Summary
The plant holobiont comprises the plant and its associated microbiota, which interact with each other and determine holobiont functioning and plant performance. We have started to understand the complexity of the involved microorganisms and their interactions, however, we need more research on plant–microbiome interactions to understand holobiont functioning. By 2020 we expect that our knowledge on these interactions will have considerably increased facilitating crop management practices based on the interactions of the plant holobiont.
Already in 1994, Richard Jefferson introduced in a symposium lecture his idea that the evolutionary selection unit is not a single organism but a macro‐organism (e.g. a plant or animal) and all its associated microorganisms that act in consortia (Jefferson, 1994). Consequently, the fitness of plants is determined by the entire suite of genes (the hologenome), consisting of the genome of the host plant as well as the genomes of all epi‐ and endophytes. Jefferson further concluded that “agriculture can only progress sustainably when balanced hologenetic combinations or holo‐alleles are present” (Jefferson, 1994).
Today, more than 20 years after Jefferson's pioneering ideas, research on the plant microbiome is blooming and the importance of the plant microbiome for plant health and development is well acknowledged (Bulgarelli et al., 2013; Hardoim et al., 2015). Those microorganisms that live in the root environment (rhizosphere and root) as well as in seeds have received most attention in this regard (Philippot et al., 2013; Barret et al., 2015; Klaedtke et al., 2016; Lareen et al., 2016). The microbial component of healthy seeds appears to be inherited between plant generations and is likely to represent an important “seed” for the microbiome build‐up, whereas soil and root‐derived microorganisms arrive later and have to compete with the already established microflora. Thus, the seed‐associated microbiota is likely to mediate different functions than root‐associated microbiota, such as effects on germination, early plant establishment and survival. In contrast to the seed microbiota, which consists of a limited range of microbial species (Truyens et al., 2014), the root environment hosts a tremendous microbial diversity due to the fact that root exudates and mucilage attract and select a high number of different soil microorganisms. The rhizosphere also represents the main source of microorganisms, which migrate into plants via the root and establish sub‐communities inside plants (Hardoim et al., 2015). Consequently, the root is a hot spot of plant–microbe interactions and the microbiome of the rhizosphere and root is especially important for plant nutrition, abiotic stress tolerance and defence against pathogen attack (Mitter et al., 2013; Ramírez‐Puebla et al., 2013). Evolution created sophisticated communication systems by which the plant influences the behaviour of microorganisms in the root environment to its own favour. For example, barley plants respond to root infection by the phytopathogenic fungi Pythium ultimum with increased exudation of phenolic and organic acids, which in turn induce the expression of the antifungal phlA in the root‐colonizing bacterium Pseudomonas fluorescens CHA0 (Jousset et al., 2011). Domestication as well as modern crop breeding together with high‐input agricultural practices have counteracted these systems and resulted in a decrease in the diversity of plant beneficial interactions and a reduction of the ability to establish such interactions in modern crop varieties (Pérez‐Jaramillo et al., 2016). In other words, conventional agriculture might have selected for unbalanced hologenetic combinations. For the successful re‐integration of microbial functions in agronomic management, we need to better understand the functioning of the holobiont plant and in particular the interactions of the plant and its microbial components as well as the interactions between different members of the microbial assemblage in and on the plant.
