Abstract
Purpose
Thirty to fifty per cent of HIV patients develop HIV associated neurocognitive disorders (HAND) despite combined antiretroviral therapy. HIV-1 infected macrophages release viral and cellular proteins that induce neuronal degeneration and death. We hypothesize that changes in the macrophage secretome of HIV-1 seropositive patients with HAND may dissect proteins related to neurotoxicity.
Experimental Design
Monocyte-derived macrophages (MDM) were isolated from the peripheral blood of 12 HIV+ and 4 HIV− women characterized for neurocognitive function. Serum-free MDM supernatants were collected for protein isolation and quantification with iTRAQ® labeling. Protein identification was performed using a LTQ Orbitrap Velos mass spectrometer and validated in MDM supernatants and in plasma using ELISA.
Results
Three proteins were different between NC and ANI, 6 between NC and HAD, and 6 between NC and HAD. Among these, S100A9 was decreased in plasma from patients with ANI, and MMP9 was decreased in the plasma of all HIV+ patients regardless of cognitive status, and was significantly reduced in supernatant of MDM isolated from patients with ANI.
Conclusions and clinical relevance
S100A9 and MMP9 have been associated with inflammation and cognitive impairment, and therefore represent potential targets for HAND treatment.
Keywords: HAND, iTRAQ, MMP9, S100A9, secretome
Introduction
Perivascular macrophages and microglia are the main cells infected by HIV in the brain. HIV can enter the brain during the acute stage of infection and this can lead to the development of HIV-associated neurocognitive disorders (HAND). HAND remains a major neurological complication even in patients adherent to the use of combined antiretroviral therapy (cART). HAND affects up to 50% of HIV+ patients [1, 2]. Although the incidence of HIV-associated dementia has decreased dramatically after cART, the prevalence of milder form of neurocognitive impairment (asymptomatic neurocognitive disorders (ANI) and mild neurocognitive disorders (MND), has increased mainly because of extended life-span of HIV+ patients.
Current diagnosis of HAND relies mainly in neuropsychological tests. There is not a therapeutic regimen for early stages in cognitive impairment, and since screening methods are not enough to diagnose ANI in all clinical settings, we are in need of biomarkers to better diagnose early stages of HAND. Therefore, all the proteins involved in pathogenesis need to be identified for the development of new therapeutic strategies. Although several studies aimed to find biomarkers for the development of HAND in varied samples such as, HIV-infected monocyte-derived macrophages (MDM) supernatant [3], monocytes cells [4, 5], CSF [5–7], and sera [8, 9], this is the first study done with MDM supernatant (secretome) from HIV+ patients characterized for cognitive status using a quantitative proteomic method. We applied isobaric tags for relative and absolute quantitation (iTRAQ®), to determine the secretome of MDM isolated from HIV+ Hispanic women characterized for neurocognitive status in search for new biomarkers for the development of HAND and potential targets for therapy.
Materials and Methods
Patients
Patient data and samples for this study were collected from Hispanic/Latino Longitudinal Women cohort from 2003 to 2009. This study had the approval of the Institutional Review Board (UPR-MSC; IRB# 0720109), and the informed consent of all patients. Patients with other infectious diseases, hepatitis C or positive toxicology were excluded. The MSK modified to include neuropsycological test performance was used for to determine cognitive function [2], which categorized cognitive performance into normal cognition (NC) or MSK of 0, Asymptomatic (ANI) or MSK of 0.5, and HIV-associated dementia (HAD) or a MSK ≥ 1.
Cell cultures
Peripheral blood mononuclear cells (PBMCs) were isolated from patients characterized for HAND, by neuropsycological tests the day of blood collection, using a Lymphosep medium (MP Biomedicals, Solon, Ohio, USA). Monocyte-derived macrophages were selected by adherence after 7 days in culture. Adherent monocytes were grown in RPMI supplemented with 20% heat-inactivated FBS, 10% heat-inactivated human sera, and 1% Pen/Strep (all from Sigma Chemical Company, St. Louis, MO) in T25 flasks at a concentration of 1.5×106cells/ml. Half of the medium was changed every 3 days for all cultures. On day 6 cells were washed twice with PBS1X, then media was replaced with serum-free media, collected 24 hrs later and stored at −80°C for further proteomics analyses.
Sample preparation for proteomics analyses
Serum free supernatants were dialyzed using distilled water for 24hrs at 4°C to remove salts. Samples were dried and resuspended in lysis buffer, and then protein quantification was performed using BCA. To precipitate samples ethanol 200 proof, in at least 10 volumes was added to each sample and incubated at −20°C overnight. Samples were centrifuged twice at 1300xg for 15 minutes at 4°C and ethanol was decanted. Finally, the samples were dried to remove any trace of ethanol. In order to reduce the protein in the samples 20ul of dissolution buffer (AB Sciex iTRAQ® Kit) and 1ul of denaturant (AB Sciex iTRAQ® Kit, Framingham, MA) were added. Samples were vortexed rigorously and 2ul of the reducing reagent was added and incubated at 60°C for 1 hour. Then 1ul of cysteine blocking reagents were added to the samples and incubated at room temperature for 10min in the dark. Finally, samples were trypsin was added at a concentration of 1ug/mL, and incubated for 12 hours.
