Abstract
Genome-wide association studies (GWASs) have linked genes to various pathological traits. However, the potential contribution of regulatory noncoding RNAs, such as microRNAs (miRNAs), to a genetic predisposition to pathological conditions has remained unclear. We leveraged GWAS meta-analysis data from >188,000 individuals to identify 69 miRNAs in physical proximity to single-nucleotide polymorphisms (SNPs) associated with abnormal levels of circulating lipids. Several of these miRNAs (miR-128-1, miR-148a, miR-130b, and miR-301b) control the expression of key proteins involved in cholesterol-lipoprotein trafficking, such as the low-density lipoprotein (LDL) receptor (LDLR) and the ATP-binding cassette A1 (ABCA1) cholesterol transporter. Consistent with human liver expression data and genetic links to abnormal blood lipid levels, overexpression and antisense targeting of miR-128-1 or miR-148a in high-fat diet–fed C57BL/6J and Apoe-null mice resulted in altered hepatic expression of proteins involved in lipid trafficking and metabolism, and in modulated levels of circulating lipoprotein-cholesterol and triglycerides. Taken together, these findings support the notion that altered expression of miRNAs may contribute to abnormal blood lipid levels, predisposing individuals to human cardiometabolic disorders.
GWASs have been extensively used to identify common variants, termed SNPs, associated with various human traits. Most of these studies have focused on genes in proximity (typically within 100 kb) to SNPs as potential causative links to traits. However, protein-coding genes account for only a very small proportion of the transcribed human genome, thus highlighting a lack of understanding of the possible contributions of noncoding RNAs to human traits and disease. Aberrant cholesterol and lipid homeostasis, as well as altered energy metabolism, are implicated in cardiometabolic diseases such as metabolic syndrome, type 2 diabetes, and coronary artery disease (CAD)1. Despite progress in the treatment (for example, with statins) of abnormal levels of cholesterol and other lipids, cardiometabolic diseases represent prominent causes of human morbidity and mortality2, thereby emphasizing the need for new therapeutic strategies. miRNAs, comprising a class of short (20–24 nt) regulatory RNAs that modulate mRNA translation and turnover, have recently emerged as crucial regulators of cholesterol, triglycerides (TAG), glucose and energy homeostasis3,4. We and others recently identified the miR-33 family of miRNAs as central regulators of mammalian cholesterol and lipid metabolism5–9. Several additional miRNAs have also been found to control aspects of cellular and in vivo cholesterol and lipid homeostasis4,10. However, the potential contribution in humans of miRNAs to derangements in circulating cholesterol and lipid levels, and to associated cardiometabolic disease, has remained unclear.
RESULTS
MicroRNAs are linked to SNPs associated with abnormal blood lipids
To explore whether miRNAs might be functionally linked to human genetic predispositions to abnormalities in circulating levels of cholesterol and (other) lipids, we carried out a systematic analysis of data from GWASs, involving >188,000 individuals, that associated common SNPs with plasma lipid levels11. We annotated these results to determine whether sequences encoding miRNAs might be located in proximity (<100 kb) to SNPs linked to aberrant lipid levels11. Our analysis identified 69 miRNAs in the vicinity of SNPs with genome-wide significant (P < 5 × 10−8) association with abnormal levels of total cholesterol (TC), HDL cholesterol (HDL-C), LDL-C, or TAGs (Supplementary Table 1). In several cases, these genomic loci harbor a nearby protein-coding gene that probably represents the main contributor to the link between the SNP and aberrant blood lipid levels (for example, fatty acid desaturases 1–3 (FADS1–FADS3) on chromosome 11 are associated with altered levels of all four blood lipid categories, and cholesteryl ester transfer protein (CETP) on chromosome 16 is associated with altered levels of HDL-C)12,13. However, many of the loci do not contain an obvious candidate gene.
We reasoned that miRNAs causally linked with abnormal blood lipids would be predicted to target genes associated with cholesterol and lipid or metabolic homeostasis, and/or with cardiometabolic disorders. We first extracted lists of predicted conserved targets for the 69 miRNAs from the widely used TargetScan 6.2 website14. We then analyzed these predicted targets using the DAVID gene ontology (GO) program15, filtered for statistically significant links with cardiometabolic disorders such as type 2 diabetes, obesity and atherosclerosis (see Online Methods). From these analyses, the top miRNAs with predicted mRNA targets involved in metabolism and metabolic control, and with significant association with cardiometabolic disorders in GO analysis, were miR-128-1 (near SNPs associated with altered TC and LDL-C levels), miR-148a (TC, LDL-C, and TAGs), and the physically closely linked and sequence-related (sharing seed sequence) miR-130b and miR-301b (TC and HDL-C) (Fig. 1a–c and Supplementary Tables 2–4). Notably, the genomic loci harboring these four miRNAs do not have clear candidate genes that might account for the genetic association of these loci with abnormal blood lipid levels. We therefore focused our follow-up studies on these miRNAs as candidate noncoding RNAs contributing to the blood lipid associations identified in GWASs.
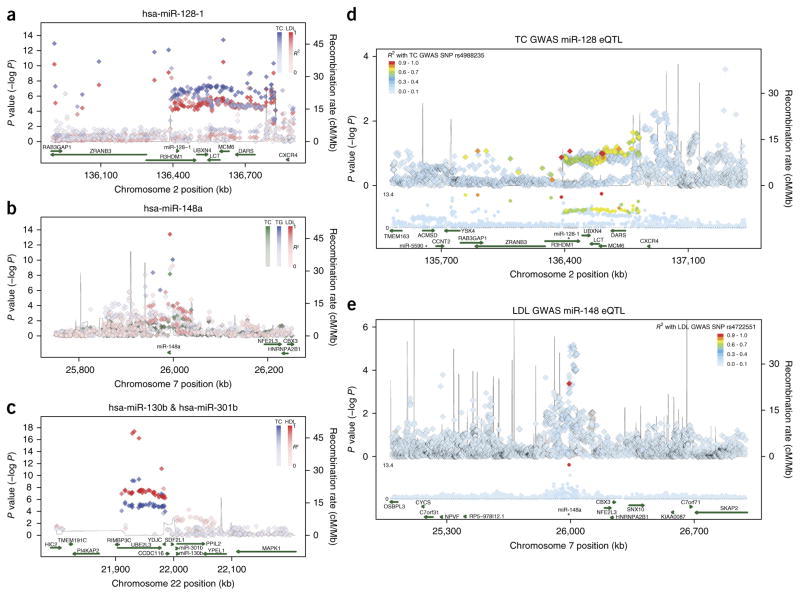
Figure 1.
Identification of miRNAs located in genomic loci that are enriched for SNPs associated with abnormal circulating total cholesterol, triglyceride, LDL-C and HDL-C levels. (a–c) Locus plots showing associations from Willer et al.11 with total cholesterol (TC), triglycerides (TG), LDL-C (LDL) and HDL-C (HDL) at the regions harboring miR-128-1 (a), miR-148a (b), and miR-130b and miR-301b (c). Color saturation levels correspond to linkage disequilibrium values (R2) to the top SNP for that particular trait in the 1000 genome EUR panel. Genetic recombination rates, estimated from the 1000 Genomes sequencing project, are plotted in blue. (d,e) Regional association plots for the eQTL analysis (top track) aligned against the Willer et al.11 joint GWAS analysis (middle track) for total cholesterol (TC) for miR-128-1 (d) and LDL cholesterol for miR-148a (e). Green arrows (bottom track) represent miRNA and protein-coding transcripts as annotated by the GENCODE Project. hsa refers to human (Homo sapiens) miRNA. The data show concordance of SNP peaks and eQTL signals, linking miR-128-1 and miR-148a hepatic expression to SNPs associated with abnormal circulating cholesterol levels.
miR-128-1 is located in intron 18 of R3HDM1, a gene on human chromosome 2 with unknown function; miR-148a is located in a gene-poor intergenic region of human chromosome 7; and miR-130b and miR-301b are found in close proximity in an inter-genic region near the UBE2L3 gene on human chromosome 22 (Fig. 1a–c). The four miRNAs are highly conserved among vertebrates (data not shown) and all four miRNAs are expressed in a number of human tissues (Supplementary Fig. 1). miR-128-1 appears to be extensively co-expressed with its host gene R3HDM1 (suggesting that they share the same primary transcript), and miR-148a is predominantly expressed in liver. Notably, the expression of miR-148a and miR-130b was significantly increased in the liver of high-fat diet (HFD)-fed mice and rhesus macaques, suggesting a potential evolutionarily conserved link to metabolic regulatory function (Supplementary Fig. 2).
