Abstract
Therapeutic delivery of small interfering RNA (siRNA) is a major challenge that limits its potential clinical application. Here, a pH-sensitive cholesterol–Schiff base–polyethylene glycol (Chol–SIB–PEG)-modified cationic liposome–siRNA complex, conjugated with the recombinant humanized anti-EphA10 antibody (Eph), was developed as an efficient nonviral siRNA delivery system. Chol–SIB–PEG was successfully synthesized and confirmed with FTIR and 1H-NMR. An Eph–PEG–SIB–Chol-modified liposome–siRNA complex (EPSLR) was prepared and characterized by size, zeta potential, gel retardation, and encapsulation efficiency. Electrophoresis results showed that EPSLR was resistant to heparin replacement and protected siRNA from fetal bovine serum digestion. EPSLR exhibited only minor cytotoxicity in MCF-7/ADR cells. The results of flow cytometry and confocal laser scanning microscopy suggested that EPSLR enhanced siRNA transfection in MCF-7/ADR cells. Intracellular distribution experiment revealed that EPSLR could escape from the endo-lysosomal organelle and release siRNA into cytoplasm at 4 hours posttransfection. Western blot experiment demonstrated that EPSLR was able to significantly reduce the levels of MDR1 protein in MCF-7/ADR cells. The in vivo study of DIR-labeled complexes in mice bearing MCF-7/ADR tumor indicated that EPSLR could reach the tumor site rather than other organs more effectively. All these results demonstrate that EPSLR has much potential for effective siRNA delivery and may facilitate its therapeutic application.
Keywords: siRNA, cationic liposome, pH sensitive, endosomal escape, anti-EphA10 antibody, gene silencing
Introduction
Cancer is a major public health problem in many parts of the world, and existing chemotherapeutic drugs are far from perfect with undesirable severe side effects, low bioavailability, or development of drug resistance.1 Worldwide, the number of clinical trials in gene therapy has increased so as to overcome serious genetic disorders, such as cancers and other types of monogenic disorders. It has been reported that RNA interference is a powerful technique that has been regarded as a potential therapeutic option for silencing target genes in various diseases. Currently, several synthetic RNAs are in clinical trials for macular degeneration,2 kidney injury,3 respiratory infection,4 and cancer.5 Small interfering RNA (siRNA) acts in a very specific manner inhibiting specific protein expression. Maximum therapeutic efficacy of siRNA can be obtained if the siRNAs are delivered to the site of action.6,7 However, a variety of physiological, cellular, and immunological barriers hinder siRNA molecules from reaching their target site.8 Physiological barriers are composed of endothelial barrier, degradation by nucleases, reticuloendothelial system uptake, etc. siRNA therapeutics also need to overcome various cellular barriers for their intracellular entry, endosomal escape, and effective protein knockdown.9 Besides this, hydrophilicity and negative charge of siRNAs are also obstacles for their intracellular delivery.
To resolve these problems, recent developments in nanotechnology have raised exciting opportunities for the design and formulation of nonviral delivery systems for siRNA therapeutics. Various liposomes, polymer micelles, and nanoparticles can be used as nonviral delivery vectors for siRNA.10 Among these vectors, cationic liposomes-based delivery systems are the most explored and promising siRNA delivery systems because of their suitable physicochemical properties and biocompatibility. However, poor serum stability and short circulation time limited the further development of cationic liposomes.
To prolong the circulation time of cationic liposomes, modification with polyethylene glycol (PEG)-conjugated lipid is preferentially employed.11,12 Although attaching PEG on the surface of liposomes presents various advantages, PEGylation hinders the interaction between liposomes and targeted cells, and may bring about lysosomal escape failure13 – referred to as the PEG dilemma.14 To circumvent this problem, labile linkages have been introduced between the hydrophilic PEG and the hydrophobic moiety15 (eg, cholesterol), which is degradable only upon exposure to relatively acidic,16 enzymatic,17 or oxidoreductive18 condition.19 Among these responsive bonds, pH-sensitive bond was mostly researched because it is independent of cellular chemical substances and does not require the exact location of tumors for triggered release.20 It is well known that the physiological pH in cancer cells is lower compared to that in blood and normal tissues, and it is about 6.0 in early endosomes, and reduces to 5.0 during the progression to late endosomes and lysosomes.21 pH-sensitive PEG-lipid can be degraded even in weak acid, in favor of endosomal escape, followed by cytoplasmic release of the liposome-incorporated drug.22 Ketal,23 vinyl ester,24 orthoester,25 hydrazone,26 and Schiff base27,28 are several examples of pH-degradable bonds that are hydrolyzed relatively rapidly at pH 6.0 but stable at neutral conditions. Schiff base is stable under neutral pH but releases drug in acidic environments.29 Recently, we reported on the production of a pH-sensitive PEG-lipid, cholesterol–Schiff base PEG (Chol–SIB–PEG) to modify doxorubicin-loaded liposome for hepatocarcinoma therapy.30 In this study, Chol–SIB–PEG was synthesized with some modification to develop an efficient siRNA delivery system.
To further enhance the transfection efficiency of PEGylated cationic liposomes, it is a good choice to develop immunoliposomes conjugated with antibody.31 Eph receptor A10, the most recent addition to the largest subfamily of receptor tyrosine kinases, is a valuable breast cancer marker that is highly expressed in breast cancer tissue in comparison to other tissues (mRNA level in testis).32 Anti-EphA10, which plays a role in tumor progression and metastasis,33 could be used as the target site for breast carcinoma therapy.34 Anti-EphA10 antibody (Eph) preferentially binds to anti-EphA10 expressed in breast cancer cells, and so when conjugated to liposomes, it enables a preferential targeting ability to cancer cells following systemic administration.
Here, we developed an approach to improve the therapeutic efficacy of siRNA-targeting MDR1 gene (MDR1-siRNA) in female breast doxorubicin-resistant MCF-7/ADR cells within one multifunctional nanoassembly carrier. The carrier is composed of siRNA–liposomes complexes coated with Eph conjugated with Chol–SIB–PEG, which is shown in Figure 1. We hypothesized that the delivery system remains relatively stable in blood system, with internalization mediated by anti-EphA10 antibody upon arrival at tumor sites, resulting in siRNA release into the cytoplasm silencing gene expression after PEG-lipid degradation and siRNA endosomal/lysosomal escape. To evaluate the hypothesis, liposome–siRNA complexes (LR), PEGylated LR (PSLR), and PSLR-conjugated anti-EphA10 antibody (EPSLR) were prepared and their physicochemical characteristics investigated. The transfection efficiency and cytotoxicity of vectors were determined on MCF-7/ADR cell line. Endosomal escape efficacy of the vectors was also investigated by confocal laser scanning microscopy (CLSM), and gene silencing was assessed using Western blot. In vivo targeting ability of EPSLR on tumor cells was evaluated using MCF-7/ADR xenograft nude mice. Our results suggest that the successful delivery and gene silencing of siRNA in vitro and targeting ability in vivo can be achieved using EPSLR.
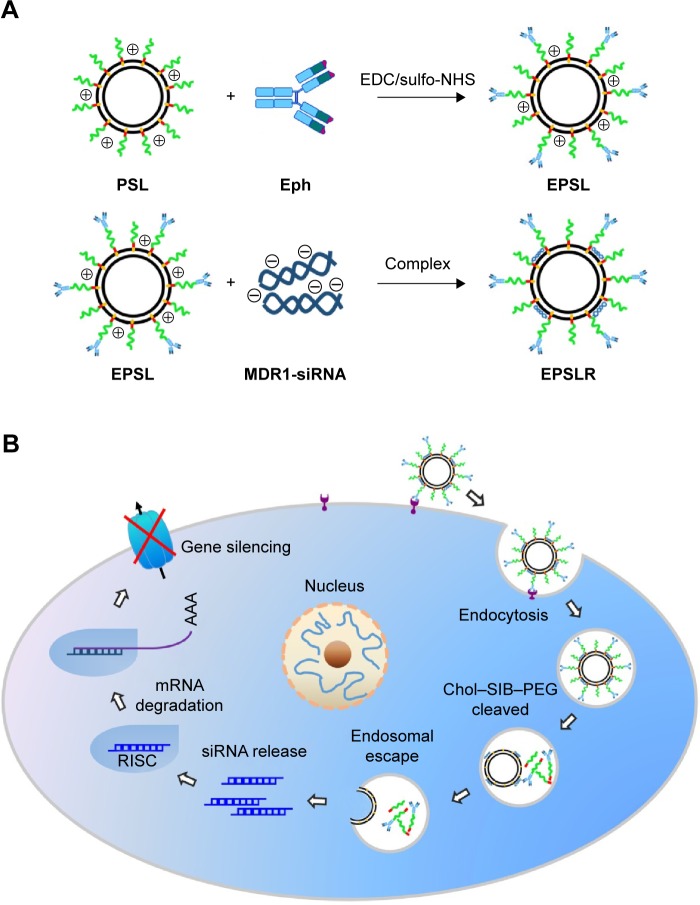
Figure 1.
A schematic diagram showing (A) the formation of EPSLR complexes and (B) the intracellular trafficking for siRNA delivery.
Notes: EPSLR is associated with MCF-7/ADR cells followed by cell internalization by anti-EphA10-mediated endocytosis. Chol–SIB–PEG–Eph degraded in acidic endosome and liposome–siRNA complexes subsequently escaped from endosomes through fusion interaction. Lastly, the siRNA was released into cytoplasm and formed RISC gene expression.
Abbreviations: LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody; RISC, RNA-induced silencing complex; EDCI, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydro chloride; sulfo-NHS, N-Hydroxysulfosuccinimide sodium salt; Eph, anti-EphA10 antibody; PSL, PEGylated lipsomes; EPSL, anti-EphA10 antibody coated PSL; Chol-SIB-PEG, cholesterol–Schiff base–polyethylene glycol.
