Abstract
Endothelial cells line the inner wall of blood vessels and play an important role in the regulation of vascular tone, vascular permeability, and new vascular formation. Endothelial cell dysfunction is implicated in the development and progression of many cardiovascular diseases including ischemic heart disease. To examine the function and characterization of coronary endothelial cells, cell isolation is the first step and it requires high purity and quantity to conduct subsequent experiments. This protocol describes an efficient method to isolate adult mouse coronary endothelial cells. The mouse heart is dissected and minced into small pieces. After the digestion of the heart using dispase and collagenase II, cells are washed and incubated with magnetic beads which are conjugated with anti-CD31 antibody. The beads with endothelial cells are washed several times and are ready to use in various applications, including imaging and molecular biological experiments. Efficient isolation yields approximately 104 cells per one heart with over 90% purity.
Keywords: Cellular Biology, Issue 113, MCEC, magnetic beads, hearts, dissection, CD31, enzymatic digestion
Introduction
Mouse models of various cardiovascular diseases and metabolic disorders bear physiological and molecular changes which are similar to those found in patients. Furthermore, genetic alteration of mice is a powerful tool that allows us to investigate the pathogenic role of specific genes in the development and progression of diseases. Nowadays, cell-type-specific gene-knock-in or -out mice are easily generated in many laboratories and the measurement of mRNA and protein levels in specific cell types is the first step in determining whether the mice were truly genetically modified.
Endothelium is a thin single layer of endothelial cells (ECs) that lines the inner vascular wall. Endothelial cells play a critical role in regulating vascular tension, vascular permeability, and new vascular formation1,2. Endothelial dysfunction is a hallmark of many pathological conditions and changes in endothelial function can lead to various cardiovascular diseases3,4. It is thus significant to study the function of endothelial cells under physiological and pathophysiological conditions.
There are several ways to isolate ECs5-8 and the isolation method has to be optimized depending on which tissues and species will be used. This is because ECs from different tissues display a high level of heterogeneity with respect to their surface markers and protein expression9. Successful isolation of endothelial cells can often be challenging and requires some degree of training and practice. Once achieved, the method of isolation proposed in this protocol proves to be stable and efficient.
The overall goal of this method is to obtain mouse coronary ECs (MCECs) of high quality and quantity. Even though the quantity of cells collected from a heart is less than cells collected from other tissues using other techniques, this technique still provides better quality. The purity of endothelial cells in the resulting population is greater than 90%. Thus, this technique would be ideal for applications that rely on purely isolated endothelial cells such as cell imaging.
Protocol
The research on mice was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Arizona and was performed according to the National Institute of Health (NIH) guidelines.
NOTE: Sections 1, 2 and 3 should be carried out the day before the experiment as setup.
1. Preparing Solutions
- CD31 buffer (50 ml)
- Add 50 ml 1× Phosphate Buffer Saline (PBS) to a 50 ml tube. Weigh 0.05 g Bovine Serum Albumin (BSA) and 0.037 g Ethylenediaminetetraacetic acid (EDTA) and add to the 50 ml tube and put the tube to rotate for about 1 h until powder dissolves. Adjust pH to 7.4, filter through a 0.2 µm filter into a new 50 ml tube, and store at 4 ˚C until use.
- Hank's Balanced Salt Solution (HBSS) with HEPES (500 ml)
- Add 1.1915 g of HEPES to make 10 mM HEPES in HBSS solution and mix well. Measure the pH and adjust to 7.4. Filter and store at 4 ˚C.
- 1x Kreb's solution (1 L)
- Add 100 ml of 10× Kreb's solution [Table 1] to double distilled water in a 1 L flask. Weigh 2.1 g Sodium Bicarbonate and 2 g D-Glucose and add to the flask. Mix and adjust the total volume to 1 L.
- Bubble solution with 95% O2/5% CO2 gas for 30 min and filter solution inside the hood. Store at 4 ˚C and keep on ice during experiment.
