ABSTRACT
Class 1 integrons are genetic systems that enable bacteria to capture and express gene cassettes. These integrons, when isolated in clinical contexts, most often carry antibiotic resistance gene cassettes. They play a major role in the dissemination of antibiotic resistance among Gram-negative bacteria. The key element of integrons is the integrase, which allows gene cassettes to be acquired and shuffled. Planktonic culture experiments have shown that integrase expression is regulated by the bacterial SOS response. In natural settings, however, bacteria generally live in biofilms, which are characterized by strong antibiotic resilience and by increased expression of stress-related genes. Here, we report that under biofilm conditions, the stringent response, which is induced upon starvation, (i) increases basal integrase and SOS regulon gene expression via induction of the SOS response and (ii) exerts biofilm-specific regulation of the integrase via the Lon protease. This indicates that biofilm environments favor integron-mediated acquisition of antibiotic resistance and other adaptive functions encoded by gene cassettes.
IMPORTANCE
Multidrug-resistant bacteria are becoming a worldwide health problem. Integrons are bacterial genetic platforms that allow the bacteria to capture and express gene cassettes. In clinical settings, integrons play a major role in the dissemination of antibiotic resistance gene cassettes among Gram-negative bacteria. Cassette capture is catalyzed by the integron integrase, whose expression is induced by DNA damage and controlled by the bacterial SOS response in laboratory planktonic cultures. In natural settings, bacteria usually grow in heterogeneous environments known as biofilms, which have very different conditions than planktonic cultures. Integrase regulation has not been investigated in biofilms. Our results showed that in addition to the SOS response, the stringent response (induced upon starvation) is specifically involved in the regulation of class 1 integron integrases in biofilms. This study shows that biofilms are favorable environments for integron-mediated acquisition/exchange of antibiotic resistance genes by bacteria and for the emergence of multidrug-resistant bacteria.
INTRODUCTION
Antibacterial drugs are one of the most important therapeutic advances in medical history, but bacterial resistance has increased dramatically over the last decade. Multidrug-resistant (MDR) Gram-negative bacteria are spreading worldwide and are becoming a major public health issue. Clinicians are now dealing with infections for which very few effective antibiotics are available. The question, therefore, is how to resist resistance and thereby preserve the effectiveness of existing antibiotics. In addition to preventing antibiotic overuse, we urgently need to better understand how bacteria acquire and disseminate determinants of antibiotic resistance (1, 2).
Along with transposons and plasmids, integrons are important genetic elements involved in the dissemination of antibiotic resistance among Gram-negative bacteria (3, 4). The integron’s functional platform is composed of a gene encoding an integron integrase, intI, a specific recombination site, attI, and a promoter, Pc, which controls the expression of promoterless genes embedded within gene cassettes (5). The integrase catalyzes gene cassette insertion and excision through site-specific RecA-independent recombination (6). Hundreds of classes of integrons have been described on the basis of the amino acid sequence of the IntI protein; they are found in all ecosystems (human, animal, and environment), providing to bacteria multiple adaptive functions (7–9). In clinical settings, five classes of integrons have been described, which mainly contain antibiotic resistance gene cassettes (7). The class 1 integrons are those most commonly encountered in human commensals and pathogens. Integrons containing antibiotic resistance gene cassettes are usually located on mobile genetic elements (plasmids or transposons) (10). More than 130 gene cassettes have been described, conferring resistance to almost all antibiotic classes (11).
Integrase expression is regulated by the bacterial SOS response (12). This coordinated response to DNA damage requires a repressor, LexA, and a sensor/activator, RecA (13, 14). During normal bacterial growth, LexA is bound at attachment sites (SOS boxes) in the promoter region of genes of the SOS regulon, which comprises at least 43 unlinked genes in Escherichia coli (15, 16). In response to DNA damage that leads to single-stranded DNA (ssDNA) formation, ssDNA-RecA nucleoprotein filaments induce LexA autoproteolysis (17), thereby releasing promoters and enabling gene expression. Among the stresses that can induce the SOS response, several antibiotics, as well as horizontal gene transfer events like transformation and conjugation, have been shown to enhance integrase expression and activity in planktonic cultures of E. coli and Vibrio cholerae (12, 18–20). In addition to SOS response regulation, the nucleoid-associated proteins FIS and H-NS were recently suggested to repress the expression of IntI1 (21). The V. cholerae integron integrase (IntIA, formerly called IntI4) was also shown to be controlled by cyclic AMP (cAMP) receptor protein (CRP)-dependent regulation (19).
All of these regulatory mechanisms have been extensively studied in planktonic culture, whereas in natural settings, bacteria mostly live in biofilms. A biofilm is a community of microbes associated with a biotic or abiotic surface, typically encased in an autoproduced extracellular matrix (22). Biofilms are characterized by high levels of antibiotic resistance/tolerance compared to those of their planktonic counterparts and represent a major health threat when they develop during chronic infections or on medical devices (23). The antibiotic resilience of bacterial biofilms results from a variety of mechanisms (24, 25). Recalcitrance (or tolerance) is mainly due to the presence of an isogenic subpopulation of nondividing, antibiotic-tolerant bacteria called persisters (26, 27). The SOS and stringent responses are the two main pathways leading to the generation of persister bacteria (24). Recently, Bernier et al. showed that starvation and SOS response induction in aging biofilms mediated bacterial tolerance to fluoroquinolones (28). Biofilms are highly heterogeneous environments with local gradients of nutrients, pH, oxygen tension, etc., creating microniches of distinct bacterial subpopulations that experience and adapt to various stresses (29, 30). Another characteristic explaining the survival of biofilm bacteria during antibiotic exposure is that biofilms facilitate the transfer of mobile genetic elements and, therefore, the spread of antibiotic resistance between bacteria (31–34). It has been shown that various environments where bacteria live in complex biofilms contain large numbers of integrons displaying a huge variety of gene cassettes (35–37).
