Abstract
To transmit signals across cellular compartments, many membrane-embedded enzymes undergo extensive conformational rearrangements. Monitoring these events in lipid bilayers by NMR at atomic resolution has been challenging due to the large size of these systems. It is further exacerbated for large mammalian proteins that are difficult to express and label with NMR-active isotopes. Here, we synthesized and engineered 13C ethyl groups on native cysteines to map the structural transitions of the sarcoplasmic reticulum Ca2+-ATPase, a 110 kDa transmembrane enzyme that transports Ca2+ into the sarcoplasmic reticulum. Using magic angle spinning NMR, we monitored the chemical shifts of the methylene and methyl groups of the derivatized cysteine residues along the major steps of the enzymatic cycle. The methylene chemical shifts are sensitive to the ATPase conformational changes induced upon nucleotide and Ca2+ ion binding and are ideal probes for active and inactive states of the enzyme. This new approach is extendable to large mammalian enzymes and signaling proteins with native or engineered cysteine residues in their amino acid sequence.
Keywords: Site-directed ethylation, Solid-state NMR, conformational transitions, membrane proteins, Ca2+-ATPase
Graphical abstract
Introduction
Cell signaling events are mediated by membrane-bound enzymes and proteins that transmit their messages across lipid membranes through chemical, structural and/or dynamical changes. Since membrane proteins are difficult to crystallize in lipid membranes, solid-state NMR (ssNMR) spectroscopy is the ideal high-resolution spectroscopic technique to monitor structural changes in liquid crystalline lipid membranes1–3. When the proteins or protein complexes exceed 100 kDa, the analysis of structural transitions becomes cumbersome as the number of structural probes (resonances) crowds the NMR spectra. For solution NMR spectroscopy, Kay and co-workers introduced recombinant protein expression protocols that enable amino acid specific 13C methyl labeling4, 5. The higher gyromagnetic ratio of 13C with respect to 15N in concert with longer relaxation times of the methyl groups makes it possible to image the methyl fingerprint region of larger proteins4, 6. For example, such labeling technique was used to monitor the conformational transitions of large enzymes by solution NMR7. For larger globular proteins containing cysteine residues, it is also possible to covalently link 13C-labeled methyl groups by site-directed labeling, incubating the protein with 13C methyl methanethiosulfonate (MMTS)8. Similar approaches have been utilized to probe the conformational landscape of membrane proteins by modification of Lys side chains9–11, or by introducing 19F probes12–15. To date, these studies have been limited to detergent micelles that can affect protein structure, topology and function compared to native lipids16–19.
Larger eukaryotic membrane proteins are very challenging to express recombinantly in amounts required for NMR spectroscopy20. Nonetheless, their structural and dynamic transitions could be still monitored by ssNMR using cysteine reductive methylation8. However, this method presents severe spectroscopic challenges as the 1H signals of non-deuterated proteins are typically broad and unresolved21. Moreover, the 13C background signals arising from natural abundance 13C from proteins and lipids complicate the appearance of the spectra. To overcome these problems, we synthesized 13C-ethyl methanethiosulfonate (13C EMTS) and used it to alkylate the native Cys side chains (Figure 1). This approach has three major merits: i) it enables one to filter out the natural abundance 13C signals using dipolar assisted rotational resonance (DARR) experiments22, making it possible to study membrane proteins in native lipid bilayers; ii) it provides better resolution of the engineered aliphatic resonances with respect to methyl groups, iii) the 13C-EMTS probe is small and relatively non-perturbing, and labeling can be carried out in aqueous milieu, retaining enzymatic activity.
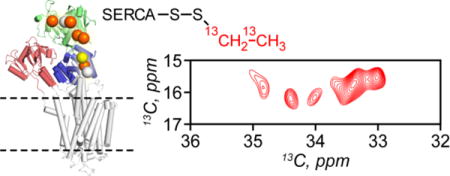
Figure 1.
a: Synthesis of 13C EMTS and its usage for the labeling of cysteines of SERCA. b: [13C, 13C]-CT-DARR spectrum of 13C-EMTS labeled SERCA. c: Normalized ATPase activity of the ethylated SERCA: reaction with 5-fold excess of EMTS did not modify the activity of the enzyme.
