Abstract
Although bioluminescence imaging (BLI) shows promise for monitoring tumor burden in animal models of cancer, these analyses remain mostly qualitative. Here we describe a method for bioluminescence imaging to obtain a semi-quantitative analysis of tumor burden and treatment response. This method is based on the calculation of a luminoscore, a value that allows comparisons of two animals from the same or different experiments. Current BLI instruments enable the calculation of this luminoscore, which relies mainly on the acquisition conditions (back and front acquisitions) and the drawing of the region of interest (manual markup around the mouse). Using two previously described mouse lymphoma models based on cell engraftment, we show that the luminoscore method can serve as a noninvasive way to verify successful tumor cell inoculation, monitor tumor burden, and evaluate the effects of in situ cancer treatment (CpG-DNA). Finally, we show that this method suits different experimental designs. We suggest that this method be used for early estimates of treatment response in preclinical small-animal studies.
Keywords: Medicine, Issue 113, Bioluminescence imaging, Quantification, Luminoscore, Murine B-cell lymphoma, Syngeneic, CpG-DNA, Cancer biology
Introduction
Early tumor cell detection remains a challenge and is crucial for enhancing cancer treatment efficacy. In vivo bioluminescence imaging (BLI) is a very sensitive, noninvasive optical technique, widely used for monitoring tumors in small animals. Firefly luciferase-expressing cells are commonly used for such experiments1,2. This oxygenase oxidizes D-luciferin with molecular oxygen but requires two cofactors - Mg2+ and adenosine triphosphate3. Firefly luciferase is more suitable for in vivo imaging than renilla luciferase4 because its quantum yield is higher.
The oxidized substrate - oxyluciferin - spontaneously emits a photon to return to its fundamental state and then becomes inactive. The emitted photons have a maximum wavelength around 530 nm. A high-sensitivity camera can detect the luminescent photons from the inside of a small animal and provide images that make it possible to locate tumor cells.
The ability to quantify tumor burden accurately by photon counts could serve as a powerful and sensitive tool for quantifying treatment efficacy. Because treatment effects can be detected sooner, this sensitivity might make it possible to determine the exact moment at which a treatment becomes effective.
Absolute quantification of total emitted photons is very complex. The number of photons gathered depends on the depth of the tumor and on the organs the photons are emitted through. Correction coefficients based on tissue absorption coefficients can be calculated5, but the absolute quantification of tumor cell numbers requires knowing the number of photons emitted by each tumor cell. Moreover, luciferase expression, like that of many reporter genes (e.g., fluorescent proteins) is not homogeneous, even in a cell population derived from a single clone6. The number of luciferase proteins in cells cannot be calculated exactly. The establishment of standardized experimental conditions thus appears crucial for a reliable semi-quantitative analysis.
We applied the luminoscore method to two different mouse lymphoma models7,8,9. In these models, syngeneic tumor cells are injected into the eye or under the skin to obtain, respectively, a primary intraocular lymphoma (PIOL) model and a subcutaneous lymphoma (SCL) model. In each of these orthotopic models, the treatment is administered in situ, seven days after the tumor inoculation for PIOL and when the tumor has reached 0.5 to 0.7 cm in its largest diameter for SCL.
We used the luminoscore method to monitor the effects of in situ CpG therapy, previously shown to be effective7,10,11. CpG is an oligonucleotide sequence and a ligand of TLR9, which in turn is an intracellular receptor expressed by numerous cells of the immune system, including dendritic cells, B lymphocytes, monocytes, and natural killer cells. CpG-DNA is a 20-mer DNA sequence that contains the CpG (CG) immunostimulatory motif; the control (ODN-control) is the same 20-mer DNA sequence, except that the immunostimulatory CG sequence is inverted (GC). TLR9 engagement in the murine lymphoma we are studying induces apoptosis10, activates the immune system12, and thereby significantly reduces tumor burden7,11.
Here we describe a standardized method for quantifying tumor burden and treatment response through bioluminescent images. This method relies on different aspects of the imaging procedure, from acquisition to analysis, to optimize reliability, reproducibility, non-user dependence, and statistical significance. A bioluminescence quantification index is attributed to each mouse; this value, which we call a luminoscore, can be compared not only between animals but also between experiments.
