Abstract
Background
Tumors have heterogeneous properties, which could be explained by the existence of hierarchically and biologically distinct tumor cells such as tumor-initiating cells (TICs). This model is clinically important, as TICs are promising targets for cancer therapies. However, TICs in spontaneous B-cell lymphoma have not been conclusively identified.
Hypothesis/Objectives
Tumor cells with a progenitor phenotype exist in B-cell lymphoma, reflecting a hierarchical organization.
Animals
Twenty-eight client-owned dogs with previously untreated B-cell lymphoma and six healthy dogs.
Methods
This was a prospective study. Flow cytometry was used to identify lymphoid progenitor cells (LPCs) that co-expressed hematopoietic progenitor antigens CD34, CD117 (KIT), and CD133, with lymphoid differentiation markers CD21 and/or CD22 in B-cell lymphoma. The polymerase chain reaction for antigen receptor rearrangements was used to analyze clonality and relatedness of tumor populations. A xenograft model using NOD/SCID/IL-2Rγ−/− mice was adapted to expand and serially transplant primary canine B-cell lymphoma.
Results
LPCs were significantly expanded in lymph node samples from 28 dogs with B-cell lymphoma compared to six healthy dogs (p=0.0022). LPCs contained a clonal antigen receptor gene rearrangement identical to that of the bulk of tumor cells. Canine B-cell lymphoma xenografts in recipient mice that maintained LPCs in the tumors were recurrently observed.
Conclusions and clinical importance
These results suggest the presence of a hierarchy of tumor cells in canine B-cell lymphoma as has been demonstrated in other cancers. These findings have the potential to impact not only the understanding of lymphoma pathogenesis but also the development of lymphoma therapies by providing novel targets for therapy.
Keywords: Flow cytometry, Dog, Xenotransplantation, CD34
Introduction
Lymphomas in dogs are a group of lymphoproliferative disorders originating from B-cells, T-cells, and NK cells. Lymphoma is the most common hematopoietic tumor in dogs, accounting for more than 20% of all canine tumors.1 Approximately 85% of dogs with lymphoma respond to standard of care CHOP-based protocols; however, durable remissions for most B-cell lymphomas are not the norm, with median survival remaining at ~9–12 months.2 A better understanding of the pathogenesis of lymphoma will assist in development of more effective treatment strategies.
It has long been known that cancer cells within tumors exhibit heterogeneity.3, 4 Although individual tumor cells initially arise from a single cell and thus share common genetic aberrations reflecting their clonal origin, single-cell analysis has demonstrated variation among tumor cells at the genetic, cellular, and molecular levels. According to the stochastic carcinogenesis model, all cells in a tumor are biologically equivalent and heterogeneity arises from random mutational or epigenetic events in individual cells. However, recent evidence supports a hierarchical model of tumor initiation where cells are not biologically equivalent, and which is the major source of tumor heterogeneity.5, 6 According to this model (also known as the cancer stem cell model), tumors are organized hierarchically into a population of cancer stem cells or tumor-initiating cells (TICs) and a population of partially differentiated tumor cells, where the TICs are the only cells able to initiate and maintain tumors. In this model, TICs are singly responsible for resistance to therapy, relapse, and metastatic spread.7, 8 A primitive population of tumor cells might be as important in lymphoma biology as it is in the biology of other hematopoietic tumors.9–13 However, TICs have not been conclusively identified in naturally occurring lymphoma of any species. Since hierarchical organization may not be a feature of every tumor type,14 it is possible that lymphoma conforms to the stochastic model. The implication that tumors organized into stochastic or hierarchical populations will have fundamentally different natural histories and response to treatment underscore the importance of this question.
In this study, we focused on B-cell lymphoma of dogs (which is more common than T-cell and NK cell lymphoma2) to test the hypothesis that lymphoid progenitor cells (LPCs) exist in lymph nodes (LNs) of dogs with B-cell lymphoma, reflecting a hierarchical organization. TICs have initially been defined in many tumors by the expression of certain lineage-specific and progenitor markers in subpopulations of enriched cells. Thus, we assessed the existence within B-cell lymphomas of cells that expressed hematopoietic progenitor markers, CD34, KIT, and CD133, in combination with lymphoid lineage markers.
Materials and Methods
Study design
This was a prospective study. The present study was designed to confirm the biological and clinical relevance of lymphoid progenitor cells (LPCs) found in lymph nodes from dogs with malignant B-cell lymphoma. LPCs, defined as cells that co-expressed CD34, Kit, and/or CD133 (“progenitor markers”) with CD45 and lymphoid lineage markers, have been identified in lymph node (LN) samples from nine dogs with diverse types of malignant lymphoma (JFM, MME, and KAH, unpublished). Here, samples from an expanded cohort of dogs with B-cell lymphoma (cohort-II) were used to prospectively analyze expression of lymphoid lineage markers in Progenitor+CD45+ cells. The initial objective was to exclude the possibility that LPCs represented resident or mobilized hematopoietic progenitor cells (HPCs) in malignant LNs. Next, the percentage of LPCs in LNs from healthy dogs vs. LNs from dogs with B-cell lymphoma, as well as the clonal relationship between LPCs and the rest of the resident malignant population in the LNs were assessed. The side population (SP) assay to determine if LPCs had the capability to exclude vital dyes was used, as has been described for normal hematopoietic stem cells15 and some cancer stem cells.16 Finally, the persistence of LPCs in tumors after xenotransplantion was analyzed.