As a proof-of-principle, we used site-directed ethyl labeling to monitor the conformational transitions of the sarcoplasmic reticulum Ca2+-ATPase (SERCA), EC: 3.6.3.8, a 110 kDa integral membrane protein that plays a key role in muscle contractility23, 24. To monitor SERCA’s conformational transitions, we labeled the most reactive cysteine residues with 13C-EMTS and followed the chemical shift changes of both CH2 and CH3 signals by ssNMR DARR experiments. To assign the resonances, we employed a combination of paramagnetic relaxation enhancements (PRE) experiments. We were able to obtain domain specific assignments for all of the resonances in the P- and N-domains of SERCA, including two unambiguous assignments for ethyl groups linked to C674 and C636 of the P-domain, defining the NMR signatures for the different conformational states of the enzyme.
Materials and Methods
The synthesis of 13C-EMTS was accomplished by refluxing equimolar amounts of 13C bromoethane and sodium methanethiosulfonate for 5 h in ethanol (Figures 1 and S1). The resulting product was filtered and the solvent was removed by evaporation. The final product was identified by solution NMR and used without further purification. To label the ATPase with EMTS, we incubated 9 μM C12E8-solubilized SERCA purified from rabbit skeletal muscle25 in reconstitution buffer (20 mM HEPES, 100 mM KCl, 1 mM Mg2+, 5% glycerol, 0.02% NaN3) with 45 μM (5-fold molar excess) of EMTS for 3 h. Enzymatic activity of labeled SERCA was measured by ATP hydrolysis assay26. The incubation time and the EMTS concentrations were varied to minimize activity loss of SERCA. Under conditions of 5-fold excess of EMTS relative to and 3 h incubation, the ATPase retains full activity (Figure 1). Additionally, we have tested SERCA labeling with 10-fold and 20-fold excess of EMTS to assess whether all the cysteine residues in the enzyme are accessible to the labeling reagent. The enzymatic assay shows only in the presence of large excess of EMTS, suggesting that the least accessible cysteines are essential for the SERCA function (Figure S2).
For SERCA reconstitution in lipid membranes, 9 mg of DMPC-d54 were solubilized in 18 mg of C12E8 and added to the ATPase. Detergent was removed by incubation with a 30-fold weight excess of biobeads for 3 h. The sample was centrifuged at 100,000×g, and the pellet was re-suspended in reconstitution buffer, dialyzed twice to remove any unreacted EMTS, and pelleted down at 100,000×g. The proteoliposome pellet was re-suspended in 1 ml reconstitution buffer with appropriate ligands, sedimented by ultracentrifugation at 350,000×g for 20 h, and the resulting hydrated pellet was transferred to a 3.2 mm MAS rotor for NMR experiments. The ligands used to induce the enzyme different SERCA conformations were obtained using the following conditions: 5 mM CaCl2 (E1-Ca2+), 2.5 mM AMPPCP (E2-ATP), 5 mM CaCl2 and 2.5 mM AMPPCP (E1-Ca2+-ATP), or 5 mM CaCl2, 2.0 mM AlCl3, 2.5 mM ADP and 20 mM NaF (E1-Ca2+~P~ADP). To increase the resolution, a constant-time version of the DARR experiment, [13C,13C]-CT-DARR, was used 22, 27. The spectrum was acquired at 4°C with a spinning rate of 10 kHz on a 600 MHz Varian spectrometer. Acquisition parameters were 4000 points with a spectral width of 100 kHz (direct dimension). The corresponding values in the indirect dimension were 36 points and 5 kHz. 1024 scans were acquired with 800 μs CP transfer, 100 ms DARR mixing time and ωRF/(2π) = 100 kHz TPPM decoupling.
