Abstract
Growing axons develop a highly motile structure at their tip, termed the growth cone. The growth cone contacts extracellular environmental cues to navigate axonal growth. Netrin, slit, semaphorin, and ephrins are known guidance molecules that can attract or repel axons upon binding to receptors and co-receptors on the axon. The activated receptors initiate various signaling molecules in the growth cone that alter the structure and movement of the neuron. Here, we describe the detailed protocol for a stripe assay to assess the ability of a guidance molecule to attract or repel neurons. In this method, dissociated hippocampal neurons from E15.5 mice are cultured on laminin-coated dishes processed with alternating stripes of ectodomain of fibronectin and leucine-rich transmembrane protein-2 (FLRT2) and control immunoglobulin G (IgG) fragment crystallizable region (Fc) protein. Both axons and cell bodies were strongly repelled from the FLRT2-coated stripe regions after 24 h of culture. Immunostaining with tau1 showed that ~90% of the neurons were distributed on the Fc-coated stripes compared to the FLRT2-Fc-coated stripes (~10%). This result indicates that FLRT2 has a strong repulsive effect on these neurons. This powerful method is applicable not only for primary cultured neurons but also for a variety of other cells, such as neuroblasts.
Keywords: Neuroscience, Issue 112, Axon guidance molecules, repulsion, attraction, FLRT, stripe assay, neurons, substrate
Introduction
Axon guidance is the process by which newly formed neurons send axons to their target during development of the nervous system1,2. Developing axons carry a highly motile structure at their tip called the growth cone. The growth cone senses extracellular cues to navigate the axon's path. Guidance molecules, such as slit, semaphorin, and ephrins, can attract or repel axons depending on their interaction with suitable receptors and co-receptors on the axon1,3,4. The activated receptors transfer signals to the growth cone that affect its cytoskeletal organization for axon and growth-cone movements.
Various methods have been developed to evaluate the action of attractant and repellent molecules. Chemo-attractants and repellents can be administered into the growth/culture medium with a gradient concentration (e.g., Dunn's chamber or µ-slides)5,6, in a highly concentrated spot by micro-pipette (e.g., turning assay)7 or at a homogenous concentration by bath application (e.g., growth cone collapse assay)8,9.
Other methods include a stripe assay or microcontact printing (µCP), in which a chemo-attractant or repellent is coated on the surface of a plate as a substrate10-12. Thestripe assay was originally developed by Bonhoeffer and colleagues in 1987 to analyze topographical mapping in the chick retino-tectal system13. The original method required a vacuum system to coat proteins onto polycarbonate nucleopore membranes using striped and meshed matrices. In later versions, the recombinant proteins were directly printed on the surface of a culture plate in a striped pattern using narrow slit silicon matrices14,15. Recently, various research groups have successfully applied this stripe assay to the analysis of axon guidance molecule activities16-21.
Here, we present the detailed protocol for a stripe assay that measures the attraction or repulsion of axon guidance molecules for dissociated hippocampal neurons. Notably, this method can be applied in minimally equipped laboratory settings. For this assay, alternating stripes of a fluorescently labeled substrate and a control protein are generated on a plastic dish using a silicon matrix with 90-µm slits and coated with laminin. In our demonstration, dissociated hippocampal neurons from E15.5 mice were cultured on alternating stripes of recombinant ectodomain of fibronectin and leucine-rich transmembrane protein-2 (FLRT2) and control Fc protein21. After 24 h of culture, both the axons and cell bodies of the neurons were strongly repelled from the FLRT2 stripes. Staining with an anti-Tau1 antibody revealed that ~90% of the neurons were distributed on the Fc-coated regions, compared to ~10% on the FLRT2-Fc, indicating that FLRT2 has a strong repulsive function for hippocampal neurons21.
Protocol
Procedures involving animal subjects have been approved by the Institutional Animal Care and Use Committee at Hamamatsu University School of Medicine.
1. Preparation of Matrices
Boil 4-8 silicone matrices in a microwave or on a hot plate for 5 min and allow them to dry completely for 1 h under laminar flow (striped side up). Note: The following procedures should be performed under laminar flow.
Blow compressed air or use transparent sticky tape to remove any dust from the striped side of the matrices. Keep the striped side clean so it can firmly adhere to the plate surface.
Carefully place the matrices onto 6-cm plastic dishes by pressing with a finger (one matrix per dish; stripe side down). Avoid getting air bubbles between the matrix and the dish. Mark the location of the stripes on the bottom side of the culture dish.
