Summary
While academic‐level studies on metabolic engineering of microorganisms for production of chemicals and fuels are ever growing, a significantly lower number of such production processes have reached commercial‐scale. In this work, we review the challenges associated with moving from laboratory‐scale demonstration of microbial chemical or fuel production to actual commercialization, focusing on key requirements on the production organism that need to be considered during the metabolic engineering process. Metabolic engineering strategies should take into account techno‐economic factors such as the choice of feedstock, the product yield, productivity and titre, and the cost effectiveness of midstream and downstream processes. Also, it is important to develop an industrial strain through metabolic engineering for pathway construction and flux optimization together with increasing tolerance to products and inhibitors present in the feedstock, and ensuring genetic stability and strain robustness under actual fermentation conditions.
Introduction
Our increasing concerns on climate change and the eventual depletion of fossil resources have directed much research towards developing renewable ways for producing chemicals and fuels. One strategy that has been under investigation for many years, beginning at the industrial‐scale with conversion of simple sugars into acetone, butanol and ethanol 100 years ago (Moon et al., 2016), and more heavily in recent years, is the utilization of microorganisms for producing chemicals and fuels from renewable non‐food biomass. This field has advanced tremendously over the past two decades, revolutionized by technological developments allowing inexpensive large‐scale genome sequencing, systems‐level gene expression profiling and other omics techniques, in silico metabolic modelling and simulation, enzyme/pathway engineering and evolution, in addition to routine and upgraded recombinant DNA techniques. Despite this, the number of examples of successful commercialization of microbial processes producing chemicals and fuels remains relatively small to date (Table 1), leaving the question of whether a large‐scale switch towards a sustainable society based on microbial biorefineries is feasible. To achieve this goal, it is critical that metabolic engineering works are designed and performed with the full techno‐economic picture in mind, considering the raw material availability, large‐scale production and downstream processes, and even applications throughout the strain development. To reach this goal, interactions between academia and industry are obviously vital (Pronk et al., 2015). In this work, we review the techno‐economic challenges for the large‐scale implementation of biorefineries with the producing microbial cell factories as the focus, and discuss metabolic engineering strategies to overcome the pending challenges. We have recently suggested 10 general and stepwise strategies to consider for strain development suitable for industrial biorefineries (Lee and Kim, 2015); thus, here we rather focused on complementary points to be considered.
Table 1.
Status of commercialization of microbial cell factories
| Product | Production organism | Status | Feed stock | Companies | Reference |
|---|---|---|---|---|---|
| Chemicals | |||||
| Acetone | Clostridium acetobutylicum | Commercialized | Corn | Green Biologics | www.greenbiologics.com |
| Citric acid | Aspergillus niger | Commercialized | |||
| Succinic acid | E. coli | Commercialized | Corn sugars | BioAmber | www.bio-amber.com |
| E. coli | Commercialized | Sucrose | Myriant | www.myriant.com | |
| S. cerevisiae | Commercialized | Starch, sugars | Reverdia | www.reverdia.com | |
| B. succiniproducens | Commercialized | Glycerol, sugars | Succinity | www.succinity.com | |
| Lactic acid | Commercialized | Corn sugars + more | NatureWorks | www.natureworksllc.com | |
| Itaconic acid | Aspergillus terreus | Commercialized | Biochemistry | Qingdao Kehai | www.kehai.info/en |
| 1,3‐PDO | E. coli | Commercialized | Corn sugars | DuPont Tate & Lyle | www.duponttateandlyle.com |
| 1,3‐BDO | Demonstration | Genomatica and Versalis | www.genomatica.com | ||
| 1,4‐BDO | E. coli | Commercialized | Sugar | Genomatica and DuPont Tate & Lyle | www.genomatica.com |
| 1,5‐PDA | Commercialized | Sugar | Cathay Industrial Biotech | www.cathaybiotech.com | |
| 3‐HP | Commercialized | Metabolix | www.metabolix.com | ||
| Demonstration | Novozymes and Cargill | www.novozymes.com | |||
| Isoprene | S. cerevisiae | Preparing | Sugar, cellulose | Amyris, Braskem, Michelin | www.amyris.com |
| Preparing | DuPont, Goodyear | www.biosciences.dupont.com | |||
| Isobutene | E. coli | Demonstration | Glucose, sucrose | Global Bioenergies | www.global-bioenergies.com |
| Adipic acid | Candida sp. | Demonstration | Plant oils | Verdezyne | www.verdezyne.com |
| Sebacic acid | Candida sp. | Demonstration | Plant oils | Verdezyne | www.verdezyne.com |
| DDDA | Candida sp. | Under commercialization | Plant oils | Verdezyne | www.verdezyne.com |
| Squalene | S. cerevisiae | Commercialized | Sugarcane | Amyris | www.amyris.com |
| PHA | E. coli | Commercialized | Metabolix | www.metabolix.com | |
| Fuels | |||||
| Ethanol | S. cerevisiae, Zymomonas mobilis, Kluyveromyces marxianus | Commercialized | Sugarcane, corn sugar, lignocellulose | Many | |
| Clostridium autoethanogenum | Demonstration | Flue gas | Lanzatech | www.lanzatech.com | |
| Farnesene | S. cerevisiae | Commercialized | Amyris | www.amyris.com | |
| Butanol | Clostridium acetobutylicum | Commercialized | Corn | Green Biologics | www.greenbiologics.com |
| Isobutanol | Yeast | Commercialized | Sugars | Gevo | www.gevo.com |
1,3‐PDO, 1,3‐propanediol; 1,3‐BDO, 1,3‐butanediol; 1,4‐BDO, 1,4‐butanediol; 1,5‐PDA, 1,5‐pentanediamine; 3‐HP, 3‐hydroxypropionic acid; DDDA, dodecanedioic acid; PHA, polyhydroxyalkanoates.
