Abstract
T cell receptor (TCR) signaling is essential in the development and differentiation of T cells in the thymus and periphery, respectively. The vast array of TCRs proves studying a specific antigenic response difficult. Therefore, TCR transgenic mice were made to study positive and negative selection in the thymus as well as peripheral T cell activation, proliferation and tolerance. However, relatively few TCR transgenic mice have been generated specific to any given antigen. Thus, studies involving TCRs of varying affinities for the same antigenic peptide have been lacking. The generation of a new TCR transgenic line can take six or more months. Additionally, any specific backcrosses can take an additional six months. In order to allow faster generation and screening of multiple TCRs, a protocol for retroviral transduction of bone marrow was established with stoichiometric expression of the TCRα and TCRβ chains and the generation of retrogenic mice. Each retrogenic mouse is essentially a founder, virtually negating a founder effect, while the length of time to generate a TCR retrogenic is cut from six months to approximately six weeks. Here we present a rapid and flexible alternative to TCR transgenic mice that can be expressed on any chosen background with any particular TCR.
Keywords: Immunology, Issue 113, T cell receptor (TCR), Major Histocompatibility Complex (MHC), Retrovirus, Bone Marrow, Transduction, Retrogenic
Introduction
T cell receptor (TCR) repertoire of humans and mice has been estimated at 1 x 108 and 2 x 106 unique TCRs respectively1,2. This large diversity allows T cells to recognize a vast array of antigen epitopes derived from self-peptides as well as from pathogens presented by the major histocompatibility complex (MHC) on antigen presenting cells (APCs). The subtle differences in the interactions of the TCRs with unique peptide-MHC complexes dictate whether a T cell will undergo apoptosis, anergy, activation, differentiation, cytokine production or cytotoxicity. However, due to the large TCR repertoire, analysis of how a specific TCR will respond to a particular antigen requires the use of single TCR systems.
Various TCR transgenic mice have been generated in order to study the function of a single TCR in an in vivo model 3-9. However, there are caveats to TCR transgenic mice including the cost, the length of time to generate a single transgenic mouse and the so called founder effect of random transgene insertion into germline DNA10. Therefore, relatively few TCR transgenic mice have been generated for any given antigen and the functional implications of high and low TCR affinity for the same epitope are rarely addressed. To address the need for a rapid approach to screen and study multiple TCRs individually or in combination, retrogenic ('retro' from retrovirus and 'genic' from transgenic) mice have been utilized as an alternative to TCR transgenic mice11-13.
The 2A peptide consensus motif found within several viruses consist of an 2A-Asp-Val/Ile-Glut-X-Asn-Pro-Gly-2B-Pro, in which cleavage occurs between the glycine of the 2A and the proline of the 2B from cis-acting hydrolase activity, resulting in ribosomal skipping during translation10,14-16. For a detailed diagram depicting the cleavage of the various 2A peptides (F2A, E2A, T2A and P2A) please refer to references 10 - 12. In this manner, 2 cistrons (TCR alpha and TCR beta) can be linked resulting in stoichiometric translation in a single vector. Utilizing this approach, we are able to express and directly compare multiple antigen specific TCRs in vivo.
Protocol
Ethics Statement: Every effort is made to keep animal discomfort or stress to a minimum during irradiation and tail vein injections. Mice are used as a source of cells in these experiments; as such there are no procedures or manipulations apart from euthanasia. Mice will be euthanized by CO2 inhalation followed by cervical dislocation to confirm death. This procedure is consistent with the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association.
1. Prepare Retroviral Construct
Sub-clone the T-cell receptor (TCR) alpha and beta chains, separated by a 2A linker, into a MSCV-based retroviral11 vector (example, pMIAII or pMIGII)11. NOTE: The 2A linker allows for stoichiometric and concordant expression of the alpha and beta TCR chains from a single open reading frame. The vector includes an IRES fluorescent protein cassette (ametrine or GFP) that is used to track transduced cells and transduction efficiency. For a detailed protocol please see Holst et al.11
Isolate Plasmid DNA using commercially obtained plasma prep kit or a similar product. Use a working concentration of 1 - 2 μg μl-1.