If the matrix fails to attach, repeat from step 1.2.
2. Stripe Generation and Laminin Coating for Neuron Culture
Prepare fluorescently labeled recombinant protein (25 µl each for injection [step 2.2] or 100 µl each for the placement method [step 2.2.1]) by mixing the Fc-tagged protein (10-50 µg/ml) and a fluorescent dye-conjugated anti-human Fc antibody (1 to 3 ratio: 30-150 µg/ml) in phosphate buffered saline (PBS). Then, incubate the mixture for 30 min at RT.
- Using a 22-G syringe, inject 25 µl of the recombinant protein prepared in the previous step (2.1) through the small hole on the side of the matrix (arrows in Figure 1A, 1C, and 1E).
- Alternatively, place 100 µl of the recombinant protein into the top slit of the matrix and aspirate approximately half of the solution from the small hole (arrows in Figure 1A, 1C, and 1E), so that the recombinant protein remains in the striped regions.
Incubate the dish for 30 min at 37 °C in a cell culture incubator.
Place 300 µl of PBS into the top slit (Figure 1B) and aspirate from the small hole located on the side of the matrix (arrows in Figure 1A, C and E) to remove unattached recombinant proteins. Do not aspirate the PBS completely, because the proteins will dry. Repeat this step twice. (3 times total)
Carefully remove the matrix and immediately place ~100 µl of control protein (10-50 µg/ml; Fc) to cover the entire striped area, creating an alternate coating. Then, incubate the dish for 30 min at 37 °C in the cell culture incubator.
After washing the dish surface three times with PBS, coat it with ~100 µl of 20 µg/ml laminin in PBS. Note: Laminin should be thawed on ice to prevent solidification.
Incubate the dish for 1 h at 37 °C in the cell culture incubator.
Wash the dish surface three times with PBS and add culture medium. Place the coated dish with culture medium in a 5% CO2 incubator at 37 °C until use.
3. Culturing Hippocampal Neurons from E15.5 Mouse Embryos
Euthanize mother via cervical dislocation and isolate three E15.5 embryos.
Cut the skin and skull sagittally at the midline of the head with scissors, and remove the brain using a small spoon. Transfer the brain carefully to a 60-mm Petri dish containing 3 ml of Hank's Balanced Salt Solution (HBSS).
Using a scalpel, cut out the hemispheres including cortex, hippocampus and striatum (Figure 2A). Then, dissect out the hippocampal region by carefully removing the meninges, and transfer the dissected hippocampal tissues to a 15-ml centrifuge tube containing 2 ml of HBSS solution, on ice (Figure 2B-C). Collect 6 hippocampi from 3 embryos in the tube, which are sufficient for culturing neurons in 8 dishes.
Replace the HBSS solution with trypsin/EDTA solution (2 ml) and incubate the tube at 37 °C for 15 min in a water bath. Note: Aspirate HBSS without centrifugation and do not decant the HBSS solution by tilting the tube.
Neutralize the trypsin activity by adding 500 µl of fetal bovine serum (FBS).
Centrifuge the mixture at 100 x g for 5 min. Remove the supernatant and wash the pellet with 2 ml of culture medium. Repeat this step once more.
Pipet the hippocampi up and down 10 times in culture medium using a 1,000-µl tip with a large hole (cut-tip) to dissociate the tissue.
Change to a regular (uncut) 1,000-µl tip and pipet the dissociated tissue up and down to obtain a single-cell suspension.
Separate the single cells by passing them through an 80-100-µm mesh cell strainer.
Centrifuge the filtered single cell suspension at 150 x g for 5 min.
Remove the supernatant by aspirating, and resuspend the pellet (neurons) in 2 ml of culture medium.
Count the cells using a hemocytometer with trypan blue staining to determine the density and viability. Then, plate 10,000 neurons in 150 µl of culture medium for each set of stripes.The suspension should be carefully placed to cover the entire stripe region.
4. Visualization of Cell Bodies and Axons by Immunofluorescence Staining using an Anti-Tau1 Antibody
After 24 h of culture, aspirate the medium without disturbing the adherent cells, and fix the neurons with pre-warmed 4% paraformaldehyde (PFA)/4% sucrose/PBS at RT for 15 min.
Wash the plate surface 3 times with PBS. If desired, use a water-repellant pen to draw a circle around the striped region to decrease the amount of antibody needed.