Labeling with iTRAQ®
iTRAQ® 4-plex labeling was performed according to manufacturer instructions (AB Sciex, Framinghan, MA). Labeling of samples was distributed as depicted in Table 1. Samples were dried and stored at −80°C.
Table 1.
Distribution of 4-plex iTRAQ® labels across samples.
| Isobaric tags for each sample | ||||
|---|---|---|---|---|
| 114 | 115 | 116 | 117 | |
| Pool #1 | HIV− (1)a) | NC (1) | ANI (1) | HAD (1) |
| Pool #2 | HAD (2) | HIV− (2) | NC (2) | ANI (2) |
| Pool #3 | ANI (3) | HAD (3) | HIV− (3) | NC (3) |
| Pool #4 | NC (4) | ANI (4) | HAD (4) | HIV− (4) |
Number in parenthesis indicates the patient ID for each classification for a total of 4 patients for each classification.
This design allows reducing technical and biological replicates. Every pool contains one sample from each group: one HIV− and one of each of the HIV+ cognition status classifications: NC, ANI, and HAD. Dye swapping was performed by assigning a different isobaric tag for each patient classification in the different pools.
Peptide Purification and Concentration
To perform the peptide purification and concentration, samples were reconstituted with 1ml of 2% formic acid at a pH<3. Water Oasis MCX cartridges (1CC 30mg Part No. 18600025) were equilibrated by carefully passing 50% methanol across the cartridge at a flow rate of 1 drop per second. On the second portion of this procedure all the washes were stored and the cartridge was washed with 1ml of wash solution (5% methanol, 0.1% formic acid in HPLC water). Column was washed with 100% methanol to remove any hydrophobic contaminants. Finally, to elute the bound peptides in the samples, 1 ml of elution buffer (50ul of NH4OH solution and 950ul of 100% methanol) was added. Each eluted peptide samples were placed in a speed vac for 6 to 8 hours and stored at −80°C.
Offgel Peptide Fractionation
Each Offgel sample was reconstituted with 0.1% formic acid. The IPG strips, frames and electrodes were assembled using Agilent 3100 Offgel fractionator manual. The electrode pads were hydrated with 95% mineral oil (Agilent Kit). The Offgel fractionator (Agilent Technologies) was programmed for the12-PE00 method (tray II) for 48hrs (at 24hrs the electropads were rehydrated to prevent overheating).
Sample Clean Up
To eliminate the impurities and the excess salts from the samples PepClean C-18 Spin Columns (Thermo Scientific) were used to optimize the quality of the samples. The columns were prepared by adding activation solution (50%Acetonitrile) and equilibration solution (0.5%Trifluoroacetic acid in 5% Acetonitrile) these steps were repeated twice on each sample and centrifuged at 1500xg for 1 minute. The samples were loaded to the top of the resin bed on a new 1.5 ml microtube and centrifuged as in the pervious steps. Thereafter samples were washed using a solution containing 0.5% Trifluoroacetic acid in 5% Acetonitrile, and subsequently eluted in 70% Acetonitrile and 0.1% Trifluoroacetic acid. Samples were centrifuged, dried in the speed vac for 12 hours and stored at −80°C. Samples were analyzed in an Orbitrap Velos (Thermo-Fisher).
iTRAQ® Protein Identification and Statistical Analysis
Protein identification was performed using a LTQ Orbitrap Velos mass spectrometer. iTRAQ labeled tryptic peptide fractions were dissolved in 40ul of 0.1% TFA, 5% Acetonitrile and 6ul was directly loaded onto a custom packed trap column (2cm × 100um C18). Peptides were then eluted and sprayed from a custom packed emitter (75uM × 25cm C18) with a linear gradient from 100% solvent A (0.1% formic acid in 5% Acetonitrile) to 35% solvent B (0.1% formic acid in Acetonitrile) in 120 minutes at a flow rate of 300 nanoliters per minute on a Proxeon Easy nanoLC system directly coupled to a LTQ Orbitrap Velos mass spectrometer. Data dependent acquisitions were set up according to an experiment where full MS scans from 350 -2000 m/z were acquired in the Orbitrap at a resolution of 30000 followed by 10 MS/MS scans acquired via HCD fragmentation at 7500 resolution in the Orbitrap. All samples were run in duplicate.
The raw data files were processed with Extract_ MSN (Thermo Scientific) into peak lists and then searched against the Human index of the SwissProt database (SwissProt_Human_09211) using the Mascot Search engine (Matrixsciences, Ltd., version 2.3.02). Parent mass tolerances were set to 10 ppm and fragment mass tolerances were set to 0.05 Da. The Fixed modifications were iTRAQ4plex (K), iTRAQ4plex (N-term), and Methylthio (C). Variable modifications were iTRAQ4plex (Y), Acetyl (protein N-term), Gln to pyro Glu (N-term Q), and oxidation (M). Mascot search results were loaded into the Scaffold viewer (Proteome Software, Inc., version 3.1) and filtered to an 80% probability for both peptide and protein identification. The iTRAQ® reporter ions were quantitated with the Proteo IQ software (Nusep, Inc., version 2.6.03) using a minimum of two peptides with a protein probability of 80%. The iTRAQ® is a relative quantitation proteomic method in which the abundance of proteins is the result of relative intensities to a reference sample. In this study the reference sample was HIV− patients. The abundance of proteins is presented as log2 ratios to HIV− patients. The ratios of all four categories (HIV−, NC, ANI, and HAD) were compared using pairwise comparison, and then the p- and t-values were obtained. The comparisons were NC vs ANI, NC vs HAD, and ANI vs HAD. All validation group samples followed a normal distribution; therefore One-way ANOVA and Tukey post-test were performed to calculate difference between groups.