We therefore studied the influence of the biofilm lifestyle on class 1 integron integrase expression by comparing the expression levels of intI1 and the recombination activities of the IntI1 integrase in planktonic and biofilm culture. We found that the stringent response acts at two levels in biofilms: it induces the SOS response, thereby increasing the basal expression level of SOS-regulated genes, and also it exerts biofilm-specific positive regulation of intI1 expression through a mechanism involving the Lon protease.
RESULTS
The SOS response and integrase expression are induced by the biofilm lifestyle.
In a continuous-culture biofilm model, we examined the expression level of the intI1 gene and that of sfiA, a gene that encodes the cell division inhibitor SulA and is known to be strongly induced by the SOS response. We used E. coli MG1656 F′ (a strain with a strong propensity to form biofilms, due to the presence of the F′ factor [38]) and plasmid pPsfiA-lacZ or pPintI1-lacZ, carrying a lacZ transcriptional fusion with, respectively, the promoter of sfiA (PsfiA) or intI1 (PintI1) (Table 1). We first compared the promoter activities by assaying β-galactosidase in MG1656 F′ cells grown for 24 h under planktonic and biofilm conditions. The MG1656 F′/pPsfiA-lacZ and MG1656 F′/pPintI1-lacZ strains exhibited, respectively, 2.2- and 3.6-fold higher β-galactosidase activity under biofilm conditions than in planktonic culture (Table 2, B/P ratio).
TABLE 1 .
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| E. coli strains | ||
| MG1656 | lacZ null derivative of E. coli MG1655 | 69 |
| MG1656 ΔrecA | Deletion of the recA gene; constitutive repression of SOS genes | 12 |
| MG1656 ΔsulA ΔlexA | Deletion of the lexA and sulA genes; constitutive expression of SOS genes | 12 |
| NEB 5-alpha F′ Iq | F′ proA+B+ lacIq Δ(lacZ)M15 zzf::Tn10 (Tetr)/fhuA2Δ(argF-lacZ)U169 phoA glnV44 φ80Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 | New England Biolabs |
| TG1 ΔrelA::KmFRT | Deletion of relA by replacement of the gene with a KmFRT cassette; Kmr | 28 |
| TG1 ΔcpxR::KmFRT | Deletion of cpxR by replacement of the gene with a KmFRT cassette; Kmr | 70 |
| TG1 ΔrpoS::KmFRT | Deletion of rpoS by replacement of the gene with a KmFRT cassette; Kmr | 28 |
| TG1 ΔluxS::KmFRT | Deletion of luxS by replacement of the gene with a KmFRT cassette; Kmr | 28 |
| TG1 Δlon::KmFRT | Deletion of lon by replacement of the gene with a KmFRT cassette; Kmr | 28 |
| MG1656 ΔcpxR | Deletion of cpxR | This study |
| MG1656 ΔrpoS | Deletion of rpoS | This study |
| MG1656 ΔluxS | Deletion of luxS | This study |
| MG1656 Δlon | Deletion of lon | This study |
| MG1656 ΔrelA ΔspoT | Deletion of relA and spoT | This study |
| Plasmids | ||
| p6851 | Cassette excision reporter pSU38::aac(6′)-Ib::attCaadA7-cat(T4)-attCVCR2; Kmr Cmr | 12 |
| pSU38ΔtotlacZ | Vector carrying the lacZ coding sequence with no translation initiation region or promoter; Kmr | 68 |
| pPsfiA-lacZ | sfiA promoter cloned into pSU38ΔtotlacZ: lacZ under the control of PsfiA; Kmr | This study |
| pPintI1 | attI site from In40 class 1 integron cloned into pSU38ΔtotlacZ: lacZ under the control of the intI1 promoter, PintI1; Kmr | 12 |
| pPintI1* | pPintI1 with PintI1 carrying the mutation LexAmut2 in the LexA box; Kmr | 12 |
| pZE1-mcs1 | Promoterless derivative of pZE12-mcs1; Ampr | 12 |
| pZE1-IntI1 | attI site + intI1 gene from In40 class I integron (integrase IntI1R32_H39 variant with the highest excision activity); Ampr | 12 |
| pZE1-IntI1* | pZE1-IntI1 carrying the mutation LexAmut2 in the LexA box of PintI1; Ampr | 12 |
| pCP20 | Vector carrying Flp gene specific to FRT sites, thermosensitive; Ampr Cmr | 66 |
| F′ | F′ conjugative plasmid allowing enhanced biofilm formation; Tetr | 38 |
| pZS*tetR11-mcs1 | Plasmid carrying PN25-tetR between the bla gene and the terminator t0 and the synthetic PLtetO-1 promoter in front of the multiple-cloning site MCS1, pSC101* origin of replication; Ampr | This study |
| pZS*tetR11-relA | Same as pZS*tetR11-mcs1 but with relA under the control of the synthetic PLtetO-1 promoter; Ampr | This study |
| pZS*tetR11-lon | lon under the control of the synthetic PLtetO-1 promoter; Ampr | This study |
TABLE 2 .
intI1 and sfiA expression under biofilm conditions versus planktonic culture
| Straina | β-Gal activity (Miller units) [mean (±SD)a or B/P ratio] in strain bearing indicated plasmid under indicated condition(s) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| pPintI1-lacZ |
pPintI1*-lacZ |
pPsfiA-lacZ |
|||||||
| P | B | B/P | P | B | B/P | P | B | B/P | |
| MG1656 F′ | 17.6 (±4.3) | 64.0 (±24.3) | 3.6# | 97.6 (±23.2) | 202.9 (±29.3) | 2.1# | 584.1 (±288.9) | 1270.6 (±469.1) | 2.2# |
| MG1656 ΔlexA F′ | 74.1 (±8.2) | 109.5 (±17.8) | 1.5# | 76.9 (±17.6) | 135.3 (±14.5) | 1.8# | 16858.7 (±4,084.4) | 15647.9 (±3,517.5) | 0.9, NS |
| MG1656 ΔrecA F′ | 6.4 (±0.7) | 8.4 (±1.7) | 1.3# | ND | ND | 1.1 (±0.1) | 2.3 (±0.1) | 2.1, NS | |
The results are from at least 12 replicates. P, planktonic culture; B, biofilm; #, significant difference at a P value of <0.001; ND, not determined; NS, not significant.