Results and Discussion
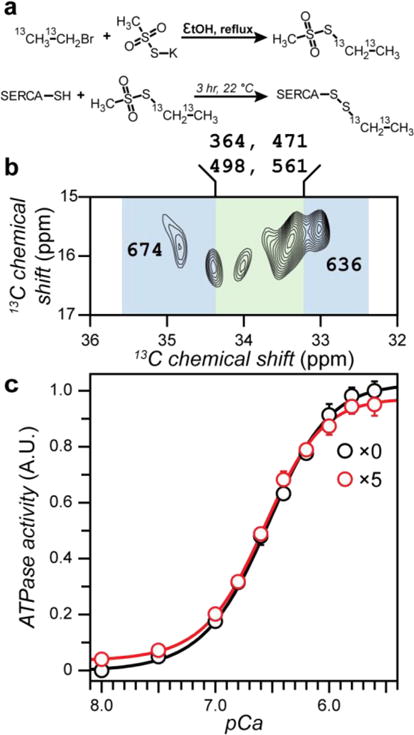
The spectrum of labeled SERCA in DMPC bilayers obtained with a 5-fold excess of EMTS is shown in Figure 1. The SERCA spectrum with non-hydrolyzable ATP analog AMPPCP without Ca2+ shows well-resolved cross peaks between CH2 and CH3 moieties of the probe. Importantly, these are the only off-diagonal signals in the spectrum, as DARR transfer occurs only between adjacent 13C nuclei, eliminating the natural abundance 13C resonances of lipids and SERCA, which dominate one-dimensional 13C spectra28. Thus, the EMTS labeling strategy enables spectral editing in 2D, enhancing the resolution. Since the transfer of magnetization in the DARR experiment is mediated by dipolar couplings, it is sensitive toward the mostly rigid groups. The differential mobility of the NMR probes can lead to distinct efficiencies of magnetization transfer, manifested as the variations in the peak intensity. The dynamics of the NMR probes can be further evaluated and quantified with the appropriate techniques29 30. The ethyl cross peaks are clearly visible in the spectrum at ~17 ppm (CH3–) and ~35 ppm (–CH2–) with much greater resolution in the –CH2– resonances. To identify the ethylated Cys residues of SERCA, we utilized trypsin digestion in combination with electrospray ionization tandem mass spectrometry (LC-MS/MS). We detected small peptides corresponding to 16 out 23 Cys residues of SERCA, covering all Cys in the cytoplasmic P-, N- and A-domains. High yield of Cys ethylation was achieved at six sites: 364, 471, 498, 561, 636 and 674 (Figures S3–S5). To estimate the labeling efficiency, a large excess of iodoacetamide was added to SERCA immediately after the ethylation reaction and prior to trypsin cleavage. For two of the modified Cys (614 and 636), we detected carbamidomethylated (CAM) peptides with higher relative abundance than of the ethylated peptides (Figure S5). Thus, we conclude that these residues have much lower percentage of ethylation and the peaks in the [13C-13C]-CT-DARR spectrum correspond to Cys 364, 471, 498, 561, 674 and 636 (Figure 2).
Figure 2.
Mass spectrometry of 13C-EMTS labeled SERCA. a: representative LC-MS chromatograms of a tryptic peptide with EMTS (+62) or CAM (+57) modifications. b: MS2 fragmentation of the indicated peaks was used to confirm the peptide identity. c: Cys in SERCA that are preferentially labeled (orange), partially labeled (yellow) and not labeled (white) with EMTS.
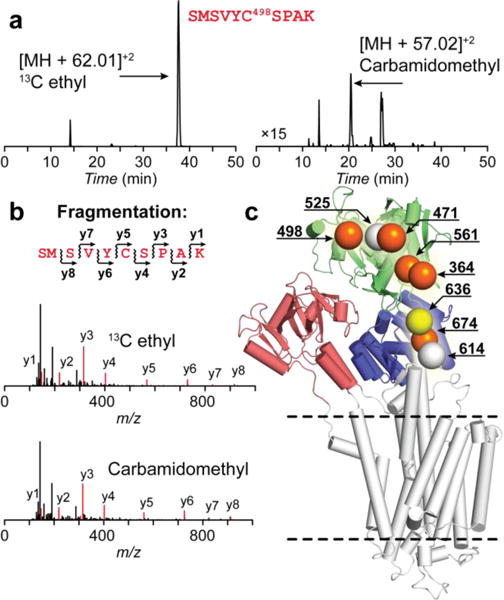
While the DARR experiment enables one to measure short-range distances between the probes22, 31, the distance separation greater than 10 Å between the labeled cysteines precludes its use for distance measurement in case of SERCA. To obtain domain specific assignments of the ethyl group fingerprint, we incubated SERCA in the E2-ATP state with various spin labels to selectively quench the NMR signals based on their proximity to the unpaired electron using paramagnetic relaxation enhancement (PRE) (Figure 1). Specifically, we engineered the TEMPO spin-label coupled to the lipid head group or to the 3′/5′ hydroxyl groups of the nucleotide (Figure S1). Additionally, we selectively attached iodoacetamide TEMPO to C674, as described previously32, and performed the experiments with the oxidized and reduced spin label. The NMR spectra of the ethyl group fingerprint and EPR spectra corresponding to the spin-labels are shown in Figure 1. Both DOPE-TEMPO and C674-MTSL are close to the lipid-water interface and quench the ethyl groups of the Cys in the P-domain. The spin labeled nucleotide, ADP-TEMPO, bound in the N-domain away from the membrane, partially quenched residues both in the P- and N-domains. Based on the quenching pattern, we assigned the resonances between 33.5 and 34.5 ppm to the ethyl groups cross-linked to the cysteines in the N-domain, whereas the resonances in the range between 34.5 and 35 ppm and 32.5 and 33.5 to those of the P-domain. Furthermore, we employed 10 mM ascorbic acid to reduce free radical of C674-MTSL to hydroxylamine. The quenched spin label does not exert any effect on NMR relaxation of the adjacent 13C nuclei, and the only missing signal can be assigned to the ethyl group on C674. Indeed, the 34.8 ppm peak is absent in the NMR spectrum, and therefore it is assigned to C674. Therefore, the remaining P-domain resonance at 33.1 ppm consequently is assigned to C636.
