ABSTRACT
Telomeres are linear guanine-rich DNA structures at the ends of chromosomes. The length of telomeric DNA is actively regulated by a number of mechanisms in highly proliferative cells such as germ cells, cancer cells, and pluripotent stem cells. Telomeric DNA is synthesized by way of the ribonucleoprotein called telomerase containing a reverse transcriptase (TERT) subunit and RNA component (TERC). TERT is highly conserved across species and ubiquitously present in their respective pluripotent cells. Recent studies have uncovered intricate associations between telomeres and the self-renewal and differentiation properties of pluripotent stem cells. Interestingly, the past decade's work indicates that the TERT subunit also has the capacity to modulate mitochondrial function, to remodel chromatin structure, and to participate in key signaling pathways such as the Wnt/β-catenin pathway. Many of these non-canonical functions do not require TERT's catalytic activity, which hints at possible functions for the extensive number of alternatively spliced TERT isoforms that are highly expressed in pluripotent stem cells. In this review, some of the established and potential routes of pluripotency induction and maintenance are highlighted from the perspectives of telomere maintenance, known TERT isoform functions and their complex regulation.
KEYWORDS: Alternative splicing, embryonic stem cells, hESC, iPSC, pluripotency, telomerase, telomere, TERT
Background
The evolution of linearly organized DNA and inability of DNA polymerases to conserve the chromosomal ends have necessitated the need for non-coding telomeric DNA to cap the chromosomes.1-3 These repeating DNA sequences protect an individual's chromosomes from fusion and erosion of coding DNA. Continued shortening of telomeric DNA due to the end-replication-problem eventually leads to critically short telomeres that induce senescence to avoid chromosomal damage.3 This effect, known as the Hayflick limit 4,5 defines the number of possible cellular replications before cells senesce, and thus limits cellular lifespan. On an organismal scale, short telomere syndromes exhibit strong correlations with certain pathologies, such as pulmonary fibrosis,6,7 arteriosclerosis,8 aplastic anemia,6,9 chronic liver disease,10 and intrauterine growth restriction (IUGR) due to trophoblast stem cell dysfunction.11-13
Landmark work published by Carol Greider and Elizabeth Blackburn 14 uncovered the telomerase protein complex needed to re-lengthen telomeric DNA and maintain proliferation of a species and its germ cells. At the core of this ribonucleoprotein complex, the telomerase reverse transcriptase (TERT) protein in conjunction with its telomerase RNA component (TERC) is sufficient to extend telomeric sequences in vitro.15 Elevated expression of TERT is found in over 90% of cancers in order to maintain proliferation, and much effort has been made in unveiling the pathogenic role of TERT in the diseased state (as previously reviewed 16,17). However, high TERT levels naturally arise in non-pathogenic settings such as in the germline and stem cells, and in particular embryonic stem cells (ESCs). Prolonged proliferative capacity of ECSs is achieved by the maintenance/re-lengthening of telomere length via high telomerase activity. As such, human (h)ESCs are capable of being passaged over 120 times with no decrease in telomerase activity, telomere length and pluripotency potential.18
Several recent reviews 19-21 have highlighted different mechanisms by which telomeres and TERT are affected by pluripotency. These discussions tend to be singularly focused with pluripotency acting as an effector. Canonically, any feedback from TERT to pluripotency occurs only indirectly through telomere-mediated effects such as DNA damage responses (DDR). It is well established that telomere length (in particular critically short telomeres), as maintained through the canonical function of TERT, affects pluripotency22 and embryonic viability.23 However, the past decade's work on TERT has revealed non-canonical, extra-telomeric functions of TERT, much of whose functional significance are yet to be elucidated in the context of stem cell biology. It is quite possible that many of the non-canonical pathway functions of TERT do affect pluripotency and stem cell maintenance. Recent studies24,25 have uncovered novel non-canonical functionality in some of the TERT isoforms that may also impact stem cell function. This review will discuss the multiple known and speculative connections to pluripotency induction and maintenance in the context of telomeres length, TERT isoforms, and their canonical and non-canonical functions.
Telomeres and pluripotent stem cells
Telomere length is a basic requirement for sustaining the replicative potential of somatic cells and its maintenance allows for unlimited self-renewal of stem cells.18,26 Both murine and human embryonic stem cells demonstrate robust telomerase expression for extended population doublings and are capable of maintaining long telomeres.27,28 Similarly, human and murine induced pluripotent stem cells (iPSCs) are capable of carrying out re-elongation of telomeres in at least some clones, although the telomere length and activity characteristics appear to be clonally unique.28-33 Conversely, the process of differentiation triggers down-regulation of telomerase and initiates telomeric attrition.34,35 Following the extended cellular division required to form adult tissues, terminally differentiated cell types typically exhibit low or undetectable levels of telomerase activity and shortened telomeres.36,37 While extensive species similarities in the regulation of TERT and telomeres exist, Table 1 highlights some significant differences that will be highlighted in subsequent sections.
Table 1.
Species-specific comparison of telomere/telomerase effects and their interactions with pluripotency regulation.
| Organism |
||||
|---|---|---|---|---|
| homo sapiens | murine | porcine | ||
| Telomere Length | Inner cell mass | 12 kb 162 | 86 kb in-vivo 50 kb in-vitro 52 | |
| ESCs | 12 kb 49 | 144 kb at p12 52 | 33.046 | |
| iPSCs | Similar to ES49 | similar to ES53 | up to 31.5kb31 | |
| Effect on ES/iPSC pluripotency | TERTknockdown* | No86/Yes 84,85 | No 45,124 /Reprogramming effects 63 | unknown |
| TERC knockdown* | No40,85 | No major effect 51/mitochondria 44, naivety 33 | unknown | |
| TERT Null Phenotype | TERT−/−* | unknown/unknown | None76,83,123, /WNT-defect 105 | unknown |
| TERC−/−* | unknown/unknown | None 77 | unknown | |
| iPSC Reprogramming | ||||
| Telomeres re-lengthen | Yes-sporadic 28 | Yes-sporadic 32 | Yes-sporadic31 | |
| Elongation period | up to passage 15 49 | continuous 52 | unknown | |
| ALT pathway | supressed 49 | active 46 | active31 | |
| iPSC TERT increase | >100x 32 | approx. 3x 32 | ||
| Knockdown SUV39h1 and SUV39h246 | Telomere shortening | Telomere elongation | Telomere elongation | |
| Non-telomere Interactions | NF-kB (RelA/p65)111,163 | Stabilizes p65 and its Nuclear Residency | mTERC−/− and mTERT−/− defective NF-kB signaling | |
| IL-6,8 & TNF-a promoter binding | ||||
| BRG1111,105 | BRG1/GNL3 | BRG1 | ||
| RNA species102,107 | RMRP | |||
| LINE1/alphoid ssRNA | ||||
| Mitochondrial membrane proteins158 | TOM20/TOM40 & TIM23 | |||
| mtDNA158 | ND1 & ND2 mtDNA | |||
Categories with mixed results are indicated with supporting references.
in absence of short telomere effects.
Both the increase and attrition of telomere length occurs gradually during S-phase 38 and requires multiple cell divisions. On the other hand, the initial stage of iPSC reprogramming is a fairly rapid process, and therefore telomere-mediated effects may not arise in the midst of reprogramming. Not surprisingly, the age of human cell donor has little effect on the ability to reprogram the cells to a pluripotent state.30,39 In fact, human iPSCs (hiPSC) derived from cells with telomerase activity defects and non-critically short telomeres still can be reprogrammed to a pluripotent state.29,40 This suggests that, while critically short telomeres significantly impact reprogramming efficiency, increasing telomere length far beyond the length needed to support initial proliferation requirements does not confer additional reprogramming advantage. This point is often masked by the use of mean telomere length, which generally has correlation with effects such as lifespan and pluripotency near short lengths where the telomere length distribution skewness is changing significantly.41 However, following reprogramming mis-localization of human TERT (hTERT), and all abnormalities leading to similar telomere attrition result in eventual loss of self-renewal and pluripotency.42 Similarly, long term self-renewal and teratoma formation in murine and porcine iPSCs with short telomeres are generally impaired.31,43 In addition, PSCs with short telomeres in a variety of species display mitochondrial dysfunction,44 impaired neuroectodermal differentiation,44 uncapping events, and reduction in H3K27me3 distal to telomeres including global hypomethylation which contributes to reactivation of pluripotency genes,45 and possibly reprogramming transgenes.31,33,46
Partially reprogrammed iPSCs often show little TERT activation and short telomeres.32 This reprogramming failure is likely the reason for the failure of the TERT promoter to be activated. Conversely, it is unlikely that lack of TERT activation causes partial reprogramming as TERT expression and/or activity does not guarantee pluripotency.28,35,47,48 However, telomere length is touted as a biomarker of reprogramming, with hiPSC telomere lengths plateauing close to the hESC telomere length of about 12 kbp despite continued high telomerase expression.49 Such stabilization of telomere length, in addition to proper downregulation of hTERT following differentiation, is a strong hallmark of non-transformed human pluripotent stem cells.30,35,49 hESCs and hiPSCs normally suppress the ALT (alternative lengthening of telomerase) pathway49 and, in the absence of hTERT, short telomeres typically lead to up-regulation of the p53-mediated apoptosis pathway, or chromosomal rearrangements.29 Thus, the aforementioned telomere maintenance characteristics in pluripotency are not preserved across vertebrates (Table 1).