2. Generation of Retroviral Producer Cell Lines
- Utilize a two-step process to produce the retroviral producer cell lines. NOTE: For a detailed protocol, please view Holst et al.11.
- Generate retrovirus by transient transfection of 293T cells with three plasmids (pVSVG, pEQ-Pam3(-E) and vector of interest11) and take the retroviral supernatant from the 293T cells to transduce the GP+E86 ecotropic retroviral producer cell line.
- Isolate the top 40% of fluorescent expressing GP+E86 producer cells by FACS and propagate the transdcuced-GP+E86 producer cells as a source of virus. NOTE: The generation of high-titer producers is critical for efficient transduction of bone marrow cells. For a complete protocol please view Holst et al.11
3. Retrovirus-mediated Stem Cell Gene Transfer (Day -5)
- Thaw out and culture 0.5 -1.0 x 106 of each GP+E86 producer cell line in complete DMEM 10% (vol/vol) FBS to allow sufficient growth for two confluent 150-mm plates by Day 0 of this protocol. NOTE: Thawing 0.5 - 1.0 x 106 of each GP+E86 producer cell line in a 10 cm plate will allow for 30 - 50% confluency at Day 5. This also allows for 5 days of culture in order to expand the GP+E86 producer cell lines to two confluent 150-mm plates by Day 0.
- Culture retroviral producer cell line in complete tissue culture media containing 5% feta calf serum, and ciprofloxacin (10 μg/ml) to avoid mycoplasma contamination. NOTE: Discontinue use of ciprofloxacin when generating the viral supernatant for bone marrow transduction. Avoid over-growth of the producer cell line, as this will negatively impact bone marrow transduction efficiency.
4. Inject Donor Mice with 5-Fluorouracil (5-FU) (Day -3)
Weigh the donor mice (5 - 12 weeks old) to determine the amount of 5-FU required. Using mice younger than 5 weeks of age results in low bone marrow yield. To ensure expression of a single TCR, use a T cell deficient mouse strain such as SCID, RAG or TCR alpha deficient mice as bone marrow donors.
Working in the tissue culture hood, dilute the 5-FU stock with sterile PBS in order to make a sufficient amount of 10 mg ml-1 working solution of 5-FU to deliver 0.15 mg 5-FU per gram of body weight.
Using a 1 ml syringe with a 37 G needle, intraperitoneal inject 0.15 mg 5-FU per gram of body weight per donor mouse. NOTE: Use 1 donor mouse per one recipient to start and adjust the number of donor mice according to the cell yield. An alternative method to the 5-FU treatment for the enrichment and isolation of bone marrow progenitors is negative selection of lineage positive cells as described in Viret et al.
5. Bone Marrow Extraction Procedure (Day 0)
Obtain dissection scissors and forceps for bone isolation. Prepare media (HBSS supplemented with 5% FCS (vol/vol)) to collect bones in a 50 ml conical tube. Keep 50 ml conical containing media on ice. Euthanize mice by CO2 inhalation followed by cervical dislocation to confirm death. Pinch paw to ensure no reflexes are detected.
Sterilize the surgical site and surgical tools with ethanol or methanol. Remove the femur, tibia, humerus, and pelvic girdle carefully by first cutting up and under the pelvic girdle. Cut back down along the femur to the tibia. Remove all surrounding tissue using scissors and forceps to obtain clean bones. Keep bones hydrated in HBSS + 5% (vol/vol) FBS.
Separate the bones by cutting at the joints, making sure to cut both ends off each bone. Insert a 25-gauge needle attached to a 10-ml syringe containing HBSS + 5% (vol/vol) FBS. Apply pressure and flush all bone marrow through a 70-μm strainer resting in a 50 ml conical.
Periodically, push the bone marrow through the 70-μm filter using the plunger from a 1 ml or 3 ml syringe. Rinse the strainer with HBSS + 5% (vol/vol) FBS. This will ensure a single cell suspension that is free of bone fragments and tissue. Keep cells sterile by performing the procedure in a sterile hood.