In this work, we focus on the bioluminescence imaging procedure as well as the image quantification by the luminoscore method. We show the effectiveness of this method for verifying the injection, monitoring tumor burden, and assessing the efficacy of in situ cancer treatment. Each of these points is illustrated in representative results from experiments using different mouse models to highlight the adaptability of the luminoscore method.
Protocol
All procedures involving mice complied with European Union guidelines, French regulations for animal experimentation (Ministry of Agriculture Act No. 2001-464, May 2001), and the guidelines of the Institut National de la Santé et de la Recherche Médicale (INSERM) Committee on Animal Research, and were approved by the relevant local committee (the Charles Darwin Ethics Committee for Animal Experiments, Paris, France; Permit Number: p3/2009/004).
1. Cell Preparation
Grow mouse B lymphoma cell line A20.IIA-luc2 in RPMI-1640 Glutamax medium supplemented with 10% fetal calf serum, 100 µg/ml penicillin, 100 µg/ml streptomycin, 10 mM sodium pyruvate, 50 µM 2-mercaptoethanol, and 0.50 mg/ml hygromycin B.
Maintain cell culture at 37 °C, 5% CO2 and change medium every two to three days. Harvest 5 ml of the cell suspension with a pipette one day after changing the medium.
Spin cells at 300 × g for 10 min and suspend the cells in 3 ml sterile phosphate-buffered saline (PBS). Repeat this step twice to wash the cells.
Mix 15 µl of cell suspension with 30 µl Trypan Blue labeling before loading a Malassez counting chamber. Calculate the cell concentration with the formula: Concentration (cells/ml) = Number of cells in the counting grid * 3 * 1000.
Spin the cells one more time at 300 x g for 10 min. Remove supernatant with a pipette. With C the concentration calculated at step 1.4), the number of cells is N = C*3.
Calculate the volume of sterile PBS 1x required to obtain cell suspension A at a concentration of 5 x 107 cells/ml with the following formula: PBS Volume (ml) = N / (5 x 107). Suspend the cell pellet (from step 1.5)) in the volume of sterile PBS 1x calculated in the previous sentence. Cell suspension A is to be used in the SCL model (in vivo injected volume is 100 µl: 5 x 106 cells).
Pipette 10 µl of cell suspension A and add 90 µl of sterile PBS in a 1.5 ml tube to obtain 100 µl of cell suspension B at 5 x 106 cells per ml. Cell suspension B is to be used in the PIOL model (in vivo injected volume is 2 µl: 1 x 104 cells).
2. Luciferin
Dilute 1 g of D-luciferin potassium salt powder in 30 ml of sterile PBS 1x in a 50-ml tube and shake for a few seconds to dissolve the aggregates. NOTE: Because luciferin is light-sensitive, prepare 500 µl aliquots in dark 1.5-ml microcentrifuge tubes.
Store the aliquots at -20 °C. NOTE: The aliquots can be stored for several months. After thawing, the aliquots must not be stored for more than 1 day at + 4 °C. Freezing-thawing cycles are preferable to storage at 4 °C.
Inject 100 µl of D-luciferin potassium salt solution intraperitoneally for each imaging assay. NOTE: This solution corresponds to 3.3 mg per mouse for a dose of 150 mg/kg.
3. Anesthetic Mixture and Anesthetization
Prepare an anesthetic solution by mixing ketamine 120 mg/kg and xylazine 6 mg/kg in sterile PBS 1x.
Inject 60 µl of anesthetic mixture intraperitoneally for each imaging assay with a 25 G needle. For surgery (with the PIOL or SCL model), inject 80 µl of the mixture to obtain a deeper anesthesia. Put the mouse back in its cage.
When the mouse appears immobile, remove it from its cage and gently squeeze the mouse's leg between fingers. If the mouse reacts with an escape reflex, wait several min. Repeat the action until the mouse does not react, which confirms satisfactory anesthetization.
Place the mouse on a warming plate or under a warming light.
Apply eye ointment to avoid eye dryness during anesthetization for the imaging assay or for SCL surgery. Apply the eye ointment after surgery for the PIOL model.
4. Surgery and Cell Inoculation
NOTE: Perform all surgical procedures on a warming plate or under a warming light, in a type 1 microbiological safety cabinet in an animal biosafety level 2 facility. All surgical tools used in this section were autoclaved before use.
- Subcutaneous Lymphoma Model:
- Prepare 100 µl of the cell suspension obtained in step 1.6 in a 1-ml syringe with a 25 G needle. Gently squeeze the mouse skin on the flank between fingers, at the injection site. Insert the needle exactly into the skin fold. To ensure subcutaneous injection, do not place the needle deep into the tissue.
