |
| O3A···H13A | 2.6507 | H3B···H17Fviii | 3.1267 |
| O3A···H15A | 2.4872 | H4A···O2Bx | 2.5858 |
| O3B···H13B | 2.6483 | H4A···C10Bv | 3.3216 |
| O3B···H15B | 2.4886 | H4A···C11Bv | 3.1694 |
| N1A···H3A | 3.2111 | H4A···C13Bv | 3.5120 |
| N1B···H3B | 3.2243 | H4A···C14Bv | 3.5312 |
| C1A···H2A | 3.2667 | H4A···C15Bv | 3.3787 |
| C1A···H4A | 3.2935 | H4A···C16Bv | 3.1988 |
| C1A···H6A | 2.5404 | H4A···H17Axi | 3.3142 |
| C1B···H2B | 3.2618 | H4B···O2A | 2.5865 |
| C1B···H4B | 3.3034 | H4B···C10Avii | 3.3067 |
| C1B···H6B | 2.5497 | H4B···C11Avii | 3.1490 |
| C2A···H4A | 3.2254 | H4B···C13Avii | 3.5271 |
| C2B···H4B | 3.2206 | H4B···C14Avii | 3.5794 |
| C3A···H1A | 3.21 (3) | H4B···C15Avii | 3.4446 |
| C3B···H1B | 3.23 (3) | H4B···C16Avii | 3.2293 |
| C4A···H2A | 3.2394 | H4B···H17Dv | 3.4945 |
| C4A···H6A | 3.3079 | H6A···C8B | 3.2615 |
| C4A···H8A1 | 2.5782 | H6A···C9B | 3.4562 |
| C4A···H8A2 | 3.3840 | H6A···H8B1 | 3.0246 |
| C4B···H2B | 3.2327 | H6A···H8B2 | 2.8674 |
| C4B···H6B | 3.3041 | H6A···H9B1 | 2.7650 |
| C4B···H8B1 | 2.5740 | H6A···H17Dv | 2.9212 |
| C4B···H8B2 | 3.3796 | H6B···C8Aiv | 3.2921 |
| C5A···H1A | 3.26 (3) | H6B···C9Aiv | 3.5559 |
| C5A···H3A | 3.2879 | H6B···H8A1iv | 3.0467 |
| C5A···H8A1 | 2.8540 | H6B···H8A2iv | 2.8645 |
| C5A···H8A2 | 3.5979 | H6B···H9A1iv | 2.8936 |
| C5B···H1B | 3.29 (3) | H6B···H15Bvi | 3.5873 |
| C5B···H3B | 3.2916 | H6B···H16Bvi | 3.5601 |
| C5B···H8B1 | 2.8515 | H6B···H17Avii | 2.9790 |
| C5B···H8B2 | 3.5983 | H8A1···O2Bx | 3.1964 |
| C6A···H4A | 2.7250 | H8A1···C17Axi | 3.2466 |
| C6A···H8A1 | 2.6674 | H8A1···H6Bx | 3.0467 |
| C6A···H8A2 | 3.0367 | H8A1···H13Axi | 3.4374 |
| C6B···H4B | 2.7201 | H8A1···H17Axi | 2.4631 |
| C6B···H8B1 | 2.6704 | H8A1···H17Bxi | 3.3408 |
| C6B···H8B2 | 3.0366 | H8A1···H17Cxi | 3.5252 |
| C7A···H4A | 2.9547 | H8A2···O1Bx | 2.9375 |
| C7A···H9A1 | 3.3699 | H8A2···O3Bv | 3.2070 |
| C7A···H9A2 | 2.8359 | H8A2···C14Bv | 3.1403 |
| C7B···H4B | 2.9446 | H8A2···C15Bv | 2.9149 |
| C7B···H9B1 | 3.3674 | H8A2···H1Biii | 3.4722 |
| C7B···H9B2 | 2.8279 | H8A2···H2Biii | 3.4956 |
| C8A···H4A | 2.5955 | H8A2···H6Bx | 2.8645 |
| C8A···H6A | 3.3662 | H8A2···H15Bv | 2.7849 |
| C8B···H4B | 2.5908 | H8B1···O2A | 3.0557 |
| C8B···H6B | 3.3663 | H8B1···C17Bv | 3.2548 |
| C9A···H13A | 2.6607 | H8B1···H6A | 3.0246 |
| C9B···H13B | 2.6681 | H8B1···H17Dv | 2.4760 |
| C10A···H8A1 | 3.3531 | H8B1···H17Ev | 3.3942 |