2. Preparing the Magnetic Beads
Add 1 ml CD31 buffer into each 1.5 ml tube (one tube per sample). Mix the sheep anti-rat IgG beads and add the appropriate amount to each sample [Table 1]. Shake the tubes vigorously 30 times and place them in the magnetic plate for 1 min of shaking. Open the tubes while on the plate and dump the solutions into a bucket.
Add 1 ml CD31 buffer into each tube. Add 2 µl Rat anti-mouse CD31 antibody to each tube and rotate tubes O/N at 4 ºC.
3. Autoclaving
Autoclave all surgery tools and pipette tips. Cut the ends of pipette tips so that there are tips with 4 different diameters, from widest to narrowest. Approximate diameters for the tips are 3 mm, 2.5 mm, 2 mm, and 1.5 mm respectively. Cut the tips and autoclave the day before with 30 min of sterilization at a temperature of 121 ºC. NOTE: Sections 4 - 11 should be carried out on the day of the experiment.
4. Preparing Solutions
Prepare washing buffer containing 2% Iron-Fortified Calf Serum (IFCS) in M199. For imaging, prepare 30 ml/mouse.
Prepare enzyme solution (10 ml/sample). Weigh 0.6 unit/ml of Dispase and 1 mg/ml of Collagenase Type II into a 50 ml tube. Add M199 and rotate the tube at 4 ºC for 15 min. Filter through 0.2 µm filter into a new 50 ml tube inside the hood and keep at 4 ˚C until use.
Prepare HBSS/HEPES with Heparin. Add 10 ml of HBSS with 10 mM HEPES in 15 ml tube. Add 100 μl of Heparin to the tube and keep at 4 ˚C until use.
5. Washing the Beads
Take out the tubes with beads from the rotator at 4 ºC and bring to the hood. Place the tubes in the magnetic plate and shake for 1 min. Dump the solution and add 1 ml CD31 buffer into each tube.
Take the tubes out of the magnetic plate and shake vigorously 30 times. Put the tubes back in the magnetic plate and repeat for a total of 3 washes. After completion, put the tubes back to 4˚C and keep rotating until use.
6. Dissection
Inject the mice intraperitoneally with 0.1 ml of Heparin to prevent blood coagulation. Wait 10 mins, then anesthetize the mice using intraperitoneal injection of 0.01 ml of Pentobarbital. Check if the mouse is feeling pain by pinching the foot.
Place the mouse on a polystyrene board with head up and hold the arms with pins. Use forceps and tough-cut-scissors to cut the skin at the upper thoracic area up to the throat.
Make a cut in the center of the rib cage just above the diaphragm. Cut the bone sideways and upwards toward the throat to make a triangle exposing the heart and lungs. Cut the aorta just above the diaphragm.
Hold the thymus with forceps without pulling and use scissors to cut any connective tissue carefully to remove lungs and heart together. Place tissues in a beaker with 1× Kreb's, shake with forceps and transfer to 6 cm dish with 1× Kreb's.
Remove the lung lobes using scissors and forceps. Insert a 20 G sterile catheter into the aorta. Flush the blood in the coronary circulatory system with HBSS/Heparin. Make a small cut in the ventricle, shake the heart and transfer to a sample tube of M199. Keep tubes on ice.
7. Tissue Digestion
Transfer the heart to a 6 cm dish filled with 10 ml of enzyme solution and mince it into small pieces using scissors. Place the dishes in the 37 ºC incubator for 15 min. Agitate digested materials by pipetting up and down 30 times through a tip with a cut end to allow the tissue to pass through easier.
Put it back in the 37 ºC incubator for another 15 min. Repeat agitation three times, each time using pipette tips from biggest openings to narrowest and 15 min incubation between each.
After last 15 min incubation, take samples out and into the hood. Pipet solution up and down 30 times using pipet tip with the narrowest opening and pass through a 40 µm cell strainer into a new 50 ml tube.
