Abstract
Improvement in activation of Rubisco by Rubisco activase can potentially enhance CO2 assimilation and photosynthetic efficiency in plants. The three homoeologous copies of TaRca2-α were identified on chromosomes 4AL, 4BS and 4DS (TaRca2-α-4AL, TaRca2-α-4BS, and TaRca2-α-4DS) in bread wheat. Expression patterns of the three copies at heading (Z55), anthesis (Z67) and grain-filling (Z73) stages were investigated through qRT-PCR analyses in a panel of 59 bread wheat genotypes and their effects on net photosynthesis rate (Pn), biomass plant-1 (BMPP) and grain yield plant-1 (GYPP) were further explored. Different but similar expression patterns were observed for the three copies of TaRca2-α at the three growth stages with highest expression at grain-filling stage. TaRca2-α-4BS expressed higher at the three stages than TaRca2-α-4AL and TaRca2-α-4DS. The 59 genotypes could be clustered into three groups as high (7 genotypes), intermediate (41 genotypes) and low (11 genotypes) expression based on the expression of the three copies of TaRca2-α at three growth stages. Significant variations (P<0.01) were observed among the three groups of bread wheat genotypes for Pn, BMPP and GYPP. Generally, the genotypes with higher TaRca2-α expression also showed higher values for Pn, BMPP and GYPP. The expressions of the three copies of TaRca2-α at heading, anthesis and grain-filling stages were positively correlated with Pn, BMPP and GYPP (P<0.01) with stronger association for TaRca2-α-4BS at grain-filling stage. These results revealed that the expression of TaRca2-α contribute substantially to Pn, BMPP and GYPP, and suggested that manipulating TaRca-α expression may efficiently improve Pn, BMPP and GYPP in bread wheat and detecting TaRca-α expression levels with emphasis on TaRca2-α-4BS may be a positive strategy for selection in improving photosynthetic efficiency and grain yield of bread wheat.
Introduction
Wheat is the cereal of choice globally and is a source of about one-fifth of the total calories consumed by the world’s population [1], and is planted over 220 Mha of land throughout the world [2]. Although, Green revolution technologies have helped to a reasonable extent to enhance overall wheat productivity [3], meeting the demand of the fast-growing global population is a challenging task [4]. In order to feed the future population, emphasis needs to be concentrated on key traits related to plant productivity in the context of prevailing environmental conditions instead of solely relying on conventional practices. Photosynthesis being the basic constituent part of plant productivity can be efficiently manipulated to improve the overall productivity of wheat crop. Furthermore, positive relationship of photosynthesis with yield [5] makes it a desirable trait to be selected and manipulated for the enhancement of wheat yield potential.
For efficient photosynthesis to occur, the central role is played by the enzyme Rubisco (Ribulose-1,5-bisphosphate carboxylase/oxygenase), which is capable of catalyzing net fixation of inorganic carbon into organic molecules [6]. In spite of Rubisco’s enormous presence on earth, it has a lower efficiency due to slow catalytic turn-over rate [7]; consequently larger quantities of the enzyme are needed to ensure optimal photosynthesis. Furthermore, some natural inhibitory sugar phosphates attach to the active sites of Rubisco, rendering it inactive and hence incapable of fixing CO2 [8]. This also results in extra investments in nitrogen with environmental implications.