Permeabilize the neurons with 0.3% Triton X-100/PBS for 5 min.
Incubate the neurons with blocking solution containing 3% donkey serum and 0.1% Triton X-100 for 30 min followed by incubation with an anti-Tau1 antibody (1:200) in 1% donkey serum/0.1% Triton X-100/PBS O/N at 4 °C.
Wash the plate surface 3 times with PBS. Incubate the neurons with a fluorophore-conjugated anti-mouse IgG antibody in 1% bovine serum albumin (BSA)/0.1% Triton X-100/PBS for 30 min.
Wash the plate surface 3 times with PBS. Cover the striped region with an 18 mm x 18 mm cover glass using 50 µl of antifade solution. avoid air bubbles, which interfere with the imaging.
Obtain fluorescent images with a fluorescence microscope using the appropriate system, e.g., x10 objective lens, filter sets for GFP and RFP, and 1,000-msec exposure time (Figure 3).
Representative Results
Dissociated hippocampal neurons from E15.5 mice were plated and cultured for 24 h on stripes of fluorescently labeled control Fc (Figure 3A-C) or FLRT2-Fc (Figure 3D-F) alternating with non-labeled control Fc. In both cases, the neurons were aggregated and extended their axons as bundles. On the control Fc/Fc stripes, the neurons were distributed evenly on the fluorescently labeled and non-labeled stripes, and they extended their axons in random directions (Figure 3A-C). In contrast, when cultured on FLRT-2Fc/Fc stripes, the axons avoided growing on the FLRT2-Fc regions. Thus, the extending axons were mainly located on the control Fc (Figure 3D-F). Notably, both the cell bodies and axons were repelled from the FLRT2-Fc. Thus, the axons extended in a direction parallel to the stripes. Figure 3D''-F'' shows that the axons grew along the border between control Fc and FLRT2-Fc but did not extend into the FLRT2 territory.
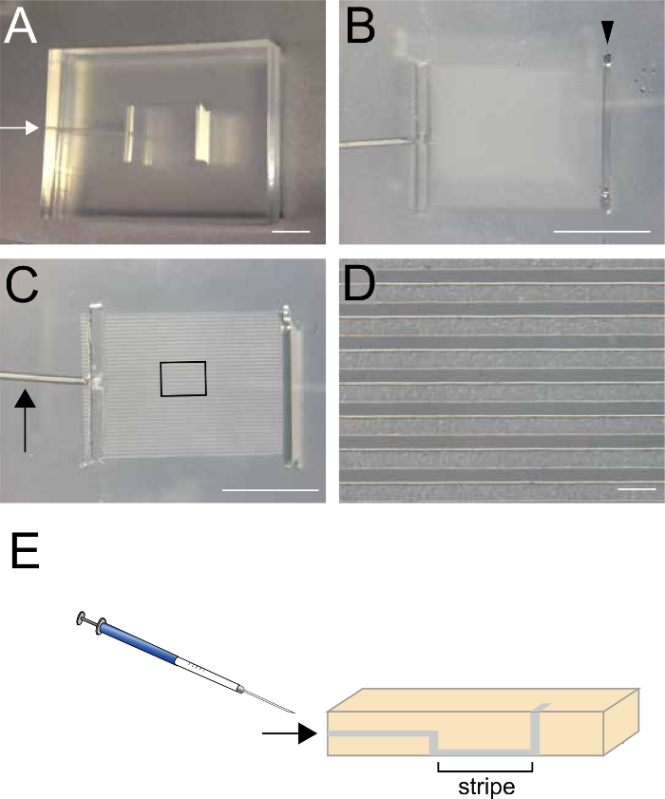 Figure 1. Silicon Matrix used to create the striped protein stamp. (A, B) Top view of the silicon matrix. The size of matrix is 30 mm x 25 mm x 5mm. White arrow in A indicates a small hole where the recombinant protein is injected (step 2.2). Arrowhead in B indicates a slit where the recombinant protein is alternatively applied (step 2.2.1). (C, D) Bottom view of the silicon matrix. The size of the striped region is 7 mm x 9 mm. The width of each stripe is 90 µm. Arrow indicates a small canal through which the recombinant protein is injected. (E) Schematic Illustration of the sagittal view at the midline of the matrix showing the connection between the small hole, the stripes, and the slit. Arrow indicates the injection hole. Scale bars are 500 µm in A-C and 200 µm in D. Please click here to view a larger version of this figure.