Most of the SNPs in our analysis are located in noncoding intergenic or intronic regions, a number of which exhibit hallmarks of enhancers and open chromatin architecture (for example, DNase I hypersensitivity; Supplementary Table 3), suggesting that some of these SNPs may alter regulatory DNA sequence elements controlling miRNA expression. To explore whether SNPs located near the four miRNAs are associated in cis with miRNA expression, we performed miRNA expression quantitative trait locus (miR-QTL) analysis using 424 liver samples from gastric bypass surgery patients (clinical information is available in Online Methods). Our analysis revealed a number of potential cis miR-QTLs (Fig. 1d,e; Supplementary Figs. 3 and 4; Supplementary Table 5), including ones near the miR-128-1 locus that broadly overlap with the GWAS TC SNP region (Fig. 1d), and to a lesser extent, the LDL-C SNP region (Supplementary Fig. 3a). In addition, there appears to be a discrete miR-QTL peak near the CXCR4 gene in this locus that is nearby an apparent enhancer, which has been shown to be bound by multiple immune cell–associated transcription factors (for example, PU.1, EBF, PBX3, IRF4; chromatin immunoprecipitation data are referenced in the University of California, Santa Cruz (UCSC) Genome Browser). We speculate that this signal may represent a contribution from immune and/or inflammatory cell infiltration into the liver in the morbidly obese patient population from which the liver samples were taken (Fig. 1d and Supplementary Fig. 3); however, this hypothesis would need to be further tested. There is also a strong miR-QTL peak near miR-148a that includes two SNPs (rs4722551 and rs4719841) in the promoter region of miR-148a (Fig. 1e; Supplementary Fig. 3b,c and Supplementary Table 5); these two SNPs are associated with TC, TAGs and LDL-C levels (Supplementary Table 3). There was no apparent overlap between liver miR-QTLs in the miR-130b and miR-301b locus and the SNPs identified by GWAS (Supplementary Fig. 4 and data not shown), suggesting either that liver is not a relevant tissue for such an association, or that the expression of these miRNAs is not linked with the GWAS SNPs in this cohort of obese individuals. Taken together, the results of this miR-QTL analysis underscore the potential of GWASs to identify genomic variants linking miRNA expression to human phenotypes.
MicroRNA regulation of lipid-related genes and cholesterol and lipoprotein trafficking
All four miRNAs are predicted to target key regulators of lipid homeostasis and lipoprotein trafficking, including the LDLR, which is important for hepatic clearance of LDL cholesterol from the circulation16,17, as well as the ATP-binding cassette transporter A1 (ABCA1), which is crucial for HDL biogenesis, cholesterol efflux from peripheral cells such as atherogenic macrophages, and reverse cholesterol transport (RCT)18–20 (Supplementary Table 4). In accordance with these target predictions, introduction of miR-128-1, miR-148a, miR-130b and miR-301b precursor oligonucleotides into human liver-derived (HepG2 hepatoma) cells decreased LDLR and ABCA1 levels, compared to precursor control-transfected cells. Conversely, transfection of antimiR oligonucleotides increased LDLR and ABCA1 levels compared to mock-treated cells (Fig. 2). The results of the antimiR experiments show that endogenous miRNAs act to repress LDLR and ABCA1 expression in HepG2 cells. These miRNAs are also predicted to target other essential factors involved in lipid and glucose homeostasis (Supplementary Table 4). We confirmed that miR-128-1 regulates expression of the insulin receptor (InsR) and the insulin receptor substrate 1 (IRS1), which acts downstream of the insulin receptor in the insulin signaling pathway21, as well as expression of SIRT1, a NAD+-dependent energy sensor and lysine deacetylase22 (Fig. 2a). Notably, of these four miRNAs, three (miR-148a, miR-130b and miR-301b) control expression of salt-inducible kinase 1 (SIK1), a regulator of hepatic lipogenesis that inhibits SREBP-1c activity by direct phosphorylation23, as well as expression of several components of the heterotrimeric AMP-activated protein kinase (AMPK; PRKAA1 and PRKAA2), a key energy stress sensor and regulator24 (Fig. 2b–d). Moreover, miR-148a regulates expression of carnitine palmitoyltransferase 1 (CPT1A), a central regulator of mitochondrial fatty acid β-oxidation25 (Fig. 2b), whereas miR-130b controls expression of PPAR gamma coactivator 1 alpha (PGC1-α), a transcriptional coactivator, and insulin induced gene 1 (INSIG1), which controls the activity of SREBPs and HMG-CoA reductase26 (Fig. 2c). Altogether, our data suggest that these miRNAs may control multiple metabolic regulatory circuits that act in concert, which could result in cumulative effects on metabolic output.
Figure 2.
Expression of LDLR and ABCA1 is regulated by miRNAs located in genomic loci associated with abnormal circulating blood lipid levels. (a–d) Immunoblots of predicted targets of miR-128-1 (a), miR-148a (b), miR-130b (c) and miR-301b (d) that are involved in cholesterol and lipid regulation (LDLR and ABCA1) and energy homeostasis (insulin receptor (InsR) precursor (Pre) and mature (Beta) forms, IRS1, INSIG1, SIRT1, AMPK-α1, AMPK-α2, SIK1, CPT1-α, PGC1-α). Human HepG2 hepatoma cells were transfected with the indicated precursor (pre-miRNA) or antisense (antimiR) oligonucleotides. β-tubulin was used as a loading control. (e) Luciferase activity in HEK293T cells transfected with a luciferase reporter with or without the LDLR 3′ UTR. (f) Luciferase activity in cells co-transfected with the indicated miRNA precursor and a wild-type LDLR 3′ UTR-luciferase reporter or a reporter harboring point mutations in the predicted miRNA target sites. (g,h) Uptake of Dil-LDL by human HepG2 hepatoma cells after treatment with miR-128-1, miR-148a, miR-130b, or miR-301b precursors (g) or antimiRs (h). (i,j) Cholesterol efflux from HepG2 cells loaded with radiolabeled cholesterol after transfection with the indicated precursors (i) or antimiRs (j). PC, precursor control. AC, antimiR control. a.u., arbitrary units. All errors bars represent mean ± s.d. of ≥3 experiments in triplicate. Statistical significance between groups is calculated by unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to cells transfected with empty vector (e), PC (f,g,i) or AC (h,j).
LDL-C uptake by the LDLR in the liver is an essential physiological process regulating circulating levels of LDL-C16. This process is of clinical importance because of the prominent role of LDLR-dependent hepatic clearance of circulating LDL-C in preventing atherosclerosis and cardiovascular disease17. Indeed, elevated hepatic LDLR expression is a key anti-atherosclerotic effect of statins27, and mutations in the LDLR gene can cause familial hypercholesterolemia (FH), a severe disease characterized by high levels of circulating LDL-C and premature death owing to accelerated cardiovascular disease28. Because of the physiological and clinical importance of the LDLR in regulating circulating LDL-C, and as LDLR expression has not been previously shown to be under miRNA control, we wished to further verify the functional effects of the four miRNAs on LDLR expression and LDL-C uptake. First, we examined whether miR-128-1, miR-148a, miR-130b, and miR-301b regulate LDLR expression directly through its 3′ untranslated region (UTR), which possesses predicted target sequences for these miRNAs (TargetScan 6.2). The 3′ UTR of the human LDLR gene, when fused to the 3′ end of a firefly luciferase reporter, resulted in decreased luciferase activity in HEK293T cells (Fig. 2e) as compared to cells transfected with an empty vector, indicating the presence of repressive elements within the LDLR 3′ UTR. Transfection of miRNA precursors caused further repression, whereas mutation of the predicted miRNA target sites in the LDLR 3′ UTR abolished the effects of the four miRNAs (Fig. 2f). Next, we evaluated whether the four miRNAs modulate LDL-C uptake into human hepatoma cells. Indeed, internalization of fluorescently labeled LDL-C (Dil-LDL) into HepG2 cells was strongly decreased upon transfection of miR-128-1, miR-148a, miR-130b, and miR-301b as compared to cells transfected with precursor control. Conversely, transfection of antisense oligonucleotides directed against these miRNAs caused a significant increase in Dil-LDL uptake, as compared to scrambled control antimiR-treated cells (Fig. 2g,h). Together, these findings demonstrate miRNA repression of the LDLR through its 3′ UTR, with functional effects on LDL-C uptake into liver-derived cells.