- Inject the cells into the skinfold. Observe whether a little liquid ball appears under the skin to confirm that the injection was performed correctly.
- PIOL model: NOTE: This procedure requires 2 operators, referred to here as operator 1 and operator 2.
- Removal of conjunctiva:
- Have Operator 1 place the mouse under a dissecting microscope. Press gently with the fingers at each side of the eye to clear it. Maintain this position.
- Have Operator 2 look through the dissecting microscope. Grip the conjunctiva with a small pair of pliers in one hand; with the other hand, cut the conjunctiva just below the pliers with a small pair of surgical scissors.
- Have Operator 1 release the eye of the mouse pressed at step 4.2.1.1)
- Cell injection:
- Prepare a 10-µl blunt precision syringe. Wash it by pumping sterile deionized water through it. Repeat twice or three times to ensure there are no bubbles in the syringe. Then prepare 2 µl of cell suspension for injection.
- Have Operator 2 press gently with the fingers of one hand at each side of the eye to clear it. Maintain this position.
- Have Operator 1 grip the edge of the eye with a small pair of pliers in one hand and pull it gently backwards to stretch the tissue.
- Have Operator 2 look through the dissecting microscope. With the other hand, make a small hole in the inferior eyeball of the mouse with a 32 G needle.
- Have Operator 2 put down the needle and pick up (with the same hand) the precision syringe and insert the needle into the hole made at step 4.2.2.4.
- Have Operator 1 push on the syringe plunger with the other hand.
- Have Operator 2 verify with the dissecting microscope that the cell suspension in the syringe has been correctly injected inside the eyeball (the liquid flow is easily observable).
- Have Operator 2 remove the precision syringe.
- Have Operator 1 release the edge of the animal eye gripped at step 4.2.2.3)
- Have Operator 2 release the sides of the eye pressed at step 4.2.2.2)
- Apply the eye ointment immediately. NOTE: If all steps were performed correctly, the mouse should not bleed at all during this procedure.
5. Bioluminescence Imaging – Day 0
NOTE: All products injected into the mouse must be at RT before injection. After the tumor cells have been inoculated, and while the animal is still anesthetized, proceed to these steps for the imaging.
Turn on the imager and open the acquisition software. Initialize the camera, the stages, and the lenses by clicking on the "Initialize" button. The initialization will take 10 to 15 min to be complete.
Inject 100 µl of D-luciferin potassium salt solution intraperitoneally with a 25 G needle. Do not administer it intravenously. If intravenous administration is required for any reason, the D-luciferin sodium salt must be used rather than D-luciferin potassium salt. NOTE: Luciferin is the excess reactant at this concentration; therefore the bioluminescence signal reaches a plateau after 3 to 7 min and persists for more than 30 min.
10 min after D-luciferin injection13, place the subject mouse in the imager. Place the mouse in its natural position, its back towards the camera, in as flat a position as possible. This position is natural and easily reproducible.
Tick the auto-exposure feature, and click on Acquire to acquire the back (posterior) image of the animal, with the auto-exposure feature. NOTE: The auto-exposure feature optimizes exposure time by calculating it from a 1-sec exposure image. If the mouse's bioluminescence signal is negative or very low, the optimal exposure time might be automatically set to more than 20 min. In this case, an exposure time of 8 to 10 min may be a good compromise. Exposure time may be set manually, but the images must not contain saturated pixels.
Turn the mouse over to expose the front of the mouse to the camera. Try to flatten the mouse and spread its anterior limbs so that they do not block the chest.
Acquire a front image of the animal. Verify that the auto-exposure checkbox is still ticked and click again on the "Acquire" button. NOTE: The front image can be acquired before the back image and vice versa. The exposure time of front and back images can be different depending on the relative intensity of each side of the mouse. It is calculated automatically when using the auto-exposure feature. Quantification uses only the photon flux (photons per sec) and does not depend on exposure time.
Place the mouse on a warming plate or under a warming light until it recovers from the anesthetic and then place it back in its cage.
6. Bioluminescence Imaging – After Day 0
Turn on the imager, initialize the camera, the stages, and the lenses as in step 5.1).
