ABSTRACT
Staphylococcus aureus is a leading cause of life-threatening infections worldwide. The MIC of an antibiotic against S. aureus, as well as other microbes, is determined by the affinity of the antibiotic for its target in addition to a complex interplay of many other cellular factors. Identifying nontarget factors impacting resistance to multiple antibiotics could inform the design of new compounds and lead to more-effective antimicrobial strategies. We examined large collections of transposon insertion mutants in S. aureus using transposon sequencing (Tn-Seq) to detect transposon mutants with reduced fitness in the presence of six clinically important antibiotics—ciprofloxacin, daptomycin, gentamicin, linezolid, oxacillin, and vancomycin. This approach allowed us to assess the relative fitness of many mutants simultaneously within these libraries. We identified pathways/genes previously known to be involved in resistance to individual antibiotics, including graRS and vraFG (graRS/vraFG), mprF, and fmtA, validating the approach, and found several to be important across multiple classes of antibiotics. We also identified two new, previously uncharacterized genes, SAOUHSC_01025 and SAOUHSC_01050, encoding polytopic membrane proteins, as important in limiting the effectiveness of multiple antibiotics. Machine learning identified similarities in the fitness profiles of graXRS/vraFG, SAOUHSC_01025, and SAOUHSC_01050 mutants upon antibiotic treatment, connecting these genes of unknown function to modulation of crucial cell envelope properties. Therapeutic strategies that combine a known antibiotic with a compound that targets these or other intrinsic resistance factors may be of value for enhancing the activity of existing antibiotics for treating otherwise-resistant S. aureus strains.
IMPORTANCE
Bacterial resistance to every major class of antibiotics has emerged, and we are entering a “post-antibiotic era” where relatively minor infections can lead to serious complications or even death. The utility of an antibiotic for a specific pathogen is limited by both intrinsic and acquired factors. Identifying the repertoire of intrinsic resistance factors of an antibiotic for Staphylococcus aureus, a leading cause of community- and hospital-acquired infections, would inform the design of new drugs as well as the identification of compounds that enhance the activity of existing drugs. To identify factors that limit the activity of antibiotics against S. aureus, we used Tn-Seq to simultaneously assess fitness of transposon mutants in every nonessential gene in the presence of six clinically important antibiotics. This work provides an efficient approach for identifying promising targets for drugs that can enhance susceptibility or restore sensitivity to existing antibiotics.
INTRODUCTION
Staphylococcus aureus is a Gram-positive pathogen with a remarkable ability to withstand antibiotics and evade the human immune system. Many factors, both intrinsic and acquired, have been shown to contribute to its ability to survive specific antibiotic stress. For example, methicillin-resistant S. aureus (MRSA) strains have acquired the mobile staphylococcal cassette chromosome mec element (SCCmec), encoding a transpeptidase, PBP2A, which is naturally resistant to β-lactams, enabling the organism to make cross-linked peptidoglycan when the native transpeptidases are inactivated by the β-lactams (1–3). Irrespective of its methicillin susceptibility status, S. aureus possesses numerous intrinsic factors that also limit the effectiveness of specific antibiotics (4). In contrast to acquired resistance factors like PBP2A, intrinsic resistance factors typically play additional roles in normal microbial physiology. For example, MprF, which modulates cell membrane charge, was initially identified in Staphylococcus xylosus as a gene that, when inactivated, increased susceptibility to the cationic peptide gallidermin (5). The activity of MprF is now known to be important for protection from other cationic antimicrobial peptides and daptomycin (6–8). TarO, which catalyzes the first step in the wall teichoic acid biosynthetic pathway (9), contributes to β-lactam resistance in MRSA, and its deletion results in cell division defects and mislocalization of cell wall biosynthetic machinery (10–12). Effective pharmacological inhibition of TarO in MRSA restores full sensitivity to β-lactams even though PBP2A is present (10, 12), highlighting the potential to mitigate antibiotic resistance by targeting intrinsic resistance factors. The most attractive candidates for targeting are those factors that hinder the activity of multiple classes of antibiotics. To identify such candidates, as well as additional factors contributing to the resistance of specific individual antibiotics, we used the massively parallel approach of transposon sequencing (Tn-Seq) (13–15) to examine large pools of S. aureus transposon mutants for fitness defects upon exposure to multiple classes of antibiotics.
Tn-Seq involves creating large transposon libraries, sequencing the transposon insertion sites with next-generation sequencing, and mapping the sequence reads to a reference genome (13, 15). This technique can be used to identify genes that contribute to fitness in a particular environment or under a particular set of growth conditions because reads mapping to these genes would be depleted compared to reads in a control. Depletion of reads in a gene implies that the mutants have reduced fitness under the test conditions. Tn-Seq can also be used to identify those insertion mutants that are highly represented in the mutant pool, indicating that inactivation of those genes increases fitness under the tested condition.
Tn-Seq has been used previously to identify antibiotic resistance factors for other organisms (16–18) but has not been used to compare multiple antibiotic classes in Staphylococcus aureus. We have performed Tn-Seq analysis using transposon libraries treated with six different antibiotics to identify genes with significantly fewer mapped reads than were seen with an untreated control. These genes, which we refer to as intrinsic resistance factors, render the bacteria more sensitive to the antibiotic tested when inactivated. The six antibiotics used in this study, ciprofloxacin, linezolid, gentamicin, oxacillin, vancomycin, and daptomycin, were chosen as clinically relevant representatives of antibiotics that target major pathways: DNA synthesis, protein synthesis, cell wall synthesis, and membrane stability (Fig. 1A and B) (19–25). In addition to previously identified factors, we have identified two hitherto-uncharacterized factors as important intrinsic resistance factors for multiple antibiotics.
FIG 1 .
Intrinsic resistance factors that contribute to antibiotic resistance can be identified by Tn-Seq. (A) The structures of the six different antibiotics used in the Tn-Seq experiments are shown. (B) The targets of the six antibiotics are shown. (C) A pooled transposon insertion library was grown with or without antibiotic and subjected to Tn-Seq to quantify the number of sequencing reads that map to each insertion location. The black lines in the columns corresponding to the three genes represent the number of reads mapping to a particular insertion location. In this example, the red gene had similar numbers of reads in the treated and untreated samples. Therefore, inactivation of this gene does not have an effect on antibiotic susceptibility. The orange gene had a lower number of reads in the treated sample than in the untreated control. Inactivation of this gene decreases bacterial fitness in the presence of the tested antibiotic. Genes of this type are known as intrinsic resistance factors. Finally, the blue gene had a higher number of reads in the treated sample than in the untreated control. Inactivation of this gene increases bacterial fitness in the presence of the test antibiotic.
RESULTS AND DISCUSSION
Experimental approach and data analysis.
Two different transposon libraries constructed in methicillin-susceptible S. aureus (MSSA) strain HG003 were used in initial screens for mutants exhibiting either enhanced resistance or enhanced susceptibility to an antibiotic (14, 26). The first transposon library was made by transformation with a temperature-sensitive plasmid and contained insertions in 71,000 unique sites (26). The second library was made using a phage-based transposition system and included transposon insertions in 126,040 unique sites (14). To identify mutants that exhibit fitness defects that are independent of the growth condition (and therefore likely to be of value in vivo), the libraries were treated with six antibiotics at concentrations below the MIC of the antibiotics and grown in different media for various numbers of generations (see Materials and Methods).
Sample preparation and Tn-Seq analysis to determine the location of the transposon insertions were performed as described previously (14, 26, 27). Subsequently, the number of reads mapping to a gene under experimental antibiotic treatment conditions was compared to the number in the untreated control to calculate the fold change in the number of reads mapping to each gene (Fig. 1C) as a surrogate measure of the representation of each mutant in the pool. Data were analyzed as previously described (14, 17), with one additional step: before comparing the number of reads/gene using the Mann-Whitney U test, the experimental condition (antibiotic treatment) was normalized to that of the untreated control using simulation-based resampling to minimize differences between the two conditions (28, 29). After all experiments for the two libraries were analyzed independently, P values and depletion/enrichment ratios for each gene in the presence of an antibiotic treatment were combined, using Fisher’s method for P values and the geometric mean of fold changes in the number of reads mapping to each gene.