| C10A···H8A2 | 2.8177 | H8B1···H17Fv | 3.4716 |
| C10A···H16A | 3.2727 | H8B2···O1A | 2.9740 |
| C10B···H8B1 | 3.3536 | H8B2···O3Avii | 3.2863 |
| C10B···H8B2 | 2.8138 | H8B2···C14Avii | 3.2033 |
| C10B···H16B | 3.2734 | H8B2···C15Avii | 3.0027 |
| C11A···H8A2 | 3.1634 | H8B2···H1Ai | 3.5253 |
| C11A···H9A1 | 3.2766 | H8B2···H2Ai | 3.4372 |
| C11A···H9A2 | 2.9629 | H8B2···H6A | 2.8674 |
| C11A···H13A | 3.2772 | H8B2···H15Avii | 2.9180 |
| C11A···H15A | 3.2692 | H9A1···O1Bx | 2.7271 |
| C11B···H8B2 | 3.1615 | H9A1···C1Bx | 2.9439 |
| C11B···H9B1 | 3.2767 | H9A1···C5Bx | 3.2986 |
| C11B···H9B2 | 2.9529 | H9A1···C6Bx | 3.3610 |
| C11B···H13B | 3.2834 | H9A1···C9Axi | 3.3310 |
| C11B···H15B | 3.2680 | H9A1···C13Axi | 3.2812 |
| C12A···H6A | 2.4573 | H9A1···H1Biii | 3.4644 |
| C12A···H8A1 | 3.3660 | H9A1···H6Bx | 2.8936 |
| C12A···H8A2 | 2.9520 | H9A1···H9A1xi | 2.9166 |
| C12A···H9A2 | 3.2682 | H9A1···H9A2xi | 2.9945 |
| C12A···H16A | 2.6312 | H9A1···H13Axi | 2.5694 |
| C12B···H6B | 2.4593 | H9A1···H17Axi | 3.2732 |
| C12B···H8B1 | 3.3639 | H9A2···N1Bvii | 2.9853 |
| C12B···H8B2 | 2.9405 | H9A2···C1Bvii | 3.2133 |
| C12B···H9B2 | 3.2609 | H9A2···C2Bvii | 2.8657 |
| C12B···H16B | 2.6379 | H9A2···C3Bvii | 2.9221 |
| C13A···H9A1 | 2.6018 | H9A2···C4Bvii | 3.1002 |
| C13A···H9A2 | 2.9143 | H9A2···C5Bvii | 3.2826 |
| C13A···H15A | 3.2690 | H9A2···C13Axi | 3.4733 |
| C13A···H17A | 2.6943 | H9A2···H1Bvii | 3.4365 |
| C13A···H17B | 2.8283 | H9A2···H2Bvii | 3.3172 |
| C13B···H9B1 | 2.6015 | H9A2···H3Bvii | 3.3936 |
| C13B···H9B2 | 2.9233 | H9A2···H9A1xi | 2.9945 |
| C13B···H15B | 3.2673 | H9A2···H13Axi | 2.5567 |
| C13B···H17D | 2.7230 | H9A2···H17Axi | 3.2760 |
| C13B···H17E | 2.7890 | H9A2···H17Bxi | 3.4983 |
| C14A···H16A | 3.2596 | H9B1···O1A | 2.6898 |
| C14A···H17A | 2.6029 | H9B1···C1A | 2.9602 |
| C14A···H17B | 2.6673 | H9B1···C5A | 3.2785 |
| C14A···H17C | 3.2037 | H9B1···C6A | 3.2624 |
| C14B···H16B | 3.2545 | H9B1···C9Bv | 3.5565 |
| C14B···H17D | 2.6222 | H9B1···C13Bv | 3.2984 |
| C14B···H17E | 2.6492 | H9B1···H1Ai | 3.5001 |
| C14B···H17F | 3.2050 | H9B1···H6A | 2.7650 |
| C15A···H13A | 3.2711 | H9B1···H9B1v | 3.1392 |
| C15B···H13B | 3.2702 | H9B1···H9B2v | 3.2489 |
| C17A···H13A | 2.5215 | H9B1···H13Bv | 2.5684 |
| C17B···H13B | 2.5167 | H9B1···H17Dv | 3.0913 |
| H1A···H2A | 2.2992 | H9B2···N1Av | 3.0336 |
| H1B···H2B | 2.2974 | H9B2···C1Av | 3.2985 |