Centrifuge bone marrow cells at 300 x g for 10 min at 4 °C.
6. Bone Marrow Culture (Day 0)
Decant or aspirate media and resuspend cell pellet in 1 ml ammonium chloride based red blood lysis buffer (or RBC lysis buffer equivalent) per 50 ml conical tube. After 1 min incubation at room temperature, quench the red cell lysis buffer by adding 10 ml of complete DMEM 20% (vol/vol) FBS.
Centrifuge cells at 300 x g for 10 min at 4 °C.
Decant or aspirate supernatant and resuspend bone marrow in 5 ml complete DMEM 20% (vol/vol) FBS. Count the cells with a Neubauer counting chamber. NOTE: General yield should be approximately 5 - 10 x 106 cells per mouse; however, the yield can vary among different strains of mice.
Plate bone marrow cells at a density of 4 x 107 cells per 150-mm plate in 30 ml of complete DMEM 20% (vol/vol) FBS supplemented with IL-3 (20 g ml-1), IL-6 (50 g ml-1) and SCF (50 g ml-1). NOTE: Use freshly thawed-aliquoted cytokines. Do not use cytokines that have been frozen and thawed more than once or kept at 4 °C for more than 2 weeks.
Incubate bone marrow overnight (approximately 24 hr) at 37 °C with 5% CO2.
7. Retroviral Producer Cell Culture (Day 0)
Plate 3 x 106 retroviral producer cells per 150-mm plate in 18 ml complete DMEM 20% (vol/vol) FBS.
Incubate retroviral producer cells at 37 °C overnight. Anticipate one 150-mm plate for 15 million bone marrow cells (approximately 2 - 3 donor mice).
8. Retroviral Supernatant (Day 1)
The next day (approximately 24 hr later), harvest the retroviral supernatant from the producer plates (set up in step 7) by drawing up the retroviral supernatant directly off of the 150-mm plate with a 10-ml syringe and filter using a 0.45-μm syringe filter.
Carefully replace the removed retroviral supernatant with 18 ml of fresh complete DMEM 20% (vol/vol) FBS.
Supplement the filtered retroviral supernatant with IL-3 (20 g ml-1), IL-6 (50 g ml-1), MSCF (50 g ml-1) and Hexadimethrine bromide (Polybrene) (6 μg ml-1). NOTE: Hexadimethrine bromide should be made fresh every 2 months to avoid a drop in efficacy of retroviral transduction.
9. Bone Marrow Harvest (Day 1)
After completing step 8.3, harvest the bone marrow cells from Step 6.4 and transfer the media containing non-adherent cells into sterile 50-ml tubes.
Wash the plates vigorously with 10 ml PBS and collect in the 50 ml conical from step 9.1. If necessary, repeat the wash step.
Centrifuge the cells at 300 x g for 10 min at 4 °C, decant the supernatant, resuspend cells in a small volume (1 - 3 ml) complete DMEM 20% (vol/vol) FBS, and count. Anticipate 50 - 75% recovery from day 0. Resuspend bone marrow at 1 x 106 cells per 1 ml of the retroviral supernatant containing cytokines and Hexadimethrine bromide.
10. Bone Marrow Transduction (Day 1)
Plate the retroviral supernatant/bone marrow mixture at a volume of 3 ml per well in a six-well tissue culture plate. Wrap the plates in plastic wrap to minimize gas exchange during the centrifugation in step 10.2.
Spin the wrapped six-well plates at 1,000 x g for 60 min a 37 °C.
After the spin is complete, remove the plastic wrap and place the plates in a 37 °C CO2 incubator for 24 hr.
11. Retroviral Supernatant and Transduction (Day 2)
After 24 hr, collect fresh viral supernatant from the viral producer cell line. Filter the retroviral supernatant using a 10-ml syringe and a 0.45-μm syringe filter. Supplement the filtered retroviral supernatant with IL-3 (20 g ml-1), IL-6 (50 g ml-1) and SCF (50 g ml-1) and Hexadimethrine bromide (6 g ml-1).
