Abstract
Background and Objectives:
This study was conducted to assess the efficacy of Mirasol pathogen reduction system for platelets aimed at preventing bacterial regrowth by spiking buffy coat pooled platelets (BCPP) with clinically relevant load of Staphylococous epidermidis.
Materials and Methods:
BCPP units were prepared using Teruflex BP-kit with Imugard III-S-PL (Terumo BCT, Tokyo, Japan). Two BCPP units were pooled, of which 40 ml of negative control (NC) was removed. The remaining volume of the platelet unit was inoculated with clinically relevant load of bacteria (total of 30 CFU of S. epidermidis in 1 ml); following this the platelet unit was split into two parts. One part served as positive control (PC) and the other part was subjected to pathogen reduction technique (Mirasol PRT, CaridianBCT Biotechnologies, Lakewood, CO, USA). Bacterial detection was performed using BacT/ALERT system, controls after day 1 and day 7 following inoculation of bacteria and on day 7 for Mirasol-treated unit.
Results:
Of the 32 treatment cycles, 28 were valid and 4 were invalid. No regrowth was observed in 96.4% (27 of 28) after treatment with Mirasol pathogen reduction system. Of four invalid tests, on two instances the NC showed growth, whereas in other 2 no regrowth was detected in 7th day PC. Bacterial screening of PCs by BacT/ALERT after 24 h of incubation was 28.6%, whereas the effectiveness increased to 100% when incubated for 7 days.
Conclusions:
Mirasol system was effective in inactivating S. epidermidis when it was deliberately inoculated into BCPP at clinically relevant concentrations. Such systems may significantly improve blood safety by inactivating traditional and emerging transfusion-transmitted pathogens.
Key words: Bacterial contamination, Mirasol, pathogen reduction system, platelets
Introduction
Platelets, derived either by apheresis or from whole blood, are stored at room temperature (20–24°C) to preserve their viability and functional status. Platelets suspended in human plasma, stored at room temperature, behave as an excellent media for bacterial growth. Although the quantity of bacteria present at the point of contamination is typically <100 CFU/product, this relatively small bacterial inoculum can grow to very high number within a short span of time.[1,2] Therefore, older units are more likely to be contaminated with a large number of organisms and can lead to septic transfusion reactions in the recipients.[3,4] The most common cause of bacterial contamination of platelets is the bacterial inoculation into the blood bags via skin plug at the time of phlebotomy. Other less common sources are asymptomatic donor bacteremia and contamination during component processing.[5,6]
The risk of bacterial contamination of platelets has been estimated to be 50–250 times higher than the combined risk of HIV, HBV, HCV, and HTLV-1/2.[7] Gram-positive skin commensals, viz., Staphylococcus epidermidis and Bacillus cereus are commonly cultured back from donated blood and implicated in bacterial contamination of platelets.[8] In the recent past improved disinfection of the venipuncture site, use of blood diversion pouches to collect 25–40 ml of the initial blood collected, along with the routine use of bacterial detection (BacT/ALERT), has decreased but not eliminated the risk of bacterial contamination. However, these interventions do not address the asymptomatic bacteremia present in the donors. The automated bacterial detection systems may miss the detection of such exceedingly low levels of bacteria in such donors. The presence of slow growing skin organisms such as S. epidermidis can be of particular concern as they may not grow to a high enough concentration at the time of bottle inoculation (approximately 24 h following collection) to be reliably detected using bacterial screening, which poses the potential risk of issuing false negative units for transfusion.[2,9,10,11,12]
Newer pathogen reduction technologies (PRTs) can reduce the levels of both bacterial and viral pathogens successfully, maintaining the adequate recovery and function of platelets.[13] Use of riboflavin and ultraviolet (UV)-light treatment of platelets and plasma (Mirasol PRT, Terumo BCT, Lakewood, CO, USA) was approved for commercial use in Europe at the end of 2007.[14] The Mirasol system is based on the ability of riboflavin (Vitamin B2) to function as a photosensitizer and selectively damage the nucleic acids of microbes upon exposure to UV light. Riboflavin is a natural component found in food (milk, beer, eggs, yeasts, leafy vegetables), and is classified as a “Generally-Recognized-As-Safe” compound by the US-Food and Drug Administration.[15] Extensive toxicological assays have shown that riboflavin and its metabolites are present in blood and are safe for use in transfusion; thus, removal of riboflavin is not required after treatment.[16]
This pilot study was undertaken for in vitro assessment of the ability and efficacy of Mirasol to inactivate clinically relevant concentrations of S. epidermidis in buffy coat pooled platelets (BCPP) suspended in “AB” group plasma and to compare it to the effectiveness of the standard BacT/ALERT microbial detection system.