Techno‐economic considerations for microbial cell factory development
To achieve the successful transition from laboratory‐scale demonstration to large‐scale commercial production, there are three key performance indicators to consider (Fig. 1): the product yield (g g−1 substrate), the productivity (g l−1 h−1) and the product titre (g l−1). In the case of bulk chemicals and fuels, the profit margins are very narrow and it is critical that these three metrics are maximized to be competitive with traditional, petrochemical processes. In many (but not all) cases, a trade‐off has to be made between productivity and yield, as maximizing the productivity often requires high density of the producing microbes, while increasing the carbon flux directed towards cell mass inevitably lowers product yield. In an industrial‐scale fermentation, often performed in a fed‐batch mode, a productivity of typically around 2–5 g l−1 h−1 (and preferably higher) needs to be achieved depending on the product. Another strategy for increasing productivity is the use of continuous cultivation, but as these are prone to costly contamination and phage infection, the process requires more expensive equipment, and places high demand of genetic stability on the production host (Croughan et al., 2015). The fed‐batch cultivation strategy should be developed based on the product formation characteristics, e.g. growth associated or non‐growth associated, and strains should be further optimized to meet the process requirements. For the production of bulk and inexpensive products, the use of rather expensive inducers is discouraged; consequently, constitutive expression of metabolic genes is often employed.
Figure 1.
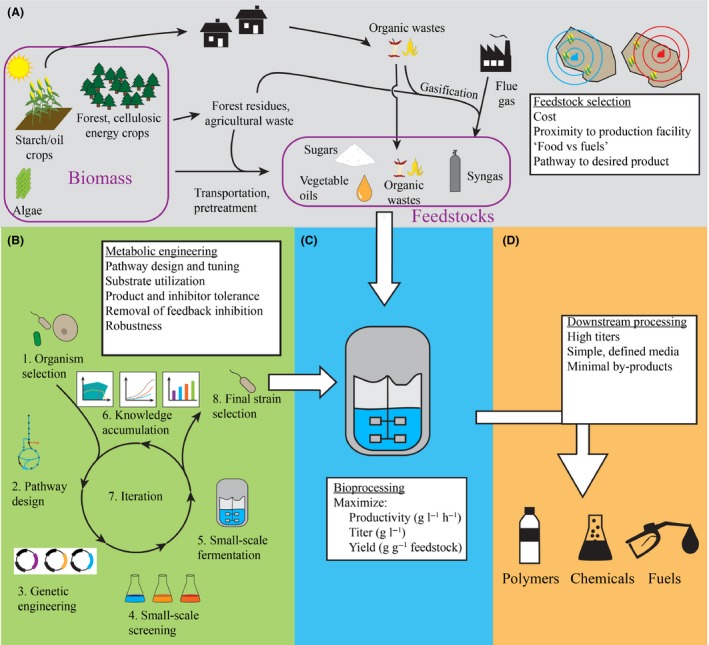
Overview of the microbial cell factory design process. For successful commercial implementation, the full picture should be considered throughout the design process. A. Considerations relating to the choice of renewable feedstock, including the location of the production facility in close proximity to the available feedstocks, as indicated by blue (correct) and red (incorrect) concentric circles. B. The metabolic engineering process, starting from selection of production organisms and iterating through design‐build‐test‐learn cycles until the process requirements are met. C and D. Critical parameters for the production process and downstream purification to final products respectively.