Pathway and Network Analysis
All identified proteins were subjected to Ingenuity Pathway Analysis software (IPA; Ingenuity System, Redwood City, CA). The relationships of the proteins identified by iTRAQ® were analyzed for direct or indirect relationship using IPA. To achieve that all proteins were connected between them, the network was expanded to include upstream or downstream proteins with direct or indirect relationships.
Results
Patients
All patients used for this iTRAQ® study were under cART except for one patient (ANI), which had no treatment. No significant differences were observed in CD4 count, plasma viral load, CSF viral load between the patients used for iTRAQ® (Table 2) or between the patients used for validation (Supplementary Table 1). The CNS penetration effectiveness (CPE) was not different, regardless of neurocognitive status (Supplementary Table 1). CPE scores were determined using the, according to the established criteria [10, 11].
Table 2.
Viral Immune parameters of HIV positive patients used for iTRAQ®.
| Classification | n | a)CD4 cells/mm3 |
a)Plasma Viral load (Log10 copies/ml) |
a)CSF Viral Load (Log10 copies/ml) |
|---|---|---|---|---|
| NC | 4 | 515 (116) | 3 (1.4) | 2.5 (0.4) |
|
|
||||
| ANI | 4 | 405 (49) | 3 (1.5) | 2 (1.3) |
|
|
||||
| HAD | 4 | 521 (558) | 1.69 (0) | 1.69 (0) |
|
|
||||
Data is presented as the mean (SD)
iTRAQ® protein quantitation
When comparing the ratios of ANI against HAD; S100A9 (Calgranulin) was differentially expressed in all four pools; and A1BG (alpha-1-B glycoprotein), APOA4 (apolipoprotein A-IV), HBB (hemoglobin beta), MMP9 (metalloproteinase 9), and TGFB1 (transforming growth factor beta) proteins were differentially expressed in at least three pools (Table 3). When comparing ANI against NC; APOE (apolipoprotein E) and S100A9 were differentially expressed in all four pools, and MMP9 was differentially expressed in at least three pools (Table 2). When comparing in NC against HAD; A1BG and HBB were differentially expressed in all four pools; and IGHG2 (Ig gamma-2 chain C region), GSN (Gelsolin), S100A9, and MMP9 (Table 2). The complete list of proteins identified and quantified is presented in Supplementary Table 2. The ratios using HIV – patients as reference value for all proteins quantified for each of the 4 pools is presented in Supplementary Table 3.
Table 3.
Statistical analysis of the peptides found in the different patient groups. All values were normalized by the values of HIV− of each pool. Protein names in bold are the only that had p-values < 0.05 in all four pools, all other have only three out of four pools with significant p-values. T-values indicated the directionality of that change.
| Protein Name |
Accession Number |
Pool #1 | Pool #2 | Pool #3 | Pool #4 | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| p-value | t-value | p-value | t-value | p-value | t-value | p-value | t-value | ||
| ANI vs HAD | |||||||||
|
| |||||||||
| A1BG | P04217 | 0.0033 | 3.54 | 0.0057 | 3.26 | 0.0012 | −4.06 | 0.0884 | 1.84 |
| S100A9 | P06702 | <.0001 | −5.76 | 0.0113 | 3.29 | 0.0012 | −4.10 | <.0001 | 23.14 |
| APOA4 | P06727 | 0.0016 | 3.90 | 0.0112 | 3.12 | 0.0774 | −1.91 | 0.5027 | −0.69 |