Materials and Methods
A prospective single-blinded randomized pilot project was planned and conducted for the evaluation of bacterial inactivation using riboflavin and UV treatment was performed with ABO and RHD type matched BCPP units by All India Institute of Medical Sciences, New Delhi, in collaboration with National Institute of Biologicals, Noida, from June 2014 to July 2014 after clearance from the Institutional Ethics Committee [Figure 1].
Figure 1.
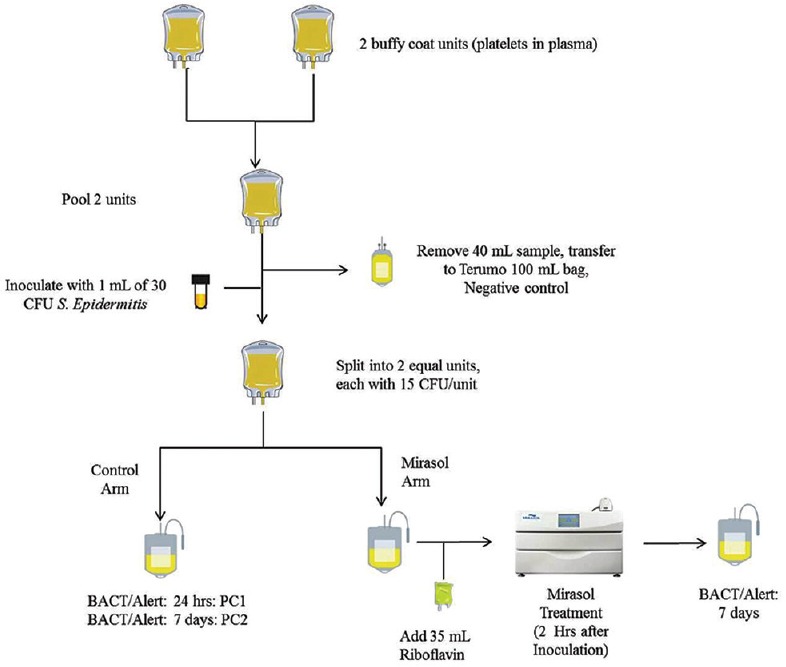
Schematic design for the Mirasol treatment
Aseptic precautions
During the entire study period, all sampling and inoculation procedures were performed using strict aseptic conditions under a biological safety cabinet (Type II A2, Thermo Scientific) and a sterile connecting device (TSCD, Terumo BCT, Lakewood, CO, USA) was used for all docking procedures.
Platelet preparation
BCPPs were prepared from 4 ABO and RHD type matched whole blood-derived buffy coat bags (collected in 450 ml quadruple top and bottom blood bags [Terumo Penpol, India]) using a dedicated set for component pooling and production (Terumo-Teruflex BP-kit with Imugard III-S-PL, Terumo BCT, Tokyo, Japan).
Bacteria propagation and storage conditions
S. epidermidis, ATCC #12228, (American Type Culture Collection, Manassas, VA, USA) was grown for 1–2 days in tryptic soya broth (OXOID, England) after which cultures were centrifuged, concentrated, and then suspended in peptone water (OXOID, England). Bacterial stock culture concentrations were determined through the use of a 10-fold end point dilution scheme plated out on tryptic soya agar (TSA) plates (Hi-Media, Mumbai, India; and Remel Inc., Lenexa, KS, USA). S. epidermidis cultures were thereafter stored at 4°C for up to 2 weeks until they were ready to be used. Before inoculation, the S. epidermidis stock culture was used to prepare a working culture of bacteria at 30 CFU/ml. Both the S. epidermidis stock cultures and working cultures were titrated on TSA plates so as to verify that the proper inoculum concentration was obtained.