Figure 1. Silicon Matrix used to create the striped protein stamp. (A, B) Top view of the silicon matrix. The size of matrix is 30 mm x 25 mm x 5mm. White arrow in A indicates a small hole where the recombinant protein is injected (step 2.2). Arrowhead in B indicates a slit where the recombinant protein is alternatively applied (step 2.2.1). (C, D) Bottom view of the silicon matrix. The size of the striped region is 7 mm x 9 mm. The width of each stripe is 90 µm. Arrow indicates a small canal through which the recombinant protein is injected. (E) Schematic Illustration of the sagittal view at the midline of the matrix showing the connection between the small hole, the stripes, and the slit. Arrow indicates the injection hole. Scale bars are 500 µm in A-C and 200 µm in D. Please click here to view a larger version of this figure.
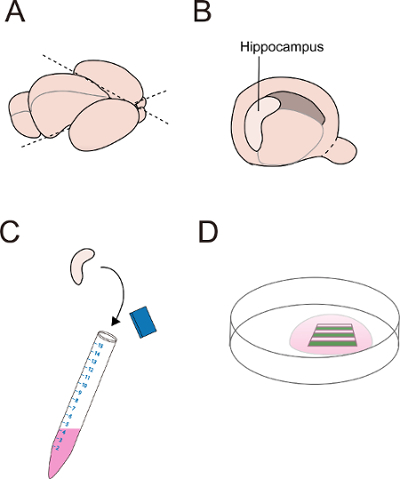 Figure 2. Dissection of mouse hippocampus. (A) Top view of mouse embryonic brain showing the cutting position (dashed line) on the isolated hemispheres including the hippocampus, the cortex, and the striatum. (B, C) The isolated hippocampus is collected in HBSS after removing the meninges. (D) Neurons are cultured on the stripes in a 6-cm dish. Please click here to view a larger version of this figure.
Figure 2. Dissection of mouse hippocampus. (A) Top view of mouse embryonic brain showing the cutting position (dashed line) on the isolated hemispheres including the hippocampus, the cortex, and the striatum. (B, C) The isolated hippocampus is collected in HBSS after removing the meninges. (D) Neurons are cultured on the stripes in a 6-cm dish. Please click here to view a larger version of this figure.
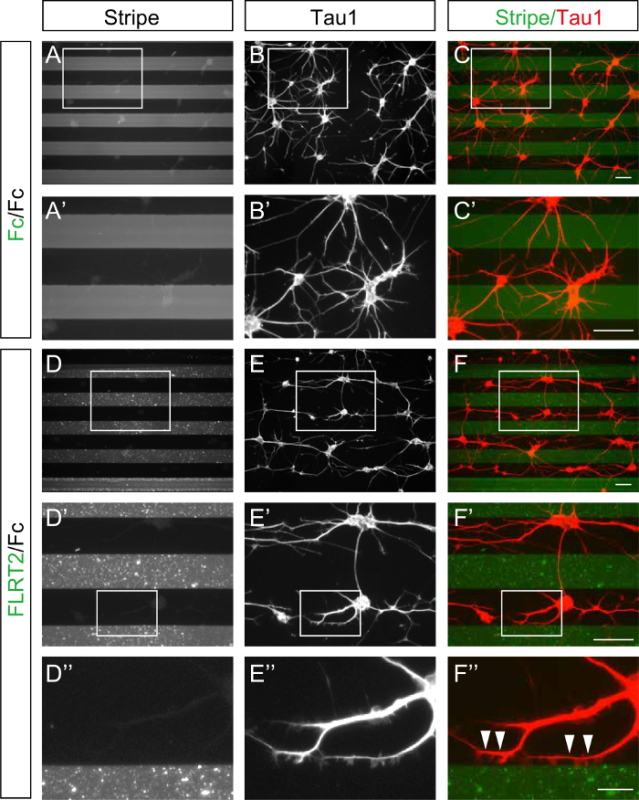 Figure 3. Stripe assays with control Fc and FLRT2-Fc and a dissociated culture of hippocampal neurons. Dissociated hippocampal neurons from E15.5 mice were cultured for 24 h on stripes of control Fc (A-C) or FLRT2-Fc (D-F). (A'-C', D'-F', D''-F'') Enlarged images from the boxed regions in (A-C, D-F, and D'-F', respectively). (A, A') Stripes generated using fluorescently labeled Fc (gray) and Fc (black) as a control experiment. (D, D', D'') Fluorescently labeled FLRT2-Fc stripes (gray) alternating with Fc (black). (B, B', E, E', E'') Anti-Tau1 antibody staining of the cell bodies and axons of the hippocampal neurons. (C, C', F, F', F'') The images of the stripes and the anti-Tau1 staining were merged. The neurons' cell bodies and axons were strongly repelled from the FLRT2-Fc (F, F', F''). Note that axons turn along the border and do not enter the FLRT2-Fc territory (arrowheads in F'').Scale bars are 100 µm in C, C', F, F' and 25 µm in F''. Please click here to view a larger version of this figure.
