Abstract
Immune checkpoint inhibitors, which unleash a patient’s own T cells to kill tumors, are revolutionizing cancer treatment. To unravel the genomic determinants of response to this therapy, we used whole-exome sequencing of non–small cell lung cancers treated with pembrolizumab, an antibody targeting programmed cell death-1 (PD-1). In two independent cohorts, higher nonsynonymous mutation burden in tumors was associated with improved objective response, durable clinical benefit, and progression-free survival. Efficacy also correlated with the molecular smoking signature, higher neoantigen burden, and DNA repair pathway mutations; each factor was also associated with mutation burden. In one responder, neoantigen-specific CD8+ T cell responses paralleled tumor regression, suggesting that anti–PD-1 therapy enhances neoantigen-specific T cell reactivity. Our results suggest that the genomic landscape of lung cancers shapes response to anti–PD-1 therapy.
Today, more than a century since the initial observation that the immune system can reject human cancers (1), immune checkpoint inhibitors are demonstrating that adaptive immunity can be harnessed for the treatment of cancer (2–7). In advanced non–small cell lung cancer (NSCLC), therapies with an antibody targeting programmed cell death-1 (anti–PD-1) demonstrated response rates of 17 to 21%, with some responses being remarkably durable (3, 8).
Understanding the molecular determinants of response to immunotherapies such as anti–PD-1 therapy is one of the critical challenges in oncology. Among the best responses have been in melanomas and NSCLCs, cancers largely caused by chronic exposure to mutagens [ultraviolet light (9) and carcinogens in cigarette smoke (10), respectively]. However, there is a large variability in mutation burden within tumor types, ranging from10s to 1000s of mutations (11–13). This range is particularly broad in NSCLCs because tumors in never-smokers generally have few somatic mutations compared with tumors in smokers (14). We hypothesized that the mutational landscape of NSCLCs may influence response to anti–PD-1 therapy. To examine this hypothesis, we sequenced the exomes of NSCLCs from two independent cohorts of patients treated with pembrolizumab, a humanized immunoglobulin G (IgG) 4-kappa isotype antibody to PD-1 (n = 16 and n = 18, respectively), and their matched normal DNA (fig. S1 and table S1) (15).
Overall, tumor DNA sequencing generated mean target coverage of 164×, and a mean of 94.5% of the target sequence was covered to a depth of at least 10×; coverage and depth were similar between cohorts, as well as between those with or without clinical benefit (fig. S2). We identified a median of 200 nonsynonymous mutations per sample (range 11 to 1192). The median number of exonic mutations per sample was 327 (range 45 to 1732). The quantity and range of mutations were similar to published series of NSCLCs (16, 17) (fig. S3). The transition/transversion ratio (Ti/Tv) was 0.74 (fig. S4), also similar to previously described NSCLCs (16–18). To ensure accuracy of our sequencing data, targeted resequencing with an orthogonal method (Ampliseq) was performed using 376 randomly selected variants, and mutations were confirmed in 357 of those variants (95%).