Anesthetize the mouse with an intraperitoneal injection of 60 µl of the ketamine (120 mg/kg) / xylazine (6 mg/kg) mixture (see steps 3.1 to 3.5). NOTE: This method of anesthetization allows imaging every 5 days. To perform daily imaging, a method that uses isoflurane as the anesthetic and a bioluminescence imager suitable for use with this anesthetic is required.
Acquire front and back images of the animal, using the auto-exposure feature, as in steps 5.5) and 5.6). Handle the mouse as in step 5.7).
7. Bioluminescence Quantification and Image Analysis
NOTE: The luminoscore method is based on image analysis. Once the images have been acquired according to the steps above, the quantification can be performed at any time (including immediately after acquisition), to associate a luminoscore to each mouse at each time point.
Display the "Tool Palette" by clicking on the "View" menu and then click on "Tool Palette" if not already displayed. Click on the "ROI Tool" tab of the Tool Palette. Choose the "Contour" button and then select "Free draw".
Manually mark the Region of Interest (ROI) around the mouse by following the edges of the mouse on the front view. Close the contour with a right click on the computer mouse.
Click on the "View" menu and then "ROI Measurements". Make sure "Radiance (Photons)" is selected in the "Measurement Types" scrolling menu on the bottom left of the "ROI Measurements" window. If not, select it. Then measure the photon flux (ph/s) by recording the value in the "Total Flux [p/s]" box. NOTE: Do not use Radiance or any other unit that is relative to the ROI surface area.
Repeat steps 7.2) and 7.3) for the back view.
Sum the two photon flux values obtained from front and back view. The result is the luminoscore.
Compare the values for each group using an appropriate statistic test (non-parametric two-tailed Mann-Whitney test, for instance)14.
Representative Results
The Luminoscore Method can Serve to Verify the Tumor Cell Injection
In models involving the injection of a small number of tumor cells or when the injection site does not permit visual verification of the injection, it can be very difficult to ensure the quality of the injection. The luminoscore method serves as a fast and convenient tool to verify the quality of the procedure and ascertain easily and instantaneously that everything went right. In both models, the tumor is detectable as early as 10 min after the injection (Figure 1A). The images alone suffice to verify that tumor cells are present in the correct location. Nonetheless, quantifying the images provides an idea of the heterogeneity of the injection. The dot plot (Figure 1B) clearly shows a difference between animals that received (black dots) and did not receive (red line) injections. Interestingly, the signal obtained in the SCL model is 100 times higher than in the PIOL model; this finding is consistent with the number of cells injected (5 x 106 and 1 x 104 cells, respectively).
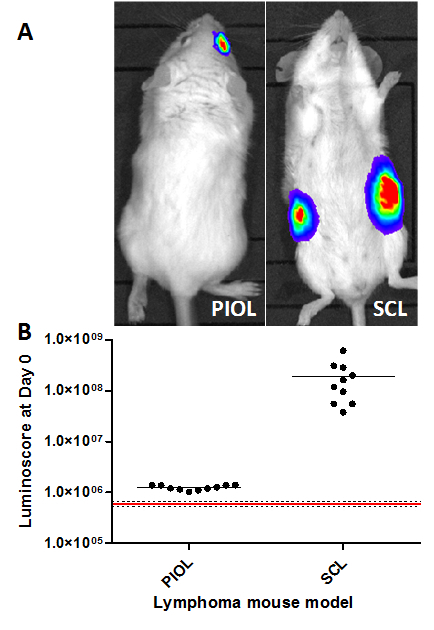 Figure 1. Verification of Injection in Different Mouse Lymphoma Models. The luminoscores measured 10 min after tumor cell inoculation in different lymphoma models. (A) Representative images of both models. (B) Luminoscore of each animal in the different models. The red line corresponds to the mean background noise measured on 10 animals before tumor cell inoculation; the dotted lines are the mean +/- standard deviation. For each model, all the animals can be distinguished from background noise. In the PIOL model, 1 x 104 cells were inoculated in the vitreous of the right eye, and in the SCL model, 5 x 106 cells subcutaneously into each flank of the mouse. Please click here to view a larger version of this figure.
Figure 1. Verification of Injection in Different Mouse Lymphoma Models. The luminoscores measured 10 min after tumor cell inoculation in different lymphoma models. (A) Representative images of both models. (B) Luminoscore of each animal in the different models. The red line corresponds to the mean background noise measured on 10 animals before tumor cell inoculation; the dotted lines are the mean +/- standard deviation. For each model, all the animals can be distinguished from background noise. In the PIOL model, 1 x 104 cells were inoculated in the vitreous of the right eye, and in the SCL model, 5 x 106 cells subcutaneously into each flank of the mouse. Please click here to view a larger version of this figure.