ABCA1 is a critical component of the RCT pathway responsible for removing cholesterol from peripheral cells (for example, atherogenic macrophages) and transporting it back to the liver for excretion18. In particular, ABCA1 facilitates the production of nascent lipid-poor HDL in the liver and intestines, and it acts in peripheral cells and tissues to pump out cholesterol that can then be ferried by HDL back to the liver. Similarly to the 3′ UTR of LDLR, we found that the 3′ UTR of ABCA1 was directly targeted by miR-128-1, miR-148a, miR-130b, and miR-301b, as revealed by miRNA overexpression and the introduction of point mutations into the miRNA target sites in the ABCA1 3′ UTR in the luciferase fusion assay (Fig. 3). Previous studies have shown that miRNAs such as miR-33a and miR-33b control ABCA1 levels and cholesterol efflux in liver cells and in macrophages, a cell type with direct relevance to atherosclerosis5–9. We found that all four miRNAs identified in the GWAS analysis were similarly able to modulate expression of ABCA1 in the human HepG2 liver cell line and in the mouse macrophage cell line J774 (Fig. 2a–d and Fig. 3c), as well as in peritoneal macrophages isolated from C57BL/6 mice, which were treated with an LXR agonist (T090) to further activate expression of Abca1 (Fig. 3e). Accordingly, miR-128-1, miR-148a, miR-130b, and miR-301b significantly affected the cholesterol efflux capacity of these cells (Fig. 2i,j and Fig. 3d,f). Ago2 PAR-CLIP analysis29 in mouse bone marrow–derived macrophages independently identified all four miRNAs as regulators of two distinct sites in the 3′ UTR of Abca1 mRNA (Fig. 3g). Taken together, our data indicate that miR-128-1, miR-148a, miR-130b, and miR-301b control expression of both LDLR and ABCA1 in functionally relevant cell types, with effects on cholesterol trafficking.
Figure 3.
miR-128-1, miR-148a, miR-130b and miR-301b regulate ABCA1 expression and cholesterol efflux in mouse macrophages. (a) Luciferase activity in HEK293T cells transfected with a luciferase reporter with or without the ABCA1 3′ UTR. (b) Luciferase activity in cells co-transfected with the indicated miRNA precursor and a wild-type ABCA1 3′ UTR-luciferase reporter or a reporter harboring point mutations in the predicted miRNA target sites. (c) Abca1 expression in the mouse macrophage cell line J774A.1 after overexpression (pre-miRNAs) or inhibition (antimiRs) of miR-128-1, miR-148a, miR-130b or miR-301b. β-tubulin was used as a loading control. (d) Cholesterol efflux from J774A.1 cells loaded with radiolabeled cholesterol after transfection with the indicated precursors (left) and antimiRs (right). (e) Abca1 expression in LXR agonist (T0901317; T090)-stimulated mouse peritoneal macrophages transfected with the indicated antimiRs. A scrambled antimiR was used as a control. Hsp90 was used as a loading control. (f) Cholesterol efflux from mouse peritoneal macrophages after transfection of the indicated antimiRs (inh-miRNAs) with or without T0901317 treatment. Cl, scrambled antimiR control. (g) Ago2 PAR-CLIP analysis of mouse BMDM. Genome browser screen shot (mouse mm9 release) of the Abca1 locus is shown. Black shadings on the top line show RNA-seq data (RPKM, reads per kilobase per million mapped reads) from BMDM. Ago2 PAR-CLIP reads, shown in blue, identified five distinct, high-confidence miRNA binding sites. The Abca1 transcript is indicated by blue boxes (the wider box indicates the coding region and the narrower box indicates the 3′ UTR) and the arrows indicate the direction of transcription of the Abca1 gene. RefSeq, reference sequence database. TargetScan 6.2 was used to identify miRNAs that interact with the miRNA binding sites identified by the Ago2 PAR-CLIP analysis. miRNAs identified in this study are labeled in red. MicroRNA expression levels in mouse BMDM are indicated in colored boxes on a log10 scale (red indicates high expression, black indicates low expression). PC, Precursor control. AC, AntimiR control. a.u., arbitrary units. All errors bars represent mean ± s.d. of ≥3 experiments in triplicate. Statistical significance between groups is calculated by unpaired t-test, *P < 0.05, **P < 0.01 compared to cells transfected with the empty vector (a), PC (b,d) or AC (d,f).
miRNA overexpression in mice inhibits hepatic Ldlr and Abca1 expression and modulates circulating cholesterol-lipoprotein levels
On the basis of the miR-QTL data linking SNPs associated with abnormal blood lipid levels to miR-128-1 and miR-148a expression, we chose to study the effects of these miRNAs on circulating blood lipids in vivo. We first investigated the effects of miR-128-1 and miR-148a overexpression in C57BL/6J mice fed an HFD (60% calories as fat). Mice were injected in the tail vein with lentivirus expressing pre-miR-128-1 or pre-miR-148a, or were injected with a control virus, and then serum and liver were harvested after two weeks. The miRNA-expressing viruses, which also encoded GFP, were active in liver, as assessed by GFP expression (Fig. 4). In accordance with the in vitro findings, miR-128-1 and miR-148a overexpression strongly reduced hepatic levels of Abca1 and Ldlr compared with control scrambled pre-miRNA expression, and it also altered the expression of other targets such as Irs1 (miR-128-1) and Cpt1a (miR-148a) (Fig. 4a,d–h and data not shown). The circulating cholesterol and lipid profile of mice overexpressing miR-148a and miR-128-1 revealed a marked reduction in plasma HDL-C levels compared with controls, as expected from the requirement of Abca1 for HDL formation18–20 (Fig. 4b,c).
Figure 4.
Overexpression of miR-128-1 and miR-148a in C57BL/6J mice decreases circulating levels of HDL-C, whereas antisense antagonism of these miRNAs increases LDL-C clearance. (a) Analysis of hepatic Ldlr and Abca1 expression in mice injected with lentivirus expressing miR-128-1 or miR-148a (n = 7 per group). β-tubulin was used as a loading control. (b) Fast protein liquid chromatography (FPLC) analysis of pooled sera from C57BL/6J mice (n = 10) injected with lentivirus expressing miR-128-1 or miR-148a and fed HFD. Control mice were injected with a control lentivirus expressing a scrambled pre-miRNA. (c) Mean area under the curve (AUC) values calculated for HDL-C fractions 33–38 isolated by FPLC from b. (d) Copepod green fluorescent protein (copGFP) expression in the liver of C57BL/6J mice infected with lentiviruses, which encoded the indicated pre-miRNA together with copGFP. (e–h). Hepatic expression of Abca1 (e), Ldlr (f), Irs1 (g), and Cpt1a (h) in C57BL/6J mice fed HFD and overexpressing the indicated miRNAs. n = 10 mice per group. (i,j) LDL clearance after inhibition of miR-128-1 (i) or miR-148a (j) in C57BL/6J mice (n = 5 per group). Control mice were injected with a scrambled LNA. Error bars represent s.e.m. Statistical significance between groups is calculated by unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001 compared to mice injected with a control lentivirus (b,c,e–h) or a scrambled LNA control (i,j). a.u., arbitrary units.
To evaluate the consequence of inhibition of endogenous miR-128-1 and miR-148a on LDL clearance, we injected locked nucleic acid (LNA) antisense oligonucleotides (antimiRs) into C57BL/6J mice, followed by retro-orbital injection of fluorescently labeled LDL (Dil-LDL), and assessed clearance of LDL in blood over the subsequent 5 h. AntimiR-128-1– and antimiR-148a–treated mice exhibited increased LDL clearance in comparison with controls (Fig. 4i,j), in keeping with elevated hepatic Ldlr expression and increased hepatic Dil-LDL levels (Supplementary Fig. 5).
