ABSTRACT
The ability for the eukaryotic cell to transcriptionally respond to various stimuli is critical for the overall homeostasis of the cell, and in turn, the organism. The human RNA polymerase II complex (Pol II), which is responsible for the transcription of protein-encoding genes and non-coding RNAs, is paused at promoter-proximal regions to ensure their rapid activation. In response to stimulation, Pol II pause release is facilitated by the action of positive transcription elongation factors such as the P-TEFb kinase. However, the majority of P-TEFb is held in a catalytically inactivate state, assembled into the 7SK small nuclear ribonucleoprotein (snRNP) complex, and must be dislodged to become catalytically active. In this review, we discuss mechanisms of 7SK snRNP recruitment to promoter-proximal regions and P-TEFb disassembly from the inhibitory snRNP to regulate ‘on site' kinase activation and Pol II pause release.
KEYWORDS: KAP1, P-TEFb, RNA polymerase II, transcription elongation, 7SK snRNP
Introduction
Proper regulation of transcription by the RNA polymerase II (Pol II) complex is essential to maintain cellular homeostasis. Checkpoints at multiple steps in the transcription cycle ensure appropriate and efficient gene activation by Pol II, both under basal conditions and in response to various stimuli.1-4 One of the major checkpoints in the transcription cycle is the pausing of Pol II ∼20–60 bp downstream of the transcription start site (TSS).5-11 The regulated release of paused Pol II into its actively elongating form is governed by the presence of negative elongation factors, restrictive nucleosome barriers directly adjacent to the pause site, and the sequestration of positive elongation factors into inhibitory complexes.10-19 Therefore, alleviation of these elongation blockades is a major rate-limiting step in gene expression, and is self-imposed by the cell to ensure transcriptional integrity.
Several transcription factors have been recently shown to play a critical role in promoting Pol II pause release.8,20-23 In the case of inducible pathways, transcription factors are recruited to target promoters to stimulate gene expression thereby allowing the cell to rapidly and efficiently respond to various stimuli.8,21-25 This is accomplished through the eviction of elongation factors from their inhibitory complexes, recruitment of chromatin-modifying enzymes, and the inactivation/eviction of negative elongation factors from early elongation complexes. At this point, paused Pol II is rapidly released into the gene body, resulting in gene activation and allowing the cell to rapidly respond to the stimulus.
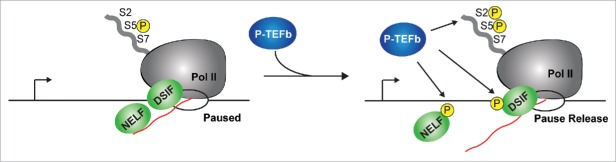
One of the best-characterized elongation factors is the positive transcription elongation factor b (P-TEFb) kinase.20,26 Upon transcriptional activation, P-TEFb phosphorylates a number of substrates at the promoter, alleviating the blockade in Pol II pause release (Fig. 1). P-TEFb inhibition blocks most Pol II regulated transcription in the cell by affecting both pause release and termination at both 5′- and 3′-ends of genes, respectively.27,28 While P-TEFb has been proposed to be inducibly recruited to gene promoters during transcriptional activation,10,20-23,26,29 several recent reports have shown that inactive P-TEFb (as part of the 7SK snRNP complex) is localized to promoter-proximal regions and enhancers genome-wide prior to, or during, gene activation.24,30-37 Recruitment of catalytically inactive P-TEFb to gene promoters and enhancers has shifted our understanding on how the cell controls transcription elongation. By localizing primed P-TEFb near paused Pol II, transcription factors can promote kinase activation to facilitate pause release (Fig. 1). This review will highlight recent advances and future challenges in the field of transcriptional regulation including the role of the 7SK snRNP complex in facilitating pause release.
Figure 1.
The P-TEFb kinase facilitates Pol II pause release. After transcription of a short (> 18 nt-long) RNA chain (red line), the negative elongation factors DSIF and NELF promote pausing by interactions with Pol II and nascent RNA. At this point, the Pol II CTD is in its hypo-phosphorylated form (S5P) by the action of the TFIIH kinase (not shown for simplicity). In response to stimulation, the P-TEFb kinase stimulates the hyper-phosphorylation of the Pol II CTD (S5P and S2P), NELF and DSIF. These phosphorylation events cause the release of NELF from the paused, early elongation complex and transforms DSIF into a positive elongation factor, stimulating the release of Pol II into productive elongation. The arrow denotes the position of the TSS, and the open circle indicates the elongation bubble.
The P-TEFb kinase promotes Pol II pause release
P-TEFb is a heterodimer composed of a regulatory cyclin subunit (primarily CycT1 and CycT2) and the cyclin dependent kinase 9 (Cdk9).38-40 During gene activation, the Cdk9 subunit of P-TEFb hyper-phosphorylates the C-terminal domain (CTD) of Pol II at Ser5 and Ser2 residues of the heptad repeats (Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7).41,42 P-TEFb also phosphorylates the multi-subunit negative elongation factor complex (NELF), particularly the NELF-E subunit, and the DRB-sensitivity inducing factor (DSIF, composed of Spt4/Spt5), which are loaded into transcription complexes after transcription initiation.12-14,43 While NELF-E phosphorylation results in its eviction from the early elongation complex, the phosphorylation of the Spt5 subunit of DSIF converts the complex into a positive elongation factor that travels with Pol II throughout the transcription unit.14,15,44-48 Collectively, these P-TEFb-mediated promoter-proximal phosphorylation events act as a molecular switch to stimulate Pol II pause release.