The interactions between the plant and its microbiota can be manifold and the positive effects of microorganisms on plant health and growth can be either directly, e.g. by the production of phytohormones, modulation of ethylene levels in the plant and inhibition of pathogen growth (Mitter et al., 2013), or indirectly, e.g. by inducing changes in the host plant gene activity (Alfano et al., 2007; Pinedo et al., 2015) or changes in microbiome composition (Ardanov et al., 2012, 2016). The plant itself also influences the composition and activity of its associated microbiota. The impact of the plant species, the plant developmental stage as well as the plant physiology on microbial assemblages in and on plants has been intensively studied (Mitter et al., 2013; Lareen et al., 2016) and plant genotype specificity of plant–microbe interactions has often been reported (Da et al., 2012; Vargas et al., 2012; Neiverth et al., 2014). More recently, researchers found out that microorganisms adjust their metabolism to adapt to the plant environment during rhizosphere or root colonization (Shidore et al., 2012; Alquéres et al., 2013; Drogue et al., 2014) or in response to plant stress reactions (Sheibani‐Tezerji et al., 2015a). All these studies point to a lively dialogue between the plant and its associated microorganisms, and in 2020 we would have obtained better understanding on the genetic determinants involved. For example, there are reports showing that inoculation with plant beneficial bacteria do not induce defence events commonly found after microbial attack in plants (Bordiec et al., 2011). But, how can the plants immune system discriminate between pathogenic and beneficial microorganisms? Or how can we explain that certain plant beneficial microorganisms speed up growth in many different genetically not related plant species but can show astonishing strong differences in their effect on different varieties within one plant species, i.e. Paraburkholderia (formerly Burkholderia) phytofirmans PsJN in potato (Da et al., 2012). To explore the full potential of plant–microbe partnerships for sustainable plant production, we need a better understanding of the plant genetic mechanisms that determine the plant's ability to interact with beneficial microorganisms. More systemic studies employing multidisciplinary research teams will allow us to reach this goal. By 2020, we should be able to provide molecular markers predicting beneficial plant–microbe interactions for plant breeding programmes with the aim to create the best plant–microbiome associations for optimum plant performance.
Numerous studies report on the complexity of plant microbiomes comprising multiple fungal and bacterial members (reviewed by Vorholt, 2012; Philippot et al., 2013; Hardoim et al., 2015) including pathogens and mutualists. It is obvious that such complex microbiomes are characterized by a dense network of interactions between individual members, which are still poorly understood. Few studies have provided evidence on such interactions not only between the plant host and its microbiome but also between different microorganisms playing a detrimental role on microbiome functioning. Known is the interaction between symbionts such as arbuscular mycorrhizal fungi (AMF) and rhizobia leading in combination to greater plant productivity than when applied alone (Larrimer et al., 2014; van der Heijden et al., 2016). Furthermore, representatives of Mollicutes and “Candidatus Glomeribacter” were shown to live in hyphae and spores of AMF (Bonfante and Anca, 2009; Naumann et al., 2010) and contribute to colonization and formation of the mycorrhizal structures in plant roots (Garbaye, 1994; Frey‐Klett et al., 2007). Several studies reported interactions between AMF and fungal endophytes including competition or antagonism (Eschen et al., 2010; Wearn et al., 2012), with alkaloids or allelopathic compounds produced by the fungal endophytes potentially interfering with AMF (Antunes et al., 2008; Mack and Rudgers, 2008). Another interesting example of multitrophic interactions is the presence of endohyphal bacteria in filamentous fungal endophytes (Hoffman and Arnold, 2010), resulting in altered functioning. For example, the endohyphal bacterium Luteibacter sp. was found to greatly enhance indole‐3‐acetic acid (IAA) production of the fungal endophyte, although the bacterium does not show IAA production when grown alone (Hoffman and Arnold, 2008). Well known is also the interaction between the phytopathogen Rhizopus microsporus, the causal agent of rice seedling blight, and its interaction with the endofungal bacterium Burkholderia endofungorum, which is responsible for toxin production (Partida‐Martinez and Hertweck, 2005). It furthermore has been demonstrated that viruses interact with plant‐associated microbiota. The geothermal grass Dichanthelium lanuginosum as well as its fungal endophyte Curvularia protuberata can tolerate temperatures of 40°C when grown separately, but in symbiosis, the combination of the host plant and the fungal endophyte infected with a virus can tolerate soil temperatures as high as 65°C (Márquez et al., 2007; Rodriguez et al., 2008).
These examples demonstrate how important it will be to shed more light on the interplay between microbiome members with regard to interacting with the plant, and a multitude of yet unknown interactions impacting plant growth and health are still to be expected. A better understanding on the type of interactions, the mechanisms and signalling cascades involved will be applicable in different directions. Defined microbial consortia comprising bacteria as well as fungi may be designed to provide a specific set of functions. Understanding the interactions between pathogens or pests with other plant‐associated microbiota may lead to novel avenues in plant disease prevention (Massart et al., 2015), which are urgently needed to reduce pesticide input and to overcome the development of resistance.
