Abstract
The p21-activated kinases (PAKs) of group I are the main effectors for the small Rho GTPases, critically involved in neurodevelopment, plasticity and maturation of the nervous system. Moreover, the neuronal complexity controlled by PAK1/PAK3 signaling determines the postnatal brain size and synaptic properties. Stress induces alterations at the level of structural and functional synaptic plasticity accompanied by reductions in size and activity of the hippocampus and the prefrontal cortex (PFC). These abnormalities are likely to contribute to the pathology of depression and, in part, reflect impaired cytoskeleton remodeling pointing to the role of Rho GTPase signaling. Thus, the present study assessed the expression of the group I PAKs and their activators in the brain of depressed subjects. Using qPCR, mRNA levels and coexpression of the group I PAKs: PAK1, PAK2, and PAK3 as well as of their activators: RAC1, CDC42 and ARHGEF7 were examined in postmortem samples from the PFC (n=25) and the hippocampus (n=23) of subjects with depression and compared to control subjects (PFC n=24; hippocampus n=21). Results demonstrated that mRNA levels of PAK1 and PAK3, are significantly reduced in the brain of depressed subjects, with PAK1 being reduced in the PFC and PAK3 in the hippocampus. No differences were observed for the ubiquitously expressed PAK2. Following analysis of gene coexpression demonstrated disruption of coordinated gene expression in the brain of subjects with depression. Abnormalities in mRNA expression of PAK1 and PAK3 as well as their altered coexpression patterns were detected in the brain of subjects with depression.
Keywords: postmortem, mRNA expression, coexpression analysis, p21-activated kinase, qPCR
Graphical abstract
It has been widely accepted that stress induces alterations at the level of structural and functional synaptic plasticity that are likely to contribute to the pathology of depression (Pittenger and Duman, 2008, Christoffel et al., 2011). For example, reductions in size and activity of the hippocampus and the prefrontal cortex (PFC) in different types of depression have been reported. The neuropathological correlates of these abnormalities include reductions in synapses or synaptic proteins, elevations in neuronal density, and reductions in neuronal size and neuropil (Rajkowska et al., 1999, Stockmeier et al., 2004, Drevets et al., 2008, Pittenger and Duman, 2008, Kang et al., 2012). Accordingly, shortening and reduced complexity of dendritic trees as well as reductions in dendritic spines and synapses has been reported also in the PFC and in the hippocampus in animal models of chronic stress (McKittrick et al., 2000, Radley et al., 2004, Drevets et al., 2008, Pittenger and Duman, 2008).
The intracellular mechanisms promoting these changes and their relevance to behavioral outcomes are poorly understood. Nevertheless, emerging evidence suggests that these structural abnormalities, in part, reflect impaired cytoskeleton remodeling and point to the role of Rho GTPase signaling as a central contributor.
The p21-activated kinases (PAKs), a family of serine/threonine protein kinases, are the main effectors for the small Rho GTPases of the RAC1 and CDC42 family. The three kinases PAK1, PAK2 and PAK3 constitute together PAKs of group I (Dan et al., 2001). It has been shown that group I PAKs have different tissue distributions: PAK1 is highly expressed in the brain and spleen, PAK2 is ubiquitously expressed and PAK3 is mainly expressed in brain (Manser et al., 1995, Teo et al., 1995, Rousseau et al., 2003). PAKs serve as key regulators of cytoskeleton dynamics and cell motility, cell cycle progression, as well as of death and survival events (Kreis and Barnier, 2009, Rane and Minden, 2014). Recent studies on neurons indicate that PAKs are important for neurite outgrowth, neuronal migration, spine morphology, and synaptic and behavioral plasticity (Daniels et al., 1998, Hayashi et al., 2002, Boda et al., 2004, Hayashi et al., 2004, Marler et al., 2005, Zhang et al., 2005, Sakakibara and Horwitz, 2006, Cobos et al., 2007, Hayashi et al., 2007, Jacobs et al., 2007, Kreis et al., 2007, Boda et al., 2008, Causeret et al., 2009). More importantly, it has been shown that the postnatal brain size and synaptic properties are determined by the neuronal complexity controlled by PAK1 and PAK3 signaling. Double knockout mice lacking both PAK1 and PAK3 exhibit reduced brain size that is accompanied by minimal changes in total cell count, due to a significant increase in cell density. Moreover, double knockout neurons have smaller soma, with markedly simplified dendritic arbors/ axons, and reduced synapse density (Huang et al., 2011).
Due to the critical involvement of PAK signaling in neuronal physiology, dysregulation of group I PAKs in brain often leads to neuronal development disorders as well as neurodegenerative pathologies such as Alzheimer’s disease, mental retardation (with PAK3 being specifically involved), and Huntington’s disease (Ma et al., 2012). On the other hand, in rats prone to depression, PAK1 was found to be downregulated in the frontal cortex when compared to animals resistant to depression (Nakatani et al., 2007).
Since the expression levels and activity of these neuroplasticity-related kinases are influenced by various factors under pathological conditions associated with neurodevelopment, neuroplasticity and maturation of the nervous system, we hypothesized that PAK signaling could be altered also in the brain of depressed subjects. Therefore, in the present study we examined the expression of PAKs of group I: PAK1, PAK2, and PAK3 as well as the expression of their regulators RAC1, CDC42 and ARHGEF7, known regulators of synaptic structure, in the hippocampus (n=23) and the PFC (n=25) of subjects with depression. Gene products that function together in common signaling cascades are expected to show greater similarities in their expression patterns than random sets of gene products (Vidal et al., 2011). Thus, correlation analyses were also conducted to determine if networks of co-expressed genes exist in control subjects and whether these networks are disrupted in the brain of subjects with depression.
2. Experimental Procedures
2.1. Subjects
Brain tissues were collected by the Brain Collection Program of the Maryland Psychiatric Research Center, Baltimore. The same cohort has been used previously in various studies published by our group (Dwivedi et al., 2008, Dwivedi et al., 2009, 2010, Pandey et al., 2014, Fuchsova et al., 2015). All the tissues from neuropsychiatric controls and depressed subjects were screened for macro- and microscopic neuropathological abnormalities by board-certified neuropathologists at the brain collection program. The presence of Alzheimer’s disease, infarcts, demyelinating diseases, or heterotopia (or clinical history of these disorders) disqualified subjects from the study. Brain samples were free of human immunodeficiency virus antibodies. Toxicology screening for alcohol and screening for antidepressant/psychoactive drugs taken prior to death were performed in blood/urine of each subject. Psychiatric diagnoses in depressed and control subjects were performed using the Diagnostic Evaluation After Death (Salzman et al., 1983) and the Structured Clinical Interview for DSM-IV (Spitzer et al., 1995). All subjects in the depression group died by suicide. The present studies were performed in the PFC in Brodmann area 9 in 25 depressed subjects and 24 nonpsychiatric control subjects. The PFC was defined as the gray matter from the most anterior 1-cm coronal slice of the cortex and was further dissected according to the Brodmann atlas. White matter was removed as much as possible, but there was still some white matter left. Hippocampi were available for 23 depressed subjects and 21 nonpsychiatric controls. This study was approved by the institutional review board of the University of Illinois at Chicago, and written informed consent was obtained from next-of-kin for each subject. Detailed demographic characteristics of subjects is provided in Table 1.
Table 1.