After relieving the elongation block, P-TEFb travels with Pol II as part of the super elongation complex (SEC), further hyper-phosphorylating both Ser5 and Ser2 residues of the Pol II CTD.49-51 In addition to P-TEFb, the SEC is composed of the 11–19 Lys-rich leukemia (ELL) family members (ELL1, ELL2, and ELL3), the AF4/FMR2 (AFF) family members AFF1 and AFF4, and the ALL1-fused gene from chromosome 9 (AF9).50,51 While the Pol II CTD is phosphorylated at other residues (including Tyr1 and Ser7),52-56 the consensus major substrates of the P-TEFb kinase (as part of the SEC) in the Pol II CTD are Ser5 and Ser2 residues. Although all CTD heptad repeats are non-redundant, phosphorylation at Ser5 and Ser2 residues takes place throughout the entire domain, as revealed through studies in budding yeast using a CTD polypeptide modified for mass spectrometry analysis.57,58
P-TEFb is utilized by a number of transcription factors to stimulate gene activation including p53, nuclear factor κB (NF-κB), heat shock factor (HSF), c-Myc, and HIV Tat, among several others.8,20-23,26,59 Previous studies have shown that these transcription factors can directly contact P-TEFb, and even modulate its kinase activity toward the Pol II CTD.8,21,23,24,59-61 It remains unclear, however, if the transcription factor does indeed need to directly bind P-TEFb at promoter-proximal regions to facilitate SEC assembly for Pol II hyper-phosphorylation, or whether the SEC assembles without participation of the transcription factor. Regardless, P-TEFb kinase activity toward the Pol II CTD is intimately linked to transcriptional stimulation of a gene by a canonical transcription factor. Future studies highlighting the potential role of transcription factors in promoting SEC assembly will be needed to clarify this issue.
The P-TEFb kinase in the 7SK snRNP complex is catalytically inactive
To ensure that P-TEFb kinase activity, and thereby transcriptional integrity of the cell remains in check, the majority of P-TEFb is held in a catalytically inactive state assembled into the 7SK snRNP complex.62-64 This ribonucleoprotein complex is composed of 7SK RNA, P-TEFb, the kinase inhibitor hexamethylene bis-acetamide inducible 1/2 (here referred as to Hexim for simplicity) and 2 RNA-binding proteins (the La-related protein 7 (Larp7) and the methylphosphate-capping enzyme (MePCE)).65-72
The human 7SK RNA is a 331 nt-long, abundant non-coding RNA synthesized by Pol III, and composed of 4 stem-loops (stems I-IV).65 MePCE stably binds to and incorporates a γ-monomethyl cap at the 5′-end of 7SK RNA stem I,73 whereas Larp7 constitutively binds stem IV and a U-rich tail at the 3′-end to protect the RNA from degradation.67,71,72 After RNA capping, MePCE and Larp7 physically bind to each other, forming a closed loop on the RNA (referred as to core 7SK snRNP), which provides further stability to the snRNP.64,66,72,74
The distal portion of 7SK stem I contains 2 structurally similar motifs, which function in the binding of Hexim1/2 homo- or hetero-dimers.70,75 The distal part of stems I (5′-end) and IV (3′-end) contain all necessary elements required for P-TEFb recruitment.76 Binding to 7SK RNA causes a conformational change in Hexim that allows it to interact with the CycT1 subunit allowing it to recruit P-TEFb, thus assembling the 7SK snRNP complex and inhibiting Cdk9 kinase activity.69,77-81 Systematic 7SK RNA mutagenesis showed that alterations in Hexim's binding abolishes P-TEFb's recruitment thereby indicating its absolute requirement in formation of the 7SK snRNP.76
Through mass spectrometry analysis, it was discovered that Cdk9 required T-loop phosphorylation (Thr186) to promote its interaction with Hexim and the 7SK RNA in vitro.68,82 In support of this model, it was found that Cdk9 T-loop mutants were incapable of associating with Hexim-7SK RNA and assembling the inhibitory snRNP complex.68,82 Therefore, Hexim physically inhibits the enzymatic activity of P-TEFb when bound to 7SK RNA and allows the cell to maintain gene expression equilibrium when the kinase is not transcriptionally engaged. Interestingly, T-loop phosphorylation is what primes the kinase for activation, similar to other Cdks,83 and this modification can be carried out by either P-TEFb's intrinsic auto-phosphorylation or by the TFIIH kinase (Cdk7).24,84,85
Collectively, it is widely accepted that the 7SK snRNP complex is the major reservoir of P-TEFb, maintaining a large fraction of P-TEFb primed until the kinase is needed for gene activation.62,63
P-TEFb is dislodged from the 7SK snRNP complex on chromatin to become catalytically active
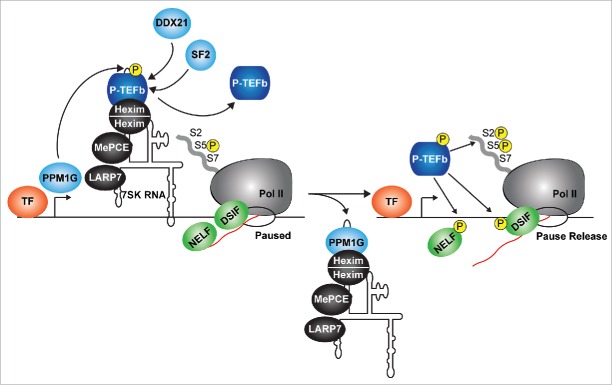
In order to accommodate the transcriptional needs of the cell, the P-TEFb kinase has to be released from the snRNP and then captured by promoter-associated factors. Given that phosphorylation of Cdk9 is a prerequisite for P-TEFb incorporation into the 7SK snRNP, it was proposed that Cdk9 T-loop dephosphorylation could trigger kinase dislodgement from Hexim-7SK RNA (thereby causing 7SK snRNP disassembly). Indeed, several Ser/Thr phosphatases (including PPM1A, PP2B ad PP1α) have been shown to dephosphorylate Cdk9 T-loop in vitro or when ectopically expressed in cells, dissociating P-TEFb from Hexim-7SK RNA.24,86,87 In addition to these enzymes, the metal-dependent Ser/Thr phosphatase PPM1G not only dephosphorylates Cdk9 T-loop in vitro, but also binds 7SK RNA and Hexim after P-TEFb is released. This binding is accomplished via direct recruitment of the phosphatase by NF-κB to the 7SK snRNP in response to various NF-κB activating stimuli to promote Pol II transcription activation24,37 (Fig. 2). Interestingly, this dephosphorylation event occurs directly on the chromatin-bound pool of 7SK snRNP to ‘locally’ activate inducible transcriptional pathways.24,37
Figure 2.