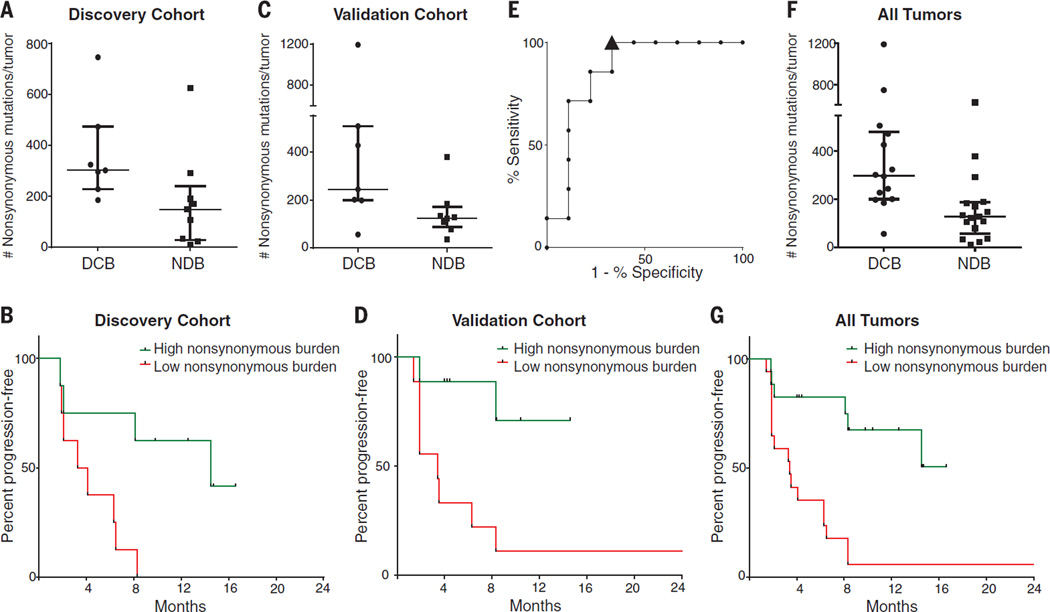
Higher somatic nonsynonymous mutation burden was associated with clinical efficacy of pembrolizumab. In the discovery cohort (n = 16), the median number of nonsynonymous mutations was 302 in patients with durable clinical benefit (DCB) (partial or stable response lasting >6 months) versus 148 with no durable benefit (NDB) (Mann-Whitney P = 0.02) (Fig. 1A). Seventy-three percent of patients with high nonsynonymous burden (defined as above the median burden of the cohort, 209) experienced DCB, compared with 13% of those with low mutation burden (below median) (Fisher’s exact P = 0.04). Both confirmed objective response rate (ORR) and progression-free survival (PFS) were higher in patients with high nonsynonymous burden [ORR 63% versus 0%, Fisher’s exact P = 0.03; median PFS 14.5 versus 3.7 months, log-rank P = 0.01; hazard ratio (HR) 0.19, 95% confidence interval (CI) 0.05 to 0.70] (Fig. 1B and table S2).
Fig. 1. Nonsynonymous mutation burden associated with clinical benefit of anti–PD-1 therapy.
(A) Nonsynonymous mutation burden in tumors from patients with DCB (n = 7) or with NDB (n = 9) (median 302 versus 148, Mann-Whitney P = 0.02). (B) PFS in tumors with higher nonsynonymous mutation burden (n = 8) compared to tumors with lower nonsynonymous mutation burden (n = 8) in patients in the discovery cohort (HR 0.19, 95% CI 0.05 to 0.70, log-rank P = 0.01). (C) Nonsynonymous mutation burden in tumors with DCB (n = 7) compared to those with NDB (n = 8) in patients in the validation cohort (median 244 versus 125, Mann-Whitney P = 0.04). (D) PFS in tumors with higher nonsynonymous mutation burden (n = 9) compared to those with lower nonsynonymous mutation burden (n = 9) in patients in the validation cohort (HR 0.15, 95% CI 0.04 to 0.59, log-rank P = 0.006). (E) ROC curve for the correlation of nonsynonymous mutation burden with DCB in discovery cohort. AUC is 0.86 (95% CI 0.66 to 1.05, null hypothesis test P = 0.02). Cut-off of ≥178 nonsynonymous mutations is designated by triangle. (F) Nonsynonymous mutation burden in patients with DCB (n = 14) compared to those with NDB (n = 17) for the entire set of sequenced tumors (median 299 versus 127, Mann-Whitney P = 0.0008). (G) PFS in those with higher nonsynonymous mutation burden (n = 17) compared to those with lower nonsynonymous mutation burden (n = 17) in the entire set of sequenced tumors (HR 0.19, 95% CI 0.08–0.47, log-rank P = 0.0004). In (A), (C), and (F), median and interquartile ranges of total nonsynonymous mutations are shown, with individual values for each tumor shown with dots.
The validation cohort included an independent set of 18 NSCLC samples from patients treated with pembrolizumab. The clinical characteristics were similar in both cohorts. The median nonsynonymous mutation burden was 244 in tumors from patients with DCB compared to 125 in those with NDB (Mann-Whitney P = 0.04) (Fig. 1C). The rates of DCB and PFS were again significantly greater in patients with a nonsynonymous mutation burden above 200, the median of the validation cohort (DCB 83% versus 22%, Fisher’s exact P = 0.04; median PFS not reached versus 3.4 months, log-rank P = 0.006; HR 0.15, 95% CI 0.04 to 0.59) (Fig. 1D and table S2).