General study design
Groups of two 48-h old BCPP units were combined using a sterile docking device. After pooling, 40 mL sample was transferred to a transfer bag and labeled as a negative control (NC). After 7 days of postincubation under standard platelet storage conditions, a 4 mL sample (NC) was taken and aseptically inoculated in an aerobic BacT/ALERT bottle. Inoculated bottles were then monitored for bacterial growth over a period of 7 days postsampling. Any NC units that were positive for bacterial growth resulted in all paired bacterial screening and riboflavin and UV light treated units to be removed from the final data analysis due to the initial product being contaminated with an unknown organism.
The remaining pooled product was inoculated with 1 ml of S. epidermidis at concentration of 30 CFU/ml. Following inoculation, the pooled platelet product was gravimetrically split into two equal-sized units. One of these paired units was tested after treatment with riboflavin and UV light, and the other half served as the positive control (PC). A total of 32 treatment cycles were performed during the study period.
Positive control
The platelet units marked as PC were incubated for 24 h at 22°C on a platelet agitator and incubator (Terumo Penpol, India). After 24 h of incubation, a 4 mL sample was withdrawn aseptically and inoculated into an aerobic BacT/ALERT bottle (PC1). After this sampling, the remaining platelet product was further incubated at 22°C for an additional 6 days. Thereafter on day 7, 4 ml of sample was removed and inoculated into an aerobic BacT/ALERT bottle to serve as the positive growth control (PC2). Both PC1 and PC2 were monitored for bacterial growth over a period of 7 days postsampling. Any negatives observed during this period also eliminated the paired bacterial screening and Mirasol-treated results from the final data analysis due to the failure of the organism to proliferate in the paired PC.
Riboflavin and ultraviolet-treated units
Each unit to be treated was incubated for a minimum of 2 h postinoculation and then treated according to the riboflavin and UV light method for platelets as has been previously described.[17] Following treatment, the unit was placed at 22°C under standard platelet storage conditions for 7 days. At the end of the 7-day period, a 4 mL sample was removed and inoculated in aerobic BacT/ALERT bottle and monitored for 7 days. Any positive culture observed during this period was considered a positive event.
Results
The BCPP unit prepared had a mean volume of 241.8 ± 29.8 ml, mean platelet yield of 2.27 ± 0.37 (×1011), mean pH of 7.0 ± 0.11, and a mean platelet concentration of 9.44 ± 1.57 (×109) per liter.
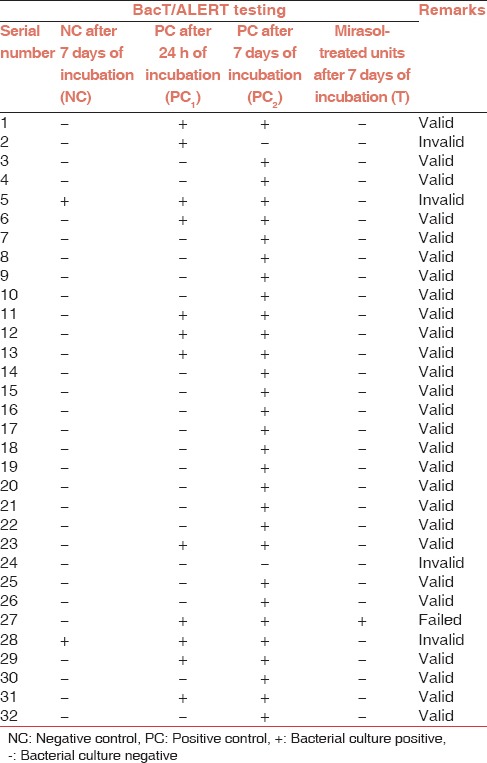
An overall summary of bacteria testing results is provided in Table 1. Of the 32 treatment cycles, 28 were valid and 4 were invalid. Of the four invalid tests, in 2 treatment cycles the NC BacT/ALERT bottle showed growth, whereas in the other 2 invalid tests no growth was detected in the PC samples at 7 days (PC2), and were thus excluded from the study. Mirasol treatment of the platelet units (T) demonstrated a 96.7% success rate at preventing bacterial growth in contaminated platelet units (1 out of 28 pooled platelet units showed growth). Bacterial screening of PCs by BacT/ALERT after 24 h incubation was 28.6%, whereas the effectiveness increased to 100% when incubated for 7 days.
Table 1.