The downstream processes for the separation and purification of a product should also be considered already at the strain development stage (Fig. 1). The final product titre has a major impact on the downstream processing costs, as a highly concentrated product is both easier to separate and allows reduction of the total volume to be handled (including during waste treatment). The use of a chemically defined medium not only allows production of a desired product under more controllable conditions but also simplifies the downstream purification by limiting the amount of contaminants. For the same reason, minimizing by‐product formation is essential during strain development to lower the downstream processing costs. The downstream processes can also be simplified by altering the fermentation conditions from typical ones together with the matching strain developed. For example, in the industrial production of carboxylic acids, it is advantageous to perform the fermentation process at a low pH as this leads to direct precipitation of the protonated acid (the desired product) rather than its salt form. This is why Cargill and Reverdia use acid‐tolerant yeast strains to produce lactic and succinic acids respectively, at low pH (Table 1).
The feedstock is another major contributor to the total bioprocess costs, especially for large volume low price products; for instance, the feedstock cost is estimated to be around 75% of the total production cost of U.S. corn‐based ethanol (FAO, 2008). Thus, selecting an appropriate feedstock is of outmost importance for cheap bulk products. So far, most commercial processes have focused on using simple sugars from sugar cane and starch (Table 1). However, a switch towards utilizing non‐edible lignocellulosic biomass is highly desirable in order to avoid conflicts with food uses. Lignocellulose is the most abundant biomass on earth with an estimated annual production of around 150–170 × 109 tons, out of which only around 2 × 109 tons are currently used for energy, timber, and paper and pulp production (Pauly and Keegstra, 2008). Nevertheless, the necessity for extensive pre‐treatment together with the presence of microbial growth inhibitors, make these feedstocks more difficult to work with compared with sugarcane sugar or starch (Jönsson et al., 2013). In fact, sugarcane is an excellent feedstock, due to its relatively low cost of production, high sugar yield and low energy input requirements for its cultivation (Renouf et al., 2008). However, it is abundantly available only in several specific regions including Brazil, Australia and Southeast Asian countries. This highlights a major consideration for feedstock selection: proximity to the chosen substrates, as shown in Fig. 1A. The costs of feedstock transportation contribute considerably to the costs of raw materials; estimated at up to 30% for locally sourced lignocellulosic feedstocks (Hess et al., 2007). Thus, it makes a lot of sense to base a Brazilian biorefinery on sugarcane, while corn has been the focus of US bioethanol production efforts (EIA, 2016). Northern countries with less favourable agricultural climates should carefully calculate the transportation costs for the use of these feedstocks, or instead focus on other feedstocks, such as the abundant forest industry residues available in Canada, Finland and Sweden.
In addition to using biomass directly as a feedstock, there have been investigations into feeding CO2 or gasified biomass in the form of synthesis gas (syngas) into biorefineries, with the former strategy bypassing the need to first fix CO2 into plant biomass. Direct CO2 utilization has been investigated using photosynthetic microalgae or cyanobacteria, either to produce oils for further conversion or for direct production of target chemicals and fuels. This technology promises superior oil yields per cultivated area compared with conventional oil crops, but much work still remains to bring costs down to levels comparable to common vegetable oils (Chisti, 2007; Hannon et al., 2010). As for syngas, it can be obtained by gasification of various biomass components or municipal organic wastes using thermochemical processes (Puigjaner, 2011). The resulting gas can in turn be fermented by acetogens such as Clostridium ljungdahlii or Clostridium carboxidivorans. These bacteria convert CO and CO2 into fuels or chemicals, including acetate, methane, ethanol and acetone, using hydrogen as a source for reducing power through the Wood–Ljungdahl pathway (Molitor et al., 2016). There is much ongoing effort on strain and bioprocess development using one‐carbon substrates for the production of chemicals and fuels, as exemplified by Lanzatech's ongoing commercialization of bioconversion of industrial off‐gases (Table 1).
Systems metabolic engineering for developing microbial cell factories
The key role of metabolic engineers in the creation of commercial microbial cell factories is engineering strains that can produce the target product efficient enough to meet the requirements for large‐scale bioprocessing as discussed above and summarized in Fig. 1. These requirements include utilization of a wide range of carbon sources, for instance the common 5‐ and 6‐carbon monosaccharides including glucose, xylose and arabinose present in lignocellulose, disaccharides including sucrose and lactose, and fatty acids, depending on feedstock availability. Furthermore, the organism should be tolerant to inhibitors present in these feedstocks and high titres of the target product, allow easy process design by growing fast in a simple defined medium, be robust in relation to heterogeneities in substrate, pH, temperature and oxygen levels typically found in large‐scale bioprocesses, be genetically stable over many divisions (at least 30 generations when scaling from 100 ml to 10 000 m3 production scale) and insensitive to infections by for instance bacteriophages. Finally, the production host should obviously possess metabolic capacities for production of the target product at high productivities and yields.