Demographic Characteristics of Subjects with Depression and Normal Control Subjects
| Age (years) | Race | Gender | PMI (hours) | Brain pH | Cause of Death | Psychotropic drugs (at the time of death) | Psychiatric Diagnosis | ||
|---|---|---|---|---|---|---|---|---|---|
| Normal Control Subjects a | |||||||||
| 1 | CONTROL | 22 | Black | Male | 19 | 6.9 | GSW | None | Normal |
| 2 | CONTROL | 42 | White | Female | 23 | 7.2 | Pneumonia | None | Normal |
| 3 | CONTROL | 37 | Black | Male | 5 | 7.1 | ASCVD | None | Normal |
| 4 | CONTROL | 31 | Black | Male | 8 | 7.2 | GSW | None | Normal |
| 5 | CONTROL | 46 | Black | Male | 9 | 7.1 | Multiple injuries | None | Normal |
| 6 | CONTROL | 33 | White | Male | 15 | 7.0 | GSW | None | Normal |
| 7 | CONTROL | 48 | White | Male | 26 | 6.9 | ASCVD | None | Normal |
| 8 | CONTROL | 40 | White | Female | 7 | 7.0 | ASCVD | None | Normal |
| 9 | CONTROL | 23 | Black | Male | 15 | 6.8 | GSW | None | Normal |
| 10 | CONTROL | 38 | Black | Male | 16 | 6.9 | Lung Sarcoidosis | None | Normal |
| 11 | CONTROL | 65 | Black | Female | 23 | 6.9 | ASCVD | None | Normal |
| 12 | CONTROL | 52 | White | Male | 30 | 7.3 | ASCVD | None | Normal |
| 13 | CONTROL | 35 | White | Male | 24 | 6.9 | Crush injury to abdomen and chest | None | Normal |
| 14 | CONTROL | 37 | White | Male | 24 | 7.0 | ASCVD | None | Normal |
| 15 | CONTROL | 45 | White | Male | 22 | 7.3 | ASCVD | None | Normal |
| 16 | CONTROL | 26 | White | Male | 12 | 6.9 | Arrhythmia | None | Normal |
| 17 | CONTROL | 72 | White | Female | 23 | 6.9 | MVA | None | Normal |
| 18 | CONTROL | 42 | White | Female | 23 | 6.9 | Mitral valve prolapse | None | Normal |
| 19 | CONTROL | 47 | White | Male | 10 | 7.0 | ASCVD | None | Normal |
| 20 | CONTROL | 31 | White | Male | 16 | 7.2 | MVA | None | Normal |
| 21 | CONTROL | 60 | White | Male | 15 | 7.1 | Accidental drowning | None | Normal |
| 22 | CONTROL | 28 | White | Male | 13 | 6.8 | Electrocution | None | Normal |
| 23 | CONTROL | 45 | White | Female | 16 | 6.9 | Cardiac arrhythmia | None | Normal |
| 24 | CONTROL | 62 | White | Male | 19 | 7.0 | Cardiac arrest | None | Normal |
|
| |||||||||
| Depressed Subjects b | |||||||||
|
| |||||||||
| 1 | SUICIDE | 27 | White | Male | 24 | 7.0 | GSW | None | MDD, Ethanol abuse |
| 2 | SUICIDE | 44 | White | Female | 11 | 7.2 | Drug OD | Nortriptyline | MDD, Ethanol abuse |
| 3 | SUICIDE | 36 | White | Female | 10 | 7.1 | GSW | None | MDD |
| 4 | SUICIDE | 24 | White | Male | 7 | 7.1 | GSW | Ethanol | MDD |
| 5 | SUICIDE | 43 | White | Male | 12 | 7.0 | Drug OD | None | MDD, Polysubstance Abuse |
| 6 | SUICIDE | 53 | White | Male | 23 | 6.9 | Jump from height | None | MDD |
| 7 | SUICIDE | 41 | White | Female | 27 | 7.1 | Drug OD | Amitriptyline, Desipramine, Nortriptyline, Ethanol | MDD, Ethanol abuse |
| 8 | SUICIDE | 22 | Black | Female | 16 | 7.3 | Drug OD | None | MDD |
| 9 | SUICIDE | 46 | White | Female | 21 | 6.9 | Drug OD | Amitriptyline, Desipramine, Ethanol | MDD |
| 10 | SUICIDE | 36 | White | Female | 18 | 7.2 | GSW | None | MDD |
| 11 | SUICIDE | 38 | White | Male | 24 | 7.0 | Drug OD, Ethanol intoxication | Ethanol | MDD, Ethanol abuse |
| 12 | SUICIDE | 46 | White | Female | 16 | 6.8 | Drug OD | Nortriptyline | MDD, Panic disorder with agoraphobia |
| 13 | SUICIDE | 23 | White | Male | 12 | 7.0 | Hanging | Paroxetine | MDD |
| 14 | SUICIDE | 30 | White | Male | 17 | 7.1 | Hanging | Venlafaxine | MDD |
| 15 | SUICIDE | 44 | White | Female | 30 | 7.2 | Drug OD, Ethanol intoxication | Fluoxetine, Ethanol | MDD, Ethanol abuse, Opioid abuse |
| 16 | SUICIDE | 74 | White | Female | 27 | 7.0 | Venlafaxine OD | Venlafaxine, Ethanol | MDD, Ethanol abuse |
| 17 | SUICIDE | 25 | White | Male | 14 | 6.8 | Hanging | Ethanol | MDD |
| 18 | SUICIDE | 23 | Black | Male | 23 | 6.9 | Hanging | None | MDD |
| 19 | SUICIDE | 63 | White | Male | 19 | 6.9 | Drug OD, Ethanol intoxication | Ethanol | MDD |
| 20 | SUICIDE | 67 | White | Male | 22 | 7.0 | GSW | Fluoxetine, Venlafaxine | MDD |
| 21 | SUICIDE | 40 | White | Female | 20 | 7.0 | Drug OD | Xanax | MDD |
| 22 | SUICIDE | 53 | White | Male | 26 | 7.1 | Stabbing | Sertraline | MDD |
| 23 | SUICIDE | 68 | White | Female | 26 | 6.8 | GSW | Amitriptyline | Bipolar Disorder |
| 24 | SUICIDE | 31 | White | Female | 31 | 6.8 | Hanging | Trazadone, Ethanol | Bipolar Disorder, Bulimia, OCD |
| 25 | SUICIDE | 51 | White | Female | 28 | 6.9 | Amitriptyline OD | Amitriptyline, Ethanol | Bipolar Disorder, Ethanol abuse |
Abbreviations: ASCVD, atherosclerotic cardiovascular disease; CO, carbon monoxide; DKA, diabetic ketoacidosis; GSW, gunshot wound; MDD, major depressive disorder; MVA, motor vehicle accident; OCD, obsessive-compulsive disorder; OD, overdose; PMI, postmortem interval
Mean ± SD age is 41.96 ± 13.19 years; PMI, 17.21 ± 6.65 h; and brain pH 7.01 ± 0.15
Mean ± SD age is 41.92 ± 15.07 years; PMI, 20.16 ± 6.68 h; and brain pH 7.00 ± 0.14
2.2. RNA isolation and reverse transcription
We used the Trizol® reagent (Invitrogen, Carlsbad, California) to extract total RNA from 100 mg of tissue according to the manufacturer’s instructions. NanoDrop®ND-1000 (NanoDrop Technologies, Montchanin, Delaware) was used to determine RNA yield and purity by absorbance ratios A260/A280 and A260/A230. All the samples had OD ratios close to 2 indicating absence of contaminants. To assess the quality of RNA, we used Agilent Bioanalyzer 2100. All samples showed RNA integrity number (RIN) above 6.6 and 28S/18S ratios >1.2. Random hexamers (2.5 μM) (Invitrogen) were employed to synthesize first-strand cDNA from 1 μg of total RNA with MMLV-reverse transcriptase (Invitrogen) according to manufacturer’s directions.
2.3. Oligonucleotide primers
We used Primer Express 3.0 software (Applied Biosystems, Foster City, California) to design primers for the amplification of human PAK1, PAK2, PAK3, RAC1, ARHGEF7, CDC42 and internal reference genes (GAPDH, CYC1, and PPIA). In all cases primers were designed to amplify all known transcript variants for each gene. Only in case of RAC1, an additional set of primers was designed to distinguish the amplification of RAC1B, a longer transcript variant of RAC1 (NM_018890.3) that includes the alternatively spliced 57 bp region (exon 3b). Primer sequence, full gene name, gene ID, function, and chromosomal localization are listed in Table 2 (reference genes) and Table 3 (target genes).
Table 2.
Primer sequences for reference genes
| Symbol | Entrez Gene ID | Name | Function | Forward primer | Reverse primer | Location |
|---|---|---|---|---|---|---|
| GAPDH | 2597 | glyceraldehyde-3- phosphate dehydrogenase, transcript variant 1 | Catalysis of oxidative phosphorylation of glyceraldehyde-3- phosphate in the presence of NAD | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | 12p13 |
| PPIA | 5478 | peptidylprolyl isomerase A (cyclophilin A) | Peptidyl-prolylcis- transisomerase | GGCAAATGCTGGACCCAACACA | TGCTGGTCTTGCCATTCCTGGA | 7p13 |
| CYC1 | 1537 | cytochrome c-1 | Electron transporter | CCAGATAGCCAAGGATGTGTGC | GACTGACCACTTGTGCCGCTTT | 8q24.3 |
Table 3.
Primer sequences for target genes
| Symbol | Entrez Gene ID | Name | Forward primer | Reverse primer | Location |
|---|---|---|---|---|---|
| PAK1 | 5058 | p21 protein (Cdc42/Rac)- activated kinase 1 | GTGAAGGCTGTGTCTGAGACTC | GGAAGTGGTTCAATCACAGACCG | 11q13-q14 |
| PAK2 | 5062 | p21 protein (Cdc42/Rac)- activated kinase 2 | CGACTCCAACACAGTGAAGCAG | TCACTACTGCGGGTGCTTCTGT | 3q29 |
| PAK3 | 5063 | p21 protein (Cdc42/Rac)- activated kinase 3 | CGCTGTCTTGAGATGGATGTGG | CAGTCTTAGCGGCTGCTGTTCT | Xq23 |
| RAC1 | 5879 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) isoforms Rac1 and Rac1b | AGTGCTCGGCGCTCACA | CGGATCGCTTCGTCAAACA | 7p22 |
| RAC1B | 5879 | ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) isoform Rac1b | CGGTGAATCTGGGCTTATGGGA | GGAGGTTATATCCTTACCGTACG | 7p22 |
| CDC42 | 998 | Cell division cycle 42 | TGACAGATTACGACCGCTGAGTT | GGAGTCTTTGGACAGTGGTGAG | 1p36.1 |
| ARHGEF7 | 8874 | Rho guanine nucleotide exchange factor 7 | CGCAAACCTGAACGGAAGCCTT | GTTTTGGCGCTGGTGCAGTAAG | 13q34 |
2.4. Quantitative polymerase chain reaction (qPCR)
Levels of mRNA were quantified by conducting qPCR reactions with SYBR®Select Master Mix (Applied Biosystems) according to the manufacturer’s directions. Stratagene Mx3005P equipped with MxPro software (Stratagene, La Jolla, California) was used to perform measurements. Plate setup always included no-RT control, no-template control and three different inter-run calibrators for each gene assay. All reactions were done in duplicate. A heat dissociation protocol to verify primer specificity was included after the final cycle of each PCR reaction. All pairs of primers showed a single peak corresponding to the melting temperature (Tm) of expected PCR product (Table 4). Reproducibility of RT reaction and inter-run reproducibility of qPCR was evaluated as described in our previous work (Fuchsova et al., 2015). We employed the relative standard curve method as described in the “Applied Biosystems Guide to Performing Relative Quantitation of Gene Expression Using Real-Time Quantitative PCR” to determine relative expression levels and report them as fold change (FC) to control. To determine the linear range and PCR reaction efficiency of each gene assay we build standard curves using 10-fold serial dilutions of cDNA derived from all subjects. We report r2≥0.988 and efficiencies between 90–110 percent for all gene assays (Table 4). qBasePLUS software (Biogazelle, Zwijnaarde, Belgium) was employed to analyze raw expression data (CT values). For further statistical analysis we used values normalized to the normalization factor calculated as a geometric mean of the expression of three reference genes (Vandesompele et al., 2002).