Instead of using murine TERT (mTERT), initial lengthening of murine telomeres during early embryo cleavage50 is largely achieved through alternative lengthening of telomerase (ALT) pathways. To a lesser extent, mESCs and miPSCs also appear to upregulate and utilize ALT mechanisms for telomere maintenance intermittently.33,51 mESCs and miPSCs also do not exhibit the in vitro plateau in telomere length that is characteristic of hESCs/hiPSCs.52 While NT-mESCs generally have completed telomere lengthening and full reprogramming by the blastocyst stage,44 miPSCs may take up to 30 passages to achieve mESC lengths 53 and only do so sporadically.32 Control of telomere length is an intricate process involving many factors. As previously reviewed,19 mESCs and miPSCs control telomere length largely by modulating telomeric epigenetics and regulation by several proteins, including ZSCAN4,51,54-56 ATRX,54,55 RIF1,56 TRF152,57 TPP1 and other shelterin complex components.52,58 Increased telomere length, as part of the establishment of the pluripotent epigenome, occurs only after multiple passages, and is accomplished through epigenetic modifications of histones and subtelomeric DNA methylation.33,59 During cellular reprogramming, hiPSC sub-telomeres become hyper-methylated with both de novo methylation and pockets of demethylation occurring.34 At the same time, histone H3.3 plays a critical role in regulating telomeric chromatin accessibility.60 Whereas during differentiation H3.3 is decreased causing telomeres and subtelomeres to take on a more heterochromatic state, complete knockdown of H3.3 leads to telomeric dysfunction.60 Early lengthening of telomeres, within the first few passages following reprogramming, is preceded by a significant reduction of H3K9/H4K20 tri-methylation.49 However, this must be followed by reestablishment of H3K9/H4K20 me3 repressive marks to stabilize telomeric length.49
While knockdown of histone methyltransferases (HMTs) SUV39h1 and SUV39h2 in mice and pigs leads to increased telomere length, decreased demethylases DNMT1/3a/3b and decreased H3K9me3 marks,46 knockdown in human cells leads to telomere length shortening.46 This disparity is likely due to species differences (Table 1) that repress ALT pathways in humans, but not murine or porcine cells.31,46 Consequently, although heterozygous mTERC−/+ miPSCs and to a lesser extent mTERC−/− miPSCs are capable of maintaining pluripotency and telomere length in mice, possibly due to the activation of the ALT pathway,33 hTERT−/+ hiPSCs show poor telomere elongation, and DKC1 (Dyskerin - a telomerase complex component) hiPSC mutants (TERC deficient) do not elongate telomeres.42
Pluripotency and canonical TERT functions
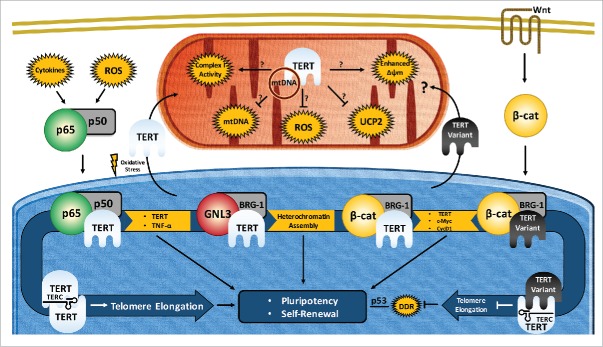
As discussed above, telomeres have strong feedback mechanisms to regulate pluripotency. TERT is the catalytic component of telomerase whose function, when combined with TERC, is to maintain telomere length and structure. This section will establish the canonical pluripotency-to-TERT links that will be contrasted with the subsequent non-canonical TERT section (below). Putative links are highlighted in Fig. 1.
Figure 1.
Proposed routes of pluripotency maintenance and self-renewal by pluripotent stem cells through functions of telomerase reverse transcriptase (TERT) isoforms. TERT expression is regulated by a number of key pluripotency/transcription factors. Conversely, TERT regulates pluripotency and self-renewal via canonical (telomeric) and non-canonical (mitochondrial, transcriptional) functions in pluripotent stem cells (PSCs). Telomere- and chromosome-mediated TERT effects lead to changes in pluripotency and cell proliferation by modulating the activities of transcription factors (c-Myc, β-Catenin, NF-κB, p53). This figure would be much more complex with additional TERT variant specific functions, as well as multiple TERT splice variants modulating each of its current roles. Common acronyms: mtDNA (mitochondrial DNA), Δψm (Mitochondrial Membrane Potential/MMP), ROS (Reactive Oxygen Species) UCP2 (Mitochondria uncoupling protein 2), β-Cat (β-Catenin/CTNNB1), DDR (DNA Damage Response). The line without arrowhead underneath p53 indicates existing evidence for interaction with lack of mechanistic understanding.
During embryogenesis and induced pluripotent stem cell (iPSC) generation, TERT and other telomerase components are upregulated, and remain so in self-renewing PSCs. TERT activation is a beneficial event for lengthening telomeres in order to stabilize them and suppressing the DNA damage response (DDR) prior to other epigenetic changes requiring extensive proliferation. Telomerase activity may thus be useful in assessing iPSC reprogramming efficiency and as a hallmark of non-transformed somatic cells, which should contain low telomerase expression and activity.61 Moreover, upregulation of pluripotency genes is strongly connected with expression of TERT, the overexpression of which enhances reprogramming efficiency.62,63 For example, porcine iPSCs can only maintain telomeres with continued expression of exogenous reprogramming factors to maintain TERT levels,31 and partially reprogrammed mouse embryonic fibroblasts (MEFs), and porcine embryonic fibroblasts (PEFs) show little TERT promoter activation and poor telomere elongation. However, while low telomerase activity is correlated with partial reprogramming, high levels of TERT alone does not induce a pluripotent state,48 nor does pluripotency strictly require high levels of TERT.28 The transcriptional control by which TERT is upregulated during reprogramming to a pluripotent state has only recently begun to be elucidated.
During reprogramming, endogenous TERT up-regulation is a late event64 that precedes endogenous upregulation of OCT4, SOX2 33 and TERC (whose promoter is bound by OCT3/4 and NANOG 40). TERT up-regulation is instead simultaneous with the overexpression of KLF4.33 Recently, KLF4 binding has been mapped directly to the TERT proximal promoter where KLF4 is able to upregulate TERT when β-catenin acts as a cofactor.49,65,66 This appears to be a key function of KLF4, as hTERT overexpression is capable of rescuing KLF4 knockdown-triggered cellular differentiation.66 Furthermore, KLF4-β-catenin in complex with TCF-4 or TCF-1 serves either to activate or to repress TERT, respectively.65,67 This interplay helps to provide a mechanism by which pluripotent and cancer stem cells are able to upregulate TERT to initiate cellular immortalization through telomere maintenance. Additionally, cell lines containing short telomeres are quite refractory to reprogramming and this inhibition is mediated by the p53 apoptosis/senescence pathway, which when removed allows the reprogramming of cell lines with critically short telomeres (albeit resulting in widespread chromosomal aberrations).68 Hence, TERT upregulation through KLF4 may serve as an additional indirect means by which the p53 pathway can be suppressed via telomere maintenance during reprogramming.69
C-MYC (one of the original Yamanaka factors 70) remains a common, albeit dispensable, cofactor during iPSC reprogramming,71 and maintains high expression following transformation to a pluripotent state. C-MYC binds to and activates the TERT promoter.65,72,73 Indirect evidence of C-MYC's importance is provided by the knockdown of SIRT1 which represses C-MYC, and accompanies a reduction in mTERT levels.47 However, C-MYC is not required for reprogramming and its expression level is generally not correlated with TERT levels post-reprogramming, raising the question of whether C-MYC actually plays a major role in maintaining TERT expression.32,33,53 It is possible that this effect is still mediated through KLF4 as SIRT1 also affects NANOG expression, which, in turn can regulate KLF4 and C-MYC through feedback loops with other pluripotency factors (for example OCT3/4 and mir145).74,75 C-MYC may have a more important role in hiPSC generation where ALT does not contribute to the initial elongation of telomeres, and thus hTERT must play a more substantial role.
While non-canonical extra-telomeric functions of TERT are becoming apparent (see following section), it appears that, at least in the murine context, it is the canonical telomeric maintenance that has the greatest effect in vitro.76,77 In agreement with this, complete abolition of telomerase function through mTERT 76 or mTERC 77 knockout in mice does not result in gross pathological symptoms for 6 to 8 generations at which point telomeres are critically shortened. Moreover, haplosufficient mTERC+/− mESCs can contribute to tetraploid embryos, whereas full-knockout mTERC−/− cells are unable to contribute despite containing unmodified mTERT.78 Collectively, it can be deduced that TERT and canonical telomerase activity is dispensable during the reprogramming process of murine cell lines with sufficient telomere length,33 but ultimately telomere maintenance is a required feature of prolonged self-renewal. In support of this, knockdown of ZSCAN4, an enhancer of chromosome stability and telomere-sister chromatid exchange (T-SCE)51 initiates cell crisis and apoptosis within 8 passages in mESCs, whereas mTERC−/− cells upregulate ZSCAN4 possibly to compensate and are subsequently sustainable for up to 450 population doublings.51
Similarly, mTERT appears to be dispensable for the in vitro differentiation of mESCs with sufficient telomere length.45 In a study by Pucci et al., mTERT −/− mESCs with both short and long telomeres appear to differentiate normally upon LIF withdrawal.45 However, differentiated cells derived from short telomere mESCs do not maintain their non-pluripotent status and begin to re-express NANOG 45 through unknown mechanisms. Prior to differentiation NANOG is also expressed at high levels in mTERC −/− mESCs with short telomeres.44 Additionally, Pucci et al. found that ESRRB, a KLF4 substitute during reprogramming 79 and a direct target of NANOG,80 is also strongly upregulated.45 Previous studies have demonstrated p53 binding to the promoters of NANOG and ESRRB 81 and its ability to either enhance or repress the pluripotency genes.80 Thus, p53 signaling in response to short telomeres of mTERT−/− mESCs may act to upregulate NANOG. Alternatively, NANOG up-regulation could be an incidental effect of DNA methyl transferase (DNMT) down-regulation leading to hypo-methylation in an attempt to upregulate the ALT pathway as previously proposed.58 Another possibility is that the short telomeres might have led to destabilization of shelterin complexes and the resultant release of RAP1. RAP1 can modulate the steady state level of E-cadherin and consequently OCT3/4 and NANOG.82 Lastly, as DKC1 regulates promoters of pluripotency genes, it is possible that removal of TERT from the telomerase complex disrupts the holoenzyme allowing DKC1 to more efficiently modulate pluripotency promoters.83 Further investigation of mechanisms of telomeric induced deregulation of pluripotency genes during differentiation should yield important information as to how short telomeres influence distal genes.
Little work has been carried out with the knockout/knockdown of telomerase subunits in human cells. Of the studies that have been carried out, conflicting results have emerged. Yang et al. 84 suggest that lentiviral knockdown of hTERT in hESCs results in their spontaneous differentiation and the inability to generate a stable ES cell line. In partial agreement, chemical inhibition of hTERT activity in hESCs leads to lowered pluripotency and increased differentiation, while curiously, similar blocking of telomerase activity through TERC steric inhibition has no effect suggesting an extra-telomeric role for TERT in maintaining self-renewal in the undifferentiated state.85 In contrast, Sexton et al. did not observe any reduction in pluripotency (as measured by sustained OCT3/4 expression only) after establishing stable hESC lines through zinc finger-mediated knockout of hTERT.86 However, Sexton et al. do observe a loss of self-renewal and differentiation beginning around 75 days post TERT-knockout, when the cells reach a critical telomere length of 2–3kb. These drastic differences likely arise from effects of the modalities used to knockout/knockdown hTERT or alternatively from telomeric differences in the cell lines used. Despite this, as discussed in the previous section (also see Table 1), humans do not normally have a substantial ALT pathway contribution and other species-specific differences exist. For example, mTERT expression is only upregulated at low (3 fold) levels during reprogramming as compared to hTERT in hiPSCs (several hundred fold), despite high promoter activation that correlates with reprogramming stage.32 TERC is typically expressed in excess, even in differentiated tissues,58 and thus may not be a limiting factor in mTERC +/− murine cells. Lastly, as laboratory mice are known to have extremely long telomeres and short lifespans, telomeric effects may be delayed compared to humans.