Because antibiotics with differing mechanisms of action exert different degrees of selective pressure on the bacterial population at fractional MIC levels, it was necessary to tailor the data analysis approach in a way that enabled comparison across different antibiotic treatments. We therefore adjusted the cutoff value for the fold change in the number of mapped reads/gene for the treated samples relative to the controls to obtain similar numbers of hits for each antibiotic. Only genes corresponding to a P value of <0.05 were considered. We used a sliding cutoff value for the number of reads/gene that resulted in a maximum of 20 genes being identified as hits for each antibiotic. This sliding fold change cutoff value ranged from 10-fold (0.1 to 10) for ciprofloxacin to 55-fold for oxacillin. The top genes contributing to susceptibility/resistance for each antibiotic included genes with fewer as well as more reads mapping to them in the treated sample than in the control. The former represent intrinsic resistance factors or impediments to antibiotic inhibition of S. aureus. The latter are also of interest as they provide information on how antibiotic resistance can arise via gene inactivation.
Intrinsic factors that decrease or increase susceptibility to antibiotics.
Of the genes implicated in antibiotic resistance via the analysis described above, 80 were unique (see Table S1 in the supplemental material). Among the 20 genes in which transposon insertion resulted in the greatest change with respect to antibiotic susceptibility/resistance, we identified few or none that, when mutated, provided a fitness advantage upon exposure to ciprofloxacin, daptomycin, linezolid, oxacillin, and vancomycin, with the notable exception of gentamicin. For gentamicin, half of the mutants exhibited a fitness advantage (Table 1; see also Fig. S1 in the supplemental material). All of these genes, the absence of which enhanced fitness in the presence of fractional MIC levels of gentamicin, occur in the oxidative phosphorylation pathway. It is known that gentamicin and other aminoglycosides rely on the membrane potential to gain entry into cells (30, 31); disrupting genes in the oxidative phosphorylation pathway therefore limits cellular penetration.
TABLE 1 .
Treatment of a pooled transposon library with six different antibiotics identified genes that contribute to fitness under antibiotic stress conditionsa
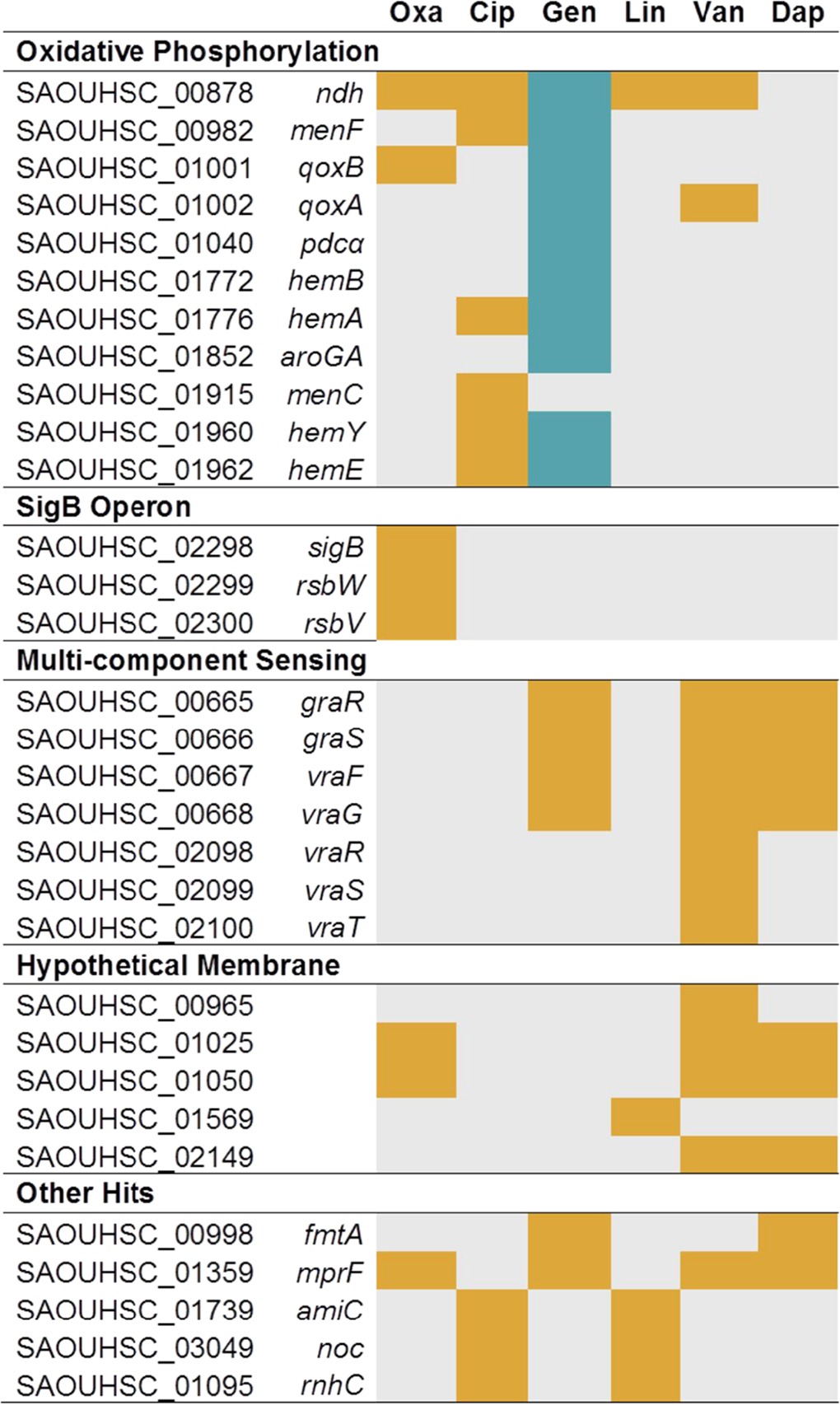
Orange rectangles indicate genes for which the numbers of reads due to transposon insertions were substantially lower than in the control, whereas blue rectangles indicate genes for which the numbers of reads due to transposon insertions were substantially higher than in the control. Gray rectangles indicate those genes which were not identified among the top 20 most affected genes. The top 20 most affected genes for each antibiotic were identified, and a subset of the 80 unique genes is shown here. The complete list is shown in Table S1 in the supplemental material. oxa, oxacillin; cip, ciprofloxacin; gen, gentamicin; lin, linezolid; van, vancomycin; dap, daptomycin.
This analysis also identified many other genes for which a contribution to resistance/susceptibility to an antibiotic had been observed previously. For example, sigB was among the hits identified with oxacillin treatment. Reads mapping to this gene, and to other components involved in the alternative sigma factor pathway, i.e., rsbV and rsbW, were significantly depleted after growth with oxacillin (>100-fold depletion in reads/gene for all three genes) (Table 1). It has been shown that overexpressing SigB causes cells to have thicker cell walls, increased transcript levels for penicillin-binding proteins, and elevated MICs to β-lactams and that deletion of sigB renders resistant S. aureus strains more sensitive to oxacillin (32, 33). Reads for pbp4, which encodes a penicillin-binding protein involved in secondary cross-linking of peptidoglycan and β-lactam resistance (34–36), were also found to be depleted under conditions of oxacillin treatment. Similarly, reads mapping to all three genes of the vraTSR operon, which encodes a multicomponent sensing (MCS) system that regulates the cell wall stress stimulon (37–41), were substantially depleted in the presence of vancomycin. norA, which encodes an efflux pump that is known to be involved in ciprofloxacin resistance (42, 43), was also identified as an important factor under conditions of ciprofloxacin treatment. In addition to these and other known intrinsic resistance factors, we identified 13 hypothetical genes that are important for resistance to these six antibiotics (see Table S1 in the supplemental material).