Table 4.
Efficiency of qPCR
| Reference Genes
| |||||
|---|---|---|---|---|---|
| Assay | Threshold (dRn) | RSq (dRn) | Slope (dRn) | Efficiency (%) | Tm (°) |
| GAPDH | 0.2 | 0.992 | −3.552 | 91.2 | 83.6 |
| PPIA | 0.2 | 0.997 | −3.353 | 98.7 | 84.4 |
| CYC1 | 0.2 | 0.988 | −3.455 | 94.7 | 86.8 |
|
| |||||
| Target Genes | |||||
|
| |||||
| Assay | Threshold (dRn) | RSq (dRn) | Slope (dRn) | Efficiency (%) | Tm (°) |
|
| |||||
| PAK1 | 0.2 | 0.988 | −3.566 | 90.7 | 83.8 |
| PAK2 | 0.2 | 0.991 | −3.351 | 98.8 | 81.8 |
| PAK3 | 0.2 | 0.997 | −3.103 | 110 | 82.8 |
| RAC1 | 0.2 | 0.995 | −3.178 | 106.4 | 83.2 |
| RAC1b | 0.2 | 0.967 | −3.147 | 107.9 | 80.4 |
| ARHGEF7 | 0.2 | 0.995 | −3.333 | 99.5 | 83.25 |
| CDC42 | 0.2 | 0.99 | −3.426 | 95.8 | 79.8 |
Efficiency data for reference genes and target genes. CT values obtained from 10-fold serial dilutions of cDNA were plotted against dilution factors and the reaction efficiency was calculated using qBasePLUS software.
RSq, r-squared; Rn, normalized reporter signal; dRn, delta Rn (Rn minus the baseline); Tm, melting temperature
2.5. Determination of reference targets for qPCR data normalization
The algorithm geNorm of qBasePLUS software was used to evaluate expression stability of reference genes used for qPCR data normalization (Vandesompele et al., 2002) as described in our previous work (Fuchsova et al., 2015). The reference targets PPIA, CYC1, and GAPDH were determined as the most stably expressed genes and were found adequate for the optimal normalization of data in our experimental setting (Table 5, see the Table 2 for full gene name, accession number, function, chromosomal localization, and primer sequences). Figure 1 shows that control and depressed subjects did not differ with respect to GAPDH, CYC1 and PPIA mRNA expression levels. The geometric mean of GAPDH, CYC1, and PPIA was further used for normalization in all relative quantitation assays.
Table 5.
Reference target stability values (M and CV)
| PFC
| ||
|---|---|---|
| Reference Target | M | CV |
| CYC1 | 0.4716 | 0.1812 |
| GAPDH | 0.6866 | 0.2651 |
| PPIA | 0.4709 | 0.2495 |
| Average | 0.5430 | 0.2319 |
|
| ||
| Hippocampus | ||
|
| ||
| Reference Target | M | CV |
|
| ||
| CYC1 | 0.5128 | 0.2437 |
| GAPDH | 0.5878 | 0.2331 |
| PPIA | 0.4601 | 0.1525 |
| Average | 0.5202 | 0.2098 |
Reference target stability values (M and CV) determined for the combination of the reference targets GAPDH, CYC1, and PPIA in human brain samples from the PFC and hippocampus. Expression stability of reference genes in human brain samples was calculated by geNorm algorithm using qBasePLUS software (Biogazelle) as described in our previous publication (Fuchsova et al., 2015). GeNorm analysis was performed on the gene expression data from PFC and hippocampal postmortem samples obtained from all depressed subjects and matched nonpsychiatric controls. The gene stability measure M is defined as the average pairwise variation of a particular gene with all other control genes (Vandesompele et al., 2002). Genes with lowest values have the most stable expression (optimal geNorm M≤0.5). To assess that the genes with the lowest M values have indeed the most stable expression, the gene-specific variation of each control gene is determined as the variation coefficient (CV) of the expression levels after normalization. Mean CV values equal or lower than 0.2 are typically observed for stably expressed reference genes in relatively homogenous sample panels. Normalization to a single control gene can lead to erroneous normalization. For this reason, in our previous publication the optimum number of three reference genes required for adequate data normalization in this experimental situation was calculated using geNorm (Fuchsova et al., 2015). Thus, in our experiments in human postmortem PFC and hippocampal samples, the optimal normalization factor was calculated as the geometric mean of reference targets PPIA, CYC1, and GAPDH.
Figure 1.
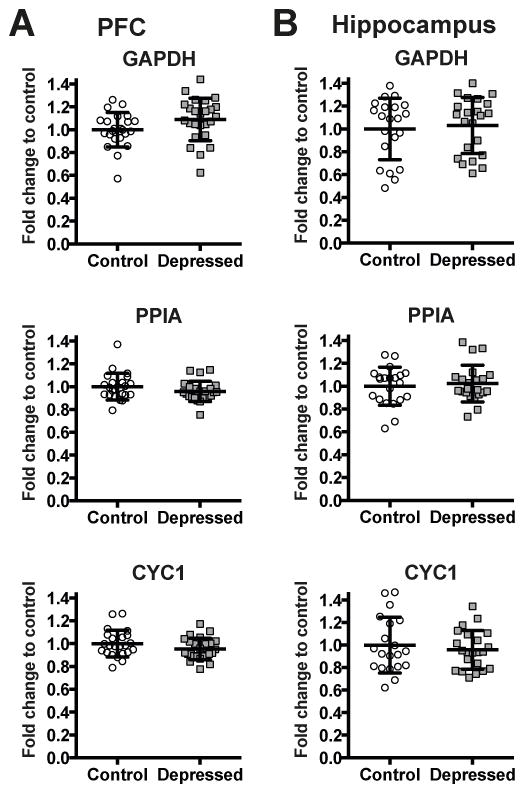
Expression levels of reference genes GAPDH, PPIA and CYC1 in the PFC (A) and in the hippocampus (B) of non-psychiatric controls (Control: PFC n=23 for GAPDH, PPIA and CYC1; Hippocampus n=21 for GAPDH, PPIA and n=20 for CYC1) and depressed subjects (Depressed: PFC n=25 for GAPDH, CYC1 and n=24 for PPIA; Hippocampus n=22 for GAPDH, PPIA and CYC1). Results are expressed as fold change in mRNA levels. Values are fold change ±SD. No significant differences between the depressed group and the control group were determined for any of the genes.
2.6. Statistical analysis
ANCOVA was used to compare the groups of controls and depressed subjects. Data analysis was done independently for each brain region and gene of interest adjusting the effects of age, postmortem interval (PMI), and brain pH. Bonferroni correction was applied for multiple comparisons to maintain alpha at 0.05 to adjust the type I error rates. Statistical outliers were identified by the ROUT method with the value Q set to 1% in order to control the false discovery rate. We report results as individual values and group means ± SD. For coexpression analysis of gene expression levels, correlation analyses were conducted to calculate pairwise Spearman correlation coefficients along with its 95% confidence intervals, and p value (two-tailed) on normalized mRNA expression levels. Cross-correlations were performed between the expression level of the 6 different genes for the depression group and for the control group independently in each brain area. This resulted in fifteen separate comparisons for each group in each area. For multiple comparisons, Spearman correlation values with Bonferroni correction to maintain alpha at 0.05 were used. To determine differences in the frequency of significant correlations between groups, Chi-square analyses were performed. GraphPad Prism software was used to do statistical analysis and graphs.
3. Results
3.1. Demographic characteristics
Detailed demography of studied subjects is reported in Table 1. The age range of control and depressed subjects was between 22–74 years without significant differences between depressed subjects and their matched controls (p=0.9925, unpaired t-test, two-tailed). The postmortem interval (PMI) was between 5–31 hours and did not differ statistically between two groups (p=0.1279, unpaired t-test, two-tailed). The mean brain pH± SD was 7.01± 0.15 in control group and 7.00± 0.14 in depressed group and did not significantly differed between groups (p=0.9163, unpaired t-test, two-tailed). In the PFC, the mean RIN± SD was 7.21± 0.56 in controls and 7.23± 0.61 in subjects with depression. The mean RIN± SD in the hippocampus was 7.16± 0.72 in controls and 7.45± 0.83 in subjects with depression. In both the PFC and the hippocampus, the RIN values did not differed between control group and depressed subjects (p=0.7 and p=0.37; respectively, unpaired t-test, two-tailed).
3.2. PAK1 expression is downregulated in the PFC of subjects with depression
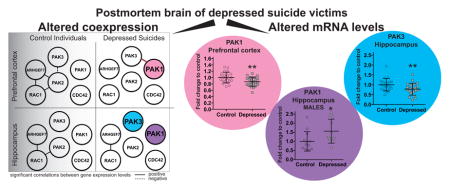
To evaluate if alteration in the expression of PAK1 is associated with pathophysiology of human depression, PAK1 mRNA level was assessed in the entire cohort using qPCR. Figures 2A and 2B show mRNA levels of PAK1 in the PFC (Figure 2A) and in the hippocampus (Figure 2B) of depressed subjects and matched non-psychiatric controls. In the hippocampus, no significant differences were observed between groups. On the other hand, F-test of ANCOVA showed that in the PFC, the mean PAK1 mRNA level was significantly decreased in the depressed group (F=23.87, **p<0.01 after Bonferroni correction, FC=0.87). The age, PMI and brain pH had no significant effects on mRNA expressions of PAK1.