Next, carefully remove with pipette or aspirate the top 2 ml of media from each well of the six-well plate containing the bone marrow cells. NOTE: Work carefully not to aspirate the bone marrow, which is usually concentrated in the middle of the well. Alternatively, the removed supernatant can be spun down to recover any lost bone marrow cells.
To each well in the bone marrow 6-well plate add 3 ml of fresh viral supernatant containing cytokines and Hexadimethrine bromide. Wrap the plates in plastic, and repeat the spin transduction at 1,000 x g for 60 min a 37 °C. Unwrap the plates, and incubate cells at 37 °C and 5% CO2 overnight (about 24 hr).
12. Addition of Fresh Supplemented DMEM (Day 3)
After 24 hr, remove the top 2 ml of media from each well of the six-well plate containing the bone marrow cells, as in step 11.2. Replace the removed media with fresh DMEM 20% (vol/vol) FBS supplemented with final concentrations of IL-3 (20 g ml-1), IL-6 (50 g ml-1) and SCF (50 g ml-1).
Incubate bone marrow plates at 37 °C CO2 incubator for another 24 hr.
13. Harvest Transduced Bone Marrow (Day 4)
- After 24 hr, collect the bone marrow cells from the six-well plates. Wash the plates vigorously with PBS to remove all non-adherent cells.
- Centrifuge the cells at 300 x g for 10 min at 4 °C and decant the supernatant.
- Resuspend the bone marrow in a small volume (1 ml) of PBS + 0.5% (vol/vol) heat-inactivated FBS. Keep cells on ice.
- Take a 10-μl sample of each bone marrow condition and count the cells using a Neubauer counting chamber. Assuming 1 - 2 donor per recipient, the yield will be sufficient to transfer at least 4 x 106 cells per recipient mouse. Resuspend the bone marrow in a sufficient volume of PBS + 0.5% (vol/vol) heat-inactivated FBS to inject 200 - 300 μl per mouse.
- Inject at least 2 x 106 cells with a transduction efficiency of 25% (can inject up to 8 x106 cells) per mouse intravenously. NOTE: We routinely inject five recipient mice per two 6-well plates of bone marrow cells. If the yield is substantially lower, more donor mice should be used until the sufficient bone marrow number can be obtained. In order to achieve optimal reconstitution, at least 5 x 105 transduced (fluorescent positive) should be injected into each recipient.
With a micro pipettor, take a 10-μl sample of each bone marrow condition and analyze transduction efficiency by flow cytometry based on expression of the fluorescent marker (Figure 1A)11. NOTE: Generally, the percentage of fluorescent positive cells is between 25% and 70%, depending on the construct and retroviral titer. The number of transduced bone marrow cells necessary to reconstitute a sub-lethally irradiate mouse can vary depending on the transduction efficiency. The lower the transduction efficiency, the more bone marrow cells required and conversely, the higher the bone marrow transduction efficiency, the lower number of cells required for reconstitution. Transduction efficiency below 25% is suboptimal and may result in insufficient reconstitution and survival. In order to ensure optimal bone marrow recovery and transduction efficiency, use freshly prepared tissue culture media and cytokines. When expressing a new TCR utilizing the retrogenic system, the selection of that TCR in vivo is unknown. Depending on each individual TCR affinity, TCR selection in the thymus and peripheral T cell expression will vary17.
14. Mouse Irradiation, Bone Marrow Injection and Care
- Irradiate mice the day before bone marrow injection (Same day as Step 13). Use the following doses: C57BL/6 mice (and other wild type strains), 900 rads; Rag1-/- mice, 500 rads; scid mice, 300 rads).
- Place recipient mice on Amoxicillin (50 mg/kg/day) or another antibiotic water treatment before irradiation, and keep on antibiotics for the first two weeks after bone marrow injection. NOTE: Optimal irradiation dose may have to be determined empirically.