Materials and methods
Materials
Para-hydroxybenzaldehyde was purchased from Meilan Co., Ltd. (Shanghai, People’s Republic of China); cholesteryl choroformate was obtained from J&K Scientific Ltd. (Karlsruhe, Germany); phenylenediamine was purchased from Bill Chemical Products Co., Ltd. (Zhengzhou, People’s Republic of China); HOOC–PEG2000–COOH was from Seebio Biotech (Shanghai, People’s Republic of China); and N,N-diisopropylethylamine (DIPEA), 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo [4,5-b] pyridinium 3-oxide hexafluorophosphate (HATU), and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDCI) were purchased from GL Biochem Ltd. (Shanghai, People’s Republic of China).
Soybean phospholipid (PC) was purchased from Shanghai Tywei Pharmacy Co. Ltd. (Shanghai, People’s Republic of China). The 1,2-Dioleoyl-3-trimethylammonium-propane (DOTAP), 1,2-dioleoyl-sn-glycero-3-phosphoethanol-amine (DOPE), and cholesterol were from A.V.T. (Shanghai, People’s Republic of China). Anti-EphA10 antibody was purchased from Bioss Biotechnology Co. Ltd. (Beijing, People’s Republic of China). Doxorubicin hydrochloride was from Beijing Huafeng United Technology Co. Ltd. (Beijing, People’s Republic of China). RPMI 1640 and fetal bovine serum (FBS) were purchased from Gibco (Thermo Fisher Scientific, Waltham, MA, USA). MDR1 rabbit mAb, β-actin rabbit mAb, and HRP-conjugated goat anti-rabbit IgG were obtained from Cell Signal Technology (Danvers, MA, USA). DIR, sulfo-NHS, EDCI, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St Louis, MO, USA). LysoTracker Red was obtained from Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA). FAM-labeled siRNA (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense, 5′-ACGUGACACGUUCGGAGAATT-3′), MDR1-siRNA (sense, 5′-CACCCAGGCAAUGAUGUAUTT-3′; antisense, 5′-AUACAUUGCCUGGGUGTT-3′), and scramble siRNA (NC; sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense, 5′-ACGUGACACGUUCGGAGAATT-3′) were synthesized by GenePharma (Shanghai, People’s Republic of China). All other reagents used were of analytical grade and were used without purification.
Human breast carcinoma doxorubicin-resistant MCF-7/ADR cells were provided by KeyGen Biotech Co. Ltd. (Nanjing, People’s Republic of China). The cells were maintained in RPMI 1640 medium containing 20% FBS (Gibco, Thermo Fisher Scientific) and 1% penicillin–streptomycin in a humidified atmosphere with 5% CO2 at 37°C.35 To maintain their multidrug resistance, MCF-7/ADR cells were cultured in a medium containing 1 μg/mL of doxorubicin.
Synthesis of Chol–SIB–PEG
To synthesize the Chol–SIB–PEG, para-hydroxybenzaldehyde and para-phenylenediamine were used to constitute pH-sensitive Schiff base bond. The synthesis of Schiff base bond can be divided into two steps:
Synthesis of Chol–CHO. Cholesteryl chloroformate was reacted with para-hydroxybenzaldehyde (molar ratio =2:1) in anhydrous dichloromethane (DCM) at room temperature under argon in the presence of N-ethyldiisopropylamine (DIPEA) for about 4 hours. After thin-layer chromatography showed the disappearance of para-hydroxybenzaldehyde, the reaction mixture was poured into distilled water and extracted using DCM. The extract was evaporated under vacuum. The residue was purified in a silica-gel chromatography column (DCM:MeOH =1:1) to get Chol–CHO.
Chol–CHO and para-phenylenediamine (molar ratio =1:2) were reacted in methylbenzene with gentle stirring at 120°C in an oil bath overnight. The mixture was evaporated under vacuum followed by dissolving in DCM and then purifying by silica-gel chromatography. The collected solution was evaporated under vacuum, and the crude product (Chol–SIB–NH2) was used in subsequent steps without further purification.
For the synthesis of the Chol–SIB–PEG conjugate, HOOC–PEG2000–COOH and Chol–SIB–NH2 (molar ratio =1.5:1) were reacted in DCM with gentle stirring at room temperature in the presence of HATU, DIPEA, and EDCI for 48 hours. The crude mixture was washed with distilled water and extracted with DCM, which was then removed using a rotary evaporator. After drying under vacuum, the residue was redissolved in chloroform and filtered again to purify the product. The Chol–SIB–PEG was obtained by removing chloroform under vacuum.
The chemical structure of Chol–SIB–PEG was determined and confirmed by 1H-NMR (Bruker ARX-300, Bruker Optik GmbH, Ettlingen, Germany) and FTIR (Bruker vector 22, Bruker Optik GmbH).
Preparation of EPSLR nanocomplexes
Liposomes were prepared by the modified film dispersion–hydration method described by Qiang.36 Briefly, the mixture of PC, DOPE, DOTAP, cholesterol, and Chol–SIB–PEG in the weight ratio of 2:2:4:2:1.5 was dissolved in 5 mL of DCM; the solvent was then evaporated at 24°C until a thin film was obtained. Then, thin films were hydrated with 3 mL of distilled water in a 60°C water bath for 20 minutes. The resultant suspension was sonicated for 4 minutes (200 W 2 minutes, 400 W 2 minutes) with a probe (Scientz-1200E, Ningbo, People’s Republic of China) to form unilamellar liposomes and filtered through a 0.22 μm microporous membrane to form Chol–SIB–PEG-modified liposomes (PSL) with a narrow-size distribution. Liposomes were prepared with PC, DOPE, DOTAP, and cholesterol as reference formulations (L).
The conjugation of anti-EphA10 antibody onto PSL was achieved by using a previously described method.37 Briefly, EDCI and sulfo-NHS dissolved in pH 7.4 phosphate-buffered saline (PBS) were added to liposome suspensions. The mixtures were incubated, with constant stirring, at room temperature for 2 hours. Then, 250 μL of anti-EphA10 antibody solution (1 mg/mL) was added and the mixture gently agitated at room temperature for 2 hours; the mixture was incubated at 4°C overnight. PSL-conjugated anti-EphA10 antibody (EPSL) was separated from free antibody by gel filtration on sepharose 4B column equilibrated in PBS.
EPSLR nanocomplexes were prepared with EPSL and siRNA at a series of N/P ratios (molar ratio of DOTAP-nitrogen atoms to siRNA-phosphate). For EPSLR formulation, various aliquots of EPSL solution were diluted to a final volume of 50 μL with RNase-free water; to this, an equal volume of siRNA solution was then added. The mixture was immediately vortexed for 5 minutes and incubated at room temperature for 20–30 minutes to assemble homogeneous EPSLR. As control, liposome-siRNA complexes (LR) and PEGylated LR (PSLR) nanocomplexes were prepared by the same procedure as described earlier.
As for biodistribution of complexes, DIR, a near-infrared fluorescence probe, was loaded into the complexes described previously except that DIR fluorescence dye was dissolved with lipid in DCM during the process of liposome preparation.
Physicochemical characteristics of EPSLR
The mean particle sizes and zeta potential of lipoplexes were determined by the dynamic light scattering (DLS) technique using a Malvern Instruments apparatus (Nano series ZS; Malvern Instruments S.A., Cedex, France) at a 90° scattering angle. The morphology of EPSL was observed on Tecnai G220 Transmission Electron Microscope (transmission electron microscopy [TEM], FEI, Hillsboro, OR, USA). Before visualization, the liposomes were placed on copper grids, dried with warm air, and then negatively stained with 2%(w/v) phosphotungstic acid for 1 minute. Finally, the images were captured with TEM using an accelerated voltage of 120 kV.
Gel retardation assay and encapsulation efficiency
The desired N/P ratios for full complexation of liposomes and siRNA were evaluated by gel electrophoresis in Tris–borate–EDTA buffer (40 mM Tris–HCl, 40 mM borate, and 1 mM EDTA). About 10 μL of lipoplexes in different N/P ratios were loaded onto 2% agarose gel stained with ethidium bromide to test the combining ability between liposomes and siRNA,38 and naked siRNA was used as a positive control. After electrophoresis, the siRNA bands were visualized using the Tannon 2500R system (Shanghai, People’s Republic of China) at 254 nm.
The encapsulation efficiency of lipoplexes was evaluated by using the same method described in the section “gel retardation assay encapsulation efficiency” except that RiboGreen (Thermo Fisher Scientific, Waltham, MA, USA) was used instead of ethidium bromide. The amount of free siRNA not encapsulated in lipoplexes was calculated by correlating band intensities to concentration. The encapsulation efficiency of siRNA was precisely evaluated by quantifying free siRNA from total siRNA. The encapsulation efficiency was calculated using the formula:
| (1) |
where Ctotal is the concentration of total siRNA and Cfree is the concentration of siRNA not encapsulated in lipoplexes.
Stability assay of the siRNA–liposome complexes
The stability of LR, PSLR, and EPSLR was evaluated using polyanion heparin7 and FBS. LR, PSLR, and EPSLR were prepared at N/P ratios of 1, 2, 3, 5, 7, 9, and 14 as in the “Preparation of EPSLR nanocomplexes” section and then incubated with heparin solution (Tianjin Biochem Pharmaceutical Co. Ltd., Tianji, People’s Republic of China) at a heparin/siRNA ratio of 5 (IU/μg) for 20 minutes at room temperature. A 5 μL sample of the mixture was electrophoresed and analyzed on agarose gel.39
For investigating stability in FBS, lipoplexes were incubated with equivoluminal FBS at 37°C, in which the N/P ratio was fixed at 7. Naked siRNA was treated as control at the same time. At predetermined time internals, samples were taken and immediately stored at −20°C. In order to release siRNA from the complexes, excessive heparin sodium solution (heparin/siRNA =20 IU/μg) was added. After another 1 hour of incubation, the samples were analyzed by agarose gel electrophoresis and visualized using the Tannon system to look for intact siRNA.