To determine whether PsfiA and PintI1 induction under biofilm conditions is linked to SOS-dependent regulation, we measured the β-galactosidase activities of both promoters in the MG1656 F′ ΔrecA (constitutive repression) and ΔlexA (constitutive expression) deletion mutant derivatives (Table 1). In biofilm culture, the PsfiA and PintI1 activities were dramatically reduced in strain MG1656ΔrecA F′ (557- and 7.6-fold, respectively) (Table 2) and increased in strain MG1656ΔlexA F′ (12.3- and 1.7-fold, respectively) (Table 2) compared to their activities in the wild-type strain. Thus, basal sfiA and intI expression (expression level in the absence of exogenous stress) was higher under biofilm conditions than in planktonic culture, nevertheless allowing both promoters to be further activated by the SOS response.
To examine the consequences of higher basal class 1 integrase expression on cassette rearrangement under biofilm conditions, we estimated the excision activity of the integrase by measuring its capacity to catalyze recombination between two attC sites located on a synthetic array of two cassettes: attCaadA7-cat(T4)-attCVCR-aac(6′)-Ib (12). When the intI1 gene was expressed from the wild-type promoter PintI1 (pZE1-intI1) (Table 1), the cassette excision frequency rose by more than 2 log under biofilm conditions compared to the frequency in planktonic culture (average values, 1.3 × 10−05 versus 6.6 × 10−07; P < 0.001) (Fig. 1).
FIG 1 .

IntI1 excision activity under biofilm conditions. IntI1 excision recombination activity was estimated by determining the frequency of emergence of tobramycin resistance as a result of recombination between the attC sites of the attCaadA7-cat(T4)-attCVCR-aac(6′)-Ib gene cassette array carried on plasmid p6851. The intI1 gene was expressed from its own promoter (pZE1-intI1). Excision frequency was estimated under biofilm conditions (B) and in planktonic culture (P) with the wild-type strain MG1656 F′ carrying both p6851 and pZE1-intI1. Assays were done at least 9 times each. The bottom and top of the box indicate the first and third quartile respectively. The median is shown as a horizontal line inside the box, and the maximum and minimum values as the ends of the whisker.
Existence of biofilm-specific PintI1 regulation.
In the ΔlexA strain, the expression of the PsfiA promoter was maximal and independent of the growth conditions (biofilm or planktonic culture), as expected for a derepressed background (Table 2). Contrary to the results for PsfiA, in the ΔlexA strain, PintI1 exhibited a significantly higher expression level under biofilm conditions than in planktonic culture (1.5-fold difference) (Table 2), suggesting either biofilm-specific regulation of PintI1 or pleiotropic effects due to lexA deletion differentially affecting the PintI1 and PsfiA promoters. To test these two possibilities, we used the plasmid pPintI1* bearing the transcriptional fusion PintI1*-lacZ, in which the LexA box within PintI1 is mutated, inhibiting LexA binding and therefore leading to constitutive expression of lacZ (Table 1) (12). This construct should therefore exhibit maximum lacZ expression independently of both growth condition (biofilm versus planktonic) and bacterial background (WT versus ΔlexA). We found that the strength of PintI1* was still higher under biofilm conditions than in planktonic culture, whatever the genetic background (MG1656 F′ or its ΔlexA derivative) (Table 2), indicating that the difference was not due to a pleiotropic effect of lexA deletion.
Together, these results suggest that the expression of the class 1 integron integrase is also subjected to biofilm-specific regulation independently of the SOS response.
Role of RelA and Lon in the regulation of intI1 expression under biofilm conditions.
Biofilms being heterogeneous environments in which various stresses are encountered, we constructed various global regulator deletion mutants of the MG1656 F′ strain, namely, rpoS (general stress response), cpxR (envelope stress response), luxS (quorum sensing), relA/spoT (stringent response), and lon (protease) mutants, in order to examine their possible involvement in the regulation of intI1 expression under biofilm conditions. We estimated the strength of the intI1 promoter in these deletion mutants of MG1656 F′ grown under planktonic and biofilm conditions. To circumvent interference with SOS regulation, we used the PintI1* promoter, which carries a mutated LexA box (Table 1).
We first tested the ability of these mutants to form a biofilm. Apart from the cpxR mutant, which, compared to the parental strain, exhibited a slightly lower capacity to form a biofilm, none of the mutants showed an altered biofilm-forming capacity (see Fig. S1 in the supplemental material). We therefore evaluated PintI1* activity in all of these mutants. The biofilm/planktonic condition ratio of β-galactosidase activities in the luxS, cpxR, and rpoS MG1656 derivatives was similar to that of the parental strain MG1656 F′. However, in the backgrounds with deletions of relA/spoT and lon, there was no longer a significant difference between the β-galactosidase activities in biofilm and planktonic culture (Table 3).
TABLE 3 .