Lymph node samples and animals
Sterile biopsy or fine needle aspirate (FNA) samples were available from 28 dogs (four dogs from cohort-I and 24 dogs in cohort-II) with a diagnosis of B-cell lymphoma at participating veterinary hospitals with owner consent under protocols approved by the University of Minnesota Institutional Animal Care and Use Committee (IACUC) and the Colorado Multiple Institutional Review Board. The 28 dogs were from 14 breeds, including six Golden Retrievers, four Labrador Retrievers, two Pembroke Welsh Corgis, two Standard Poodles, and one each of 10 other breeds, as well as four mixed breed dogs. The gender distribution was 50:50, and the average age was 8.6 years (median = 9 years, range 3 - 14). Normal tissue controls were obtained at the time of euthanasia from six healthy, purpose-bred, one-year-old female Beagle dogs that were part of an unrelated project. Collection of normal tissues was done under a protocol approved by the University of Minnesota IACUC. Sample preparation for histopathology, cytology, and flow cytometry was performed as described.17 The final histopathological classification was determined by one author (VEV) with a consensus of three additional authors (DI, TDO, JFM) upon review of the specimens. Tumors were classified according to the modified WHO classification.18 They included 13 diffuse large B-cell lymphomas, four Burkitt-like lymphomas, two marginal zone lymphomas, two low-grade B-cell lymphomas of intermediate centrocytic cells with cleaved nuclei, one follicular lymphoma, and one anaplastic large cell lymphoma. Sufficient sample material was not available for histopathological analysis of five cases. An uncommon phenotype of canine B-cell lymphomas that express CD34 was previously reported.19 Because it is unclear if this represents a unique subtype with distinct biology, inclusion criteria for samples was set as <5% CD34+ cells in the tumor populations to limit the confounding effects of this variable. NOD/SCID/IL-2Rγ−/− (NSG) mice were purchased from Jackson Laboratories (Bar Harbor, ME) and maintained under specific pathogen-free conditions according to institutional guidelines. Lymph node collection from dogs and xenotransplantation of canine lymphoma cells into conditioned, immunocompromised (NSG) mice were conducted with approval of the University of Minnesota IACUC.
Flow cytometry
We used flow cytometry to prospectively analyze the existence of LPCs in single cell suspensions from lymphoma biopsies as described.17 Primary antibodies are listed in Supplementary Table 1. LPCs were classified based on the expression of hematopoietic progenitor markers CD34, KIT, and CD133 (detected by a cocktail of antibodies against these antigens where the mix was designated as “Progenitor”), the common leukocyte antigen CD45, and lymphoid lineage antigens CD21 and CD22. The frequency of Progenitor+ cells and Progenitor+CD22+ cells was normalized to the total number of CD45+ cells. Samples also were stained with a separate antibody (labeled by different fluorescent) for each antigen to analyze the specific antigens expressed in Progenitor+ cells. Side population (SP) assay was performed as described using Hoechst 33342 (Sigma-Aldrich, St. Louis, MO), except that exposure to Hoechst dye was limited to 30 minutes to minimize toxicity.15 To set the SP gate, paired aliquots of cells were pre-incubated with or without verapamil (50 µM), which acts as an ABC transporter inhibitor, for 15 minutes at 37°C. Flow cytometry was performed using Coulter Cytomics FC500 (Beckman Coulter, Fullerton, CA), FACSCalibur, or LSRII cytometers (BD Immunocytometry Systems, San Jose, CA), and results were analyzed using Winlist (Verity Software, Topsham, ME) and FlowJo (Tree Star, Ashland, OR).
Polymerase chain reaction for clonal antigen receptor rearrangement (PARR)
LPCs were enriched from lymphoma samples using immunomagnetic selection (EasySep PE selection kit; STEMCELL Technologies, Vancouver, Canada) with a cocktail of PE-conjugated “Progenitor” antibodies. DNA samples were prepared from enriched Progenitor+ cells and from Progenitor-depleted cells using QIAamp DNA mini kit (Qiagen, Valencia, CA), and PCR was run to detect immunoreceptor gene rearrangements using primers for Ig heavy chain (IgH), T-cell receptor gamma chain (TCRγ), and the constant region of IgM (Cμ; positive control) as described.20 IgH, TCRγ, and Cμ primers labeled with VIC, PET, and 6FAM dyes, respectively, were obtained from Applied Biosystems (Foster City, CA) and used for PCR reactions and capillary electrophoresis. The size of PCR products was analyzed with the GeneScan 600 LIZ size standard (Applied Biosystems) using the Genetic Analyzer 3130xl (Applied Biosystems). For gel electrophoresis analysis, PCR products were separated on 15% acrylamide gels, visualized by ethidium bromide staining using an AlphaImager HP imaging system (Alpha Innotech, San Leandro, CA). Semi-quantitative PARR with serial dilutions of input DNA from 1 to 0.01 ng was performed to confirm the assay did not simply detect clonal IgH gene rearrangement from residual Progenitor− cells in the “Progenitor+” sample. This assay can be used to infer the relative number of clonal lymphoma cells in a population based on the relationship between the extinction slopes obtained in the serial dilution from antigen receptor and germline Cμ control reactions.