Cellular uptake studies
For uptake analysis, the cells were cultured in six-well plates containing glass coverslips for 24 hours with a density of 3×105 cells/well. The cells were treated with lipoplexes prepared with FAM-labeled siRNA. After 4 hours of incubation, the cells were fixed with 4% paraformaldehyde followed by staining with Hoechst 33258 to stain the nucleus. The cell imaging was performed by a CLSM (FV1000-IX81; Olympus, Tokyo, Japan).
The quantitative measurement of cellular uptake siRNA was determined using a fluorescence-activated cell sorter (FACS; BD FACSCalibur; BD Biosciences, San Jose, CA, USA). MCF-7/ADR cells were seeded in six-well plates with a density of 1×106 cells/well and incubated overnight to allow adhesion of cells. The original culture medium was then replaced by fresh supplemented medium, and then cells were incubated with FAM-labeled siRNA–liposome complexes, LR, PSLR, and EPSLR-loaded 50 nM FAM-siRNA. After 4 hours of incubation, MCF-7/ADR cells were washed twice with cold PBS and dissociated with 0.25% trypsin–EDTA, and then harvested and resuspended in PBS solution. The cell suspension obtained was further analyzed by FACS. The level of cellular uptake was quantified based on FAM-siRNA fluorescence determined in FL1H, in which a total of 10,000 events were analyzed. For free ligand competition study, cells were coincubated with 10 μg/mL of anti-EphA10 antibody with formulations.
Intracellular distribution
Approximately 3×105 MCF-7/ADR cells were seeded per well (with cover slips). After 24 hours of culture, the original medium was replaced with fresh RPMI 1640 containing FAM-labeled siRNA (100 nM), FAM-labeled siRNA–liposome complexes for 4 hours at 37°C, followed by staining with LysoTracker Red for 30 minutes. The medium was removed, and cells were washed three times with cold PBS and then fixed with 4% paraformaldehyde for 30 minutes. Cell nucleus was stained with 2 μg/mL Hoechst 33258 for 10 minutes. The fluorescence images were captured using a CLSM (FV1000-IX81).
Cytotoxicity assay
The cytotoxicity of complexes was evaluated using the MTT method in MCF-7/ADR cells. MCF-7/ADR cells were seeded in 96-well plates with a density of 10,000 cells per well. The cells were incubated for 24 hours to allow for attachment to the culture vessel before they were washed with prewarmed sterile PBS (pH 7.4), followed by exposure to free siRNA and different siRNA–liposome complexes at different N/P ratios diluted with a culture medium to the same siRNA concentration (50 nM) at 37°C for 24, 48, and 72 hours, separately. Then, cell viability was evaluated using the MTT assay. The amounts of MTT formazan products were analyzed spectrophotometrically at 490 nm using a flash multimode reader (Varioskan™; Thermo Fisher Scientific). The concentrations of all complexes were tested in six replicates. The percentage of cell viability was calculated using the following equation:
| (2) |
where A1 and A2 are the absorbance of control and sample wells, respectively, and A0 is the absorbance of blank wells without samples and cells.
Western blotting analysis of MDR1 expression
To observe the MDR1 gene silencing, MCF-7/ADR cells were seeded in six-well plates at a density of 3×105 and cultivated for 24 hours. Then, cells were transfected with MDR1-siRNA (50 nM)-loaded different formulations. After 4 hours of incubation, the supernatant was discarded, and then cells were incubated with a fresh complete culture medium for 72 hours. MCF-7/ADR cells were lysed in 50 μL of RIPA lysis buffer with 1% PMSF, and the lysates were centrifuged at 14,000 g for 10 minutes at 4°C to gather total protein. Protein extracts were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis under reducing conditions and transferred onto polyvinylidene fluoride films (Invitrogen), which were then allowed to block with 5% skimmed milk for 2 hours. MDR1 and β-actin protein were detected using the MDR1/ABCB1 rabbit monoclonal antibody (1:300; Cell Signal Technology) and the rabbit anti-β-actin antibody at a dilution of 1:1,000, respectively. The secondary antibody was HRP-conjugated goat anti-rabbit IgG (1:2,000). Signals were detected by adding ECL chemical substrates (Thermo Fisher Scientific) according to manufacturer’s instructions.
Biodistribution
Female BALB/c nude mice aged 5–6 weeks were obtained from Beijing HFK Bioscience Co. Ltd. (Beijing, People’s Republic of China). Mice were housed in the SPF II laboratories under natural light/night conditions and allowed free access to food and water. All animal procedures were approved by the Animal study committee of Shenyang Pharmaceutical University and followed the Guide for the Care and Use of Laboratory Animals Published by the National Institutes of Health.
Tumor-bearing mice were established by inoculating a suspension of 1×107 MCF-7/ADR cells (subcutaneously injected) into the right axillary fossa. When the tumor volume reached approximately 50–100 mm3, DIR-loaded LR, PSLR, and EPSLR were administrated via the tail vein of xenograft mice. The distribution of nanocomplexes in MCF-7/ADR tumor-bearing nude mice was analyzed using a near-infrared fluorescence imaging system (Carestream Health, Inc., Rochester, NY, USA). Then, the mice were sacrificed and tumor and the main organs, including heart, liver, spleen, lung, kidney, and brain, removed. Each organ or tumor was immersed in saline solutions followed by measurement of fluorescence intensity.
Statistical analysis
All the experiments were repeated at least three times. Results are presented as mean ± SD. Statistical comparisons were performed using a one-way analysis of variance. Pairwise comparisons between treatments were made using Student’s t-test (two-tailed) at a confidence level of P<0.05 (*) or P<0.01 (**).
Results and discussions
Synthesis and characteristics of Chol–SIB–PEG
To construct siRNA delivery carrier targeting cancer cells, a pH-sensitive cholesterol–PEG derivative (Chol–SIB–PEG) was synthesized according to the procedure illustrated in Figure 2. The terminal acyl chloride group of commercial Chol–COCl was activated with para-hydroxybenzaldehyde to synthesize Chol–CHO. Chol–SIB–NH2 was synthesized by conjugating para-phenylenediamine to the aldehyde reactive group of Chol–CHO through aldimine condensation reaction. The method of the aldimine condensation reaction is well understood, and an important advantage of this method is that the byproducts and excess reactant can be easily separated from the reaction product. Then, the resulting PEG was reacted with Chol–SIB–NH2 to produce a good overall yield of the detachable PEG–lipid (Chol–SIB–PEG). The Chol–SIB–PEG was characterized by FTIR and 1H-NMR. The FTIR data for Chol–SIB–PEG showed several bands at 1,623 cm−1 (ν–CH=N– in Chol–SIB–PEG), 2,889 cm−1 (ν–C–H– in PEG), and 1,110 cm−1 (ν–C–O–C in PEG). 1H-NMR spectra (300 MHz, CDCl3) of Chol–SIB–PEG showed a singlet proton at δ=8.81(1H, s, –CH=N–), 3.71 (t, J=7.0 Hz, –O–CH2CH2– of PEG), and 2.03–1.07 (m, cholesterol) and confirmed the successful synthesis of Chol–SIB–PEG.
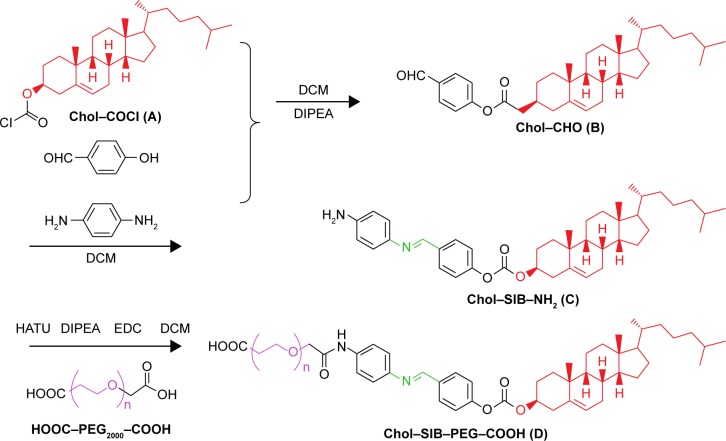
Figure 2.
Synthesis of Chol–SIB–PEG conjugates.
Notes: The synthesis of Chol–SIB–PEG can be divided into three steps. Chol–COCl (A) was reacted with para-hydroxy benzaldehyde in anhydrous dichloromethane at room temperature under argon in the presence of N-Ethyldiisopropylamine to synthesize Chol–CHO (B). (II) Chol–CHO (B) and para-phenylenediamine were reacted in the methylbenzene with gentle stirring at 120°C in oil bath overnight to synthesize Chol–SIB–NH2 (C). Finally, Chol–SIB–PEG (D) was synthesized by conjugating C with HOOC–PEG–COOH in the presence of HATU, DIPEA, and EDC.
Abbreviations: Chol, cholesterol; DIPEA, N,N-diisopropylethylamine; DCM, dichloromethane; HATU, 1-[bis(dimethylamino)methylene]-1H-1,2,3-triazolo [4,5-b]pyridinium 3-oxide hexafluorophosphate; SIB, Schiff base; EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride; PEG, polyethylene glycol.