While no inactivated gene altered susceptibility to all six antibiotics, we did identify 21 genes that were hits with two or more antibiotics, and 8 of these were hits with more than two antibiotics (see Table S1 in the supplemental material). These 8 genes included mprF, ndh, fmtA, components of the graRS and vraFG (graRS/vraFG) multicomponent sensing system, and two genes of unknown function, SAOUHSC_01025 and SAOUHSC_01050.
Our ability to detect numerous previously identified resistance factors using this massively parallel fitness profiling approach served as a validation of the method. To further confirm the results and to determine whether these resistance factors were also important in other S. aureus strains, we examined the fitness of mutants in genes identified as hits against all six antibiotics under two or more sets of conditions using an agar spot dilution assay (Fig. 2; see also Fig. S1 in the supplemental material). Mutants were chosen based on whether they were identified as hits under two or more sets of conditions. In general, the agreement between Tn-Seq results and the spot dilution assay results was excellent. Given that the spot dilutions did not involve competition between thousands of mutants and that the assays were performed at a single concentration (chosen so that wild-type [WT] growth would be relatively unaffected; see Materials and Methods) and involved mutants from different genetic backgrounds (Newman or USA300 instead of HG003), the high validation rate is remarkable and supports the choice of the approach described here.
FIG 2 .

Tn-Seq results were validated by testing mutant fitness in spot dilution assays. Tn-Seq results and validation for selected genes are shown. ndh encodes an NADH dehydrogenase involved in oxidative phosphorylation; fmtA is a cell surface protein of undetermined function; graR is a member of a multicomponent sensing system (MCS); mprF and dltA are members of the regulon of this MCS. Asterisks indicate those conditions under which the pertinent gene was among the top 20 hits for that antibiotic. (A) Bar graph depicting fold change in reads per gene relative to the untreated control for each of the six antibiotics. As very few insertions in dltA were present in the untreated control, changes in fitness could not be detected by Tn-Seq. (B) Bar graph depicting fitness of mutant strains in which the indicated genes are inactivated compared to that of the WT. Fitness was assessed by spotting 10-fold dilutions of WT and mutant strains on antibiotic plates and comparing the highest dilutions that resulted in growth (see Materials and Methods). Because the data are shown on a logarithmic scale, the lack of a bar indicates that no change (fold change value of 1) was observed for that mutant under the relevant treatment condition. Fitness assessed by spot dilution validated the Tn-Seq results for the list of the top 20, with the exception of ndh with vancomycin treatment. Strain backgrounds were USA300 LAC JE2 for the ndh, fmtA, and mprF transposon mutants and Newman for the graR and dltA deletion mutants.
Intrinsic factors that impact multiple classes of antibiotics.
Of the eight genes found to impact multiple classes of antibiotics, ndh (NADH dehydrogenase) is the only one that, when inactivated, promotes resistance to some antibiotics while promoting sensitization to others. Ndh is a component of the electron transport chain. The electron transport chain creates a membrane potential, which is required for penetration of gentamicin through the cell membrane (44). We show here that, in addition to conferring resistance to gentamicin when inactivated, ndh is an intrinsic resistance factor for oxacillin, linezolid, and ciprofloxacin. ndh is in the same pathway as several genes that are commonly found to be inactivated in small colony variants (SCVs), a phenotype correlated with persistent infections that are resistant to β-lactams as well as aminoglycosides (45–48). Our data confirmed the importance of the oxidative phosphorylation pathway in antibiotic resistance, identified new genes of importance, and suggest that a better understanding of how S. aureus modulates this system could increase our understanding of antibiotic resistance.
FmtA is a cell surface protein of uncertain function. It was identified as a factor involved in methicillin resistance and has since been proposed to act as a carboxypeptidase and a teichoic acid d-ala esterase (49–51). We did not find that inactivation of fmtA resulted in increased sensitivity to oxacillin in the Tn-Seq analysis or in the agar spot dilution assay. In fact, in these tests, fitness was enhanced compared to that of other mutants under conditions of oxacillin selection. However, we did observe increased sensitivity of fmtA mutants to daptomycin and vancomycin. The impact of fmtA mutations showed that fmtA met the threshold for inclusion among the top 20 genes affecting daptomycin resistance but was not among the top 20 for vancomycin selection. Nevertheless, these data support the idea of an important role for FmtA in withstanding cell envelope stress for at least some classes of antibiotics (52).
MprF catalyzes formation of lyslyphosphatidylglycerol, a membrane modification that confers protection against cationic antibiotics, which are repelled by the increased cell surface positive charge (6, 8, 53, 54). MprF is also known to contribute to methicillin resistance in MRSA strains (55), and point mutations increasing the activity of MprF are a mechanism for daptomycin resistance (7, 56, 57). In our studies, mprF was found to be an intrinsic resistance factor for all antibiotics tested, although it occurred among the top 20 for only four of the six (Table 1 and Fig. 2A). Enhanced susceptibility of mprF mutants to these four antibiotics was also found in the agar spot dilution assay performed using mutants in the USA300 (MRSA) background (Fig. 2B) (58). Since mprF inactivation potently promotes sensitization to daptoymcin, vancomycin, and ciprofloxacin, which are not rich in positive charges, the positively charged product of MprF, lysylphosphatidylglycerol, likely plays biophysical roles in membrane stability or organization. This has been suggested previously for daptomycin (59, 60).
GraRS/VraFG is the most important MCS across tested antibiotics.
Multicomponent sensing systems (MCSs) allow bacteria to sense and respond to their environments. These systems typically include a membrane-anchored extracellular sensory domain fused to an intracellular kinase domain and a separate, cytosolic response regulator, but they can also include additional elements. A stimulus sensed by the sensory domain results in a change in the phosphorylation state of the response regulator, which in turn modulates the expression of downstream targets (61). S. aureus contains many multicomponent sensing systems, and we identified multiple components of three of these systems, agrABCD, vraTSR, and graXRS/vraFG, among the top hits under conditions of selection with at least one antibiotic (Table 1; see also Table S1 in the supplemental material).
AgrABCD is involved in quorum sensing and regulation of virulence factors and autolysin expression (62–64). Sequencing reads from transposon insertions mapping to components agrA, agrB, and agrD were depleted under conditions of oxacillin or daptomycin treatment, suggesting that factors regulated by this MCS are involved in the response to these antibiotics, possibly due to its regulation of the autolysin lytM and the penicillin-binding proteins (64–66). While reads mapping to agrC were also depleted under these treatment conditions, agrC did not meet the threshold for inclusion in the top 20 hits. The vraTSR system is known for its crucial role in withstanding vancomycin treatment (38), and all three components were among the top genes identified under conditions of vancomycin treatment. This sensing system regulates expression of cell wall biosynthetic genes, and it has also been implicated in β-lactam resistance (37, 39, 67). Although reads mapping to these genes were also depleted under conditions of oxacillin treatment, they were not depleted enough to be included among the top 20 genes contributing to oxacillin resistance.
The single most important MCS across all the six antibiotics tested is graXRS/vraFG. Four components of this system met our cutoffs under conditions of gentamicin, daptomycin, and vancomycin treatment (Table 1). Moreover, compared to that of the wild type, we found the fitness of a ΔgraR mutant plated on these antibiotics to be reduced by 4 to 5 orders of magnitude (Fig. 2B). This mutant was also sensitive to ciprofloxacin, although less so than to the other antibiotics tested. The graXRS/vraFG regulon includes other global regulators such as agr and walKR, and its function has been linked to numerous stress response and virulence genes (68). However, its best-characterized role is that of regulation of mprF and the dlt operon, both of which are involved in modulating cell surface charge. The dlt operon attaches d-alanine to lipoteichoic and wall teichoic acids (69), increasing the positive charge of the cell surface. The dlt pathway has been shown to modulate resistance to cationic antimicrobial peptides, aminoglycosides, and other positively charged antibiotics (70). Whereas mprF was identified as a top hit under several treatment conditions, transposon insertions in the dlt genes were poorly represented in the control libraries, because dlt mutants exhibit substantial fitness defects even in the absence of antibiotic selection and so do not compete well against the other mutants in the library. However, upon direct testing, we found the fitness of a dltA mutant plated on three of the six tested antibiotics—vancomycin, ciprofloxacin, and gentamicin—to be greatly reduced compared to that of the wild type (Fig. 2B). Binding of the zwitterionic fluoroquinolones to the cell surface is known to be antagonized by calcium or magnesium ions, and perhaps the presence of d-alanylation of cell wall components is similarly antagonistic (71). The sensitivity of dltA mutants to vancomycin has been previously reported and was suggested to be due to increased binding of vancomycin to the cell surface (72). It was previously shown that the number of positive charges on aminoglycosides correlates with activity against the dltA mutant (73), but in our tests, we did not observe a strong correlation between the number of positive charges and the fitness of the dltA mutant for different classes of antibiotics. As d-alanylation of the cell envelope results in pleiotropic effects, the fitness of the dltA mutant in the presence of different antibiotics likely reflects its different cellular roles. Nevertheless, our results suggest that the importance of the graRS/vraFG MCS can be explained in part by the combined action of two members of its regulon, dltA and mprF, which modify cell envelope charge.