Rubisco needs to be converted from an inactive to catalytically active state for the efficient catalysis of photosynthetic assimilation of inorganic CO2 into organic compounds, and Rubsico activase (Rca) is the enzyme facilitating this conversion [9, 10]. Rca belongs to an extended AAA+ superfamily of ATPases, which is involved in various cellular activities [11] and regulates Rubisco’s activity by removing inhibitory sugar phosphates from Rubisco active sites using energy from ATP hydrolysis [12, 13]. Resultantly, the active sites of Rubisco are spontaneously carbamylated by CO2 and normal photosynthesis is supported. In most of plants, there are two isoforms of Rca, i.e. a large α isoform and a small β isoform with differences at the carboxy terminus [12] and also differing in maximal activity [14, 15] with Rca-α showing higher expression under most of the growth conditions [16]. The importance of Rca-α is also evident from the results of previous studies in different crops due its positive effect on plant productivity traits [16, 17] and therefore can also identify genotypes with improved phenotype under prevailing crop growth conditions. The success of crop plants depends on their final performance in the field where plants experience unpredictable changes in environmental conditions e.g. fluctuating irradiance, water deficit etc. Most studies concerning the effect of Rca on plant phenotype in wheat are conducted under preset conditions. Therefore, understanding the effect of Rca-α on wheat photosynthesis, biomass and grain yield under natural field conditions may provide a better strategy for improving overall productivity. Furthermore, it may also facilitate to detect the existing genetic variability among different wheat genotypes for photosynthesis, biomass and yield rated traits.
Being hexaploid, wheat has a very complex genome, each individual gene is potentially present in triplicate (A, B and D), and each homoeologue may express differentially and affect the phenotype in different manner. Investigation on the difference among the three copies not only helps in understanding the effect of a specific copy of a gene, but also reveals sequence diversity and facilitates to develop gene-based functional markers for marker-assisted breeding [18].
Based on the potential role of TaRca2-α on wheat photosynthesis and therefore grain yield especially under fluctuating environmental conditions, the present study was designed to investigate the expression patterns of TaRca2-α in flag leaves at three main growth stages (heading, anthesis and mid grain-filling) of bread wheat in a panel of 59 bread wheat genotypes grown under natural field conditions, and to test whether the expression levels of TaRca2-α in flag leaves associated with Pn, and BMPP and GYPP, and the contributions of the three individual copies of TaRca2-α.
Materials and Methods
Plant material and sowing
A panel of 59 winter wheat genotypes from two major wheat growing regions of China was used in the present study (Table 1), among those, 29 genotypes each were from the Northern and Huang-Huai Winter Wheat Regions, respectively and one genotype from Southwestern Winter Wheat Region. The genotypes were sown during 2013–14 and 2014–15 crop seasons on the experimental field at the Northwest A&F University, Yangling, Shaanxi, China (N 34°10′, E 108°10′, 526 m elevation). The experiment was laid-out in Randomized Complete Block Design with two replications. Each genotype was planted in 3 rows of 2 m length with row-to-row and plant-to-plant distance of 25 cm and 6.7 cm, respectively. All genotypes were sown under natural field conditions solely dependent on the soil moisture and the natural rainfall in season.
Table 1. Details of the genotypes used in the current study.