Construction of siRNA-liposome complexes
This study aimed to develop multifunctional liposomes formulated for enhancing siRNA delivery. The system consisted of the multifunctional components including a core of siRNA–cationic liposome complexes, pH-sensitive-based lipid envelope Chol–SIB–PEG, and active targeting ligand anti-EphA10 antibody. Since first introduced by Felgner, cationic liposomes have been proven to be promising gene delivery systems to cells in culture for gene therapy.40 To pack and deliver siRNA, DOTAP, cholesterol, DOPE, and PC were employed to prepare cationic liposomes, which were used to interact electrostatically with siRNA to form the siRNA–liposome complex. In the composition of liposomes, DOTAP was a major contributor to the surface charge of cationic liposomes, which promoted complete complexation with siRNA by electrostatic interaction.41 DOPE also caused transition of lipid bilayers from lamellar phase to hexagonal phase in low pH, enabling endosomal escape, and therefore cargo was released into cellular plasma.42 PC and cholesterol were basic lipids, which were essential for membrane stability, especially for the incorporation of Chol–SIB–PEG.
Chol–SIB–PEG was added to the bilayers of the positively charged complexes to form a coating on the surface. In this siRNA delivery system, the important lipid was Chol–SIB–PEG, which, because of PEG chains, could prevent aggregation and extend circulation time, and then degrade in acidic pH condition.
Active targeting liposomal formulations were nanoscale vesicles modified with a ligand that can be used as drug-carrying vesicles. Here, we coupled the anti-EphA10 antibody onto carboxyl terminal of Chol–SIB–PEG to prepare the immunoliposomes. Anti-EphA10 antibody was attached on the surface of Chol–SIB–PEG using the primary amines of anti-EphA10 and Chol–SIB–PEG–COOH by amide reaction, and successful conjugation was confirmed with sodium dodecyl sulfate polyacrylamide gel electrophoresis (Figure S1).
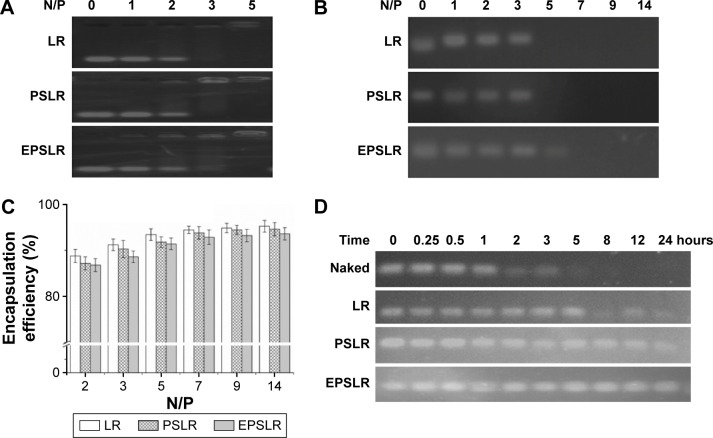
Gel retardation assay
RiboGreen is a sensitive fluorescence nuclear stain for determining RNA concentration (as little as 1 ng/mL) and is generally used for siRNA encapsulation efficiency evaluation.31 Here, siRNA encapsulation efficiency of LR, PSLR, and EPSLR formed at different N/P ratios was detected with the gel retardation assay (Figure 3). As shown in Figure 3C, the encapsulation efficiency of siRNA–liposome complexes was N/P ratio dependent and exhibited increased encapsulation efficiency as N/P ratio increased. When the N/P ratio was above 5, the encapsulation efficiencies of LR, PSLR, and EPSLR were all above 85%. When the N/P ratio increased to 19, the encapsulation efficiency of LR, PSLR, and EPSLR was almost the same at about 94%. Compared with the LR, there were relatively low encapsulation efficiencies of PSLR and EPSLR. It could be due to steric hindrance of PEGylation, electric hindrance, or the adverse effect of antibody modification on encapsulation efficiency.43 The results of the gel retardation assay (Figure 3A) showed that the siRNA in EPSLR was fully complexed at an N/P ratio of 5, which was consistent with the encapsulation efficiency results.
Figure 3.
Encapsulation efficiency and protection by EB (RiboGreen) interaction and gel retardation assay.
Notes: (A) Gel retardation of LR, PSLR, and EPSLR at different N/P ratios; (B) heparin-resistant assay with different N/P ratios; (C) encapsulation efficiency of LR, PSLR, and EPSLR at various N/P ratios; (D) stability in serum of naked siRNA and liposome–siRNA.
Abbreviations: LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody; EB, ethidium bromide; N/P ratios, molar ratio of DOTAP-nitrogen atoms to siRNA-phosphate.
To study the electrostatic interaction of the siRNA with liposomes, EPSLR was subjected to heparin displacement. Heparin is an anionic polysaccharide and a major component of extracellular matrix that can compete with siRNA for binding to disrupt EPSLR stability.44 siRNA displaced by heparin can bind to EB, where fluorescence emission is captured when excited.7 As shown in Figure 3B, no siRNA released from LR or PSLR was observed when the N/P ratio reached 5, whereas a fractional release occurred from EPSLR at this ratio. The results indicated that the interactions between L or PSL and siRNA were stronger than EPSLR, thereby resisting dissociation, and this might be due to the high negative charge on the anti-EphA10 antibody diminishing siRNA–liposome interaction to some degree. However, no siRNA released from EPSLR when the N/P ratio reached 7, which suggested that an increase in the N/P ratio caused greater stability of EPSLR. These results showed that EPSL was not only involved in the formation of EPSLR but also protected the siRNA to some extent. The N/P ratio was fixed at 7 in the following experiments if not mentioned.
In therapeutic application, siRNA is vulnerable to degradation by nucleases.45 We investigated whether complexation with liposome could protect siRNA from degradation in the presence of FBS. The results showed that the amount of intact siRNA decreased along with increase in the time of exposure (Figure 3D). Naked siRNA incubated with FBS was stable within 1 hour and completely degraded after 5 hours (disappearance of band in gel). Meanwhile, serum incubation with LR complexes resulted in a significantly lower siRNA degradation ratio (stable for 5 hours), which was attributed to the siRNA that was only bound to the outer surface of liposomes, and would immediately release and be degraded by nucleases.43 The siRNA in EPSLR and PSLR showed the best integrity, which was visualized even after 24 hours incubation, suggesting that EPSLR and PSLR complexes provided a significant protection for siRNA against RNase degradation compared to LR. The serum stability assay demonstrated that EPSLR could significantly increase siRNA stability, which was likely due to the presence of the PEGylated shell and the anti-EphA10 antibody that sterically hampered the access of nucleases to siRNA.
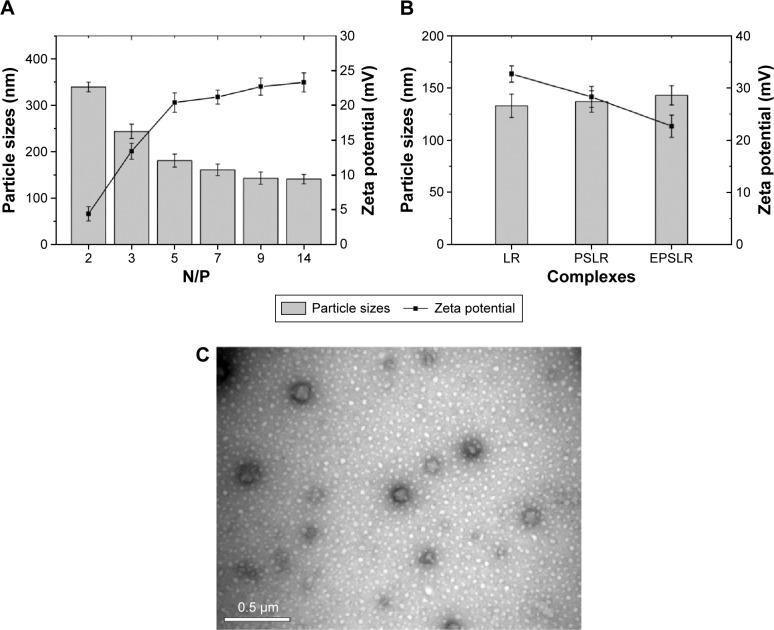
Physicochemical characteristics of lipoplexes
The characteristics of siRNA–liposome complexes on the particle sizes and zeta potential were investigated. Figure 4A shows that the particle size and zeta potential of complexes were N/P ratio dependent. As the N/P ratio increased from 2 to 14, the particle size of EPSLR decreased (from 339.7 to 141.1 nm), whereas the zeta potential increased (from +4.41 to +23.3 mV). This suggested that siRNA was condensed by cationic liposomes and that the condensation efficiency was dependent on the N/P ratio in the formulation, which was consistent with a previous report.46 Similar results were obtained in PSLR and LR (data not shown). As shown in Figure 4B, when the N/P ratio was 9, the diameter increased from 133 nm of PSLR to 143 nm of EPSLR, whereas the zeta potential decreased from +32.7 mV of PSLR to +22.7 mV of EPSLR, which indicated that the particle size and zeta potential of complexes were dependent on the ligand moiety in the vector. In general, the zeta potential of gene-loaded complexes was +20 mV or more. The positive potential could induce nonspecific interactions of these complexes with blood components,47 which could severely reduce the half-life and targeting ability of the complexes. Complexes coated with PEG and ligand can reduce the uptake of particles by the reticuloendothelial system, resulting in a longer circulation in vivo, and this subsequently facilitates tumor targeting.48 In this study, anti-EphA10 antibody-conjugated Chol–SIB–PEG was decorated on complexes to achieve zeta potential reduction and prolonged circulation time.
Figure 4.
Physicochemical characteristics of complexes.