| H2A···H3A | 2.3127 | H9B2···C2Av | 2.8904 |
| H2B···H3B | 2.3064 | H9B2···C3Av | 2.9447 |
| H3A···H4A | 2.3474 | H9B2···C4Av | 3.1532 |
| H3B···H4B | 2.3525 | H9B2···C5Av | 3.3625 |
| H4A···H8A1 | 1.9890 | H9B2···C13Bv | 3.5161 |
| H4A···H8A2 | 2.6223 | H9B2···H1Av | 3.4543 |
| H4B···H8B1 | 1.9888 | H9B2···H2Av | 3.3194 |
| H4B···H8B2 | 2.6163 | H9B2···H3Av | 3.3943 |
| H6A···H8A1 | 3.5813 | H9B2···H9B1v | 3.2489 |
| H6B···H8B1 | 3.5854 | H9B2···H13Bv | 2.5824 |
| H8A1···H9A1 | 2.4195 | H9B2···H17Dv | 3.0969 |
| H8A1···H9A2 | 2.3025 | H9B2···H17Ev | 3.3127 |
| H8A2···H9A1 | 2.3013 | H13A···O1Biii | 3.5115 |
| H8A2···H9A2 | 2.8577 | H13A···N1Bx | 3.4617 |
| H8B1···H9B1 | 2.4135 | H13A···C9Axi | 2.9967 |
| H8B1···H9B2 | 2.3030 | H13A···H1Bx | 3.2562 |
| H8B2···H9B1 | 2.3020 | H13A···H1Biii | 3.3828 |
| H8B2···H9B2 | 2.8566 | H13A···H8A1xi | 3.4374 |
| H9A1···H13A | 2.4265 | H13A···H9A1xi | 2.5694 |
| H9A2···H13A | 2.9957 | H13A···H9A2xi | 2.5567 |
| H9B1···H13B | 2.4275 | H13B···O1A | 3.5000 |
| H9B2···H13B | 3.0136 | H13B···O1Ai | 3.5656 |
| H13A···H17A | 2.2374 | H13B···N1A | 3.4371 |
| H13A···H17B | 2.3898 | H13B···C1A | 3.5271 |
| H13A···H17C | 3.4914 | H13B···C9Bv | 3.0242 |
| H13B···H17D | 2.2690 | H13B···H1A | 3.2363 |
| H13B···H17E | 2.3465 | H13B···H1Ai | 3.3392 |
| H13B···H17F | 3.4883 | H13B···H9B1v | 2.5684 |
| H15A···H16A | 2.3198 | H13B···H9B2v | 2.5824 |
| H15B···H16B | 2.3119 | H15A···O1Avii | 3.3651 |
| O1A···H1Ai | 1.82 (3) | H15A···C17Bviii | 2.9481 |
| O1A···H2Ai | 3.5344 | H15A···H2Aviii | 2.9548 |
| O1A···H8B2 | 2.9740 | H15A···H8B2vii | 2.9180 |
| O1A···H9B1 | 2.6898 | H15A···H17Dviii | 3.4425 |
| O1A···H13B | 3.5000 | H15A···H17Eviii | 2.4894 |
| O1A···H13Bi | 3.5656 | H15A···H17Fviii | 2.5707 |
| O1A···H15Avii | 3.3651 | H15B···O1Bvi | 3.1705 |
| O1A···H17Ei | 2.4719 | H15B···C17Axii | 2.9788 |
| O1B···H1Bii | 1.80 (3) | H15B···H1Bix | 3.5086 |
| O1B···H2Bii | 3.5356 | H15B···H2Bix | 2.9363 |
| O1B···H8A2iv | 2.9375 | H15B···H6Bvi | 3.5873 |
| O1B···H9A1iv | 2.7271 | H15B···H8A2v | 2.7849 |
| O1B···H13Aiii | 3.5115 | H15B···H17Axii | 3.5094 |
| O1B···H15Bvi | 3.1705 | H15B···H17Bxii | 2.3967 |
| O1B···H17Biii | 2.4912 | H15B···H17Cxii | 2.7158 |
| O2A···H3B | 2.9423 | H16A···O2Avii | 3.4235 |
| O2A···H3Bvii | 3.3296 | H16A···C3B | 3.1966 |
| O2A···H4B | 2.5865 | H16A···H3B | 2.3163 |
| O2A···H8B1 | 3.0557 | H16A···H17Fviii | 2.7900 |