By 2020, we should have acquired better knowledge on the ecology and functioning of plant–microbiome interactions in different plant compartments. Of main interest are the root environment and reproductive organs such as seeds (Fig. 1). Our understanding on which microbial traits are needed to preferentially colonize seeds, roots or other plant tissues is still limited. One bottleneck is that (cultivation‐independent) microbiome analysis is mostly based on the analysis of 16S rRNA genes, which provides information at the species or genus level. However, various important characteristics with regard to interaction with the plant such as plant tissue or plant genotype preferences, mutualism or competitive ability are manifested at the strain rather than at the species level. This becomes evident when investigating different isolated strains belonging to the same species (Idris et al., 2004; Sheibani‐Tezerji et al., 2015b). Therefore, it will be important that in 2020, we will have strain‐dependent tools available to monitor microbiome members, and to have a better understanding on the determinants for establishment in a particular tissue. Current sequencing efforts, genome comparison, improved tools in genome annotation, combined with classical microbial genetics and detailed functional analysis will lead to advanced knowledge on which microorganisms will perform particular plant growth‐promoting functions and will thrive/compete in a particular environment. From an application point of view, this information will lead to tailored microbial applications for crop enhancement, which will improve the reliability and field success of such applications. Depending on the target microbial trait (e.g. improvement of seed germination or antagonism of soil‐borne pathogens), it will be possible to select appropriate strains showing these functions with the desired plant genotype(s), to establish well in the target environment (e.g. in/on seeds for seed germination, in the root environment for pathogen antagonism), and to also express the target traits in the relevant environment.
Figure 1.
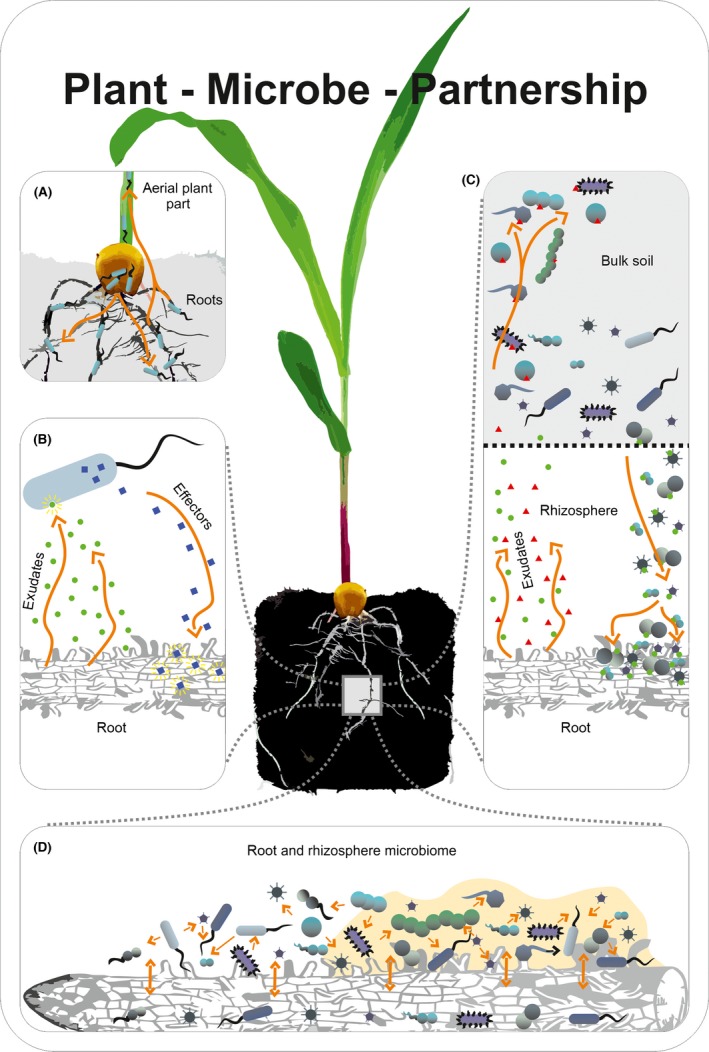
Close‐up view of plant–microbe partnership. (A) Plants are colonized initially by microbes originating from the seed. This seed‐derived microbiota is complemented and partly substituted gradually by rhizosphere microorganisms migrating into the plant via roots. Plant–microbe partnerships occur at different levels of complexity. (B) The plant interacts with single organisms. It responds to the presence of a microbe and its metabolites and vice versa the microbe is affected by the plant environment and reacts to plant metabolism and physiology. (C) The plant interacts with the microbiota in the soil and rhizosphere. Plant exudates attract microbes in the soil thereby directing a subset of them to the root zone. In turn, the activity of the microbiota in the root zone has strong impact on plant growth and health. (D) The microorganisms within the root and rhizosphere microbiota dynamically interact with each other and the microbiota in the root.