Monitoring Tumor Burden and Treatment Response
The luminoscore is also a powerful tool for studying tumor growth and treatment efficacy. Figure 2A shows representative images of the monitoring of tumor growth in the control and the CpG treated PIOL groups. The mice were inoculated with 1 x 104 tumor cells each, and treatment was administered in situ on Day 7. The images show that after a sufficient time (28 days), metastases began to appear in the control group. Although the primary tumor did not appear sensitive to treatment, fewer metastases were observed in the CpG-treated group (Table 1). The quantitative analysis of the tumor burden in each group (Figure 2B) shows that CpG slowed tumor development, producing a statistically significant difference between the treated and the control group after 28 days (non-parametric two-tailed Mann-Whitney test p = 0.0079). Nonetheless, the tumors still grew in the treated mice, and none of them survived (data not shown). This finding is consistent with previous reports: CpG has no significant effects on primary ocular tumors10. The significant effect that we have here between the ODN and the CpG group comes from the metastasis growth inhibition.
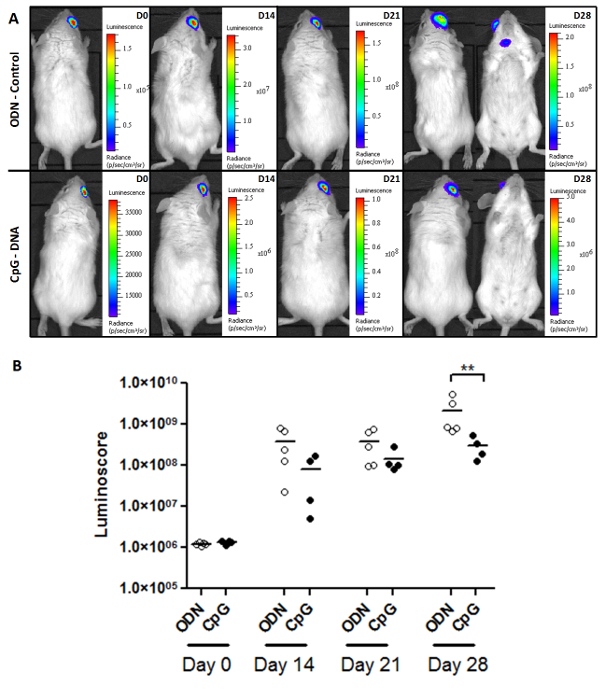 Figure 2. Monitoring Tumor Burden and Treatment Response. (A) Representative images of the two groups (ODN-control and CpG-treated) of PIOL-bearing mice. (B) Monitoring tumor burden with the luminoscore. As seen on the dot plot, the effect of CpG on the luminoscore is significant on day 28. In those two groups, this method made it possible to monitor tumor burden and to measure the slight but significant (p = 0.0079) effects of CpG-DNA, which could have not been detected by images alone. Please click here to view a larger version of this figure.
Figure 2. Monitoring Tumor Burden and Treatment Response. (A) Representative images of the two groups (ODN-control and CpG-treated) of PIOL-bearing mice. (B) Monitoring tumor burden with the luminoscore. As seen on the dot plot, the effect of CpG on the luminoscore is significant on day 28. In those two groups, this method made it possible to monitor tumor burden and to measure the slight but significant (p = 0.0079) effects of CpG-DNA, which could have not been detected by images alone. Please click here to view a larger version of this figure.
| Groups | Mouse | Presence of metastases | Location | Photon flux (ph/s) |
| CpG | 1 | No | X | X |
| 2 | Yes | Eye-draining lymp node | 9.32E+05 | |
| 3 | No | X | X | |
| 4 | No | X | X | |
| 5 | Yes | Eye-draining lymp node | 2.21E+07 | |
| ODN | 1 | Yes | Eye-draining lymp node | 1.06E+07 |
| 2 | Yes | Eye-draining lymp node | 7.25E+07 | |
| 3 | Yes | Eye-draining lymp node + contralateral | 7.64E+08 | |
| 4 | Yes | Eye-draining lymp node + contralateral | 1.74E+09 | |
| 5 | Yes | Eye-draining lymp node + contralateral | 9.76E+07 |
Table 1. The Luminoscore Method can be Adapted to Different Approaches
The luminoscore method is not limited to a single type of mouse model. Figure 3 shows that it can be adapted to different mouse models. In the SCL model, two tumors are grafted on each side of the mouse. The treatment is administered in situ to a single side on Day 0, and the contralateral tumor serves as its control (Figure 3A). Therefore, two ROI were drawn per mouse: one for each side (Figure 3C). The ratio between the luminoscore on the treated and control sides describes the relative course of each tumor. This ratio was set to 1 on Day 0, when the treatment was administered. We observed a significant decrease in this ratio 13 days after treatment administration (Figure 3D; non-parametric two-tailed Mann-Whitney test p = 0.004). This decrease reveals that the treated-side tumor has been resorbed, as shown on the representative bioluminescence images (Figure 3B).