| O2A···H16Avii | 3.4235 | H16B···O2Bvi | 3.1853 |
| O2A···H17Dv | 2.6679 | H16B···C3Aiv | 3.2609 |
| O2A···H17Fv | 3.1116 | H16B···C17Axii | 3.5415 |
| O2B···H3Aiv | 2.8622 | H16B···H3Aiv | 2.3352 |
| O2B···H3Av | 3.4629 | H16B···H6Bvi | 3.5601 |
| O2B···H4Aiv | 2.5858 | H16B···H17Bxii | 3.4993 |
| O2B···H8A1iv | 3.1964 | H16B···H17Cxii | 2.7607 |
| O2B···H16Bvi | 3.1853 | H17A···O2Bvii | 2.6864 |
| O2B···H17Avii | 2.6864 | H17A···C6Bvii | 3.2234 |
| O2B···H17Cvii | 3.1185 | H17A···C7Bvii | 3.2767 |
| O3A···H2Aviii | 2.3436 | H17A···C8Axi | 3.3019 |
| O3A···H8B2vii | 3.2863 | H17A···C9Axi | 3.4878 |
| O3B···H2Bix | 2.3441 | H17A···C12Bvii | 3.0581 |
| O3B···H8A2v | 3.2070 | H17A···H4Axi | 3.3142 |
| N1A···H1Ai | 2.93 (3) | H17A···H6Bvii | 2.9790 |
| N1A···H9B2v | 3.0336 | H17A···H8A1xi | 2.4631 |
| N1A···H13B | 3.4371 | H17A···H9A1xi | 3.2732 |
| N1A···H17E | 3.3986 | H17A···H9A2xi | 3.2760 |
| N1B···H1Bii | 2.92 (3) | H17A···H15Bxiii | 3.5094 |
| N1B···H9A2vii | 2.9853 | H17B···O1Biii | 2.4912 |
| N1B···H13Aiv | 3.4617 | H17B···N1Bx | 3.5354 |
| N1B···H17Biv | 3.5354 | H17B···C1Biii | 3.2559 |
| C1A···H1Ai | 2.71 (3) | H17B···C15Bxiii | 3.2749 |
| C1A···H9B1 | 2.9602 | H17B···H1Bx | 2.9558 |
| C1A···H9B2v | 3.2985 | H17B···H8A1xi | 3.3408 |
| C1A···H13B | 3.5271 | H17B···H9A2xi | 3.4983 |
| C1A···H17Ei | 3.2226 | H17B···H15Bxiii | 2.3967 |
| C1B···H1Bii | 2.70 (3) | H17B···H16Bxiii | 3.4993 |
| C1B···H9A1iv | 2.9439 | H17C···O2Bvii | 3.1185 |
| C1B···H9A2vii | 3.2133 | H17C···C2Aviii | 3.5271 |
| C1B···H17Biii | 3.2559 | H17C···C3Aviii | 3.5886 |
| C2A···H9B2v | 2.8904 | H17C···C12Bvii | 3.4482 |
| C2A···H17Cix | 3.5271 | H17C···C15Bxiii | 3.2706 |
| C2B···H9A2vii | 2.8657 | H17C···C16Bxiii | 3.2954 |
| C2B···H17Fviii | 3.5513 | H17C···H2Aviii | 2.9064 |
| C3A···H9B2v | 2.9447 | H17C···H3Aviii | 3.0415 |
| C3A···H16Bx | 3.2609 | H17C···H8A1xi | 3.5252 |
| C3A···H17Cix | 3.5886 | H17C···H15Bxiii | 2.7158 |
| C3B···H9A2vii | 2.9221 | H17C···H16Bxiii | 2.7607 |
| C3B···H16A | 3.1966 | H17D···O2Av | 2.6679 |
| C4A···H9B2v | 3.1532 | H17D···C6Av | 3.1614 |
| C4B···H9A2vii | 3.1002 | H17D···C7Av | 3.2187 |
| C5A···H9B1 | 3.2785 | H17D···C8Bv | 3.2661 |
| C5A···H9B2v | 3.3625 | H17D···C9Bv | 3.3347 |
| C5B···H9A1iv | 3.2986 | H17D···C12Av | 3.0137 |
| C5B···H9A2vii | 3.2826 | H17D···H4Bv | 3.4945 |
| C6A···H9B1 | 3.2624 | H17D···H6Av | 2.9212 |
| C6A···H17Dv | 3.1614 | H17D···H8B1v | 2.4760 |