P-TEFb is released from the 7SK snRNP on chromatin. The 7SK snRNP complex (containing P-TEFb phosphorylated in the T-loop, (P)) occupies promoter-proximal regions of genes containing paused Pol II, DSIF and NELF. In response to stimulation, PPM1G is recruited by the canonical transcription factor (TF) to dephosphorylate the P-TEFb T-loop, disrupting its interaction with Hexim-7SK RNA and facilitating its release. Other P-TEFb releasing factors include DDX21 (which directly unwinds the 7SK RNA), and the splicing regulator SF2 (which liberates P-TEFb through interactions with exonic splicing sequences). Through T-loop re-phosphorylation, P-TEFb is re-activated to phosphorylate paused Pol II and the negative elongation factors to promote pause release. The arrow denotes the position of the TSS. TF, transcription factor.
Another enzyme, the DDX21 DEAD-box RNA helicase, has also been implicated in the release of P-TEFb from the 7SK snRNP complex, but using a different enzymatic activity as compared with PPM1G33 (Fig. 2). DDX21 is recruited to the promoters of Pol II-transcribed genes encoding ribosomal proteins and snoRNAs and promotes their activation by releasing P-TEFb from the 7SK snRNP through direct unwinding of 7SK RNA (by virtue of its helicase activity).33 In addition, the splicing regulator SRSF2 (also known as SC35, an SR-splicing factor) was also shown to mediate P-TEFb release from the 7SK snRNP upon recognition of exonic splicing enhancer sequences on target RNAs32 (Fig. 2).
Taken together, while P-TEFb release from the 7SK snRNP can be accomplished through the activity of various enzymes/factors, all of these observations point toward a unifying model of 7SK snRNP disassembly and P-TEFb release/activation on chromatin, where transcription and pre-mRNA processes occur.24,31-33,35-37,88 Therefore, activation of P-TEFb can occur ‘on-site', in direct proximity to the paused Pol II complex.
Recruitment of 7SK RNA and the 7SK snRNP complex to chromatin
The deposition of the 7SK snRNP complex on chromatin was initially demonstrated at the HIV promoter, and was dependent on the presence of the general transcription machinery.31,88 Since then, it has become increasingly apparent that there exists an intimate association of 7SK snRNP components with chromatin at many cellular genes, both at promoters and enhancers.24,30-34,37 However, a conceptual understanding of how the 7SK snRNP complex is recruited to chromatin has been murky.
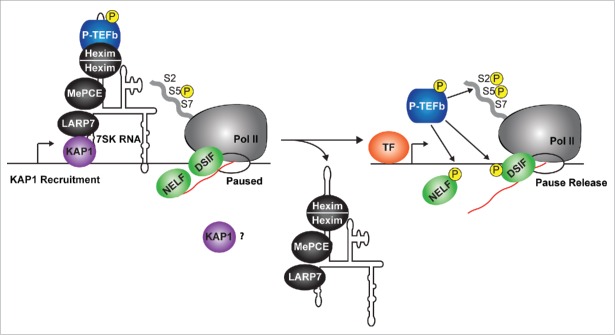
One recent genome-wide study has provided us with insight into how the 7SK snRNP complex is recruited to chromatin.30 Using ChIP-seq of 3 7SK snRNP components (Larp7, Hexim, and Cdk9), McNamara et al. recently shown that the 7SK snRNP occupies promoter-proximal regions of over 70% of Pol II paused genes in human cells.30 This placement of the 7SK snRNP was attributed to the Kruppel-associated box-protein 1 (KAP1, also referred as to TRIM28 and TIF1β),89,90 which physically interacts with Larp7 and recruits the 7SK snRNP to the promoter-proximal sites of transcriptionally active genes30 (Fig. 3). Moreover, KAP1 appears to shuttle in more P-TEFb (through 7SK snRNP) to promoters upon gene activation.30 This model is supported by the observation that antagonizing 7SK snRNP recruitment to promoter-proximal regions through KAP1 knockdown sharply reduces P-TEFb recruitment and Pol II pause release thereby blunting activation of inducible genes in response to stimulation.