Starting strain selection
The metabolic engineering process begins with the selection of the base strain for modification. As it is unlikely that a single wild‐type organism will have a phenotype covering all the requirements for the production of diverse products, the selection of a starting strain considering metabolic capacity towards the desired product, bioprocess compatibility, easiness of metabolic and genetic engineering, ability to utilize inexpensive feedstocks and others. So far, many studies have focused on engineering simple model organisms, including Escherichia coli and S. cerevisiae, as these microorganisms have been relatively more studied, have well‐developed tools for various genetic manipulations and have well‐validated genome‐scale metabolic models available for genome‐scale metabolic simulations. Several other strains that have also been employed for biorefineries include Corynebacterium glutamicum Clostridium sp., Bacillus sp. and Pseudomonas sp. Nevertheless, with recent developments in inexpensive genome sequencing, more rapid construction of genome‐scale metabolic models, new genetic and genomic manipulation tools, most notably the use of clustered regularly interspersed short palindromic repeats (CRISPR) for metabolic engineering of microbes (Li et al., 2015; Tong et al., 2015; Blin et al., 2016), metabolic engineering of microorganisms including non‐conventional host strains has become easier than in the past.
One good example of using a non‐model organism to achieve efficient chemical production is the metabolic engineering of Mannheimia succiniproducens for succinic acid production. This organism was isolated from the rumen of a Korean cow, an environment with a high CO2 partial pressure. The reason for screening the cow rumen was based on the hypothesis that there might be a bacterium capable of efficiently performing phosphoenolpyruvate carboxylation in this CO2‐rich environment, which was indeed the case. After genome sequencing and generation of a validated genome‐scale model, development of expression vectors and genetic manipulation tools, and systems metabolic engineering, a final strain capable of producing succinic acid at titres, yields and productivities comparable to or exceeding those of the best current industrial producers could be developed (Choi et al., 2016b).
In silico genome‐scale metabolic modelling and simulation can be a great tool in selecting an appropriate production organism, by allowing evaluation of the metabolic capacities of different organisms. One example is a recent extensive evaluation of the E. coli capacities for biosynthesis of a large range of chemicals (Zhang et al., 2016). This study found that up to 1777 non‐native products, out of which 279 have known commercial applications, could be derived from the E. coli metabolism by introducing heterologous enzymes.
Pathway design for novel products
One of the greatest challenges faced by metabolic engineers is the creation of pathways for target products with no known natural producer. For these products, new enzymes need to be developed, starting from ones catalysing similar reactions as the desired one. Identification of such suitable enzyme candidates is no easy task, but recent developments in computer tools are very helpful for this. For instance, the pathway for 1,4‐butanediol production in E. coli could efficiently designed and tested by using the SimPheny Biopathway Predictor (www.genomatica.com; Yim et al., 2011). A number of such tools exist for prediction of pathways and selection of enzyme candidates, as reviewed elsewhere (Shin et al., 2013). Additionally, cell‐free pathway assembly tools more recently reported promises rapid screening of heterologous pathways in vitro (Karim and Jewett, 2016).
Removing negative regulatory circuits
When overproducing some natural metabolites, a common problem encountered is feedback inhibition and transcriptional attenuation control of the production pathway caused by accumulation of the desired product. Such negative regulations can occur both at the transcriptional level and be caused by allosteric regulation of pathway enzymes, and should be removed at the early stage of strain development. In the case of transcriptional regulation, the strategy is straightforward. Modern DNA‐manipulation tools allow relatively simple editing of chromosomal transcription regions to introduce desired changes. Alternatively, the transcription factors involved in this regulation may also be knocked out. Examples of these strategies include the production of different amino acids (pathways that are heavily affected by such negative regulatory circuits); a number of examples are available, for instance knocking out the aromatic amino acid biosynthesis pathway regulator tyrosine‐activated repressor (tyrR) to allow production of L‐tyrosine (Lütke‐Eversloh and Stephanopoulos, 2007) and its derivative phenol (Kim et al., 2014), and replacing the native promoters of the L‐valine and L‐threonine operons with a strong, inducible promoter for more efficient production of L‐valine and L‐threonine, respectively, in E. coli (Lee et al., 2007; Park et al., 2007).
Dealing with allosteric feedback inhibition is more difficult compared with transcriptional regulation. The most straightforward solution is to look for heterologous enzymes that lack the allosteric regulation. Alternatively, enzyme engineering can be employed to generate feedback‐resistant mutants. This was exemplified for production of L‐threonine using E. coli, where previously identified single point mutations in the thrA and lysC genes (encoding aspartokinase I and III respectively) were used to remove allosteric feedback regulation of this pathway (Lee et al., 2007), and for the production of L‐tyrosine, where a single mutation removed the allosteric regulation of aroG and two mutations removed the regulation of tyrA (Lütke‐Eversloh and Stephanopoulos, 2007; Kim et al., 2014).