The labeled Cys in the P and N domains of SERCA are ideally positioned to probe the allosteric communication between Ca2+ binding in the TM domain and nucleotide binding in the cytoplasmic domain (Figures S6–S7). It has been proposed that the enzymatic cycle of the ATPase comprises several different conformational states24, 33. For example, the structures of SERCA in the Ca2+-free nucleotide-bound (E2-ATP)34, Ca2+- and nucleotide-bound (E1-Ca2+-ATP)35 and Ca2+-bound nucleotide-free (E1-Ca2+)36 states show distinct conformations in the cytoplasmic domains37–39. To test whether the 13C-EMTS labeled sites are sensitive to the conformational transitions of SERCA, we mimicked these major states of the enzymatic cycle40, saturating the enzyme with AMPPCP, ADP/AlF4, or Ca2+. The ethyl group signatures of the enzyme’s states display pronounced differences. In the Ca2+-free form, SERCA populates the E2 conformational state, while addition of Ca2+ shifts the equilibrium to the E1 state33, 40. Upon Ca2+ addition, the P domain peaks of the E2 state spectrum change substantially (Figure 3). These peaks are sensitive probes for tracing the enzyme’s interconversion between the E1/E2 states.
Figure 3.
a: Crystal structure of SERCA with the spin label positions indicated. b–e: NMR and EPR spectra of the ethyl labeled enzyme with the spin labels indicated in panel a. Residues in the proximity of the spin label are absent in NMR spectra due to paramagnetic relaxation enhancement. EPR acquisition conditions were identical to the ones described previously28.
The center of mass of the cytoplasmic domains has the largest separation in the E1-Ca2+ structure (Figure 3), which has been predicted to be highly dynamic37, 38. It is notable that the resonance pattern and signal intensity of SERCA in this state differ significantly from the rest of the spectra. We speculate this may be attributed to the distinct motion time scale of this state. During the E1/E2 transition, the N-domain undergoes rotation and translation, with the ATP molecule bridging the N- and P-domains, stabilizing the enzyme. Accordingly, in the E2-ATP state we observed all of the Cys ethyl groups, suggesting reduced conformational dynamics. The crystal structures of the E1-Ca2+-ATP and E1-Ca2+~P~ADP states are nearly identical (1.2 Å RMSD), and one can expect the ethyl fingerprints to be similar. Nevertheless, the pattern differs in both N-domain signals as well as C636, indicating structural and/or dynamic changes of SERCA going from ATP-bound to the transition mimic ADP·P-bound state.
The activity of SERCA is regulated by several factors: lipids41, other proteins42, 43, and post-translational modifications44. The associated structural changes have been monitored using site-directed spin labeling of SERCA’s Cys residues32, 45–53. Although most of the chemical modifications of SERCA’s Cys residues did not completely inactivate this enzyme, their size and mobility represent a problem for the accurate distance measurement, or monitoring allosteric coupling between ATP hydrolysis and Ca2+ transport47–49, 51, 54, 55. These problems are even more apparent when small molecules inhibitors or activators are screened for drug discovery56. In such cases, the combination of ethylation and ssNMR can provide a powerful approach to monitor ligand-induced conformational changes with SERCA function in a native membrane environment.