Pathogen inactivation efficacy of Mirasol technology
Discussion
Bacterial contamination of blood components is the second most common cause of transfusion-associated mortality (posttransfusion errors).[18] The British Haemovigilance system Serious Hazards of Transfusions reports of 2011 states that since 1996, 40 bacterial transfusion transmitted infection incidents have been confirmed, involving a total of 43 recipients. Of the 40 reported incidents, 33 were related to the transfusion of platelets, whereas only 7 incidents were related to transfusion of red cells.[19] Similarly in the United States, the Bacon study also showed that infection risk from platelet transfusion is higher compared with that from red blood cells, and overall the risk of infection from bacterial contamination now may exceed that from viral agents.[20] Almost all the published studies show that the most common organisms isolated from contaminated platelet units are Gram-positive bacteria from the donor's skin.[21,22,23] Hemovigilance data from German blood establishments indicates that the most frequently identified pathogenic agents are S. epidermidis followed by S. aureus.[24]
This study evaluated the in vitro effectiveness of the Mirasol system for bacterial inactivation when whole blood-derived BCPP units were challenged with concentration of S. epidermidis (15 CFU/product) to mimic conditions seen in a routine clinical setting. The system showed no growth in 96.7% of challenged sample even after 7 days incubation in a platelet agitator at 22°C. In a similar study, Jocic et al. reported that complete inactivation of all bacterial species with CFU concentrations of 100 and 1000 per platelet product throughout storage or investigation period.[25] Goodrich et al. spiked 20–100 CFU per platelet product and demonstrated that riboflavin and UV light treatment was 91% effective against broad spectrum of bacteria. They reported that the Mirasol system may provide up to 98% protection of bacterially contaminated units at the most of clinically relevant contamination levels (20 CFUs/product).[26] In our study, one Mirasol treated unit showed positive growth, the cause for which could not be ascertained as we could not subject the BacT/ALERT bottle or original unit for further culture and identification. The possibility of bacterial re-growth following Mirasol treatment might be attributed to the breach in aseptic technique while sampling or during the inoculation of the BacT/ALERT bottle. Although, two of our tests that were invalid due to the presence of bacterial growth in NCs, after 7 days of incubation, no bacterial re-growth was detected after Mirasol treatment.
Culture studies performed on platelets early after collection may miss bacterial contaminated units that can achieve dangerous levels of overgrowth during the storage of blood components.[1] In this study, after a 24-h hold after inoculation, only 32.1% of samples were detected using the BacT/ALERT with a 4 ml inoculum. The low detection rate was probably due to the slow growth rate of the S. epidermidis. Since lag-phase of the growth of organisms may depend on the conditions used to culture microorganisms; slow-growing bacteria like S. epidermidis may grow in an idiosyncratic manner which may allow the slow-growing organism to escape such a detection scheme. Wagner et al. reported that average time to the first detection of S. epidermidis was 23.8 h, whereas a minimum time of 37 h was required for detection of all organisms in the experiment using the BacT/ALERT.[27] Increasing the hold time before sampling for bacterial screening can increase the successful detection rate of contaminated units. However, the limited shelf life of platelet concentrates would lead to less time available for platelets to be transfused and a high rate of expired platelet units in an already scarce inventory. Given the rate of 0.2–3.2% false positive alarms in 3 days BacT/ALERT incubation, another threat to inventory is an unnecessary discarding of precious platelet units.[28]
Conclusion
Use of bacterial screening of platelets in India is done mainly for quality control purposes as high cost and limited resources preclude the routine use of sophisticated, sensitive, and expensive technologies for the screening of blood products.[29,30] Again, the problem is further compounded in developing nations where hemovigilance systems are still in their infancy with no nation-wide reporting of transfusion-associated bacterial infections. This is a first prospective pilot study from India regarding pathogen inactivation in blood components. Considering high morbidity and mortality associated with bacterial infections compounded with the high cost of treatment, more studies from India are required to evaluate other viral and bacterial strains as well as to check the cost-effectiveness of bacterial inactivation in the developing countries.
Limitations
This study was limited by the fact that we relied on the NC and assumed that the growth seen in PC was due to the spiked S. epidermidis only. Furthermore, we were not able to test the quantitative reduction of bacterial load in terms of “log reduction.” Assessment of the quality parameters and function of the treated platelets were beyond the scope of the current pilot study.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
The authors would like to thank Mr. Anupam J. Lall and Mr. Mohammed Zafar who assisted in execution of the study.