In the discovery cohort, there was high concordance between nonsynonymous mutation burden and DCB, with an area under the receiver operator characteristic (ROC) curve (AUC) of 87% (Fig. 1E). Patients with nonsynonymous mutation burden ≥178, the cut point that combined maximal sensitivity with best specificity, had a likelihood ratio for DCB of 3.0; the sensitivity and specificity of DCB using this cut point was 100% (95% CI 59 to 100%) and 67% (29 to 93%), respectively. Applying this cut point to the validation cohort, the rate of DCB in patients with tumors harboring ≥178 mutations was 75% compared to 14% in those with <178, corresponding to a sensitivity of 86% and a specificity of 75%.
There were few but important exceptions. Five of 18 tumors with ≥178 nonsynonymous mutations had NDB, and one tumor with a very low burden (56 nonsynonymous mutations) responded to pembrolizumab. However, this response was transient, lasting 8 months. Across both cohorts, this was the only patient with a tumor mutation burden <178 and confirmed objective response. Notably, although higher nonsynonymous mutation burden correlated with improved ORR, DCB, and PFS (Fig. 1, F and G), this correlation was less evident when examining total exonic mutation burden (table S2).
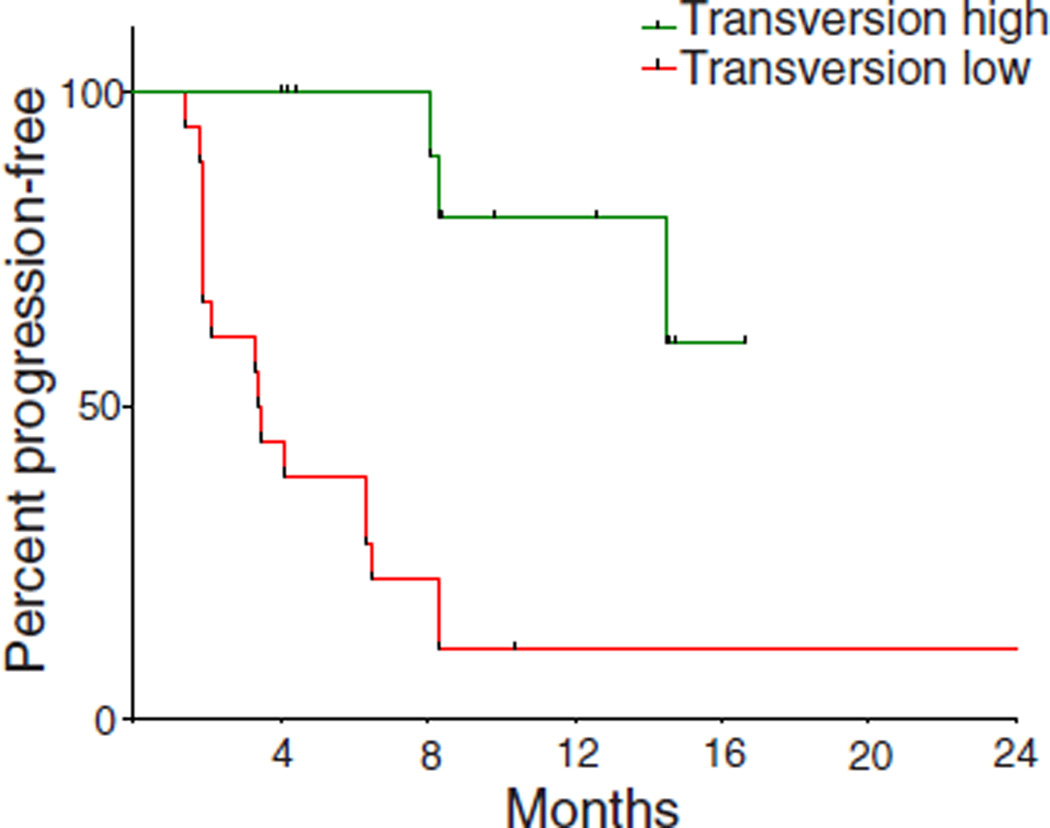
We next examined all 34 exomes collectively to determine how patterns of mutational changes were associated with clinical benefit to pembrolizumab (tables S4 and S5). C-to-A transversions were more frequent, and C-to-T transitions were less frequent, in patients with DCB compared to NDB (Mann-Whitney P = 0.01 for both) (fig. S5). A previously validated binary classifier to identify the molecular signature of smoking (17) was applied to differentiate transversion-high (TH, smoking signature) from transversion-low (TL, never-smoking signature) tumors. Efficacy was greatest in patients with tumors harboring the smoking signature. The ORR in TH tumors was 56% versus 17% in TL tumors (Fisher’s exact P = 0.03); the rate of DCB was 77% versus 22% (Fisher’s exact P = 0.004); the PFS was also significantly longer in TH tumors (median not reached versus 3.5 months, log-rank P = 0.0001) (Fig. 2A). Self-reported smoking history did not significantly discriminate those most likely to benefit from pembrolizumab. The rates of neither DCB nor PFS were significantly different in ever-smokers versus never-smokers (Fisher’s exact P = 0.66 and log-rank P = 0.29, respectively) or heavy smokers (median pack-years >25) versus light/never smokers (pack-years ≤25) (Fisher’s exact P = 0.08 and log-rank P = 0.15, respectively). The molecular smoking signature correlated more significantly with nonsynonymous mutation burden than smoking history (fig. S6, A and B).