Figure 3. Stripe assays with control Fc and FLRT2-Fc and a dissociated culture of hippocampal neurons. Dissociated hippocampal neurons from E15.5 mice were cultured for 24 h on stripes of control Fc (A-C) or FLRT2-Fc (D-F). (A'-C', D'-F', D''-F'') Enlarged images from the boxed regions in (A-C, D-F, and D'-F', respectively). (A, A') Stripes generated using fluorescently labeled Fc (gray) and Fc (black) as a control experiment. (D, D', D'') Fluorescently labeled FLRT2-Fc stripes (gray) alternating with Fc (black). (B, B', E, E', E'') Anti-Tau1 antibody staining of the cell bodies and axons of the hippocampal neurons. (C, C', F, F', F'') The images of the stripes and the anti-Tau1 staining were merged. The neurons' cell bodies and axons were strongly repelled from the FLRT2-Fc (F, F', F''). Note that axons turn along the border and do not enter the FLRT2-Fc territory (arrowheads in F'').Scale bars are 100 µm in C, C', F, F' and 25 µm in F''. Please click here to view a larger version of this figure.
Discussion
This protocol describes a stripe assay that uses recombinant protein and dissociated neurons from the E15.5 mouse hippocampus. This assay allows the efficient observation of repulsive, attractive, or neutral responses of neurons to a recombinant protein of interest placed in a striped pattern. A major advantage of this protocol is the simple method for generating the stripes, in which the protein is directly printed onto the surface of a plastic dish, compared to the traditional method, which requires special matrices, a vacuum system, and a nucleopore membrane20,21. To make the protein stripes, the labeled recombinant protein can be injected through a small hole, or a larger sample (~100 µl) can be placed in the slit on top of the matrix, followed by the careful aspiration of excess buffer and unbound proteins through the hole.
Another advantage of this method is that a large number of neurons can be visualized in a single experiment, and many growth cones can be analyzed simultaneously21. In contrast, in a conventional turning assay, only a single neuron and single growth cone is visualized. The repulsive molecule FLRT2 is sensed by neurons, which migrate away from FLRT2-Fc stripes21. A reporter molecule like green fluorescent protein (GFP) or red fluorescent protein (RFP) can be expressed in the neurons to visualize them against the carpet color and allow real-time observation without the need for immunostaining. Other tags like FLAG, Myc, and His can be used instead of the Fc tag, and identified with the appropriate fluorescently labeled anti-tag antibody (step 2.1). Some high hydrophobic proteins cannot make proper stripe because of self-aggregation. Furthermore, the relative action of attraction and repulsion between two different target proteins can be compared by substituting a second target protein for the control Fc stripe20.
To increase the viability of the neurons, adjustments in the enzymatic digestion time and trypsin inhibition may be needed. If the survival ratio of neurons is low, dilute the concentration of trypsin or shorten the enzymatic digestion time. The humidity level of the incubator should be maintained to avoid evaporation-induced osmotic pressure in the complete system throughout the culture period.
The adhesion of the matrix to the dish is an important factor for preparing good stripes. Improper adherence to the dish surface results in a disorganized stripe pattern. However, the density of cells and concentration of substrate are also critical for a successful experiment. A higher protein concentration in the stripe is recommended for analyzing a molecule whose repulsive or attractive activity is unknown. Although we have demonstrated this protocol using dissociated primary neurons, it is also applicable for organotypic explant cultures15.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (23700412, 25122707 and 26670090 to S.Y.).