Figure 3.
The 7SK snRNP complex is present at the promoters of most Pol II transcribed genes. The 7SK snRNP is recruited onto promoter-proximal regions through KAP1. Here, the P-TEFb kinase is directly released from the 7SK snRNP complex (by the mechanisms described in Fig. 2) and ‘on-site' activation (Pol II, DSIF, and NELF phosphorylation) occurs at the targeted gene. It remains unclear what the fate of KAP1 and the core 7SK snRNP is once P-TEFb has been released from the promoter-proximal region. TF, transcription factor.
Interestingly, the Rosenfeld lab provided recent evidence that components of the 7SK snRNP complex (Cdk9 and Hexim) appear to be recruited to distal gene enhancer sites in the human genome.34,91 These enhancers participate in the activation of gene expression through a specialized set of chromatin-modifying enzymes and through long-range interactions between promoters and enhancers.34 One of these enzymes (JMJD6) demethylates H4R3me2 (which is directly read by the 7SK RNA and the capping enzyme MePCE), ensuring eviction of the 7SK snRNP complex for gene activation. Thus, in this particular case it appears that 7SK RNA plays a key role in recruiting the 7SK snRNP to promoter-proximal regions through gene looping and that histone modifications communicate with the 7SK snRNP to promote its disassembly.
In a separate study, using ChIRP-seq, the Chang lab demonstrated that the scaffold of the 7SK snRNP (the 7SK RNA) occupies both enhancers and super enhancers genome-wide in mouse embryonic stem cells.35 This study found higher levels of 7SK RNA occupancy at traditional and super enhancers compared to promoters, and that deposition of the non-coding RNA at enhancers directly influenced transcriptional activity (enhancer RNA synthesis). Interestingly, the disproportionately high levels of 7SK RNA found at enhancers in mouse embryonic stem cells were not to be mirrored by levels of the Hexim protein. This lends further support to the notion that 7SK's role in nuclear events could extend beyond serving as a reservoir for P-TEFb. It is important to note that Hexim can be incorporated into 7SK complexes without P-TEFb,37 and that core 7SK snRNP complexes lacking P-TEFb-Hexim and containing other RNA-binding factors exist.92-94 In fact, the authors found high co-occupancy between 7SK RNA and the chromatin remodeling complex BAF, and that 7SK RNA is required to limit enhancer RNA initiation and synthesis in a manner that appears to be distinct from its effects on Pol II pausing at promoters.35 It is thus imperative to conduct further genome-wide studies to precisely determine whether the “canonical” 7SK snRNP is present or not at enhancers, and what additional roles 7SK RNA plays at these genomic domains.
These previous studies demonstrate examples of 7SK snRNP recruitment to chromatin. However, it is possible that other mechanisms of 7SK recruitment to chromatin exist and that 7SK RNA plays a key role in controlling nuclear dynamics and partitioning of the inhibitory snRNP between different nuclear territories such as chromatin and speckles.95,96 Collectively, there is much still to be learned about how the ‘on-site' P-TEFb activation through the 7SK snRNP influences basal and inducible gene expression, and how this process controls cellular homeostasis, cell cycle progression, and cell fate from promoters and enhancers.
Conclusions and future challenges
The elongation step of transcription has continued to receive considerable attention over the past decade. While major advances in the field have occurred during this time, there still remains much to be understood about this intricately evolved pathway in metazoans. While it has long been accepted that the major function of the 7SK snRNP is to regulate transcription elongation, recent advances have changed our view of how the inhibitory complex accomplishes this function. From its initial observation at the HIV promoter,31 the proposed model of 7SK snRNP recruitment to promoters has been the subject of considerable debate. However, gene-by-gene and global genome-wide analysis of the 7SK snRNP has greatly strengthened the support for this model. These previous studies highlighted the importance of 7SK RNA recruitment to chromatin and its critical role in controlling transcription from promoters and enhancers. However, several key points remain to be addressed.
One outstanding question in the field is how P-TEFb is released from the 7SK snRNP complex at promoter-proximal regions of genes belonging to distinct signaling pathways. The observation that KAP1 co-localizes with the 7SK snRNP at most Pol II regulated genes points toward a model where KAP1 initially recruits P-TEFb to promoter-proximal regions prior to or simultaneously with the recruitment of an inducible transcription factor (such as NF-κB). Upon recruitment of the inducible transcription factor, P-TEFb would be released and able to directly phosphorylate Pol II CTD and the negative elongation factors (DSIF and NELF), allowing for rapid release of Pol II into productive elongation. However, additional genome-wide analyses are needed to test the model that KAP1 recruits the 7SK snRNP to most promoter-proximal regions in the genome and that this recruitment event results in the positioning of P-TEFb at target genomic domains for ‘on-site' kinase activation to promote Pol II pause release.97 In contrast to the promoter case, the mechanism of 7SK snRNP recruitment to enhancers remains poorly understood. Flynn et al. showed that 7SK RNA recruitment was sensitive to JQ1, which is an acetylated histone mimic that blocks recruitment of the bromodomain-containing protein BRD4 to chromatin.10,35,46 This is in contrast to previously held models in which BRD4 was implicated in P-TEFb recruitment without other 7SK snRNP components.34,98-101 Therefore, further investigation is needed to clarify this discrepancy.