| HBB | P68871 | 0.0012 | 4.04 | <.0001 | 9.70 | 0.1357 | −1.58 | 0.0044 | −3.52 |
| MMP9 | P14780 | 0.3223 | −1.03 | 0.0033 | −3.72 | <.0001 | −7.87 | <.0001 | 6.88 |
| TGFB1 | Q15582 | 0.2595 | −1.18 | 0.0025 | −3.73 | 0.0107 | −2.94 | 0.0009 | 4.18 |
|
| |||||||||
| ANI vs NC | |||||||||
|
| |||||||||
| APOE | P02649 | 0.0146 | −2.79 | 0.0176 | −2.75 | 0.0249 | −2.68 | 0.0011 | 4.49 |
| S100A9 | P06702 | <.0001 | −18.63 | 0.0009 | −4.44 | 0.0042 | −3.57 | <.0001 | 20.87 |
| MMP9 | P14780 | <.0001 | −7.02 | 0.0021 | −3.78 | 0.0002 | −5.86 | 0.8845 | 0.15 |
|
| |||||||||
| NC vs HAD | |||||||||
|
| |||||||||
| IGHG2 | P01859 | 0.0300 | −2.43 | 0.0432 | −2.30 | 0.0212 | 2.83 | 0.8324 | −0.22 |
| A1BG | P04217 | <.0001 | −5.89 | 0.0107 | −2.96 | <.0001 | 7.43 | 0.0192 | 2.65 |
| GSN | P06396 | 0.0012 | −4.07 | 0.1066 | −1.75 | 0.0280 | 2.58 | 0.0536 | −2.11 |
| S100A9 | P06702 | <.0001 | −16.99 | 0.0006 | −5.12 | 0.1152 | 1.72 | 0.0004 | −4.74 |
| MMP9 | P14780 | <.0001 | −5.96 | 0.5309 | −0.64 | 0.0015 | 4.45 | <.0001 | −6.15 |
| HBB | P68871 | 0.0054 | −3.29 | 0.0259 | −2.50 | 0.0005 | 5.39 | 0.0006 | 4.37 |
Network of proteins elucidated by IPA
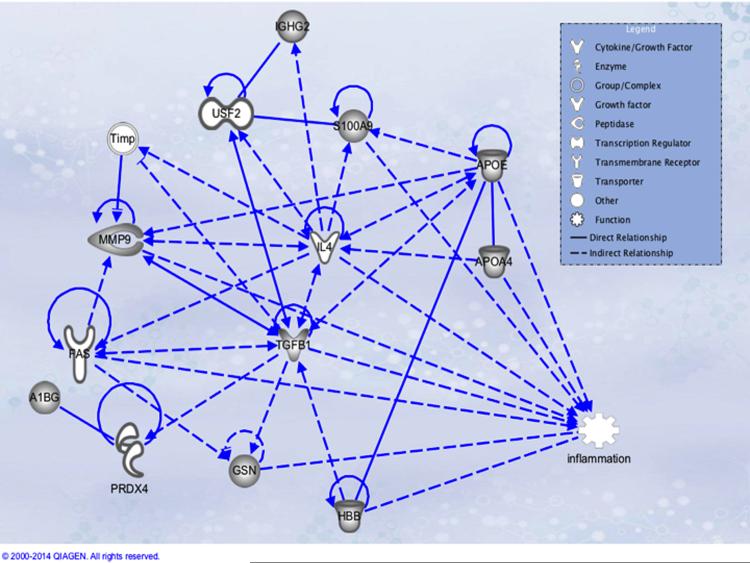
According to IPA analysis most of the proteins identified relate with inflammation, cell proliferation, and migration (Figure 1). TGFB1 seems to be a hub protein, it modulates MMP9 activity and expression [12–16], also it decreases expression of GSN mRNA and protein [17]. TGFB1 a increases APOE expression [18] and secretion [19]. Knockout of APOE is involved in the upregulation of S100A9 expression [20], and an increase in MMP9 activity and expression [21]. MMP9 expression is also increased after stimulation of FAS [22].
Figure 1. Ingenuity Pathway Analysis with all proteins found.
Molecules with gray filling are the proteins identified by iTRAQ®, all other were added by the IPA software beacause they have either a direct or indirect relationship with the found proteins. A1BG = alpha -1- beta glycoprotein; APOA4 = Apolipoprotein A IV; APOE = Apolipoprotein E; GSN = Gelsolin; HBB = Hemoblogin Beta; TGFB = Transforming Growth Factor Beta. S100A9 (Calgranulin B). IL4 = Interleukin-4; FAS = apoptosis antigen 1; PRDX4 = Peroxiredoxin-4; IGHG2 = Ig gamma-2 chain C region; Timp = Tissue inhibitor of metalloproteinases; USF2 = Upstream transcription factor 2.
Validation of proteins
Proteins found significantly different (p<0.05) in MDM secretome by iTRAQ® were analyzed from the MDM supernatants and plasma of patients with HAND using ELISA. Secretion of S100A9 in MDM did not differ between patients with different neurocognitive status (Figure 2a). However, S100A9 was found to be significantly decreased in plasma from HIV+ patients with ANI, when compared with patients with NC or HAD (Figure 2b). Secretion of MMP9 was found to be significantly reduced in supernatants of MDM isolated from patients with ANI (Figure 3a), and expression of MMP9 was decreased in plasma from all patients with HIV+, independently of cognitive status (Figure 3a).
Figure 2. Validation of S100A9 in MDM supernatants and plasma of patients with HAND.