References
- 1.Brecher ME, Holland PV, Pineda AA, Tegtmeier GE, Yomtovian R. Growth of bacteria in inoculated platelets: Implications for bacteria detection and the extension of platelet storage. Transfusion. 2000;40:1308–12. doi: 10.1046/j.1537-2995.2000.40111308.x. [DOI] [PubMed] [Google Scholar]
- 2.Pearce S, Rowe GP, Field SP. Screening of platelets for bacterial contamination at the Welsh Blood Service. Transfus Med. 2011;21:25–32. doi: 10.1111/j.1365-3148.2010.01037.x. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz DH, Young VM, Friedman NR, Reilly JA, Mardiney MR., Jr Detection and quantitation of bacteria in platelet products stored at ambient temperature. Transfusion. 1973;13:268–75. doi: 10.1111/j.1537-2995.1973.tb05488.x. [DOI] [PubMed] [Google Scholar]
- 4.Dodd RY. Bacterial contamination and transfusion safety: Experience in the United States. Transfus Clin Biol. 2003;10:6–9. doi: 10.1016/s1246-7820(02)00277-x. [DOI] [PubMed] [Google Scholar]
- 5.Anderson KC, Lew MA, Gorgone BC, Martel J, Leamy CB, Sullivan B. Transfusion-related sepsis after prolonged platelet storage. Am J Med. 1986;81:405–11. doi: 10.1016/0002-9343(86)90290-1. [DOI] [PubMed] [Google Scholar]
- 6.Puckett A. Bacterial contamination of blood for transfusion: A study of the growth characteristics of four implicated organisms. Med Lab Sci. 1986;43:252–7. [PubMed] [Google Scholar]
- 7.Ribault S, Harper K, Grave L, Lafontaine C, Nannini P, Raimondo A, et al. Rapid screening method for detection of bacteria in platelet concentrates. J Clin Microbiol. 2004;42:1903–8. doi: 10.1128/JCM.42.5.1903-1908.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brecher ME, Hay SN. Bacterial contamination of blood components. Clin Microbiol Rev. 2005;18:195–204. doi: 10.1128/CMR.18.1.195-204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumont LJ, Kleinman S, Murphy JR, Lippincott R, Schuyler R, Houghton J, et al. Screening of single-donor apheresis platelets for bacterial contamination: The passport study results. Transfusion. 2010;50:589–99. doi: 10.1111/j.1537-2995.2009.02460.x. [DOI] [PubMed] [Google Scholar]
- 10.Larsen CP, Ezligini F, Hermansen NO, Kjeldsen-Kragh J. Six years' experience of using the BacT/ALERT system to screen all platelet concentrates, and additional testing of outdated platelet concentrates to estimate the frequency of false-negative results. Vox Sang. 2005;88:93–7. doi: 10.1111/j.1423-0410.2005.00596.x. [DOI] [PubMed] [Google Scholar]
- 11.Murphy WG, Foley M, Doherty C, Tierney G, Kinsella A, Salami A, et al. Screening platelet concentrates for bacterial contamination: Low numbers of bacteria and slow growth in contaminated units mandate an alternative approach to product safety. Vox Sang. 2008;95:13–9. doi: 10.1111/j.1423-0410.2008.01051.x. [DOI] [PubMed] [Google Scholar]
- 12.Walther-Wenke G, Wirsing von König CH, Däubener W, Heiden M, Hoch J, Hornei B, et al. Monitoring bacterial contamination of blood components in Germany: Effect of contamination reduction measures. Vox Sang. 2011;100:359–66. doi: 10.1111/j.1423-0410.2010.01432.x. [DOI] [PubMed] [Google Scholar]
- 13.Seltsam A, Müller TH. Update on the use of pathogen-reduced human plasma and platelet concentrates. Br J Haematol. 2013;162:442–54. doi: 10.1111/bjh.12403. [DOI] [PubMed] [Google Scholar]
- 14.Goodrich RP, Edrich RA, Li J, Seghatchian J. The Mirasol PRT system for pathogen reduction of platelets and plasma: An overview of current status and future trends. Transfus Apher Sci. 2006;35:5–17. doi: 10.1016/j.transci.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 15.CFR. Code of Federal Regulations, Title 21, Food and Drug Administration. U.S.: FDA; 2014. [Google Scholar]