In summary, we demonstrated that site-directed ethyl labeling in combination with paramagnetic spin-labeling and solid-state NMR is a powerful method to study SERCA in lipid bilayers under fully functional conditions. Paramagnetic relaxation enhancement strategy enabled domain-specific assignments of the spectra, with two resonances unambiguously assigned to the individual cysteine residues. Importantly, the ethyl chemical shifts are sensitive to conformational changes of the enzyme, giving distinct signatures for the major structural states and will constitute specific reporters for active and inhibited states of the ATPase. This method will be widely applicable to monitor the conformational states of large proteins or protein complexes, purified from natural sources. It can be easily extended to smaller proteins and receptors expressed recombinantly, including GPCRs, where site-directed mutagenesis can make the assignments unambiguous. Moreover, it can be utilized to monitor dynamic changes via nuclear spin relaxation experiments. Finally, spectral overlap can be resolved by introducing new probes in the 13C-EMTS groups, such as 19F, enabling multidimensional NMR spectroscopy.
Supplementary Material
Figure 4.
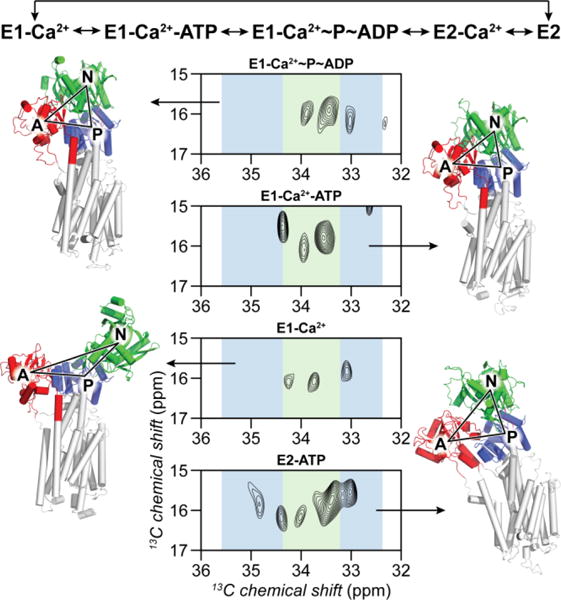
Enzymatic cycle57 and conformations of SERCA by ethyl fingerprints. The crystal structures are Ca2+-free E2 state (PDB ID: 1IWO), E1-Ca2+ (1SU4), E1-Ca2+-ATP (1T5S) and E1-Ca2+~P~ADP (2ZBD). Triangle vertices correspond to the centers of mass of the three cytoplasmic domains.
Acknowledgments
Technical assistance of L. Higgins and T. Markowski in LC-MS/MS data acquisition is gratefully acknowledged.
Funding Sources
This research was supported by grants from National Institute of Health: GM64742 to G.V. and training grants 5T32AR007612 and 13POST14670054 to V.V.
Abbreviations
- DARR
Dipolar assisted rotational resonance
- EMTS
ethyl methanethiosulfonate
- SERCA
sarco(endo)plasmic reticulum Ca2+-ATPase
- ssNMR
solid state NMR
- TM
transmembrane
Footnotes
Supporting Information.
Identification of the 13C-EMTS labeled sites. Synthesis of the NMR and spin labels. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors.