Activity of PintI1* under biofilm and planktonic conditions for the deletion mutants
| Strain | β-Gal activity (Miller units) [mean (±SD)a or B/P ratio] under indicated condition(s) of strain bearing pPintI1*-lacZ |
||
|---|---|---|---|
| P | B | B/P | |
| MG1656 F′ | 97.6 (±23.2) | 202.9 (±29.3) | 1.8# |
| MG1656 ΔluxS F′ | 65.0 (±20.1) | 138.0 (±38.5) | 2.1† |
| MG1656 ΔcpxR F′ | 46.0 (±4.7) | 81.3 (±6.2) | 1.8† |
| MG1656 ΔrpoS F′ | 89.3 (±28.6) | 182.6 (±38.0) | 2.0† |
| MG1656 ΔrelA/spoT F′ | 65.5 (±8.2) | 88.0 (±17.6) | 1.3, NS |
| MG1656 Δlon F′ | 70.0 (±15.9) | 92.8 (±22.4) | 1.3, NS |
Results shown are from at least 6 replicates. P, planktonic culture; B, biofilm; # and †, significant difference at a P value of <0.001 or <0.01, respectively; NS, not significant.
To confirm the role of the stringent response and of the Lon protease in the biofilm-specific regulation of intI1, we complemented the MG1656ΔrelA/spoT F′ and MG1656Δlon F′ mutants with RelA and Lon, respectively (Table 1) (39). As shown by the results in Fig. 2, the induction of RelA and Lon protein production, respectively, in the relA/spoT and lon deletion mutants restored the wild-type phenotype in both mutants, i.e., higher lacZ expression from PintI1* under biofilm conditions than in planktonic culture (1.7- and 2.1-fold differences, respectively). These results showed that PintI1* induction under biofilm conditions resulted from a direct and/or indirect effect of the stringent response and that Lon protein also played a role in this regulation.
FIG 2 .
Complementation experiment with relA and lon mutants. The activity level of the derepressed integrase promoter PintI1* was estimated by β-galactosidase assay. (A) Results for complementation of relA. MG1656ΔrelA/spoT F′/pPintI1*-lacZ also carried pZS*tetR11-mcs1 (pZS-mcs1) or pZS*tetR11-relA (pZS-relA). (B) Results for complementation of lon. MG1656Δlon F′/pPintI1*-lacZ also carried pZS*tetR11-mcs1 or pZS*tetR11-lon (pZS-lon); Strains were grown for 24 h under biofilm conditions or planktonic culture in the absence or presence of 0.2 mM anhydrotetracycline (ATc; induction of RelA or Lon protein synthesis). Error bars indicate the standard deviations of the results from 6 different assays.
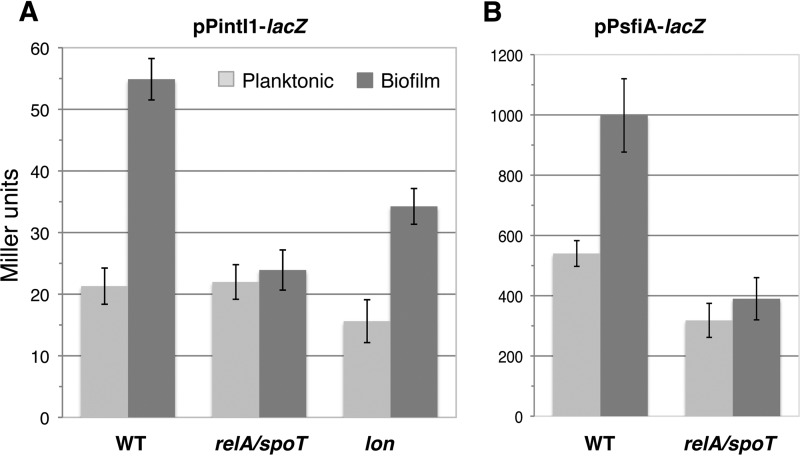
We then examined whether the biofilm-specific regulation observed with PintI1* also affected the wild-type SOS-regulated PintI1 promoter by estimating the β-galactosidase activities from PintI1-lacZ in strains MG1656ΔrelA/spoT F′ and MG1656Δlon F′. As observed with PintI1*, there was no longer any difference between the β-galactosidase activities under planktonic and biofilm conditions with the MG1656ΔrelA/spoT PintI1-lacZ strain (Fig. 3). This result was surprising, as we knew from the above-described PsfiA experiments that the increase of PsfiA basal activity in biofilm compared to that in planktonic culture was SOS dependent. Thus, we expected that the increased activity of PintI1 in biofilm should also be at least partially dependent on the SOS response (Table 2). We therefore examined whether RelA was also responsible for the higher basal expression level of sfiA under biofilm conditions than in planktonic culture. As observed with PintI1, the β-galactosidase activity from PsfiA-lacZ in strain MG1656ΔrelA/spoT F′ was similar under biofilm and planktonic conditions (Fig. 3). These results thus suggested that the stringent response might somehow induce the SOS response, which would in turn increase basal intI1 and sfiA expression under biofilm conditions. In the MG1656Δlon F′ background, contrary to what was observed with PintI1*, the β-galactosidase activity from PintI1-lacZ was 2.2-fold higher (P < 0.01) under biofilm conditions than in planktonic culture (Fig. 3).
FIG 3 .
Effect of the stringent response on intI1 and sfiA expression under biofilm conditions. The activity levels of the PintI1 and PsfiA promoters, expressed as Miller units, were estimated by β-galactosidase assay in 24-h planktonic and biofilm cultures of the wild-type (WT) strain MG1656 F′ and its ΔrelA/spoT and Δlon derivatives, as indicated. The bacteria carried plasmid pPintI1-lacZ (A) or PsfiA-lacZ (B). Error bars indicate the standard deviations of the results from at least 6 different assays.