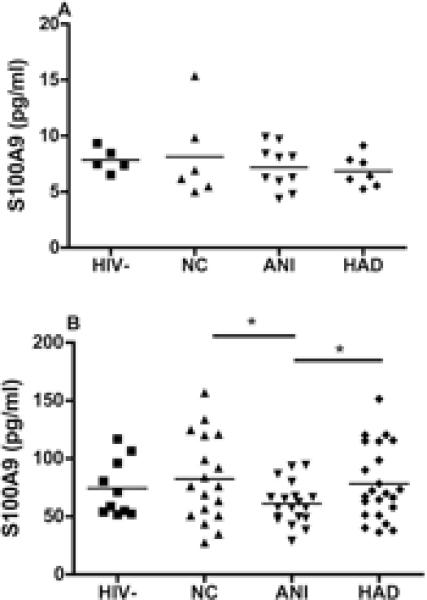
Concentration of S100A9 was determined by sandwich ELISA (BioVendor, Asheville, NC). (A) PBMCs were isolated from HIV+ Hispanic women characterized for cognitive function, and then macrophages were selected by adherence. Supernatant were collected after 6 days of macrophage differentiation. Macrophage supernatants were from HIV− donors (n=5) and HIV+ patients characterized for neurocognitive status of normal cognition (NC, n=6), asymptomatic neurocognitive impairment (ANI, n=10), and HIV-associated dementia (HAD, n=7) were analyzed for S100A9 expression. (B) Plasma from HIV− donors (n=10) and HIV+ patients characterized for neurocognitive status of NC (n=10), ANI (n=19), and HAD (n=23) were analyzed for S100A9 expression. Data were analyzed by one-way ANOVA and Tukey post-test. *p<0.05.
Figure 3. Validation of MMP9 in MDM supernatants and plasma of patients with HAND.
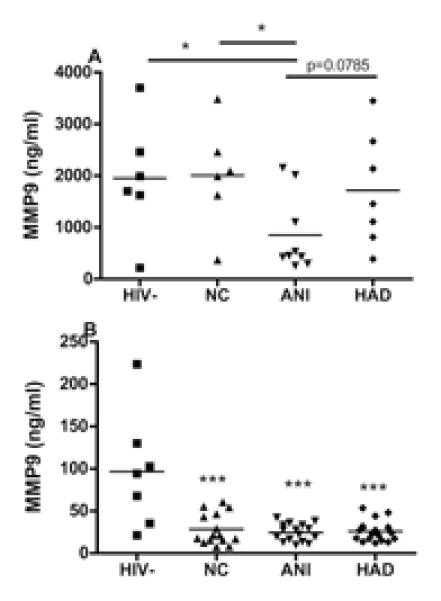
Concentration of MMP9 was determined by sandwich ELISA (R&D, Minneapolis, MN). (A) PBMCs were isolated from HIV+ Hispanic women characterized for cognitive function. Supernatant were collected after 6 days of macrophage differentiation. Macrophage supernatants were from HIV− donors (n=5) and HIV+ patients characterized for neurocognitive status of normal cognition (NC, n=6), asymptomatic neurocognitive impairment (ANI, n=9), and HIV-associated dementia (HAD, n=7) were analyzed for MMP9 expression. Macrophages supernatant from ANI had lower levels of MMP9. (B) Plasma from HIV− donors (n=10) and HIV+ patients characterized for neurocognitive status of NC (n=18), ANI (n=19), and HAD (n=23) were analyzed for MMP9 expression. Data were analyzed by one-way ANOVA and Tukey post-test. HIV+ patients, regardless of cognitive status, had lower levels of MMP9 when compared to HIV− donors. *p<0.05; *** p<0.001.
Discussion
In this study, we analyzed the macrophage secretome from Hispanic women characterized for cognitive function in search for biomarkers that could be used for diagnosis and treatment of early stages of HAND. Using iTRAQ® and tandem mass spectrometry, we found that S100A9 and MMP9 were differentially expressed in several HAND stages (NC, ANI and HAD). These proteins were validated in plasma and in supernatant from MDM isolated from HIV+ patients. S100A9 is also known as myeloid inhibitory factor-related protein 14 (MRP-14) or calgranulin B. This protein is constitutively expressed in macrophages and has been related with acute and chronic inflammation [23]. A S100A9 knockout decreases memory impairment in a Tg2576 mice model [24]. Previously, we identified by mass spectrometry that S100A9 and its partner S100A8 as present in HIV-infected MDM and placental macrophages, and absent in uninfected cells. Although S100A8 has been associated with increasing HIV replication in cervico-vaginal secretion [25], no validation was done for this protein since the scope of that project was to compare proteins differentially expressed in placental macrophages and MDM [26].
In the present study, S100A9 was significantly decreased in the plasma of patients with ANI when compared to NC and HAD patients, but we did not found differences in MDM supernatants. This could suggest that other blood cells besides macrophages, including other myeloid cells or platelets, could be secreting S100A9 into the plasma, or that number of samples were small to reach for significance.
MMP9 degrades extracellular matrix proteins and is involved in many processes including inflammation [27–29]. Overexpression of MMP9 prevents cognitive impairment seen in 5xRAD mice, an Alzheimer’s disease model [30]. It has been observed that injection into the caudate putamen of mice gp120 alone is able to increase the expression of MMP2 and MMP9 and its inhibitors, TIMP-1 and -2 in the region [31]. Interestingly antiretroviral therapy was demonstrated to inhibit secretion of MMP2 and MMP9 in activated astrocytes and microglia [32], and in patient’s PBMCs [33]. MMP9 expression is also enhanced in astrocytes treated with gp120, and this expression is exacerbated if Nrf2 siRNA is added to cells [34]. Nrf2 is a transcription factor that regulates the expression of proteins involved in antioxidant response. MMP9 was found to be downregulated in plasma of all HIV+ patients, regardless of neurocognitive status (Figure 3b). As for the MDM supernatant, we found that MMP9 was significantly reduced in ANI patients (Figure 3a). We tested MMP9 in CSF from HIV+ patients but it resulted in undetectable levels. Our results agree with another study in which MMP9 secretion was reduced after HIV infection of MDM [35].