Antisense inhibition of miR-128-1 and miR-148a in Apoe−/− mice alters cholesterol and metabolism
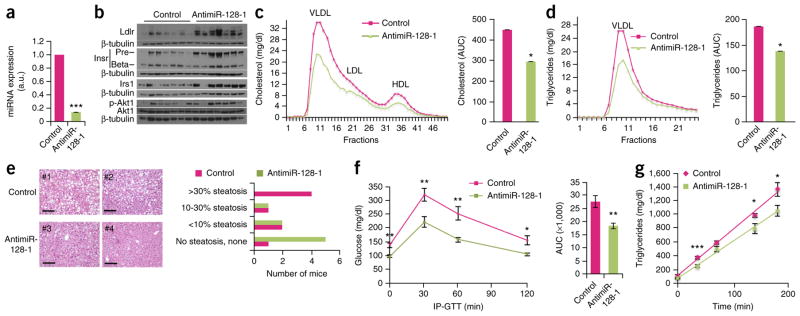
Next, we investigated the effects of antimiR-128-1 and antimiR-148a treatment in Apoe−/− mice fed a Western-type diet. Apoe−/− mice recapitulate some aspects of the abnormal serum lipid profiles observed in humans with dyslipidemia (for example, high VLDL-C and LDL-C levels and low HDL-C levels), and they exhibit accelerated atherosclerosis when fed a Western-type diet30,31. Short-term studies (5 d) with tail vein injection of 20 mg/kg antimiR-128-1, antimiR-148a, or control nonspecific LNA antimiR on days 1 and 3, revealed strongly decreased hepatic miR-128-1 and miR-148a levels, indicating potent on-target effects of both the miR-128-1– and miR-148a– targeting antimiRs (Figs. 5 and 6). As miR-128-1 exhibits some sequence similarity with the miR-27 family and shares a number of targets with those miRNAs, we verified that antimiR-128-1 LNA did not affect hepatic miR-27 levels (Supplementary Fig. 6a). In accordance with the in vitro studies and in vivo data in C57BL/6J mice, antisense inhibition of miR-128-1 and miR-148a increased hepatic levels of Ldlr as compared with control LNA-treated mice (Figs. 5b and 6b). AntimiR-148a treatment also markedly elevated hepatic Abca1 levels at both the mRNA and protein levels (Fig. 5b,c). Although we did not observe consistent effects of antimiR-128-1 treatment on Abca1 expression across the treated animals in the short-term study (data not shown), we found potent de-repression of other targets such as the insulin signaling components Irs1 and the insulin receptor (Insr), with a concomitant increase in insulin signaling output, as evidenced by elevated phosphorylated Akt (Fig. 6b). These results suggest that abnormal expression of miR-128-1 might contribute to insulin resistance, a hallmark of the metabolic syndrome1, as well as to other effects of insulin dysregulation, such as hepatic steatosis and dyslipidemia.
Figure 5.
Antisense inhibition of miR-148a in Apoe−/− mice fed a Western-type diet results in altered circulating lipoprotein/lipid levels. (a) Hepatic miR-148a levels determined by qRT-PCR in Apoe−/− mice injected with a LNA targeting miR-148a (n = 10 per group). (b) Ldlr and Abca1 expression in liver from Apoe−/− mice treated with antimiR-148a. (c) Hepatic expression of Ldlr, Abca1, and Cpt1a mRNA after short-term (5 d) antimiR-148a treatment of Apoe−/− mice fed a Western-type diet (n = 10 mice per group). (d) FPLC analysis of pooled sera from Apoe−/− mice (n = 10) fed a Western-type diet after treatment with antimiR-148a for 5 d. (e,f) Mean AUC values calculated for the LDL (e) and HDL (f) fractions (17 to 24 and 31 to 37, respectively) isolated by FPLC from d. (g) FPLC analysis of pooled sera from 10 mice after longitudinal (16 weeks) antimiR-148a treatment of Apoe−/− mice fed a Western-type diet. (h,i) Mean AUC values calculated for the VLDL/LDL (h) and HDL (i) fractions (15 to 24 and 31 to 36, respectively) isolated by FPLC from g. Control mice were injected with a scrambled LNA. AUC, area under the curve. Error bars represent s.e.m. Statistical significance between groups is calculated by unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001. a.u., arbitrary units.
Figure 6.
Antisense inhibition of miR-128-1 in Apoe−/− mice fed a Western-type diet results in altered circulating lipoprotein/lipid levels, improved glucose homeostasis/insulin signaling and decreased hepatic steatosis. (a) Hepatic miR-128-1 levels determined by quantitative RT-PCR in Apoe−/− mice injected with an LNA targeting miR-128-1. n = 10 per group. (b) Immunoblots in liver tissue from Apoe−/− mice treated with antimiR-128-1 showing expression levels of Ldlr, insulin receptor (InsR) precursor (Pre) and mature (Beta) forms, Irs1, phosphorylated (activated) and total Akt1. β-tubulin was used as a loading control. (c,d) Levels of cholesterol (c) and TAGs in the VLDL fraction (d) isolated by FPLC fractionation from pooled sera of Apoe−/− mice (n = 10) fed a Western-type diet. Histograms represent the mean AUC values for cholesterol (c, right) and triglyceride (d, right). (e) H&E staining (left) and steatosis quantification (right) in liver of Apoe−/− mice treated with antimiR-128-1 over 16 weeks. Micrographs from two control LNA–treated (#1 and #2) and two antimiR-128-1 LNA–treated (#3 and #4) mice are shown. Scale bars, 1 mm. (f) IP-GTT performed after 16 weeks of treatment with antimiR-128-1 in Apoe−/− mice (n = 10 per group). (g) VLDL-TAG secretion in Apoe−/− mice treated with antimiR-128-1 after inhibition of lipoprotein lipase with Triton WR1339 (n = 10 per group). Error bars represent s.e.m. Statistical significance between groups is calculated by unpaired t-test, *P < 0.05, **P < 0.01, ***P < 0.001. a.u., arbitrary units.
The circulating cholesterol and lipid profile of Apoe−/− mice treated with antimiR-148a revealed an increase of HDL-C levels, as expected from the involvement of ABCA1 in HDL-C generation as compared to controls, but little effect on VLDL-associated TAGs (Fig. 5d,f and Supplementary Fig. 6b). Interestingly, we also observed a decrease in LDL-C levels after inhibition of miR-148a in Apoe−/− mice (Fig. 5d,e). In this context, because in mice ApoE function is required for LDL-C clearance via LDLR, the decrease in circulating LDL-C levels observed after inhibition of miR-148a must result from LDLR-independent processes. AntimiR-128-1–treated Apoe−/− mice exhibited modestly decreased blood levels of VLDL-C and LDL-C and a shift of HDL-C to a smaller and denser form, presumably nascent HDL-C (Supplementary Fig. 6c). The miRNA-targeting LNA injections did not cause liver toxicity, as assessed by blood levels of alanine transaminase (ALT) and asparagine transaminase (AST) (Supplementary Fig. 6d,e). Together, these findings show that miR-148a and miR-128-1, which are located in genomic regions harboring SNPs associated with abnormal blood lipid levels, regulate circulating lipoprotein-cholesterol levels in vivo in an experimental model of metabolic disease, probably through multiple mechanisms acting in concert (for example, regulation of cholesterol and lipid transporters, hepatic insulin signaling, and cholesterol and TAG production and clearance).
To further test the ability of miR-128-1 and miR-148a to regulate lipid metabolism, as well as the ability of miR-128-1 to affect glucose homeostasis and insulin sensitivity, we carried out longitudinal studies (16 weeks) using once-weekly subcutaneous injection of antimiR-128-1, antimiR-148a (each 20 mg/kg) or control scrambled LNA oligonucleotide in Apoe−/− mice fed a Western-type diet. Long-term antisense antagonism of miR-148a maintained the strong elevation of HDL-C levels seen in the short-term study, whereas the LDL-C–lowering effect found in the short-term treatment was no longer apparent (Fig. 5g–i). In antimiR-128-1–treated animals, we found strongly decreased circulating total cholesterol levels (~35%), reflected primarily in a reduction of VLDL-C/LDL-C, as well as a marked decrease in VLDL-associated TAGs (~25%), as compared with control LNA-injected mice (Fig. 6c,d). Gene expression analysis by DNA microarray profiling and quantitative RT-PCR analysis of livers from antimiR-128-1–treated mice showed that the mRNA levels of many predicted miR-128-1 targets (TargetScan 6.2 mouse), including Ldlr and Abca1, were significantly de-repressed (Supplementary Fig. 7a and Supplementary Table 6). Pathological examination of the livers, as assessed by H&E and Oil Red O staining, showed significantly (P < 0.05) decreased hepatic steatosis in animals treated with antimiR-128-1 (Fig. 6e and Supplementary Fig. 7c,d) as compared to controls. We speculate that this effect on steatosis may in part be due to decreased lipogenesis, as Fasn, a rate-limiting enzyme in fatty acid biosynthesis, was downregulated (Supplementary Fig. 7a). AntimiR-128-1–treated mice also showed significantly improved glucose homeostasis and a modest effect on insulin sensitivity, as revealed by intra-peritoneal glucose tolerance test (IP-GTT) and insulin tolerance test (ITT) (Fig. 6f and Supplementary Fig. 7b). The potent effect of antimiR-128-1 treatment on circulating cholesterol and TAGs in Apoe−/− mice cannot be explained by changes in hepatic Ldlr and Abca1 levels, but may relate to the decreased VLDL secretion that could accompany improved hepatic insulin sensitivity. Accordingly, we observed a modest but significant decrease in VLDL secretion and lowered circulating apoB-100 levels in Apoe−/− mice treated with antimiR-128-1 (Fig. 6g and Supplementary Fig. 8).