For injection, load the cells from Step 13.1 into a 1-ml syringe using blunt needles immediately before injection, then change the needle to 27-gauge for injection, expel any air to avoid air embolism. Heat mice under a heating lamp and inject 0.2 ml of bone marrow per mouse intravenously. NOTE: Do not let cells sit in the syringe for any length of time or the cells will settle.
Monitor the mice weekly to ensure that they remain healthy.
After 5 - 6 weeks, bleed the mice to check for reconstitution on the basis of GFP+/ametrine+ and TCR co-expression (Figure 1B)11.
Representative Results
Bone marrow transduction efficiency is checked at step 13.3 of the protocol before the bone marrow is injected into tail vein i.v. In the representative bone marrow transduction figure (Figure 1A), approximately 10-μl of the harvested bone marrow was added to 100-μl of PBS and analyzed for ametrine expression. Generally the percentage of fluorescent positive cells is between 25% and 70%, depending on the construct and retroviral titer. After 6 weeks bone marrow injection, the mice are bled to determine bone marrow reconstitution (Figure 1B). Approximately 25 μl of peripheral blood was red cell lysed and stained with anti-TCRβ. In Figure 2B all TCR positive cells concordantly express the ametrine fluorescent protein.
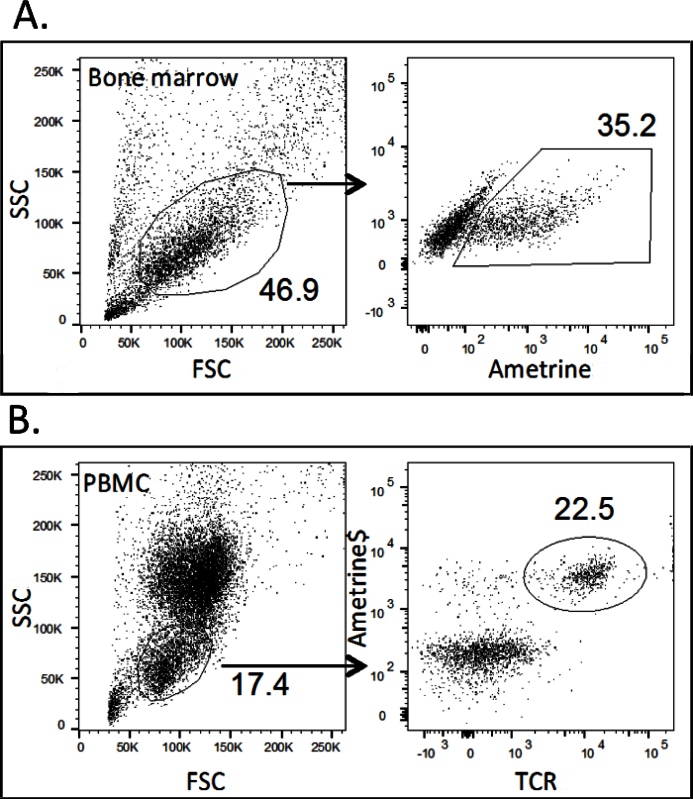 Figure 1.Example of Bone Marrow Transduction and Peripheral TCR Retrogenic T Cells.A) Transduction efficiency of bone marrow cells is analyzed based on the fluorescent marker (ametrine) on the day of bone marrow transfer. Approximately 10-μl of the harvested bone marrow was added to 100-μl of PBS and analyzed for ametrine expression. A successful bone marrow transduction will yield a percentage of fluorescent positive cells between 25% and 70%. B) Example of TCR retrogenic T cells in peripheral blood 6 weeks post bone marrow reconstitution. Peripheral blood was obtained and red blood cells were lysed with RBC lysis buffer. The remaining cells were stained for TCRβ. Representative analysis for ametrine expression and anti-TCRβ staining 6 weeks post bone marrow reconstitution. Please click here to view a larger version of this figure.