To our knowledge, this is the first study that determined the secretome of MDM isolated from HIV+ patients characterized for cognitive status. Our study is also unique because it used samples from a cohort that is composed of only women followed longitudinally.
Blood samples were collected for MDM isolation on the day of evaluation for their cognitive status, therefore the amount of MDM cultures obtained for each category (HIV−, NC, ANI, and HAD) were limited. In our study, MDM were isolated in absence of growth factors such as M-CSF or GM-CSF that are used in several in vitro studies [36]. These growth factors can switch the macrophage into M1 (GM-CSF) or M2 (M-CSF) phenotypes. Therefore, the samples analyzed in this iTRAQ® study reflect the macrophage intrinsic phenotype.
iTRAQ® is a quantitative proteomic technique that has its drawbacks, among them, the quantitation of proteins is based on the relative abundance against a reference sample and therefore is not an absolute quantification. It is also an expensive technique and this may limit the number of samples to analyze. After identifying the proteins, we noticed that some of the proteins identified were originated from the serum, indicating that one or two additional washings with PBS prior of adding serum-free media could improve the removal of serum-derived proteins. However these additional washings may also result in loss of cells due to detachment.
All these results suggest that S100A9 and MMP9 may represent a new important biomarker for early stages of HAND, since S100A9 and MMP9 lower levels in plasma and MDM supernatant, respectively, was found in patients with ANI. Further studies in brain tissue, animal models, and patients are required to further understand their mechanisms of action, its potential for therapy, and if they serve as possible biomarkers for the development of HAND.
Supplementary Material
Statement of clinical relevance.
The prevalence of HIV-associated neurocognitive disorders (HAND) has increased mainly because the HIV-infected population is living longer, and therefore aging also comes into play. The more common forms of HAND are the milder forms, mostly asymptomatic neurocognitive disorders (ANI). The diagnosis of ANI is mainly through neuropsychological tests, not available in all clinical settings. Hence, the establishment of biomarkers is of great need for the early detection of ANI and for effective treatment. The potential clinical significance of this study relies on the identification of possible candidate biomarkers for the development of HAND. This study is important as the field lacks biomarkers to reliably identify HIV-seropositive patients that will eventually develop HAND. Additionally the proteins identified could serve as potential targets for more effective treatment of HIV-infected patients.
Acknowledgments
This work was supported in part by NIH grants R01-MH08316-01, RCMI Translational Proteomics Center NIMHH (8G12-MD007600), SNRP-NINDS-1-U54NS43011, IDeA Networks of Biomedical Research Excellence (INBRE) P20RR016470, NeuroID-1R25GM097635, RISE R25GM061838 and CRC-NCRR-P20RR11126. We appreciate the contribution of Dr. Pawel Ciborowski in the implementation of iTRAQ® at the Translational Proteomics Center.
Abbreviations
- HAND
HIV-associated neurocognitive disorders
- NC
normal cognition
- ANI
asymptomatic neurocognitive disorders
- HAD
HIV-associated dementia
- PBMC
perivascular mononuclear cells
- MDM
monocyte-derived macrophages
- CSF
cerebral spinal fluid
- CPE
CSF penetration index
Footnotes
Conflict of interests
Authors declare that they have no financial or commercial conflict of interest.
References
- 1.Heaton RK, Clifford DB, Franklin DR, Woods SP, Ake C, Vaida F, Ellis RJ, Letendre SL, Marcotte TD, Atkinson JH, Rivera-Mindt M, Vigil OR, Taylor MJ, Collier a C, Marra CM, Gelman BB, McArthur JC, Morgello S, Simpson DM, McCutchan J a, Abramson I, Gamst a, Fennema-Notestine C, Jernigan TL, Wong J, Grant I. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology. 2010;75:2087–96. doi: 10.1212/WNL.0b013e318200d727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wojna V, Skolasky RL, Hechavarría R, Mayo R, Selnes O, McArthur JC, Meléndez LM, Maldonado E, Zorrilla CD, García H, Kraiselburd E, Nath A. Prevalence of human immunodeficiency virus-associated cognitive impairment in a group of Hispanic women at risk for neurological impairment. Journal of neurovirology. 2006;12:356–64. doi: 10.1080/13550280600964576. [DOI] [PubMed] [Google Scholar]