Two previously uncharacterized genes are broadly important under cell envelope stress conditions.
Among the novel genes identified as hits under one or more sets of treatment conditions, two genes, SAOUHSC_01025 and SAOUHSC_01050, encoding polytopic membrane proteins, stood out as particularly important because both were identified as hits for three of the six antibiotics: oxacillin, vancomycin, and daptomycin. These genes are conserved in S. aureus. SAOUHSC_01025 is predicted to have 10 transmembrane domains, with a 93-amino-acid extracellular domain between helices six and seven, while SAOUHSC_01050 is predicted to have 3 transmembrane domains and a 191-amino-acid C-terminal extracellular domain. BLAST and PSI-BLAST searches performed with SAOUHSC_01025 and SAOUHSC_01050 did not reveal extensive amino acid identity with proteins from any other source or of any known function.
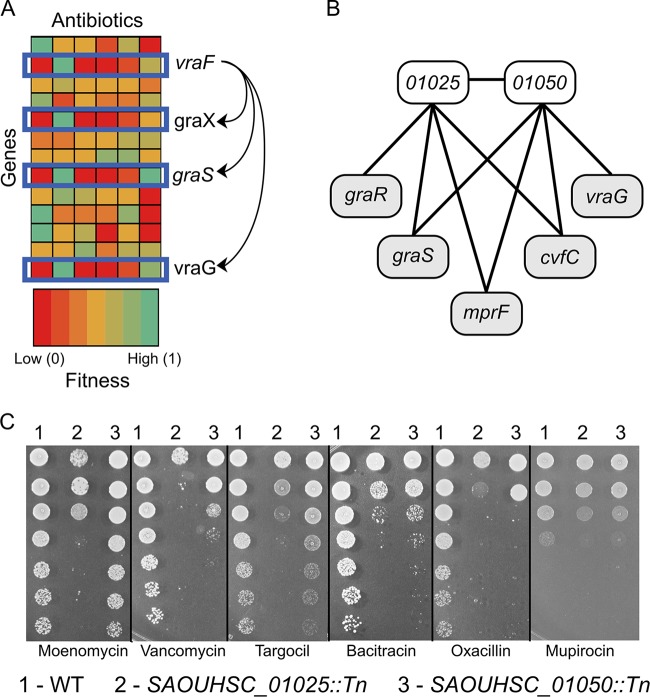
Taking advantage of the data generated via the Tn-Seq experiments, we used a machine learning approach to identify the genes with the fitness profiles (i.e., representing fitness of an inactivation insertion mutation in that gene under each set of antibiotic conditions) most similar to those of SAOUHSC_01025 and SAOUHSC_01050 (see Materials and Methods). We first tested the K-nearest neighbors algorithm by using graX, graS, vraF, or vraG as the query gene and then searched across all nonessential genes for those with the most similar fitness profiles for all six antibiotic selections. For each of these genes, at least two of the other components of the graXRS/vraFG MCS system were among the five genes that most closely paralleled their contribution to fitness in these tests. This result was expected because the individual components of the graXRS/vraFG system would be expected to exhibit similar fitness profiles (Fig. 3A). We then applied the algorithm to identify the five genes with the profiles most similar to those of mutants with insertions in SAOUHSC_01025 and SAOUHSC_01050. Transposon mutations in SAOUHSC_01025 and SAOUHSC_01050 were most similar in their effect to one another, and they shared three of four additional nearest neighbors, graS, mprF, and cvfC (Fig. 3B). In addition, graR and vraG were identified as similar to SAOUHSC_01025 and SAOUHSC_01050, respectively. These data collectively indicate that SAOUHSC_01025 and SAOUHSC_01050 are new factors involved in maintaining membrane integrity and withstanding envelope stress.
FIG 3 .
Two genes encoding polytopic membrane proteins were found to be important for withstanding antibiotics that target the cell envelope. (A) Schematic depicting the fitness of a subset of genes upon treatment with different antibiotics. Each column represents an antibiotic, and each row represents a gene. Genes with related functions, such as the components of the graRS-vraFG MCS, have similar fitness profiles across a panel of antibiotics. Therefore, for any given test gene, it is possible to identify the five genes with the most similar fitness profiles, and it is inferred that these genes are involved in pathways related to the test gene. (B) The K-nearest neighbors algorithm predicted that SAOUHSC_01025 and SAOUHSC_01050, which encode polytopic membrane proteins of unknown function, were most similar to one another and also shared similarity with three of four other identified neighbors. As these neighbors play an important role in protecting S. aureus from certain classes of antibiotics, we predict that SAOUHSC_01025 and SAOUHSC_01050 are important for cell envelope integrity. (C) Spot dilution assays showing fitness of inactivation mutants in SAOUHSC_01025 and SAOUHSC_01050 upon plating on the indicated antibiotics compared to that of the WT (for data representing additional antibiotics, see Fig. S2 in the supplemental material). The first five antibiotics target the cell envelope, and at least one of the two mutant strains is highly sensitive at an antibiotic concentration that permits growth of the WT at all dilutions. The sixth antibiotic targets protein translation, and the mutants show a decrease in fitness of only 1 log compared to that of the WT.
Using the agar spot dilution assay, we tested the fitness of mutants with transposon insertions in SAOUHSC_01025 (SAOUHSC_01025::Tn) and SAOUHSC_01050 (SAOUHSC_01050::Tn) against a panel of 12 antibiotics with a greater range of different targets. In addition to the six antibiotics originally used, we added moenomycin, targocil, bacitracin, fosfomycin, mupirocin, and rifampin (Fig. 3C; see also Fig. S2 in the supplemental material). Moenomycin inhibits peptidoglycan synthesis by binding to the extracellular transglycosylases that polymerize lipid II (74–76), while bacitracin inhibits the same pathway by binding to undecaprenylpyrophosphate released during lipid II polymerization, thereby preventing lipid II recycling and new lipid II synthesis (77). Fosfomycin inhibits peptidoglycan synthesis by inhibiting an intracellular enzyme, MurA (78). Targocil inhibits wall teichoic acid biosynthesis, resulting in depletion of peptidoglycan precursors and, therefore, inhibition of peptidoglycan synthesis (36, 79, 80). Mupirocin inhibits protein translation by targeting an acyl-tRNA synthetase (81), and rifampin inhibits RNA polymerase (82, 83). Against cell wall-active antibiotics, SAOUHSC_01025::Tn and SAOUHSC_01050::Tn generally showed large reductions in fitness, although SAOUHSC_01025::Tn was typically more susceptible than SAOUHSC_01050::Tn. Moenomycin provides a striking example of this: the fitness of the SAOUHSC_01025::Tn mutant decreased by 4 orders of magnitude whereas the fitness of the SAOUHSC_01050::Tn mutants did not change. Both the SAOUHSC_01025::Tn and SAOUHSC_01050::Tn mutants plated on non-cell wall-related antibiotics showed a more modest reduction in fitness. The one exception was that SAOUHSC_01050::Tn was found to be far more sensitive to gentamicin than SAOUHSC_01025::Tn, a unique behavior of SAOUHSC_01050::Tn not observed with any other antibiotic. The greatly decreased fitness of both SAOUHSC_01025::Tn and SAOUHSC_01050::Tn mutants with a wide range of cell wall-active antibiotics verifies that they play an important role in cell envelope integrity.