DISCUSSION
The aim of this study was to assess the expression/activity of the class 1 integron integrase IntI1 under biofilm conditions. In agreement with Bernier et al., who showed that the SOS response is gradually induced in aging static biofilm culture in minimal medium (up to twofold after 96 h) (28), we found that both the SOS response and class 1 integron integrase expression were induced more than twofold (up to 3.6-fold for intI1) in 24-h continuous biofilm culture in LB medium compared to their expression level in planktonic culture. We also found that the expression of sfiA and intI1 was enhanced under biofilm conditions in the lexA deletion mutant background compared to their levels in the parental strain (up to 12.3-fold for sfiA). This indicates that, although the SOS response is a signature of the biofilm lifestyle, its level of induction under biofilm conditions varies with the growth conditions and does not reach its fully derepressed level, providing bacteria with some leeway to cope with exogenous stresses.
We also observed that, under derepressed conditions (lexA deletion mutant background or with PintI1*), the intI1 expression level was still higher under biofilm conditions than in planktonic culture. This suggests the existence of unexpected biofilm-specific regulation of intI1 expression, indicating that the regulation of integron integrase is more complex than previously thought. It was recently shown that, besides its regulation via the SOS response, the V. cholerae integron integrase IntIA is also subject to positive CRP-dependent regulation, likely fully independent of SOS regulation (19). CRP, the c-AMP receptor protein, has been implicated in the regulation not only of the catabolic pathway but also of genes involved in adaptation and survival in the environment and virulence (40, 41). Using the virtual footprint tool PRODORIC (http://www.prodoric.de), we found no CRP binding site within the In40 class 1 integron attI site that encompasses the intI1 promoter (42), suggesting that PintI1 is not regulated by CRP.
In bacteria, various nucleotides [c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp, etc.] have emerged as important second messengers in the regulation of key processes required for adaptation and biofilm formation (43, 44). The E. coli stringent response, mediated by the alarmone (p)ppGpp, is responsible for reorganizing cellular transcription in response to nutritional starvation and other stresses, ultimately reducing the growth rate (45, 46). The concentration of (p)ppGpp [denoting both ppGpp and (p)ppGpp] is governed by the two synthases RelA and SpoT, the latter protein also acting as a hydrolase. Surprisingly, the deletion of relA and spoT abrogated the induction of both PintI1 and PsfiA under biofilm conditions (Table 3; Fig. 3). (p)ppGpp regulates replication, transcription and translation (47). It induces pausing of transcription elongation at some positions, which can hamper replication (48, 49) and lead to R loop formation (reviewed in reference 50). R loop formation has been shown to induce the SOS response (51). Furthermore, transcription profiling showed that the stringent response in E. coli induces the SOS response (52). It is thus conceivable that the stringent response is activated under biofilm conditions, leading to mild induction of the SOS response and, thus, to the observed increases in intI1 and sfiA expression compared to their expression in planktonic culture.
Our results showed that the stringent response also regulates intI1 expression in biofilms independently of the SOS response (Table 3). RelA is a global regulator of the stringent response and cannot act directly on PintI1. (p)ppGpp also plays a role in regulating the acid stress response, facilitates the use of alternative sigma factors (such as σS, σE, and σN), and stabilizes σS, the sigma factor that is encoded by rpoS and controls the general stress response (for recent reviews, see references 53 and 54). The MG1656ΔrpoS F′ mutant exhibited higher expression from PintI1* under biofilm conditions than in planktonic culture (Table 3), indicating that σS is not the missing link between RelA/(p)ppGpp and biofilm-specific intI1 regulation.
The stringent response also represses the activity of exopolyphosphatase (PPX), resulting in the accumulation of polyphosphate (poly-P), which binds to Lon, stimulating its protease activity toward proteins such as free ribosomal proteins and antitoxins (55, 56). Poly-P also reduces Lon activity in vitro (57, 58). Interestingly, lon deletion had an effect similar to that of relA/spoT deletion on PintI1* activity under biofilm conditions, i.e., no induction compared to that in planktonic culture (Table 3). Our results thus suggest that, in biofilms, by activating the stringent response through RelA, the poly-P–Lon complex would control the amount of a biofilm-specific PintI1 regulator. Things may not be so simple, however, as lon deletion had no effect on the PintI1 expression level under our biofilm conditions (Fig. 3), suggesting that Lon-mediated regulation is not active when LexA is bound to PintI1. This implies that an unknown regulator of PintI1, the stability of which would be controlled by the poly-P–Lon complex, might display steric interference with bound LexA.
As biofilms are heterogeneous environments, only bacteria within certain microniches might experience nutrient starvation (59) and therefore be subject to (p)ppGpp regulation. In this case, PintI1 expression in a fraction of the biofilm population might be even higher than the global level found here. (p)ppGpp has been shown to be important for the formation of E. coli and Pseudomonas aeruginosa persisters in both planktonic and biofilm culture (60–63). Maisonneuve et al. demonstrated that the degradation of antitoxins by the poly-P–Lon complex in type II toxin-antitoxin (TA) modules is pivotal to E. coli persistence (62). These and our results raise the possibility that biofilm-specific induction of integrase expression might take place in persister cells.
This study demonstrates that the regulation of class 1 integron integrase expression is more complex than previously thought, as summarized in Fig. 4. In the ubiquitous and natural settings represented by biofilms, some bacteria experience nutrient starvation that triggers a stringent response. The resulting increase in the (p)ppGpp concentration induces (i) a moderate increase in the SOS response, leading to increased basal expression of LexA-regulated genes, and (ii) biofilm-specific positive regulation of class 1 integron integrase expression through the poly-P–Lon complex.
FIG 4 .
Proposed mechanism of class 1 integron regulation via stringent response under biofilm conditions. In biofilms, upon nutrient starvation, the alarmone (p)ppGpp, synthesized by the RelA and SpoT proteins, would mediate the inhibition of replication initiation and transcription of specific genes, thereby stalling the RNA polymerase and leading to the generation of single-stranded DNA (ssDNA) and, thus, to mild induction of the SOS response, resulting in autoproteolysis of the LexA dimer bound to the PintI1 promoter and expression of intI1 (a) and the inhibition of the exopolyphosphatase (PPX) activity, resulting in the accumulation of inorganic polyphosphate (poly-P) via the polyphosphate kinase (PPK), whereupon poly-P would bind the Lon protease to form the poly-P–Lon complex that would regulate the degradation of an unknown regulator of derepressed PintI1 (b).