Figure 2.
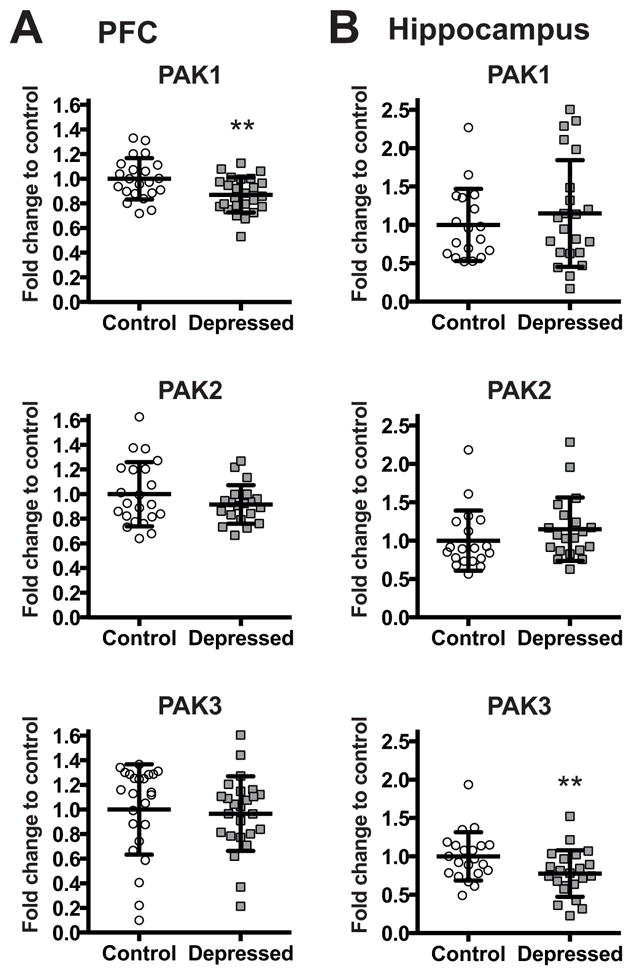
mRNA levels of PAK1, PAK2, and PAK3 in the PFC (A) and in the hippocampus (B) of non-psychiatric controls (Control: PFC n=22 for PAK1, PAK2, and n=24 for PAK3; Hippocampus n=18 for PAK1, n=19 for PAK2, and n=21 for PAK3) and depressed subjects (Depressed: PFC n=25 for PAK1, PAK3, and n=20 for PAK2; Hippocampus n=22 for PAK1, PAK3, and n=20 for PAK2) normalized to the geometric mean of the three reference targets (GAPDH, CYC1, PPIA). Results are expressed as fold change in mRNA levels. Values are fold change ±SD. Significant differences between the subjects with depression and the control group were determined for PAK1 (F=23.87, **p<0.01) in the PFC and PAK3 (F=17.27, **p<0.01) in the hippocampal tissue. Significant effects are marked by asterisks showing Bonferroni corrected significance thresholds.
3.3. Altered expression of PAK3 in the hippocampus of subjects with depression
We next examined whether the expression of two other members of group I PAKs, PAK2 and PAK3, is altered in the brain of subjects with depression. PAK2 and PAK3 mRNA levels were measured by qPCR in the PFC (Figure 2A) and in the hippocampus (Figure 2B) of subjects with depression and matched non-psychiatric controls. In the PFC, we did not detect any differences between groups for PAK3. In contrast, F-test of ANCOVA revealed that in the hippocampus, the mean PAK3 mRNA level was significantly reduced in the depressed subjects (F=17.27, **p<0.01 after Bonferroni correction, FC=0.78). The age, PMI and brain pH had no significant effects on mRNA expression of PAK3. Expression of PAK2 mRNA did not differ in neither of the tissues.
3.4. Expression of RAC1, CDC42, and ARHGEF7 in the postmortem brain of subjects with depression
We next assessed if the expression of the genes encoding PAKs regulators: the cell division cycle 42 (CDC42), the Ras-related C3 botulinum toxin substrate (RAC1) and ARHGEF7 (also known as betaPIX) is altered in the postmortem brain of subjects with depression. Figure 3 shows mRNA levels of RAC1, CDC42, and ARHGEF7 in the PFC (Figure 3A) and in the hippocampus (Figure 3B) of depressed subjects and matched control individuals.
Figure 3.
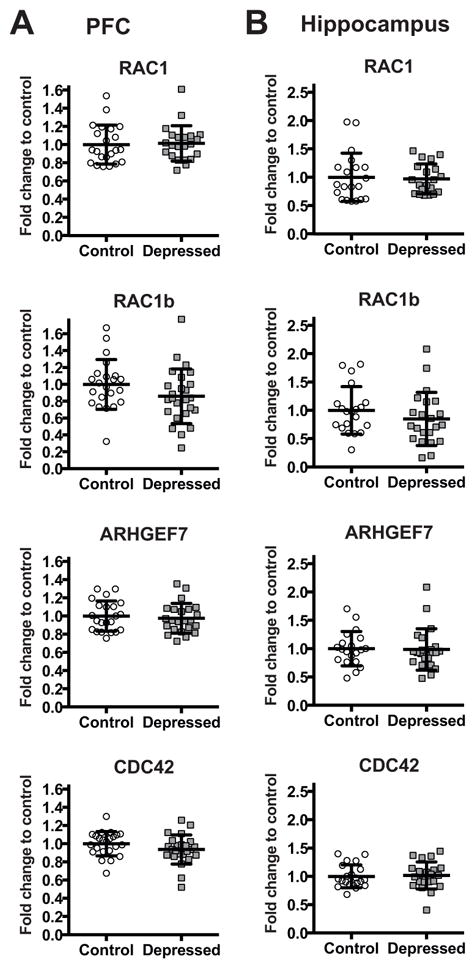
mRNA levels of RAC1, RAC1B, CDC42, and ARHGEF7 in the PFC (A)and in the hippocampus (B) of non-psychiatric controls (Control: PFC n=22 for RAC1, RAC1B, and ARHGEF7, n=23 for CDC42; Hippocampus n=20 for RAC1, RAC1B, and ARHGEF7, n=21 for CDC42) and depressed subjects (Depressed: PFC n=22 for RAC1, n=25 for RAC1B, n=23 for ARHGEF7, and n=24 for CDC42; Hippocampus n=20 for RAC1, and n=22 for RAC1B, ARHGEF7, and CDC42) normalized to the geometric mean of the three reference targets (GAPDH, CYC1, PPIA). Results are expressed as fold change in mRNA levels. Values are fold change ±SD. No significant differences between the depressed group and the control group were determined for any of the genes.
No significant changes were observed for RAC1, CDC42, and ARHGEF7 in neither of the tissues. Furthermore, we evaluated separately the expression of RAC1B, a constitutively active splice variant of RAC1, whose increased expression has been found to contribute to neurodegeneration seen in Alzheimer’s disease (Perez et al., 2012). Similarly to the previous cases, the expression of RAC1B mRNA did not differ in neither of the tissues.
3.5. Coordinated expression
To assess interrelationships in the expression levels of PAK1, PAK2, PAK3, RAC1, CDC42, and ARHGEF7, cross-correlation analyses were conducted in the control group and in the depressed subjects group independently for each brain region. Normalized mRNA expression levels of all studied genes have been correlated to each other and Spearman correlation coefficients and p values were calculated.
Figure 4A shows correlation matrices with Spearman correlation coefficients and statistically significant Spearman correlation p values indicated by asterisks. In the control PFC, 4 of 15 possible correlations were statistically significant. Within the hippocampus of control individuals, 2 of 15 correlations were statistically significant and these were the same 2 positive correlations as detected in the control PFC (correlations between PAK2 and RAC1, and RAC1 and ARHGEF7). In the PFC in of subjects with depression, the number of significant correlations declined to one, which in fact was a new positive correlation between PAK1 and PAK3. The difference in the frequency of significant correlations between groups was statistically significant (chi-square==6, *p=0.0143).
Figure 4. Coordinated expression is altered in the brain of subjects with depression.