References
- 1.Baldus M. Molecular interactions investigated by multi-dimensional solid-state NMR. Curr Opin Struct Biol. 2006;16:618–623. doi: 10.1016/j.sbi.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 2.McDermott AE. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu Rev Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 3.Hong M, Zhang Y, Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annu Rev Phys Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tugarinov V, Kay LE. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. Chembiochem. 2005;6:1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- 5.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 6.Ruschak AM, Kay LE. Methyl groups as probes of supra-molecular structure, dynamics and function. J Biomol NMR. 2010;46:75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- 7.Religa TL, Kay LE. Optimal methyl labeling for studies of supra-molecular systems. J Biomol NMR. 2010;47:163–169. doi: 10.1007/s10858-010-9419-7. [DOI] [PubMed] [Google Scholar]
- 8.Religa TL, Ruschak AM, Rosenzweig R, Kay LE. Site-directed methyl group labeling as an NMR probe of structure and dynamics in supramolecular protein systems: applications to the proteasome and to the ClpP protease. J Am Chem Soc. 2011;133:9063–9068. doi: 10.1021/ja202259a. [DOI] [PubMed] [Google Scholar]
- 9.Bokoch MP, Zou Y, Rasmussen SG, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi HJ, Thian FS, Kobilka TS, Puglisi JD, Weis WI, Pardo L, Prosser RS, Mueller L, Kobilka BK. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–112. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larda ST, Bokoch MP, Evanics F, Prosser RS. Lysine methylation strategies for characterizing protein conformations by NMR. J Biomol NMR. 2012;54:199–209. doi: 10.1007/s10858-012-9664-z. [DOI] [PubMed] [Google Scholar]
- 11.Xie Q, Fulton DB, Andreotti AH. A selective NMR probe to monitor the conformational transition from inactive to active kinase. ACS Chem Biol. 2015;10:262–268. doi: 10.1021/cb5004702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. Biased signaling pathways in β2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–1110. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shekhawat SS, Pham GH, Prabakaran J, Strieter ER. Simultaneous detection of distinct ubiquitin chain topologies by 19F NMR. ACS Chem Biol. 2014;9:2229–2236. doi: 10.1021/cb500589c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W, Manglik A, Venkatakrishnan AJ, Laeremans T, Feinberg EN, Sanborn AL, Kato HE, Livingston KE, Thorsen TS, Kling RC, Granier S, Gmeiner P, Husbands SM, Traynor JR, Weis WI, Steyaert J, Dror RO, Kobilka BK. Structural insights into μ-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manglik A, Kim TH, Masureel M, Altenbach C, Yang Z, Hilger D, Lerch MT, Kobilka TS, Thian FS, Hubbell WL, Prosser RS, Kobilka BK. Structural Insights into the Dynamic Process of β2-Adrenergic Receptor Signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. Eur Biophys J. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal A, Hoop CL, DeLucia M, Kodali R, Kagan VE, Ahn J, van der Wel PC. Structural Changes and Proapoptotic Peroxidase Activity of Cardiolipin-Bound Mitochondrial Cytochrome c. Biophys J. 2015;109:1873–1884. doi: 10.1016/j.bpj.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rankenberg JM, Vostrikov VV, DuVall CD, Greathouse DV, Koeppe RE, 2nd, Grant CV, Opella SJ. Proline kink angle distributions for GWALP23 in lipid bilayers of different thicknesses. Biochemistry. 2012;51:3554–3564. doi: 10.1021/bi300281k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sani MA, Whitwell TC, Separovic F. Lipid composition regulates the conformation and insertion of the antimicrobial peptide maculatin 1.1. Biochim Biophys Acta. 2012;1818:205–211. doi: 10.1016/j.bbamem.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 20.Weingarth M, Baldus M. Solid-state NMR-based approaches for supramolecular structure elucidation. Acc Chem Res. 2013;46:2037–2046. doi: 10.1021/ar300316e. [DOI] [PubMed] [Google Scholar]
- 21.Lewandowski JR, Dumez JN, Akbey U, Lange S, Emsley L, Oschkinat H. Enhanced Resolution and Coherence Lifetimes in the Solid-State NMR Spectroscopy of Perdeuterated Proteins under Ultrafast Magic-Angle Spinning. J Phys Chem Lett. 2011;2:2205–2211. [Google Scholar]