Non-canonical functions of telomerase and pluripotency
In recent years, the discovery of telomere-independent effects of TERT on proliferation87-95 and perturbations of resistance to reactive oxygen species damage 96-100 have led to a burgeoning body of evidence that indicates that TERT has many functions outside it's canonical role in telomere length maintenance.101 Some of these extra-telomeric effects include acting as an RNA-dependent RNA polymerase,102 a mitochondrial reverse transcriptase,103 interacting with nucleostemin (GNL3),104 WNT, BRG1, β-catenin,87,105 NF-κB,106 and modulating endogenous siRNA 102,107 and miRNA profiles.108 Although the non-canonical effects have been observed mainly in non-pluripotent cell types, many of the affected pathways play key roles in pluripotency. This section will discuss extra-telomeric roles and possible direct or indirect connections to pathways also involved in the self-renewal and maintenance of stem cells. Some of these confirmed and speculative pathways are summarized in Fig. 1.
TERT, BRG1 and Wnt
A number of effects point to TERT (with or without TERC) possessing non-canonical functions. TERT is capable of aiding the maintenance of self-renewal in human limbal and mesenchymal stem cells, increasing the efficiency of directed differentiation,48,99 enhancing iPSC reprogramming efficiency 40 and activating resting hair follicle stem cells.90 Several recent reports have shown that TERT is able to interact with BRG1 in a WNT/β-catenin dependent manner. The canonical WNT pathway is extensively involved in differentiation of cell lineages and BRG1 loss of function is embryonic lethal and a critical developmental transcription factor.109-113 Despite demonstrating TERT-BRG1 interaction in both murine cellular and tissue ChIP assays,105 and the TERT-BRG1-GNL3 complex in specific human cell lines,107 the widespread applicability of TERT-BRG1 interactions has been called into question.114 Of 4 cell lines investigated, Listerman et al. could not show a consistent interaction,114 and other studies have not found BRG1 interactions.106,115 However, supportive evidence has been offered in several other studies.105,107,108 MicroRNA (miRNA) expression alterations during the suppression of TERT in THP-1 and HeLa cells are strikingly similar to microRNA changes during BRG1 suppression.108 Though mostly inferred from correlative data, it is likely that these changes are mediated through TERT's interaction with transcriptional factors BRG1, GNL3 and NF-κB that regulate miRNAs and not by direct miRNA interaction.108 Additionally, axis formation defects following TERT knockdown in Xenopus are consistent with a role for TERT in the Wnt pathway.105 In contrast, in human cells, association of GNL3 with TERT-BRG1 produce RNA dependent RNA polymerase activity (RdRP), forming siRNA targeted to regulate centromeric alphoid domains, and non-centromeric LINE1 elements, thereby modulating heterochromatin during mitosis.107 Thus, TERT-BRG1 may play varying roles depending on which cofactors are present, and these associations if present during pluripotency and differentiation would directly link TERT to a developmental role.
Unfortunately, despite in vitro evidence for TERT-BRG1 interactions, in vivo defects observed in TERT-null mice (with sufficient telomeres) have only been seen in a single mouse strain by one research group.105 It is possible that interaction may be strongly context dependent and pathological transformed lines may have additional, or conversely lack, compensatory mechanisms for TERT-BRG1 interactions. On the other hand, multiple groups have indicated possible TERT interaction with β-catenin in both humans and mice 87,105,115 and the ability to activate resting hair and epithelial stem cells lends credence to an in vivo interaction possibly involving β-catenin.87,90 Since even catalytically inactive TERT is able to enhance iPSC reprogramming 63 requiring an mesenchymal-to-epithelial transition (MET), it is initially curious that the TERT-β-catenin association supports the reverse epithelial-to-mesenchymal transition (EMT) in transformed cells.115 The upregulation of the EMT inhibitor KLF7 in TERT containing iPSCs 63 and nuclear export of β-catenin by E-cadherin 116 may mitigate this effect . However, recent evidence suggests that early activation of an EMT pathway prior to a delayed EMT event actually enhances reprogramming.117 Ultimately, the enhancement of iPSC reprogramming may be supported by more than one of TERT's non-canonical effects, such as enhanced proliferation, ROS mitigation, NF-κB interaction in addition to its canonical telomere stabilizing effect.
TERT and mitochondria
Mitochondrial localization of TERT is extensively documented, however, its role in mitigating ROS (Reactive Oxygen Species), and acting as a TERC independent reverse transcriptase are only beginning to be understood.96-100,118,119 It is difficult to unravel the contributions from non-canonical TERT effects versus telomeric effects. The canonical telomere shortening effects generally occur after extended proliferation, and are mediated by telomeric dysfunction. On the other hand, non-canonical effects of TERT should appear immediately upon knockout. For example, removal of the canonical function of mTERT by mTERC knockout leads to mitochondrial dysfunction after iPSC and SCNT reprogramming of cells with short telomeres.44 However, this mitochondrial dysfunction is likely a p53 mediated effect due to the poorly maintained telomeres. The donor cells came from late-generation Terc−/− mice (hence short telomeres to begin with) and were allowed to grow/differentiate under telomerase-activity-deficient conditions, which may have led to mitochondrial damage.44 As there is evidence that most mitochondrial localized TERT may not carry TERC at all and instead act as a reverse transcriptase for mitochondrial RNA 103 it would be informative to additionally knockdown mTERT 44 to look for decreased ROS scavenging.
To add to the complexity, TERT is capable of binding alternative RNA's including the RNA component of mitochondrial RNA processing endoribonuclease (RMRP), with which it can form dsRNA that is processed into siRNA directed against RMRP.102 RMRP is required for development, and aside from its involvement in mitochondrial DNA replication,120 it is also processed into siRNAs that targets several developmentally relevant genes.121,122 TERT also appears capable of altering miRNA levels, possibly through a BRG1 associated mechanism,108 although in rat cardiomyocytes the effect is solely on mature miRNAs and seems likely to be mediated through non-specific effects such as increased ROS.123 It should be interesting to further investigate if these RMRP related effects are present in pluripotent stem cells and whether they are capable of altering cellular differentiation pathways.
Aside from the highlighted telomeric effects, TERT and TERC knockouts are not lethal in mice, nor do they result in phenotypic/transcriptional WNT pathway defects 124 or typical RMRP-related defects.124 The extensive homology of TERT across species and in-particular vertebrates, and the ability to make at least one hTERT-null pluripotent human line 86 makes it difficult to imagine embryonic lethality in hTERT-null humans. On the other hand, as discussed in the previous sections, species differences exist and humans appear to suppress ALT pathways and rely more heavily on hTERT during development.49 Alternatively, it is feasible that extra-telomeric effects may be redundant, appearing in sporadic situations where compensatory mechanisms are absent. As functional redundancy may, on a much larger scale, play a significant role in survival of a species, these effects still warrant a significant amount of attention. Additionally, as TERT has abundant isoforms, most of which are catalytically inactive at telomeres, these extra-telomeric roles may be significant.
Alternative splice forms of telomerase
Telomerase expression and activity is highly regulated by a number of mechanisms, one of which is alternative splicing.125-129 Alternative splicing is a method utilized by most mammalian cells to increase the transcriptome and protein diversity for extra means of gene regulation.130 To date, 22 and 31 unique alternative splice variants of TERT have been identified for human and chicken, respectively.25,131-134
The human TERT gene is approximately 42 kb in length and contains 15 introns and 16 exons.135 In most cases, exons are assembled together by joining of evolutionarily conserved 5′-splice donor and 3′-splice acceptor sequences in introns through long-range RNA interaction.136 Alternative splicing of TERT is achieved by exon-skipping, intron-retention, and use of alternate splice acceptor sequences,136 and is regulated by SRSF and hnRNP protein families.24 However, the mechanism of regulation remains largely unclear. Notably, multiple splicing events can occur simultaneously in a single transcript, resulting in very diverse population of TERT isoforms.137 In many cases, early splicing events within 5′-region of the transcript will introduce premature termination codons (PTCs) nullifying the effect of splicing 3′ to these,25 causing many protein isoforms to be degenerate and/or targeted for non-sense mediated decay (NMD).138 TERT has several evolutionarily conserved functional domains. The N-terminal end harbours a mitochondrial targeting sequence (MTS), nuclear localization signals (NLSs), and RNA-interacting domains, while the C-terminal half contains multiple reverse-transcriptase-like (RT-like) motifs.135,139-145 These critical elements are often disrupted in a majority of TERT isoforms to varying extents.25 Due to this, no currently identified alternative splice variant of hTERT exhibits reverse-transcriptase activity and only the full-length hTERT (containing all 16 exons) retains catalytic activity.25 Nevertheless, catalytically inactive isoforms are highly expressed throughout human development in a tissue-specific and development-stage dependent manner.126,128 Moreover, some hTERT isoforms are constitutively generated independently of the full-length hTERT levels.24,125,127,128,146 Accumulating evidence points to potential roles of hTERT isoforms in the regulation of telomerase functions.24,128,147-151
An early attempt to characterize hTERT revealed two of the best-characterized hTERT isoforms, α- and β-variants.131,135 The α-variant results from a partial in-frame deletion (36bp) of exon 6, leading to the partial loss of RT-motif A. This catalytically inactive form of hTERT is a dominant negative inhibitor of telomerase activity when overexpressed in telomerase positive immortal cells.137,152 Considering the relatively low transcript abundance of the α-variant,146,147,151 the level of inhibition may be insufficient for it to have significant impact on telomere maintenance of indefinitely dividing cells. Shortly after the discovery of the α-variant, Hisatomi et al. discovered a new in-frame splicing event that deletes two RT-motifs, rendering catalytic inactivity.132 Like the α-variant, the γ-variant was suggested as a negative regulator of telomerase activity. However, γ-transcript levels, relative to those of α- and β-variants, are extremely low or nearly undetectable in tested cell types.132,150 Despite low expression levels, the α- and γ-variants hold much promise in micro-regulation of non-canonical TERT functions, as they are both in-frame deletion variants with good structural and sequence homology to full-length TERT.