Fine‐tuning pathway expression and minimizing metabolic burden
After establishment of a deregulated production pathway to the target product, the next step is fine‐tuning of the pathway to maximize the flux to the target product while minimizing the metabolic burden. As the number of pathway enzymes increase, the task of tuning the expression of each one quickly becomes very large, as shown in a recent work describing optimization of violacein production in E. coli (Jones et al., 2015). In this work, a five‐step pathway for violacein production was cloned combinatorially under five different promoter sequences, resulting in a randomized library of 3125 variants. By screening a fraction of this library, a strain with a 62‐fold improvement in violacein production over a control having only strong promoter variants was obtained.
As an alternative to promoter engineering, the influence of the Shine–Dalgarno sequence was systematically investigated in E. coli. A tool named Empirical Modeling and Oligos for Protein Expression Changes (EMOPEC, http://emopec.biosustain.dtu.dk) was developed for generating oligo sequences for tuning the translation of chromosomal genes in E. coli (Bonde et al., 2016). This tool promises efficient generation of combinatorial libraries for high‐throughput tuning of chromosomal pathways. Another recent contribution of interest describes the use of a chromosomally integrated green fluorescent protein (GFP) under a constitutive promoter as a reporter for examining the protein synthesis capacity of an engineered cell (Ceroni et al., 2015). By comparing the GFP expression in strains containing different plasmid constructs, they were able to select for constructs that produced the optimum level of the target protein, while minimizing the reduction in capacity for synthesis of other proteins. Combining this method with the above‐mentioned combinatorial library techniques could greatly assist in future pathway‐tuning efforts.
Furthermore, inducer‐free expression systems are preferable in industry to remove the cost of the inducer. Traditionally, this has been achieved using constitutive promoters, but this prevents separation of cell growth and product formation. It is envisioned that the progress in design of synthetic circuits, not least the recent report of a software for automatic design of biological circuits with predictable output (Nielsen et al., 2016), will lead to the development of advanced, dynamically regulated auto‐induction strategies in the future.
Substrate utilization engineering
Not only should the production organism contain efficient, well‐tuned production pathways as discussed above but it also needs to efficiently utilize the inexpensive feedstock of choice. A classic example of this problem is that S. cerevisiae is incapable of growing on xylose (Jeffries and Jin, 2004). As xylose typically makes up around 15–23% of carbohydrates in lignocellulose (Huang et al., 2009), this is a critical issue that needs to be solved to obtain higher yields from this feedstock. Consequently, this was one of the most heavily studied topics, and there are now a number of reports describing engineered S. cerevisiae for this purpose, as reviewed elsewhere (Laluce et al., 2012). Other organisms, including for instance E. coli, can natively utilize the five‐carbon carbohydrates arabinose and xylose. However, in E. coli a strict substrate utilization hierarchy is maintained, with a preference for glucose utilization before other carbohydrates. In order for efficient fed‐batch processing of mixed lignocellulosic sugars, this hierarchy needs to be removed to enable simultaneous sugar utilization. One such example is the deletion of the E. coli glucose phosphotransferase system domain IIC (encoded by ptsG) that removes this substrate preference from E. coli. This deletion was recently used to enable synthesis of poly(lactate‐co‐glycolate) through simultaneous utilization of glucose and xylose (Choi et al., 2016a). Another example is the inability of the commonly used E. coli K12 and B strains to utilize sucrose as a carbon source; simple overexpression of a single β‐fructofuranosidase gene from M. succiniproducens could confer the sucrose‐utilization phenotype to an engineered E. coli K12 strain, allowing direct production of L‐threonine from sucrose (Lee et al., 2010).
Engineering cells to tolerate target products and inhibitors present in feedstock
For utilization of complex substrates such as lignocellulosic biomass and accumulation of target products to high titres, it is necessary that industrial strains should be resistant towards both the target product and inhibitors present in the feedstock. However, inhibitor tolerance is one of the most complex phenotypes to engineer as a wide number of hard‐to‐predict genes are often involved. Thus, this phenotype is commonly achieved by one of two main strategies. First, adaptive laboratory evolution can be used, as described for production of D‐lactate in E. coli (Utrilla et al., 2012) and L‐arginine in C. glutamicum (Park et al., 2014). Following the genome sequencing of evolved strains, the contributing alterations from adaptive laboratory evolution can then be introduced into the production strain. Second, system‐wide genomic or transcriptomic engineering followed by screening and selection can also be used. For this, various methods including transcription factor engineering (Alper et al., 2006), and gene knockdown libraries based on sRNA (Na et al., 2013) or CRISPR interference (CRISPRi; Qi et al., 2013) can be employed.