Figure 1.Example of Bone Marrow Transduction and Peripheral TCR Retrogenic T Cells.A) Transduction efficiency of bone marrow cells is analyzed based on the fluorescent marker (ametrine) on the day of bone marrow transfer. Approximately 10-μl of the harvested bone marrow was added to 100-μl of PBS and analyzed for ametrine expression. A successful bone marrow transduction will yield a percentage of fluorescent positive cells between 25% and 70%. B) Example of TCR retrogenic T cells in peripheral blood 6 weeks post bone marrow reconstitution. Peripheral blood was obtained and red blood cells were lysed with RBC lysis buffer. The remaining cells were stained for TCRβ. Representative analysis for ametrine expression and anti-TCRβ staining 6 weeks post bone marrow reconstitution. Please click here to view a larger version of this figure.
Discussion
In the protocol, we detail several critical steps to ensure optimal bone marrow health, transduction efficiency and reconstitution. First critical step is the generation and proper maintenance of the GP+E86 viral producer cells. Use early passage producer cell lines and maintain at 80% confluency or lower prior to use. When making fresh GP+E86 viral producer cells, ensure the 293T cells are early passage and growing in culture for 24 - 48 hr. Plating too many GP+E86 cells during the transduction step will lower viral titer. Determine viral titer as described in Holst et al.11 to ensure GP+E86 are robust viral producers. Additionally, the removal of red blood cells from the harvested bone marrow improves transduction efficiency. Ensuring the health of the bone marrow progenitors is critical to the success of the protocol. To this end, always use fresh media (less than 4 weeks at 4 °C after supplemented with FCS) and fresh cytokines (less than 2 weeks when kept at 4 °C). Hexadimethrine bromide is also sensitive to degradation and should be made fresh, once it is in solution, every 2 months. Other details that will ensure optimal bone marrow health and transduction include pre-warming the centrifuge before the spin transduction. Spin-transduce at 37 °C and wrap plates with plastic wrap to lessen gas exchange as much as possible. Also, do not let the bone marrow sit outside of the 37 °C incubator or on ice for extended periods of time as this will increase cell death and decrease bone marrow transduction and reconstitution. After the bone marrow cells have been harvested at Day 4 (step 13), only load the syringes immediately before injecting the mice. Finally, ensure that mice are placed on antibiotics the day before or the day of the irradiation.
Utilization of this retroviral mediated transduction protocol allows for the transduction of bone marrow progenitor cells resulting in the generation of T cell receptor retrogenic mice. Retrogenic mice are faster to generate than TCR transgenic mice and cost considerably less than generating a single TCR transgenic. However, retrogenic mice cannot be bred and must generated anew with each experiment10. The number of peripheral T cells are lower in retrogenic mice than seen in TCR transgenic mice however the regulation of the TCR is comparable between TCR retrogenic and TCR transgenics10. The TCR retrogenic system allows for the expression of multiple TCRs within the same colony and even within the same experiment. Utilizing the TCR retrogenic protocol described here allows for an investigator to test and screen the development and functionality of numerous newly identified MHC class I or II restricted TCRs. With the development of the "humanized mouse" model18,19, the TCR retrogenic could further be expanded utilizing cloned human TCRs. Additionally, the 2A linked constructs can also be used to study other multi-proteins including the CD3 chains, interleukins and chimeric antigen receptors20-24.
Disclosures
No conflicts of interest declared.
Acknowledgments
This work was supported by grants from NIH (5K22A1119151-01 and 1R56DK104903-01) to M.L.B, Pilot/Feasibility Program of the Diabetes Research Center (P30-DK079638) at BCM, JDRF 1-FAC-2014-243-A-N APF, ADA 1-15-JF-07, AAI Careers in Immunology Fellowship to M.B., and The Robert and Janice McNair Foundation.