This work confirms that biofilms are environments favorable to integron-mediated acquisition/exchange of antibiotic resistance determinants through specific regulation of class 1 integron integrase. Moreover, metagenomics studies have shown that class 1 integrons may also be found on the chromosomes of environmental bacteria (64). These class 1 integrons contain a huge diversity of gene cassettes, mostly of unknown function, potentially providing adaptive functions to bacteria (37, 65). Besides antibiotic resistance, our study thus indicates that biofilms are ideal niches for shaping bacterial evolution through the exchange of gene cassettes.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The F′Tet factor, designated F′ for convenience, was introduced into E. coli MG1656 and its derivatives by conjugation, using the commercial strain NEB 5-alpha F′ as the donor.
Cells were grown under planktonic or biofilm conditions at 37°C in Luria-Bertani (LB) medium supplemented when necessary with kanamycin (Km; 25 µg ⋅ ml−1), ampicillin (Amp; 100 µg ⋅ ml−1), tetracycline (Tet; 7.5 µg ⋅ ml−1), zeocyn (Zeo; 30 µg ⋅ ml−1), or chloramphenicol (Cm; 25 µg ⋅ ml−1).
Biofilm and planktonic culture.
Biofilms were produced by culturing bacteria at 37°C in LB medium for 24 h in a continuous-flow glass microfermentor containing a removable spatula, as described in reference 38. The microfermentors were inoculated by dipping the removable glass slides for 2 min into 15 ml of bacterial culture containing 1× 109 cells/ml, followed by a brief rinse in LB medium before insertion in the microfermentor. After 24 h of growth under nonbubbling conditions, the biofilm that formed on the removable glass slide was resuspended in 10 ml of ice-cold LB by vortexing. Biofilm biomass was estimated by determining the optical density at 600 nm (OD600).
For planktonic culture, 100 µl of the culture used to inoculate the microfermentors was diluted in 10 ml of LB and grown for 24 h at 37°C with shaking.
Mutant construction.
MG1656Δgene::KmFRT strains [“gene” denotes relA, cpxR, rpoS, luxS, or lon, and KmFRT is the resistance cassette used to replace a gene of interest, composed of the aph(3′)-II gene (Km resistance) flanked on each side by a FRT site (specific recombination site of the FLP recombinase of Saccharomyces cerevisiae)] were created by P1vir transduction from strain TG1Δgene::KmFRT into MG1656 F′. The Km resistance gene was then removed by flippase action (66) to obtain strain MG1656Δgene F′.
Constructs were verified by PCR and sequencing (Applied Biosystems 3130XL Genetic Analyser). All primers are listed in Table S1 in the supplemental material.
Plasmid construction.
pPsfiA-lacZ was constructed by amplifying the sfiA promoter, PsfiA, from MG1656 genomic DNA with PCR using primers psulA-3 and psulA-EcoRI-5 and cloning the product into pSU38ΔtotlacZ at the EcoRI/BamHI sites.
pZS*tetR11-relA was constructed as follows: relA was amplified from the MG1656 genome by using primers relA-KpnI-5 and relA-HindIII-3 and cloned via KpnI/HindIII into pZS*21mcs1 (39), yielding pZS*21-relA. The neo gene (kanamycin resistance) from pZS*21-relA was replaced by the bla gene of pZE1-mcs1 by XhoI/SacI cloning, yielding pZS*11-relA. tetR was amplified with its PN25 promoter from pZEtetR21-gfp (67) using primers tetR-SacIinfu-3 and tetR-SacIIinfu-5 and cloned at the SacI site of pZS*11-relA by using the In-Fusion method (In-Fusion HD cloning kit, Clontech), following the manufacturer’s instructions, to yield pZS*tetR11-relA.
pZS*tetR11-lon was constructed by using the in-Fusion approach to replace relA with lon. The lon fragment was amplified from MG1656 by using lon-infusion-3′ and lon-infusion-5′ primers and cloned with linearized pZS*tetR11-relA (KpnI/HindIII), following the manufacturer’s instructions.
pZS*tetR11-mcs1 was constructed as follows: the XhoI/SacI fragment from pZS*tetR11-relA containing tetR was cloned into pZS*21-mcs1, replacing the neo gene with the tetR-bla fragment.
All constructs were verified by sequencing. All primers are listed in Table S1 in the supplemental material.
β-Galactosidase assay.
The β-galactosidase assay was performed with 0.5-ml aliquots of planktonic culture or 0.5 ml of resuspended biofilm, as described in reference 68.
Cassette excision assay.
A synthetic array of two cassettes [attCaadA7-cat(T4)-attCVCR-aac(6′)-Ib], preceded by the lac promoter, Plac, and conferring chloramphenicol resistance [cat(T4)], is carried on plasmid p6851. The excision assay is described in reference 12. Briefly, MG1656 F′/p6851 cells electroporated with pZE1-IntI1 or pZE1-IntI1* (Table 1) were grown overnight in LB medium. These cultures were used to inoculate both planktonic and biofilm cultures, which were then grown for 24 h. Dilutions of resuspended biofilm or planktonic culture were plated on LB-Amp-Km plates (total population) and LB-tobramycin (Tobra) plates (recombinants only). The excision frequency was calculated by determining the ratio of Tobrar to Ampr Kmr colonies (CFU/ml). Experiments were performed at least 9 times.
Statistical analysis.