Notes: The particle sizes (columns) and zeta potentials (lines) of EPSLR at different N/P ratios (A) and its reference formulations at an N/P ratio of 9 (B). Morphology of Eph-modified liposomes was characterized with TEM after staining with phosphotungstic acid (C).
Abbreviations: LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody; TEM, transmission electron microscopy; N/P ratios, molar ratio of DOTAP-nitrogen atoms to siRNA-phosphate; Eph, anti-EphA10 antibody.
TEM showed that EPSL were spherical in shape and had a narrow size distribution (Figure 4C). The mean nanoparticle sizes observed by TEM were smaller than that measured by the dynamic light scattering technique because of the shrinkage of nanoparticles owing to the drying process in the TEM preparation.49
In vitro cellular uptake of EPSLR
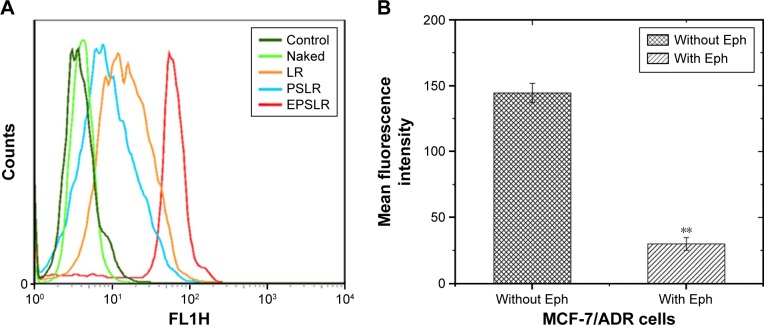
The in vitro cellular uptake of EPSLR was investigated by CLSM and flow cytometry. FAM-labeled siRNA was chosen as a fluorescence probe in place of siRNA. The CLSM study showed that a very weak green fluorescence signal was obtained for siRNA, indicating that only small amounts of siRNA were internalized into the cells (Figure 5A). In contrast, MCF-7/ADR cells treated with LR, PSLR, and EPSLR showed significant internalization as shown by intensive green fluorescence (Figure 5). LR resulted in a higher amount of internalization (Figure 5B) than PSLR (as shown in Figure 5C), indicating that the cellular uptake of PSLR was inhibited in the presence of PEG. The cells treated with EPSLR exhibited intensive green fluorescence, indicating that the EPSLR had significantly increased transfection efficiency in MCF7/ADR cells in comparison to PSLR and even LR (Figure 5D). It has to be noted that the zeta potential of LR was much higher than that of EPSLR, and higher zeta potential would contribute to nonspecific internalization. These results firmly demonstrated that anti-EphA10 conjugation could specifically enhance the binding affinity of EPSLR in breast cancer of anti-EphA10 expression.
Figure 5.
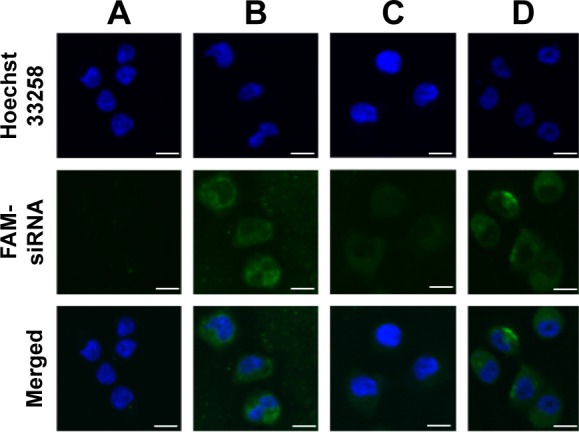
CLSM images of MCF-7/ADR cells incubated with naked siRNA (A), FAM-labeled LR (B), PSLR (C), and EPSLR (D).
Notes: Blue and green colors indicate Hoechst 33258 and FAM-siRNA, respectively. Scale bar =10 μm.
Abbreviations: CLSM, confocal laser scanning microscopy; LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody.
In order to evaluate the cellular uptake of siRNA–liposome complexes, MCF-7/ADR cells were incubated with LR, PSLR, and EPSLR prepared with FAM-labeled siRNA for 4 hours at 37°C. Fluorescence intensity was quantitatively analyzed by flow cytometry (Figure 6A). The amount of cellular uptake of naked siRNA after 4 hours incubation was significantly lower than that of siRNA–liposome complexes. The EPSLR resulted in 4.42-fold higher cellular uptake than PSLR, and thus 1.67-fold higher cellular uptake than the LR after 4 hours incubation, respectively. The cells incubated with PSLR demonstrated less fluorescence intensity in contrast to LR, which indicated that the PEG modification led to a decrease in the complexes–cell interaction as a result of the steric and electrostatic hindrance to entry into the target cell caused by PEG.50 The high amount of cellular uptake of EPSLR in MCF-7/ADR cells may be attributed to the anti-EphA10-mediated targeting, and this result is consistent with the findings of the fluorescence intensity study as shown in Figure 5. Surface-functionalized liposomes with antibodies decreased steric hindrance during targeting, facilitating binding of liposomes to cancer cells.51 Anti-EphA10 is expressed in all subsets of breast cancer but is not expressed in normal breast tissue.52 Nagano et al53 revealed that the administration of an anti-EphA10 antibody significantly suppressed tumor growth in a xenograft mouse model, indicating anti-EphA10 as the target for breast cancer. To improve the targeting ability of loaded siRNA, cationic liposomes can be modified with anti-EphA10 antibody that selectively binds to anti-EphA10 preferentially expressed on the surface of breast cancer cells. The uptake experiments demonstrated that anti-EphA10 antibody modification facilitated siRNA–liposome complexes internalization in MCF-7/ADR cells.
Figure 6.
Flow cytometry measurement of intracellular uptake of FAM-siRNA-loaded complexes (A) and anti-EphA10 antibody competitive assay (B) in MCF-7/ADR cells (P<0.01).
Note: **P<0.01.
Abbreviations: LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody; Eph, anti-EphA10 antibody.
To further investigate the targeting ability of EPSLR, competitive inhibition of free Eph was performed. MCF-7/ADR cells were incubated with EPSLR in medium with or without Eph. The result revealed that uptake of EPSLR without Eph was significantly higher than that with anti-EphA10 (Figure 6B, P<0.01). The aforementioned results indicated that the introduction of anti-EphA10 antibody may greatly enhance the affinity of EPSLR to MCF-7/ADR cells through Eph-mediated endocytosis, which is highly Eph dependent, and free antibody can competitively bind to anti-EphA10 on MCF-7/ADR surface. Although the endocytosis mechanism mediated by Eph is not clear and needs further research, Eph conjugation significantly increased EPSLR accumulation in the breast cancer cells.
Intracellular distribution
To further explore the intracellular distribution of siRNA in EPSLR, subcellular localization of FAM-labeled siRNA was observed using CLSM with respect to LysoTracker Red for lysosomes and Hoechst 33258 for nucleus. As shown in Figure 7A, naked siRNA displayed poor green fluorescence, based on which it is difficult to judge successful endosomal escape. The separation of green and red fluorescence was significant when cells were incubated with LR (Figure 7B); siRNA in LR indicating a successful endosomal escape and an efficient release into cytoplasm. Comparatively, FAM-siRNA formulated in PSLR and EPSLR exhibited a diffuse distribution after 4 hours incubation (Figure 7C and D). The results could be due to the fact that PEG–SIB–Chol or Eph–PEG–SIB–Chol was sensitive to H+ and degraded to remove the PEG or Eph–PEG long chains, contributing to membrane–membrane fusion between liposome and plasma.24 In addition, stronger green fluorescence was found on incubation with EPSLR than PSLR as well as LR. The results were consistent with flow cytometry results, confirming facilitated internalization by Eph.
Figure 7.
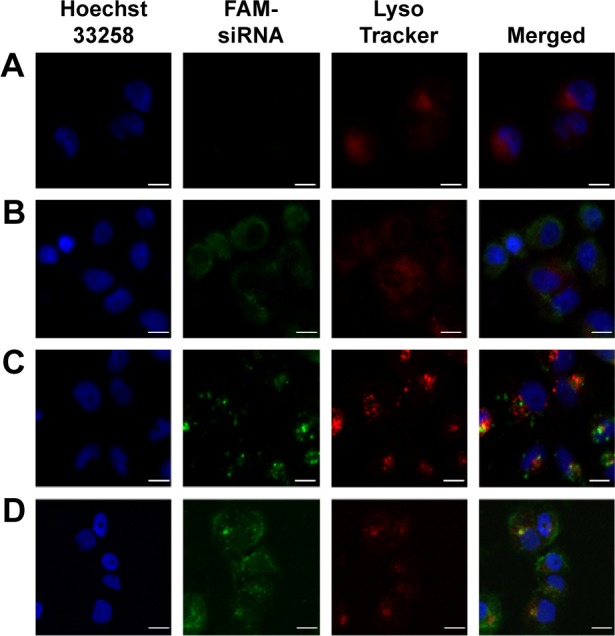
CLSM images of the intracellular distribution of FAM-siRNA.
Notes: For each panel, images from left to right show nuclei stained by Hoechst 33258 (blue), FAM-siRNA (green), lysosome stained with LysoTracker (red) and merged images. MCF-7/ADR cells were incubated with naked siRNA (A), LR (B), PSLR (C), and EPSLR (D) for 4 hours. Scale bar =10 μm.
Abbreviations: CLSM, confocal laser scanning microscopy; LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody.