References
- Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnitsyna VI, Evavold BD, Schoettle LN, Blattman JN, Antia R. Estimating the diversity, completeness, and cross-reactivity of the T cell repertoire. Frontiers in immunology. 2013;4:485. doi: 10.3389/fimmu.2013.00485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisielow P, Bluthmann H, Staerz UD, Steinmetz M, von Boehmer H. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature. 1988;333:742–746. doi: 10.1038/333742a0. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Verdaguer J, et al. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J Exp Med. 1997;186:1663–1676. doi: 10.1084/jem.186.10.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JD, Wang B, Haskins K, Benoist C, Mathis D. Following a diabetogenic T cell from genesis through pathogenesis. Cell. 1993;74:1089–1100. doi: 10.1016/0092-8674(93)90730-e. [DOI] [PubMed] [Google Scholar]
- Pauza ME, et al. T-cell receptor transgenic response to an endogenous polymorphic autoantigen determines susceptibility to diabetes. Diabetes. 2004;53:978–988. doi: 10.2337/diabetes.53.4.978. [DOI] [PubMed] [Google Scholar]
- Jasinski JM, et al. Transgenic insulin (B:9-23) T-cell receptor mice develop autoimmune diabetes dependent upon RAG genotype, H-2g7 homozygosity, and insulin 2 gene knockout. Diabetes. 2006;55:1978–1984. doi: 10.2337/db06-0058. [DOI] [PubMed] [Google Scholar]
- Kersh GJ, et al. TCR transgenic mice in which usage of transgenic alpha- and beta-chains is highly dependent on the level of selecting ligand. Journal of immunology. 1998;161:585–593. [PubMed] [Google Scholar]
- Bettini ML, Bettini M, Vignali DA. TCR retrogenic mice: A rapid, flexible alternative to TCR transgenic mice. Immunology. 2012;136(3):265–272. doi: 10.1111/j.1365-2567.2012.03574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst J, et al. Generation of T-cell receptor retrogenic mice. Nat Protoc. 2006;1:406–417. doi: 10.1038/nprot.2006.61. [DOI] [PubMed] [Google Scholar]
- Holst J, Vignali KM, Burton AR, Vignali DA. Rapid analysis of T-cell selection in vivo using T cell-receptor retrogenic mice. Nat Methods. 2006;3:191–197. doi: 10.1038/nmeth858. [DOI] [PubMed] [Google Scholar]
- Bettini ML, Bettini M, Nakayama M, Guy CS, Vignali DA. Generation of T cell receptor-retrogenic mice: improved retroviral-mediated stem cell gene transfer. Nat Protoc. 2013;8:1837–1840. doi: 10.1038/nprot.2013.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly ML, et al. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Atkins JF, et al. A case for "StopGo": reprogramming translation to augment codon meaning of GGN by promoting unconventional termination (Stop) after addition of glycine and then allowing continued translation (Go) RNA. 2007;13:803–810. doi: 10.1261/rna.487907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doronina VA, et al. Site-specific release of nascent chains from ribosomes at a sense codon. Mol Cell Biol. 2008;28:4227–4239. doi: 10.1128/MCB.00421-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettini M, et al. TCR affinity and tolerance mechanisms converge to shape T cell diabetogenic potential. Journal of immunology. 2014;193:571–579. doi: 10.4049/jimmunol.1400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm MA, Wiles MV, Greiner DL, Shultz LD. Generation of improved humanized mouse models for human infectious diseases. J Immunol Methods. 2014;410:3–17. doi: 10.1016/j.jim.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm MA, Shultz LD, Greiner DL. Humanized mouse models to study human diseases. Curr Opin Endocrinol Diabetes Obes. 2010;17:120–125. doi: 10.1097/MED.0b013e328337282f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin PJ, et al. Production of interleukin-12 as a self-processing 2A polypeptide. J Interferon Cytokine Res. 1999;19:235–241. doi: 10.1089/107999099314162. [DOI] [PubMed] [Google Scholar]
- Collison LW, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- Holst J, et al. Scalable signaling mediated by T cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nature immunology. 2008;9:658–666. doi: 10.1038/ni.1611. [DOI] [PubMed] [Google Scholar]
- Kalos M, et al. T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med. 2011;3:95ra73. doi: 10.1126/scitranslmed.3002842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanSeggelen H, et al. T Cells Engineered With Chimeric Antigen Receptors Targeting NKG2D Ligands Display Lethal Toxicity in Mice. Mol Ther. 2015;23:1600–1610. doi: 10.1038/mt.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]