Fig. 2. Molecular smoking signature is significantly associated with improved PFS in NSCLC patients treated with pembrolizumab.
PFS in tumors characterized as TH by molecular smoking signature classifier (n = 16) compared to TL tumors (n = 18) (HR 0.15, 95% 0.06 to 0.39, log-rank P = 0.0001).
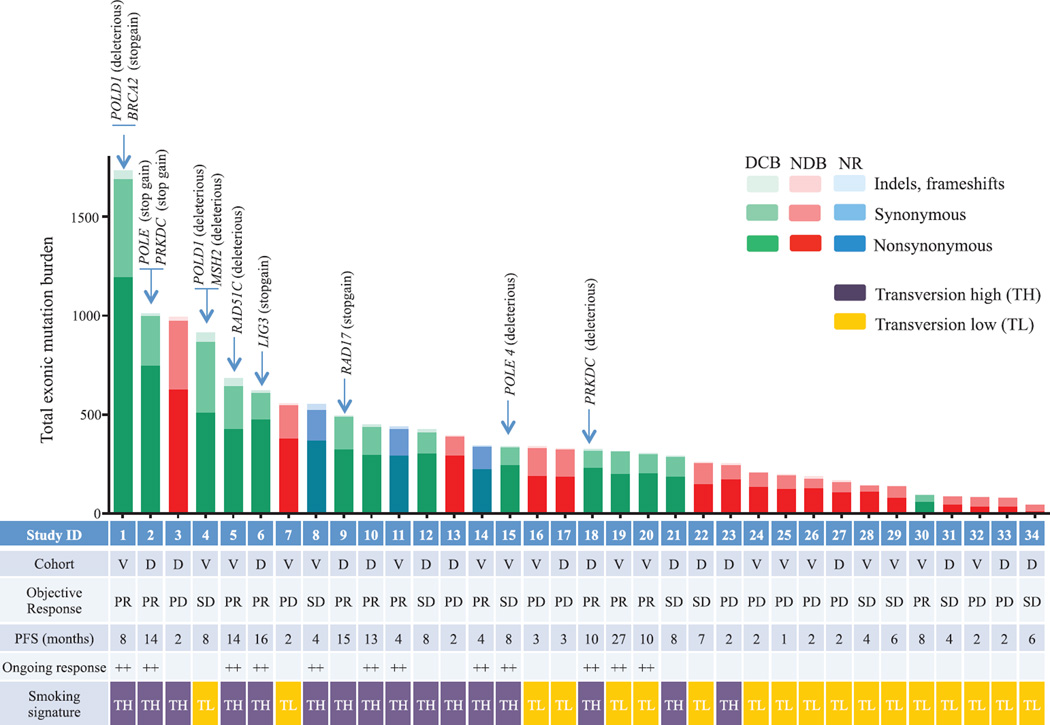
Although carcinogens in tobacco smoke are largely responsible for the mutagenesis in lung cancers (19), the wide range of mutation burden within both smokers and never-smokers implicates additional pathways contributing to the accumulation of somatic mutations. We found deleterious mutations in a number of genes that are important in DNA repair and replication. For example, in three responders with the highest mutation burden, we identified deleterious mutations in POLD1, POLE, and MSH2 (Fig. 3). Of particular interest, a POLD1 E374K mutation was identified in a never-smoker with DCB whose tumor harbored the greatest nonsynonymous mutation burden (n = 507) of all never-smokers in our series. POLD1 Glu374 lies in the exonuclease proofreading domain of Pol δ (20), and mutation of this residue may contribute to low-fidelity replication of the lagging DNA strand. Consistent with this hypothesis, this tumor exome had a relatively low proportion of C-to-A transversions (20%) and predominance of C-to-T transitions (51%), similar to other POLD1 mutant, hypermutated tumors (21) and distinct from smoking-related lung cancers. Another responder, with the greatest mutation burden in our series, had a C284Y mutation in POLD1, which is also located in the exonuclease proofreading domain. We observed nonsense mutations in PRKDC, the catalytic subunit of DNA-dependent protein kinase (DNA-PK), and RAD17. Both genes are required for proper DNA repair and maintenance of genomic integrity (22, 23).