DISCUSSION
Previous GWASs have uncovered associations of genes with various human traits; however, to our knowledge, no systematic analysis has been carried out to evaluate potential associations of regulatory noncoding RNAs, such as miRNAs, with such traits. We leveraged GWAS meta-analysis in >188,000 individuals to identify 69 miRNAs located in genomic regions associated with abnormal blood lipid levels. Our work identifies four miRNAs (miR-128-1, miR-148a, miR-130b and miR-301b) that regulate LDL-C uptake and cholesterol efflux by controlling the expression of the LDLR and ABCA1 proteins, respectively, in physiologically relevant cell types such as liver and macrophage-derived cells. miR-QTL studies using liver tissue from 424 morbidly obese individuals revealed an association of miR-128-1 and miR-148a expression with SNPs linked to abnormal human blood lipid levels, suggesting the relevance of these miRNAs, identified by GWASs, to human cardiometabolic disorders. In vivo studies in C57BL/6J mice and the Apoe−/− mouse metabolic disease model verified that miR-128-1 and miR-148a can indeed regulate cholesterol/lipid and energy homeostasis. Moreover, the effects of antimiR-128-1 treatment on TAGs, as well as on insulin signaling and sensitivity and glucose homeostasis, in Apoe−/− mice suggests that miR-128-1 dysregulation could potentially contribute to other aspects of the metabolic syndrome.
Taken together, our studies suggest that noncoding RNAs, such as miRNAs, uncovered in GWAS analyses could play key roles in contributing to human disease states. The therapeutic tractability of potent and specific antisense technologies targeting single or multiple miRNAs implicated in human cardiometabolic diseases may thus have important clinical ramifications for the treatment of these diseases.
ONLINE METHODS
Mouse lentivirus and antimiR studies
Lentivirus
Lentiviruses encoding miR-128-1 (miRZip-128-1) and miR-148a (miRZip-148a) precursors, as well as a scrambled control (miRZip-Scramble), were obtained from System Biosciences, Inc. (no. CS970MZ-Z). 12-week-old male C57BL/6J mice were purchased from the Jackson Laboratory, Bar Harbor, ME, and fed an HFD supplemented with 60% kcal from milk fat (D12492, Research Diets) during treatment. Mice were injected in the tail vein with 1 × 109 pfu/mouse diluted in 100 μl PBS on day 1, and serum and liver were harvested on day 14.
AntimiRs
LNA antimiRs directed against miR-128-1 (TTCACTGTG), miR-148a (TTCTGTAGTGCACTG), and a scrambled control LNA oligonucleotide (TCATACTA) were purchased from Exiqon, Denmark. Experiments were performed using 12-week-old male Apoe-null mice. Mice (no. 002052) were purchased from the Jackson Laboratory, Bar Harbor, ME and fed a Western-type diet supplemented with 45% kcal from milk fat (D12451, Research Diets) for 6 weeks before and during treatment. For the short-term (5 d) studies, miR-128-1 and miR-148a LNA antimiRs (20 mg/kg) or control scrambled LNA oligonucleotide were dissolved in PBS (total volume of 200 μl) and administered through tail vein injections on days 1 and 3 at the same time of day on each day of treatment. Mice were sacrificed 48 h after the last tail vein injection. The longitudinal studies were performed in Apoe-null mice fed with a Western-type diet for 16 weeks, with once-weekly subcutaneous injection of the miR-128-1 or miR-148a LNA antimiRs (20 mg/kg) or control scrambled LNA oligonucleotide. Upon sacrifice, livers were collected and ~1 ml of blood was obtained from each mouse by right ventricular puncture. Blood was centrifuged at 8,000 rpm for 5 min to obtain serum, which was frozen at −80 °C. Total serum cholesterol and triglyceride levels were determined with a Heska Dri-Chem 4000 Chemistry Analyzer (Heska, Loveland, CO) at Massachusetts General Hospital, Center for Comparative Medicine, Diagnostic Laboratory. FPLC analysis of pooled serum was carried out as described5. All mouse procedures were approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee.
GWAS analysis
The GWAS analysis was performed by identifying miRNAs located in proximity (<100 kb) of SNPs (P-value <5 × 10−8) associated with total cholesterol (TC), low-density lipoprotein-cholesterol (LDL-C), high- density lipoprotein-cholesterol (HDL-C) and triglyceride (TAG) levels11. The list of miRNAs was extracted from miRBase (release 19). The predicted target genes were extracted using the TargetScan (6.2, Human) and gene ontology/functional annotation was performed with the DAVID software based on two diseases categories: GENETIC_ASSOCIATION_DB_DISEASE (http://geneticassociationdb.nih.gov/) and OMIM_DISEASE (http://www.ncbi.nlm.nih.gov/omim). To represent the SNP (P-value <10−4) density in proximity of miRNAs shown in Figure 1, we extracted the list of SNPs associated with TC, LDL-C, HDL-C and TAG levels previously published by the Global Lipids Genetics Consortium11,32. The recombination values shown in Figure 1b–d were obtained as pre-calculated values from the 1000 Genomes consortium and were accessed from (http://mathgen.stats.ox.ac.uk/impute/ALL_1000G_phase1integrated_feb2012_impute/). The R2 calculations were produced using Plink with the 1000 Genomes phase 1 variants for the EUR (CEU, TSI, FIN, GBR, IBS) populations.
miR-QTL analysis using a RYGB patient cohort
For this study, we were interested in finding cis QTLs for four miRNAs: miR-128-1 (chromosome 2), miR-148a (chromosome 7), and the physically closely linked and related miR-130b and miR-301b (chromosome 22 – their seed sequences are identical). The full data set including QTL and expression analysis of all miRNAs and SNPs profiled will be published separately.
From February 2000 until April 2007, consent was obtained from 1,018 individuals undergoing Roux-en-Y gastric bypass (RYGB) at the Massachusetts General Hospital to collect and analyze tissue samples removed at the time of surgery. Surgical procedures and the study population have been described previously33–35. Briefly, intraoperative liver samples were collected in RNAlater (Ambion) and stored at −80 °C. Genomic DNA was extracted from liver samples, and 950 samples were successfully genotyped with the Illumina HumanHap 650Y BeadChip array at the Gene Expression Laboratory of Rosetta Inpharmatics. MicroRNAs were extracted from liver tissues and RT-PCR was performed to derive miRNA expression from the samples using a previously published approach36. Standard curves were created for all miRNAs profiled, allowing for the characterization of miRNA abundance as copies/10 pg of total RNA, with a typical mammalian cell containing 10–30 pg of total RNA. All human studies were approved by and conducted in accordance with the ethical standards of the Partners Human Research Committee, and written informed consent was obtained from all participants.
Samples were processed in three separate batches over the course of several years. Because measured miRNA expression may be affected by batch effects as well as by other nongenetic factors, we first determined potential factors that may need to be accounted for in our miR-QTL analysis. For a given factor, we tested for its association with every miRNA using a simple linear regression model. We then compared the resulting association P-value distribution against the uniform distribution; a uniform P-value distribution would be expected if the factor tested does not systematically contribute to the variability of the miRNA expression measurements. This procedure identified that 1) sample processing batch, 2) surgery year, 3) surgery type, and 4) two qPCR controls are significant contributors to the variability of miRNA expression. We therefore corrected for the effects of these factors for all miRNAs using mixed-effect modeling37 and retained the fitted residuals for miR-QTL analysis. The model we used, expressed using R/lme4 notation, is
where E is the measured expression; here we treat batch, surgery year, and surgery type as random effects and the two control probes (X1 and X2) as fixed effects.