Xenotransplantation assay
A limitation in studies of primary B-cell lymphomas in dogs and humans is the absence of established methods to analyze the cells using in vivo models. The conditioned NSG mouse model, which is robust for hematopoietic cell transplantation,21 was adapted to serially xenotransplant T-cell depleted canine B-cell tumors in vivo. Immunomagnetic depletion using a PE-conjugated anti-CD5 antibody was used to eliminate residual T-cells from primary B-cell lymphomas. Serially diluted malignant B-cells (3×107, 1×107, or 2×106) were transplanted intraperitoneally into conditioned NSG mice (by 200 cGy irradiation) using an X-RAD 320 (Precision X-Ray, North Branford, CT). Tumor growth was assessed daily, and tumor-bearing mice were sacrificed after 12–14 weeks. Tumor engraftment in LNs, spleen, lung, liver, and kidney was analyzed by histopathology. Single cell suspensions of spleen cells were prepared and blocked with dog IgG and 2.4G2 antibody (BD Biosciences, San Jose, CA). The phenotype of tumor cells and the presence of LPCs in spleen samples were analyzed using routine immunohistochemistry and flow cytometry as described above. Engrafted tumor cells were serially transplanted into conditioned NSG recipients twice more to confirm tumor propagation potential.
Statistical analysis
The Mann-Whitney Test (Prism 4, GraphPad Software, Inc., La Jolla, CA) was used to determine significance between LPC numbers in LNs from dogs with lymphoma and LNs from unaffected dogs.
Results
Lymphoid progenitor cells reside in lymph nodes of dogs with lymphoma
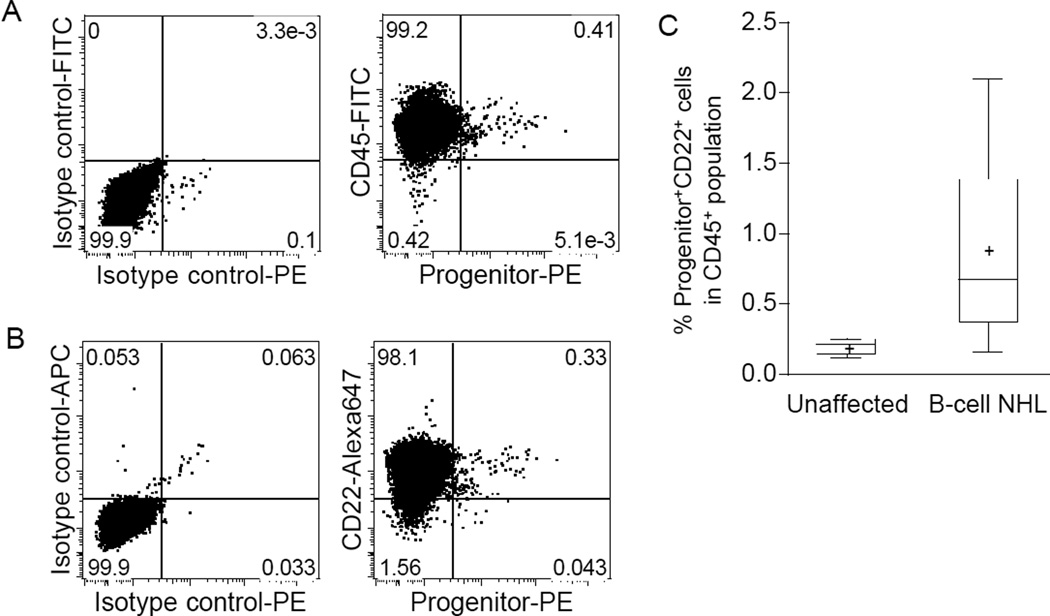
The first cohort included 4 dogs with B-cell lymphoma, each of which contained Progenitor+ cells in LN biopsies (0.11, 0.13, 0.29, and 1.51% of the total CD45+ cells, respectively); the second independent cohort included 24 dogs with B-cell lymphoma. Figure 1A and 1B show two-dimensional flow cytometry dot plots from a representative sample of cohort-II, showing >88% of Progenitor+ cells co-expressed CD45 and the B-cell marker, CD22. The percent of CD22+ cells in the total Progenitor+ population from the 24 samples in cohort-II was 90.8 ± 7.4% (mean, SD), and these Progenitor+CD22+ cells comprised 0.16–2.10% (mean = 0.89%) of all LN lymphocytes (Fig. 1C). Two samples tested also showed co-expression of the B-cell antigen CD21 (data not shown). Most of the Progenitor+ cells in each sample CD34-single positive (0.63 ± 0.58%, mean, SD, n = 23), with fewer KIT-single positive (0.18 ± 0.12%, mean, SD, n = 22) and CD133-single positive cells (0.19 ± 0.21%, mean, SD, n =22). There were less than 0.05% CD34- and KIT-double-positive cells and CD34- and CD133-double-positive cells in all the samples. The percent Progenitor+CD22+ cells in LNs from six healthy dogs (without lymphoma) ranged from 0.12 to 0.25% (mean=0.20%, Fig. 1C). The percentage of Progenitor+CD22+ cells in B-cell lymphomas was statistically significantly different (p = 0.0022, two-tailed Mann-Whitney test) from that in the control group (Fig. 1C).