The β-splice variant results from a 182 bp out-of-frame deletion and the subsequent loss of exons 7 and 8.131,135 Unlike the α-deletion, the β-deletion introduces a premature termination codon (PTC) causing it to be truncated and a potential NMD target.138 However, Listerman et al. and our group have recently shown that the β-variant transcript associates with polyribosomes, escapes NMD, and is likely translated.24,85 Surprisingly, the β-deletion transcript not only survives degradation but is one of the most highly expressed TERT transcripts in various cell types including human embryonic stem cells.24,128,132,147,151 A recent discovery suggests that the β-variant inhibits telomerase activity by sequestering TERC.24 However, TERC is expressed at very high levels in most immortal cell types and is rarely a limiting factor for telomerase activity.125,134,153,154 Despite the truncation, the β-variant retains the MTS, RNA-interaction sites and NLSs. Its abilities to interact with different RNA species and translocate between different cell compartments 24 are strongly suggestive of possible roles in regulation of RNA-mediated extra-telomeric functions of hTERT. Maida et al. was the first group to have detected RdRP activity of full-length TERT and to have characterized catalytically active hTERT as a negative regulator of RMRP via siRNA mediated-knockdown.102,122 This was later confirmed by work of Mukherjee et al. in which they also observed reduced proliferation of human mammary epithelial cells upon shRNA mediated-RMRP knockdown.155 Since the production of siRNA by TERT-RMRP requires catalytic activity, any catalytically inactive forms of hTERT with an intact RNA-binding domain such as β-variant may inhibit cell proliferation by down-regulating siRNA production. Overexpression of the β-variant in HeLa cells showed its ability to associate with mitochondria, the significance of which is not fully understood.24 In the same study, overexpressed β-variant exhibited anti-apoptotic effects in cisplatin-treated human breast cancer cells by significantly reducing caspase 3/7 activity. Human TERT overexpression studies have shown that full-length hTERT augments mitochondrial function and maintains the integrity of mitochondria DNA under oxidative stress in a context dependent manner.156-158 Remarkably, overexpression of dominant-negative mutant TERT abrogated or reversed some of these mitochondrial changes, pointing to a potential counter-regulatory role of hTERT alternative splice variants against full-length hTERT in regulating mitochondrial function.158 This putative interplay between hTERT isoforms hold much significance in settings such as embryonic stem cells where mitochondria (and metabolism) likely play critical roles in governing the stress response, cell fate decisions, and pluripotency. Known factors that affect hTERT-splicing patterns are TGF-β1 and hypoxia.159,160
TGF-β1 in human epidermal cells shifts hTERT splicing patterns toward β-variant transcript at the expense of the full-length transcript,159 whereas hypoxic conditions causes it to shift in the other direction.160 TGF-β1 is an important cytokine involved in cell proliferation, stem cell maintenance and differentiation through SMAD signaling.1,2 In addition, hypoxia increases hESC resistance to spontaneous differentiation and improves cell pluripotency through stabilization of hypoxia-inducible factors and the expression of their target genes.161 Together, this data suggests that the preferential shift of the TERT splicing pattern is a highly regulated process, which can promptly respond to extra-cellular stimuli that ultimately affect stem cell identity. Reverse-transcriptase function of mTERT may be dispensable for modulation of Wnt-β-catenin signaling and its targeted gene expression.105 This indicates the likelihood that naturally occurring catalytically inactive TERT isoforms regulate Wnt signaling possibly through disruption and/augmentation of the Brg1-TCF-TERT transcription complex formation.
More recently, a novel hTERT isoform (now termed Δ4–13 variant) was discovered in both telomerase-positive and negative human cells.25 Remarkably, this in-frame alternatively spliced isoform is probably the most promising candidate to help us discern TERT functions that require catalytic activity from the ones that do not, since the alternative splicing almost cleanly excises the RT-domain while maintaining an open reading frame and sequence homology with its full-length counterpart. Overexpression of this variant in telomerase-negative cell lines enhanced cell proliferation and LiCl-induced Wnt signaling.25 However, the response was not as strong as that seen in the full-length hTERT overexpression controls, suggesting that the isoform may merely be carrying out a redundant role of its full-length counterpart.
Lastly, four independent alternative splicing events within the 5′ region of the transcript result in the partial to complete deletion of exon 2 and introduce early PTCs.25,134 Exon 2 contains portions of the telomerase essential N-terminal domain and RNA-interaction domain, which are lost, along with the catalytic core, in the process of splicing. Translation of the transcripts harboring exon 2 deletions through the wild-type reading frame would render all other potential splicing events downstream of exon 3 defunct due to the PTCs. Of note, the PTC introduced by exon 2 splicing may be bypassed if translation starts at an alternative start site downstream of the wild-type start codon. However, no evidence exists to confirm such a translation pattern to date. Interestingly, Wither et al. showed that exon 2-deletion (termed e2) is a common splice event in primates and that the e2 transcripts are highly abundant.134 In agreement, our preliminary data indicates that the e2 transcripts are present in hESCs and whose amount varies in cells cultured under different oxygen tensions (Betts, unpublished data). Despite the abundance of e2 transcripts, its translational competency is yet to be investigated. Regardless, its naturally high transcript abundance implies that exon 2-deletion may be a non-functional by-product of TERT regulation by alternative splicing, serving as a powerful switch to control telomerase activity and/or non-canonical functions of TERT.
Lack of consistency in methods of comparing isoform transcript abundance and poor antibody availability for TERT and its variants have greatly limited the progress of characterizing alternate splice isoforms of TERT in various species. Nonetheless, newly emerging pieces of evidence indicating the extra-telomeric roles of TERT (Fig. 1) and cell-type specific biases in splicing patterns are supportive of the functional importance of TERT and its variants in pluripotent stem cells. Fig. 1 may be much more complex with additional isoform specific functions, as well as multiple isoforms modulating each of its current roles.
Conclusion
Once regarded as a unipartite protein whose sole purpose is to re-lengthen telomeres, TERT is now considered a multi-functional protein, capable of modulating functions beyond telomere maintenance in various cell compartments. It is well accepted that TERT is upregulated in pluripotent stem cells, presumably to support the indefinite cell self-renewal by maintaining telomere lengths. Yet, this understanding lacks appreciation of TERT's possible contribution to pluripotency through its cross talk with the pluripotency network and non-canonical roles. The non-canonical roles of TERT as an active regulator of major signaling pathways, histone modulation, and mitochondrial function have been characterized mostly in a non-PSC context and in different species, making it challenging to gauge the true significance of TERT in PSCs based on our current knowledge of the field. Nonetheless, these functions of TERT, including telomere maintenance, closely relate to a number of highly regulated cell mechanisms of PSCs such as epigenetic and metabolic modulation. Here, we propose ways by which TERT may govern pluripotency and self-renewal in pluripotent stem cells. TERT's canonical and non-canonical functions collectively have a significant impact on pluripotency and self-renewal in pluripotent stem cells.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Dr. Andrew J. Watson and Nicole Edwards for their careful review of the manuscript.
Funding
This research is supported by operating grants to D.H.B. from the Canadian Institutes of Health Research (CIHR), the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Children's Health Research Institute (CHRI).
References
- 1.Muller HJ. The Remaking of Chromosomes. Collecting Net - Woods Hole 1938; 13:181-98 [Google Scholar]
- 2.McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics 1941; 26:234-82; PMID:17247004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan SR, Blackburn EH. Telomeres and telomerase. Philos Trans R Soc Lond B Biol Sci 2004; 359:109-21; PMID:15065663; http://dx.doi.org/ 10.1098/rstb.2003.1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayflick L. The Limited in Vitro Lifetime of Human Diploid Cell Strains. Exp Cell Res 1965; 37:614-36; PMID:14315085; http://dx.doi.org/ 10.1016/0014-4827(65)90211-9 [DOI] [PubMed] [Google Scholar]
- 5.Hayflick L, Moorhead PS. The serial cultivation of human diploid cell strains. Exp Cell Res 1961; 25:585-621; PMID:13905658; http://dx.doi.org/ 10.1016/0014-4827(61)90192-6 [DOI] [PubMed] [Google Scholar]
- 6.Armanios M. Telomerase and idiopathic pulmonary fibrosis. Mutat Res 2012; 730:52-58; http://dx.doi.org/ 10.1016/j.mrfmmm.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA 3rd, et al.. Telomerase Mutations in Families with Idiopathic Pulmonary Fibrosis. New England Journal of Medicine 2007; 356:1317-26; PMID:17392301; http://dx.doi.org/ 10.1056/NEJMoa066157 [DOI] [PubMed] [Google Scholar]
- 8.Aviv A. Genetics of leukocyte telomere length and its role in atherosclerosis. Mutat Res 2012; 730:68-74; http://dx.doi.org/ 10.1016/j.mrfmmm.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi H, Calado RT, Ly H, Kajigaya S, Baerlocher GM, Chanock SJ, Lansdorp PM, Young NS. Mutations in TERT, the Gene for Telomerase Reverse Transcriptase, in Aplastic Anemia. New England Journal of Medicine 2005; 352:1413-24; PMID:15814878; http://dx.doi.org/ 10.1056/NEJMoa042980 [DOI] [PubMed] [Google Scholar]