A different, rational engineering strategy to improve both productivity and product tolerance is to express product efflux pumps that enhance the export of the product. One example of this strategy is the screening of a library of 43 efflux pumps for improving the tolerance of E. coli against seven different biofuels, resulting in the identification of efflux pumps improving the tolerance against five of seven of the tested biofuels (Dunlop et al., 2011). Another recent work describes the screening of 16 ABC transporters for the ability to enhance secretion of carotenoids from E. coli (Doshi et al., 2013). Using this strategy, up to 5.4‐fold increase in carotenoid secretion was achieved while maintaining cell viability.
Genetic stability and strain robustness
Industrial strains need to be stable over a great number of generations to allow scaling up to production scale. This is no small challenge, as cost issues and environmental concerns prohibit the use of antibiotics at production scale. Thus, it is preferable to use chromosomal pathway integration over plasmid‐based expression if possible. The invention of the CRISPR system has greatly simplified this, as exemplified by a recent report describing the integration of a five‐gene pathway in a single step using CRISPR (Bassalo et al., 2016). Furthermore, pathway tuning to minimize the metabolic burden of the production pathway is necessary to avoid preferential selection of non‐producers. To combat this, biosensors are developed to couple target products to an essential gene for cell growth (Xiao et al., 2016). In this design, cells with a high production rate obtain a growth advantage compared with low‐producers, leading to the highest fatty acids titre reported to date in E. coli (21.5 g l−1). Such approaches will be more frequently employed as synthetic biology research advances rapidly.
Finally, the production host must also be robust towards the stresses experienced in large‐scale bioprocesses, such as inhomogeneities resulting from insufficient mixing. This is why pilot plant testing (and even domo‐plant testing) is important. It has also been suggested to repeatedly test strains under process‐like conditions during the strain development process, for instance using scale‐down bioreactor systems (Neubauer and Junne, 2016). Biotechnology companies have accumulated much unpublished knowledge on scaling‐up process, and thus academia‐industry collaboration will speed up the successful scale‐up process.
Conclusion
In this work, we have briefly reviewed the challenges associated with the establishing industrial‐scale microbial biorefineries for the production of chemicals and fuels. As has been discussed, many different aspects need to be taken into account during the strain development by metabolic engineering. The most important lesson for successful cell factory development is thus to have an all‐encompassing view during strain development, including everything from feedstock selection, through the bioprocessing steps and to the downstream processes. It is clear that such rapid advances in the field promise successful establishment of microbial biorefineries producing chemicals and fuels from renewable non‐food biomass, allowing us to move towards a bio‐based economy.
Conflict of interest
The authors declare no conflicts of interest.
Microbial Biotechnology (2016) 9(5), 610–617
Funding Information
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science, ICT and Future Planning (MSIP) through the National Research Foundation (NRF) of Korea (NRF‐2012M1A2A2026556, NRF‐2012M1A2A2026557). Martin Gustavsson was additionally supported by the Swedish Research Council Formas project “Microbial conversion of fat from food waste into renewable biochemicals” (Formas 2014‐1620).
References
- Alper, H. , Moxley, J. , Nevoigt, E. , Fink, G.R. , and Stephanopoulos, G.N. (2006) Engineering yeast transcription machinery for improved ethanol tolerance and production. Science 314: 1565–1568. [DOI] [PubMed] [Google Scholar]
- Bassalo, M.C. , Garst, A.D. , Halweg‐Edwards, A.L. , Grau, W.C. , Domaille, D.W. , Mutalik, V.K. , et al (2016) Rapid and efficient one‐step metabolic pathway integration in E. coli . ACS Synth Biol (in press). DOI: 10.1021/acssynbio.5b00187 [DOI] [PubMed] [Google Scholar]
- Blin, K. , Pedersen, L.E. , Weber, T. , and Lee, S.Y. (2016) CRISPy‐web: an online resource to design sgRNAs for CRISPR applications. Synth Syst Biotechnol 1: 118–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonde, M.T. , Pedersen, M. , Klausen, M.S. , Jensen, S.I. , Wulff, T. , Harrison, S. , et al (2016) Predictable tuning of protein expression in bacteria. Nat Methods 13: 233–236. [DOI] [PubMed] [Google Scholar]
- Ceroni, F. , Algar, R. , Stan, G.‐B. , and Ellis, T. (2015) Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods 12: 415–418. [DOI] [PubMed] [Google Scholar]
- Chisti, Y. (2007) Biodiesel from microalgae. Biotechnol Adv 25: 294–306. [DOI] [PubMed] [Google Scholar]
- Choi, S.Y. , Park, S.J. , Kim, W.J. , Yang, J.E. , Lee, H. , Shin, J. , and Lee, S.Y. (2016a) One‐step fermentative production of poly (lactate‐co‐glycolate) from carbohydrates in Escherichia coli . Nat Biotechnol 34: 435–440. [DOI] [PubMed] [Google Scholar]
- Choi, S. , Song, H. , Lim, S.W. , Kim, T.Y. , Ahn, J.H. , Lee, J.W. , et al (2016b) Highly selective production of succinic acid by metabolically engineered Mannheimia succiniciproducens and its efficient purification. Biotechnol Bioeng (in press). DOI: 10.1002/bit.25988 [DOI] [PubMed] [Google Scholar]
- Croughan, M.S. , Konstantinov, K.B. , and Cooney, C. (2015) The future of industrial bioprocessing: batch or continuous? Biotechnol Bioeng 112: 648–651. [DOI] [PubMed] [Google Scholar]
- Doshi, R. , Nguyen, T. , and Chang, G. (2013) Transporter‐mediated biofuel secretion. Proc Natl Acad Sci USA 110: 7642–7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop, M.J. , Dossani, Z.Y. , Szmidt, H.L. , Chu, H.C. , Lee, T.S. , Keasling, J.D. , et al (2011) Engineering microbial biofuel tolerance and export using efflux pumps. Mol Syst Biol 7: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EIA (2016) U.S. ethanol exports exceed 800 million gallons for second year in a row [WWW document]. URL www.eia.gov/todayinenergy/detail.cfm?id=25312.