| Code | Name | Origin | Region | Code | Name | Origin | Region |
|---|---|---|---|---|---|---|---|
| 1 | Luohan 2 | Henan | HHWWR | 31 | Aifeng 3 | Shaanxi | HHWWR |
| 2 | Shijiazhuang 8 | Hebei | NWWR | 32 | Bainong 160 | Henan | HHWWR |
| 3 | Jinmai 47 | Shanxi | NWWR | 33 | Shaanhan 187 | Shaanxi | HHWWR |
| 4 | Linhan51329 | Shanxi | NWWR | 34 | Shijiazhuang 54 | Hebei | NWWR |
| 5 | Shaan 229 | Shaanxi | HHWWR | 35 | Luomai 21 | Henan | HHWWR |
| 6 | Xiaoyan 6 | Shaanxi | HHWWR | 36 | Lunxuan 061 | Beijing | NWWR |
| 7 | Pubing 143 | Shaanxi | HHWWR | 37 | Luo 9908 | Henan | HHWWR |
| 8 | Zhonghan 110 | Beijing | NWWR | 38 | Heng95Guan26 | Hebei | NWWR |
| 9 | Liken 2 | Shaanxi | HHWWR | 39 | Jinmai 33 | Shanxi | NWWR |
| 10 | Changwu135 | Shaanxi | HHWWR | 40 | Kedong 81 | Beijing | NWWR |
| 11 | Linfen 10 | Shanxi | NWWR | 41 | Shaanken 81 | Shaanxi | HHWWR |
| 12 | Luohan 3 | Henan | HHWWR | 42 | Han 6172 | Hebi | NWWR |
| 13 | Linhan536 | Shanxi | NWWR | 43 | Huaimai 21 | Jiangsu | HHWWR |
| 14 | Jing 411 | Beijing | NWWR | 44 | Yunong 982 | Henan | HHWWR |
| 15 | Tongmai 3 | Shaanxi | HHWWR | 45 | Xifeng 20 | Gansu | HHWWR |
| 16 | Mianyang 11 | Sichuan | SWWWR | 46 | Lunxuan 715 | Beijing | NWWR |
| 17 | Xinyuan 958 | Henan | HHWWR | 47 | Nongda 198 | Beijing | NWWR |
| 18 | Linfen 10 | Shanxi | NWWR | 48 | Fengkang 5 | Beijing | NWWR |
| 19 | Taishan 5 | Shandong | NWWR | 49 | Luohan 6 | Henan | HHWWR |
| 20 | Jining 18 | Shandong | NWWR | 50 | Jingwang 9 | Beijing | NWWR |
| 21 | Xinmai 13 | Henan | HHWWR | 51 | Jingdong 1 | Beijing | NWWR |
| 22 | Youmai 2 | Shandong | NWWR | 52 | Jinmai 21 | Shanxi | NWWR |
| 23 | Xinmai 18 | Henan | HHWWR | 53 | Jimai 23 | Hebei | NWWR |
| 24 | Xinong 2000–7 | Shaanxi | HHWWR | 54 | Jinan 18 | Shandong | NWWR |
| 25 | Shaanmai 150 | Shaanxi | HHWWR | 55 | Hanxuan 1 | Shanxi | NWWR |
| 26 | Zhoumai 16 | Henan | HHWWR | 56 | Lumai 1 | Shandong | NWWR |
| 27 | Yuanfeng 139 | Shaanxi | HHWWR | 57 | Wenmai 6 | Henan | HHWWR |
| 28 | Fengchan 3 | Shaanxi | HHWWR | 58 | Yunhan 618 | Shanxi | NWWR |
| 29 | Xinong 979 | Shaanxi | HHWWR | 59 | Hanxuan 10 | Shanxi | NWWR |
| 30 | Zhongyu 8 | Henan | HHWWR |
Note: HHWWR: Huang-huai Winter Wheat Region; NWWR: Northern Winter Wheat Region; SWWWR: Southwestern Winter Wheat Region.
Identification and sequence analysis of TaRca2-α
DNA sequence of wheat Rubisco activase (TaRca2-α, accession No. LM992845) was used to search the homoeologous copies through BLAST against wheat chromosome sequence survey database (http://wheatgenome.org). The exon/intron distribution of the three copies was predicted using spidey tool in NCBI database (http://ncbi.nlm.nig.gov/spidey/). The predicted amino acid sequences of the three copies were fruther determined through ExPASy (http://web.expasy.org/translate/). The conserved domains were analyzed using Conserved Domain Search of NCBI database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
Primer designing for expression analysis of TaRca2-α by qRT-PCR
Multiple alignments of the 3′ untranslated regions (UTRs) at the C-terminal extension on the three copies of TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS were carried-out using ClustalW program in Bioedit 7.0 [19]. Primer pairs were hand-picked based on sequence polymorphisms at the 3′ ends of their forward and reverse sequences. General properties of primers picked were further checked using PrimerPREMIER version 5.0 (PREMIER Biosoft International). The genome-specificities of the primer pairs were tested with RT-PCR using RNA from three nulli-tetrasomic (NT) lines (N4AT4B, N4BT4D and N4DT4A) of Chinese Spring. Three reference genes (TaActin, TaSand and TaCell) were used for background standardization in wheat [20]. The details of the primers used are given in Table 2.