- 10.Carulli L, Anzivino C. Telomere and telomerase in chronic liver disease and hepatocarcinoma. World Journal of Gastroenterology : WJG 2014; 20:6287-92; PMID:24876749; http://dx.doi.org/20350645 10.3748/wjg.v20.i20.6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biron-Shental T, Sukenik-Halevy R, Sharon Y, Goldberg-Bittman L, Kidron D, Fejgin MD, Amiel A. Short telomeres may play a role in placental dysfunction in preeclampsia and intrauterine growth restriction. American journal of obstetrics and gynecology 2010; 202:381-e1; PMID:20350645; http://dx.doi.org/ 10.1016/j.ajog.2010.01.036 [DOI] [PubMed] [Google Scholar]
- 12.Biron-Shental T, Sukenik Halevy R, Goldberg-Bittman L, Kidron D, Fejgin MD, Amiel A. Telomeres are shorter in placental trophoblasts of pregnancies complicated with intrauterine growth restriction (IUGR). Early human development 2010; 86:451-6; PMID:20619976; http://dx.doi.org/ 10.1016/j.earlhumdev.2010.06.002 [DOI] [PubMed] [Google Scholar]
- 13.Biron-Shental T, Kidron D, Sukenik-Halevy R, Goldberg-Bittman L, Sharony R, Fejgin MD, Amiel A. TERC telomerase subunit gene copy number in placentas from pregnancies complicated with intrauterine growth restriction. Early human development 2011; 87:73-5; PMID:21168289; http://dx.doi.org/ 10.1016/j.earlhumdev.2010.08.024 [DOI] [PubMed] [Google Scholar]
- 14.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 1985; 43:405-13; PMID:3907856; http://dx.doi.org/ 10.1016/0092-8674(85)90170-9 [DOI] [PubMed] [Google Scholar]
- 15.Blackburn EH, Collins K. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 2011; 3:a003558; PMID:20660025; http://dx.doi.org/ 10.1101/cshperspect.a003558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis 2010; 31:9-18; PMID:19887512; http://dx.doi.org/ 10.1093/carcin/bgp268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Autexier C, Greider CW. Telomerase and cancer: revisiting the telomere hypothesis. Trends Biochem Sci 1996; 21:387-91; PMID:8918193; http://dx.doi.org/ 10.1016/S0968-0004(96)10042-6 [DOI] [PubMed] [Google Scholar]
- 18.Xie X, Hiona A, Lee AS, Cao F, Huang M, Li Z, Cherry A, Pei X, Wu JC. Effects of long-term culture on human embryonic stem cell aging. Stem Cells Dev 2010; 20:127-38; PMID:20629482; http://dx.doi.org/ 10.1089/scd.2009.0475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang Y, Liang P, Liu D, Huang J, Songyang Z. Telomere regulation in pluripotent stem cells. Protein Cell 2014; 5:194-202; PMID:24563217; http://dx.doi.org/ 10.1007/s13238-014-0028-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalmbach K, Robinson LG Jr, Wang F, Liu L, Keefe D. Telomere length reprogramming in embryos and stem cells. Biomed Res Int 2014; 2014:925121; PMID:24719895; http://dx.doi.org/ 10.1155/2014/925121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gourronc FA, Klingelhutz AJ. Therapeutic opportunities: Telomere maintenance in inducible pluripotent stem cells. Mutat Res 2012. 730:98-105; PMID:21605571; http://dx.doi.org/21283131 10.1016/j.mrfmmm.2011.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang J, Wang F, Okuka M, Liu N, Ji G, Ye X, Zuo B, Li M, Liang P, Ge WW, et al.. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res 2011; 21:779-92; PMID:21283131; http://dx.doi.org/ 10.1038/cr.2011.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keefe DL, Franco S, Liu L, Trimarchi J, Cao B, Weitzen S, Agarwal S, Blasco MA. Telomere length predicts embryo fragmentation after in vitro fertilization in women–toward a telomere theory of reproductive aging in women. Am J Obstet Gynecol 2005; 192:1256-60; discussion 1260–1; PMID:15846215; http://dx.doi.org/ 10.1016/j.ajog.2005.01.036 [DOI] [PubMed] [Google Scholar]
- 24.Listerman I, Sun J, Gazzaniga FS, Lukas JL, Blackburn EH. The major reverse transcriptase–incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis. Cancer Res 2013; 73:2817-28; PMID:23610451; http://dx.doi.org/ 10.1158/0008-5472.CAN-12-3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hrdlickova R, Nehyba J, Bose HR Jr. Alternatively spliced telomerase reverse transcriptase variants lacking telomerase activity stimulate cell proliferation. Mol Cell Biol 2012; 32:4283-96; PMID:22907755; http://dx.doi.org/ 10.1128/MCB.00550-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Telomere length predicts replicative capacity of human fibroblasts. Proc Natl Acad Sci U S A 1992; 89:10114-8; PMID:1438199; http://dx.doi.org/ 10.1073/pnas.89.21.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 2000; 227:271-278; PMID:11071754; http://dx.doi.org/ 10.1006/dbio.2000.9912 [DOI] [PubMed] [Google Scholar]
- 28.Vaziri H, Chapman KB, Guigova A, Teichroeb J, Lacher MD, Sternberg H, Singec I, Briggs L, Wheeler J, Sampathkumar J, et al.. Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Regenerative Medicine 2010; 5:345-63; PMID:20230312; http://dx.doi.org/ 10.2217/rme.10.21 [DOI] [PubMed] [Google Scholar]
- 29.Winkler T, Hong SG, Decker JE, Morgan MJ, Wu C, Hughes WM 5th, Yang Y, Wangsa D, Padilla-Nash HM, Ried T, et al.. Defective telomere elongation and hematopoiesis from telomerase-mutant aplastic anemia iPSCs. J Clin Investig 2013; 123:1952-63; http://dx.doi.org/ 10.1172/JCI67146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, Murthy S, Cibelli JB. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS ONE 2009; 4:e8124; PMID:19956585; http://dx.doi.org/ 10.1371/journal.pone.0008124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji G, Ruan W, Liu K, Wang F, Sakellariou D, Chen J, Yang Y, Okuka M, Han J, Liu Z, et al.. Telomere reprogramming and maintenance in porcine iPS cells. PLoS ONE 2013; 8:e74202; PMID:24098638; http://dx.doi.org/20354136 10.1371/annotation/f5e4554b-18cc-46ef-ac39-73ac4d6750ae [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathew R, Jia W, Sharma A, Zhao Y, Clarke LE, Cheng X, Wang H, Salli U, Vrana KE, Robertson GP, et al.. Robust activation of the human but not mouse telomerase gene during the induction of pluripotency. FASEB J 2010; 24:2702-15; PMID:20354136; http://dx.doi.org/ 10.1096/fj.09-148973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Yin Y, Ye X, Liu K, Zhu H, Wang L, Chiourea M, Okuka M, Ji G, Dan J, et al.. Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res 2012; 22:757-68; PMID:22184006; http://dx.doi.org/ 10.1038/cr.2011.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yehezkel S, Rebibo-Sabbah A, Segev Y, Tzukerman M, Shaked R, Huber I, Gepstein L, Skorecki K, Selig S. Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics 2011; 6:63-75; PMID:20861676; http://dx.doi.org/ 10.4161/epi.6.1.13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park KD, Seong SK, Park YM, Choi Y, Park JH, Lee SH, Baek DH, Kang JW, Choi KS, Park SN, et al.. Telomerase reverse transcriptase related with telomerase activity regulates tumorigenic potential of mouse embryonic stem cells. Stem Cells Dev 2010; 20:149-57; PMID:20486780; http://dx.doi.org/ 10.1089/scd.2009.0523 [DOI] [PubMed] [Google Scholar]
- 36.Holt SE, Wright WE, Shay JW. Regulation of telomerase activity in immortal cell lines. Mol Cell Biol 1996; 16:2932-9; PMID:8649404; http://dx.doi.org/ 10.1128/MCB.16.6.2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet 1996; 18:173-9; PMID:8934879; http://dx.doi.org/ 10.1002/(SICI)1520-6408(1996)18:2%3c173::AID-DVG10%3e3.0.CO;2-3 [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, Wright WE. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 2009; 138:463-75; PMID:19665970; http://dx.doi.org/ 10.1016/j.cell.2009.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez JM, De Vos J, et al.. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Gen Dev 2011; 25:2248-53; PMID:22056670; http://dx.doi.org/ 10.1101/gad.173922.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal S, Loh YH, McLoughlin EM, Huang J, Park IH, Miller JD, Huo H, Okuka M, Dos Reis RM, Loewer S, et al.. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature 2010; 464:292-6; PMID:20164838; http://dx.doi.org/ 10.1038/nature08792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hemann MT, Strong MA, Hao L.-Y, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107:67-77; PMID:11595186; http://dx.doi.org/ 10.1016/S0092-8674(01)00504-9 [DOI] [PubMed] [Google Scholar]
- 42.Batista LFZ, Pech MF, Zhong FL, Nguyen HN, Xie KT, Zaug AJ, Crary SM, Choi J, Sebastiano V, Cherry A, et al.. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 2011; 474:399-402; PMID:21602826; http://dx.doi.org/ 10.1038/nature10084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J, Wang F, Okuka M, Liu N, Ji G, Ye X, Zuo B, Li M, Liang P, Ge WW, et al.. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res 2011; 21:779-92; PMID:21283131; http://dx.doi.org/ 10.1038/cr.2011.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le R, Kou Z, Jiang Y, Li M, Huang B, Liu W, Li H, Kou X, He W, Rudolph KL, et al.. Enhanced telomere rejuvenation in pluripotent cells reprogrammed via nuclear transfer relative to induced pluripotent stem cells. Cell Stem Cell 2014; 14:27-39; PMID:24268696; http://dx.doi.org/ 10.1016/j.stem.2013.11.005 [DOI] [PubMed] [Google Scholar]
- 45.Pucci F, Gardano L, Harrington L. Short Telomeres in ESCs Lead to Unstable Differentiation. Cell Stem Cell 2013; 12:479-86; PMID:23561444; http://dx.doi.org/ 10.1016/j.stem.2013.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dang-Nguyen TQ, Haraguchi S, Furusawa T, Somfai T, Kaneda M, Watanabe S, Akagi S, Kikuchi K, Tajima A, Nagai T. Downregulation of histone methyltransferase genes SUV39H1 and SUV39H2 increases telomere length in embryonic stem-like cells and embryonic fibroblasts in pigs. J Reprod Dev 2013; 59:27-32; PMID:23018532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Bonis ML, Ortega S, Blasco MA. SIRT1 is necessary for proficient telomere elongation and genomic stability of induced pluripotent stem cells. Stem cell reports 2014; 2:690-706; PMID:24936455; http://dx.doi.org/ 10.1016/j.stemcr.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai C-C, Chen CL, Liu HC, Lee YT, Wang HW, Hou LT, Hung SC. Overexpression of hTERT increases stem-like properties and decreases spontaneous differentiation in human mesenchymal stem cell lines. J Biomed Sci 2010; 17:10-1186; PMID:20152059; http://dx.doi.org/ 10.1186/1423-0127-17-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng S, Liu L, Sun Y, Xie P, Hu L, Yuan D, Chen D, Ouyang Q, Lin G, Lu G. Telomerase-mediated telomere elongation from human blastocysts to embryonic stem cells. J Cell Sci 2014; 127:752-62; PMID:24338368; http://dx.doi.org/ 10.1242/jcs.131433 [DOI] [PubMed] [Google Scholar]