Our understanding of higher organisms is changing fundamentally. Plants are no longer understood as monogenetic individuals but as polygenetic entities, in which the microbiota play a central role in fitness, adaption and diversification of the holobiont (Bordenstein and Theis, 2015). By 2020, we should have developed a better understanding of the functioning of the holobiont plant on the basis of fundamental knowledge on plant–microbiome interactions. This will allow realizing Jefferson's vision of a sustainable agriculture based on balanced hologenetic combinations in future.
Conflict of interest
Authors have no conflict of interest to declare.
Microbial Biotechnology (2016) 9(5), 635–640
Funding Information
This work was supported by a grant provided by the FWF (Austrian Science Foundation) (grant P26203‐B22) to A.S.
References
- Aleklett, K. , and Hart, M. (2013) The root microbiota – a fingerprint in the soil? Plant Soil 370: 671–686. [Google Scholar]
- Alfano, G. , Lewis Ivey, M.L. , Cakir, C. , Bos, J.I.B. , Miller, S.A. , Madden, L.V. , et al (2007) Systemic modulation of gene expression in tomato by Trichoderma hamatum 382. Phytopathology 97: 429–437. [DOI] [PubMed] [Google Scholar]
- Alquéres, S. , Meneses, C. , Rouws, L. , Rothballer, M. , Baldani, I. , Schmid, M. , and Hartmann, A. (2013) The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol. Plant‐Microb. Interact. 26: 937–945. [DOI] [PubMed] [Google Scholar]
- Antunes, P.M. , Miller, J. , Carvalho, L.M. , Klironomos, J.N. , and Newman, J.A. (2008) Even after death the endophytic fungus of Schedonorus phoenix reduces the arbuscular mycorrhizas of other plants. Funct Ecol 22: 912–918. [Google Scholar]
- Ardanov, P. , Sessitsch, A. , Häggman, H. , Kozyrovska, N. , and Pirttilä, A.M. (2012) Methylobacterium‐induced endophyte community changes correspond with protection of plants against pathogen attack. PLoS ONE 7: e46802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardanov, P. , Lyastchenko, S. , Karppinen, K. , Häggman, H. , Kozyrovska, N. , and Pirttilä, A.M. (2016) Effects of Methylobacterium sp. on emergence, yield, and disease prevalence in three cultivars of potato (Solanum tuberosum L.) were associated with the shift in endophytic microbial community. Plant Soil [In press]. doi:10.1007/s11104‐015‐2500‐y. [Google Scholar]
- Barret, M. , Briand, M. , Bonneau, S. , Préveaux, A. , Valière, S. , Bouchez, O. , et al (2015) Emergence shapes the structure of the seed microbiota. Appl Environ Microbiol 81: 1257–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante, P. , and Anca, I.‐A. (2009) Plants, mycorrhizal fungi, and bacteria: a network of interactions. Annu Rev Microbiol 63: 363–383. [DOI] [PubMed] [Google Scholar]
- Bordenstein, S.R. , and Theis, K.R. (2015) Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol 13: e1002226. doi:10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordiec, S. , Paquis, S. , Lacroix, H. , Dhondt, S. , Ait Barka, E. , Kauffmann, S. , et al (2011) Comparative analysis of defence responses induced by the endophytic plant growth‐promoting rhizobacterium Burkholderia phytofirmans strain PsJN and the non‐host bacterium Pseudomonas syringae pv. pisi in grapevine cell suspensions. J Exp Bot 62: 595–602. [DOI] [PubMed] [Google Scholar]
- Bulgarelli, D. , Schlaeppi, K. , Stijn Spaepen, S. , van Themaat, E.V.L. , and Schulze‐Lefert, P. (2013) Structure and functions of the bacterial microbiota of plants. Ann. Rev. Plant Biol. 64: 807–838. [DOI] [PubMed] [Google Scholar]
- Da, K. , Nowak, J. , and Flinn, B. (2012) Potato cytosine methylation and gene expression changes induced by a beneficial bacterial endophyte, Burkholderia phytofirmans strain PsJN. Plant Physiol Biochem 50: 24–34. [DOI] [PubMed] [Google Scholar]