Table 2. Primer pairs used for qRT-PCR analysis in 59 bread wheat genotypes.
| Name of primer | Primer sequence (5′ - 3′) | Usage |
|---|---|---|
| TaRca2- α _AF | GGTGTCTGCAAGGGTATCTTC | TaRca2-α-4AL |
| TaRca2- α _AR | TCGACTGTCATCTTTGGCTG | |
| TaRca2- α _BF | ACGCCGACCAACTTCCTT | TaRca2-α-4BS |
| TaRca2- α _BR | CAAGACCCTTCCACTTGTCC | |
| TaRca2- α _DF | GACGAGAAGAGGAACACC | TaRca2-α-4DS |
| TaRca2- α _DR | TGGCTGACGTACTCGTAT | |
| TaActin_F | TTGCTGACCGTATGAGCAAG | Reference gene TaActin |
| TaActin_R | ACCCTCCAATCCAGACACTG | |
| TaSand_F | TGCCTTGCCCATAAGAAATC | Reference gene TaSand |
| TaSand_R | GTGCGGACCAGTTGCTTTAT | |
| TaCell_F | GAGGAGGATGAGGTGGATGA | Reference gene TaCell |
| TaCell_R | CCTGGTACTTGCGGATGTCT |
Total RNA isolation and cDNA synthesis
Fully expanded flag leaf samples of five randomly chosen plants from each of the 59 experimental genotypes were taken and pooled together, respectively at heading (Z55), anthesis (Z65) and grain-filling (Z73) stages. A three-step RNA extraction was carried out using modified hot phenol method [21, 22]. Initial extraction was carried out in 1 mL (80°C) 1:1 Phenol/Extraction buffer (0.1 M Tris-HCL, pH 8.0, 0.1 M LiCl, 1% (w/v) SDS and 10 mM EDTA). Afterwards, two phenol/chloroform/IAA (25:24:1) extractions were conducted. Genomic DNA contamination was removed with DNaseI (TAKARA, Dalian) treatment according to the manufacturer’s instruction. The first strand cDNA was synthesized from 10 μg of total RNA from each template with PrimScriptIII RT-PCR kit (TAKARA, Dalian) using oligo (dT)18 primer according to the manufacturer’s instructions. The cDNA samples were stored at -20°C for subsequent analysis.
Expression analysis by qRT-PCR
cDNA sample from each genotype was replicated three times as per specifications of the SYBER Premix ExTaq Kit (Takara, Dalian), qRT-PCR were conducted using ABI7300 real time PCR system (Applied Bio Systems, USA). The reaction mixture was consisted of a total volume of 20μl including 10μl 2X SYMBER MIX, 0.3μl of each of the forward and reverse primer (0.6 μM), 1.5μl template cDNA (100 ng), ddH2O was added to get the final volume of 20μl. The qRT-PCR reaction was programmed as initial denaturation at 95°C for 20s, followed by 40 cycles at 95°C for 5s, 60°C for 30s. Relative expression of the target gene was calculated as under:
Where NE is the relative expression of target gene, E is the primer efficiency, Ct value is collected where the fluorescence is above the thresh-hold value, X indicates values from the target gene, R indicates the geometric mean of values from the three reference genes [23, 24].
Phenotypic evaluation
Net photosynthesis rate (Pn) was determined on fully expanded flag leaves of 5 randomly selected plants in each plot of each replication at heading (Z55), anthesis (Z65) and grain-filling stages(Z73), respectively, using portable photosynthesis system (LI6400XT, USA). The leaf chamber’s conditions were as reference CO2 concentration = 400 μmol mol-1, PPFD = 1800 μmol m-2 s-1, relative humidity = 50–70% and block temperature = 20°C. The measurements were taken between 9:00 and 11:00 am in sunny and windless conditions.
At maturity, 10 plants from each plot and each replication were randomly selected; the above-ground plant parts were harvested, dried and weighed for biomass plant-1 (g) using electronic balance. The same 10 plants were then threshed separately to record grain yield plant-1 (g).