Fig. 3. Mutation burden, clinical response, and factors contributing to mutation burden.
Total exonic mutation burden for each sequenced tumor with nonsynonymous (dark shading), synonymous (medium shading), and indels/frameshift mutations (light shading) displayed in the histogram. Columns are shaded to indicate clinical benefit status: DCB, green; NDB, red; not reached 6 months follow-up (NR), blue. The cohort identification (D, discovery; V, validation), best objective response (PR, partial response; SD, stable disease; PD, progression of disease), and PFS (censored at the time of data lock) are reported in the table. Those with ongoing progression-free survival are labeled with ++. The presence of the molecular smoking signature is displayed in the table with TH cases (purple) and TL cases (orange). The presence of deleterious mutations in specific DNA repair/replication genes is indicated by the arrows.
Genes harboring deleterious mutations common to four or more DCB patients and not present in NDB patients included POLR2A, KEAP1, PAPPA2, PXDNL, RYR1, SCN8A, and SLIT3. Mutations in KRAS were found in 7 of 14 tumors from patients with DCB compared to 1 of 17 in the NDB group, a finding that may be explained by the association between smoking and the presence of KRAS mutations in NSCLC (24). There were no mutations or copy-number alterations in antigen-presentation pathway–associated genes or CD274 [encoding programmed cell death ligand-1 (PD-L1)] that were associated with response or resistance.
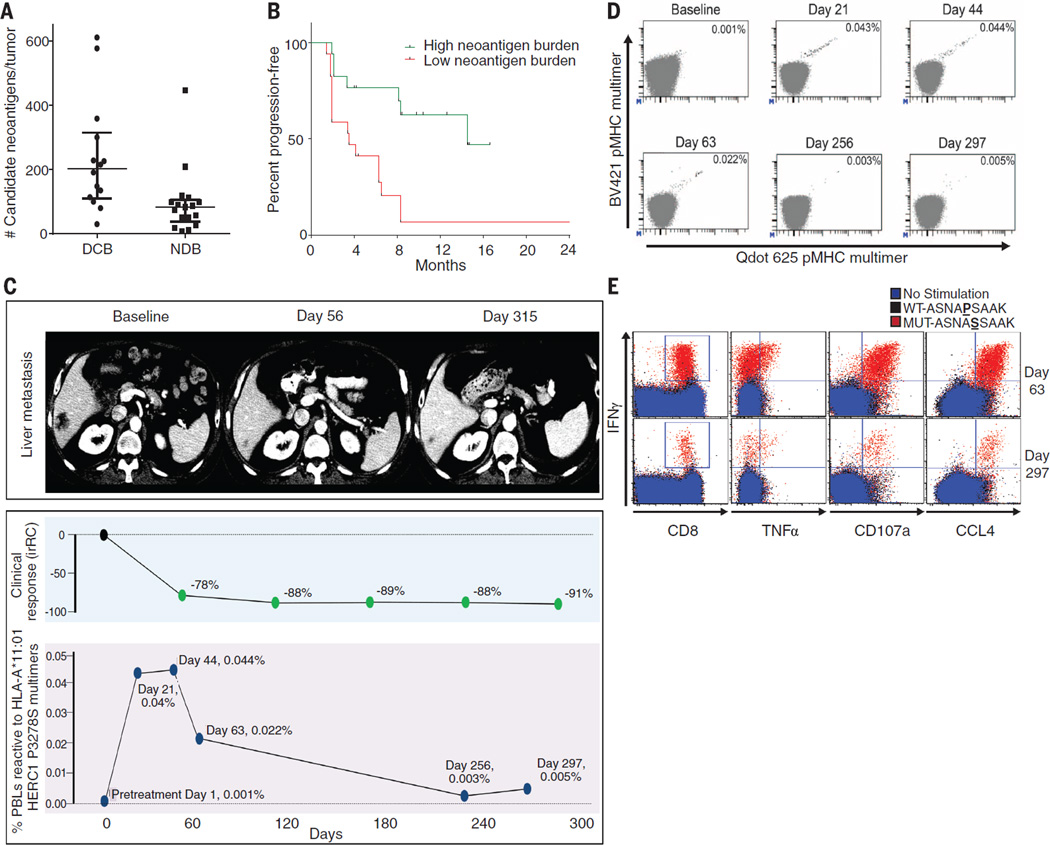
How does increased mutation burden affect tumor immunogenicity? The observation that nonsynonymous mutation burden is associated with pembrolizumab efficacy is consistent with the hypothesis that recognition of neoantigens, formed as a consequence of somatic mutations, is important for the activity of anti–PD-1 therapy. We examined the landscape of neoantigens using our previously described methods (25) (fig. S7). Briefly, this approach identifies mutant nonamers with ≤500 nM binding affinity for patient-specific class I human lymphocyte antigen (HLA) alleles (26, 27), which are considered candidate neoantigens (table S6). We identified a median of 112 candidate neoantigens per tumor (range 8 to 610), and the quantity of neoantigens per tumor correlated with mutation burden (Spearman ρ 0.91, P < 0.0001), similar to the correlation recently reported across cancers (28). Tumors from patients with DCB had significantly higher candidate neoantigen burden compared to those with NDB (Fig. 4A), and high candidate neoantigen burden was associated with improved PFS (median 14.5 versus 3.5 months, log-rank P = 0.002) (Fig. 4B). The presence of specific HLA alleles did not correlate with efficacy (fig. S8). The absolute burden of candidate neoantigens, but not the frequency per nonsynonymous mutation, correlated with response (fig. S9).
Fig. 4. Candidate neoantigens, neoantigen-specific T cell response, and response to pembrolizumab.