- 3.Ciborowski P, Kadiu I, Rozek W, Smith L, Bernhardt K, Fladseth M, Ricardo-Dukelow M, Gendelman HE. Investigating the human immunodeficiency virus type 1-infected monocyte-derived macrophage secretome. Virology. 2007;363:198–209. doi: 10.1016/j.virol.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kraft-Terry S, Gerena Y, Wojna V, Plaud-Valentin M, Rodriguez Y, Ciborowski P, Mayo R, Skolasky R, Howard E, Meléndez LM, Gendelman HE. Proteomic analyses of monocytes obtained from Hispanic women with HIV-associated dementia show depressed antioxidants. Proteomics Clinical applications. 2010;4:706–14. doi: 10.1002/prca.201000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velázquez I, Plaud M, Wojna V, Skolasky R, Laspiur JP, Meléndez LM. Antioxidant enzyme dysfunction in monocytes and CSF of Hispanic women with HIV-associated cognitive impairment. Journal of neuroimmunology. 2009;206:106–11. doi: 10.1016/j.jneuroim.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laspiur JP, Anderson ER, Ciborowski P, Wojna V, Rozek W, Duan F, Mayo R, Rodríguez E, Plaud-Valentín M, Rodríguez-Orengo J, Gendelman HE, Meléndez LM. CSF proteomic fingerprints for HIV-associated cognitive impairment. Journal of neuroimmunology. 2007;192:157–70. doi: 10.1016/j.jneuroim.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rozek W, Ricardo-Dukelow M, Holloway S, Gendelman HE, Wojna V, Melendez LM, Ciborowski P. Cerebrospinal fluid proteomic profiling of HIV-1-infected patients with cognitive impairment. Journal of proteome research. 2007;6:4189–99. doi: 10.1021/pr070220c. [DOI] [PubMed] [Google Scholar]
- 8.Rozek W, Horning J, Anderson J, Ciborowski P. Sera proteomic biomarker profiling in HIV-1 infected subjects with cognitive impairment. Proteomics Clinical applications. 2008;2:1498–507. doi: 10.1002/prca.200780114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiederin J, Rozek W, Duan F, Ciborowski P. Biomarkers of HIV-1 associated dementia: proteomic investigation of sera. Proteome science. 2009;7:8. doi: 10.1186/1477-5956-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Letendre S, Marquie-Beck J, Capparelli E, Best B, Clifford D, Collier AC, Gelman BB, McArthur JC, McCutchan JA, Morgello S, Simpson D, Grant I, Ellis RJ. Validation of the CNS Penetration-Effectiveness rank for quantifying antiretroviral penetration into the central nervous system. Archives of neurology. 2008;65:65–70. doi: 10.1001/archneurol.2007.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smurzynski M, Wu K, Letendre S, Robertson K, Bosch RJ, Clifford DB, Evans S, Collier AC, Taylor M, Ellis R. Effects of central nervous system antiretroviral penetration on cognitive functioning in the ALLRT cohort. AIDS (London, England) 2011;25:357–65. doi: 10.1097/QAD.0b013e32834171f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Yang K, Mao Q, Zheng X, Kong D, Xie L. Inhibition of TGF-beta receptor I by siRNA suppresses the motility and invasiveness of T24 bladder cancer cells via modulation of integrins and matrix metalloproteinase. International urology and nephrology. 2010;42:315–23. doi: 10.1007/s11255-009-9620-3. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen J, Knapnougel P, Lesavre P, Bauvois B. Inhibition of matrix metalloproteinase-9 by interferons and TGF-beta1 through distinct signalings accounts for reduced monocyte invasiveness. FEBS letters. 2005;579:5487–93. doi: 10.1016/j.febslet.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 14.Ogawa K, Chen F, Kuang C, Chen Y. Suppression of matrix metalloproteinase-9 transcription by transforming growth factor-beta is mediated by a nuclear factor-kappaB site. The Biochemical journal. 2004;381:413–22. doi: 10.1042/BJ20040058. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W, Wang X, Chun J, Vilaysane A, Clark S, French G, Bracey NA, Trpkov K, Bonni S, Duff HJ, Beck PL, Muruve DA. Inflammasome-independent NLRP3 augments TGF-β signaling in kidney epithelium. Journal of immunology (Baltimore, Md : 1950) 2013;190:1239–49. doi: 10.4049/jimmunol.1201959. [DOI] [PubMed] [Google Scholar]
- 16.Sehgal I, Thompson TC. Novel regulation of type IV collagenase (matrix metalloproteinase-9 and -2) activities by transforming growth factor-beta1 in human prostate cancer cell lines. Molecular biology of the cell. 1999;10:407–16. doi: 10.1091/mbc.10.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Park JS, Chu JSF, Krakowski A, Luo K, Chen DJ, Li S. Proteomic profiling of bone marrow mesenchymal stem cells upon transforming growth factor beta1 stimulation. The Journal of biological chemistry. 2004;279:43725–34. doi: 10.1074/jbc.M407368200. [DOI] [PubMed] [Google Scholar]
- 18.Roes J, Choi BK, Cazac BB. Redirection of B cell responsiveness by transforming growth factor beta receptor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7241–6. doi: 10.1073/pnas.0731875100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter CJ. Convergence of genes implicated in Alzheimer’s disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochemistry international. 2007;50:12–38. doi: 10.1016/j.neuint.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Farris SD, Hu JH, Krishnan R, Emery I, Chu T, Du L, Kremen M, Dichek HL, Gold E, Ramsey SA, Dichek DA. Mechanisms of urokinase plasminogen activator (uPA)-mediated atherosclerosis: role of the uPA receptor and S100A8/A9 proteins. The Journal of biological chemistry. 2011;286:22665–77. doi: 10.1074/jbc.M110.202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemdahl A-L, Gabrielsen A, Zhu C, Eriksson P, Hedin U, Kastrup J, Thorén P, Hansson GK. Expression of neutrophil gelatinase-associated lipocalin in atherosclerosis and myocardial infarction. Arteriosclerosis, thrombosis, and vascular biology. 2006;26:136–42. doi: 10.1161/01.ATV.0000193567.88685.f4. [DOI] [PubMed] [Google Scholar]