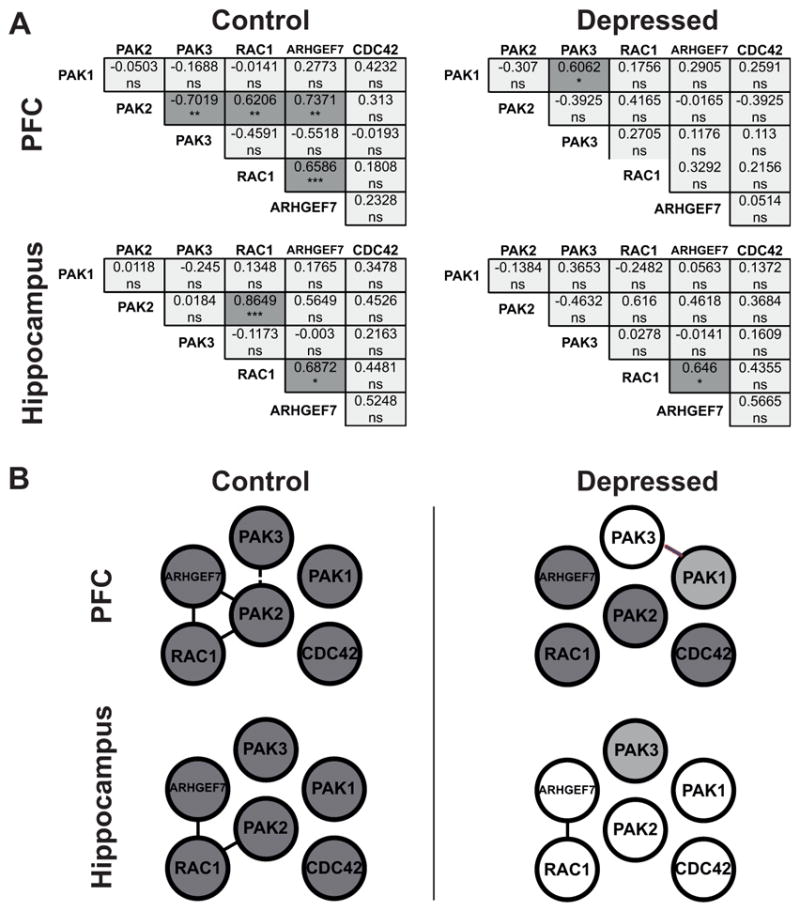
(A) Correlation analysis of mRNA expression levels in the PFC and in the hippocampus In the correlation matrices all pairwise Spearman correlation coefficients calculated on normalized mRNA expression levels are shown. Cross-correlations were performed between the expression level of the 6 different genes for the depression group and for the control group independently in each brain area. This resulted in fifteen separate comparisons for each group in each area. Statistically significant Spearman correlation p values (two-tailed) are indicated by asterisks (Bonferroni corrected significance threshold, after correction: *p<0.05, **p<0.01, ***p<0.001, ns=no significant; dark grey indicates a statistically significant correlation, light grey indicates a non significant correlation). In the PFC and in the hippocampus of depressed subjects, significant correlations appear less frequently than in the control PFC and the control hippocampus. In the PFC, decrease in the frequency of significant correlations between controls and depressed subjects is statistically significant (chi-square=6, *p=0.0143). No statistical difference is detected in the hippocampus (chi-square=1.034, p=0.3091). (B) Network of the gene coexpression pattern in the PFC and in the hippocampus in control and depressed group. Gene pairs that show a significant correlation are indicated: a positive significant correlation (full line); a negative significant correlation (dashed line). Dark circles indicate differentially expressed genes. Differences between control and depressed groups are observed in both tissues.
In the hippocampus, only one significant correlation evident also in controls was detected in the brain of subjects with depression. The frequency of significant correlations between the depressed group and the control group in the hippocampus did not statistically differ (chi-square=1.034, p=0.3091).
Figure 4B depicts the networks of the gene coexpression patterns in the control and depressed groups to illustrate our findings. Significant positive or negative correlation between two genes is shown with a full or dashed line respectively. In addition, genes showing differential expression between depressed subjects and their non-psychiatric matched controls (namely PAK1 in the PFC and PAK3 in the hippocampus) are shaded in gray. In both tissues of the control group, correlated genes connect suggesting that they function in common signaling cascades. If the pathway is affected by disease, this type of analysis is expected to reveal changes in the coordinated gene expression. Indeed, in the depressed group, this coordinated gene expression is slightly altered in the hippocampus while it almost disappears in the PFC, where the only correlation detected was a new correlation between PAK3 and PAK1.
3.6. Effect of psychotropic drugs, ethanol and gender
Since several of the subjects in the depressed group were on antidepressant medication at the time of death and/or suffered from comorbid substance or ethanol abuse, we examined if the presence of psychotropic drugs or ethanol had any influence on the mRNA expression or on the coordinated gene expression of studied genes. For this purpose, depressed subjects were separated in three groups: 1) the depressed group free of any psychotropic medication (7 subjects), 2) the group taking different types of antidepressants (14 subjects), and 3) the group with ethanol consumption (4 subjects).
When we compared the mRNA expression, we did not find any significant effect of antidepressants or ethanol on the expression of any of the studied genes in the hippocampus and on the expression of PAK1, PAK3, RAC1, RAC1B, CDC42, and ARHGEF7 in the PFC. On the other hand, F-test of ANCOVA determined a statistically significant effect of drugs and ethanol on the expression of PAK2 in the PFC (F=3.98, p=0.0449). However, subsequent post hoc tests for multiple comparisons using the Bonferroni correction revealed no significant differences among the three groups. Moreover, ANCOVA analysis on each of the three groups compared to control group did not reveal any significant differences in the PAK2 expression in the PFC. We conclude that the differences in the PAK1 and PAK3 expression in the PFC and the hippocampus between the depressed group and normal controls do not appear to be related to the presence of psychotropic drugs and ethanol.
To evaluate potential effects of antidepressants and ethanol on coordinated gene expressions correlation analyses were performed. Spearman correlation coefficients and p values with Bonferroni correction for multiple comparisons were calculated for all genes on normalized mRNA expression levels independently in each brain region. No significant correlations were detected in any of the three groups. Consequently, no differences were detected between subjects with depression who were free of any psychotropic medication and those who had been taking antidepressants or ethanol. This would suggest that the presence of antidepressants or ethanol might not have a significant influence on coordinated gene expressions. However, this observation is limited by the small n of analyzed samples (7 in the group 1, 14 in the group 2, and 4 in the group 3).
To assess the effect of gender on gene expression we analyzed the data by two way ANOVA independently for each brain region with diagnosis and gender as categorical independent variables adjusting the effects of age, PMI, and brain pH. In the PFC, a gender effect with tendency toward significance was detected for PAK3 (F=4.04, p=0.0510) but no significant gender by diagnosis interaction was revealed (F=0.72, p=0.3999). In accordance, ANCOVA analysis on males and females separately did not reveal any significant differences in the PAK3 expression in the PFC of depressed male or female subjects when compared to control males or females. No gender effect on PAK3 expression was detected in the hippocampus. On the other hand, in the hippocampus, a significant gender effect was detected for PAK1 (F=5.19, *p=0.0294). Here, also a significant gender by diagnosis interaction was revealed (F=4.47, *p=0.0422). ANCOVA analysis on males and females separately revealed a significant difference in the PAK1 expression in the hippocampus of depressed males when compared to control males (F=16.48, **p<0.01 after Bonferroni correction, FC=1.56). No differences were detected in depressed females (Fig. 5). No gender effect on PAK1 expression was detected in the PFC.
Figure 5.
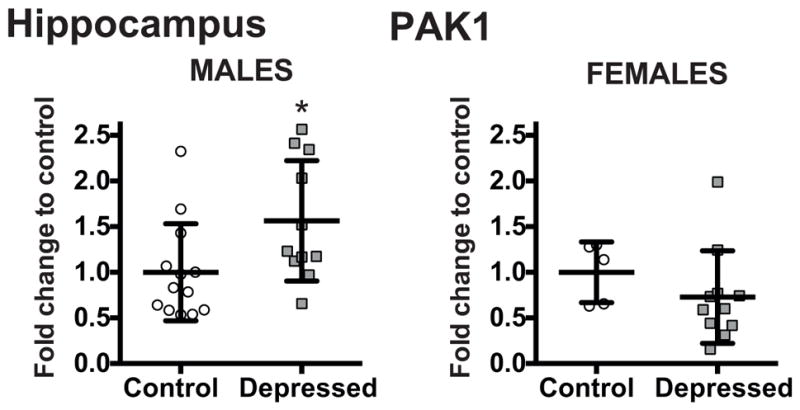
Effect of gender on mRNA levels of PAK1 in the hippocampus of non-psychiatric controls (Control: Males n=13; Females n=5) and depressed subjects (Depressed: Males n=11; Females n=11) normalized to the geometric mean of the three reference targets (GAPDH, CYC1, PPIA). Results are expressed as fold change in mRNA levels. Values are fold change ±SD. Significant difference between the subjects with depression and the control group was determined in the PAK1 expression in the hippocampual tissue of depressed males when compared to control males (F=16.48, **p<0.01). No differences were detected in depressed females. Significant effects are marked by asterisks showing Bonferroni corrected significance thresholds.
4. Discussion
The highly conserved PAK family of serine/threonine kinases participates in the regulation of cytoskeletal dynamics, and therefore is implicated in a wide range of physiological and pathological processes (Kreis and Barnier, 2009, Ma et al., 2012, Ba et al., 2013, Rane and Minden, 2014). In the present study, we show that in the brain of subjects with depression mRNA levels of two members of group I PAK predominantly expressed in neurons, PAK1 and PAK3, are significantly diminished in the PFC and in the hippocampus respectively. Moreover, a significant gender effect was detected for PAK1 expression in the hippocampus, where a significant difference was detected in depressed males but not females. No differences were observed for the ubiquitously expressed PAK2.
PAK3 is the only group I member whose expression is relatively restricted to neurons whereas PAK1 is highly expressed in the nervous system but also in other tissues (Manser et al., 1995, Teo et al., 1995, Rousseau et al., 2003). Changes in their activity have been detected in pathology of some cognitive and neurodegenerative disorders (Ma et al., 2012). For example, X-linked mental retardation can be caused by mutations in PAK3 that disrupt actin dynamics in dendritic spines (Allen et al., 1998). On the other hand, in Alzheimer’s disease, both PAK1 and PAK3 are significantly decreased in the hippocampus and PAK3 also in the temporal cortex (Zhao et al., 2006). Moreover, PAK1 is involved in the pathology of Huntington’s disease (Luo et al., 2008). The observed dysregulation of PAK1 and PAK3 in the brain of subjects with depression in the present study suggests that disruption in the signaling of both PAKs might also play a role in the pathology of human depression.