Figure 1.
Flow cytometry analysis of malignant LNs from dogs with B-cell lymphoma for LPCs. (A) A representative example of two-dimensional flow cytometry dot plots of tumor cells analyzed to identify hematopoietic progenitor cells. The left panel shows staining with isotype control antibodies. The right panel shows expression of “Progenitor” antigens CD34, KIT, and CD133 (using an antibody cocktail) on the X-axis and expression of CD45 on the Y-axis. (B) Analysis of the same sample showing expression of the lymphoid lineage commitment marker, CD22, in Progenitor+ cells. (C) Significantly greater numbers of Progenitor+CD22+ cells are detectable in LN samples from dogs with B-cell lymphoma as compared to LNs from healthy (unaffected control) dogs. The bottom and top edges of the boxes correspond to the sample 25th and 75th percentiles, and the box lengths represent one interquartile range.
Lymphoid progenitor cells are related to lymphoma cells
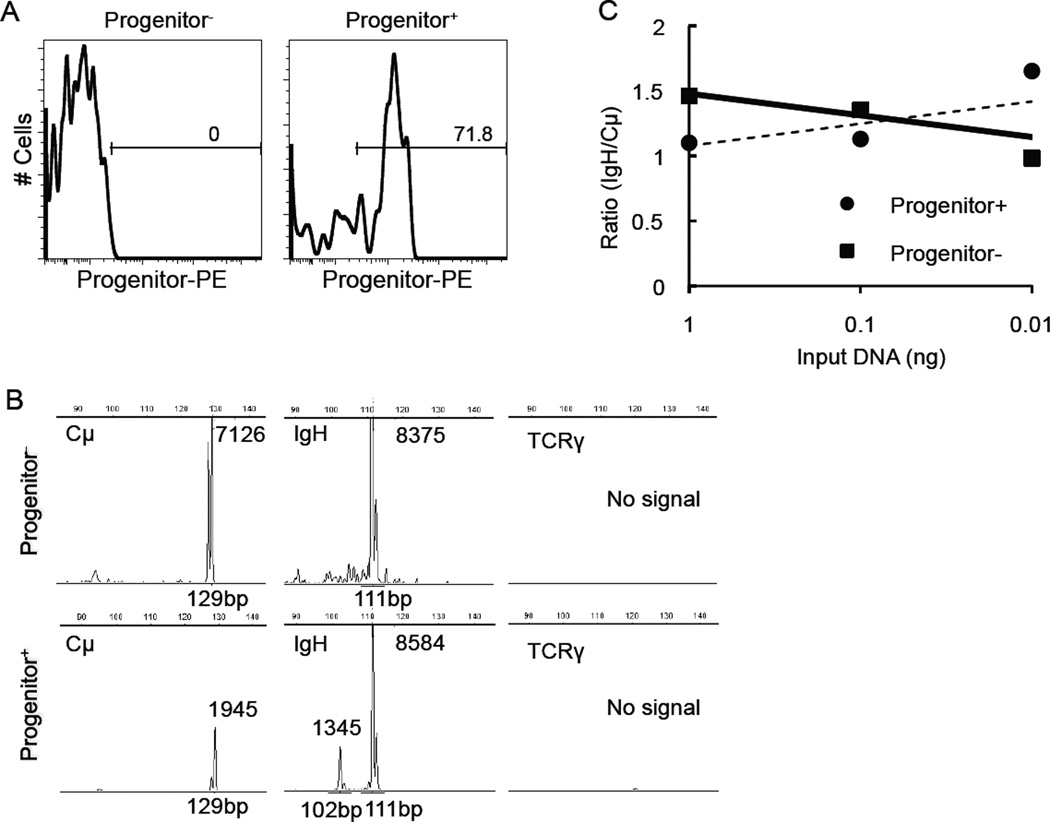
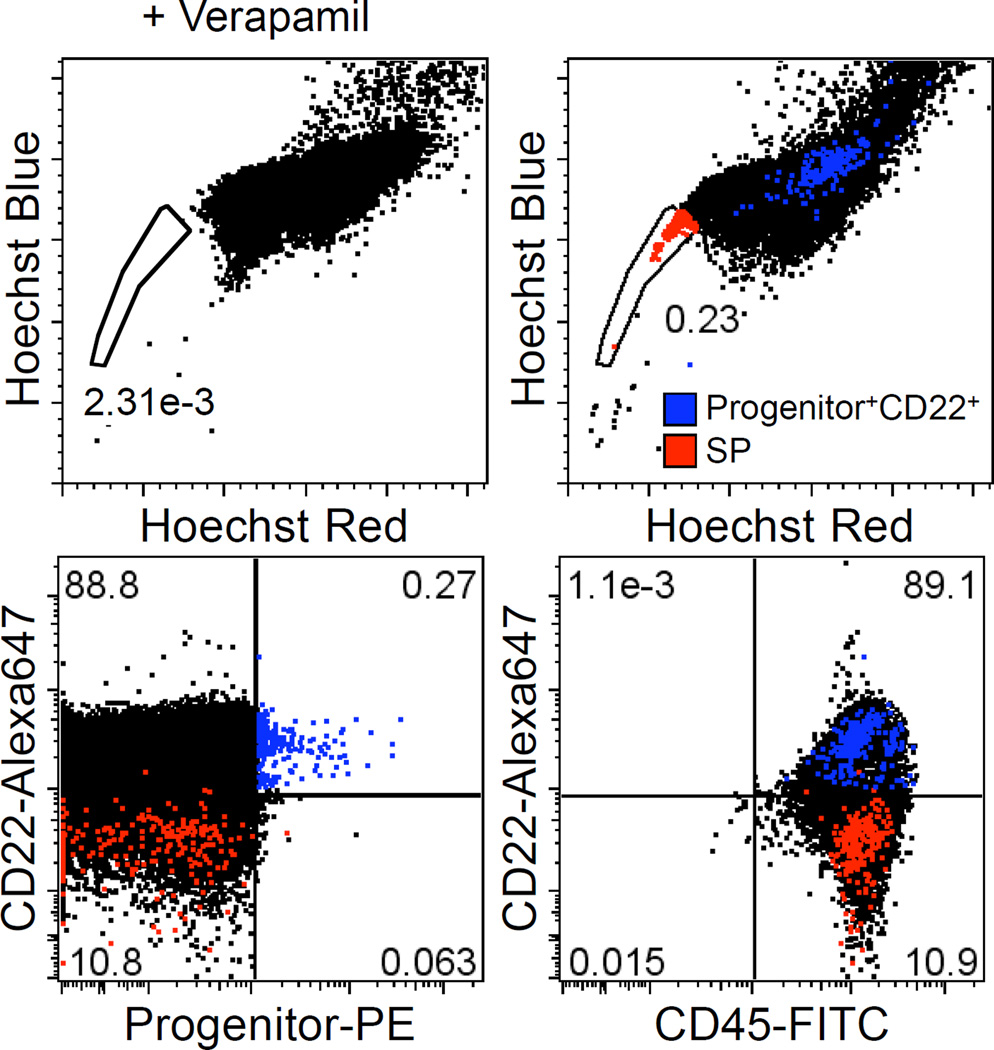
Progenitor+ cells were enriched by ~2 orders of magnitude (from 0.86% to 71.8%) by immunomagnetic selection. Figure 2A shows the percentage of Progenitor+ cells in Progenitor-enriched (Progenitor+) and Progenitor-depleted (Progenitor−) fractions from a representative B-cell lymphoma sample as determined by flow cytometry. Clonal IgH gene rearrangements of the same size were found in the Progenitor+ and Progenitor− populations (Fig. 2B). By semi-quantitative PARR, there was no difference in the extinction rate between Progenitor+ and Progenitor− samples (Fig. 2C). Amplified IgH genes were further