There are a number of ways in which SAOUHSC_01025 and SAOUHSC_01050 could affect cell envelope integrity. Because these genes have fitness profiles similar to those of graRS/vraFG and mprF, it is tempting to suggest that their expression is regulated by this MCS. However, this system has been very well studied, and these two genes have not been identified as members of the graRS/vraFG regulon (68, 84). Another possibility is that these genes are part of a parallel cell envelope stress pathway, responding to stresses similar to those responded to by the graRS/vraFG MCS. Both genes are predicted to encode proteins with extracellular domains. It is possible that these extracellular domains could serve a variety of functions, including acting as sensory domains which respond to a small molecule or other metabolites produced as a result of cell envelope stress. The possibility that these proteins could be acting as scaffolding proteins, coordinating cell envelope synthesis or repair, cannot be ruled out. The heightened sensitivity of SAOUHSC_01050::Tn to gentamicin suggests that this protein plays a role, directly or indirectly, in regulating cell membrane potential or cell envelope positive charge. Future work will provide insight into the cellular functions of these highly important intrinsic resistance factors.
Conclusion.
Treating transposon libraries with subinhibitory concentrations of antibiotics using a format that involves massively parallel competition between mutants allows the robust identification of factors that contribute to resistance. Mutant fitness based on Tn-seq profiles correlated well among multiple approaches, including assessment of fitness of individual mutants plated on antibiotics. Previously characterized intrinsic resistance factors were identified, validating the method, but the use of saturating mutant pools in a massively parallel assay also identified novel intrinsic resistance factors, including those contributing to resistance under conditions of treatment with antibiotics in multiple classes. We also used a machine learning approach to obtain insights into the physiological roles of genes annotated as hypothetical. Two novel intrinsic resistance factors, SAOUHSC_01025 and SAOUHSC_01050, were found to be important in withstanding cell envelope damage, but elucidation of the mechanism by which they do so will require further direct characterization. These or other intrinsic resistance factors may be of considerable value as targets for the development of small-molecule potentiators that extend the utility of existing and new antibiotics for treating S. aureus infections.
MATERIALS AND METHODS
Library 1 antibiotic treatment and DNA preparation.
Library 1 was constructed by transformation of a temperature-sensitive plasmid as previously described (26). Briefly, a 100-µl aliquot of this initial S. aureus HG003 transposon library freezer stock, containing 108 CFU, was used to inoculate 100 ml of Mueller-Hinton (MH) cation-adjusted broth and incubated for 15 h at 37°C with shaking at 200 rpm. A 10-µl aliquot (106 CFU) of this input culture was then inoculated into a final volume of 200 µl in a 96-well plate broth microdilution format and incubated at 37°C for 8 h, representing approximately 5.5 generations (5 × 107 CFU/200 µl). The 0.5×, 0.25×, and 0.125× MIC wells for the library pool were determined on the basis of the MIC of a small mutant pool (consisting of 10 innocuous transposon mutants). This small pool was used to determine MICs in order to compensate for resistant mutants potentially present in the library pool. The chosen wells (0.5×, 0.25×, and 0.125×) were then subcultured (3 × 105 CFU) into a second iteration of serial dilutions of antibiotics as described above and incubated for 15 h at 37°C, representing approximately 9 generations (2 × 108 CFU/200 µl). The 0.5×, 0.25×, and 0.125× MIC wells were determined based on the small pool, and these wells were transferred to 10 ml of brain heart infusion (BHI) broth and incubated for 4 h at 37°C with shaking at 200 rpm. Biological replicates were conducted for each growth condition. Genomic DNA was harvested using a DNeasy blood and tissue kit (Qiagen, Valencia, CA) following the manufacturer’s instructions. Library 1 was prepared for next-generation sequencing using the shearing method as described previously (27).
Library 2 antibiotic treatment and DNA preparation.
Library 2 was constructed by phage-based transposition of six different transposon constructs as previously described (14). Tryptic soy broth (TSB) supplemented with 25 mg/liter Ca2+ and 12.5 mg/liter Mg2+ was used for all antibiotics except oxacillin, which was tested using cation-adjusted Mueller-Hinton broth (MHB). For all antibiotics, an untreated control was prepared in the same media as was used for the tested antibiotic. A stock of the complete library was thawed and diluted to an optical density at 600 nm (OD600) of 0.2 and grown to an OD600 of ~0.4 to minimize changes in library composition prior to treatment. The culture was then diluted to 4 × 105 CFU/ml and added to 1 ml of media with 2× the desired concentration of the antibiotic to give a final starting inoculum of 2 × 105 CFU/ml in 2-ml culture volumes. A 107 CFU/2 ml starting inoculum was used for vancomycin- and daptomycin-treated samples. Samples were grown at 37°C and harvested when they reached stationary phase (~1 × 109 CFU/2 ml). The samples were treated with 2×, 1×, 0.5×, and 0.25× the MIC of the antibiotic. Then, we identified antibiotic concentrations that caused the transposon library to reach stationary phase with a few hours of delay compared to the untreated control. These antibiotic concentrations were prepared for sequencing following the protocol described by Santiago et al. (14). Samples from at least two of the concentrations of library 2 were sent for sequencing. Illumina sequencing was completed at the Harvard Biopolymers Facility or the Tufts Genomic DNA Sequencing Core Facility on a HiSeq 2500 sequencing system.
Data analysis of both libraries.
We identified datasets from library 2 where reads mapped to approximately 25% to 40% of the TA dinucleotide sites with hits in the untreated control (with the exception of the vancomycin treatment, which hit 67% of the TA dinucleotide sites hit in the untreated control). These were processed for further analysis. This percentage of decrease was chosen so that we could identify genes with an increase and a decrease in the number of reads mapping to them. Library 2 contains transposon constructs with outward-facing promoters that can upregulate proximal genes in addition to the traditional construct which can only insert into and inactivate genes. For these experiments, we considered only data from the inactivation constructs. Data for both library 1 and library 2 were analyzed as described previously (14), with the following modifications using in-house python scripts. Data from biological replicates were combined, and before the numbers of reads/gene were compared using the Mann-Whitney U test, the experimental data were normalized to the control data using simulation-based resampling (28, 29). Data for each antibiotic treatment from each of the library 1 experiments were then combined with the data from the library 2 experiments using the geometric mean of the ratios of reads in the antibiotic-treated sample compared to the control and Fisher’s method for combining corrected P values. Top hits were identified first by filtering for genes with a P value of less than 0.05 and then by increasing the fold change cutoff value by integers until 20 genes or fewer were left.
Spot dilution assays.
Identified hits qoxA, qoxB, ndh, fmtA, SAOUHSC_01025, and SAOUHSC_01050 were validated using transposon mutants from the Nebraska library in the USA300 LAC JE2 background (58). dltA and graR deletion mutants were tested in methicillin-susceptible S. aureus (MRSA) strain Newman (17). Agar plates were prepared with TSB supplemented with Ca2+ (25 mg/liter) and Mg2+ (12.5 mg/liter) and the six antibiotics at concentrations below the MIC. Overnight cultures of mutants were diluted 1:100 in fresh TSB and grown to an OD600 of 1. They were then serially diluted 10-fold, spotted onto an agar plate, and incubated at 37°C overnight. The concentration of the antibiotic at which growth of the WT was severely inhibited, showing growth in only the highest 1 or 2 dilutions on agar plates under these conditions, was determined. This was considered to be the MIC under these conditions. The spot dilution assays were then set up using three different antibiotic concentrations. The concentration closest to the MIC at which the WT was at most 3 logs more depleted than the control was used to calculate fitness. This concentration was used so that reduced fitness of any mutants could be observed. The exception to this was the use of the MIC for gentamicin to evaluate the resistance of inactivation mutants in the oxidative phosphorylation pathway (see Fig. S1 in the supplemental material). Control plates with no antibiotic were set up for all strains assayed, and under these conditions, the mutant and WT strains showed equal levels of growth (not shown). Fitness was assessed by determining the highest dilution for which growth was observed for a mutant and the WT strain. The highest dilution showing full growth for the mutant was then divided by the highest dilution showing full growth for the WT to calculate the fitness of the mutant compared to that of the WT, and the results were plotted on a log scale. Those spots that showed hazy growth indicative of cell lysis, those that showed mixed populations of colonies of different sizes that suggested the possible presence of suppressors and reduced fitness relative to spots with homogenous colonies, and those that had fewer than 10 individual colonies were regarded as not representative of full growth.