Moreover, it remains unclear the mechanism by which KAP1 interacts with promoter-proximal regions. Thus, this opens the question: is KAP1 directly recruited to the template DNA, modified histones tails and/or the transcription machinery (e.g., Pol II)? Further biochemical and genetic studies are needed to answer these critical questions. Given the interaction of both KAP1 and other bromodomain-containing factors with the 7SK snRNP, it will also be interesting to determine if there is any interplay between these 7SK snRNP anchoring complexes and various P-TEFb releasing factors (e.g. PPM1G and DDX21). This would provide evidence that these anchoring complexes act as central scaffolds for factors involved in P-TEFb recruitment to chromatin as well as release from the inhibitory 7SK snRNP complex.
Is KAP1 the ubiquitous anchoring factor to all promoter-proximal regions containing paused Pol II, or are there other cell-type specific factors involved in this process? Recent evidence supports the latter since it was found that TIF1γ, a highly homologous protein to KAP1, interacted with P-TEFb to regulate transcription elongation of a small subset of erythroid specific genes.89,102 Whether this interaction is bridged by 7SK RNA (as is the case for the KAP1–P-TEFb complex) or not has yet to be addressed. However, given the strikingly high level of protein identity between KAP1 and TIF1γ, a TIF1γ-mediated 7SK snRNP recruitment model to target genes could be envisioned.
Future studies on how promoter and enhancer localized 7SK snRNP complexes serve to control various transcriptional pathways will need to be addressed, as well as other functions of the 7SK snRNP in pluripotent and committed cells, and in the responses to various stimuli in different cell types. With the role these transcriptional regulatory factors may play in diseases such as cancer and HIV infection, further understanding of this process will not only increase our knowledge about biology in general, but will also pave the way for drug development against a number of diseases.
Abbreviations
- ASF/SF2
alternative splicing factor/Serine-arginine-rich splicing factor 2
- AFF
AF4/FMR2 family
- AF9
ALL1-fused gene from chromosome 9
- BAF
BRG1 associated factor
- c-Myc
Myc proto-oncogene protein
- Cdk9
Cyclin dependent kinase 9
- ChIP
chromatin immunoprecipitation
- ChiRP
chromatin isolation by RNA immunoprecipitation
- CTD
C-terminal domain
- DRB
5,6-dichloro-1-β-D-ribofuranosyl-benzimidazole
- DSIF
DRB-sensitivity inducing factor
- DDX21
Nucleolar RNA helicase 21
- Hexim
hexamethylene bis-acetamide inducible protein 1
- ELL
Eleven-19 Lys-rich leukemia
- HSF
heat shock factor
- KAP1
Kruppel-associated protein 1
- Larp7
La-related protein 7
- MePCE
methylphosphate-capping enzyme
- NELF
Negative elongation factor complex
- NF-κB
Nuclear factor κB
- Pol II
RNA polymerase II
- PPM1G
Protein phosphatase 1G
- P-TEFb
positive transcription elongation factor b
- SEC
super elongation complex
- snRNP
small nuclear ribonucleoprotein
- TIF1
transcription intermediary factor 1
- TRIM28
tripartite motif-containing protein 28
- TSS
transcription start site
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Research reported in this publication was supported by an intramural grant from the American Cancer Society (ACS-IRG-02-196); the National Cancer Institute (NCI) of the NIH under award number 5P30CA142543; the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH under award numbers R01AI114362; and Welch Foundation grant I-1782 to Iván D'Orso. C.W.B. work was supported by training grant 5T32GM8203.
References
- [1].Ljungman M. The transcription stress response. Cell Cycle 2007; 6:2252-7; PMID:17700065; http://dx.doi.org/ 10.4161/cc.6.18.4751. [DOI] [PubMed] [Google Scholar]
- [2].Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle 2008; 7:1539-44; PMID:18469524; http://dx.doi.org/ 10.4161/cc.7.11.6006 [DOI] [PubMed] [Google Scholar]
- [3].Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 2013; 50:212-22; PMID:23523369; http://dx.doi.org/ 10.1016/j.molcel.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 2013; 52:517-28; PMID:24184211; http://dx.doi.org/ 10.1016/j.molcel.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet 2007; 39:1507-11; PMID:17994021; http://dx.doi.org/ 10.1038/ng.2007.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 2007; 39:1512-6; PMID:17994019; http://dx.doi.org/ 10.1038/ng.2007.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science 2009; 324:92-4; PMID:19251593; http://dx.doi.org/ 10.1126/science.1169628 [DOI] [PubMed] [Google Scholar]
- [8].Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell 2010; 141:432-45; PMID:20434984; http://dx.doi.org/ 10.1016/j.cell.2010.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 2012; 13:720-31; PMID:22986266; http://dx.doi.org/ 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem 2012; 81:119-43; PMID:22404626; http://dx.doi.org/ 10.1146/annurev-biochem-052610-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 2015; 16:167-77; PMID:25693130; http://dx.doi.org/ 10.1038/nrm3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 1999; 97:41-51; PMID:10199401; http://dx.doi.org/ 10.1016/S0092-8674(00)80713-8 [DOI] [PubMed] [Google Scholar]