sequenced and confirmed to be identical between Progenitor+ and Progenitor− samples (data not shown). A small number of verapamil-sensitive SP cells were detectable in B-cell lymphoma samples (Fig. 3), but these SP cells did not express Progenitor markers or CD22.
Figure 2.
Characterization of LPCs by PARR. (A) Flow cytometry analysis was used to determine the efficiency of Progenitor+ cell enrichment and depletion in a B-cell lymphoma sample after immunomagnetic separation. (B) PCR analysis of Progenitor+ and Progenitor− samples using primers for Cμ, IgH, and TCRγ primers. PCR product size was analyzed by capillary electrophoresis. The number next to the peak represents the fluorescence intensity. (C) Serial dilutions of Progenitor+ and Progenitor− cell DNA were used for PARR analysis. Data show the ratio of scanned pixel intensity from the IgH product (band) over the Cμ product. For DNA from LPC-enriched cells, the ratios of Cμ/IgH were 1.1, 1.1, and 1.6. For DNA from LPC-depleted cells, the values were 1.5, 1.4, and 1.0 (1 ng to 0.01 ng DNA input samples, respectively). Similar results were seen in two dogs.
Figure 3.
SP assay of B-cell lymphoma cells. The SP assay was performed to detect cells with stem cell-like characteristics of high dye-efflux activity. B-cell lymphoma cells were incubated with Hoechst 33342 in the presence or absence of verapamil. Cells were stained with antibodies as indicated and analyzed on a LSRII flow cytometer. SP population is displayed in two-dimensional dot plots for Hoechst-blue versus Hoechst-red fluorescence. Red and blue dots represent SP cells and Progenitor+CD22+ cells, respectively.