Significance was determined using the nonparametric Mann-Whitney U test to compare the results under the two experimental conditions (biofilm and planktonic) and for the wild-type and mutant strains. P values of <0.05 were considered to indicate statistical significance.
SUPPLEMENTAL MATERIAL
Biofilm formation by mutants. Download
List of primers and probes used in this study.
ACKNOWLEDGMENTS
We are grateful to Jérémy Mounier for his help with construction of the deletion strains and to Elena Bülow for critical reading of the manuscript. We also thank Christophe Beloin and Jean-Marc Ghigo (Institut Pasteur, Paris) for the kind gift of strains and Benoît Marin and François Dalmay (UMR Inserm U1094, Limoges, France) for their help with statistical analysis.
This work was supported by grants from Ministère de l’Enseignement Supérieur et de la Recherche, Conseil Régional du Limousin, Institut National de la Santé et de la Recherche Médicale (Inserm), Agence Nationale pour la Recherche (ANR-08-MIE-016 and ANR-12-BSV3-0015-01), and Direction de la Recherche et Innovation du CHU de Limoges (APREL 2014). The funders had no role in study design, data collection and analysis, or the decision to submit the work for publication.
Footnotes
Citation Strugeon E, Tilloy V, Ploy M-C, Da Re S. 2016. The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. mBio 7(4):e00868-16. doi:10.1128/mBio.00868-16.
REFERENCES
- 1.Davies J, Davies D. 2010. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev 74:417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalan L, Wright GD. 2011. Antibiotic adjuvants: multicomponent anti-infective strategies. Expert Rev Mol Med 13:e5. doi: 10.1017/S1462399410001766. [DOI] [PubMed] [Google Scholar]
- 3.Cambray G, Guerout AM, Mazel D. 2010. Integrons. Annu Rev Genet 44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 4.Gillings MR. 2014. Integrons: past, present, and future. Microbiol Mol Biol Rev 78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stokes HW, Hall RM. 1989. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol 3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 6.Collis CM, Hall RM. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol 174:1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazel D. 2006. Integrons: agents of bacterial evolution. Nat Rev Microbiol 4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 8.Boucher Y, Labbate M, Koenig JE, Stokes HW. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol 15:301–309. doi: 10.1016/j.tim.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Cury J, Jové T, Touchon M, Néron B, Rocha EP. 2016. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res 44:4539–4550. doi: 10.1093/nar/gkw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domingues S, da Silva GJ, Nielsen KM. 2012. Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mob Genet Elements 2:211–223. doi: 10.4161/mge.22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev 33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 12.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbé J, Ploy MC, Mazel D. 2009. The SOS response controls integron recombination. Science 324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 13.Erill I, Campoy S, Barbé J. 2007. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 31:637–656. doi: 10.1111/j.1574-6976.2007.00082.x. [DOI] [PubMed] [Google Scholar]
- 14.Walker GC. 1984. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Res 48:60–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández De Henestrosa AR, Ogi T, Aoyagi S, Chafin D, Hayes JJ, Ohmori H, Woodgate R. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol Microbiol 35:1560–1572. doi: 10.1046/j.1365-2958.2000.01826.x. [DOI] [PubMed] [Google Scholar]
- 17.Little JW. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411–421. doi: 10.1016/0300-9084(91)90108-D. [DOI] [PubMed] [Google Scholar]
- 18.Baharoglu Z, Bikard D, Mazel D. 2010. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet 6:e00868-16. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baharoglu Z, Krin E, Mazel D. 2012. Connecting environment and genome plasticity in the characterization of transformation-induced SOS regulation and carbon catabolite control of the Vibrio cholerae integron integrase. J Bacteriol 194:1659–1667. doi: 10.1128/JB.05982-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baharoglu Z, Mazel D. 2011. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother 55:2438–2441. doi: 10.1128/AAC.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cagle CA, Shearer JE, Summers AO. 2011. Regulation of the integrase and cassette promoters of the class 1 integron by nucleoid-associated proteins. Microbiology 157:2841–2853. doi: 10.1099/mic.0.046987-0. [DOI] [PubMed] [Google Scholar]
- 22.Monds RD, O’Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol 17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Stewart PS, Costerton JW. 2001. Antibiotic resistance of bacteria in biofilms. Lancet 358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 24.Lebeaux D, Ghigo JM, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Acker H, Van Dijck P, Coenye T. 2014. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 27.Shah D, Zhang Z, Khodursky A, Kaldalu N, Kurg K, Lewis K. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol 6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernier SP, Lebeaux D, DeFrancesco AS, Valomon A, Soubigou G, Coppée JY, Ghigo JM, Beloin C. 2013. Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genet 9:e00868-16. doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boles BR, Singh PK. 2008. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A 105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart PS, Franklin MJ. 2008. Physiological heterogeneity in biofilms. Nat Rev Microbiol 6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 31.Maeda S, Ito M, Ando T, Ishimoto Y, Fujisawa Y, Takahashi H, Matsuda A, Sawamura A, Kato S. 2006. Horizontal transfer of nonconjugative plasmids in a colony biofilm of Escherichia coli. FEMS Microbiol Lett 255:115–120. doi: 10.1111/j.1574-6968.2005.00072.x. [DOI] [PubMed] [Google Scholar]
- 32.Molin S, Tolker-Nielsen T. 2003. Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr Opin Biotechnol 14:255–261. doi: 10.1016/S0958-1669(03)00036-3. [DOI] [PubMed] [Google Scholar]