- FAO (2008) The State of Food and Agriculture 2008 [WWW document]. URL www.fao.org/docrep/011/i0100e/i0100e00.htm.
- Hannon, M. , Gimpel, J. , Tran, M. , Rasala, B. , and Mayfield, S. (2010) Biofuels from algae: challenges and potential. Biofuels 1: 763–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, J.R. , Wright, C.T. , and Kenney, K.L. (2007) Cellulosic biomass feedstocks and logistics for ethanol production. Biofuels Bioprod Bioref 1: 181–190. [Google Scholar]
- Huang, H.J. , Ramaswamy, S. , Al‐Dajani, W. , Tschirmer, U. , and Cairncross, R.A. (2009) Effect of biomass species and plant size on cellulosic ethanol: a comparative process and economic analysis. Biomass Bioenerg 33: 234–246. [Google Scholar]
- Jeffries, T.W. , and Jin, Y.S. (2004) Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol 63: 495–509. [DOI] [PubMed] [Google Scholar]
- Jones, J.A. , Vernacchio, V.R. , Lachance, D.M. , Lebovich, M. , Fu, L. , Shirke, A.N. , et al (2015) ePathOptimize: a combinatorial approach for transcriptional balancing of metabolic pathways. Sci Rep 5: 11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson, L.J. , Alriksson, B. , and Nilvebrant, N.‐O. (2013) Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim, A.S. , and Jewett, M.C. (2016) A cell‐free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab Eng 36: 116–126. [DOI] [PubMed] [Google Scholar]
- Kim, B. , Park, H. , Na, D. , and Lee, S.Y. (2014) Metabolic engineering of Escherichia coli for the production of phenol from glucose. Biotechnol J 9: 621–629. [DOI] [PubMed] [Google Scholar]
- Laluce, C. , Schenberg, A.C.G. , Gallardo, J.C.M. , Coradello, L.F.C. , and Pombeiro‐Sponchiado, S.R. (2012) Advances and developments in strategies to improve strains of Saccharomyces cerevisiae and processes to obtain the lignocellulosic ethanol ‐ A review. Appl Biochem Biotechnol 166: 1908–1926. [DOI] [PubMed] [Google Scholar]
- Lee, S.Y. , and Kim, H.U. (2015) Systems strategies for developing industrial microbial strains. Nature Biotechnol 33: 1061–1072. [DOI] [PubMed] [Google Scholar]
- Lee, K.H. , Park, J.H. , Kim, T.Y. , Kim, H.U. , and Lee, S.Y. (2007) Systems metabolic engineering of Escherichia coli for L‐threonine production. Mol Syst Biol 3: 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.W. , Choi, S. , Park, J.H. , Vickers, C.E. , Nielsen, L.K. , and Lee, S.Y. (2010) Development of sucrose‐utilizing Escherichia coli K‐12 strain by cloning β‐fructofuranosidases and its application for L‐threonine production. Appl Microbiol Biotechnol 88: 905–913. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Lin, Z. , Huang, C. , Zhang, Y. , Wang, Z. , Tang, Y.J. , et al (2015) Metabolic engineering of Escherichia coli using CRISPR‐Cas9 meditated genome editing. Metab Eng 31: 13–21. [DOI] [PubMed] [Google Scholar]
- Lütke‐Eversloh, T. , and Stephanopoulos, G. (2007) L‐tyrosine production by deregulated strains of Escherichia coli . Appl Microbiol Biotechnol 75: 103–110. [DOI] [PubMed] [Google Scholar]
- Molitor, B. , Richter, H. , Martin, M.E. , Jensen, R.O. , Juminaga, A. , Mihalcea, C. , and Angenent, L.T. (2016) Carbon recovery by fermentation of CO‐rich off gases ‐ Turning steel mills into biorefineries. Bioresource Technol 215: 386–396. [DOI] [PubMed] [Google Scholar]
- Moon, H.G. , Jang, Y.‐S. , Cho, C. , Lee, J. , Binkley, R. , and Lee, S.Y. (2016) One hundred years of clostridial butanol fermentation. FEMS Microbiol Lett 363: 1–15. [DOI] [PubMed] [Google Scholar]