LPCs remain after xenotransplantation into immunocompromised laboratory mice
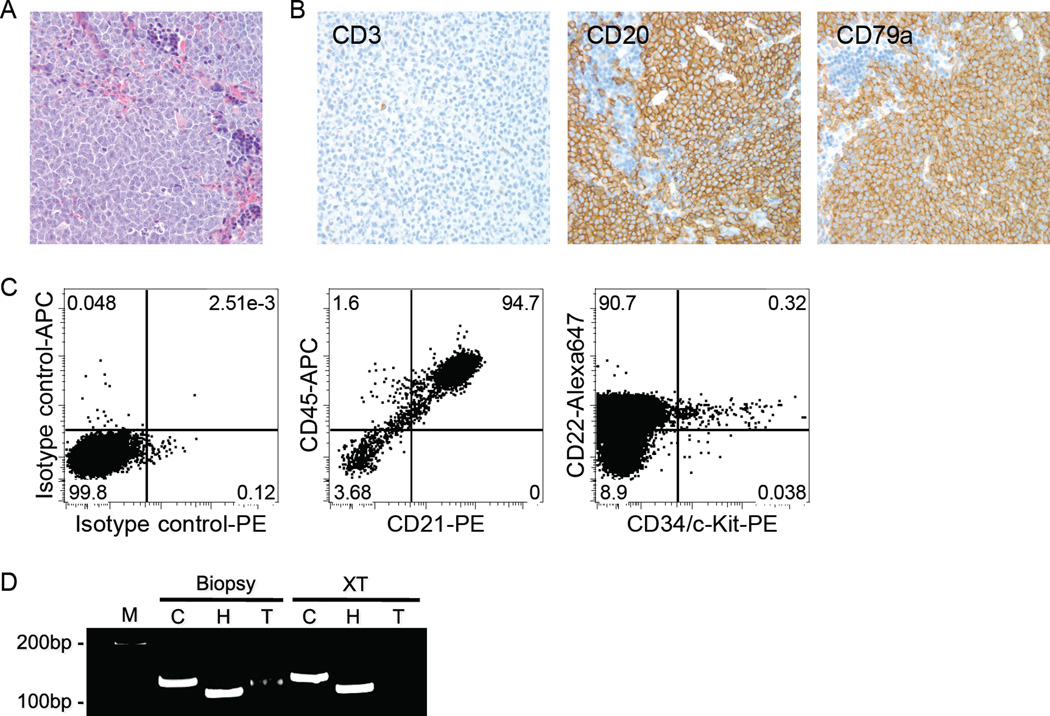
Three different primary canine B-lymphomas (of three attempted in this model) were successfully transplanted. Engrafted canine B-cell lymphoma cells became detectable within 12–14 weeks after transplant. Figure 4A shows diffuse infiltration of tumor cells in spleen in a representative recipient mouse. Lymphoma cells also were present in LNs, liver, kidneys, lungs, and pancreas (data not shown). The tumor cells expressed (canine) CD20 and CD79a, but not CD3 (Fig. 4A), and >90% of the spleen cells were found to be canine-derived large B-cells by flow cytometry (Fig. 4B). A population of Progenitor+ cells persisted recurrently in the tumor xenografts (Fig. 4B). The PARR analysis showed an identical IgH gene rearrangement in a biopsy sample and its associated xenograft (Fig. 4C). Success was not limited to primary transplants, as canine B-lymphoma cells derived from spleens of recipient mice and containing the LPCs were serially transplanted twice more into NSG mice (data not shown).
Figure 4.
Tumor engraftment of primary canine B-cell lymphoma in NSG mice. (A) Photomicrograph of diffuse infiltration of canine tumor cells in a recipient mouse spleen (hematoxylin-eosin (HE) stain, magnification ×200) and immunostaining of the donor (dog) cells for expression of canine CD3, CD20 and CD79a (magnification ×200). (B) Analysis of canine B-cell lymphoma in the spleen of a recipient mouse by multi-parameter flow cytometry. More than 94% of the cells were canine B-cells expressing CD21 and CD45. Similar data were obtained when cells were stained using anti-CD22 and anti-CD45. A small population of Progenitor+ cells was observed within the CD22+ population. (C) PARR performed for Cμ (C), IgH (H), and TCRγ (T) on genomic DNA from a biopsy sample of a dog LN and its corresponding xenograft in a NSG mouse spleen (XT). An identical clonal IgH gene rearrangement was present in both samples. M represents the molecular weight marker.