Genotyped SNPs were initially filtered to have a minor allele frequency (MAF) >0.01, SNP call rate >95%, individual missing rate <1%, and Hardy-Weinberg equilibrium (HWE) test P >1e-6. SNPs were then imputed using MaCH-Admix38 with 1000 Genomes haplotype data (Phase I release version 3) as reference. Imputed SNPs were filtered based on imputation quality (Rsq >0.3). After imputation, the combined genotyped and imputed SNPs were filtered again to ensure MAF >0.03, SNP call rate >95%, individual missing rate <1%, and HWE P >1e-6.
To reduce the potentially confounding effect of race, only Caucasian patients’ samples were selected for analysis by excluding the small number of non-Caucasians. Furthermore, to account for population stratification, we computed principal components of SNP genotypes using Eigenstrat39 and included them in our association model (see below). A total of 424 samples were used to perform miR-QTL analysis.
We performed association between each of the four miRNAs and the SNPs located within 1 Mb on either side of the transcription start site of the miRNAs using a linear, additive model. We included the first 3 principal components of SNP genotypes as covariates in the miR-QTL analysis. Gender, age, and pre-surgery BMI were also included as covariates. The “Matrix eQTL” package40 was used to perform the association analysis in R. The “dosage” output (.dose files) from MaCH-Admix was used for association analysis.
Bone marrow-derived macrophage (BMDM) culture and Ago2 PAR-CLIP analysis
Mice were euthanized using carbon dioxide, and bone marrow cells from the femur and tibia were collected by flushing through a 23-gauge needle with PBS. Bone marrow cells were cultured with complete growth media containing DMEM and 20% L929 cell conditioned medium for 6 d (ref. 41),
BMDM for the PAR-CLIP experiment were incubated with 100 μM 4-thio-uridine (Sigma) for 16 h, washed with PBS and UV-crosslinked (0.15 J/cm2 total energy using 365 nm UV light in a Stratalinker (Invitrogen)). BMDM were then harvested and frozen at −80 °C. The details of the PAR-CLIP protocol were described previously42,43 with several minor modifications as follows. We used mouse monoclonal anti-mouse AGO2 antibody (Clone 2D4 WAKO) for Ago2 IP. Immunoprecipitated RNA-protein complex was labeled with γ-[32P]-ATP and polynucleotide kinase, purified by PAGE and the cDNA library was prepared for deep sequencing. We used 3′ adapters, 5′ adapters, RT primers, and PCR primers from the Illumina TruSeq Small RNA Sample Preparation Kit. PAR-CLIP small RNA libraries from 2 samples were sequenced for 45 cycles on the Illumina HiSeq 2000 platform (Illumina).
We used PARalyzer software to identify miRNA binding sites, as described previously44. Briefly, reads were aligned to the mouse genome sequence, version MM9. Ago2 PAR-CLIP clusters were defined as having at least 25 reads, as excluding genomic repeat regions, and as meeting the T to C conversion criteria. We only considered overlapping sequence reads that exceeded a threshold of ≥ 50% T to C conversion frequency. We selected sites with more than 100 reads for further analysis, such as miRNA prediction. Clusters that overlapped with predicted miRNA binding sites from the TargetScan database (TargetScan 6.2, mouse non-conserved and conserved predictions) were identified using custom scripts. Since TargetScan 6.2 uses canonical seed match sites (≥7mer1A, i.e., nucleotide 2–7 match with an A across from position one of the mature miRNA), when no TargetScan 6.2 predictions were found for a given cluster, we also used the PITA algorithm (Segal Lab of Computational Biology) to search for miRNA target sites; this algorithm allows either 1 G:U wobble or 1 mismatch in the 7–8 nt seed site. Detailed information on PAR-CLIP data, miRNA sequencing data and mRNA sequencing data were published previously29, and sequencing data have been submitted to the GEO database with the following accession number: GSE63199.
Hepatic miRNA expression in mice and rhesus macaques fed chow versus high-fat diets
Male rhesus monkeys (age 7–13 years old) were fed a standard (TestDiet no. 5038; Purina Mills) or high-fat, high-sucrose diet (42% kcal in fat, Custom formula no. 07802; Harlan, Teklad, Madison, WI) for 2 years and maintained as previously described45. Animal procedures were approved by the Animal Care and Use Committee of the National Institute on Aging (NIA) Intramural Research Program. After animal euthanasia, liver RNA was isolated using the Bullet Blender Homogenizer (Next Advance) in TRIzol. For miRNA quantification, 1 μg of total RNA was reverse transcribed using the miScript II RT Kit (Qiagen). Primers specific for miR-128-1, miR-148a, miR-130b and miR-301b (Qiagen) were used and values were normalized to SNORD68 (Qiagen) as a housekeeping gene.
Male C57BL/6J mice were purchased from the Jackson Laboratories (Bar Harbor, ME) and kept under constant temperature and humidity in a 12 h controlled dark/light cycle. For HFD studies, 8-week old male mice were placed on a chow diet or HFD containing 0.3% cholesterol and 21% (wt/wt) fat (Dyets, Inc.) for 3 weeks. Liver samples were collected as previously described46 and stored at −80 °C until total RNA was harvested for miRNA expression analysis, as described above.
Mouse peritoneal macrophage miRNA antisense treatment and cholesterol efflux
Thioglycollate-elicited peritoneal mouse macrophages (1 × 106 cells per well) were transfected either with a control inhibitor (IN-001005-01-20, Dharmacon) or an anti-miRNA inhibitor (#Inh-mmu-miR-128-3p: #IH-310398-08-0005, Inh-mmu-miR-148a-3p: #IH-310496-07-0005, Inh-mmu-miR-130a-3p: #IH-310399-07-0005, Inh-mmu-miR-301a-3p: #IH-310482-07-0005, Dharmacon) 48 h before loading with 0.5 μCi/ml 3H-cholesterol for 24 h with or without the LXR agonist T0901317 (Sigma-Aldrich; 3 μmol/l) for 12 h. Then, cells were washed twice with PBS and incubated in RPMI supplemented with 2 mg/ml fatty-acid free BSA (FAFA-media) in the presence of ACAT inhibitor (Sandoz 58-035, Sigma-Aldrich; 2 μmol/l) for 2 h before addition of 50 μg/ml human ApoA1 in FAFA-media with or without the indicated treatments. Supernatants were collected after 6 h and expressed as a percentage of total cell 3H-cholesterol content (total effluxed 3H-cholesterol + cell-associated 3H-cholesterol).
Quantitative RT-PCR
Total RNA and miRNA were extracted from cells using TRIzol (Life Technologies/Invitrogen) and the mirVana miRNA Isolation Kit (Life Technologies/Invitrogen), respectively, according to the manufacturer’s instructions. After extraction, total RNA and miRNA were reverse transcribed using the High Capacity cDNA Reverse Transcription Kit and the TaqMan MicroRNA Reverse Transcription Kit (Life Technologies/Invitrogen). RT products were quantified by real-time qPCR (Lightcycler, Roche) using the TaqMan Universal PCR Master Mix. The amount of the indicated mRNA or miRNA was normalized to the amount of GAPDH mRNA and U6 RNA, respectively. Total RNA for miRNA expression levels analysis in human tissues was purchased from Ambion (FirstChoice Human Total RNA Survey Panel, AM6000). To quantify pri-miRNA-128-1 levels, RNAs were reverse transcribed using the High Capacity cDNA Reverse Transcription Kit and quantified with the specific TaqMan Pri-miRNA assay for pri-miRNA-128-1 (Life Technologies/Invitrogen). To quantify miR-148a, miR-130b and miR-301b levels, RNAs were reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit and quantified with the miRNA-specific TaqMan miRNA assays (Life Technologies/Invitrogen). PCR primer sequences are available from A.M.N. upon request.