References
- Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298(5600):1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Klein R. Signaling from axon guidance receptors. Cold Spring Harb Perspect Biol. 2010;2(5):001941. doi: 10.1101/cshperspect.a001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Nishiyama M, Henley J, Tessier-Lavigne M, Poo M. Calcium signalling in the guidance of nerve growth by netrin-1. Nature. 2000;403(6765):93–98. doi: 10.1038/47507. [DOI] [PubMed] [Google Scholar]
- Ming G-l, Henley J, Tessier-Lavigne M, Song H-j, Poo M-m. Electrical Activity Modulates Growth Cone Guidance by Diffusible Factors. Neuron. 2001;29(2):441–452. doi: 10.1016/s0896-6273(01)00217-3. [DOI] [PubMed] [Google Scholar]
- Dudanova I, et al. Genetic evidence for a contribution of EphA:ephrinA reverse signaling to motor axon guidance. J Neurosci. 2012;32(15):5209–5215. doi: 10.1523/JNEUROSCI.5707-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BQ, Geng ZH, Ma L, Geng JG. Slit2 regulates attractive eosinophil and repulsive neutrophil chemotaxis through differential srGAP1 expression during lung inflammation. J Immunol. 2010;185(10):6294–6305. doi: 10.4049/jimmunol.1001648. [DOI] [PubMed] [Google Scholar]
- Ly A, et al. DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin-1. Cell. 2008;133(7):1241–1254. doi: 10.1016/j.cell.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Kaibuchi K, Inagaki S, Yamashita T. Unc5B associates with LARG to mediate the action of repulsive guidance molecule. J Cell Biol. 2009;184(5):737–750. doi: 10.1083/jcb.200807029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea J, et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron. 2005;47(4):515–528. doi: 10.1016/j.neuron.2005.06.029. [DOI] [PubMed] [Google Scholar]
- von Philipsborn AC, Lang S, Jiang Z, Bonhoeffer F, Bastmeyer M. Substrate-Bound Protein Gradients for Cell Culture Fabricated by Microfluidic Networks and Microcontact Printing. Sci Signal. 2007. [DOI] [PubMed]
- Jackman R, Wilbur J, Whitesides G. Fabrication of submicrometer features on curved substrates by microcontact printing. Science. 1995;269(5224):664–666. doi: 10.1126/science.7624795. [DOI] [PubMed] [Google Scholar]
- Mrksich M, Whitesides GM. Using self-assembled monolayers to understand the interactions of man-made surfaces with proteins and cells. Annu Rev Biophys Biomol Struct. 1996;25:55–78. doi: 10.1146/annurev.bb.25.060196.000415. [DOI] [PubMed] [Google Scholar]
- Walter J, Kern-Veits B, Huf J, Stolze B, Bonhoeffer F. Recognition of position-specific properties of tectai cell membranes by retinal axons in vitro. Development. 1987;101:685–696. doi: 10.1242/dev.101.4.685. [DOI] [PubMed] [Google Scholar]
- Knoll B, Weinl C, Nordheim A, Bonhoeffer F. Stripe assay to examine axonal guidance and cell migration. Nat Protoc. 2007;2(5):1216–1224. doi: 10.1038/nprot.2007.157. [DOI] [PubMed] [Google Scholar]
- Weschenfelder M, Weth F, Knoll B, Bastmeyer M. The stripe assay: studying growth preference and axon guidance on binary choice substrates in vitro. Methods Mol Biol. 2013;1018:229–246. doi: 10.1007/978-1-62703-444-9_22. [DOI] [PubMed] [Google Scholar]
- Seiradake E, et al. Structure and functional relevance of the Slit2 homodimerization domain. EMBO Rep. 2009;10(7):736–741. doi: 10.1038/embor.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt C, Bastmeyer M, Weth F. Balancing of ephrin/Eph forward and reverse signaling as the driving force of adaptive topographic mapping. Development. 2012;139(2):335–345. doi: 10.1242/dev.070474. [DOI] [PubMed] [Google Scholar]
- Atapattu L, et al. Antibodies binding the ADAM10 substrate recognition domain inhibit Eph function. J Cell Sci. 2012;125:6084–6093. doi: 10.1242/jcs.112631. (Pt 24) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark DA, Karvas RM, Siegel AL, Cornelison DD. Eph/ephrin interactions modulate muscle satellite cell motility and patterning. Development. 2011;138(24):5279–5289. doi: 10.1242/dev.068411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiradake E, et al. FLRT Structure: Balancing Repulsion and Cell Adhesion in Cortical and Vascular Development. Neuron. 2014;84(2):370–385. doi: 10.1016/j.neuron.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi S, et al. FLRT2 and FLRT3 act as repulsive guidance cues for Unc5-positive neurons. EMBO J. 2011;30(14):2920–2933. doi: 10.1038/emboj.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]