- 50.Liu L, Bailey SM, Okuka M, Muñoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, et al.. Telomere lengthening early in development. Nat Cell Biol 2007; 9:1436-41; PMID:17982445; http://dx.doi.org/ 10.1038/ncb1664 [DOI] [PubMed] [Google Scholar]
- 51.Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, et al.. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 2010; 464:858-63; PMID:20336070; http://dx.doi.org/ 10.1038/nature08882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varela E, Schneider RP, Ortega S, Blasco MA. Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc Natl Acad Sci 2011; 108(37):15207-12; PMID: 21873233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, Serrano M, Blasco MA. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 2009; 4:141-54; PMID:19200803; http://dx.doi.org/ 10.1016/j.stem.2008.12.010 [DOI] [PubMed] [Google Scholar]
- 54.Jiang J, Lv W, Ye X, Wang L, Zhang M, Yang H, Okuka M, Zhou C, Zhang X, Liu L, et al.. Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell research 2013; 23:92-106; PMID:23147797; http://dx.doi.org/ 10.1038/cr.2012.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee K, Gollahon L. Abstract 2047: The interaction of ZSCAN4 with TRF1: Effects on regulation of telomere elongation in cancer cells. Cancer research 2012; 72:2047; PMID:26403970; http://dx.doi.org/24735877 10.1158/1538-7445.AM2012-2047 [DOI] [Google Scholar]
- 56.Dan J, Liu Y, Liu N, Chiourea M, Okuka M, Wu T, Ye X, Mou C, Wang L, Wang L, et al.. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Developmental cell 2014; 29:7-19; PMID:24735877; http://dx.doi.org/ 10.1016/j.devcel.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider RP, Garrobo I, Foronda M, Palacios JA, Marión RM, Flores I, Ortega S, Blasco MA. TRF1 is a stem cell marker and is essential for the generation of induced pluripotent stem cells. Nature communications 2013; 4:1946; PMID: 23735977 [DOI] [PubMed] [Google Scholar]
- 58.Ozturk S, Sozen B, Demir N. Telomere length and telomerase activity during oocyte maturation and early embryo development in mammalian species. Molecular human reproduction 2014; 20:15-30; PMID:23928157; http://dx.doi.org/ 10.1093/molehr/gat055 [DOI] [PubMed] [Google Scholar]
- 59.Blasco MA. Telomere length, stem cells and aging. Nature chemical biology 2007; 3:640-649; PMID:17876321; http://dx.doi.org/ 10.1038/nchembio.2007.38 [DOI] [PubMed] [Google Scholar]
- 60.Wong LH, Ren H, Williams E, McGhie J, Ahn S, Sim M, Tam A, Earle E, Anderson MA, Mann J, et al.. Histone H3. 3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome research 2009; 19:404-14; PMID:19196724; http://dx.doi.org/ 10.1101/gr.084947.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y, Zhou P, Xin Y, Wang J, Zhu Z, Hu J, Wei S, Ma H. Trend of telomerase activity change during human iPSC self-renewal and differentiation revealed by a quartz crystal microbalance based assay. Sci Rep 2014; 4:6978; PMID:25381797; http://dx.doi.org/ 10.1038/srep06978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dashinimaev EB, et al.. Induction of telomerase activity increase reprogramming efficiency of human dermal fibroblasts. Moscow University Biological Sciences Bulletin 2012; 67:6-12; http://dx.doi.org/ 10.3103/S0096392512010038 [DOI] [Google Scholar]
- 63.Kinoshita T, Nagamatsu G, Saito S, Takubo K, Horimoto K, Suda T. Telomerase Reverse Transcriptase Has an Extratelomeric Function in Somatic Cell Reprogramming. Journal of Biological Chemistry 2014; 289:15776-87; PMID:24733392; http://dx.doi.org/ 10.1074/jbc.M113.536037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining Molecular Cornerstones during Fibroblast to iPS Cell Reprogramming in Mouse. Cell Stem Cell 2008;2:230-40; PMID:18371448; http://dx.doi.org/ 10.1016/j.stem.2008.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R, Kemler R. Wnt/β-Catenin Signaling Regulates Telomerase in Stem Cells and Cancer Cells. Science 2012; 336:1549-54; PMID:22723415; http://dx.doi.org/ 10.1126/science.1218370 [DOI] [PubMed] [Google Scholar]
- 66.Wong C-W, Hou PS, Tseng SF, Chien CL, Wu KJ, Chen HF, Ho HN, Kyo S, Teng SC. Krüppel-Like Transcription Factor 4 Contributes to Maintenance of Telomerase Activity in Stem Cells. Stem Cells 2010; 28:1510-17; PMID:20629177; http://dx.doi.org/ 10.1002/stem.477 [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y, Toh L, Lau P, Wang X. Human telomerase reverse transcriptase (hTERT) is a novel target of the Wnt/β-catenin pathway in human cancer. Journal of Biological Chemistry 2012; 287:32494-511; PMID:22854964; http://dx.doi.org/ 10.1074/jbc.M112.368282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009; 460:1149-53; PMID:19668189; http://dx.doi.org/ 10.1038/nature08287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol 2005; 7:1074-82; PMID:16244670; http://dx.doi.org/ 10.1038/ncb1314 [DOI] [PubMed] [Google Scholar]
- 70.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 126:663-76; PMID:16904174; http://dx.doi.org/ 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 71.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc Is Dispensable for Direct Reprogramming of Mouse Fibroblasts. Cell Stem Cell 2:10-12; PMID:18371415; http://dx.doi.org/ 10.1016/j.stem.2007.12.001 [DOI] [PubMed] [Google Scholar]
- 72.Wu K-J, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet 1999; 21:220-4; PMID:9988278; http://dx.doi.org/ 10.1038/6010 [DOI] [PubMed] [Google Scholar]
- 73.Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Gen Dev 1998; 12:1769-74; PMID:9637678; http://dx.doi.org/ 10.1101/gad.12.12.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bourillot PY, Aksoy I, Schreiber V, Wianny F, Schulz H, Hummel O, Hubner N, Savatier P. Novel STAT3 target genes exert distinct roles in the inhibition of mesoderm and endoderm differentiation in cooperation with Nanog. Stem Cells 2009; 27:1760-71; PMID:19544440; http://dx.doi.org/ 10.1002/stem.110 [DOI] [PubMed] [Google Scholar]
- 75.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 Regulates OCT4, SOX2, and KLF4 and Represses Pluripotency in Human Embryonic Stem Cells. Cell 137:647-58; PMID:19409607; http://dx.doi.org/ 10.1016/j.cell.2009.02.038 [DOI] [PubMed] [Google Scholar]
- 76.Erdmann N, Liu Y, Harrington L. Distinct dosage requirements for the maintenance of long and short telomeres in mTert heterozygous mice. Proc Natl Acad Sci of the United States of America 2004; 101:6080-5; PMID:15079066; http://dx.doi.org/ 10.1073/pnas.0401580101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blasco MA, Lee HW, Hande MP, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell 1997; 91:25-34; PMID:9335332; http://dx.doi.org/ 10.1016/S0092-8674(01)80006-4 [DOI] [PubMed] [Google Scholar]
- 78.Sung L-Y, Chang WF, Zhang Q, Liu CC, Liou JY, Chang CC, Ou-Yang H, Guo R, Fu H, Cheng WT, et al.. Telomere Elongation and Naive Pluripotent Stem Cells Achieved from Telomerase Haplo-Insufficient Cells by Somatic Cell Nuclear Transfer. Cell reports 2014; 9:1603-9; PMID:25464850; http://dx.doi.org/ 10.1016/j.celrep.2014.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al.. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol 2009; 11:197-203; PMID:19136965; http://dx.doi.org/ 10.1038/ncb1827 [DOI] [PubMed] [Google Scholar]
- 80.Festuccia N, Osorno R, Halbritter F, Karwacki-Neisius V, Navarro P, Colby D, Wong F, Yates A, Tomlinson SR, Chambers I. Esrrb is a direct nanog target gene that can substitute for nanog function in pluripotent cells. Cell Stem Cell 2012; 11:477-90; PMID:23040477; http://dx.doi.org/ 10.1016/j.stem.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li M, He Y, Dubois W, Wu X, Shi J, Huang J. Distinct Regulatory Mechanisms and Functions for p53-Activated and p53-Repressed DNA Damage Response Genes in Embryonic Stem Cells. Molecular Cell 2012; 46:30-42; PMID:22387025; http://dx.doi.org/ 10.1016/j.molcel.2012.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li L, Wang S, Jezierski A, Moalim-Nour L, Mohib K, Parks RJ, Retta SF, Wang L. A unique interplay between Rap1 and E-cadherin in the endocytic pathway regulates self-renewal of human embryonic stem cells. Stem Cells 2010; 28:247-57; PMID:20039365; http://dx.doi.org/ 10.1002/stem.532 [DOI] [PubMed] [Google Scholar]
- 83.Fong YW, Ho JJ, Inouye C, Tjian R. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells (ed Blencowe BJ.) Elife 3; e03573 (2014); PMID:2540768018203676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang C, Przyborski S, Cooke MJ, Zhang X, Stewart R, Anyfantis G, Atkinson SP, Saretzki G, Armstrong L, et al.. A key role for telomerase reverse transcriptase unit in modulating human embryonic stem cell proliferation, cell cycle dynamics, and in vitro differentiation. Stem Cells 2008; 26:850-63; PMID:18203676; http://dx.doi.org/ 10.1634/stemcells.2007-0677 [DOI] [PubMed] [Google Scholar]
- 85.Radan L, Hughes CS, Teichroeb JH, Vieira Zamora FM, Jewer M, Postovit LM, Betts DH. Microenvironmental regulation of telomerase isoforms in human embryonic stem cells. Stem Cells Dev 2014; 23:2046-66; PMID:24749509; http://dx.doi.org/ 10.1089/scd.2013.0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sexton AN, Regalado SG, Lai CS, Cost GJ, O'Neil CM, Urnov FD, Gregory PD, Jaenisch R, Collins K, Hockemeyer D. Genetic and molecular identification of three human TPP1 functions in telomerase action: recruitment, activation, and homeostasis set point regulation. Gen Dev 2014; 28:1885-99; PMID:25128433; http://dx.doi.org/ 10.1101/gad.246819.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Choi J, Southworth LK, Sarin KY, Venteicher AS, Ma W, Chang W, Cheung P, Jun S, Artandi MK, Shah N, et al.. TERT Promotes Epithelial Proliferation through Transcriptional Control of a Myc- and Wnt-Related Developmental Program. PLoS Genet 2008; 4:e10; PMID:18208333; http://dx.doi.org/ 10.1371/journal.pgen.0040010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cong Y, Shay JW. Actions of human telomerase beyond telomeres. Cell Res 2008; 18:725-32; PMID:18574498; http://dx.doi.org/ 10.1038/cr.2008.74 [DOI] [PubMed] [Google Scholar]