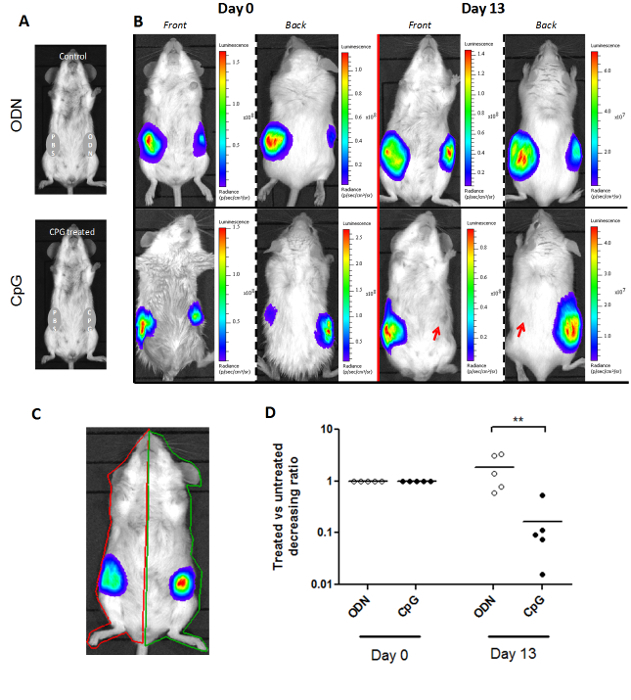 Figure 3. Adaptation of the Luminoscore Method to a Model with Two Primary Tumor Sites. (A) Experimental design of an SCL assay. The mice were injected with two primary tumors in each flank. One side is injected with either CpG-DNA or ODN-control, and the other side with PBS, to serve as its control. (B) Representative images of CpG-treated and control mice at day 0 and day 13. The treatment was administered on day 0. Tumor growth was inhibited on the CpG-treated side of the mouse. (C) The manually marked up regions of interest per animal for two primary tumor sites. (D) The ratio between the luminoscores of the CpG-treated or ODN-control side and the PBS control side is an index that reflects the relative growth of each tumor within the same animal. This ratio was set to 1 on day 0. On day 13, the ratio of the CpG-treated group had decreased significantly (p = 0.004), revealing inhibition of tumor growth by in situ administration of CpG-DNA. Please click here to view a larger version of this figure.
Figure 3. Adaptation of the Luminoscore Method to a Model with Two Primary Tumor Sites. (A) Experimental design of an SCL assay. The mice were injected with two primary tumors in each flank. One side is injected with either CpG-DNA or ODN-control, and the other side with PBS, to serve as its control. (B) Representative images of CpG-treated and control mice at day 0 and day 13. The treatment was administered on day 0. Tumor growth was inhibited on the CpG-treated side of the mouse. (C) The manually marked up regions of interest per animal for two primary tumor sites. (D) The ratio between the luminoscores of the CpG-treated or ODN-control side and the PBS control side is an index that reflects the relative growth of each tumor within the same animal. This ratio was set to 1 on day 0. On day 13, the ratio of the CpG-treated group had decreased significantly (p = 0.004), revealing inhibition of tumor growth by in situ administration of CpG-DNA. Please click here to view a larger version of this figure.
Discussion
Light absorption by organs and tissues remains a limitation of bioluminescence imaging, although this limitation is intrinsic to any optical imaging modality. In the context of our approach, the effects of anatomic structures on the luminoscore are expected to have low variability provided that the studies are performed in a given model (location and mouse strand), allowing then comparisons. Bioluminescence does not need excitation light and thus is more adapted to whole body in vivo imaging than fluorescence.