Data analysis
Analysis of variance (ANOVA) was conducted for the expressions of TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS at heading (Z55), anthesis (Z67) and grain-filling (Z73) stages. Separate analysis at heading (Z55), anthesis (Z67) and grain-filling (Z73) stages were also carried-out for Pn, whereas ANOVA for the final BMPP and GYPP was conducted after harvest. Hierarchical cluster analyses were performed to classify the 59 bread wheat genotypes on the basis of average expression of TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS across the three growth stages. Correlation coefficients between the expressions of the three copies at the three stages with the measured traits at the respective stages were determined using Pearson Product Moment Correlation test. All statistical analyses were carried-out using SPSS statistical software version 19.0 (IBM SPSS Statistics, USA).
Results
Characterization of the TaRca2-αgene in wheat
BLAST search using DNA sequence of TaRca2-α (accession No. LM992845) against wheat chromosome sequence survey database (http://wheatgenome.org) revealed that there were three homoeologous copies of TaRca2-α located on long arm of chromosome 4A, and short arms of chromosome 4B and 4D, respectively, and designated as TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS. The predicted size of TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS was 1719 bp, 1749 bp, and 1735 bp in length, respectively. Analysis using spidey tool in NCBI database (http://ncbi.nlm.nih.gov/spidey/) predicted five exons and four introns for each copy, and the coding sequences (CDS) of the three copies were 1193 bp (TaRca2-α-4AL), 1195 bp (TaRca2-α-4BS) and 1195 bp (TaRca2-4DS), respectively. Multiple sequence alignment of CDS of the three copies revealed 26 nucleotide difference between TaRca2-α-4AL and TaRca2-α-4BS, 29 nucleotide difference between TaRca2-α-4BS and TaRca2-α-4DS, and 19 nucleotide difference between TaRca2-α-4AL and TaRca2-α-4DS (S1 Fig). The three copies encoded 397, 397 and 396 amino acids for TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS, respectively (S2 Fig). High similarities were observed in the conserved protein (AAA) domains of the three copies (S2 Fig).
Expression of TaRca2-α in 59 bread wheat genotypes
The specificity of individual primer pairs for the three copies of TaRca2-α was tested in three nulli-tetrasomic lines (N4ATB, N4BT4D and N4DT4A) of Chinese Spring. The single RT-PCR product for TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS was 233, 151 and 164 bp, respectively. The 4AL-specific primer pair amplified a single PCR product from N4BT4D and N4DT4A, but not from N4ATB. The amplicon produced by the 4BS-specific primer pair was absent in N4BT4D, but present in N4AT4D and N4DT4A. The 4DS-specific primer set generated a single PCR product from N4AT4B and N4BT4D, but not from N4DT4A (S3 Fig).
Expression analysis by qRT-PCR showed that the three copies of TaRca2-α were highly expressed at grain-filling stage (0.68) than at heading (0.45) and anthesis (0.54) stages (Table 3). Significantly different expression patterns at the three growth stages and among those genotypes were observed for TaRca2-α-4AL with a range of 0.18 to 0.83 at heading, 0.32 to 1.18 at anthesis and 0.45 to 1.22 at grain-filling stages (P<0.05) (Table 3). Similar results were observed for TaRca2-α-4BS expression, which ranged between 0.25 and 1.07 at heading, 0.30 to 1.26 at anthesis and 0.49 to 1.35 at grain-filling stages (Table 3). No significant (P<0.05) differences for TaRca2-α-4DS expression were found between heading and anthesis stages, whereas it was significantly higher at grain-filling stage with a range of 0.41 to 1.29 (Table 3). Overall, TaRca2-α-4BS showed higher expression at the three growth stages than TaRca2-α-AL and TaRca2-α-4DS (Table 3). The expressions of three copies of TaRca2-α at the three growth stages were significantly (P<0.01) and positively correlated with each other, with the highest correlations at grain-filling stage. The correlation coefficient (r) between TaRca2-α-4AL and TaRca2-α-4BS, TaRca2-α-4AL and TaRca2-α-4DS, TaRca2-α-4BS and TaRca2-α-4DS at grain-filling stage was 0.916, 0.928, and 0.954, respectively (S1 Table).