Antibodies
Rabbit anti-LDLR (ab30532), mouse anti-ABCA1 (ab18180), rabbit anti-AMPKα1 (ab32047), rabbit anti-AMPKα2 (ab3760), rabbit anti-INSIG1 (ab-70784), rabbit anti-CPT1α (ab83862), and mouse anti-Sik1 (ab64428) were purchased from Abcam Research Products, Cambridge, UK. Mouse anti-β tubulin (T7816) was purchased from Sigma Aldrich. Rabbit anti-SIRT1 (CY-P1016) was purchased from Cyclex. Rabbit anti-IRS1 (2382) was purchased from Cell Signaling Technology. Rabbit anti-IR (sc-711) was purchased from Santa Cruz Biotechnology. Rabbit anti-AKT (9272) and rabbit anti-phospho-AKT (9271) were purchased from Cell Signaling, mouse anti-PGC1α (ST1202-1SET) from Millipore, and mouse anti-HSP90 (no. 610418) from BD Bioscience.
Plasmids, miRNA antisense and precursor oligonucleotide transfection
Luciferase plasmids harboring the ABCA1 (HmiT004727) and LDLR (HmiT010577) 3′ UTRs were purchased from GeneCopoeia. Mutagenesis of the 3′ UTRs was performed with the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. Primer sequences used to mutate the three miR-128-1 binding sites in the LDLR 3′ UTR are the following (all are in 5′–3′ order): (816; 867): CACTTTCTCAGTTCAGAGTTG TACCCTATGTACATTTGGCATTTGTGTTATT; (2046; 2078): GTCCCCAGG GACAAAACCCTATGTCCCCCCCAG; (2444; 2506): TAGAAGGTTTTTGTA GCCTGAATGTCTTATTGTTATCAATTAAATTTCTTAAATGAACCAATT. The two binding sites in the ABCA1 3′ UTRs were mutated with the following primers: (2255; 2329): CAAGGCCATATTTTAAAAAATCAAAAGGCAATGT AAACTATTTTGAAGAAAACACAACATTTTAA; (2563; 2626): TTTGGGAC ACCTCAGAAAACTTATTAACAAATGTAAATATGAGAAATACAGAAGAA AATAATA. Mutagenesis of the two miR-148a binding sites in the LDLR 3′ UTR were performed with the following primers: (840; 893): ACTGTGTACATTTG GCATTTGTGTTATTATTTTACAATGTTTTCTGTCGTGTGT; (2547; 2587): GAAATCGCCGTGTTACTGTTACAATGATGTCCGGAGAGACA. The four miR-130b and miR-301b binding sites in the LDLR 3′ UTR were mutated using the following primers: (796; 836): CCATTCCCGTGGTCTCCTTACCCTTT CTCAGTTCAGAGTTG; (841;892): CTGTGTACATTTGGCATTTGTGTTA TTATTTTACCCTGTTTTCTGTCGTGTG; (1948; 1987): TTCTGAAATCG CCGTGTTACTGTTACCCTGATGTCCGGAG; (2136; 2192): AGATAGTGG GGATTTTTTGTTATGTTTACCCTTTGTATATTGGTTGAAACTGTTATC. Mutagenesis of both the miR-128-1 and miR-130b/miR-301b binding sites in the ABCA1 3′ UTR was performed using a single primer (the two binding sites for these three miRNAs overlap): (3081; 3131): CTTTACGCTGTCTGTAATG GGATCTATTTTTACAATGGAATATCTGAGAAT. The mutated residues are indicated in bold. Antisense (no. 4464084) and precursor (no. 4464066) oligo-nucleotides for miR-128-1, miR-148a, miR-130b and miR-301b for cell culture studies were purchased from Life Technologies/Invitrogen.
Cell culture transfection
HepG2, J774A.1 and HEK293T cells were obtained from the ATCC and propagated according to ATCC’s instructions. Transfection of antisense and precursor oligonucleotides (35 nM final concentration) were performed in HepG2 cells using Lipofectamine RNAiMAX Reagent (Life Technologies/Invitrogen), and in J774A.1 cells using TransIT TKO Transfection Reagent (Mirus Bio LLC) according to the manufacturers’ instructions. Plasmid transfection in HEK293T cells was performed with Lipofectamine 2000 (Life Technologies/Invitrogen) according to the manufacturer’s instructions.
Luciferase assays and immunoblotting
Luciferase activity was measured according to the manufacturer’s protocol (Promega, Madison, WI) and was normalized to β-galactosidase activity. Immunoblotting was performed by standard procedures.
Efflux of cholesterol from mouse J774A.1 macrophages
J774A.1 mouse macrophage cells were seeded in 24-well plates at 2 × 105 cells per well. After 24 h, cells were incubated in 3H-cholesterol and transfected with antisense or precursor oligonucleotides for 24 h. Then, cells were washed twice with PBS and incubated with fresh 5% lipoprotein deficient bovine calf serum (BT-907, Biomedical Technology) and ApoA1 protein (BT 927, Biomedical Technology) at 25 μg/ml final concentration. After 2 h, 3H-cholesterol present in medium and in alkaline lysed cells was determined by liquid scintillation counting.
Dil-LDL binding
HepG2 cells were transfected in 12-well plates at 5 × 105 cells/well. 24 h after antisense or precursor oligonucleotide transfection, Dil-LDL labeling procedure was performed according to the manufacturer’s protocol (BT-904, Biomedical Technology).
Intra-peritoneal glucose tolerance test and insulin tolerance test
Experiments were performed in 8 week-old Apoe null male mice purchased from Jackson Laboratory and treated once-weekly by subcutaneous injection with an LNA antimiR-128-1 (20 mg/kg) over 16 weeks. Mice were fed a Western-type diet supplemented with 45% kcal from milk fat (D12451, Research Diets) during the treatment. IP-GTT was performed in mice fasted for 6 h. Mice were injected with 1.5 mg/g D-(+)-glucose (G7528, Sigma Aldrich). Glucose levels were measured at 0 (before injection), 30, 60 and 120 min post-injection. Insulin tolerance test (ITT) was performed in 4 h fasted mice. Mice were injected with 0.00075 U/g insulin (I9278-5 ML, Sigma Aldrich). Glucose levels were measured at 0 (before injection), 15, 45 and 60 min after injection.
Lipid extraction from liver tissue
Each mouse liver was homogenized and incubated in a hexane:isopropanol (3:2) mixture for 16 h. After centrifugation at 3,000g for 10 min at 4 °C, the organic phase with lipids was removed and the pellet was used for protein quantification using the Bradford assay. To quantify the levels of triglycerides and cholesterol in the organic fraction, solvents in the lipid extract were removed under vacuum for 30 min and the remaining lipids were dissolved in DMSO. Colorimetric assays to measure triglycerides and cholesterol were performed according to the manufacturer’s instructions (Wako diagnostics). Lipid levels were normalized to protein concentrations and expressed as μg of lipid/mg of protein.
Liver histology
Liver tissue from each study animal was sectioned for histology47,48. Frozen and paraffin-embedded sections were prepared for staining for fat content with Oil Red O and H&E, respectively. Metamorph NX software (Molecular Devices, Sunnyvale, CA) was used to calculate the percent Oil Red O staining from digital images obtained at 20 × magnification. A paired t-test was used to compare the percentage of Oil Red O staining in the control LNA versus antimiR-128-1–treated mice. H&E slides were assigned an ordinal value on the basis of the amount of macrovesicular and microvesicular steatosis identified in a representative 20× image: 0, none; 1, <10%; 2, 10–30%; 3, >30%.
Statistics
In vitro experiments were repeated at least three times and histograms represent mean ± s.d. The number of animals used in each mouse study was determined on the basis of pertinent literature for comparable studies in which the desired effect sizes were admitted to be significant. No mice were excluded in the analyses. Data analyses from mouse experiments show mean ± s.e.m. The scoring of liver steatosis was performed in a blinded manner. The area under the curve (AUC) was calculated using the linear trapezoidal method based on a linear interpolation between data points49. The AUC was calculated as follows: AUC = 1/2 (C1+C2), where C indicates the concentration of lipids in two incremental FPLC fractions. The average AUC value for the VLDL, LDL or HDL fractions was calculated and indicated with the s.e.m. and P value. Statistical differences were measured using an unpaired two-sided Student’s t-test. P ≤ 0.05 was considered as statistically significant. Data analysis was performed using Excel Software Version 14.4.5. Statistical tests for miR-QTL and liver steatosis analyses are indicated in the corresponding Online Methods sections.