| C6B···H9A1iv | 3.3610 | H17D···H9B1v | 3.0913 |
| C6B···H17Avii | 3.2234 | H17D···H9B2v | 3.0969 |
| C7A···H3Bvii | 3.4330 | H17D···H15Aix | 3.4425 |
| C7A···H17Dv | 3.2187 | H17E···O1Ai | 2.4719 |
| C7B···H3Av | 3.5482 | H17E···N1A | 3.3986 |
| C7B···H17Avii | 3.2767 | H17E···C1Ai | 3.2226 |
| C8A···H6Bx | 3.2921 | H17E···C15Aix | 3.3979 |
| C8A···H17Axi | 3.3019 | H17E···H1A | 2.8349 |
| C8B···H6A | 3.2615 | H17E···H2A | 3.5542 |
| C8B···H17Dv | 3.2661 | H17E···H8B1v | 3.3942 |
| C9A···H6Bx | 3.5559 | H17E···H9B2v | 3.3127 |
| C9A···H9A1xi | 3.3310 | H17E···H15Aix | 2.4894 |
| C9A···H13Axi | 2.9967 | H17F···O2Av | 3.1116 |
| C9A···H17Axi | 3.4878 | H17F···C2Bix | 3.5513 |
| C9B···H6A | 3.4562 | H17F···C12Av | 3.4151 |
| C9B···H9B1v | 3.5565 | H17F···C15Aix | 3.2020 |
| C9B···H13Bv | 3.0242 | H17F···C16Aix | 3.3113 |
| C9B···H17Dv | 3.3347 | H17F···H2Bix | 2.9233 |
| C10A···H1Biii | 3.36 (3) | H17F···H3Bix | 3.1267 |
| C10A···H4Bvii | 3.3067 | H17F···H8B1v | 3.4716 |
| C10B···H1Ai | 3.36 (3) | H17F···H15Aix | 2.5707 |
| C10B···H4Av | 3.3216 | H17F···H16Aix | 2.7900 |
| C14A—O3A—C17A | 117.87 (17) | C1A—N1A—H1A | 116.4 (16) |
| C14B—O3B—C17B | 118.04 (17) | C2A—N1A—H1A | 118.6 (16) |
| C1A—N1A—C2A | 124.77 (18) | C1B—N1B—H1B | 117.6 (16) |
| C1B—N1B—C2B | 124.09 (19) | C2B—N1B—H1B | 118.2 (16) |
| O1A—C1A—N1A | 119.19 (18) | N1A—C2A—H2A | 120.047 |
| O1A—C1A—C5A | 124.94 (19) | C3A—C2A—H2A | 120.062 |
| N1A—C1A—C5A | 115.9 (2) | N1B—C2B—H2B | 119.577 |
| O1B—C1B—N1B | 119.22 (18) | C3B—C2B—H2B | 119.570 |
| O1B—C1B—C5B | 124.91 (19) | C2A—C3A—H3A | 120.562 |
| N1B—C1B—C5B | 115.87 (19) | C4A—C3A—H3A | 120.571 |
| N1A—C2A—C3A | 119.9 (2) | C2B—C3B—H3B | 120.798 |
| N1B—C2B—C3B | 120.9 (2) | C4B—C3B—H3B | 120.786 |
| C2A—C3A—C4A | 118.9 (3) | C3A—C4A—H4A | 118.985 |
| C2B—C3B—C4B | 118.4 (3) | C5A—C4A—H4A | 118.981 |
| C3A—C4A—C5A | 122.0 (2) | C3B—C4B—H4B | 118.977 |
| C3B—C4B—C5B | 122.0 (2) | C5B—C4B—H4B | 118.991 |
| C1A—C5A—C4A | 118.51 (19) | C5A—C6A—H6A | 114.725 |
| C1A—C5A—C6A | 115.3 (2) | C7A—C6A—H6A | 114.724 |
| C4A—C5A—C6A | 126.10 (19) | C5B—C6B—H6B | 114.869 |
| C1B—C5B—C4B | 118.65 (19) | C7B—C6B—H6B | 114.869 |
| C1B—C5B—C6B | 115.41 (19) | C7A—C8A—H8A1 | 108.953 |
| C4B—C5B—C6B | 125.82 (19) | C7A—C8A—H8A2 | 108.949 |
| C5A—C6A—C7A | 130.6 (2) | C9A—C8A—H8A1 | 108.951 |
| C5B—C6B—C7B | 130.3 (2) | C9A—C8A—H8A2 | 108.949 |