Machine learning algorithm optimization.
We used the machine learning algorithm K-nearest nearest neighbors to identify other genes with similar resistance and sensitization patterns in an unsupervised manner using the Scikit-learn python library (85). However, because of the different selective pressures exerted by each antibiotic, the ratio of reads under the experimental treatment conditions versus the control conditions that map to each gene could not be used as the metric for classification. In addition, we wanted to distinguish between the two following conditions: (i) a ratio change of 0.1 due to 100 reads in the control and 10 reads in the experiment and (ii) a ratio change of 0.1 due to 1,000 reads in the control and 100 reads in the experiment. For two genes of same length, option 1 is much less relevant than option 2, as 100 reads/gene and 10 reads/gene both correspond to a gene with a significant fitness defect whereas a change from 1,000 reads/gene to 100 reads/gene is more likely to be a significant change. Therefore, we converted the ratios to a more appropriate fitness measurement value by first choosing a value for the minimum number of reads per gene that could be considered interesting. Based on empirical observations, we noticed that essential/fitness-defective genes tended to have <1/10,000 of the total number of reads in the sample, so the value for any gene with fewer reads mapping to it was converted to this value. Then, ratios were recalculated. Next, the new modified ratio was multiplied by the number of reads mapping to that gene under the treatment conditions and was normalized to the length of the gene. Genes were ordered from lowest to highest “fitness” level. To place all the samples on the same scale, the gene with the lowest “fitness” was given a value of 0, and the gene with the highest “fitness” was given a value of 1. All other genes were placed in order between these values, in increments that increased by 1/(total number of genes). This final value, which we call the “normalized fitness value,” was subsequently used in the machine learning analysis. Essential genes were removed to reduce bias in the data set, and the K-nearest neighbors algorithm was further optimized by adjusting the Minkowski distance metric to output the genes with the resistance/sensitization patterns most similar to those of the test gene. We identified the five genes (the five nearest neighbors) with the most similar patterns of “normalized fitness values.”
Data availability.
All raw next-generation sequencing data as well as the python scripts used in the analysis are available on the publically accessible Harvard Dataverse Network at https://dataverse.harvard.edu/dataverse/intrinsicresistancefactordata.
SUPPLEMENTAL MATERIAL
Inactivation of the oxidative phosphorylation pathway confers resistance to gentamicin. (A) Schematic of the oxidative phosphorylation pathway is depicted here. Numbers of reads due to transposon insertions were greatly increased under conditions of exposure to gentamicin for the 11 genes illustrated. Inactivation of the oxidative phosphorylation pathway is a known mechanism of resistance to gentamicin, which depends on the membrane potential for cell entry. (B) Results representative of the fold change in the number of reads/gene for a subset of genes involved in oxidative phosphorylation that were tested to determine if inactivation confers resistance to gentamicin. (C) Fitness comparison of the WT to mutant strains in which the indicated genes were inactivated. Spot dilutions of WT and mutant strains were plated on gentamicin, and fitness was calculated as the ratio of the highest dilution that allowed growth of the WT to highest dilution that allowed growth of the mutant (see Materials and Methods). Download
SAOUHSC_01025::Tn and SAOUHSC_01050::Tn are particularly sensitive to antibiotics that damage the cell envelope. (A) Data representative of results of analysis of the targeted pathways of the additional antibiotics tested against SAOUHSC_01025::Tn and SAOUHSC_01050::Tn mutants are shown. The abbreviations used here are as follows: mup, mupirocin; lin, linezolid; rif, rifampin; cip, ciprofloxacin; gen, gentamicin; van, vancomycin; bac, bacitracin; tar, targocil; fos, fosfomycin; moe, moenomycin A; dap, daptomycin. (B) A summary of the fitness of these mutants relative to that of the WT was assessed by spot dilution against the various antibiotics tested. (C) Spot dilution assay plates for these mutants and all antibiotics tested are shown here. The results obtained with the first six antibiotics are reproduced from Fig. 3 for comparison. Download
A total of 80 unique genes were identified as important for fitness by treatment of pooled transposon libraries with six antibiotics. The top 20 genes with the greatest fold change in numbers of mapped reads are shown for each antibiotic. Fold change in the number of mapped reads is indicated by colored rectangles. Orange rectangles indicate genes for which the numbers of reads due to transposon insertions were substantially lower than in the control, whereas blue rectangles indicate genes for which the numbers of reads due to transposon insertions were substantially higher than in the control. Gray rectangles indicate that they were not identified as a hit using that antibiotic treatment.
ACKNOWLEDGMENTS
Portions of this research performed in the Gilmore and Walker laboratories were supported by the Harvard-wide Program on Antibiotic Resistance (P01 AI083214). Work in the laboratory of M.S.G. was also supported by a grant from Institut Merieux. Work in the laboratory of S.W. was also supported by a Center for Excellence in Translational research grant (U19 AI109764).
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Rajagopal M, Martin MJ, Santiago M, Lee W, Kos VN, Meredith T, Gilmore MS, Walker S. 2016. Multidrug intrinsic resistance factors in Staphylococcus aureus identified by profiling fitness within high-diversity transposon libraries. mBio 7(4):e00950-16. doi:10.1128/mBio.00950-16.