Wash the dish with 10 ml washing buffer using a 10 ml pipette and transfer onto cell strainer. Put the 50 ml tubes in the centrifuge and spin at 400 g for 10 min at RT.
8. Washing and Conjugation with Beads
Remove supernatant without disrupting the pellet. Wash cells with 5 ml washing buffer twice.
Get one of the previously prepared 1.5 ml tubes with beads and work on one tube at a time. Put one 1.5 ml tube in the magnetic plate, shake 3 times and open it.
After last centrifuge, remove as much supernatant as possible without disrupting the pellet. Disassociate the cell clumps by flicking vigorously.
Add 1 ml washing buffer to the 50 ml tube and mix the cells with a 1 ml pipette. Dump the solution from the 1.5 ml tube and add 1 ml of cell suspension from the 50 ml tube to the beads. Flick the tubes 30 times then rotate at 4 ºC for 30 min.
9. Preparing for Cell Culture/Imaging
Prepare chambers for plating cells. Add gelatin solution (5% w/v, 300 µl) to the chamber well to coat the surface and incubate for 30 min at 37 ºC. After incubation, remove gelatin and dry the surface.
Take out the tubes with beads/cells after 30 min of rotation and put in the magnetic plate inside the hood. Shake for 2 min, dump and add 1 ml washing buffer.
Take tubes out of the plate, shake vigorously 30 times then flick 30 times. Put them back in magnetic plate and shake for 1 min. Repeat 4 times for a total of 5 washes, but with 1 min shaking on the plate.
After the last wash, take out one tube and work on each tube one at a time. Shake 30 times then flick 30 times. Put it on the magnetic plate and shake for 1 min. Dump and add 1 ml 20% IFCS EC media (pre-warmed at 37 ºC).
Shake vigorously 30 times then flick 30 times. Adjust pipet to 500 µl and pipet up and down 10 times. Transfer 500 µl to each chamber.
Put at 37 ºC incubator and leave O/N. Next day, wash cells with 10% FBS EC media.
10. Preparing for Western Blot (WB) Samples
Take out the tubes with beads/cells after 1 hr of rotation. Put them in the magnetic plate and shake for 2 min. Dump supernatant and add 1 ml cold HBSS. Shake vigorously 30 times then flick 30 times. Repeat twice for 3 washes, but with 1 min shaking on the plate.
After last wash, put tubes on magnetic plate and shake for 1 min. Remove buffer using pipette and lyse the cells for WB using a method previously described11,12. Collected lysate usually contains around 30 µg of protein.
11. Imaging
Wash the cells 3 times with media, with final volume of 250 µl for a chamber. Add 1.5 µl of Dil-acLDL to the cells. From this point, keep the cells in the dark.
Incubate for 3.5 hr at 37 ˚C. Add 1.5 µl of Hoechst, mix well and incubate for 30 min at 37 ˚C.Wash cells with PBS and fix with 4% Paraformaldehyde (PFA) for 20 min at RT.
Wash twice with PBS and block with 5% BSA-PBS for 30 min at RT. Wash twice with 1% BSA-PBS. Dilute BS-lectin-FITC using 1% BSA-PBS to a concentration of 2 µg/ml and add 300 µl to each chamber.
Put the chamber on the orbital shaker at RT for 30 min. Wash with PBS 3 times.
Visualize cells under the fluorescent microscope with exposure time of 200 msec for FITC (Lectin staining, an EC marker, excitation/emission 488 nm/519 nm), 30 msec for DAPI (Hoechst, a nuclear staining, excitation/emission at 358 nm/461 nm), and 300 msec for TRITC (acLDL staining, an EC marker, excitation/emission at 557 nm/576 nm). Acquire images using a 20X objective lens.