- [13].Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, Winston F, et al.. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev 1998; 12:343-56; PMID:9450929; http://dx.doi.org/ 10.1101/gad.12.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J 1998; 17:7395-403; PMID:9857195; http://dx.doi.org/ 10.1093/emboj/17.24.7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 2003; 17:1402-14; PMID:12782658; http://dx.doi.org/ 10.1101/gad.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol 2006; 7:557-67; PMID:16936696; http://dx.doi.org/ 10.1038/nrm1981 [DOI] [PubMed] [Google Scholar]
- [17].Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 2009; 461:186-92; PMID:19741698; http://dx.doi.org/ 10.1038/nature08449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 2010; 143:540-51; PMID:21074046; http://dx.doi.org/ 10.1016/j.cell.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev 2008; 22:1921-33; PMID:18628398; http://dx.doi.org/ 10.1101/gad.1643208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, et al.. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev 1997; 11:2633-44; PMID:9334326; http://dx.doi.org/ 10.1101/gad.11.20.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 2001; 8:327-37; PMID:11545735; http://dx.doi.org/ 10.1016/S1097-2765(01)00314-8 [DOI] [PubMed] [Google Scholar]
- [22].Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev 2006; 20:601-12; PMID:16510875; http://dx.doi.org/ 10.1101/gad.1398206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev 2000; 14:792-803; PMID:10766736 [PMC free article] [PubMed] [Google Scholar]
- [24].McNamara RP, McCann JL, Gudipaty SA, D'Orso I. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep 2013; 5:1256-68; PMID:24316072; http://dx.doi.org/ 10.1016/j.celrep.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zobeck KL, Buckley MS, Zipfel WR, Lis JT. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell 2010; 40:965-75; PMID:21172661; http://dx.doi.org/ 10.1016/j.molcel.2010.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 2006; 23:297-305; PMID:16885020; http://dx.doi.org/ 10.1016/j.molcel.2006.06.014 [DOI] [PubMed] [Google Scholar]
- [27].Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 2001; 276:31793-9; PMID:11431468; http://dx.doi.org/ 10.1074/jbc.M102306200 [DOI] [PubMed] [Google Scholar]
- [28].Laitem C, Zaborowska J, Isa NF, Kufs J, Dienstbier M, Murphy S. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat Struct Mol Biol 2015; 22:396-403; PMID:25849141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 2009; 138:129-45; PMID:19596240; http://dx.doi.org/ 10.1016/j.cell.2009.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].McNamara RP, Reeder JE, McMillan EA, Bacon CW, McCann JL, D'Orso I. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol Cell 2016; 61:39-53; PMID:26725010; http://dx.doi.org/ 10.1016/j.molcel.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 2010; 17:815-21; PMID:20562857; http://dx.doi.org/ 10.1038/nsmb.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 2013; 153:855-68; PMID:23663783; http://dx.doi.org/ 10.1016/j.cell.2013.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Calo E, Flynn RA, Martin L, Spitale RC, Chang HY, Wysocka J. RNA helicase DDX21 coordinates transcription and ribosomal RNA processing. Nature 2015; 518:249-53; PMID:25470060; http://dx.doi.org/ 10.1038/nature13923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 2013; 155:1581-95; PMID:24360279; http://dx.doi.org/ 10.1016/j.cell.2013.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Flynn RA, Do BT, Rubin AJ, Calo E, Lee B, Kuchelmeister H, Rale M, Chu C, Kool ET, Wysocka J, et al.. 7SK-BAF axis controls pervasive transcription at enhancers. Nat Struct Mol Biol 2016; 23:231-8; PMID:26878240; http://dx.doi.org/ 10.1038/nsmb.3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cherrier T, Le Douce V, Eilebrecht S, Riclet R, Marban C, Dequiedt F, Goumon Y, Paillart JC, Mericskay M, Parlakian A, et al.. CTIP2 is a negative regulator of P-TEFb. Proc Natl Acad Sci U S A 2013; 110:12655-60; PMID:23852730; http://dx.doi.org/ 10.1073/pnas.1220136110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gudipaty SA, McNamara RP, Morton EL, D'Orso I. PPM1G Binds 7SK RNA and Hexim1 To Block P-TEFb Assembly into the 7SK snRNP and Sustain Transcription Elongation. Mol Cell Biol 2015; 35:3810-28; PMID:26324325; http://dx.doi.org/ 10.1128/MCB.00226-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 1998; 12:755-62; PMID:9499409; http://dx.doi.org/ 10.1101/gad.12.5.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cho S, Schroeder S, Ott M. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle 2010; 9:1697-705; PMID:20436276; http://dx.doi.org/ 10.4161/cc.9.9.11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998; 92:451-62; PMID:9491887; http://dx.doi.org/ 10.1016/S0092-8674(00)80939-3 [DOI] [PubMed] [Google Scholar]
- [41].McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 1997; 385:357-61; PMID:9002523; http://dx.doi.org/ 10.1038/385357a0 [DOI] [PubMed] [Google Scholar]
- [42].Eick D, Geyer M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem Rev 2013; 113:8456-90; PMID:23952966; http://dx.doi.org/ 10.1021/cr400071f [DOI] [PubMed] [Google Scholar]
- [43].Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, Inukai N, et al.. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol Cell Biol 2003; 23:1863-73; PMID:12612062; http://dx.doi.org/ 10.1128/MCB.23.6.1863-1873.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell 2006; 21:227-37; PMID:16427012; http://dx.doi.org/ 10.1016/j.molcel.2005.11.024 [DOI] [PubMed] [Google Scholar]