Discussion
In this study, an expanded population of LPCs that expressed hematopoietic progenitor antigens and lymphoid markers was identified in primary LN samples of B-cell lymphoma. In addition, it was demonstrated that LPCs harbored tumor specific IgH gene rearrangements, which take place along a highly ordered B-cell developmental pathway, and thus they act as “genetic fingerprints” to identify clonally derived B-cells.22 Because the PARR analysis might be sensitive enough to detect clonal IgH gene rearrangement in residual Progenitor− cells from the Progenitor+ sample,20 the result was further confirmed using semi-quantitative PARR with serial dilutions of input DNA. There was no difference in the extinction rate of IgH normalized to the germline Cμ control between Progenitor+ and Progenitor− samples, suggesting the rearrangements were not present exclusively in residual Progenitor− cells. Taken together, these results suggest LPCs represent an atypical population that is expanded in malignant LNs of dogs with B-cell lymphoma. The LPCs were evident in different grades and types of canine B-cell lymphoma, suggesting they might be a common feature required to maintain and expand these various subtypes. In addition, these cells persist in serial xenotransplantation, suggesting they are pathogenetically involved in tumor formation.
To our knowledge, reports of resident LPCs in normal and malignant LNs are limited. There was an independent subpopulation of cells in malignant LNs that could differentiate to myeloid lineage cells in long-term in vitro cultures, but these cells were clonally unrelated to the tumor (DI and JFM, unpublished data). A population of cells that can exclude vital dyes (SP cells), which were similarly distinct from the LPCs, was also found in this study. These suggest that normal HPCs co-exist with LPCs in malignant LNs. Recent work reviewed in detail by Caligiuri identified a small population of CD34+ cells (<0.05% of total lymphoid cells) in normal human LNs.23 This population is comprised almost entirely of lineage-committed NK cell progenitors. While the cells characterized by Caliguiri may retain multipotency, additional work will be necessary to document their significance in lymphoma.
Although there is anecdotal evidence of spontaneous regression of lymphoma in humans associated with immune responses against the causative Epstein-Barr virus,24 lymphoma in dogs is not associated with a viral etiology.2 Tumor reactive LPCs might give rise to expanded clones that mediate anti-tumor immunity elicited by active immunization through the use of idiotype vaccines, dendritic cell therapy, and other strategies,25, 26 and infiltrating regulatory T cells have been shown to limit anti-tumor responses.27 However, the peculiar tumor-like phenotype (i.e. CD22+ in the B-cell lymphomas) and the tumor-related immunoreceptor gene rearrangement of the Progenitor+CD45+ cells indicate these cells are probably not tumor-specific or tumor-infiltrating lymphocytes. Likewise, the absence of clonal TCR rearrangements excludes the possibility these are regulatory T-cells. We therefore conclude that the atypical, expanded population of LPCs represents a potential pathogenetically significant subset of cells in lymphoma.
It has been reported that tumor cells with progenitor phenotypes are prognostically significant in some types of cancer.28, 29 While sample size in this study was large enough to establish the recurrent presence of LPCs, it was too small and too diverse to determine their prognostic significance. Additional work will be necessary to assess if LPCs have prognostic or predictive value.
In summary, there was an expanded population of LPCs related to malignant cells in LNs of dogs with B-cell lymphoma, suggesting that B-cell lymphoma in dogs might conform to the hierarchical progression model found in other types of cancer. Further, we have defined a reliable xenotransplantation model that can be used to test the biological behavior of these LPCs, as well as the potential benefits of treatment strategies targeting these cells.
Supplementary Material
Acknowledgments
The authors thank all the dog owners who allowed their pets to participate in this study, as well as Drs. Scott Sunbury (Sunbury Veterinary Hospital, Austin, TX), Benn Doyle (Westfield Veterinary Group and Wellness Center, Westfield, NJ), and Kaye Fuller (Anderson Mill Animal Clinic, Austin TX), for assistance collecting biopsy samples and for providing follow-up information, Dr. Roberto Novo for providing samples from unaffected dogs, and Drs. Matthew Breen and Rachael Thomas (North Carolina State University, Raleigh, NC) for assistance with histopathologic analyses. The authors would like to thank Veterinary Cancer Associates, P.A. for defraying costs of sample collection from dogs treated at Gulf Coast Veterinary Oncology, and Ms. Mitzi Lewellen for editorial assistance.
This work was supported by AKC Canine Health Foundation grants 615a and 1113, NIH grants P30CA046934, and P30CA077598, the University of Minnesota Animal Cancer Care and Research Program, and the Starlight Fund.
List of abbreviations
- NSG
NOD/SCID/IL-2Rγ−/−
- TICs
tumor-initiating cells
- LPCs
lymphoid progenitor cells
- SP
Side population
- PI
propidium iodide
- IgH
immunoglobulin heavy chain
- TCRγ
T-cell receptor gamma chain
- Cμ
the constant region of IgM
- HPCs
hematopoietic progenitor cells
- PARR
polymerase chain reaction for clonal antigen receptor rearrangement
Footnotes
The main part of this work was done at the Masonic Cancer Center, University of Minnesota, Minneapolis, MN, USA.
A part of the study was presented at AACR 100th Annual Meeting, Washington, DC, USA, April 2010.