| C6A—C7A—C8A | 126.42 (19) | H8A1—C8A—H8A2 | 107.768 |
| C6A—C7A—C12A | 116.3 (2) | C7B—C8B—H8B1 | 108.959 |
| C8A—C7A—C12A | 117.25 (17) | C7B—C8B—H8B2 | 108.961 |
| C6B—C7B—C8B | 126.74 (19) | C9B—C8B—H8B1 | 108.958 |
| C6B—C7B—C12B | 116.4 (2) | C9B—C8B—H8B2 | 108.957 |
| C8B—C7B—C12B | 116.84 (18) | H8B1—C8B—H8B2 | 107.760 |
| C7A—C8A—C9A | 113.13 (19) | C8A—C9A—H9A1 | 109.091 |
| C7B—C8B—C9B | 113.11 (19) | C8A—C9A—H9A2 | 109.092 |
| C8A—C9A—C10A | 112.54 (19) | C10A—C9A—H9A1 | 109.085 |
| C8B—C9B—C10B | 112.72 (19) | C10A—C9A—H9A2 | 109.087 |
| C9A—C10A—C11A | 120.40 (18) | H9A1—C9A—H9A2 | 107.832 |
| C9A—C10A—C13A | 119.5 (2) | C8B—C9B—H9B1 | 109.041 |
| C11A—C10A—C13A | 119.94 (19) | C8B—C9B—H9B2 | 109.049 |
| C9B—C10B—C11B | 120.00 (18) | C10B—C9B—H9B1 | 109.055 |
| C9B—C10B—C13B | 119.72 (19) | C10B—C9B—H9B2 | 109.048 |
| C11B—C10B—C13B | 120.15 (18) | H9B1—C9B—H9B2 | 107.811 |
| C10A—C11A—C12A | 121.74 (18) | C10A—C13A—H13A | 120.007 |
| C10A—C11A—C16A | 119.34 (19) | C14A—C13A—H13A | 120.000 |
| C12A—C11A—C16A | 118.8 (2) | C10B—C13B—H13B | 120.131 |
| C10B—C11B—C12B | 121.83 (18) | C14B—C13B—H13B | 120.134 |
| C10B—C11B—C16B | 118.97 (19) | C14A—C15A—H15A | 120.013 |
| C12B—C11B—C16B | 119.2 (2) | C16A—C15A—H15A | 120.010 |
| O2A—C12A—C7A | 121.79 (19) | C14B—C15B—H15B | 119.841 |
| O2A—C12A—C11A | 120.46 (19) | C16B—C15B—H15B | 119.843 |
| C7A—C12A—C11A | 117.75 (19) | C11A—C16A—H16A | 119.693 |
| O2B—C12B—C7B | 121.47 (19) | C15A—C16A—H16A | 119.690 |
| O2B—C12B—C11B | 120.49 (18) | C11B—C16B—H16B | 119.642 |
| C7B—C12B—C11B | 118.04 (19) | C15B—C16B—H16B | 119.641 |
| C10A—C13A—C14A | 120.0 (2) | O3A—C17A—H17A | 109.475 |
| C10B—C13B—C14B | 119.7 (2) | O3A—C17A—H17B | 109.477 |
| O3A—C14A—C13A | 124.7 (2) | O3A—C17A—H17C | 109.471 |
| O3A—C14A—C15A | 115.17 (19) | H17A—C17A—H17B | 109.471 |
| C13A—C14A—C15A | 120.1 (2) | H17A—C17A—H17C | 109.464 |
| O3B—C14B—C13B | 124.3 (2) | H17B—C17A—H17C | 109.469 |
| O3B—C14B—C15B | 115.61 (19) | O3B—C17B—H17D | 109.470 |
| C13B—C14B—C15B | 120.1 (2) | O3B—C17B—H17E | 109.468 |
| C14A—C15A—C16A | 120.0 (3) | O3B—C17B—H17F | 109.473 |
| C14B—C15B—C16B | 120.3 (2) | H17D—C17B—H17E | 109.466 |
| C11A—C16A—C15A | 120.6 (3) | H17D—C17B—H17F | 109.473 |
| C11B—C16B—C15B | 120.7 (3) | H17E—C17B—H17F | 109.478 |
| C17A—O3A—C14A—C13A | −0.9 (4) | C8B—C7B—C12B—O2B | −169.3 (2) |
| C17A—O3A—C14A—C15A | 179.70 (19) | C8B—C7B—C12B—C11B | 10.0 (3) |