REFERENCES
- 1.Chambers HF. 1988. Methicillin-resistant staphylococci. Clin Microbiol Rev 1:173–186. doi: 10.1128/CMR.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Utsui Y, Yokota T. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother 28:397–403. doi: 10.1128/AAC.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol 158:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger-Bächi B. 1995. Factors affecting methicillin resistance in Staphylococcus aureus. Int J Antimicrob Agents 6:13–21. doi: 10.1016/0924-8579(95)00021-Y. [DOI] [PubMed] [Google Scholar]
- 5.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med 193:1067–1076. doi: 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishi H, Komatsuzawa H, Fujiwara T, McCallum N, Sugai M. 2004. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob Agents Chemother 48:4800–4807. doi: 10.1128/AAC.48.12.4800-4807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman L, Alder JD, Silverman JA. 2006. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother 50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang SJ, Xiong YQ, Dunman PM, Schrenzel J, François P, Peschel A, Bayer AS. 2009. Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. Antimicrob Agents Chemother 53:2636–2637. doi: 10.1128/AAC.01415-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soldo B, Lazarevic V, Karamata D. 2002. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148:2079–2087. doi: 10.1099/00221287-148-7-2079. [DOI] [PubMed] [Google Scholar]
- 10.Campbell J, Singh AK, Santa Maria JP Jr, Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S. 2011. Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosyntheses in Staphylococcus aureus. ACS Chem Biol 6:106–116. doi: 10.1021/cb100269f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. 2010. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci U S A 107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann PA, Müller A, Wolff KA, Fischmann T, Wang H, Reed P, Hou Y, Li W, Müller CE, Xiao J, Murgolo N, Sher X, Mayhood T, Sheth PR, Mirza A, Labroli M, Xiao L, McCoy M, Gill CJ, Pinho MG, Schneider T, Roemer T. 2016. Chemical genetic analysis and functional characterization of staphylococcal wall teichoic acid 2-epimerases reveals unconventional antibiotic drug targets. PLoS Pathog 12:e00950-16. doi: 10.1371/journal.ppat.1005585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santiago M, Matano LM, Moussa SH, Gilmore MS, Walker S, Meredith TC. 2015. A new platform for ultra-high density Staphylococcus aureus transposon libraries. BMC Genomics 16:252. doi: 10.1186/s12864-015-1361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Opijnen T, Bodi KL, Camilli A. 2009. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00950-16. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santa Maria JP Jr, Sadaka A, Moussa SH, Brown S, Zhang YJ, Rubin EJ, Gilmore MS, Walker S. 2014. Compound-gene interaction mapping reveals distinct roles for Staphylococcus aureus teichoic acids. Proc Natl Acad Sci U S A 111:12510–12515. doi: 10.1073/pnas.1404099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan Y, Lazinski D, Rowe S, Camilli A, Lewis K. 2015. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 6:e00950-16. doi: 10.1128/mBio.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman JA, Perlmutter NG, Shapiro HM. 2003. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother 47:2538–2544. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol 194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drlica K. 1999. Mechanism of fluoroquinolone action. Curr Opin Microbiol 2:504–508. doi: 10.1016/S1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 22.Barna JC, Williams DH. 1984. The structure and mode of action of glycopeptide antibiotics of the vancomycin group. Annu Rev Microbiol 38:339–357. doi: 10.1146/annurev.mi.38.100184.002011. [DOI] [PubMed] [Google Scholar]
- 23.Frère J. 1977. Mechanism of action of beta-lactam antibiotics at the molecular level. Biochem Pharmacol 26:2203–2210. doi: 10.1016/0006-2952(77)90280-5. [DOI] [PubMed] [Google Scholar]
- 24.Kotra LP, Haddad J, Mobashery S. 2000. Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother 44:3249–3256. doi: 10.1128/AAC.44.12.3249-3256.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinabarger D. 1999. Mechanism of action of the oxazolidinone antibacterial agents. Expert Opin Investig Drugs 8:1195–1202. doi: 10.1517/13543784.8.8.1195. [DOI] [PubMed] [Google Scholar]
- 26.Valentino MD, Foulston L, Sadaka A, Kos VN, Villet RA, Santa Maria J Jr, Lazinski DW, Camilli A, Walker S, Hooper DC, Gilmore MS. 2014. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 5:e00950-16. doi: 10.1128/mBio.01729-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein BA, Tenorio EL, Lazinski DW, Camilli A, Duncan MJ, Hu LT. 2012. Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics 13:578. doi: 10.1186/1471-2164-13-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chao MC, Pritchard JR, Zhang YJ, Rubin EJ, Livny J, Davis BM, Waldor MK. 2013. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nucleic Acids Res 41:9033–9048. doi: 10.1093/nar/gkt654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e00950-16. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mates SM, Eisenberg ES, Mandel LJ, Patel L, Kaback HR, Miller MH. 1982. Membrane potential and gentamicin uptake in Staphylococcus aureus. Proc Natl Acad Sci U S A 79:6693–6697. doi: 10.1073/pnas.79.21.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryan LE, Van Den Elzen HM. 1977. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob Agents Chemother 12:163–177. doi: 10.1128/AAC.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morikawa K, Maruyama A, Inose Y, Higashide M, Hayashi H, Ohta T. 2001. Overexpression of sigma factor, sigma(B), urges Staphylococcus aureus to thicken the cell wall and to resist beta-lactams. Biochem Biophys Res Commun 288:385–389. doi: 10.1006/bbrc.2001.5774. [DOI] [PubMed] [Google Scholar]
- 33.Singh VK, Schmidt JL, Jayaswal RK, Wilkinson BJ. 2003. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int J Antimicrob Agents 21:256–261. doi: 10.1016/S0924-8579(02)00359-X. [DOI] [PubMed] [Google Scholar]
- 34.Łeski TA, Tomasz A. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol 187:1815–1824. doi: 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. 2008. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother 52:3955–3966. doi: 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiao Y, Lebar MD, Schirner K, Schaefer K, Tsukamoto H, Kahne D, Walker S. 2014. Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J Am Chem Soc 136:14678–14681. doi: 10.1021/ja508147s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuroda M, Kuroda H, Oshima T, Takeuchi F, Mori H, Hiramatsu K. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49:807–821. doi: 10.1046/j.1365-2958.2003.03599.x. [DOI] [PubMed] [Google Scholar]
- 38.Qureshi NK, Yin S, Boyle-Vavra S. 2014. The role of the staphylococcal VraTSR regulatory system on vancomycin resistance and vanA operon expression in vancomycin-resistant Staphylococcus aureus. PLoS One 9:e00950-16. doi: 10.1371/journal.pone.0085873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardete S, Wu SW, Gill S, Tomasz A. 2006. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob Agents Chemother 50:3424–3434. doi: 10.1128/AAC.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin S, Daum RS, Boyle-Vavra S. 2006. VraSR two-component regulatory system and its role in induction of pbp2 and vraSR expression by cell wall antimicrobials in Staphylococcus aureus. Antimicrob Agents Chemother 50:336–343. doi: 10.1128/AAC.50.1.336-343.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, Singh VK, Jayaswal RK, Wilkinson BJ. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719–2732. doi: 10.1099/mic.0.26426-0. [DOI] [PubMed] [Google Scholar]
- 42.Ubukata K, Itoh-Yamashita N, Konno M. 1989. Cloning and expression of the norA gene for fluoroquinolone resistance in Staphylococcus aureus. Antimicrob Agents Chemother 33:1535–1539. doi: 10.1128/AAC.33.9.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neyfakh AA, Borsch CM, Kaatz GW. 1993. Fluoroquinolone resistance protein NorA of Staphylococcus aureus is a multidrug efflux transporter. Antimicrob Agents Chemother 37:128–129. doi: 10.1128/AAC.37.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schurig-Briccio LA, Yano T, Rubin H, Gennis RB. 2014. Characterization of the type 2 NADH:menaquinone oxidoreductases from Staphylococcus aureus and the bactericidal action of phenothiazines. Biochim Biophys Acta 1837:954–963. doi: 10.1016/j.bbabio.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 46.Proctor RA, Kahl B, von Eiff C, Vaudaux PE, Lew DP, Peters G. 1998. Staphylococcal small colony variants have novel mechanisms for antibiotic resistance. Clin Infect Dis 27(Suppl 1):S68–S74. doi: 10.1086/514906. [DOI] [PubMed] [Google Scholar]
- 47.Bulger RJ. 1969. In vitro studies on highly resistant small colony variants of Staphylococcus aureus resistant to methicillin. J Infect Dis 120:491–494. doi: 10.1093/infdis/120.4.491. [DOI] [PubMed] [Google Scholar]
- 48.Schnitzer RJ, Camagni LJ, Buck M. 1943. Resistance of small colony variants (G-forms) of a staphylococcus towards the bacteriostatic activity of penicillin. Exp Biol Med 53:75–78. doi: 10.3181/00379727-53-14192. [DOI] [Google Scholar]