Supplementary Material
Acknowledgments
This work was supported by the following grants: US National Institutes of Health (NIH) grants R21DK084459 and R01DK094184 (A.M.N); R37DK048873 and R01DK056626 to D.E.C.; K24DK078772 to R.T.C.; and R01HL107953 and R01HL106063 to C.F.-H. A.M.N. was supported by a scholar award from the Massachusetts General Hospital (MGH). R.E.G. was supported by an Established Investigator Award from the American Heart Association. C.F.-H. was supported by a Fondation Leducq Transatlantic Network of Excellence in Cardiovascular Research Award. T.H. acknowledges the support of NIH grant R01HL49094 and a Fondation Leducq Transatlantic Network of Excellence in Cardiovascular Research Award. N.S. and J.S.T. were supported by the Intramural Research Program of the US National Institute of Allergy and Infectious Diseases. C.M.R. was supported by the American Heart Association SDG Grant 15SDG23000025. A.W. was supported by a fellowship from the MGH Executive Committee on Research. We thank C. Molony of Merck Research Laboratories for help with genotyping data quality assessment. We thank the Harvard Medical School Neurobiology Department and the Neurobiology Imaging Facility for consultation and instrument availability that supported this work. This facility is supported in part by the Neural Imaging Center as part of a National Institutes on Neurological Disorders and Stroke P30 Core Center grant no. NS072030, and by the Harvard Digestive Diseases Center (P30 DK034854).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
S.H.N.-S. and A.M.N. conceived and carried out the initial miR-128-1 studies that provided support for the expanded project. A.W. and A.M.N. conceived and designed the expanded, published studies, interpreted the data, and wrote the manuscript, which was commented on by all authors. A.W., S.H.N.-S., L.W., S.S., Y. Li, F.K., N.P., D.E.C. and R.E.G. performed experiments and analyzed data in Apoe deficient mice and in C57BL/6 mice. L.G., C.M.R. and C.F.-H. quantified miRNAs in Rhesus monkeys and mice fed with different diets and performed efflux experiments in mouse peritoneal macrophages. N.S., Y. Lu, J.S.T., E.S., R.T., I.H., P.C.S. and L.M.K. contributed to the human liver miR-QTL analysis. Y.-C.L. and T.H. performed the Ago2 PAR-CLIP analysis in BMDMs. A.S.d. and R.T.C. performed the liver histology analysis. V.V. injected the lentivirus in C57BL/6 mice. J.C.B. and J.W. performed DNA microarray analysis from mouse liver samples prepared by A.W. S.K. and A.B. analyzed the GWAS data. R.d.C. carried out the non-human primate studies.
References
- 1.Cornier MA, et al. The metabolic syndrome. Endocr Rev. 2008;29:777–822. doi: 10.1210/er.2008-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go AS, et al. Heart disease and stroke statistics–2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Najafi-Shoushtari SH, et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayner KJ, et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci USA. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horie T, et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci USA. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rayner KJ, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goedeke L, Aranda JF, Fernandez-Hernando C. microRNA regulation of lipoprotein metabolism. Curr Opin Lipidol. 2014;25:282–288. doi: 10.1097/MOL.0000000000000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willer CJ, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merino DM, Ma DW, Mutch DM. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids Health Dis. 2010;9:63. doi: 10.1186/1476-511X-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charles MA, Kane JP. New molecular insights into CETP structure and function: a review. J Lipid Res. 2012;53:1451–1458. doi: 10.1194/jlr.R027011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 15.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 16.Ishibashi S, et al. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown MS, Goldstein JL. How LDL receptors influence cholesterol and atherosclerosis. Sci Am. 1984;251:58–66. doi: 10.1038/scientificamerican1184-58. [DOI] [PubMed] [Google Scholar]
- 18.Tall AR, Yvan-Charvet L, Terasaka N, Pagler T, Wang N. HDL, ABC transporters, and cholesterol efflux: implications for the treatment of atherosclerosis. Cell Metab. 2008;7:365–375. doi: 10.1016/j.cmet.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Navab M, Reddy ST, Van Lenten BJ, Fogelman AM. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat Rev Cardiol. 2011;8:222–232. doi: 10.1038/nrcardio.2010.222. [DOI] [PubMed] [Google Scholar]
- 20.Duffy D, Rader DJ. Update on strategies to increase HDL quantity and function. Nat Rev Cardiol. 2009;6:455–463. doi: 10.1038/nrcardio.2009.94. [DOI] [PubMed] [Google Scholar]
- 21.Sparks JD, Sparks CE, Adeli K. Selective hepatic insulin resistance, VLDL overproduction, and hypertriglyceridemia. Arterioscler Thromb Vasc Biol. 2012;32:2104–2112. doi: 10.1161/ATVBAHA.111.241463. [DOI] [PubMed] [Google Scholar]
- 22.Stein S, Matter CM. Protective roles of SIRT1 in atherosclerosis. Cell Cycle. 2011;10:640–647. doi: 10.4161/cc.10.4.14863. [DOI] [PubMed] [Google Scholar]
- 23.Yoon YS, Seo WY, Lee MW, Kim ST, Koo SH. Salt-inducible kinase regulates hepatic lipogenesis by controlling SREBP-1c phosphorylation. J Biol Chem. 2009;284:10446–10452. doi: 10.1074/jbc.M900096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hardie DG. Organismal carbohydrate and lipid homeostasis. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a006031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnefont JP, et al. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol Aspects Med. 2004;25:495–520. doi: 10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 27.Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soutar AK, Naoumova RP. Mechanisms of disease: genetic causes of familial hypercholesterolemia. Nat Clin Pract Cardiovasc Med. 2007;4:214–225. doi: 10.1038/ncpcardio0836. [DOI] [PubMed] [Google Scholar]
- 29.Lu YC, et al. ELAVL1 modulates transcriptome-wide miRNA binding in murine macrophages. Cell Rep. 2014;9:2330–2343. doi: 10.1016/j.celrep.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang SH, Reddick RL, Piedrahita JA, Maeda N. Spontaneous hypercholesterolemia and arterial lesions in mice lacking apolipoprotein E. Science. 1992;258:468–471. doi: 10.1126/science.1411543. [DOI] [PubMed] [Google Scholar]
- 31.Nakashima Y, Plump AS, Raines EW, Breslow JL, Ross R. ApoE-deficient mice develop lesions of all phases of atherosclerosis throughout the arterial tree. Arterioscler Thromb. 1994;14:133–140. doi: 10.1161/01.atv.14.1.133. [DOI] [PubMed] [Google Scholar]
- 32.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatoum IJ, et al. Melanocortin-4 receptor signaling is required for weight loss after gastric bypass surgery. J Clin Endocrinol Metab. 2012;97:E1023–E1031. doi: 10.1210/jc.2011-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenawalt DM, et al. A survey of the genetics of stomach, liver, and adipose gene expression from a morbidly obese cohort. Genome Res. 2011;21:1008–1016. doi: 10.1101/gr.112821.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatoum IJ, et al. Weight loss after gastric bypass is associated with a variant at 15q26.1. Am J Hum Genet. 2013;92:827–834. doi: 10.1016/j.ajhg.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bates D, et al. Fitting linear mixed-effects models using lme4. 2014 arXiv http://arxiv.org/abs/1406.5823.
- 38.Liu EY, Li M, Wang W, Li Y. MaCH-admix: genotype imputation for admixed populations. Genet Epidemiol. 2013;37:25–37. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 40.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weischenfeldt J, Porse B. Bone marrow-derived macrophages (BMM): isolation and applications. CSH Protoc 2008. 2010 doi: 10.1101/pdb.prot5080. [DOI] [PubMed] [Google Scholar]
- 42.Hafner M, et al. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corcoran DL, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattison JA, et al. Resveratrol prevents high fat/sucrose diet-induced central arterial wall inflammation and stiffening in nonhuman primates. Cell Metab. 2014;20:183–190. doi: 10.1016/j.cmet.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rayner KJ, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lillie RD, Fulmer HM. Histopathologic Technique and Practical Histochemistry. McGraw-Hill; 1976. [Google Scholar]
- 48.Ljungberg O, Tibblin S. Peroperative fat staining of frozen sections in primary hyperparathyroidism. Am J Pathol. 1979;95:633–641. [PMC free article] [PubMed] [Google Scholar]
- 49.Galabova G, et al. Peptide-based anti-PCSK9 vaccines - an approach for long-term LDLc management. PLoS ONE. 2014;9:e114469. doi: 10.1371/journal.pone.0114469. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.