Both PAK1 and PAK3 have been shown to be critically involved in biological mechanisms associated with neurodevelopment, neuroplasticity and maturation of the nervous system such as neurite outgrowth, spine morphogenesis and manteinance, and synapse formation (Rashid et al., 2001, Hayashi et al., 2002, Boda et al., 2004, Bryan et al., 2004, Zhang et al., 2005, Cobos et al., 2007, Hayashi et al., 2007, Kreis et al., 2007, Dubos et al., 2012). They both share important homologies with regard to structure and possibly also functions (Hofmann et al., 2004). For example, in spine morphogenesis, PAK1 and PAK3 share distinct but also compensatory roles and their activity may overlap allowing the compensation of the PAK3 deficit by PAK1 (Boda et al., 2008). Furthermore, some PAK functions may depend on a crosstalk between PAK1 and PAK3 during the activation process since PAK3 proteins were shown to form heterodimers with PAK1 (Combeau et al., 2012).
Knockout of either PAK1 or PAK3 alone does not cause a robust morphological phenotype in mice (Meng et al., 2005, Asrar et al., 2009, Kelly and Chernoff, 2012). On the other hand, double PAK1 and PAK3 knockout are severely impaired in postnatal brain growth. Reduced brain size is observed due to simplified neuronal dendrites/axons, neurons with smaller soma and significant increase in cell density without global neuron loss. As a consequence, synapse development and function is impaired in such a way that these mice exhibit reduced synapse density, defects in dendritic spines, LTD, LTP, as well as learning and memory deficits, hyperactivity and increased anxiety (Huang et al., 2011).
Remarkably, in depressed patients structural and functional alterations have been observed in the PFC and hippocampus, where reduced volume is inversely correlated with length of illness, time of treatment, and severity of depression (Drevets et al., 2008, MacQueen and Frodl, 2011). In patients, characterization of the cellular determinants underlying these changes is limited. Nevertheless, postmortem studies demonstrate decreased neuronal cell body size, atrophy of dendritic processes, as well as decreased synapse number (Rajkowska et al., 1999, Stockmeier et al., 2004, Drevets et al., 2008, Pittenger and Duman, 2008, Kang et al., 2012). Thus, we hypothesize that neuronal size, complexity and synaptic properties regulated by PAK1/PAK3 dependent signaling may be a key factor responsible for volumetric abnormalities of hippocampus and PFC in depression resulting in altered connectivity of these regions.
Regulation of PAK activity is a complex process that apart from the homo/heterodimer formation and phosphorylation events is also dependent on activators. In particular, the small GTPases RAC1 and CDC42 play a central role in the activation of PAK1 and PAK3 (Manser et al., 1994). Here, we observed that neither RAC1 nor CDC42 expression is altered in the hippocampus or in the PFC of subjects with depression. Similarly, we did not observe any significant changes in neither of the tissues for RAC1B, a constitutively active splice variant of RAC1, whose increased expression has been found to contribute to neurodegeneration seen in Alzheimer’s disease (Perez et al., 2012).
A number of other signaling proteins can also play key roles in PAK activation. In fact, PAK1 can even lie upstream to RAC, by binding to the ARHGEF7 (also known as betaPIX for PAK-interacting exchange factor), which is a guanine nucleotide exchange factor (GEF) for RAC (Manser et al., 1998, Obermeier et al., 1998, Rane and Minden, 2014). Evidence shows that the GIT1/ARHGEF7/PAK complex plays a crucial role in neuronal signaling during neurite outgrowth and spine morphogenesis (Obermeier et al., 1998, Zhang et al., 2005). Alterations in this complex has been suggested to associate with decreased neuronal connectivity and cognitive defects such as those seen in nonsyndromic mental retardation (Zhang et al., 2005). Furthermore, GIT1/ARHGEF7/RAC1/PAK pathway has been also recently shown to be required for GABAA receptor synaptic stability and inhibitory neurotransmission (Smith et al., 2014). In the present work, when ARHGEF7 mRNA expression was measured in the hippocampus or in the PFC of subjects with depression, we did not observed any alterations. The same was true for the mRNA expression of GIT1 assessed in our previous study (Fuchsova et al., 2015).
Our observations are in line with findings by Golden et al (Golden et al., 2013). In their recent work, transcriptional profiling for Rho GTPase–related genes revealed that RAC1 is transcriptionally downregulated through an epigenetic mechanism in the NAc of subjects with depression and after chronic social defeat stress in animal model of depression-like behaviors. However, they did not observe downregulation of RAC1 in other brain structures in susceptible mice. The expression levels of neither PAK1 nor ARHGEF7 were affected in the NAc by social defeat (Golden et al., 2013). We suggest that observed differences in the present work reflect region specific effects of depression in agreement with rodent studies that demonstrate reductions in dendrite complexity and spine density in the hippocampus and PFC but increases in the basolateral amygdala and NAc (Duman and Duman, 2015). We hypothesize that depression is associated with a shift toward synaptic instability through region specific dysregulation of different Rho GTPase–related mechanisms.
Depression may lead to the downregulation of PAK1 and PAK3 through different mechanisms. Epigenetic mechanisms could be a possible explanation since both genes have been identified as targets of posttranscriptional regulation by microRNAs. PAK1 and PAK3 are validated targets of miR-320 and miR-335 respectively in humans (Tavazoie et al., 2008, Helwak et al., 2013). miR-320 and miR-335 were found to be differentially expressed in blood of depressed patients (Camkurt et al., 2015, Li et al., 2015). Whether these microRNAs regulate PAK1/PAK3 expression in the hippocampus and PFC remains to be established. Another possible mechanism could involve Erk5 activation. In a recent study, Komaravolu et al showed that endothelial Erk5 activation decreased PAK1 mRNA levels and that this process is mediated by the Krüppel-like transcription factor KLF2 (Komaravolu et al., 2015). It would be interesting to evaluate association of Erk5/KLF2 with depression.
Gene products functioning along in common signaling pathways are supposed to display higher coordination in their expression levels than random sets of gene products (Vidal et al., 2011). In accordance, in our investigation we detected significant correlations between PAK2 and RAC1, and ARHGEF7 and RAC1 in the control hippocampus. In the control PFC, we detect the same positive significant correlations. Moreover, a positive correlation between ARHGEF7 and PAK1 and a negative correlation between PAK3 and PAK2 are observed indicating that co-regulation of some of the studied gene products exists in the hippocampus and the PFC of humans.
However, the coordination we observe is disrupted or reorganized in subjects with depression. In the PFC, the only significant correlation is a new positive correlation between PAK3 and PAK1. In the hippocampus, one correlation between the expression levels of ARHGEF7 and RAC1 detected also in controls remains significant. Different authors have provided important insights into the pathways disbalanced in psychiatric disease by using the analysis of gene coexpression networks (Merali et al., 2004, Poulter et al., 2010, Zhurov et al., 2012, Gaiteri et al., 2014). Our observations fit well with the conclusions that pathological mechanisms resulting in depression affect the coordination of gene expression in the brain (Merali et al., 2004, Gaiteri et al., 2010, Poulter et al., 2010, Zhurov et al., 2012, Gaiteri et al., 2014). The observed changes may reflect loss of coordinated transcriptional response or may result from other regulatory processes such as altered mRNA stability or microRNA actions.
Since PAK3 and PAK1 signaling are linked together, and PAK1 and PAK3 share distinct but also compensatory roles, it is tempting to speculate that as a consequence of their differential expression as well as altered coexpression in the brain of subjects with depression, their function in neurite outgrowth, spine morphogenesis and synaptic plasticity would be affected with consequences for neuronal complexity and connectivity. This would fit with the observations that dendritic complexity and synapse density are reduced in depression (Drevets et al., 2008, Kang et al., 2012, Duman and Duman, 2015). Observed differences in the expression of PAK1 and PAK3 between control and subjects with depression in the present work are low. Nevertheless, major depressive disorder and other neuropsychiatric disorders are complex diseases implicating large number of genes. Each of them confers a small and incremental risk that potentially converges in dysregulated biological pathways, cellular functions, local circuit changes, and, eventually scales up to brain region pathophysiology (Belmaker and Agam, 2008). We suppose that the collective effect of the changes in the balance between mRNA levels as a result of altered expression as well as altered coexpression of these genes in a genetically complex disease such as depression can lead to alterations in the molecular interactions of cellular pathways, affect neuronal connectivity, and, thus, contribute to the behavioral symptoms of depression.
One of the limitations of the present study is that the samples originate from suicide victims. Thus, the present observations cannot be automatically generalized to depression without suicidal intent, even though all subjects that had died by suicide had a history of depression (depressed phase in case of bipolar disorder). Moreover, we cannot say that suicide itself was without effect (van Heeringen, 2001). Gene expression patterns associated with suicidality have been reported (Sequeira et al., 2007, Sequeira et al., 2009), although some overlap exists with those associated with major depression. Furthermore, additional studies to evaluate the effect on the protein levels of studied genes are required. Nevertheless, despite these limitations, we think that the reported changes in mRNA levels and disruption of the coordinated gene expression of the PAK1/PAK3 signaling signify an important contribution to the efforts of identifying potential candidates associated with mental illness.
Since PAKs are common downstream substrates of a number of signaling pathways, including those mediated by the Rho/Ras GTPases, neurotrophins, and glutamate receptors (Kreis and Barnier, 2009), and many molecules involved in these pathways have been shown to be important for activity-dependent neuronal morphogenesis in vitro (Kreis and Barnier, 2009), it is possible that the PAK1/PAK3-dependent actin reorganization may represent a converged mechanism dysregulated in the pathology of depression.
5. Conclusion
Abnormalities in the PAK1 and PAK3 mRNA levels as well as their altered coexpression patterns were observed in the postmortem brain of subjects with depression. We hypothesize that dysregulated PAK1/PAK3 dependent signaling may be a key factor responsible for volumetric abnormalities observed in the hippocampus and in the PFC in depression resulting in altered connectivity of these regions.
Highlights.
Group I PAKs are critical for development, plasticity, and maturation of the nervous system.