- [45].Pirngruber J, Shchebet A, Johnsen SA. Insights into the function of the human P-TEFb component CDK9 in the regulation of chromatin modifications and co-transcriptional mRNA processing. Cell Cycle 2009; 8:3636-42; PMID:19844166; http://dx.doi.org/ 10.4161/cc.8.22.9890 [DOI] [PubMed] [Google Scholar]
- [46].Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol 2013; 33:2497-507; PMID:23589332; http://dx.doi.org/ 10.1128/MCB.01180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 2004; 24:787-95; PMID:14701750; http://dx.doi.org/ 10.1128/MCB.24.2.787-795.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A 2010; 107:11301-6; PMID:20534440; http://dx.doi.org/ 10.1073/pnas.1000681107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].He N, Liu M, Hsu J, Xue Y, Chou S, Burlingame A, Krogan NJ, Alber T, Zhou Q. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Mol Cell 2010; 38:428-38; PMID:20471948; http://dx.doi.org/ 10.1016/j.molcel.2010.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 2010; 37:429-37; PMID:20159561; http://dx.doi.org/ 10.1016/j.molcel.2010.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 2012; 13:543-7; PMID:22895430; http://dx.doi.org/ 10.1038/nrm3417 [DOI] [PubMed] [Google Scholar]
- [52].Hsin JP, Li W, Hoque M, Tian B, Manley JL. RNAP II CTD tyrosine 1 performs diverse functions in vertebrate cells. Elife 2014; 3:e02112; PMID:24842995; http://dx.doi.org/ 10.7554/eLife.02112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Descostes N, Heidemann M, Spinelli L, Schuller R, Maqbool MA, Fenouil R, Koch F, Innocenti C, Gut M, Gut I, et al.. Tyrosine phosphorylation of RNA polymerase II CTD is associated with antisense promoter transcription and active enhancers in mammalian cells. Elife 2014; 3:e02105; PMID:24842994; http://dx.doi.org/ 10.7554/eLife.02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mayer A, Heidemann M, Lidschreiber M, Schreieck A, Sun M, Hintermair C, Kremmer E, Eick D, Cramer P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012; 336:1723-5; PMID:22745433; http://dx.doi.org/ 10.1126/science.1219651 [DOI] [PubMed] [Google Scholar]
- [55].Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell 2009; 36:541-6; PMID:19941815; http://dx.doi.org/ 10.1016/j.molcel.2009.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Czudnochowski N, Bosken CA, Geyer M. Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun 2012; 3:842; PMID:22588304; http://dx.doi.org/ 10.1038/ncomms1846 [DOI] [PubMed] [Google Scholar]
- [57].Suh H, Ficarro SB, Kang UB, Chun Y, Marto JA, Buratowski S. Direct Analysis of Phosphorylation Sites on the Rpb1 C-Terminal Domain of RNA Polymerase II. Mol Cell 2016; 61:297-304; PMID:26799764; http://dx.doi.org/ 10.1016/j.molcel.2015.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Schuller R, Forne I, Straub T, Schreieck A, Texier Y, Shah N, Decker TM, Cramer P, Imhof A, Eick D. Heptad-Specific Phosphorylation of RNA Polymerase II CTD. Mol Cell 2016; 61:305-14; PMID:26799765; http://dx.doi.org/ 10.1016/j.molcel.2015.12.003 [DOI] [PubMed] [Google Scholar]
- [59].Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J Biol Chem 2002; 277:40156-62; PMID:12177005; http://dx.doi.org/ 10.1074/jbc.M207441200 [DOI] [PubMed] [Google Scholar]
- [60].Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature 2010; 465:747-51; PMID:20535204; http://dx.doi.org/ 10.1038/nature09131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol 2000; 20:5077-86; PMID:10866664; http://dx.doi.org/ 10.1128/MCB.20.14.5077-5086.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 2001; 414:322-5; PMID:11713533; http://dx.doi.org/ 10.1038/35104581 [DOI] [PubMed] [Google Scholar]
- [63].Yang Z, Zhu Q, Luo K, Zhou Q. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 2001; 414:317-22; PMID:11713532; http://dx.doi.org/ 10.1038/35104575 [DOI] [PubMed] [Google Scholar]
- [64].Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci U S A 2009; 106:7798-803; PMID:19416841; http://dx.doi.org/ 10.1073/pnas.0903188106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol 1991; 11:3432-45; PMID:1646389; http://dx.doi.org/ 10.1128/MCB.11.7.3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Muniz L, Egloff S, Kiss T. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res 2013; 41:4686-98; PMID:23471002; http://dx.doi.org/ 10.1093/nar/gkt159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep 2008; 9:569-75; PMID:18483487; http://dx.doi.org/ 10.1038/embor.2008.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem 2005; 280:28819-26; PMID:15965233; http://dx.doi.org/ 10.1074/jbc.M502712200 [DOI] [PubMed] [Google Scholar]
- [69].Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell 2003; 12:971-82; PMID:14580347; http://dx.doi.org/ 10.1016/S1097-2765(03)00388-5 [DOI] [PubMed] [Google Scholar]
- [70].Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J Biol Chem 2005; 280:16368-76; PMID:15713661; http://dx.doi.org/ 10.1074/jbc.M500912200 [DOI] [PubMed] [Google Scholar]