Spatial resolution is also a limitation of bioluminescence imaging, and precise location of the organ from which bioluminescence photons are emitted remains difficult. However, good knowledge of the model can help in the qualitative location of the tumor sites. The only output in this bioluminescence-based method is the luminoscore. Location does not influence the luminoscore because the manual mark-up Region of Interest (ROI) covers the whole mouse. Finally, firefly luciferase requires oxygen. Accordingly, bioluminescence imaging usually underestimates necrotic tumors. Once again, a good knowledge of this aspect of the model is required.
In this paper, we are not aiming at assessing the efficacy of CpG as an anti-tumor drug (which has already been demonstrated7,9,11) but to describe a method allowing comparison of bioluminescence datasets.We indeed describe a method to quantify tumor burden intended to help standardize the acquisition protocol for comparing different assays in different places at different times and that does not require computer calculations. To ensure reproducibility and correct photon flux quantification, the imaging device must be calibrated with a light reference for home-made devices or as recommended by the provider for commercial devices.
Our protocol requires careful attention to several critical points: (i) First, the quality of the mouse anesthesia is crucial for obtaining clear images, especially in cases with long exposure times. (ii) The quantification unit must be always be the photon flux because radiance depends on the surface area of each mouse and might be irrelevant for comparing different mice. (iii) The bioluminescence images must not contain saturated pixels, because these would bias the luminoscore. (iv) Back and front acquisition are required to collect all photons emitted from the tumor site (i.e., front acquisition may not necessarily detect photons from the back of the tumor). Various different ROI drawings were tested during the development of the luminoscore method. Only the manual mark-up yielded satisfactory results that are more likely to be statistically significant (data not shown).
Inoue et al. recommend a luciferin dose of 75 mg/kg13. By using a dose of 150 mg/kg instead, the timing of imaging after the luciferin administration remains unchanged and we ensure that the plateau lasts throughout a long exposure acquisition. Luciferin must be the excess reactant throughout the acquisition process. Depending on the model, the region of interest can be adapted, as we showed in the SCL model where we drew two ROI per animal. In the SCL model, the treatment is injected when the tumor reaches 0.5 cm in its largest diameter to limit variability of engraftment. Depending on the mice, the tumor might have grown differently. To standardize and compare mice, we decided to use a ratio between treated and non-treated side that reveals the relative growth of both flanks tumors.
If no signal is observed from an injected mouse expected to be positive, either (i) the number of cells is very low and the signal is below the detection threshold; or (ii) the mouse lacks oxygen and requires immediate care.
Several of the quantitative bioluminescence analyses described by various authors require complex calculations and instruments (e.g., 3D bioluminescence tomography) to approach the absolute quantification of emitted bioluminescence photons5,15. There is no consensus on a method for quantifying bioluminescence reproducibly, especially in tumor models, with a 2D bioluminescence imager. Our aim was to standardize the image acquisition protocol to limit user-dependence.
The injection of CpG in situ after tumor inoculation reduced tumor burden in both models. The luminoscore method can serve as a tool to monitor tumor burden in other models of tumor immunotherapies. Monitoring tumor burden provides a noninvasive method that can improve our understanding of tumor growth and metastasizing mechanisms without interfering with the tumor microenvironment. The identification of animals that do and do not respond to treatment with this method is reinforced by the verification of successful tumor cell injection at the beginning of the experiment.
We here showed the adaptability of the luminoscore method in a SCL model using A20.IIA-luc2 cells. However, this method can be adapted to any other tumor model using other cell lines provided that they express luciferase, or in the context of T-cell studies (tumor specific cytotoxic T-cells, regulatory T-cells, etc.). The monitoring of in vivo gene transfer in the context of gene therapy of rare disease could also be done using the luminoscore method.
Finally the data shows that the bioluminescence-based luminoscore method enables comparisons between experiments, offers flexibility and adaptability to specific experimental needs, and is a useful tool for noninvasive longitudinal preclinical studies.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank the animal facilities of the Cordelier Research Center (CEF, Paris, France), Genethon (Evry, France) and Genopole (CERFE, Evry, France). We thank Jo Ann Cahn for her careful reading of the manuscript. This research was supported by the Institut National de la Santé Et de la Rechercher Médicale, Paris Descartes University, Pierre & Marie Curie University, the Association pour la Recherche contre le Cancer, the Tunisian Direction Générale de la Recherche Scientifique, the Franco-Tunisian CMCU project, and the Actions Thématiques Incitatives de Genopole (ATIGE) funds. J.C was supported by the Frontiers in Life Science doctoral school and by a fellowship from the INCA (Institut National du Cancer). R.B.A. was a recipient of grants from the DGRS-INSERM and the CMCU. S.D. received a grant from the Institut National du Cancer.