Representative Results
The successful isolation of coronary endothelial cells from a mouse heart typically yields around 104 cells with over 90% purity with a cobblestone shape. For a pure population of endothelial cells, caution should be taken throughout the protocol to ensure a sterile environment free from any contamination. The appearance of elongated cells or enlarged cells within the population indicates contamination with other cell types, smooth muscle cells and fibroblasts. Endothelial cell purity test is performed by staining cells with an endothelial cell surface marker, BS-Lectin and acLDL (Figure 1). The percentage of positive cells which exhibit both colors was over 90% in MCEC. Such a result indicates that the technique was performed successfully.
| PBS (10x) (1 L) | |||
| Material | MW | Amount (g) | |
| NaCl | 58.44 | 80 | |
| KCl | 74.55 | 2 | |
| Na2HPO4 | 141.96 | 6.1 | |
| KH2PO4 | 136.08 | 2 | |
| Krebs (10x) (1 L) | |||
| Material | MW | Conc (mM) | Amount (g) |
| NaCl | 58.44 | 118 | 69 |
| KCl | 74.56 | 4.7 | 3.5 |
| CaCl2•2H2O | 147 | 1.8 | 2.6 |
| MgSO4•7H2O | 246.5 | 1.2 | 2.96 |
| NaH2PO4 | 120 | 1.2 | 1.44 |
| EC Media (20%) (500 ml) | |||
| Material | Amount | ||
| M199 | 444.5 ml | ||
| 10 - 20 U/ml Heparin | 500 µl | ||
| D-Valine | 25 mg | ||
| IFCS | 100 ml | ||
| EGS | 10 mg | ||
| Penicillin/Streptomycin | 5 ml | ||
| Amount of beads to be used for one mouse | |||
| For imaging | 5 µl | ||
| For WB/PCR | 10 µl |
Table 1. Chemical Compositions of Solutions Used.
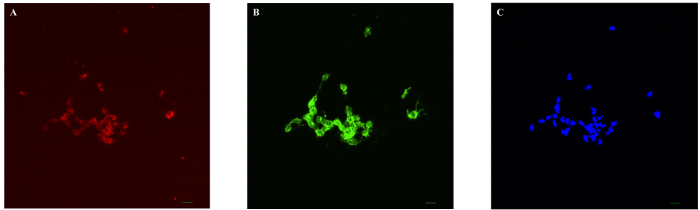 Figure 1. Representative Images of MCECs. Fluorescence images show that mouse coronary ECs (MCEC) isolated from hearts exhibit high purity of the EC population. Purity was tested by both acLDL uptake of red color (A) and lectin staining in green color (B) in MCEC. The nucleus was stained by Hoechst in blue color (C). The percentage of positive cells which exhibit both colors was over 90% in MCEC. Bar is representative of 20 µm. Please click here to view a larger version of this figure.
Figure 1. Representative Images of MCECs. Fluorescence images show that mouse coronary ECs (MCEC) isolated from hearts exhibit high purity of the EC population. Purity was tested by both acLDL uptake of red color (A) and lectin staining in green color (B) in MCEC. The nucleus was stained by Hoechst in blue color (C). The percentage of positive cells which exhibit both colors was over 90% in MCEC. Bar is representative of 20 µm. Please click here to view a larger version of this figure.
Discussion
The most widely used endothelial cells in research are human umbilical vein endothelial cells (HUVECs) because of their convenience and ease of culturing. However, a lot of research requires the availability of a physiological model attained by the isolation of endothelial cells from other specific organs10. Endothelial cells make up a very minor population of cells in the heart tissue; their isolation and culturing can prove to be very difficult.
There are three critical steps within this protocol. The first is to flush blood well. This is necessary, as leukocytes within blood also express CD31 as a cell surface marker, and will compete against ECs for the binding of the CD31 antibody. As other endothelial cells may also be present in the blood, flushing is essential to prevent contamination by such cells. The second critical step is to maintain a sterile environment throughout the isolation process. Any contamination results in a dramatic decrease in the number of cells attained. Finally, the third critical step is to adjust the pH of the CD31 buffer, as failure to do so will decrease the efficiency of the isolation.