(A) Neoantigen burden in patients with DCB (n = 14) compared to NDB (n = 17) across the overall set of sequenced tumors (median 203 versus 83, Mann-Whitney P = 0.001). (B) PFS in tumors with higher candidate neoantigen burden (n = 17) compared to tumors with lower candidate neoantigen burden (n = 17) (HR 0.23, 95%CI 0.09 to 0.58, log-rank P = 0.002). (C) (Top) Representative computed tomography (CT) images of a liver metastasis before and after initiation of treatment. (Middle) Change in radiographic response. (Bottom) Magnitude of the HERC1 P3278S reactive CD8+ T cell response measured in peripheral blood. (D) The proportion of CD8+ T cell population in serially collected autologous PBLs recognizing the HERC1 P3278S neoantigen (ASNASSAAK) before and during pembrolizumab treatment. Each neoantigen is encoded by a unique combination of two fluorescently labeled peptide-MHC complexes (represented individually on each axis); neoantigen-specific T cells are represented by the events in the double positive position indicated with black dots. Percentages indicate the number of CD8+ MHC multimer+ cells out of total CD8 cells. (E) Autologous T cell response to wild-type HERC1 peptide (black), mutant HERC1 P3278S neoantigen (red), or no stimulation (blue), as detected by intracellular cytokine staining. T cell costains for IFNγ and CD8, TNFα, CD107a, and CCL4, respectively, are displayed for the Day 63 and Day 297 time points.
We next sought to assess whether anti–PD-1 therapy can alter neoantigen-specific T cell reactivity. To directly test this, identified candidate neoantigens were examined in a patient (Study ID no. 9 in Fig. 3 and table S3) with exceptional response to pembrolizumab and available peripheral blood lymphocytes (PBLs). Predicted HLA-A–restricted peptides were synthesized to screen for ex vivo autologous T cell reactivity in serially collected PBLs (days 0, 21, 44, 63, 256, and 297, where day 0 is the first date of treatment) using a validated high-throughput major histocompatibility complex (MHC) multimer screening strategy (29, 30). This analysis revealed a CD8+ T cell response against a neoantigen resulting from a HERC1 P3278S mutation (ASNASSAAK) (Fig. 4C). Notably, this T cell response could only be detected upon the start of therapy (level of detection 0.005%). Three weeks after therapy initiation, the magnitude of response was 0.040% of CD8+ T cells, and this response was maintained at Day 44. This rapid induction of T cell reactivity correlated with tumor regression, and this T cell response returned to levels just above background in the subsequent months as tumor regression plateaued (Fig. 4D). HERC1 P3278S-multimer–reactive T cells from PBLs collected on day 44 were characterized by a CD45RA-CCR7-HLA-DR+LAG-3 phenotype, consistent with an activated effector population (fig. S10). These data reveal autologous T cell responses against cancer neoantigens in the context of a clinical response to anti–PD-1 therapy.