- Drogue, B. , Sanguin, H. , Borland, S. , Prigent‐Combaret, C. , and Wisniewski‐Dyé, F. (2014) Genome wide profiling of Azospirillum lipoferum 4B gene expression during interaction with rice roots. FEMS Microbiol Ecol 87: 543–555. [DOI] [PubMed] [Google Scholar]
- Eschen, R. , Hunt, S. , Mykura, C. , Gange, A.C. , and Sutton, B.C. (2010) The foliar endophytic fungal community composition in Cirsium arvense is affected by mycorrhizal colonization and soil nutrient content. Fungal Biol. 114: 991–998. [DOI] [PubMed] [Google Scholar]
- Frey‐Klett, P. , Garbaye, J. , and Tarkka, M. (2007) The mycorrhiza helper bacteria revisited. New Phytol 176: 22–36. [DOI] [PubMed] [Google Scholar]
- Garbaye, J. (1994) Helper bacteria ‐ a new dimension to the mycorrhizal symbiosis. New Phytol 128: 197–210. [DOI] [PubMed] [Google Scholar]
- Hardoim, P.R. , van Overbeek, L.S. , Berg, G. , Pirttilä, A.M. , Compant, S. , Campisano, A. , et al (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79: 293–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden, M.G.A. , de Bruin, S. , Luckerhoff, L. , van Logtestijn, R.S.P. , and Schlaeppi, K. (2016) A widespread plant‐fungal‐bacterial symbiosis promotes plant biodiversity, plant nutrition and seedling recruitment. ISME J 10: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, M.T. , and Arnold, A.E. (2008) Geographic locality and host identity shape fungal endophyte communities in cupressaceous trees. Mycol Res 112: 331–344. [DOI] [PubMed] [Google Scholar]
- Hoffman, M.T. , and Arnold, A.E. (2010) Diverse bacteria inhabit living hyphae of phylogenetically diverse fungal endophytes. Appl Environ Microbiol 76: 4063–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idris, R. , Trifonova, R. , Puschenreiter, M. , Wenzel, W.W. , and Sessitsch, A. (2004) Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense . Appl Environ Microbiol 70: 2667–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R. (1994) The Hologenome. Agriculture, Environment and the Developing World: A Future of PCR. Cold Spring Harbor, NY: Cold Spring Harbor or in fiull Cold Spring Harbor Laboratory Press. [Google Scholar]
- Jousset, A. , Rochat, L. , Lanoue, A. , Bonkowski, M. , Keel, C. , and Scheu, S. (2011) Plants respond to pathogen infection by enhancing the antifungal gene expression of root‐associated bacteria. Mol. Plant‐Microbe Interact. 24: 352–358. [DOI] [PubMed] [Google Scholar]
- Klaedtke, S. , Jacques, M.‐A. , Raggi, L. , Préveaux, A. , Bonneau, S. , Negri, V. , et al (2016) Terroir is a key driver of seed‐associated microbial assemblages. Environ Microbiol 18: 1792–1804. [DOI] [PubMed] [Google Scholar]
- Lareen, A. , Burton, F. , and Schäfer, P. (2016) Plant root‐microbe communication is shaping root microbiomes. Plant Mol Biol 90: 575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrimer, A.L. , Clay, K. , and Bever, J.D. (2014) Synergism and context dependency of interactions between arbuscular mycorrhizal fungi and rhizobia with prairie legumes. Ecology 95: 1045–1054. [DOI] [PubMed] [Google Scholar]
- Mack, K.M.L. , and Rudgers, J.A. (2008) Balancing multiple mutualists: asymmetric interactions among plants, arbuscular mycorrhizal fungi, and fungal endophytes. Oikos 117: 310–320. [Google Scholar]
- Márquez, L.M. , Redman, R.S. , Rodriguez, R.J. , and Roossinck, M.J. (2007) A virus in a fungus in a plant: three‐way symbiosis required for thermal tolerance. Science 315: 513–515. [DOI] [PubMed] [Google Scholar]
- Massart, S. , Martinez‐Medina, M. , and Jijakli, H. (2015) Biological control in the microbiome era: challenges and opportunities. Biol Control 89: 98–108. [Google Scholar]