- 89.Gu B-W, Bessler M, Mason PJ. A pathogenic dyskerin mutation impairs proliferation and activates a DNA damage response independent of telomere length in mice. Proc Natl Acad Sci 2008; 105:10173-8; PMID:18626023; http://dx.doi.org/16107853 10.1073/pnas.0803559105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sarin KY. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 2005; 436:1048-52; PMID:16107853; http://dx.doi.org/ 10.1038/nature03836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stewart SA, Hahn WC, O'Connor BF, Banner EN, Lundberg AS, Modha P, Mizuno H, Brooks MW, Fleming M, Zimonjic DB, et al.. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc Natl Acad Sci 2002; 99:12606-11; PMID:12193655; http://dx.doi.org/12717449 10.1073/pnas.182407599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol 2003; 5:474-9; PMID:12717449; http://dx.doi.org/ 10.1038/ncb985 [DOI] [PubMed] [Google Scholar]
- 93.Flores I, Cayuela ML, Blasco MA. Effects of Telomerase and Telomere Length on Epidermal Stem Cell Behavior. Science 2005; 309:1253-6; PMID:16037417; http://dx.doi.org/ 10.1126/science.1115025 [DOI] [PubMed] [Google Scholar]
- 94.Martínez P, Blasco MA. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat Rev Cancer 2011; 11:161-76; PMID:21346783; http://dx.doi.org/12154355 10.1038/nrc3025 [DOI] [PubMed] [Google Scholar]
- 95.Blasco MA. Telomerase beyond telomeres. Nat Rev Cancer 2002; 2:627-33; PMID:12154355; http://dx.doi.org/ 10.1038/nrc862 [DOI] [PubMed] [Google Scholar]
- 96.Singhapol C, Pal D, Czapiewski R, Porika M, Nelson G, Saretzki GC. Mitochondrial telomerase protects cancer cells from nuclear DNA damage and apoptosis. PLoS ONE 2013; 8:9; PMID:23326372; http://dx.doi.org/21071633 10.1371/journal.pone.0052989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Indran IR, Hande MP, Pervaiz S. hTERT Overexpression Alleviates Intracellular ROS Production, Improves Mitochondrial Function, and Inhibits ROS-Mediated Apoptosis in Cancer Cells. Cancer Res 2011; 71:266-76; PMID:21071633; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-1588 [DOI] [PubMed] [Google Scholar]
- 98.Nitta E, Yamashita M, Hosokawa K, Xian M, Takubo K, Arai F, Nakada S, Suda T. Telomerase reverse transcriptase protects ATM-deficient hematopoietic stem cells from ROS-induced apoptosis through a telomere-independent mechanism 2011. [DOI] [PubMed] [Google Scholar]
- 99.Liu Z, Wan P, Duan H, Zhou J, Tan B, Liu Y, Zhou Q, Zhou C, Huang Z, Tian B, et al.. ES micro-environment enhances stemness and inhibits apoptosis in human limbal stem cells via the maintenance of telomerase activity. PLoS ONE 2013; 8:e53576; PMID:23326460; http://dx.doi.org/ 10.1371/journal.pone.0053576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mattiussi M, Tilman G, Lenglez S, Decottignies A. Human telomerase represses ROS-dependent cellular responses to Tumor Necrosis Factor-α without affecting NF-κB activation. Cellular Signalling 2012; 24:708-17; PMID:22108091; http://dx.doi.org/ 10.1016/j.cellsig.2011.11.004 [DOI] [PubMed] [Google Scholar]
- 101.Kong F, Zheng C, Xu D. Telomerase as a “stemness” enzyme. Science China Life Sciences 2014; 57:564-70; PMID:24829107; http://dx.doi.org/ 10.1007/s11427-014-4666-6 [DOI] [PubMed] [Google Scholar]
- 102.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009; 461:230-5; PMID:19701182; http://dx.doi.org/ 10.1038/nature08283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sharma NK, Reyes A, Green P, Caron MJ, Bonini MG, Gordon DM, Holt IJ, Santos JH. Human telomerase acts as a hTR-independent reverse transcriptase in mitochondria. Nucleic Acids Res 2011. 40:712-25; PMID: 2193751321730156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Okamoto N, Yasukawa M, Nguyen C, Kasim V, Maida Y, Possemato R, Shibata T, Ligon KL, Fukami K, Hahn WC, et al.. Maintenance of tumor initiating cells of defined genetic composition by nucleostemin. Proc Natl Acad Sci U S A 2011; 108:20388-93; PMID:21730156; http://dx.doi.org/ 10.1073/pnas.1015171108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, et al.. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 2009; 460:66-72; PMID:19571879; http://dx.doi.org/ 10.1038/nature08137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, Wong M, Zhang Z, Li G, Sung WK, et al.. Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol 2012; 14:1270-1281; PMID:23159929; http://dx.doi.org/ 10.1038/ncb2621 [DOI] [PubMed] [Google Scholar]
- 107.Maida Y, Yasukawa M, Okamoto N, Ohka S, Kinoshita K, Totoki Y, Ito TK, Minamino T, Nakamura H, Yamaguchi S, et al.. Involvement of telomerase reverse transcriptase in heterochromatin maintenance. Mol Cell Biol 2014; 34:1576-93; PMID:24550003; http://dx.doi.org/ 10.1128/MCB.00093-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lassmann T, Maida Y, Tomaru Y, Yasukawa M, Ando Y, Kojima M, Kasim V, Simon C, Daub CO, Carninci P, et al.. Telomerase Reverse Transcriptase Regulates microRNAs. Int J Mol Sci 2015; 16:1192-208; PMID:25569094; http://dx.doi.org/ 10.3390/ijms16011192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hang CT, Yang J, Han P, Cheng HL, Shang C, Ashley E, Zhou B, Chang CP. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010; 466:62-67; PMID:20596014; http://dx.doi.org/ 10.1038/nature09130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Griffin CT, Brennan J, Magnuson T. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development 2008; 135:493-500; PMID:18094026; http://dx.doi.org/ 10.1242/dev.010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development 2005; 132:105-15; PMID:15576411; http://dx.doi.org/ 10.1242/dev.01548 [DOI] [PubMed] [Google Scholar]
- 112.Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al.. A Brg1 Null Mutation in the Mouse Reveals Functional Differences among Mammalian SWI/SNF Complexes. Mol Cell 2000; 6:1287-95; PMID:11163203; http://dx.doi.org/ 10.1016/S1097-2765(00)00127-1 [DOI] [PubMed] [Google Scholar]
- 113.Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Gen Dev 2006; 20:1744-54; PMID:16818606; http://dx.doi.org/ 10.1101/gad.1435106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Listerman I, Gazzaniga FS, Blackburn EH. An investigation of the effects of the core protein telomerase reverse transcriptase on wnt signaling in breast Cancer cells. Mol Cell Biol 2014; 34:280-9; PMID:24216762; http://dx.doi.org/ 10.1128/MCB.00844-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu Z, Li Q, Li K, Chen L, Li W, Hou M, Liu T, Yang J, Lindvall C, Björkholm M, et al.. Telomerase reverse transcriptase promotes epithelial–mesenchymal transition and stem cell-like traits in cancer cells. Oncogene 2013; 32:4203-13; PMID:23045275; http://dx.doi.org/ 10.1038/onc.2012.441 [DOI] [PubMed] [Google Scholar]
- 116.Orsulic S, Huber O, Aberle H, Arnold S, Kemler R. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J Cell Sci 1999; 112 (Pt 8), 1237-45; PMID:10085258 [DOI] [PubMed] [Google Scholar]
- 117.Liu X, Sun H, Qi J, Wang L, He S, Liu J, Feng C, Chen C, Li W, Guo Y, et al.. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT–MET mechanism for optimal reprogramming. Nat Cell Biol 2013; 15:829-38; PMID:23708003; http://dx.doi.org/ 10.1038/ncb2765 [DOI] [PubMed] [Google Scholar]
- 118.Moslehi J, DePinho RA, Sahin E. Telomeres and Mitochondria in the Aging Heart. Circulation Res 2012; 110:1226-37; PMID:22539756; http://dx.doi.org/ 10.1161/CIRCRESAHA.111.246868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saretzki G. Extra-telomeric functions of human telomerase: cancer, mitochondria and oxidative stress. Curr Pharmaceutical Design 2014; 20:6386-403; PMID:24975608; http://dx.doi.org/ 10.2174/1381612820666140630095606 [DOI] [PubMed] [Google Scholar]
- 120.Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. The EMBO Journal 1987; 6:409-17; PMID:3582365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rosenbluh J, Nijhawan D, Chen Z, Wong KK, Masutomi K, Hahn WC. RMRP Is a Non-Coding RNA Essential for Early Murine Development. PLoS ONE 2011; 6:e26270; PMID:22039455; http://dx.doi.org/ 10.1371/journal.pone.0026270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Maida Y, Kyo S, Lassmann T, Hayashizaki Y, Masutomi K. Off-Target Effect of Endogenous siRNA Derived from RMRP in Human Cells. Int J Mol Sci 2013; 14:9305-18; PMID:23629666; http://dx.doi.org/ 10.3390/ijms14059305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Drevytska TI, Nagibin VS, Gurianova VL, Kedlyan VR, Moibenko AA, Dosenko VE. Silencing of TERT decreases levels of miR-1, miR-21, miR-29a and miR-208a in cardiomyocytes. Cell Biochem Funct 2014; 32:565-70; PMID:25156787; http://dx.doi.org/ 10.1002/cbf.3051 [DOI] [PubMed] [Google Scholar]
- 124.Strong MA, Vidal-Cardenas SL, Karim B, Yu H, Guo N, Greider CW. Phenotypes in mTERT(+)/(−) and mTERT(−)/(−) mice are due to short telomeres, not telomere-independent functions of telomerase reverse transcriptase. Mol Cell Biol 2011; 31:2369-79; PMID:21464209; http://dx.doi.org/ 10.1128/MCB.05312-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yi X, Shay JW, Wright WE. Quantitation of telomerase components and hTERT mRNA splicing patterns in immortal human cells. Nucleic Acids Res 2001; 29:4818-25; PMID:11726691; http://dx.doi.org/ 10.1093/nar/29.23.4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ulaner GA, Giudice LC. Developmental regulation of telomerase activity in human fetal tissues during gestation. Mol Hum Reprod 1997; 3:769-73; PMID:9358002; http://dx.doi.org/ 10.1093/molehr/3.9.769 [DOI] [PubMed] [Google Scholar]
- 127.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res 1998; 58:4168-72; PMID:9751630 [PubMed] [Google Scholar]
- 128.Ulaner GA, Hu J.-F, Vu TH, Giudice LC, Hoffman AR. Tissue-specific alternate splicing of human telomerase reverse transcriptase (hTERT) influences telomere lengths during human development. Int J Cancer 2001; 91:644-9; PMID:11267974; http://dx.doi.org/11517292 10.1002/1097-0215(200002)9999:9999%3c::AID-IJC1103%3e3.0.CO;2-V [DOI] [PubMed] [Google Scholar]