- [71].He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 2008; 29:588-99; PMID:18249148; http://dx.doi.org/ 10.1016/j.molcel.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al.. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res 2008; 36:2219-29; PMID:18281698; http://dx.doi.org/ 10.1093/nar/gkn061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al.. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 2007; 27:262-74; PMID:17643375; http://dx.doi.org/ 10.1016/j.molcel.2007.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res 2010; 38:360-9; PMID:19906723; http://dx.doi.org/ 10.1093/nar/gkp977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog 2010; 6:e1001152; PMID:20976203; http://dx.doi.org/ 10.1371/journal.ppat.1001152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol 2006; 26:630-42; PMID:16382153; http://dx.doi.org/ 10.1128/MCB.26.2.630-642.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, et al.. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J 2004; 23:2608-19; PMID:15201869; http://dx.doi.org/ 10.1038/sj.emboj.7600275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J 2005; 24:4291-303; PMID:16362050; http://dx.doi.org/ 10.1038/sj.emboj.7600883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res 2005; 33:7000-10; PMID:16377779; http://dx.doi.org/ 10.1093/nar/gki997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Schulte A, Czudnochowski N, Barboric M, Schonichen A, Blazek D, Peterlin BM, Geyer M. Identification of a cyclin T-binding domain in Hexim1 and biochemical analysis of its binding competition with HIV-1 Tat. J Biol Chem 2005; 280:24968-77; PMID:15855166; http://dx.doi.org/ 10.1074/jbc.M501431200 [DOI] [PubMed] [Google Scholar]
- [81].Dames SA, Schonichen A, Schulte A, Barboric M, Peterlin BM, Grzesiek S, Geyer M. Structure of the Cyclin T binding domain of Hexim1 and molecular basis for its recognition of P-TEFb. Proc Natl Acad Sci U S A 2007; 104:14312-7; PMID:17724342; http://dx.doi.org/ 10.1073/pnas.0701848104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem 2004; 279:4153-60; PMID:14627702; http://dx.doi.org/ 10.1074/jbc.M310044200 [DOI] [PubMed] [Google Scholar]
- [83].Fisher RP. The CDK Network: Linking Cycles of Cell Division and Gene Expression. Genes Cancer 2012; 3:731-8; PMID:23634260; http://dx.doi.org/ 10.1177/1947601912473308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol Cell Biol 2000; 20:6958-69; PMID:10958691; http://dx.doi.org/ 10.1128/MCB.20.18.6958-6969.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 2012; 19:1108-15; PMID:23064645; http://dx.doi.org/ 10.1038/nsmb.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, et al.. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev 2008; 22:1356-68; PMID:18483222; http://dx.doi.org/ 10.1101/gad.1636008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wang Y, Dow EC, Liang YY, Ramakrishnan R, Liu H, Sung TL, Lin X, Rice AP. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J Biol Chem 2008; 283:33578-84; PMID:18829461; http://dx.doi.org/ 10.1074/jbc.M807495200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].D'Orso I, Jang GM, Pastuszak AW, Faust TB, Quezada E, Booth DS, Frankel AD. Transition step during assembly of HIV Tat:P-TEFb transcription complexes and transfer to TAR RNA. Mol Cell Biol 2012; 32:4780-93; PMID:23007159; http://dx.doi.org/ 10.1128/MCB.00206-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 2011; 286:26267-76; PMID:21652716; http://dx.doi.org/ 10.1074/jbc.R111.252569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Iyengar S, Ivanov AV, Jin VX, Rauscher FJ 3rd, Farnham PJ. Functional analysis of KAP1 genomic recruitment. Mol Cell Biol 2011; 31:1833-47; PMID:21343339; http://dx.doi.org/ 10.1128/MCB.01331-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kim TK, Shiekhattar R. Architectural and Functional Commonalities between Enhancers and Promoters. Cell 2015; 162:948-59; PMID:26317464; http://dx.doi.org/ 10.1016/j.cell.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J 2007; 26:3570-80; PMID:17611602; http://dx.doi.org/ 10.1038/sj.emboj.7601783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol Cell Biol 2007; 27:6996-7006; PMID:17709395; http://dx.doi.org/ 10.1128/MCB.00975-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hogg JR, Collins K. RNA-based affinity purification reveals 7SK RNPs with distinct composition and regulation. RNA 2007; 13:868-80; PMID:17456562; http://dx.doi.org/ 10.1261/rna.565207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Mizutani T, Ishizaka A, Suzuki Y, Iba H. 7SK small nuclear ribonucleoprotein complex is recruited to the HIV-1 promoter via short viral transcripts. FEBS Lett 2014; 588:1630-6; PMID:24607481; http://dx.doi.org/ 10.1016/j.febslet.2014.01.067 [DOI] [PubMed] [Google Scholar]
- [96].Prasanth KV, Camiolo M, Chan G, Tripathi V, Denis L, Nakamura T, Hubner MR, Spector DL. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell 2010; 21:4184-96; PMID:20881057; http://dx.doi.org/ 10.1091/mbc.E10-02-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G, et al.. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol 2014; 21:876-83; PMID:25173174; http://dx.doi.org/ 10.1038/nsmb.2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005; 19:535-45; PMID:16109377; http://dx.doi.org/ 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- [99].Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol 2008; 28:967-76; PMID:18039861; http://dx.doi.org/ 10.1128/MCB.01020-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 2005; 19:523-34; PMID:16109376; http://dx.doi.org/ 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- [101].Itzen F, Greifenberg AK, Bosken CA, Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res 2014; 42:7577-90; PMID:24860166; http://dx.doi.org/ 10.1093/nar/gku449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, et al.. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 2010; 142:133-43; PMID:20603019; http://dx.doi.org/ 10.1016/j.cell.2010.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]