PAK1 and PAK3 but not PAK2 mRNA levels alter in the PFC and the hippocampus of depressed suicides respectively.
mRNA levels of PAKs activators: RAC1, CDC42, and ARHGEF7 are not affected in these brain regions.
Coexpression of the group I PAKs and of their activators alters in the brain of depressed suicides.
Dysregulated PAK1/PAK3 signaling associates with human depression and suicide.
Acknowledgments
This study was supported by grants to GNP (RO1 MH 48153 and RO1 MH 98554) from the National Institute of Mental Health, Rockville, MD. This work was also supported by grants to ACF and to BF from the ANPCyT and by grants to BF from the UNSAM, CONICET, and Fulbright Commission. AAJ is recipient of a doctoral fellowship from CONICET. BF and ACF are researchers from CONICET. The funding sources had no role in study design, acquisition, and interpretation of data or writing of the report.
Abbreviations
- ANCOVA
analysis of covariance
- PAK
p21-activated kinase
- PFC
prefrontal cortex
- PMI
postmortem interval
- qPCR
quantitative polymerase chain reaction
- RT
reverse transcription
Footnotes
Conflict of interest
The authors report no biomedical financial interests or potential conflicts of interest. Dr. Pandey reports grants from USA: National Institute of Health, during the conduct of the study.
Author contributions
Conceived and designed the experiments: BF. Performed the experiments: BF AAJ HR. Analyzed the data: BF AAJ. Interpretation of data: BF AAJ ACF GNP. Wrote the paper: BF.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- Asrar S, Meng Y, Zhou Z, Todorovski Z, Huang WW, Jia Z. Regulation of hippocampal longterm potentiation by p21-activated protein kinase 1 (PAK1) Neuropharmacology. 2009;56:73–80. doi: 10.1016/j.neuropharm.2008.06.055. [DOI] [PubMed] [Google Scholar]
- Ba W, van der Raadt J, Nadif Kasri N. Rho GTPase signaling at the synapse: implications for intellectual disability. Exp Cell Res. 2013;319:2368–2374. doi: 10.1016/j.yexcr.2013.05.033. [DOI] [PubMed] [Google Scholar]
- Belmaker RH, Agam G. Major depressive disorder. The New England journal of medicine. 2008;358:55–68. doi: 10.1056/NEJMra073096. [DOI] [PubMed] [Google Scholar]
- Boda B, Alberi S, Nikonenko I, Node-Langlois R, Jourdain P, Moosmayer M, Parisi-Jourdain L, Muller D. The mental retardation protein PAK3 contributes to synapse formation and plasticity in hippocampus. J Neurosci. 2004;24:10816–10825. doi: 10.1523/JNEUROSCI.2931-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boda B, Jourdain L, Muller D. Distinct, but compensatory roles of PAK1 and PAK3 in spine morphogenesis. Hippocampus. 2008;18:857–861. doi: 10.1002/hipo.20451. [DOI] [PubMed] [Google Scholar]
- Bryan B, Kumar V, Stafford LJ, Cai Y, Wu G, Liu M. GEFT, a Rho family guanine nucleotide exchange factor, regulates neurite outgrowth and dendritic spine formation. J Biol Chem. 2004;279:45824–45832. doi: 10.1074/jbc.M406216200. [DOI] [PubMed] [Google Scholar]
- Camkurt MA, Acar S, Coskun S, Gunes M, Gunes S, Yilmaz MF, Gorur A, Tamer L. Comparison of plasma MicroRNA levels in drug naive, first episode depressed patients and healthy controls. J Psychiatr Res. 2015;69:67–71. doi: 10.1016/j.jpsychires.2015.07.023. [DOI] [PubMed] [Google Scholar]
- Causeret F, Terao M, Jacobs T, Nishimura YV, Yanagawa Y, Obata K, Hoshino M, Nikolic M. The p21-activated kinase is required for neuronal migration in the cerebral cortex. Cereb Cortex. 2009;19:861–875. doi: 10.1093/cercor/bhn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffel DJ, Golden SA, Russo SJ. Structural and synaptic plasticity in stress-related disorders. Rev Neurosci. 2011;22:535–549. doi: 10.1515/RNS.2011.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobos I, Borello U, Rubenstein JL. Dlx transcription factors promote migration through repression of axon and dendrite growth. Neuron. 2007;54:873–888. doi: 10.1016/j.neuron.2007.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combeau G, Kreis P, Domenichini F, Amar M, Fossier P, Rousseau V, Barnier JV. The p21- activated kinase PAK3 forms heterodimers with PAK1 in brain implementing trans-regulation of PAK3 activity. J Biol Chem. 2012;287:30084–30096. doi: 10.1074/jbc.M112.355073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- Daniels RH, Hall PS, Bokoch GM. Membrane targeting of p21-activated kinase 1 (PAK1) induces neurite outgrowth from PC12 cells. EMBO J. 1998;17:754–764. doi: 10.1093/emboj/17.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC, Price JL, Furey ML. Brain structural and functional abnormalities in mood disorders: implications for neurocircuitry models of depression. Brain Struct Funct. 2008;213:93–118. doi: 10.1007/s00429-008-0189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos A, Combeau G, Bernardinelli Y, Barnier JV, Hartley O, Gaertner H, Boda B, Muller D. Alteration of synaptic network dynamics by the intellectual disability protein PAK3. J Neurosci. 2012;32:519–527. doi: 10.1523/JNEUROSCI.3252-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman CH, Duman RS. Spine synapse remodeling in the pathophysiology and treatment of depression. Neurosci Lett. 2015;601:20–29. doi: 10.1016/j.neulet.2015.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Teppen T, Zhang H, Mondal A, Roberts RC, Conley RR, Pandey GN. Lower phosphoinositide 3-kinase (PI 3-kinase) activity and differential expression levels of selective catalytic and regulatory PI 3-kinase subunit isoforms in prefrontal cortex and hippocampus of suicide subjects. Neuropsychopharmacology. 2008;33:2324–2340. doi: 10.1038/sj.npp.1301641. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN. Aberrant extracellular signal-regulated kinase (ERK)1/2 signalling in suicide brain: role of ERK kinase 1 (MEK1) Int J Neuropsychopharmacol. 2009;12:1337–1354. doi: 10.1017/S1461145709990575. [DOI] [PubMed] [Google Scholar]
- Dwivedi Y, Rizavi HS, Zhang H, Roberts RC, Conley RR, Pandey GN. Modulation in activation and expression of phosphatase and tensin homolog on chromosome ten, Akt1, and 3-phosphoinositide-dependent kinase 1: further evidence demonstrating altered phosphoinositide 3-kinase signaling in postmortem brain of suicide subjects. Biol Psychiatry. 2010;67:1017–1025. doi: 10.1016/j.biopsych.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsova B, Alvarez Julia A, Rizavi HS, Frasch AC, Pandey GN. Altered expression of neuroplasticity-related genes in the brain of subjects with depression. Neuroscience. 2015;299:1–17. doi: 10.1016/j.neuroscience.2015.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Ding Y, French B, Tseng GC, Sibille E. Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014;13:13–24. doi: 10.1111/gbb.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri C, Guilloux JP, Lewis DA, Sibille E. Altered gene synchrony suggests a combined hormone-mediated dysregulated state in major depression. PloS one. 2010;5:e9970. doi: 10.1371/journal.pone.0009970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden SA, Christoffel DJ, Heshmati M, Hodes GE, Magida J, Davis K, Cahill ME, Dias C, Ribeiro E, Ables JL, Kennedy PJ, Robison AJ, Gonzalez-Maeso J, Neve RL, Turecki G, Ghose S, Tamminga CA, Russo SJ. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat Med. 2013;19:337–344. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Ohshima T, Hashimoto M, Mikoshiba K. Pak1 regulates dendritic branching and spine formation. Dev Neurobiol. 2007;67:655–669. doi: 10.1002/dneu.20363. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Ohshima T, Mikoshiba K. Pak1 is involved in dendrite initiation as a downstream effector of Rac1 in cortical neurons. Mol Cell Neurosci. 2002;20:579–594. doi: 10.1006/mcne.2002.1144. [DOI] [PubMed] [Google Scholar]
- Hayashi ML, Choi SY, Rao BS, Jung HY, Lee HK, Zhang D, Chattarji S, Kirkwood A, Tonegawa S. Altered cortical synaptic morphology and impaired memory consolidation in forebrain- specific dominant-negative PAK transgenic mice. Neuron. 2004;42:773–787. doi: 10.1016/j.neuron.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Shepelev M, Chernoff J. The genetics of Pak. J Cell Sci. 2004;117:4343–4354. doi: 10.1242/jcs.01392. [DOI] [PubMed] [Google Scholar]
- Huang W, Zhou Z, Asrar S, Henkelman M, Xie W, Jia Z. p21-Activated kinases 1 and 3 control brain size through coordinating neuronal complexity and synaptic properties. Mol Cell Biol. 2011;31:388–403. doi: 10.1128/MCB.00969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs T, Causeret F, Nishimura YV, Terao M, Norman A, Hoshino M, Nikolic M. Localized activation of p21-activated kinase controls neuronal polarity and morphology. J Neurosci. 2007;27:8604–8615. doi: 10.1523/JNEUROSCI.0765-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, Licznerski P, Lepack A, Majik MS, Jeong LS, Banasr M, Son H, Duman RS. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly ML, Chernoff J. Mouse models of PAK function. Cellular logistics. 2012;2:84–88. doi: 10.4161/cl.21381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaravolu RK, Adam C, Moonen JR, Harmsen MC, Goebeler M, Schmidt M. Erk5 inhibits endothelial migration via KLF2-dependent down-regulation of PAK1. Cardiovasc Res. 2015;105:86–95. doi: 10.1093/cvr/cvu236. [DOI] [PubMed] [Google Scholar]