References
- 1.Hansen K, Khanna C. Spontaneous and genetically engineered animal models; use in preclinical cancer drug development. Eur J Cancer. 2004;40:858–880. doi: 10.1016/j.ejca.2003.11.031. [DOI] [PubMed] [Google Scholar]
- 2.Modiano J, Breen M, Valli V, et al. Predictive value of p16 or Rb inactivation in a model of naturally occurring canine non-Hodgkin's lymphoma. Leukemia. 2007;21:184–187. doi: 10.1038/sj.leu.2404392. [DOI] [PubMed] [Google Scholar]
- 3.Fidler I, Hart I. Biological diversity in metastatic neoplasms: origins and implications. Science. 1982;217:998–1003. doi: 10.1126/science.7112116. [DOI] [PubMed] [Google Scholar]
- 4.Park C, Bergsagel D, McCulloch E. Mouse myeloma tumor stem cells: a primary cell culture assay. J Natl Cancer Inst. 1971;46:411–422. [PubMed] [Google Scholar]
- 5.Dick J. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 6.Clarke MF, Dick JE, Dirks PB, et al. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer research. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 7.Vermeulen L, Sprick M, Kemper K, et al. Cancer stem cells--old concepts, new insights. Cell Death Differ. 2008;15:947–958. doi: 10.1038/cdd.2008.20. [DOI] [PubMed] [Google Scholar]
- 8.Visvader J, Lindeman G. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 9.Jamieson C, Ailles L, Dylla S, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 10.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 11.Bonnet D, Dick J. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 12.Matsui W, Huff C, Wang Q, et al. Characterization of clonogenic multiple myeloma cells. Blood. 2004;103:2332–2336. doi: 10.1182/blood-2003-09-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones R, Gocke C, Kasamon Y, et al. Circulating clonotypic B cells in classic Hodgkin lymphoma. Blood. 2009;113:5920–5926. doi: 10.1182/blood-2008-11-189688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quintana E, Shackleton M, Sabel M, et al. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodell M, Brose K, Paradis G, et al. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hadnagy A, Gaboury L, Beaulieu R, et al. SP analysis may be used to identify cancer stem cell populations. Exp Cell Res. 2006;312:3701–3710. doi: 10.1016/j.yexcr.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 17.Ito D, O'Brien TD, Modiano JF. Exclusion of cytoplasmic fragments in flow cytometric analysis of lymph node samples from dogs with lymphoma using membrane-permeable violet laser-excitable DNA-binding fluorescent dye (DyeCycle Violet) Vet Clin Pathol. 2010;39:494–498. doi: 10.1111/j.1939-165X.2010.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valli VEJR, Parodi AL, Vernau W, Moore PF. Classification of Hematopoietic Tumors of Domestic Animals. 2. Washington DC: AFIP American Registry of Pathology; 2002. [Google Scholar]
- 19.Wilkerson M, Dolce K, Koopman T, et al. Lineage differentiation of canine lymphoma/leukemias and aberrant expression of CD molecules. Vet Immunol Immunopathol. 2005;106:179–196. doi: 10.1016/j.vetimm.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 20.Burnett R, Vernau W, Modiano J, et al. Diagnosis of canine lymphoid neoplasia using clonal rearrangements of antigen receptor genes. Vet Pathol. 2003;40:32–41. doi: 10.1354/vp.40-1-32. [DOI] [PubMed] [Google Scholar]
- 21.Shultz L, Lyons B, Burzenski L, et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J Immunol. 2005;174:6477–6489. doi: 10.4049/jimmunol.174.10.6477. [DOI] [PubMed] [Google Scholar]
- 22.Jung D, Giallourakis C, Mostoslavsky R, et al. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 23.Freud A, Becknell B, Roychowdhury S, et al. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Abe R, Ogawa K, Maruyama Y, et al. Spontaneous regression of diffuse large B-cell lymphoma harbouring Epstein-Barr virus: a case report and review of the literature. J Clin Exp Hematop. 2007;47:23–26. doi: 10.3960/jslrt.47.23. [DOI] [PubMed] [Google Scholar]
- 25.Kofler D, Mayr C, Wendtner C. Current status of immunotherapy in B cell malignancies. Curr Drug Targets. 2006;7:1371–1374. doi: 10.2174/138945006778559120. [DOI] [PubMed] [Google Scholar]
- 26.Houot R, Levy R. Vaccines for lymphomas: idiotype vaccines and beyond. Blood Rev. 2009;23:137–142. doi: 10.1016/j.blre.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Qin F. Dynamic behavior and function of Foxp3+ regulatory T cells in tumor bearing host. Cell Mol Immunol. 2009;6:3–13. doi: 10.1038/cmi.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Rhenen A, Feller N, Kelder A, et al. High stem cell frequency in acute myeloid leukemia at diagnosis predicts high minimal residual disease and poor survival. Clin Cancer Res. 2005;11:6520–6527. doi: 10.1158/1078-0432.CCR-05-0468. [DOI] [PubMed] [Google Scholar]
- 29.Pallini R, Ricci-Vitiani L, Banna G, et al. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14:8205–8212. doi: 10.1158/1078-0432.CCR-08-0644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.