As endosomal escape is a well-known barrier to efficient nucleic acid delivery, vectors are generally entrapped in endosomes after being internalized, and tend to either recycle their contents back to the cell surface or fuse with the acidic lysosomes later, resulting in sequestration followed by degradation of the cargo plasmid by the lysosomal enzymes, with no access to the cytoplasm or nucleus. As a potential long circulation candidate for siRNA delivery, pH-sensitive PEG–lipid has been proven to assist to reduce liposome surface protein absorption and aggregation to extend half-life and release siRNA into cytoplasm.54 High transfection efficiency does not always mean superior gene silencing activity because enough siRNA escape from endosome/lysosome is required. Upon release of FAM-siRNA from EPSLR into cytoplasm, the cellular fluorescence is expected to increase significantly (Figure 7D). The results confirmed that the novel PEG–cholesterol derivative could degrade in mild acidic environment, assisting in siRNA endosomal escape and release into cytoplasm through membrane fusion between liposome and endosome/lysosome.
Cytotoxicity assay
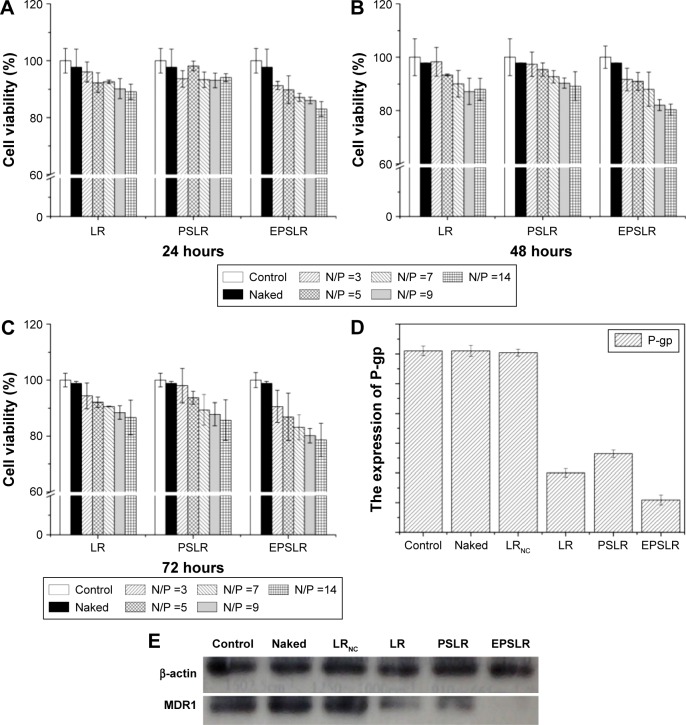
Cytotoxicity is one of the most important issues in biological application of the siRNA delivery system, and cytotoxicity may be related to factors including stabilizer, structure, and zeta potential.55 As for cationic liposomes, there are two factors contributing to cytotoxicity: the positive zeta potential and inherent biotoxicity of ligands conjugation (eg, HER2 antibody). To assess the cytotoxicity of siRNA-loaded complexes, the MTT assay was used to compare the viability of MCF-7/ADR cells after incubation with different formulations (naked siRNA, LR, PSLR, and EPSLR) at different N/P ratios for 24, 48, and 72 hours. As shown in Figure 8A, LR and PSLR did not exhibit significant cytotoxicity at different N/P ratios against MCF-7/ADR cells at 24 hours. However, a significant difference was observed when the N/P ratio was above 9 after 48 hours, indicating that LR and PSLR were slightly more cytotoxic at 48 hours (Figure 8B) and N/P dependent, and the same trend was observed at 72 hours (Figure 8C). In comparison with LR and PSLR, the viability of cells adapted for EPSLR with N/P ratios above 7 in MCF-7/ADR cells reduced significantly after 24 hours incubation. These results indicate that anti-EphA10 antibody-mediated targeting increases the cytotoxicity of EPSLR against MCF-7/ADR cells. It has been reported that anti-EphA10 was specifically expressed in various subtypes of breast cancer tissues, but not within most normal tissues, indicating that anti-EphA10 is a promising drug target potentially useful for breast cancers.53 Therefore, the cytotoxicity of EPSLR is probably attributed to the fact that anti-EphA10 antibody decoration can facilitate EPSLR internalization into carcinoma cells. Furthermore, anti-EphA10 promoted cell proliferation following ligand stimulation. Anti-EphA10 mAb as a therapeutic tool can accumulate in breast tumor tissues and is capable of mediating significant tumor growth suppression both in vitro and in vivo.56 These results confirm that anti-EphA10 modification plays an essential role in increasing the cytotoxicity of EPSLR because of receptor-mediated endocytosis. In addition, no significant difference was found at different time points in MCF-7/ADR cells treated with EPSLR at an N/P ratio of 14 (Figure 8A–C). It is important to note that the cell viability remained at over 75% even after exposure to EPSLR at an N/P ratio of 14 at 72 hours, suggesting that EPSLR can serve as a safe gene vector with low cytotoxicity.
Figure 8.
In vitro cytotoxicity studies of LR, PSLR, and EPSLR.
Notes: Studies of LR, PSLR, and EPSLR for 24 (A), 48 (B), and 72 hours (C) (P<0.05) and Western blot. Quantification of Western blot bands using ImageJ (D). MDR1 silencing mediated by control, naked siRNA, LRNC, LR, PSLR, and EPSLR (E).
Abbreviations: LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody; LRNC, Liposome–negative control siRNA complexes; N/P ratios, molar ratio of DOTAP-nitrogen atoms to siRNA-phosphate; P-gp, P-glycoprotein.
Gene silencing
In order to demonstrate the effectiveness of EPSLR delivery siRNA into MCF-7/ADR cells, the cells were treated with different formulations for 4 hours and MDR1 expression was analyzed using the Western blot assay after 72 hours. As shown in Figure 8D and E, the level of MDR1 protein expression in MCF-7/ADR cells significantly decreased after the delivery of MDR1-siRNA using siRNA–liposome complexes in comparison with naked MDR1-siRNA. This is probably due to the fact that the negative charge and the large molecule weight hamper naked siRNA cross cellular membrane. Upon examination of the primary siRNA delivery system in the study, MDR1-siRNA-loaded EPSLR showed a higher gene-silencing efficiency in MCF-7/ADR cells compared to both MDR1-siRNA-loaded LR and MDR1-siRNA-loaded PSLR at the protein level. It has been proven that cationic liposomes have great potential for siRNA delivery. It is a promising strategy to deliver therapeutic agents using nanoparticles conjugated with antibody for tumor-specific antigens in tumor therapy,57 and in vitro targeting ability experiments suggested that anti-EphA10 antibody conjugation is highly efficient in facilitating the cellular uptake of siRNA in MCF-7/ADR cells. Furthermore, Chol–SIB–PEG as a pH-sensitive PEG–lipid has reportedly been used to develop a pH-sensitive liposome for cancer therapy.30 As a result of these advantages coupled with the ability to escape from endosome/lysosome (Figure 7), EPSLR achieved more efficient MDR1 gene silencing. Besides, MDR1 protein expression in LR was lower than that of PSLR, which further confirmed that PEG-modified nanoparticles hinder its endocytosis into cells. It is noteworthy that there was no obvious change in the expression of MDR1 protein in MCF-7/ADR cells treated with LRNC in comparison to control, suggesting that gene silencing was attributed to a specific sequence of siRNA inducing mRNA degradation. Gene silencing experiments suggested that EPSLR was significantly internalized with the assistance of anti-EphA10 antibody, escaped from endo-lysosome and released siRNA into cellular plasma, and finally siRNA formed RNA-induced silencing complex (RISC) and silenced target gene.
In vivo tumor-targeting of EPSLR
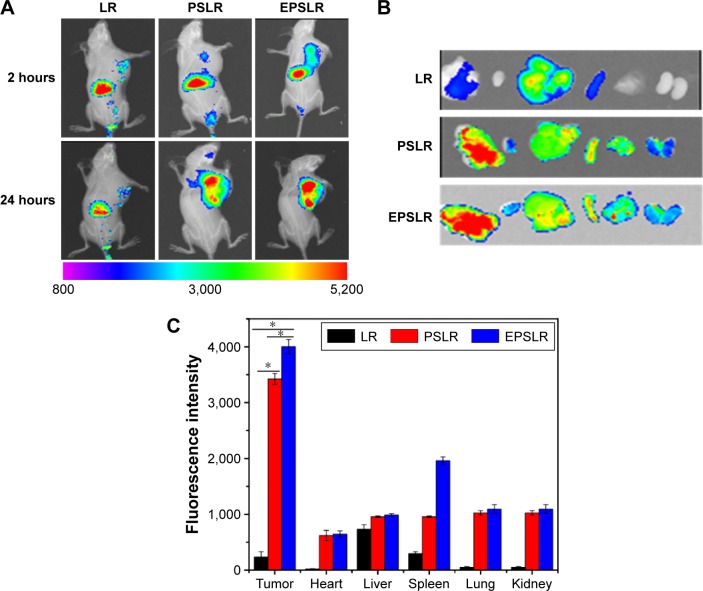
The in vivo biodistribution and targeting ability of EPSLR in tumor-bearing mice were evaluated using a near infrared fluorescence image system. The tumor-bearing mice were injected with DIR-loaded LR, PSLR, and EPSLR. Figure 9A shows the real-time images in nude mice at 1 and 24 hours after injection of the complexes. Most of the DIR accumulated in liver at 1 hour postadministration of LR, PSLR, and EPSLR, and then there was a decrease in the fluorescence intensity after 24 hours. The targeted EPSLR revealed a distinct uptake, with the highest accumulation in a MCF-7/ADR xenograft tumor model, which is consistent with anti-EphA10 antibody-mediated accumulation of EPSLR in breast cancer cells. Injection of PSLR resulted in significantly lower fluorescence intensities in tumor at 24 hours compared to mice that received EPSLR, revealing EPSLR affinity to MCF-7/ADR tumor. Compared to LR, the fluorescence intensity of PSLR or EPSLR in the tumor region was significantly increased at 24 hours postinjection, which was attributed to enhanced permeability and retention effect58 or a combination effect with Eph-mediated endocytosis,59 indicating that PEGylation is capable of prolonging circulation times of complexes in blood. Numerous reports showed that the nanoparticles with sizes ranging from 100 to 200 nm could take a longer circulation time than larger diameters.60 It should be noted that our prepared EPSLR and PSLR at N/P ratio possess a particle size of ~170 nm and ~160 nm (data not shown), which is partly responsible for accumulation of liposome-siRNA complexes in tumor site. The ex vivo analysis of fluorescence intensity (Figures 9B and C) also supported the realization that EPSLR could be delivered to tumor rather than other organs, providing the substantial evidence of tumor targeting ability of EPSLR as an effective delivery system for siRNA delivery.