- 22.Palao G, Santiago B, Galindo MA, Rullas JN, Alcamí J, Ramirez JC, Pablos JL. Fas activation of a proinflammatory program in rheumatoid synoviocytes and its regulation by FLIP and caspase 8 signaling. Arthritis and rheumatism. 2006;54:1473–81. doi: 10.1002/art.21768. [DOI] [PubMed] [Google Scholar]
- 23.Foell D, Frosch M, Sorg C, Roth J. Phagocyte-specific calcium-binding S100 proteins as clinical laboratory markers of inflammation. Clinica chimica acta; international journal of clinical chemistry. 2004;344:37–51. doi: 10.1016/j.cccn.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, Chang K-A, Ha T-Y, Kim J, Ha S, Shin K-Y, Moon C, Nacken W, Kim H-S, Suh Y-H. S100A9 knockout decreases the memory impairment and neuropathology in crossbreed mice of Tg2576 and S100A9 knockout mice model. PloS one. 2014;9:e88924. doi: 10.1371/journal.pone.0088924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashemi FB, Mollenhauer J, Madsen LD, Sha BE, Nacken W, Moyer MB, Sorg C, Spear GT. Myeloid-related protein (MRP)-8 from cervico-vaginal secretions activates HIV replication. AIDS (London, England) 2001;15:441–9. doi: 10.1097/00002030-200103090-00002. [DOI] [PubMed] [Google Scholar]
- 26.Luciano-Montalvo C, Ciborowski P, Duan F, Gendelman HE, Meléndez LM. Proteomic analyses associate cystatin B with restricted HIV-1 replication in placental macrophages. Placenta. 2008;29:1016–23. doi: 10.1016/j.placenta.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leifler KS, Svensson S, Abrahamsson A, Bendrik C, Robertson J, Gauldie J, Olsson A-K, Dabrosin C. Inflammation induced by MMP-9 enhances tumor regression of experimental breast cancer. Journal of immunology (Baltimore, Md : 1950) 2013;190:4420–30. doi: 10.4049/jimmunol.1202610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks WC, Wilson CL, López-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nature reviews Immunology. 2004;4:617–29. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 29.Moore BA, Manthey CL, Johnson DL, Bauer AJ. Matrix metalloproteinase-9 inhibition reduces inflammation and improves motility in murine models of postoperative ileus. Gastroenterology. 2011;141:1283–92. doi: 10.1053/j.gastro.2011.06.035. 1292.e1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fragkouli A, Tsilibary EC, Tzinia AK. Neuroprotective role of MMP-9 overexpression in the brain of Alzheimer’s 5xFAD mice. Neurobiology of disease. 2014;70:179–89. doi: 10.1016/j.nbd.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 31.Louboutin J-P, Reyes B a S, Agrawal L, Van Bockstaele EJ, Strayer DS. HIV-1 gp120 upregulates matrix metalloproteinases and their inhibitors in a rat model of HIV encephalopathy. The European journal of neuroscience. 2011;34:2015–23. doi: 10.1111/j.1460-9568.2011.07908.x. [DOI] [PubMed] [Google Scholar]
- 32.Liuzzi GM, Mastroianni CM, Latronico T, Mengoni F, Fasano A, Lichtner M, Vullo V, Riccio P. Anti-HIV drugs decrease the expression of matrix metalloproteinases in astrocytes and microglia. Brain : a journal of neurology. 2004;127:398–407. doi: 10.1093/brain/awh049. Pt 2. [DOI] [PubMed] [Google Scholar]
- 33.Latronico T, Liuzzi GM, Riccio P, Lichtner M, Mengoni F, D’Agostino C, Vullo V, Mastroianni CM. Antiretroviral therapy inhibits matrix metalloproteinase-9 from blood mononuclear cells of HIV-infected patients. AIDS (London, England) 2007;21:677–84. doi: 10.1097/QAD.0b013e328018751d. [DOI] [PubMed] [Google Scholar]
- 34.Reddy PVB, Agudelo M, Atluri VSR, Nair MP. Inhibition of nuclear factor erythroid 2-related factor 2 exacerbates HIV-1 gp120-induced oxidative and inflammatory response: role in HIV associated neurocognitive disorder. Neurochemical research. 2012;37:1697–706. doi: 10.1007/s11064-012-0779-0. [DOI] [PubMed] [Google Scholar]
- 35.Ciborowski P, Enose Y, Mack A, Fladseth M, Gendelman HE. Diminished matrix metalloproteinase 9 secretion in human immunodeficiency virus-infected mononuclear phagocytes: modulation of innate immunity and implications for neurological disease. Journal of neuroimmunology. 2004;157:11–6. doi: 10.1016/j.jneuroim.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 36.Lacey DC, Achuthan a, Fleetwood a J, Dinh H, Roiniotis J, Scholz GM, Chang MW, Beckman SK, Cook a D, Hamilton J a. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188:5752–5765. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.