Table 3. Expression of the three copies of TaRca2-α at three growth stages in flag leaves of 59 bread wheat genotypes.
| TaRca2- α gene | Item | Heading (Z55) | Anthesis (Z67) | Grain-filling (Z73) |
|---|---|---|---|---|
| TaRca2- α -4AL | Mean | 0.41±0.018 C | 0.57±0.020 B | 0.67±0.021 A |
| Range | 0.18–0.83 | 0.32–1.18 | 0.45–1.22 | |
| TaRca2- α -4BS | Mean | 0.54±0.024 C | 0.63±0.029 B | 0.76±0.029 A |
| Range | 0.25–1.07 | 0.30–1.26 | 0.49–1.35 | |
| TaRca2- α -4DS | Mean | 0.38±0.019 B | 0.42±0.020 B | 0.61±0.022 A |
| Range | 0.15–0.94 | 0.11–0.84 | 0.41–1.29 | |
| Overall | Mean | 0.45±0.019 C | 0.54±0.0.21 B | 0.68±0.023 A |
| Range | 0.22–0.93 | 0.27–1.02 | 0.45–1.29 |
Data are shown as the mean ± SE (standard error) of all genotypes; uppercase letters represent differences significant among the three growth stages (P≤0.01).
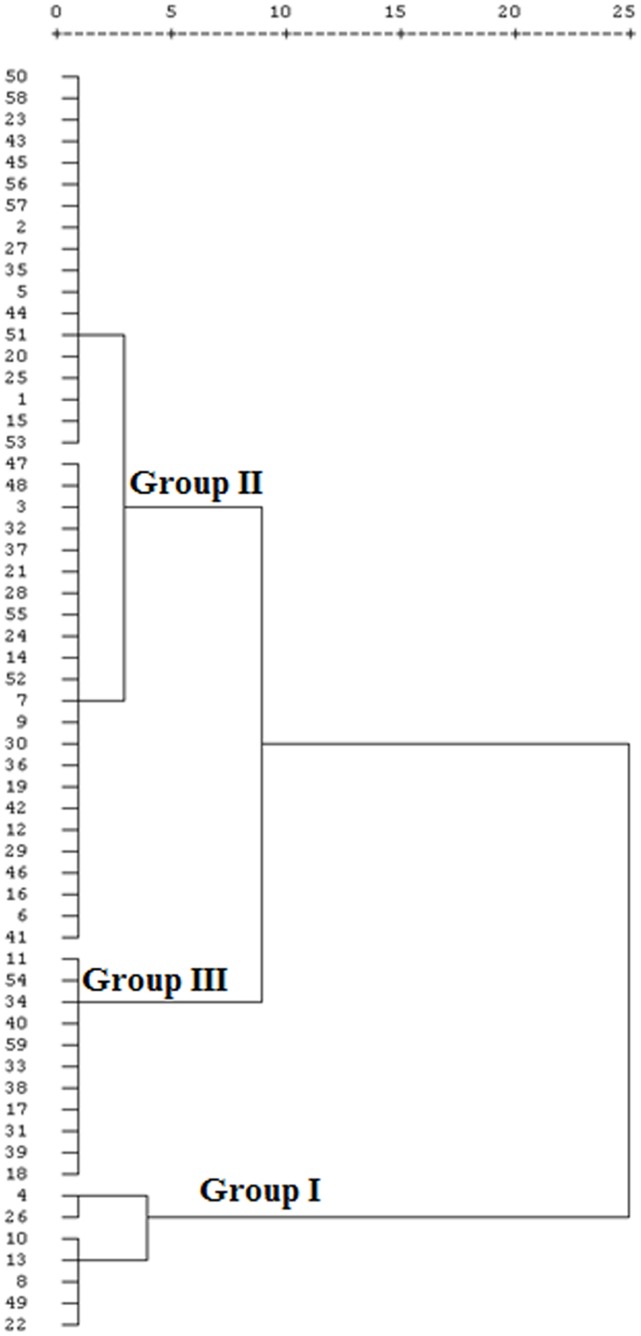
Based on the relative expression of the three homoeologous copies of TaRca2-α at the three growth stages, the 59 wheat genotypes could be clustered into three groups (Fig 1). The group I was comprised of 7 genotypes with high expression, Group II contained 41 genotypes with intermediate expression, whereas group III was consisted of 11 genotypes with low expression.