- Mitter, B. , Brader, G. , Afzal, M. , Compant, S. , Naveed, M. , Trognitz, F. , and Sessitsch, A. (2013) Advances in elucidating beneficial interactions between plants, soil and bacteria. Adv. Agronomy. 121: 381–445. [Google Scholar]
- Naumann, M. , Schussler, A. , and Bonfante, P. (2010) The obligate endobacteria of arbuscular mycorrhizal fungi are ancient heritable components related to the Mollicutes. ISME J 4: 862–871. [DOI] [PubMed] [Google Scholar]
- Neiverth, A. , Delai, S. , Garcia, D.M. , Saatkamp, K. , Souza, E.M. , Pedrosa, F.O. , et al (2014) Performance of different wheat genotypes inoculated with the plant growth promoting bacterium Herbaspirillum seropedicae . Eur. J. Soil Biol. 64: 1–5. [Google Scholar]
- Partida‐Martinez, L.P. , and Hertweck, C. (2005) Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature 437: 884–888. [DOI] [PubMed] [Google Scholar]
- Pérez‐Jaramillo, J. , Mendes, R. , and Raaijmakers, J.M. (2016) Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol 90: 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot, L. , Raaijmakers, J.M. , Lemanceau, P. , and van der Putten, W.H. (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11: 789–799. [DOI] [PubMed] [Google Scholar]
- Pinedo, I. , Ledger, T. , Greve, M. , and Poupin, M.J. (2015) Burkholderia phytofirmans PsJN induces long‐term metabolic and transcriptional changes involved in Arabidopsis thaliana salt tolerance. Front. Plant Sci. 6: 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez‐Puebla, S.T. , Servín‐Garcidueñas, L.E. , Jiménez‐Marín, B. , Bolaños, L.M. , Rosenblueth, M. , Martínez, J. , et al (2013) Gut and root microbiota commonalities. Appl Environ Microbiol 79: 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, R.J. , Henson, J. , van Volkenburgh, E. , Hoy, M. , Wright, L. , Beckwith, F. , et al (2008) Stress tolerance in plants via habitat‐adapted symbiosis. ISME J 2: 404–416. [DOI] [PubMed] [Google Scholar]
- Sheibani‐Tezerji, R. , Rattei, T. , Sessitsch, A. , Trognitz, F. , and Mitter, B. (2015a) Transcriptome profiling of the endophyte Burkholderia phytofirmans PsJN indicates sensing of the plant environment and drought stress. mBio 6: e00621–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheibani‐Tezerji, R. , Naveed, M. , Jehl, M.‐A. , Sessitsch, A. , Rattei, T. , and Mitter, B. (2015b) The genomes of closely related Pantoea ananatis maize seed endophytes having different effects on the host plant differ in secretion system genes and mobile genetic elements. Front Microbiol 6: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shidore, T. , Dinse, T. , Öhrlein, J. , Becker, A. , and Reinhold‐Hurek, B. (2012) Transcriptomic analysis of response to exudates reveals genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ Microbiol 14: 2775–2787. [DOI] [PubMed] [Google Scholar]
- Truyens, S. , Weyens, N. , Cuypers, A. , and Vangronsveld, J. (2014) Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7: 40–50. [Google Scholar]
- Vargas, L. , de Carvalho, T.L.G. , Ferreira, P.C.G. , Baldani, V.L.D. , Baldani, J.I. , and Hemerly, A.S. (2012) Early responses of rice (Oryza sativa L.) seedlings to inoculation with beneficial diazotrophic bacteria are dependent on plant and bacterial genotypes. Plant Soil 356: 127–137. [Google Scholar]
- Vorholt, J. (2012) Microbial life in the phyllosphere. Nat Rev Microbiol 10: 828–840. [DOI] [PubMed] [Google Scholar]
- Wearn, J.A. , Sutton, B.C. , Morley, N.J. , and Gange, A.C. (2012) Species and organ specificity of fungal endophytes in herbaceous grassland plants. J Ecol 100: 1085–1092. [Google Scholar]