To validate the specificity of the neoantigen-reactive T cells, PBLs from days 63 and 297 were expanded in vitro in the presence of mutant peptide and subsequently restimulated with either mutant or wild-type peptide (ASNASSAAK versus ASNAPSAAK), and intracellular cytokines were analyzed. At both time points, a substantial population of polyfunctional CD8+ T cells [characterized by production of the cytokines interferon (IFN) γ and tumor necrosis factor (TNF) α, the marker of cytotoxic activity CD107a, and the chemokine CCL4] was detected in response to mutant but not wild-type peptide (Fig. 4E and fig. S11).
In the current study, we show that in NSCLCs treated with pembrolizumab, elevated nonsynonymous mutation burden strongly associates with clinical efficacy. Additionally, clinical efficacy correlates with a molecular signature characteristic of tobacco carcinogen–related mutagenesis, certain DNA repair mutations, and the burden of neoantigens. The molecular smoking signature correlated with efficacy, whereas self-reported smoking status did not, highlighting the power of this classifier to identify molecularly related tumors within a heterogeneous group.
Previous studies have reported that pretreatment PD-L1 expression enriches for response to anti–PD-1 therapies (3, 8, 31), but many tumors deemed PD-L1 positive do not respond, and some responses occur in PD-L1–negative tumors (8, 31). Semiquantitative PD-L1 staining results were available for 30 of 34 patients, where strong staining represented ≥50% PD-L1 expression, weak represented 1 to 49%, and negative represented <1% [clone 22C3, Merck (8)]. As this trial largely enrolled patients with PD-L1 tumor expression, most samples had some degree of PD-L1 expression (24 of 30, 80%) (table S3), limiting the capacity to determine relationships between mutation burden and PD-L1 expression. Among those with high nonsynonymous mutation burden (>200, above median of overall cohort) and some degree of PD-L1 expression (weak/strong), the rate of DCB was 91% (10 of 11, 95% CI 59 to 99%). In contrast, in those with low mutation burden and some degree of PD-L1 expression, the rate of DCB was only 10% (1 of 10, 95% CI 0 to 44%). When exclusively examining patients with weak PD-L1 expression, high nonsynonymous mutation burden was associated with DCB in 75% (3 of 4, 95% CI 19 to 99%), and low mutation burden was associated with DCB in 11% (1 of 9, 0 to 48%). Large-scale studies are needed to determine the relationship between PD-L1 intensity and mutation burden. Additionally, recent data have demonstrated that the localization of PD-L1 expression within the tumor microenvironment [on infiltrating immune cells (32), at the invasive margin, tumor core, and so forth (33)] may affect the use of PD-L1 as a biomarker.
T cell recognition of cancers relies upon presentation of tumor-specific antigens on MHC molecules (34). A few preclinical (35–41) and clinical reports have demonstrated that neoantigen-specific effector T cell response can recognize (25, 42–45) and shrink established tumors (46). Our finding that nonsynonymous mutation burden more closely associates with pembrolizumab clinical benefit than total exonic mutation burden suggests the importance of neoantigens in dictating response.
The observation that anti–PD-1–induced neoantigen-specific T cell reactivity can be observed within the peripheral blood compartment may open the door to development of blood-based assays to monitor response during anti–PD-1 therapy. We believe that our findings have an important impact on our understanding of response to anti–PD-1 therapy and on the application of these agents in the clinic.