| C17B—O3B—C14B—C13B | −1.2 (4) | C12B—C7B—C8B—C9B | −38.7 (3) |
| C17B—O3B—C14B—C15B | 179.56 (19) | C7A—C8A—C9A—C10A | 48.0 (3) |
| C1A—N1A—C2A—C3A | 1.6 (4) | C7B—C8B—C9B—C10B | 48.6 (3) |
| C2A—N1A—C1A—O1A | −178.9 (2) | C8A—C9A—C10A—C11A | −30.5 (3) |
| C2A—N1A—C1A—C5A | −0.2 (4) | C8A—C9A—C10A—C13A | 153.73 (18) |
| C1B—N1B—C2B—C3B | 1.2 (4) | C8B—C9B—C10B—C11B | −31.3 (3) |
| C2B—N1B—C1B—O1B | −178.9 (2) | C8B—C9B—C10B—C13B | 152.79 (18) |
| C2B—N1B—C1B—C5B | 0.6 (4) | C9A—C10A—C11A—C12A | 0.7 (4) |
| O1A—C1A—C5A—C4A | 176.6 (2) | C9A—C10A—C11A—C16A | −176.02 (19) |
| O1A—C1A—C5A—C6A | −0.5 (4) | C9A—C10A—C13A—C14A | 175.01 (18) |
| N1A—C1A—C5A—C4A | −2.1 (3) | C11A—C10A—C13A—C14A | −0.8 (4) |
| N1A—C1A—C5A—C6A | −179.12 (17) | C13A—C10A—C11A—C12A | 176.46 (19) |
| O1B—C1B—C5B—C4B | 176.5 (2) | C13A—C10A—C11A—C16A | −0.3 (4) |
| O1B—C1B—C5B—C6B | 0.4 (4) | C9B—C10B—C11B—C12B | 2.2 (4) |
| N1B—C1B—C5B—C4B | −2.9 (3) | C9B—C10B—C11B—C16B | −175.18 (18) |
| N1B—C1B—C5B—C6B | −179.10 (18) | C9B—C10B—C13B—C14B | 174.82 (18) |
| N1A—C2A—C3A—C4A | −0.5 (4) | C11B—C10B—C13B—C14B | −1.0 (4) |
| N1B—C2B—C3B—C4B | −0.7 (4) | C13B—C10B—C11B—C12B | 178.08 (19) |
| C2A—C3A—C4A—C5A | −1.9 (4) | C13B—C10B—C11B—C16B | 0.7 (4) |
| C2B—C3B—C4B—C5B | −1.8 (4) | C10A—C11A—C12A—O2A | −169.6 (2) |
| C3A—C4A—C5A—C1A | 3.2 (4) | C10A—C11A—C12A—C7A | 11.0 (4) |
| C3A—C4A—C5A—C6A | 179.8 (2) | C10A—C11A—C16A—C15A | 0.7 (4) |
| C3B—C4B—C5B—C1B | 3.6 (4) | C12A—C11A—C16A—C15A | −176.1 (2) |
| C3B—C4B—C5B—C6B | 179.3 (2) | C16A—C11A—C12A—O2A | 7.2 (4) |
| C1A—C5A—C6A—C7A | −163.1 (2) | C16A—C11A—C12A—C7A | −172.2 (2) |
| C4A—C5A—C6A—C7A | 20.2 (4) | C10B—C11B—C12B—O2B | −171.6 (2) |
| C1B—C5B—C6B—C7B | −162.5 (2) | C10B—C11B—C12B—C7B | 9.2 (3) |
| C4B—C5B—C6B—C7B | 21.6 (4) | C10B—C11B—C16B—C15B | 0.0 (4) |
| C5A—C6A—C7A—C8A | 1.5 (4) | C12B—C11B—C16B—C15B | −177.4 (2) |
| C5A—C6A—C7A—C12A | −177.9 (2) | C16B—C11B—C12B—O2B | 5.8 (4) |
| C5B—C6B—C7B—C8B | 0.6 (4) | C16B—C11B—C12B—C7B | −173.4 (2) |
| C5B—C6B—C7B—C12B | −178.0 (2) | C10A—C13A—C14A—O3A | −178.1 (2) |
| C6A—C7A—C8A—C9A | 142.6 (3) | C10A—C13A—C14A—C15A | 1.3 (4) |
| C6A—C7A—C12A—O2A | 8.7 (4) | C10B—C13B—C14B—O3B | −178.5 (2) |
| C6A—C7A—C12A—C11A | −171.92 (19) | C10B—C13B—C14B—C15B | 0.7 (4) |
| C8A—C7A—C12A—O2A | −170.8 (2) | O3A—C14A—C15A—C16A | 178.6 (2) |
| C8A—C7A—C12A—C11A | 8.6 (3) | C13A—C14A—C15A—C16A | −0.9 (4) |