Figure 9.
In vivo fluorescence images of nude mice.
Notes: (A) Nude mice bearing MCF-7/ADR cells after tail vein administration of DIR-loaded LR, PSLR and EPSLR. (B) Ex vivo fluorescence images of tumors and organs collected at 24 hours postinjection of LR, PSLR, and EPSLR. (C) Quantification of excised organs and tumor uptake characteristics of nanocomplexes. Uptake expressed as fluorescence per mm2 of tumor and organs. Data expressed as mean values ± SD (n=3, *P<0.05).
Abbreviations: LR, liposome–siRNA complexes; siRNA, small interfering RNA; PSLR, PEGylated LR; EPSLR, PSLR-conjugated anti-EphA10 antibody; SD, standard deviation; DIR, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindotricarbocyanine iodide.
Conclusion
A multifunctional siRNA-loaded liposomal nanocarrier, decorated with the monoclonal antibody anti-EphA10 and a pH-sensitive PEG shield, was designed and characterized. This carrier could act as a stimulus-sensitive carrier targeted with antitumor antibody, with systemic long circulation characteristics and with pH sensitivity to lower pH. The cytotoxicity assays showed that the anti-EphA10 antibody coating could slightly increase the cytotoxicity of siRNA–liposome complexes against MCF-7/ADR cells. The results of the cellular uptake experiments demonstrated that anti-EphA10 antibody coating could improve the transfection efficiency of EPSLR in MCF-7/ADR cells by Ephrin receptor-mediated endocytosis. The gene expression studies in MCF-7/ADR cells clearly showed that MDR1 protein expression was significantly downregulated using EPSLR. Furthermore, the results of CLSM indicated that EPSLR could facilitate in siRNA endosomal escape and efficient release into cytoplasm. Biodistribution of EPSLR results demonstrated that PEGylation and anti-EphA10 antibody modification increased tumor targeting in vivo. Thus, the formation of the EPSLR complex provides a facile approach to constructing a multifunctional delivery system for gene drug targeting. These EPSLR complexes capable of efficiently delivering siRNA into cancer cells are highly promising for cancer therapy.
Supplementary material
SDS–PAGE of free (1–2) and conjugated anti-EphA10 antibody (3–4).
Abbreviation: SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Acknowledgments
The authors thank the financial support from the National Natural Science Foundation of China (81202483 and 81302721) as well as Liaoning Natural Science Foundation for Excellent Talents in University (LR 2015020543), Scientific Research Foundation for the Returned Overseas Chinese Scholars by State Education Ministry (201303003), and Science and Technology Project of Shenyang (F15-199-1-24, F15-139-9-06).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wang S, Chen R, Morott J, Repka MA, Wang Y, Chen M. mPEG-b-PCL/TPGS mixed micelles for delivery of resveratrol in overcoming resistant breast cancer. Expert opin Drug Deliv. 2015;12(3):361–373. doi: 10.1517/17425247.2014.951634. [DOI] [PubMed] [Google Scholar]
- 2.London NR, Whitehead KJ, Li DY. Endogenous endothelial cell signaling systems maintain vascular stability. Angiogenesis. 2009;12(2):149–158. doi: 10.1007/s10456-009-9130-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457(7228):426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo P, Coban O, Snead NM, et al. Engineering RNA for targeted siRNA delivery and medical application. Adv Drug Deliv Rev. 2010;62(6):650–666. doi: 10.1016/j.addr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandhi NS, Tekade RK, Chougule MB. Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release. 2014;194:238–256. doi: 10.1016/j.jconrel.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mokhtarieh AA, Cheong S, Kim S, Chung BH, Lee MK. Asymmetric liposome particles with highly efficient encapsulation of siRNA and without nonspecific cell penetration suitable for target-specific delivery. Biochim Biophys Acta. 2012;1818(7):1633–1641. doi: 10.1016/j.bbamem.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Li Y, Wang X, Lee RJ, Teng L. Fatty acid modified octaarginine for delivery of siRNA. Int J Pharm. 2015;495(1):527–535. doi: 10.1016/j.ijpharm.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 8.Corbet C, Ragelle H, Pourcelle V, et al. Delivery of siRNA targeting tumor metabolism using non-covalent PEGylated chitosan nanoparticles: identification of an optimal combination of ligand structure, linker and grafting method. J Control Release. 2016;223:53–63. doi: 10.1016/j.jconrel.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 9.Tyagi N, Arora S, Deshmukh SK, Singh S, Marimuthu S, Singh AP. Exploiting nanotechnology for the development of microRNA-based cancer therapeutics. J Biomed Nanotechnol. 2016;12(1):28–42. doi: 10.1166/jbn.2016.2172. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira C, Ribeiro AJ, Veiga F, Silveira I. Recent advances in nucleic acid-based delivery: from bench to clinical trials in genetic diseases. J Biomed Nanotechnol. 2016;12(5):841–862. doi: 10.1166/jbn.2016.2245. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura K, Yamashita K, Itoh Y, Yoshino K, Nozawa S, Kasukawa H. Comparative studies of polyethylene glycol-modified liposomes prepared using different PEG-modification methods. Biochim Biophys Acta. 2012;1818(11):2801–2807. doi: 10.1016/j.bbamem.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi N, Ghosh PC. Folate receptor mediated targeted delivery of ricin entrapped into sterically stabilized liposomes to human epidermoid carcinoma (KB) cells: effect of monensin intercalated into folate-tagged liposomes. Eur J Pharm Sci. 2011;43(4):343–353. doi: 10.1016/j.ejps.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Xu H, Hu M, Yu X, et al. Design and evaluation of pH-sensitive liposomes constructed by poly(2-ethyl-2-oxazoline)-cholesterol hemisuccinate for doxorubicin delivery. Eur J Pharm Biopharm. 2015;91:66–74. doi: 10.1016/j.ejpb.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Hatakeyama H, Akita H, Ito E, et al. Systemic delivery of siRNA to tumors using a lipid nanoparticle containing a tumor-specific cleavable PEG-lipid. Biomaterials. 2011;32(18):4306–4316. doi: 10.1016/j.biomaterials.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 15.Deshpande PP, Biswas S, Torchilin VP. Current trends in the use of liposomes for tumor targeting. Nanomedicine (Lond) 2013;8(9):1509–1528. doi: 10.2217/nnm.13.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Ren W, Zhong T, et al. Tumor-specific pH-responsive peptide-modified pH-sensitive liposomes containing doxorubicin for enhancing glioma targeting and anti-tumor activity. J Control Release. 2016;222:56–66. doi: 10.1016/j.jconrel.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 17.Veiman KL, Kunnapuu K, Lehto T, et al. PEG shielded MMP sensitive CPPs for efficient and tumor specific gene delivery in vivo. J Control Release. 2015;209:238–247. doi: 10.1016/j.jconrel.2015.04.038. [DOI] [PubMed] [Google Scholar]
- 18.Lin D, Jiang Q, Cheng Q, et al. Polycation-detachable nanoparticles self-assembled from mPEG-PCL-g-SS-PDMAEMA for in vitro and in vivo siRNA delivery. Acta Biomater. 2013;9(8):7746–7757. doi: 10.1016/j.actbio.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 19.Mei L, Fu L, Shi K, et al. Increased tumor targeted delivery using a multistage liposome system functionalized with RGD, TAT and cleavable PEG. Int J Pharm. 2014;468(1–2):26–38. doi: 10.1016/j.ijpharm.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 20.Qiu L, Li Z, Qiao M, et al. Self-assembled pH-responsive hyaluronic acid-g-poly((L)-histidine) copolymer micelles for targeted intracellular delivery of doxorubicin. Acta Biomater. 2014;10(5):2024–2035. doi: 10.1016/j.actbio.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 21.Malamas AS, Gujrati M, Kummitha CM, Xu R, Lu ZR. Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J Controlled Release. 2013;171(3):296–307. doi: 10.1016/j.jconrel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Y, Sun D, Wang GL, et al. An efficient PEGylated liposomal nanocarrier containing cell-penetrating peptide and pH-sensitive hydrazone bond for enhancing tumor-targeted drug delivery. Int J Nanomedicine. 2015;10:6199–6214. doi: 10.2147/IJN.S92519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, Zhao Y. Controlled release from cleavable polymerized liposomes upon redox and pH stimulation. Bioconjug Chem. 2011;22(4):523–528. doi: 10.1021/bc1003197. [DOI] [PubMed] [Google Scholar]
- 24.Shin J, Shum P, Thompson DH. Acid-triggered release via dePEGylation of DOPE liposomes containing acid-labile vinyl ether PEG–lipids. J Control Release. 2003;91(1–2):187–200. doi: 10.1016/s0168-3659(03)00232-3. [DOI] [PubMed] [Google Scholar]