- 129.Yokoyama Y, Wan X, Takahashi Y, Shinohara A, Tamaya T. Alternatively spliced variant deleting exons 7 and 8 of the human telomerase reverse transcriptase gene is dominantly expressed in the uterus. Mol Hum Reprod 2001; 7:853-7; PMID:11517292; http://dx.doi.org/ 10.1093/molehr/7.9.853 [DOI] [PubMed] [Google Scholar]
- 130.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet 2001; 17:100-7; PMID:11173120; http://dx.doi.org/ 10.1016/S0168-9525(00)02176-4 [DOI] [PubMed] [Google Scholar]
- 131.Kilian A, Bowtell DD, Abud HE, Hime GR, Venter DJ, Keese PK, Duncan EL, Reddel RR, Jefferson RA. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet 1997; 6:2011-9; PMID:9328464; http://dx.doi.org/ 10.1093/hmg/6.12.2011 [DOI] [PubMed] [Google Scholar]
- 132.Hisatomi H, Ohyashiki K, Ohyashiki JH, Nagao K, Kanamaru T, Hirata H, Hibi N, Tsukada Y. Expression profile of a gamma-deletion variant of the human telomerase reverse transcriptase gene. Neoplasia 2003; 5:193-7; PMID:12869302; http://dx.doi.org/ 10.1016/S1476-5586(03)80051-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sæbøe-Larssen S, Fossberg E, Gaudernack G. Characterization of novel alternative splicing sites in human telomerase reverse transcriptase (hTERT): analysis of expression and mutual correlation in mRNA isoforms from normal and tumour tissues. BMC Mol Biol 2006; 7:26; PMID: 1693964123110161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Withers JB, Ashvetiya T, Beemon KL. Exclusion of exon 2 is a common mRNA splice variant of primate telomerase reverse transcriptases. PLoS One 2012; 7:e48016; PMID:23110161; http://dx.doi.org/ 10.1371/journal.pone.0048016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wick M, Zubov D, Hagen G. Genomic organization and promoter characterization of the gene encoding the human telomerase reverse transcriptase (hTERT). Gene 1999; 232:97-106; PMID:10333526; http://dx.doi.org/ 10.1016/S0378-1119(99)00108-0 [DOI] [PubMed] [Google Scholar]
- 136.Wong Mandy S, Chen L, Foster C, Kainthla R, Shay JW, Wright WE. Regulation of Telomerase Alternative Splicing: A Target for Chemotherapy. Cell Rep 2013; 3:1028-35; PMID:23562158; http://dx.doi.org/ 10.1016/j.celrep.2013.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yi X, White DM, Aisner DL, Baur JA, Wright WE, Shay JW. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia 2000; 2:433-40; PMID:11191110; http://dx.doi.org/ 10.1038/sj.neo.7900113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Mühlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci 2010; 67:677-700; PMID:19859661; http://dx.doi.org/ 10.1007/s00018-009-0177-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Santos JH, Meyer JN, Skorvaga M, Annab LA, Van Houten B. Mitochondrial hTERT exacerbates free-radical-mediated mtDNA damage. Aging Cell 2004; 3:399-411; PMID:15569357; http://dx.doi.org/ 10.1111/j.1474-9728.2004.00124.x [DOI] [PubMed] [Google Scholar]
- 140.Chung J, Khadka P, Chung IK. Nuclear import of hTERT requires a bipartite nuclear localization signal and Akt-mediated phosphorylation. J Cell Sci 2012; 125:2684-97; PMID:22366458; http://dx.doi.org/ 10.1242/jcs.099267 [DOI] [PubMed] [Google Scholar]
- 141.Xia J, Peng Y, Mian IS, Lue NF. Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol Cell Biol 2000; 20:5196-207; PMID:10866675; http://dx.doi.org/ 10.1128/MCB.20.14.5196-5207.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Seimiya H, Tanji M, Oh-hara T, Tomida A, Naasani I, Tsuruo T. Hypoxia up-regulates telomerase activity via mitogen-activated protein kinase signaling in human solid tumor cells. Biochem Biophys Res Commun 1999; 260:365-70; PMID:10403776; http://dx.doi.org/ 10.1006/bbrc.1999.0910 [DOI] [PubMed] [Google Scholar]
- 143.Haendeler J, Hoffmann J, Brandes RP, Zeiher AM, Dimmeler S. Hydrogen peroxide triggers nuclear export of telomerase reverse transcriptase via Src kinase family-dependent phosphorylation of tyrosine 707. Mol Cell Biol 2003; 23:4598-610; PMID:12808100; http://dx.doi.org/ 10.1128/MCB.23.13.4598-4610.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Arai K, Masutomi K, Khurts S, Kaneko S, Kobayashi K, Murakami S. Two independent regions of human telomerase reverse transcriptase are important for its oligomerization and telomerase activity. J Biol Chem 2002; 277:8538-44; PMID:11751869; http://dx.doi.org/ 10.1074/jbc.M111068200 [DOI] [PubMed] [Google Scholar]
- 145.Jakob S, Schroeder P, Lukosz M, Büchner N, Spyridopoulos I, Altschmied J, Haendeler J. Nuclear protein tyrosine phosphatase Shp-2 is one important negative regulator of nuclear export of telomerase reverse transcriptase. J Biol Chem 2008; 283:33155-61; PMID:18829466; http://dx.doi.org/ 10.1074/jbc.M805138200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mavrogiannou E, Strati A, Stathopoulou A, Tsaroucha EG, Kaklamanis L, Lianidou ES. Real-time RT-PCR quantification of human telomerase reverse transcriptase splice variants in tumor cell lines and non-small cell lung cancer. Clin Chem 2007; 53:53-61; PMID:17130181; http://dx.doi.org/ 10.1373/clinchem.2006.073015 [DOI] [PubMed] [Google Scholar]
- 147.Lincz LF, Mudge LM, Scorgie FE, Sakoff JA, Hamilton CS, Seldon M. Quantification of hTERT splice variants in melanoma by SYBR green real-time polymerase chain reaction indicates a negative regulatory role for the beta deletion variant. Neoplasia 2008; 10:1131-7; PMID:18813352; http://dx.doi.org/ 10.1593/neo.08644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ohyashiki JH, Hisatomi H, Nagao K, Honda S, Takaku T, Zhang Y, Sashida G, Ohyashiki K. Quantitative relationship between functionally active telomerase and major telomerase components (hTERT and hTR) in acute leukaemia cells. Br J Cancer 2005; 92:1942-7; PMID:15827550; http://dx.doi.org/ 10.1038/sj.bjc.6602546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Villa R, Porta CD, Folini M, Daidone MG, Zaffaroni N. Possible regulation of telomerase activity by transcription and alternative splicing of telomerase reverse transcriptase in human melanoma. J Invest Dermatol 2001; 116:867-73; PMID:11407973; http://dx.doi.org/ 10.1046/j.1523-1747.2001.01343.x [DOI] [PubMed] [Google Scholar]
- 150.Liu Y, Wu BQ, Zhong HH, Tian XX, Fang WG. Quantification of alternative splicing variants of human telomerase reverse transcriptase and correlations with telomerase activity in lung cancer. PLoS One 2012; 7:e38868; PMID:22723897; http://dx.doi.org/ 10.1371/journal.pone.0038868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rha SY, Jeung HC, Park KH, Kim JJ, Chung HC. Changes of telomerase activity by alternative splicing of full-length and beta variants of hTERT in breast cancer patients. Oncol Res 2009; 18:213-20; PMID:20225759; http://dx.doi.org/ 10.3727/096504009X12596189659123 [DOI] [PubMed] [Google Scholar]
- 152.Colgin LM, Wilkinson C, Englezou A, Kilian A, Robinson MO, Reddel RR. The hTERTalpha splice variant is a dominant negative inhibitor of telomerase activity. Neoplasia 2000; 2:426-32; PMID:11191109; http://dx.doi.org/ 10.1038/sj.neo.7900112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR. Protein composition of catalytically active human telomerase from immortal cells. Science 2007; 315:1850-3; PMID:17395830; http://dx.doi.org/ 10.1126/science.1138596 [DOI] [PubMed] [Google Scholar]
- 154.Yi X, Tesmer VM, Savre-Train I, Shay JW, Wright WE. Both transcriptional and posttranscriptional mechanisms regulate human telomerase template RNA levels. Mol Cell Biol 1999; 19:3989-97; PMID:10330139; http://dx.doi.org/ 10.1128/MCB.19.6.3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mukherjee S, Firpo EJ, Wang Y, Roberts JM. Separation of telomerase functions by reverse genetics. Proc Natl Acad Sci U S A 2011; 108:E1363-71; PMID:21949400; http://dx.doi.org/ 10.1073/pnas.1112414108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Santos JH, Meyer JN, Van Houten B. Mitochondrial localization of telomerase as a determinant for hydrogen peroxide-induced mitochondrial DNA damage and apoptosis. Hum Mol Genet 2006; 15:1757-68; PMID:16613901; http://dx.doi.org/ 10.1093/hmg/ddl098 [DOI] [PubMed] [Google Scholar]
- 157.Buchner N, Zschauer TC, Lukosz M, Altschmied J, Haendeler J. Downregulation of mitochondrial telomerase reverse transcriptase induced by H2O2 is Src kinase dependent. Exp Gerontol 2010; 45:558-62; PMID:20211239; http://dx.doi.org/ 10.1016/j.exger.2010.03.003 [DOI] [PubMed] [Google Scholar]
- 158.Haendeler J, Dröse S, Büchner N, Jakob S, Altschmied J, Goy C, Spyridopoulos I, Zeiher AM, Brandt U, Dimmeler S. Mitochondrial telomerase reverse transcriptase binds to and protects mitochondrial DNA and function from damage. Arterioscler Thromb Vasc Biol 2009; 29:929-35; PMID:19265030; http://dx.doi.org/ 10.1161/ATVBAHA.109.185546 [DOI] [PubMed] [Google Scholar]
- 159.Cerezo A, Kalthoff H, Schuermann M, Schafer B, Boukamp P. Dual regulation of telomerase activity through c-Myc-dependent inhibition and alternative splicing of hTERT. J Cell Sci 2002; 115:1305-12; PMID:11884529 [DOI] [PubMed] [Google Scholar]
- 160.Anderson CJ, Hoare SF, Ashcroft M, Bilsland AE, Keith WN. Hypoxic regulation of telomerase gene expression by transcriptional and post-transcriptional mechanisms. Oncogene 2006; 25:61-9; PMID:16170363 [DOI] [PubMed] [Google Scholar]
- 161.Forristal CE, Wright KL, Hanley NA, Oreffo RO, Houghton FD. Hypoxia inducible factors regulate pluripotency and proliferation in human embryonic stem cells cultured at reduced oxygen tensions. Reproduction 2010; 139:85-97; PMID:19755485; http://dx.doi.org/ 10.1530/REP-09-0300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Turner S, Wong HP, Rai J, Hartshorne GM. Telomere lengths in human oocytes, cleavage stage embryos and blastocysts. Molecular Human Reproduction 2010; 16:685-694; PMID:20573647; http://dx.doi.org/ 10.1093/molehr/gaq048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Akiyama M, Yamada O, Hideshima T, Yanagisawa T, Yokoi K, Fujisawa K, Eto Y, Yamada H, Anderson KC. TNFalpha induces rapid activation and nuclear translocation of telomerase in human lymphocytes. Biochem Biophys Res Commun 2004; 316:528-32; PMID:15020249; http://dx.doi.org/ 10.1016/j.bbrc.2004.02.080 [DOI] [PubMed] [Google Scholar]