- 16.Hardwick CC, Herivel TR, Hernandez SC, Ruane PH, Goodrich RP. Separation, identification and quantification of riboflavin and its photoproducts in blood products using high-performance liquid chromatography with fluorescence detection: A method to support pathogen reduction technology. Photochem Photobiol. 2004;80:609–15. doi: 10.1562/2004-04-14-TSN-139. [DOI] [PubMed] [Google Scholar]
- 17.Ruane PH, Edrich R, Gampp D, Keil SD, Leonard RL, Goodrich RP. Photochemical inactivation of selected viruses and bacteria in platelet concentrates using riboflavin and light. Transfusion. 2004;44:877–85. doi: 10.1111/j.1537-2995.2004.03355.x. [DOI] [PubMed] [Google Scholar]
- 18.Hillyer CD, Josephson CD, Blajchman MA, Vostal JG, Epstein JS, Goodman JL. Bacterial contamination of blood components: Risks, strategies, and regulation: Joint ASH and AABB educational session in transfusion medicine. Hematology Am Soc Hematol Educ Program. 2003:575–89. doi: 10.1182/asheducation-2003.1.575. [DOI] [PubMed] [Google Scholar]
- 19.Bolton-Maggs P, Cohen H, Watt A, Poles D. SHOT Annual Report 2011. 2012. [last accessed on 2015 April 15]. Available from: http://www.shotuk.org/wp-content/uploads/SHOT-ANNUL-REPowpsANNRT_FinalWebVersionBookmarked_2012_06_22.pdf .
- 20.Kuehnert MJ, Roth VR, Haley NR, Gregory KR, Elder KV, Schreiber GB, et al. Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion. 2001;41:1493–9. doi: 10.1046/j.1537-2995.2001.41121493.x. [DOI] [PubMed] [Google Scholar]
- 21.Blajchman MA. Incidence and significance of the bacterial contamination of blood components. Dev Biol (Basel) 2002;108:59–67. [PubMed] [Google Scholar]
- 22.Ness P, Braine H, King K, Barrasso C, Kickler T, Fuller A, et al. Single-donor platelets reduce the risk of septic platelet transfusion reactions. Transfusion. 2001;41:857–61. doi: 10.1046/j.1537-2995.2001.41070857.x. [DOI] [PubMed] [Google Scholar]
- 23.Engelfriet CP, Reesink HW, Blajchman MA, Muylle L, Kjeldsen-Kragh J, Kekomäki R, et al. Bacterial contamination of blood components. Vox Sang. 2000;78:59–67. doi: 10.1159/000031151. [DOI] [PubMed] [Google Scholar]
- 24.Funk MB, Lohmann A, Guenay S, Henseler O, Heiden M, Hanschmann KM, et al. Transfusion-Transmitted Bacterial Infections – Haemovigilance Data of German Blood Establishments (1997-2010) Transfus Med Hemother. 2011;38:266–271. doi: 10.1159/000330372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jocic M, Trkuljic M, Jovicic D, Borovcanin N, Todorovic M, Balint B. Mirasol PRT system inactivation efficacy evaluated in platelet concentrates by bacteria-contamination model. Vojnosanit Pregl. 2011;68:1041–6. [PubMed] [Google Scholar]
- 26.Goodrich RP, Gilmour D, Hovenga N, Keil SD. A laboratory comparison of pathogen reduction technology treatment and culture of platelet products for addressing bacterial contamination concerns. Transfusion. 2009;49:1205–16. doi: 10.1111/j.1537-2995.2009.02126.x. [DOI] [PubMed] [Google Scholar]
- 27.Wagner SJ, Friedman LI, Dodd RY. Transfusion-associated bacterial sepsis. Clin Microbiol Rev. 1994;7:290–302. doi: 10.1128/cmr.7.3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hundhausen T, Müller TH. GERMS Group. False-positive alarms for bacterial screening of platelet concentrates with BacT/ALERT new-generation plastic bottles: A multicenter pilot study. Transfusion. 2005;45:1267–74. doi: 10.1111/j.1537-2995.2005.00194.x. [DOI] [PubMed] [Google Scholar]
- 29.Saran RK. Transfusion Medicine: Technical Manual. 2nd ed. India, New Delhi: Directorate General of Health Services, Ministry of Health and Family Welfare, Govt. of India; 2003. [Google Scholar]
- 30.Marwaha N, Singh S, Bisht A. Setting up haemovigilance from the very first step. The Indian perspective. ISBT Sci Ser. 2014;9:178–83. [Google Scholar]