- 25.Chen H, Zhang H, Thor D, Rahimian R, Guo X. Novel pH-sensitive cationic lipids with linear ortho ester linkers for gene delivery. Eur J Med Chem. 2012;52:159–172. doi: 10.1016/j.ejmech.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang T, Li YM, Lv Y, Cheng YJ, He F, Zhuo RX. Amphiphilic polycarbonate conjugates of doxorubicin with pH-sensitive hydrazone linker for controlled release. Colloids Surf B Biointerfaces. 2013;111:542–548. doi: 10.1016/j.colsurfb.2013.06.054. [DOI] [PubMed] [Google Scholar]
- 27.Torchilin VP. Targeted pharmaceutical nanocarriers for cancer therapy and imaging. AAPS J. 2007;9(2):E128–E147. doi: 10.1208/aapsj0902015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao R, Liu L, Deng S, Ren W. Preparation of carboxymethylchitosan nanoparticles with Acid-sensitive bond based on solid dispersion of 10-hydroxycamptothecin. ISRN Pharm. 2011:624–704. doi: 10.5402/2011/624704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Lü S, Gao C, et al. Polymeric micelle-coated mesoporous silica nanoparticle for enhanced fluorescent imaging and pH-responsive drug delivery. Chem Eng J. 2015;279:851–860. [Google Scholar]
- 30.Chen Q, Ding H, Zhou J, et al. Novel glycyrrhetinic acid conjugated pH-sensitive liposomes for the delivery of doxorubicin and its antitumor activities. RSC Adv. 2016;6(22):17782–17791. [Google Scholar]
- 31.Gao J, Sun J, Li H, et al. Lyophilized HER2-specific PEGylated immunoliposomes for active siRNA gene silencing. Biomaterials. 2010;31(9):2655–2664. doi: 10.1016/j.biomaterials.2009.11.112. [DOI] [PubMed] [Google Scholar]
- 32.Imai S, Nagano K, Yoshida Y, et al. Development of an antibody proteomics system using a phage antibody library for efficient screening of biomarker proteins. Biomaterials. 2011;32(1):162–169. doi: 10.1016/j.biomaterials.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 33.Nagano K, Kanasaki S, Yamashita T, et al. Expression of Eph receptor A10 is correlated with lymph node metastasis and stage progression in breast cancer patients. Cancer Med. 2013;2(6):972–977. doi: 10.1002/cam4.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lisle JE, Mertens-Walker I, Rutkowski R, Herington AC, Stephenson SA. Eph receptors and their ligands: promising molecular biomarkers and therapeutic targets in prostate cancer. Biochim Biophys Acta. 2013;1835(2):243–257. doi: 10.1016/j.bbcan.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Qiu L, Qiao M, Chen Q, et al. Enhanced effect of pH-sensitive mixed copolymer micelles for overcoming multidrug resistance of doxorubicin. Biomaterials. 2014;35(37):9877–9887. doi: 10.1016/j.biomaterials.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Feng Q, Yu MZ, Wang JC, et al. Synergistic inhibition of breast cancer by co-delivery of VEGF siRNA and paclitaxel via vapreotide-modified core–shell nanoparticles. Biomaterials. 2014;35(18):5028–5038. doi: 10.1016/j.biomaterials.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Mizoue T, Horibe T, Maruyam K, et al. Targetability and intracellular delivery of anti-BCG antibody-modified, pH-sensitive fusogenic immunoliposomes to tumor cells. Int J Pharm. 2002;237(1–2):129–137. doi: 10.1016/s0378-5173(02)00044-3. [DOI] [PubMed] [Google Scholar]
- 38.Luvino D, Khiati S, Oumzil K, Rocchi P, Camplo M, Barthelemy P. Efficient delivery of therapeutic small nucleic acids to prostate cancer cells using ketal nucleoside lipid nanoparticles. J Control Release. 2013;172(3):954–961. doi: 10.1016/j.jconrel.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Qi R, Liu S, Chen J, et al. Biodegradable copolymers with identical cationic segments and their performance in siRNA delivery. J Control Release. 2012;159(2):251–260. doi: 10.1016/j.jconrel.2012.01.015. [DOI] [PubMed] [Google Scholar]
- 40.Almofti MR, Harashima H, Shinohara Y, Almofti A, Baba Y, Kiwada H. Cationic liposome-mediated gene delivery: biophysical study and mechanism of internalization. Arch Biochem Biophys. 2003;410(2):246–253. doi: 10.1016/s0003-9861(02)00725-7. [DOI] [PubMed] [Google Scholar]
- 41.Terp MC, Bauer F, Sugimoto Y, et al. Differential efficacy of DOTAP enantiomers for siRNA delivery in vitro. Int J Pharm. 2012;430(1–2):328–334. doi: 10.1016/j.ijpharm.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yousefi A, Bourajjaj M, Babae N, et al. Anginex lipoplexes for delivery of anti-angiogenic siRNA. Int J Pharm. 2014;472(1–2):175–184. doi: 10.1016/j.ijpharm.2014.06.028. [DOI] [PubMed] [Google Scholar]
- 43.Gao J, Liu W, Xia Y, et al. The promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomes. Biomaterials. 2011;32(13):3459–3470. doi: 10.1016/j.biomaterials.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 44.Ambardekar VV, Han HY, Varney ML, Vinogradov SV, Singh RK, Vetro JA. The modification of siRNA with 3′ cholesterol to increase nuclease protection and suppression of native mRNA by select siRNA polyplexes. Biomaterials. 2011;32(5):1404–1411. doi: 10.1016/j.biomaterials.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gary DJ, Puri N, Won YY. Polymer-based siRNA delivery: perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J Control Release. 2007;121(1–2):64–73. doi: 10.1016/j.jconrel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 46.Shahbazi B, Taghipour M, Rahmani H, Sadrjavadi K, Fattahi A. Preparation and characterization of silk fibroin/oligochitosan nanoparticles for siRNA delivery. Colloids Surf B Biointerfaces. 2015;136:867–877. doi: 10.1016/j.colsurfb.2015.10.044. [DOI] [PubMed] [Google Scholar]
- 47.Ogris M, Brunner S, Schüller S, Kircheis R, Wagner E. PEGylated DNA/transferrin-PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther. 1999;6(4):595–605. doi: 10.1038/sj.gt.3300900. [DOI] [PubMed] [Google Scholar]
- 48.Xia Y, Tian J, Chen X. Effect of surface properties on liposomal siRNA delivery. Biomaterials. 2016;79:56–68. doi: 10.1016/j.biomaterials.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Qiu L, Chen Q, et al. pH-sensitive nanoparticles of poly (L-histidine)-poly(lactide-co-glycolide)-tocopheryl polyethylene glycol succinate for anti-tumor drug delivery. Acta Biomater. 2015;11:137–150. doi: 10.1016/j.actbio.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Cheng Q, Jiang Q, et al. Enhanced endosomal/lysosomal escape by distearoyl phosphoethanolamine-polycarboxybetaine lipid for systemic delivery of siRNA. J Control Release. 2014;176:104–114. doi: 10.1016/j.jconrel.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Brown BS, Patanam T, Mobli K, et al. Etoposide-loaded immunoliposomes as active targeting agents for GD2-positive malignancies. Cancer Biol Ther. 2014;15(7):851–861. doi: 10.4161/cbt.28875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagano K. Challenge to the Development of Molecular Targeted Therapy against a Novel Target Candiate Identified by Antibody Proteomics Technology. J Pharm Soc Japan. 2016;136(2):145–149. doi: 10.1248/yakushi.15-00226-1. [DOI] [PubMed] [Google Scholar]
- 53.Nagano K, Maeda Y, Kanasaki S, et al. Ephrin receptor A10 is a promising drug target potentially useful for breast cancers including triple negative breast cancers. J Control Release. 2014;189:72–79. doi: 10.1016/j.jconrel.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Simard P, Leroux JC. pH-sensitive immunoliposomes specific to the CD33 cell surface antigen of leukemic cells. Int J Pharm. 2009;381(2):86–96. doi: 10.1016/j.ijpharm.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 55.Zhu H, Zhang S, Ling Y, Meng G, Yang Y, Zhang W. pH-responsive hybrid quantum dots for targeting hypoxic tumor siRNA delivery. J Control Release. 2015;220(Pt A):529–544. doi: 10.1016/j.jconrel.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 56.Taki S, Kamada H, Inoue M, et al. A novel bispecific antibody against human CD3 and ephrin receptor A10 for breast cancer therapy. PloS one. 2015;10(12):e0144712. doi: 10.1371/journal.pone.0144712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li J, Wang Y, Liang R, et al. Recent advances in targeted nanoparticles drug delivery to melanoma. Nanomedicine. 2015;11(3):769–794. doi: 10.1016/j.nano.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 58.Kono K, Takashima M, Yuba E, et al. Multifunctional liposomes having target specificity, temperature-triggered release, and near-infrared fluorescence imaging for tumor-specific chemotherapy. J Control Release. 2015;216:69–77. doi: 10.1016/j.jconrel.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 59.Ho EA, Osooly M, Strutt D, et al. Characterization of long-circulating cationic nanoparticle formulations consisting of a two-stage PEGylation step for the delivery of siRNA in a breast cancer tumor model. J Pharm Sci. 2013;102(1):227–236. doi: 10.1002/jps.23351. [DOI] [PubMed] [Google Scholar]
- 60.Gao J, Yu Y, Zhang Y, et al. EGFR-specific PEGylated immunoliposomes for active siRNA delivery in hepatocellular carcinoma. Biomaterials. 2012;33(1):270–282. doi: 10.1016/j.biomaterials.2011.09.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SDS–PAGE of free (1–2) and conjugated anti-EphA10 antibody (3–4).
Abbreviation: SDS–PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.