- Koth AP, Oliveira BR, Parfitt GM, de Buonocore JQ, Barros DM. Participation of group I p21-activated kinases in neuroplasticity. Journal of physiology, Paris. 2014;108:270–277. doi: 10.1016/j.jphysparis.2014.08.007. [DOI] [PubMed] [Google Scholar]
- Kreis P, Barnier JV. PAK signalling in neuronal physiology. Cellular signalling. 2009;21:384–393. doi: 10.1016/j.cellsig.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Kreis P, Thevenot E, Rousseau V, Boda B, Muller D, Barnier JV. The p21-activated kinase 3 implicated in mental retardation regulates spine morphogenesis through a Cdc42- dependent pathway. J Biol Chem. 2007;282:21497–21506. doi: 10.1074/jbc.M703298200. [DOI] [PubMed] [Google Scholar]
- Li J, Meng H, Cao W, Qiu T. MiR-335 is involved in major depression disorder and antidepressant treatment through targeting GRM4. Neurosci Lett. 2015;606:167–172. doi: 10.1016/j.neulet.2015.08.038. [DOI] [PubMed] [Google Scholar]
- Luo S, Mizuta H, Rubinsztein DC. p21-activated kinase 1 promotes soluble mutant huntingtin self-interaction and enhances toxicity. Hum Mol Genet. 2008;17:895–905. doi: 10.1093/hmg/ddm362. [DOI] [PubMed] [Google Scholar]
- Ma QL, Yang F, Frautschy SA, Cole GM. PAK in Alzheimer disease, Huntington disease and X-linked mental retardation. Cellular logistics. 2012;2:117–125. doi: 10.4161/cl.21602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen G, Frodl T. The hippocampus in major depression: evidence for the convergence of the bench and bedside in psychiatric research? Mol Psychiatry. 2011;16:252–264. doi: 10.1038/mp.2010.80. [DOI] [PubMed] [Google Scholar]
- Manser E, Chong C, Zhao ZS, Leung T, Michael G, Hall C, Lim L. Molecular cloning of a new member of the p21-Cdc42/Rac-activated kinase (PAK) family. J Biol Chem. 1995;270:25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Marler KJ, Kozma R, Ahmed S, Dong JM, Hall C, Lim L. Outgrowth of neurites from NIE-115 neuroblastoma cells is prevented on repulsive substrates through the action of PAK. Mol Cell Biol. 2005;25:5226–5241. doi: 10.1128/MCB.25.12.5226-5241.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayberg HS. Targeted electrode-based modulation of neural circuits for depression. J Clin Invest. 2009;119:717–725. doi: 10.1172/JCI38454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick CR, Magarinos AM, Blanchard DC, Blanchard RJ, McEwen BS, Sakai RR. Chronic social stress reduces dendritic arbors in CA3 of hippocampus and decreases binding to serotonin transporter sites. Synapse. 2000;36:85–94. doi: 10.1002/(SICI)1098-2396(200005)36:2<85::AID-SYN1>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Meng J, Meng Y, Hanna A, Janus C, Jia Z. Abnormal long-lasting synaptic plasticity and cognition in mice lacking the mental retardation gene Pak3. J Neurosci. 2005;25:6641–6650. doi: 10.1523/JNEUROSCI.0028-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merali Z, Du L, Hrdina P, Palkovits M, Faludi G, Poulter MO, Anisman H. Dysregulation in the suicide brain: mRNA expression of corticotropin-releasing hormone receptors and GABA(A) receptor subunits in frontal cortical brain region. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:1478–1485. doi: 10.1523/JNEUROSCI.4734-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani N, Ohnishi T, Iwamoto K, Watanabe A, Iwayama Y, Yamashita S, Ishitsuka Y, Moriyama K, Nakajima M, Tatebayashi Y, Akiyama H, Higuchi T, Kato T, Yoshikawa T. Expression analysis of actin-related genes as an underlying mechanism for mood disorders. Biochem Biophys Res Commun. 2007;352:780–786. doi: 10.1016/j.bbrc.2006.11.101. [DOI] [PubMed] [Google Scholar]
- Obermeier A, Ahmed S, Manser E, Yen SC, Hall C, Lim L. PAK promotes morphological changes by acting upstream of Rac. EMBO J. 1998;17:4328–4339. doi: 10.1093/emboj/17.15.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey GN, Rizavi HS, Ren X, Bhaumik R, Dwivedi Y. Toll-like receptors in the depressed and suicide brain. J Psychiatr Res. 2014;53:62–68. doi: 10.1016/j.jpsychires.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez SE, Getova DP, He B, Counts SE, Geula C, Desire L, Coutadeur S, Peillon H, Ginsberg SD, Mufson EJ. Rac1b increases with progressive tau pathology within cholinergic nucleus basalis neurons in Alzheimer’s disease. Am J Pathol. 2012;180:526–540. doi: 10.1016/j.ajpath.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C, Duman RS. Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacology. 2008;33:88–109. doi: 10.1038/sj.npp.1301574. [DOI] [PubMed] [Google Scholar]
- Poulter MO, Du L, Zhurov V, Palkovits M, Faludi G, Merali Z, Anisman H. Altered Organization of GABA(A) Receptor mRNA Expression in the Depressed Suicide Brain. Frontiers in molecular neuroscience. 2010;3:3. doi: 10.3389/neuro.02.003.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radley JJ, Sisti HM, Hao J, Rocher AB, McCall T, Hof PR, McEwen BS, Morrison JH. Chronic behavioral stress induces apical dendritic reorganization in pyramidal neurons of the medial prefrontal cortex. Neuroscience. 2004;125:1–6. doi: 10.1016/j.neuroscience.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Rajkowska G, Miguel-Hidalgo JJ, Wei J, Dilley G, Pittman SD, Meltzer HY, Overholser JC, Roth BL, Stockmeier CA. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry. 1999;45:1085–1098. doi: 10.1016/s0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- Rane CK, Minden A. P21 activated kinases: structure, regulation, and functions. Small GTPases. 2014:5. doi: 10.4161/sgtp.28003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid T, Banerjee M, Nikolic M. Phosphorylation of Pak1 by the p35/Cdk5 kinase affects neuronal morphology. J Biol Chem. 2001;276:49043–49052. doi: 10.1074/jbc.M105599200. [DOI] [PubMed] [Google Scholar]
- Rousseau V, Goupille O, Morin N, Barnier JV. A new constitutively active brain PAK3 isoform displays modified specificities toward Rac and Cdc42 GTPases. J Biol Chem. 2003;278:3912–3920. doi: 10.1074/jbc.M207251200. [DOI] [PubMed] [Google Scholar]
- Sakakibara A, Horwitz AF. Mechanism of polarized protrusion formation on neuronal precursors migrating in the developing chicken cerebellum. J Cell Sci. 2006;119:3583–3592. doi: 10.1242/jcs.03080. [DOI] [PubMed] [Google Scholar]
- Salzman S, Endicott J, Clayton P, Winokur G. Diagnostic Evaluation After Death (DEAD) Rockville, MD: National Institute of Mental Health, Neuroscience Research Branch; 1983. [Google Scholar]
- Savitz J, Drevets WC. Bipolar and major depressive disorder: neuroimaging the developmental-degenerative divide. Neurosci Biobehav Rev. 2009;33:699–771. doi: 10.1016/j.neubiorev.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira A, Klempan T, Canetti L, ffrench-Mullen J, Benkelfat C, Rouleau GA, Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- Sequeira A, Mamdani F, Ernst C, Vawter MP, Bunney WE, Lebel V, Rehal S, Klempan T, Gratton A, Benkelfat C, Rouleau GA, Mechawar N, Turecki G. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS One. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Davenport EC, Wei J, Li X, Pathania M, Vaccaro V, Yan Z, Kittler JT. GIT1 and betaPIX are essential for GABA(A) receptor synaptic stability and inhibitory neurotransmission. Cell reports. 2014;9:298–310. doi: 10.1016/j.celrep.2014.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer R, Williams J, Gibbon M, First M. Structural Clinical Interview for DSM-IV (SCID) New York, NY: New York State Psychiatric Institute Biometrics Research; 1995. [Google Scholar]
- Stockmeier CA, Mahajan GJ, Konick LC, Overholser JC, Jurjus GJ, Meltzer HY, Uylings HB, Friedman L, Rajkowska G. Cellular changes in the postmortem hippocampus in major depression. Biol Psychiatry. 2004;56:640–650. doi: 10.1016/j.biopsych.2004.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo M, Manser E, Lim L. Identification and molecular cloning of a p21cdc42/rac1- activated serine/threonine kinase that is rapidly activated by thrombin in platelets. J Biol Chem. 1995;270:26690–26697. doi: 10.1074/jbc.270.44.26690. [DOI] [PubMed] [Google Scholar]
- van Heeringen C. Suicide, serotonin, and the brain. Crisis. 2001;22:66–70. doi: 10.1027//0227-5910.22.2.66. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ma QL, Calon F, Harris-White ME, Yang F, Lim GP, Morihara T, Ubeda OJ, Ambegaokar S, Hansen JE, Weisbart RH, Teter B, Frautschy SA, Cole GM. Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease. Nat Neurosci. 2006;9:234–242. doi: 10.1038/nn1630. [DOI] [PubMed] [Google Scholar]
- Zhurov V, Stead JD, Merali Z, Palkovits M, Faludi G, Schild-Poulter C, Anisman H, Poulter MO. Molecular pathway reconstruction and analysis of disturbed gene expression in depressed individuals who died by suicide. PloS one. 2012;7:e47581. doi: 10.1371/journal.pone.0047581. [DOI] [PMC free article] [PubMed] [Google Scholar]