| C12A—C7A—C8A—C9A | −37.9 (3) | O3B—C14B—C15B—C16B | 179.3 (2) |
| C6B—C7B—C8B—C9B | 142.7 (3) | C13B—C14B—C15B—C16B | 0.0 (4) |
| C6B—C7B—C12B—O2B | 9.5 (4) | C14A—C15A—C16A—C11A | −0.2 (4) |
| C6B—C7B—C12B—C11B | −171.26 (19) | C14B—C15B—C16B—C11B | −0.4 (4) |
Symmetry codes: (i) −x+3, −y+1, −z; (ii) −x+1, −y+2, −z+1; (iii) −x+1, −y+1, −z+1; (iv) x, y+1, z; (v) −x+2, −y+1, −z; (vi) −x+2, −y+2, −z; (vii) −x+2, −y+1, −z+1; (viii) x−1, y, z+1; (ix) x+1, y, z−1; (x) x, y−1, z; (xi) −x+2, −y, −z+1; (xii) x+1, y+1, z−1; (xiii) x−1, y−1, z+1.
(II) 3-[(E)-(6-Methoxy-1-oxo-1,2,3,4-tetrahydronaphthalen-2-ylidene)methyl]pyridin-2(1H)-one . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1A—H1A···O1Ai | 0.96 (3) | 1.82 (3) | 2.778 (3) | 178 (3) |
| N1B—H1B···O1Bii | 0.98 (3) | 1.80 (3) | 2.778 (3) | 176 (3) |
Symmetry codes: (i) −x+3, −y+1, −z; (ii) −x+1, −y+2, −z+1.
References
- Bandgar, B. P., Gawande, S. S., Bodade, R. G., Totre, J. V. & Khobragade, C. N. (2010). Bioorg. Med. Chem. 18, 1364–1370. [DOI] [PubMed]
- Dimmock, J. R., Zello, G. A., Oloo, E. O., Quail, J. W., Kraatz, H. B., Perjési, P., Aradi, F., Takács-Novák, K., Allen, T. M., Santos, C. L., Balzarini, J., De Clercq, E. & Stables, J. P. (2002). J. Med. Chem. 45, 3103–3111. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst B72, 171–179. [DOI] [PMC free article] [PubMed]
- Katsori, A. M. & Hadjipavlou-Litina, D. (2011). Expert Opin. Ther. Pat. 21, 1575–1596. [DOI] [PubMed]
- Nowakowska, Z. (2007). Eur. J. Med. Chem. 42, 125–137. [DOI] [PubMed]
- Prasad, Y. R., Kumar, P. P., Kumar, P. R. & Rao, A. S. (2008). E-J. Chem. 5, 144–148.
- Rigaku (1998). REQAB. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2011). Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Shenvi, S., Kumar, K., Hatti, K. S., Rijesh, K., Diwakar, L. & Reddy, G. C. (2013). Eur. J. Med. Chem. 62, 435–442. [DOI] [PubMed]
- Yee, S. W., Jarno, L., Gomaa, M. S., Elford, C., Ooi, L. L., Coogan, M. P., McClelland, R., Nicholson, R. I., Evans, B. A. J., Brancale, A. & Simons, C. (2005). J. Med. Chem. 48, 7123–7131. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) General, I, II. DOI: 10.1107/S2056989016009300/bg2587sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989016009300/bg2587Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S2056989016009300/bg2587IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report