References
- Rehemtulla A, et al. Neoplasia. 6. Vol. 2. New York, N.Y: 2000. Rapid and Quantitative Assessment of Cancer Treatment Response Using In Vivo Bioluminescence Imaging; pp. 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger M, et al. European Journal of Cancer. 16. Vol. 38. Oxford, England: 2002. Advancing animal models of neoplasia through in vivo bioluminescence imaging; pp. 2128–2136. [DOI] [PubMed] [Google Scholar]
- Hastings JW, Gibson QH. The Role of Oxygen in the Photoexcited Luminescence of Bacterial Luciferase. Journal of Biological Chemistry. 1967;242(4):720–726. [PubMed] [Google Scholar]
- Inouye S, Shimomura O. The Use of Renilla Luciferase, Oplophorus Luciferase, and Apoaequorin as Bioluminescent Reporter Protein in the Presence of Coelenterazine Analogues as Substrate. Biochemical and Biophysical Research Communications. 1997;233(2):349–353. doi: 10.1006/bbrc.1997.6452. [DOI] [PubMed] [Google Scholar]
- Pesnel S, et al. Quantitation in Bioluminescence Imaging by Correction of Tissue Absorption for Experimental Oncology. Molecular Imaging and Biology. 2010;13(4):646–652. doi: 10.1007/s11307-010-0387-9. [DOI] [PubMed] [Google Scholar]
- Corre G, et al. Stochastic Fluctuations and Distributed Control of Gene Expression Impact Cellular Memory. PLoS ONE. 2014;9(12) doi: 10.1371/journal.pone.0115574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Abdelwahed R, et al. Lymphoma B-cell responsiveness to CpG-DNA depends on the tumor microenvironment. Journal of Experimental & Clinical Cancer Research CR. 2013;32(1):18. doi: 10.1186/1756-9966-32-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelwahed RB, et al. Preclinical Study of Ublituximab, a Glycoengineered Anti-Human CD20 Antibody, in Murine Models of Primary Cerebral and Intraocular B-Cell Lymphomas. Investigative Ophthalmology & Visual Science. 2013;54(5):3657–3665. doi: 10.1167/iovs.12-10316. [DOI] [PubMed] [Google Scholar]
- Donnou S, Galand C, Touitou V, Sautès-Fridman C, Fabry Z, Fisson S. Murine Models of B-Cell Lymphomas: Promising Tools for Designing Cancer Therapies. Advances in Hematology. 2012;2012 doi: 10.1155/2012/701704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X-F, et al. CpG oligodeoxynucleotide induces apoptosis and cell cycle arrest in A20 lymphoma cells via TLR9-mediated pathways. Molecular Immunology. 2013;54(3-4):327–337. doi: 10.1016/j.molimm.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Houot R, Levy R. T-cell modulation combined with intratumoral CpG cures lymphoma in a mouse model without the need for chemotherapy. Blood. 2009;113(15):3546–3552. doi: 10.1182/blood-2008-07-170274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg AM. Toll-like receptor 9 (TLR9) agonists in the treatment of cancer. Oncogene. 2008;27(2):161–167. doi: 10.1038/sj.onc.1210911. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Kiryu S, Watanabe M, Tojo A, Ohtomo K. Timing of Imaging after D-Luciferin Injection Affects the Longitudinal Assessment of Tumor Growth Using In Vivo Bioluminescence Imaging. International Journal of Biomedical Imaging. 2010;2010 doi: 10.1155/2010/471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann HB, Whitney DR. On a Test of Whether one of Two Random Variables is Stochastically Larger than the Other. Annals of Mathematical Statistics. 1947;18(1):50–60. [Google Scholar]
- Darne C, Lu Y, Sevick-Muraca EM. Small animal fluorescence and bioluminescence tomography: a review of approaches, algorithms and technology update. Physics in Medicine and Biology. 2014;59(1):1. doi: 10.1088/0031-9155/59/1/R1. [DOI] [PubMed] [Google Scholar]