- Na, D. , Yoo, S.M. , Chung, H. , Park, H. , Park, J.H. , and Lee, S.Y. (2013) Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol 31: 170–174. [DOI] [PubMed] [Google Scholar]
- Neubauer, P. , and Junne, S. (2016) Scale‐Up and scale‐down methodologies for bioreactors In Bioreactors: Design, Operation and Novel Applications. Mandenius C.‐F. (ed). Chichester: John Wiley & Sons, pp. 323–354. [Google Scholar]
- Nielsen, A.A.K. , Der, B.S. , Shin, J. , Vaidyanathan, P. , Paralanov, V. , Strychalski, E.A. , et al (2016) Genetic circuit design automation. Science 352: aac7341. [DOI] [PubMed] [Google Scholar]
- Park, J.H. , Lee, K.H. , Kim, T.Y. , and Lee, S.Y. (2007) Metabolic engineering of Escherichia coli for the production of L‐valine based on transcriptome analysis and in silico gene knockout simulation. Proc Nat Acad Sci USA 104: 7797–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.H. , Kim, H.U. , Kim, T.Y. , Park, J.S. , Kim, S.‐S. , and Lee, S.Y. (2014) Metabolic engineering of Corynebacterium glutamicum for L‐arginine production. Nat Commun 5: 4618. [DOI] [PubMed] [Google Scholar]
- Pauly, M. , and Keegstra, K. (2008) Cell‐wall carbohydrates and their modification as a resource for biofuels. Plant J 54: 559–568. [DOI] [PubMed] [Google Scholar]
- Pronk, J.T. , Lee, S.Y. , Lievense, J. , Pierce, J. , Palsson, B. , Uhlén, M. , and Nielsen, J. (2015) How to set up collaborations between academia and industrial biotech companies. Nature 33: 237–240. [DOI] [PubMed] [Google Scholar]
- Puigjaner, L. (2011) Introduction In Syngas from Waste, Green Energy and Technology. Puigjaner L. (ed). London: Springer Verlag, pp. 1–9. [Google Scholar]
- Qi, L.S. , Larson, M.H. , Gilbert, L.A. , Doudna, J.A. , Weissman, J.S. , Arkin, A.P. , and Lim, W.A. (2013) Repurposing CRISPR as an RNA‐guided platform for sequence‐specific control of gene expression. Cell 152: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renouf, M.A. , Wegener, M.K. , and Nielsen, L.K. (2008) An environmental life cycle assessment comparing Australian sugarcane with US corn and UK sugar beet as producers of sugars for fermentation. Biomass Bioenerg 32: 1144–1155. [Google Scholar]
- Shin, J.H. , Kim, H.U. , Kim, D.I. , and Lee, S.Y. (2013) Production of bulk chemicals via novel metabolic pathways in microorganisms. Biotechnol Adv 31: 925–935. [DOI] [PubMed] [Google Scholar]
- Tong, Y. , Charusanti, P. , Zhang, L. , Weber, T. , and Lee, S.Y. (2015) CRISPR‐Cas9 based engineering of actinomycetal genomes. ACS Synth Biol 4: 1020–1029. [DOI] [PubMed] [Google Scholar]
- Utrilla, J. , Licona‐Cassani, C. , Marcellin, E. , Gosset, G. , Nielsen, L.K. , and Martínez, A. (2012) Engineering and adaptive evolution of Escherichia coli for D‐lactate fermentation reveals GatC as a xylose transporter. Metab Eng 14: 469–476. [DOI] [PubMed] [Google Scholar]
- Xiao, Y. , Bowen, C.H. , Liu, D. , and Zhang, F. (2016) Exploiting nongenetic cell‐to‐cell variation for enhanced biosynthesis. Nat Chem Biol 12: 339–344. [DOI] [PubMed] [Google Scholar]
- Yim, H. , Haselbeck, R. , Niu, W. , Pujol‐Baxley, C. , Burgard, A. , Boldt, J. , et al (2011) Metabolic engineering of Escherichia coli for direct production of 1,4‐butanediol. Nat Chem Biol 7: 445–452. [DOI] [PubMed] [Google Scholar]
- Zhang, X. , Tervo, C.J. , and Reed, J.L. (2016) Metabolic assessment of E. coli as a biofactory for commercial products. Metab Eng 35: 64–74. [DOI] [PubMed] [Google Scholar]