Cell quality is most important during this isolation. Smooth muscle cell growth can be inhibited by the addition of an appropriate amount of heparin to the culture media. Fibroblast proliferation could be slowed down by adding D-Valine to the media. Endothelial cells can be characterized by the uptake of Dil-AcLDL and co-staining with BS-Lectin in order to test for the purity of the population.The endothelial cell phenotype is unstable and their behavior can change rapidly once taken out of the original tissue and cultured. A limitation of the technique is that cells should not be used after 5 days of culturing to ensure that they still retain their phenotype. It is strongly recommended not to pass the cells. The primary cells will provide a consistent result.
Overall, the technique results in a good number of cells of outstanding purity. With the required skills, the isolation process may be completed within 6 hr. It provides a great method for obtaining mouse coronary endothelial cells for either cell culture or molecular biological experiment.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by the grant from the National Institutes of Health (HL115578 to A. Makino).
References
- Michiels C. Endothelial cell functions. J Cell Physiol. 2003;196:430–443. doi: 10.1002/jcp.10333. [DOI] [PubMed] [Google Scholar]
- Sumpio BE, Riley JT, Dardik A. Cells in focus: endothelial cell. Int J Biochem Cell Biol. 2002;34:1508–1512. doi: 10.1016/s1357-2725(02)00075-4. [DOI] [PubMed] [Google Scholar]
- Cines DB, et al. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–3561. [PubMed] [Google Scholar]
- Deanfield JE, Halcox JP, Rabelink TJ. Endothelial function and dysfunction: testing and clinical relevance. Circulation. 2007;115:1285–1295. doi: 10.1161/CIRCULATIONAHA.106.652859. [DOI] [PubMed] [Google Scholar]
- Jin Y, Liu Y, Antonyak M, Peng X. Isolation and characterization of vascular endothelial cells from murine heart and lung. Methods Mol Biol. 2012;843:147–154. doi: 10.1007/978-1-61779-523-7_14. [DOI] [PubMed] [Google Scholar]
- Makino A, Platoshyn O, Suarez J, Yuan JX, Dillmann WH. Downregulation of connexin40 is associated with coronary endothelial cell dysfunction in streptozotocin-induced diabetic mice. Am J Physiol Cell Physiol. 2008;295:C221–C230. doi: 10.1152/ajpcell.00433.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli-Berg FM, Peek E, Lidington EA, Stauss HJ, Lechler RI. Isolation of endothelial cells from murine tissue. J Immunol Methods. 2000;244:205–215. doi: 10.1016/s0022-1759(00)00258-1. [DOI] [PubMed] [Google Scholar]
- van Beijnum JR, Rousch M, Castermans K, vander Linden E, Griffioen AW. Isolation of endothelial cells from fresh tissues. Nat Protoc. 2008;3:1085–1091. doi: 10.1038/nprot.2008.71. [DOI] [PubMed] [Google Scholar]
- Nolan DJ, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak M, Dargatz J, Chrzanowska-Wodnicka M. Isolation and culture of pulmonary endothelial cells from neonatal mice. J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Makino A, Dai A, Han Y, Youssef KD, Wang W, Donthamsetty R, Scott BT, Wang H, Dillmann WH. O-GlcNAcase overexpression reverses coronary endothelial cell dysfunction in type 1 diabetic mice. Am J Physiol Cell Physiol. 2015;309:C593–C599. doi: 10.1152/ajpcell.00069.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Donthamsetty R, Heldak M, Cho Y, Scott BT, Makino A. VDAC: old protein with new roles in diabetes. Am J Physiol Cell Physiol. 2012;303:C1055–C1060. doi: 10.1152/ajpcell.00087.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]