- 33.Hennequin C, Aumeran C, Robin F, Traore O, Forestier C. 2012. Antibiotic resistance and plasmid transfer capacity in biofilm formed with a CTX-M-15-producing Klebsiella pneumoniae isolate. J Antimicrob Chemother 67:2123–2130. doi: 10.1093/jac/dks169. [DOI] [PubMed] [Google Scholar]
- 34.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol Med Microbiol 65:183–195. doi: 10.1111/j.1574-695X.2012.00960.x. [DOI] [PubMed] [Google Scholar]
- 35.Gaze WH, Zhang L, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, Brown H, Davis S, Kay P, Boxall AB, Wellington EM. 2011. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 5:1253–1261. doi: 10.1038/ismej.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stalder T, Barraud O, Casellas M, Dagot C, Ploy MC. 2012. Integron involvement in environmental spread of antibiotic resistance. Front Microbiol 3:119. doi: 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stalder T, Barraud O, Jové T, Casellas M, Gaschet M, Dagot C, Ploy MC. 2014. Quantitative and qualitative impact of hospital effluent on dissemination of the integron pool. ISME J 8:768–777. doi: 10.1038/ismej.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 39.Lutz R, Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res 25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Microbiol 6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 41.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 42.Ploy MC, Courvalin P, Lambert T. 1998. Characterization of In40 of Enterobacter aerogenes BM2688, a class 1 integron with two new gene cassettes, cmlA2 and qacF. Antimicrob Agents Chemother 42:2557–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalia D, Merey G, Nakayama S, Zheng Y, Zhou J, Luo Y, Guo M, Roembke BT, Sintim HO. 2013. Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem Soc Rev 42:305–341. doi: 10.1039/c2cs35206k. [DOI] [PubMed] [Google Scholar]
- 44.Landini P. 2009. Cross-talk mechanisms in biofilm formation and responses to environmental and physiological stress in Escherichia coli. Res Microbiol 160:259–266. doi: 10.1016/j.resmic.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 46.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 47.Srivatsan A, Wang JD. 2008. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol 11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Kingston RE, Nierman WC, Chamberlin MJ. 1981. A direct effect of guanosine tetraphosphate on pausing of Escherichia coli RNA polymerase during RNA chain elongation. J Biol Chem 256:2787–2797. [PubMed] [Google Scholar]
- 49.Krohn M, Wagner R. 1996. Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. J Biol Chem 271:23884–23894. doi: 10.1074/jbc.271.39.23884. [DOI] [PubMed] [Google Scholar]
- 50.McGlynn P, Savery NJ, Dillingham MS. 2012. The conflict between DNA replication and transcription. Mol Microbiol 85:12–20. doi: 10.1111/j.1365-2958.2012.08102.x. [DOI] [PubMed] [Google Scholar]
- 51.Gan W, Guan Z, Liu J, Gui T, Shen K, Manley JL, Li X. 2011. R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes Dev 25:2041–2056. doi: 10.1101/gad.17010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. 2008. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol 190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gaca AO, Colomer-Winter C, Lemos JA. 2015. Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol 197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuroda A. 2006. A polyphosphate-lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci Biotechnol Biochem 70:325–331. doi: 10.1271/bbb.70.325. [DOI] [PubMed] [Google Scholar]
- 56.Tsilibaris V, Maenhaut-Michel G, Van Melderen L. 2006. Biological roles of the Lon ATP-dependent protease. Res Microbiol 157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 57.Nomura K, Kato J, Takiguchi N, Ohtake H, Kuroda A. 2004. Effects of inorganic polyphosphate on the proteolytic and DNA-binding activities of Lon in Escherichia coli. J Biol Chem 279:34406–34410. doi: 10.1074/jbc.M404725200. [DOI] [PubMed] [Google Scholar]
- 58.Osbourne DO, Soo VW, Konieczny I, Wood TK. 2014. Polyphosphate, cyclic AMP, guanosine tetraphosphate, and c-di-GMP reduce in vitro Lon activity. Bioengineered 5:264–268. doi: 10.4161/bioe.29261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang CT, Xu KD, McFeters GA, Stewart PS. 1998. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl Environ Microbiol 64:1526–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amato SM, Brynildsen MP. 2014. Nutrient transitions are a source of persisters in Escherichia coli biofilms. PLoS One 9:e00868-16. doi: 10.1371/journal.pone.0093110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amato SM, Orman MA, Brynildsen MP. 2013. Metabolic control of persister formation in Escherichia coli. Mol Cell 50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 62.Maisonneuve E, Castro-Camargo M, Gerdes K. 2013. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 63.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gillings M, Boucher Y, Labbate M, Holmes A, Krishnan S, Holley M, Stokes HW. 2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J Bacteriol 190:5095–5100. doi: 10.1128/JB.00152-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koenig JE, Boucher Y, Charlebois RL, Nesbø C, Zhaxybayeva O, Bapteste E, Spencer M, Joss MJ, Stokes HW, Doolittle WF. 2008. Integron-associated gene cassettes in Halifax Harbour: assessment of a mobile gene pool in marine sediments. Environ Microbiol 10:1024–1038. doi: 10.1111/j.1462-2920.2007.01524.x. [DOI] [PubMed] [Google Scholar]
- 66.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
- 67.Da Re S, Le Quéré B, Ghigo JM, Beloin C. 2007. Tight modulation of Escherichia coli bacterial biofilm formation through controlled expression of adhesion factors. Appl Environ Microbiol 73:3391–3403. doi: 10.1128/AEM.02625-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jové T, Da Re S, Denis F, Mazel D, Ploy MC. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 6:e00868-16. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Espéli O, Moulin L, Boccard F. 2001. Transcription attenuation associated with bacterial repetitive extragenic BIME elements. J Mol Biol 314:375–386. doi: 10.1006/jmbi.2001.5150. [DOI] [PubMed] [Google Scholar]
- 70.Beloin C, Valle J, Latour-Lambert P, Faure P, Kzreminski M, Balestrino D, Haagensen JA, Molin S, Prensier G, Arbeille B, Ghigo JM. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol Microbiol 51:659–674. doi: 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biofilm formation by mutants. Download
List of primers and probes used in this study.