- 22.Takegoshi K, Nakamura S, Terao T. 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 23.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 24.Brini M, Carafoli E. Calcium pumps in health and disease. Physiol Rev. 2009;89:1341–1378. doi: 10.1152/physrev.00032.2008. [DOI] [PubMed] [Google Scholar]
- 25.Stokes DL, Green NM. Three-dimensional crystals of CaATPase from sarcoplasmic reticulum. Symmetry and molecular packing. Biophys J. 1990;57:1–14. doi: 10.1016/S0006-3495(90)82501-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madden TD, Chapman D, Quinn PJ. Cholesterol modulates activity of calcium-dependent ATPase of the sarcoplasmic reticulum. Nature. 1979;279:538–541. doi: 10.1038/279538a0. [DOI] [PubMed] [Google Scholar]
- 27.Chen L, Olsen RA, Elliott DW, Boettcher JM, Zhou DH, Rienstra CM, Mueller LJ. Constant-time through-bond 13C correlation spectroscopy for assigning protein resonances with solid-state NMR spectroscopy. J Am Chem Soc. 2006;128:9992–9993. doi: 10.1021/ja062347t. [DOI] [PubMed] [Google Scholar]
- 28.Gustavsson M, Verardi R, Mullen DG, Mote KR, Traaseth NJ, Gopinath T, Veglia G. Allosteric regulation of SERCA by phosphorylation-mediated conformational shift of phospholamban. Proc Natl Acad Sci U S A. 2013;110:17338–17343. doi: 10.1073/pnas.1303006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cady SD, Goodman C, Tatko CD, DeGrado WF, Hong M. Determining the orientation of uniaxially rotating membrane proteins using unoriented samples: a 2H, 13C, and 15N solid-state NMR investigation of the dynamics and orientation of a transmembrane helical bundle. J Am Chem Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 30.Sengupta I, Nadaud PS, Helmus JJ, Schwieters CD, Jaroniec CP. Protein fold determined by paramagnetic magic-angle spinning solid-state NMR spectroscopy. Nat Chem. 2012;4:410–417. doi: 10.1038/nchem.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vostrikov VV, Mote KR, Verardi R, Veglia G. Structural dynamics and topology of phosphorylated phospholamban homopentamer reveal its role in the regulation of calcium transport. Structure. 2013;21:2119–2130. doi: 10.1016/j.str.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wawrzynow A, Collins JH, Coan C. An iodoacetamide spin-label selectively labels a cysteine side chain in an occluded site on the sarcoplasmic reticulum Ca2+-ATPase. Biochemistry. 1993;32:10803–10811. doi: 10.1021/bi00091a035. [DOI] [PubMed] [Google Scholar]
- 33.Toyoshima C, Inesi G. Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Annu Rev Biochem. 2004;73:269–292. doi: 10.1146/annurev.biochem.73.011303.073700. [DOI] [PubMed] [Google Scholar]
- 34.Jensen AM, Sorensen TL, Olesen C, Moller JV, Nissen P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 2006;25:2305–2314. doi: 10.1038/sj.emboj.7601135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Toyoshima C, Mizutani T. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 2004;430:529–535. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- 36.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 37.Winters DL, Autry JM, Svensson B, Thomas DD. Interdomain fluorescence resonance energy transfer in SERCA probed by cyan-fluorescent protein fused to the actuator domain. Biochemistry. 2008;47:4246–4256. doi: 10.1021/bi702089j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinoza-Fonseca LM, Thomas DD. Atomic-level characterization of the activation mechanism of SERCA by calcium. PLoS ONE. 2011;6:e26936. doi: 10.1371/journal.pone.0026936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou Z, Hu Z, Blackwell DJ, Miller TD, Thomas DD, Robia SL. 2-Color calcium pump reveals closure of the cytoplasmic headpiece with calcium binding. PLoS ONE. 2012;7:e40369. doi: 10.1371/journal.pone.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moller JV, Olesen C, Winther AM, Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q Rev Biophys. 2010;43:501–566. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- 41.Gustavsson M, Traaseth NJ, Veglia G. Activating and deactivating roles of lipid bilayers on the Ca2+-ATPase/phospholamban complex. Biochemistry. 