- 49.Komatsuzawa H, Sugai M, Ohta K, Fujiwara T, Nakashima S, Suzuki J, Lee CY, Suginaka H. 1997. Cloning and characterization of the fmt gene which affects the methicillin resistance level and autolysis in the presence of Triton X-100 in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 41:2355–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qamar A, Golemi-Kotra D. 2012. Dual roles of FmtA in Staphylococcus aureus cell wall biosynthesis and autolysis. Antimicrob Agents Chemother 56:3797–3805. doi: 10.1128/AAC.00187-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rahman MM, Hunter HN, Prova S, Verma V, Qamar A, Golemi-Kotra D. 2016. The Staphylococcus aureus methicillin resistance factor FmtA is a d-amino esterase that acts on teichoic acids. mBio 7:e00950-16. doi: 10.1128/mBio.02070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Komatsuzawa H, Ohta K, Labischinski H, Sugai M, Suginaka H. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 43:2121–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. 2004. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 150:45–51. doi: 10.1099/mic.0.26706-0. [DOI] [PubMed] [Google Scholar]
- 54.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 5:e00950-16. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komatsuzawa H, Ohta K, Fujiwara T, Choi GH, Labischinski H, Sugai M. 2001. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 203:49–54. doi: 10.1111/j.1574-6968.2001.tb10819.x. [DOI] [PubMed] [Google Scholar]
- 56.Julian K, Kosowska-Shick K, Whitener C, Roos M, Labischinski H, Rubio A, Parent L, Ednie L, Koeth L, Bogdanovich T, Appelbaum PC. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob Agents Chemother 51:3445–3448. doi: 10.1128/AAC.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones T, Yeaman MR, Sakoulas G, Yang SJ, Proctor RA, Sahl HG, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother 52:269–278. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00950-16. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mishra NN, Bayer AS, Weidenmaier C, Grau T, Wanner S, Stefani S, Cafiso V, Bertuccio T, Yeaman MR, Nast CC, Yang SJ. 2014. Phenotypic and genotypic characterization of daptomycin-resistant methicillin-resistant Staphylococcus aureus strains: relative roles of mprF and dlt operons. PLoS One 9:e00950-16. doi: 10.1371/journal.pone.0107426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mishra NN, Yang SJ, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:2312–2318. doi: 10.1128/AAC.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 62.Arvidson S, Tegmark K. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int J Med Microbiol 291:159–170. doi: 10.1078/1438-4221-00112. [DOI] [PubMed] [Google Scholar]
- 63.George EA, Muir TW. 2007. Molecular mechanisms of agr quorum sensing in virulent staphylococci. Chembiochem 8:847–855. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- 64.Singh VK, Carlos MR, Singh K. 2010. Physiological significance of the peptidoglycan hydrolase, LytM, in Staphylococcus aureus. FEMS Microbiol Lett 311:167–175. doi: 10.1111/j.1574-6968.2010.02087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lioliou E, Fechter P, Caldelari I, Jester BC, Dubrac S, Helfer AC, Boisset S, Vandenesch F, Romby P, Geissmann T. 2016. Various checkpoints prevent the synthesis of Staphylococcus aureus peptidoglycan hydrolase LytM in the stationary growth phase. RNA Biol 13:427–440 doi: 10.1080/15476286.2016.1153209:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Píriz Durán S, Kayser FH, Berger-Bächi B. 1996. Impact of sar and agr on methicillin resistance in Staphylococcus aureus. FEMS Microbiol Lett 141:255–260. [DOI] [PubMed] [Google Scholar]
- 67.Boyle-Vavra S, Yin S, Jo DS, Montgomery CP, Daum RS. 2013. VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother 57:83–95. doi: 10.1128/AAC.01651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Falord M, Mäder U, Hiron A, Débarbouillé M, Msadek T. 2011. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 6:e00950-16. doi: 10.1371/journal.pone.0021323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev 67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 71.Kotera Y, Watanabe M, Yoshida S, Inoue M, Mitsuhashi S. 1991. Factors influencing the uptake of norfloxacin by Escherichia coli. J Antimicrob Chemother 27:733–739. doi: 10.1093/jac/27.6.733. [DOI] [PubMed] [Google Scholar]
- 72.Peschel A, Vuong C, Otto M, Götz F. 2000. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob Agents Chemother 44:2845–2847. doi: 10.1128/AAC.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pasquina L, Santa Maria JP Jr, McKay Wood B, Moussa SH, Matano LM, Santiago M, Martin SE, Lee W, Meredith TC, Walker S. 2016. A synthetic lethal approach for compound and target identification in Staphylococcus aureus. Nat Chem Biol 12:40–45. doi: 10.1038/nchembio.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ostash B, Walker S. 2010. Moenomycin family antibiotics: chemical synthesis, biosynthesis, and biological activity. Nat Prod Rep 27:1594–1617. doi: 10.1039/c001461n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rebets Y, Lupoli T, Qiao Y, Schirner K, Villet R, Hooper D, Kahne D, Walker S. 2014. Moenomycin resistance mutations in Staphylococcus aureus reduce peptidoglycan chain length and cause aberrant cell division. ACS Chem Biol 9:459–467. doi: 10.1021/cb4006744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wallhausser KH, Nesemann G, Prave P, Steigler A. 1965. Moenomycin, a new antibiotic. I. Fermentation and isolation. Antimicrob Agents Chemother (Bethesda) 5:734–736. [PubMed] [Google Scholar]
- 77.Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C(55)-isoprenyl pyrophosphate. Proc Natl Acad Sci U S A 68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci 235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 79.Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S. 2009. Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Staphylococcus aureus. ACS Chem Biol 4:875–883. doi: 10.1021/cb900151k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee K, Campbell J, Swoboda JG, Cuny GD, Walker S. 2010. Development of improved inhibitors of wall teichoic acid biosynthesis with potent activity against Staphylococcus aureus. Bioorg Med Chem Lett 20:1767–1770. doi: 10.1016/j.bmcl.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hughes J, Mellows G. 1978. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coli by pseudomonic acid. Biochem J 176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hartmann G, Honikel KO, Knüsel F, Nüesch J. 1967. The specific inhibition of the DNA-directed RNA synthesis by rifamycin. Biochim Biophys Acta 145:843–844. doi: 10.1016/0005-2787(67)90147-5. [DOI] [PubMed] [Google Scholar]
- 83.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. 2001. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104:901–912. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 84.Falord M, Karimova G, Hiron A, Msadek T. 2012. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother 56:1047–1058. doi: 10.1128/AAC.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. 2011. Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inactivation of the oxidative phosphorylation pathway confers resistance to gentamicin. (A) Schematic of the oxidative phosphorylation pathway is depicted here. Numbers of reads due to transposon insertions were greatly increased under conditions of exposure to gentamicin for the 11 genes illustrated. Inactivation of the oxidative phosphorylation pathway is a known mechanism of resistance to gentamicin, which depends on the membrane potential for cell entry. (B) Results representative of the fold change in the number of reads/gene for a subset of genes involved in oxidative phosphorylation that were tested to determine if inactivation confers resistance to gentamicin. (C) Fitness comparison of the WT to mutant strains in which the indicated genes were inactivated. Spot dilutions of WT and mutant strains were plated on gentamicin, and fitness was calculated as the ratio of the highest dilution that allowed growth of the WT to highest dilution that allowed growth of the mutant (see Materials and Methods). Download
SAOUHSC_01025::Tn and SAOUHSC_01050::Tn are particularly sensitive to antibiotics that damage the cell envelope. (A) Data representative of results of analysis of the targeted pathways of the additional antibiotics tested against SAOUHSC_01025::Tn and SAOUHSC_01050::Tn mutants are shown. The abbreviations used here are as follows: mup, mupirocin; lin, linezolid; rif, rifampin; cip, ciprofloxacin; gen, gentamicin; van, vancomycin; bac, bacitracin; tar, targocil; fos, fosfomycin; moe, moenomycin A; dap, daptomycin. (B) A summary of the fitness of these mutants relative to that of the WT was assessed by spot dilution against the various antibiotics tested. (C) Spot dilution assay plates for these mutants and all antibiotics tested are shown here. The results obtained with the first six antibiotics are reproduced from Fig. 3 for comparison. Download
A total of 80 unique genes were identified as important for fitness by treatment of pooled transposon libraries with six antibiotics. The top 20 genes with the greatest fold change in numbers of mapped reads are shown for each antibiotic. Fold change in the number of mapped reads is indicated by colored rectangles. Orange rectangles indicate genes for which the numbers of reads due to transposon insertions were substantially lower than in the control, whereas blue rectangles indicate genes for which the numbers of reads due to transposon insertions were substantially higher than in the control. Gray rectangles indicate that they were not identified as a hit using that antibiotic treatment.
Data Availability Statement
All raw next-generation sequencing data as well as the python scripts used in the analysis are available on the publically accessible Harvard Dataverse Network at https://dataverse.harvard.edu/dataverse/intrinsicresistancefactordata.