Supplementary Material
Acknowledgments
We thank the members of the Thoracic Oncology Service and the Chan and Wolchok laboratories at Memorial Sloan Kettering Cancer Center (MSKCC) for helpful discussions. We thank the Immune Monitoring Core at MSKCC, including L. Caro, R. Ramsawak, and Z. Mu, for exceptional support with processing and banking peripheral blood lymphocytes. We thank P. Worrell and E. Brzostowski for help in identifying tumor specimens for analysis. We thank A. Viale for superb technical assistance. We thank D. Philips, M. van Buuren, and M. Toebes for help performing the combinatorial coding screens. This work was supported by the Geoffrey Beene Cancer Research Center (M.D.H., N.A.R., T.A.C., J.D.W., and A.S.), the Society for Memorial Sloan Kettering Cancer Center (M.D.H.), Lung Cancer Research Foundation (W.L.), Frederick Adler Chair Fund (T.A.C.), The One Ball Matt Memorial Golf Tournament (E.B.G.), Queen Wilhelmina Cancer Research Award (T.N.S.), The STARR Foundation (T.A.C. and J.D.W.), the Ludwig Trust (J.D.W.), and a Stand Up To Cancer-Cancer Research Institute Cancer Immunology Translational Cancer Research Grant (J.D.W., T.N.S., and T.A.C.). Stand Up To Cancer is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research.
Footnotes
The data presented in this paper are tabulated in the main paper and in the supplementary materials.
Data are publicly available at the Cancer Genome Atlas (TCGA) cBio portal and database (www.cbioportal.org; study ID: Rizvi lung cancer). All genotype and phenotype data are deposited at dpGAP under accession no. phs000980.v1.p1. T.A.C. is the inventor on a patent (provisional application number 62/083,088). The application is directed toward methods for identifying patients who will benefit from treatment with immunotherapy.
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/348/6230/124/suppl/DC1
Materials and Methods
Figs. S1 to S12
Tables S1 to S6
References (47–68)
REFERENCES AND NOTES
- 1.Coley WB. Clin. Orthop. Relat. Res. 1991;1991(262):3–11. [PubMed] [Google Scholar]
- 2.Hodi FS, et al. N Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Topalian SL, et al. N Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolchok JD, et al. N Engl. J. Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert C, et al. Lancet. 2014;384:1109–1117. doi: 10.1016/S0140-6736(14)60958-2. [DOI] [PubMed] [Google Scholar]
- 6.Powles T, et al. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 7.Ansell SM, et al. N Engl. J. Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garon EB, et al. Ann. Oncol. 2014;25:LBA43. [Google Scholar]
- 9.Pfeifer GP, You YH, Besaratinia A. Mutat. Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 10.Pfeifer GP, et al. Oncogene. 2002;21:7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence MS, et al. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, et al. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein B, et al. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govindan R, et al. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.See supplementary text available on Science Online.
- 16.Hammerman PS, et al. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cancer Genome Atlas Research Network. Nature. 2014;511:543–550. [Google Scholar]
- 18.Abaan OD, et al. Cancer Res. 2013;73:4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann D, Hoffmann I, El-Bayoumy K. Chem. Res. Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- 20.Hindges R, Hübscher U. Biol. Chem. 1997;378:345–362. doi: 10.1515/bchm.1997.378.5.345. [DOI] [PubMed] [Google Scholar]
- 21.Palles C, et al. Nat. Genet. 2013;45:136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodwin JF, Knudsen KE. Cancer Discov. 2014;4:1126–1139. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, et al. Genes Dev. 2003;17:965–970. doi: 10.1101/gad.1065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dogan S, et al. Clin. Cancer Res. 2012;18:6169–6177. doi: 10.1158/1078-0432.CCR-11-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snyder A, et al. N Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen M, et al. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundegaard C, et al. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. (Web Server) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodenko B, et al. Nat. Protoc. 2006;1:1120–1132. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- 30.Andersen RS, et al. Nat. Protoc. 2012;7:891–902. doi: 10.1038/nprot.2012.037. [DOI] [PubMed] [Google Scholar]
- 31.Taube JM, et al. Clin. Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbst RS, et al. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tumeh PC, et al. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schreiber RD, Old LJ, Smyth MJ. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 35.Matsutake T, Srivastava PK. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3992–3997. doi: 10.1073/pnas.071523398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsushita H, et al. Nature. 2012;482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castle JC, et al. Cancer Res. 2012;72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 38.Schumacher T, et al. Nature. 2014;512:324–327. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 39.Gubin MM, et al. Nature. 2014;515:577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav M, et al. Nature. 2014;515:572–576. doi: 10.1038/nature14001. [DOI] [PubMed] [Google Scholar]
- 41.Duan F, et al. J Exp. Med. 2014;211:2231–2248. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Rooij N, et al. J Clin. Oncol. 2013;31:e439–e442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robbins PF, et al. Nat. Med. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rajasagi M, et al. Blood. 2014;124:453–462. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linnemann C, et al. Nat. Med. 2015;21:81–85. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 46.Tran E, et al. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.