2011;50:10367–10374. doi: 10.1021/bi200759y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacLennan DH, Kranias EG. Phospholamban: a crucial regulator of cardiac contractility. Nat Rev Mol Cell Biol. 2003;4:566–577. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 43.Gustavsson M, Traaseth NJ, Karim CB, Lockamy EL, Thomas DD, Veglia G. Lipid-mediated folding/unfolding of phospholamban as a regulatory mechanism for the sarcoplasmic reticulum Ca2+-ATPase. J Mol Biol. 2011;408:755–765. doi: 10.1016/j.jmb.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, Cohen RA. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. 2004;10:1200–1207. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 45.Murphy AJ. Sulfhydryl group modification of sarcoplasmic reticulum membranes. Biochemistry. 1976;15:4492–4496. doi: 10.1021/bi00665a025. [DOI] [PubMed] [Google Scholar]
- 46.Coan CR, Inesi G. Ca2+-dependent effect of ATP on spin-labeled sarcoplasmic reticulum. J Biol Chem. 1977;252:3044–3049. [PubMed] [Google Scholar]
- 47.Kawakita M, Yasuoka K, Kaziro Y. Selective modification of functionally distinct sulfhydryl groups of sarcoplasmic reticulum Ca2+,Mg2+-adenosine triphosphatase with N-ethylmaleimide. J Biochem. 1980;87:609–617. doi: 10.1093/oxfordjournals.jbchem.a132785. [DOI] [PubMed] [Google Scholar]
- 48.Kawakita M, Yamashita T. Reactive sulfhydryl groups of sarcoplasmic reticulum ATPase. III Identification of cysteine residues whose modification with N-ethylmaleimide leads to loss of the Ca2+-transporting activity. J Biochem. 1987;102:103–109. doi: 10.1093/oxfordjournals.jbchem.a122021. [DOI] [PubMed] [Google Scholar]
- 49.Saito-Nakatsuka K, Yamashita T, Kubota I, Kawakita M. Reactive sulfhydryl groups of sarcoplasmic reticulum ATPase. I Location of a group which is most reactive with N-ethylmaleimide. J Biochem. 1987;101:365–376. doi: 10.1093/oxfordjournals.jbchem.a121921. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki H, Obara M, Kuwayama H, Kanazawa T. A conformational change of N-iodoacetyl-N′-(5-sulfo-1-naphthyl)ethylenediamine-labeled sarcoplasmic reticulum Ca2+-ATPase upon ATP binding to the catalytic site. J Biol Chem. 1987;262:15448–15456. [PubMed] [Google Scholar]
- 51.Yamashita T, Kawakita M. Reactive sulfhydryl groups of sarcoplasmic reticulum ATPase. II Site of labeling with iodoacetamide and its fluorescent derivative. J Biochem. 1987;101:377–385. doi: 10.1093/oxfordjournals.jbchem.a121922. [DOI] [PubMed] [Google Scholar]
- 52.Bishop JE, Squier TC, Bigelow DJ, Inesi G. (Iodoacetamido)fluorescein labels a pair of proximal cysteines on the Ca2+-ATPase of sarcoplasmic reticulum. Biochemistry. 1988;27:5233–5240. doi: 10.1021/bi00414a043. [DOI] [PubMed] [Google Scholar]
- 53.Rice WJ, Green NM, MacLennan DH. Site-directed disulfide mapping of helices M4 and M6 in the Ca2+ binding domain of SERCA1a, the Ca2+ ATPase of fast twitch skeletal muscle sarcoplasmic reticulum. J Biol Chem. 1997;272:31412–31419. doi: 10.1074/jbc.272.50.31412. [DOI] [PubMed] [Google Scholar]
- 54.Viner RI, Williams TD, Schoneich C. Peroxynitrite modification of protein thiols: oxidation, nitrosylation, and S-glutathiolation of functionally important cysteine residue(s) in the sarcoplasmic reticulum Ca-ATPase. Biochemistry. 1999;38:12408–12415. doi: 10.1021/bi9909445. [DOI] [PubMed] [Google Scholar]
- 55.Sharov VS, Dremina ES, Galeva NA, Williams TD, Schoneich C. Quantitative mapping of oxidation-sensitive cysteine residues in SERCA in vivo and in vitro by HPLC-electrospray-tandem MS: selective protein oxidation during biological aging. Biochem J. 2006;394:605–615. doi: 10.1042/BJ20051214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornea RL, Gruber SJ, Lockamy EL, Muretta JM, Jin D, Chen J, Dahl R, Bartfai T, Zsebo KM, Gillispie GD, Thomas DD. High-throughput FRET assay yields allosteric SERCA activators. J Biomol Screen. 2013;18:97–107. doi: 10.1177/1087057112456878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clausen JD, Bublitz M, Arnou B, Montigny C, Jaxel C, Moller JV, Nissen P, Andersen JP, le Maire M. SERCA mutant E309Q binds two Ca2+ ions but adopts a catalytically incompetent conformation. EMBO J. 2013;32:3231–3243. doi: 10.1038/emboj.2013.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