Fig 1. Cluster analysis of 59 bread wheat genotypes based on averaged expressions of three copies of TaRca2- α.
Group I: high expression, Group II: intermediate expression; Group III: low expression. The numbers on Y axis represent the codes of 59 wheat genotypes as in Table 1. X axis represents the squared Euclidean distance.
Significant differences among the three groups of bread wheat genotypes were observed on the expressions of the three copies of TaRca2-α at heading, anthesis and grain-filling stages (Table 4). For all the three copies of TaRca2-α, the group I genotypes always revealed the highest mean expression, and the group III genotypes showed the lowest, while the group II genotypes expressed the intermediate mean expression (Table 4). At each growth stage, the group I genotypes showed significantly higher expressions (0.67, 0.67 and 0.81 at heading, anthesis and grain-filling stages, respectively), whereas the group III genotypes observed the lowest expressions (0.22, 0.21 and 0.43 at heading, anthesis and grain-filling stages respectively).
Table 4. Expression of the three copies of TaRca2-α at heading (Z755), anthesis (Z67) and grain-filling (Z73) stages in the three groups of 59 bread wheat genotypes.
| TaRca2-α gene | Growth Stage | Items | Group I | Group II | Group III |
|---|---|---|---|---|---|
| TaRca2- α -4AL | Heading (Z55) | Mean | 0.66±0.042 A | 0.42±0.011 B | 0.22±0.006 C |
| Range | 0.60–0.83 | 0.30–0.53 | 0.18–0.24 | ||
| Anthesis (Z67) | Mean | 0.86±0.058 A | 0.57±0.012 B | 0.38±0.006 C | |
| Range | 0.75–1.18 | 0.45–0.73 | 0.32–0.39 | ||
| Grain-filling (Z73) | Mean | 1.02±0.068 A | 0.66±0.009 B | 0.51±0.008 C | |
| Range | 0.80–1.22 | 0.57–0.74 | 0.45–0.53 | ||
| TaRca2- α -4BS | Heading (Z55) | Mean | 0.97±0.026 A | 0.53±0.007 B | 0.31±0.014 C |
| Range | 0.90–1.07 | 0.49–0.60 | 0.25–0.40 | ||
| Anthesis (Z67) | Mean | 1.18±0.028 A | 0.59±0.005 B | 0.42±0.019 C | |
| Range | 1.09–1.26 | 0.53–0.71 | 0.30–0.48 | ||
| Grain-filling (Z73) | Mean | 1.26±0.027 A | 0.74±0.014 B | 0.50±0.003 C | |
| Range | 1.15–1.35 | 0.60–0.90 | 0.49–0.53 | ||
| TaRca2- α -4DS | Heading (Z55) | Mean | 0.67±0.067 A | 0.37±0.011 B | 0.22±0.008 C |
| Range | 0.55–0.94 | 0.24–0.49 | 0.15–0.24 | ||
| Anthesis (Z67) | Mean | 0.67±0.044 A | 0.43±0.014 B | 0.21±0.020 C | |
| Range | 0.59–0.84 | 0.29–0.54 | 0.11–0.30 | ||
| Grain-filling (Z73) | Mean | 0.98±0.063 A | 0.59±0.015 B | 0.43±0.004 C | |
| Range | 0.81–1.29 | 0.45–0.77 | 0.49–0.53 |
Data are shown as the mean ± SE (standard error) of the genotypes in each group; Group I: high expression; Group II: intermediate expression; Group III: low expression. Uppercase letters represent differences significant among the three groups (P≤0.01).
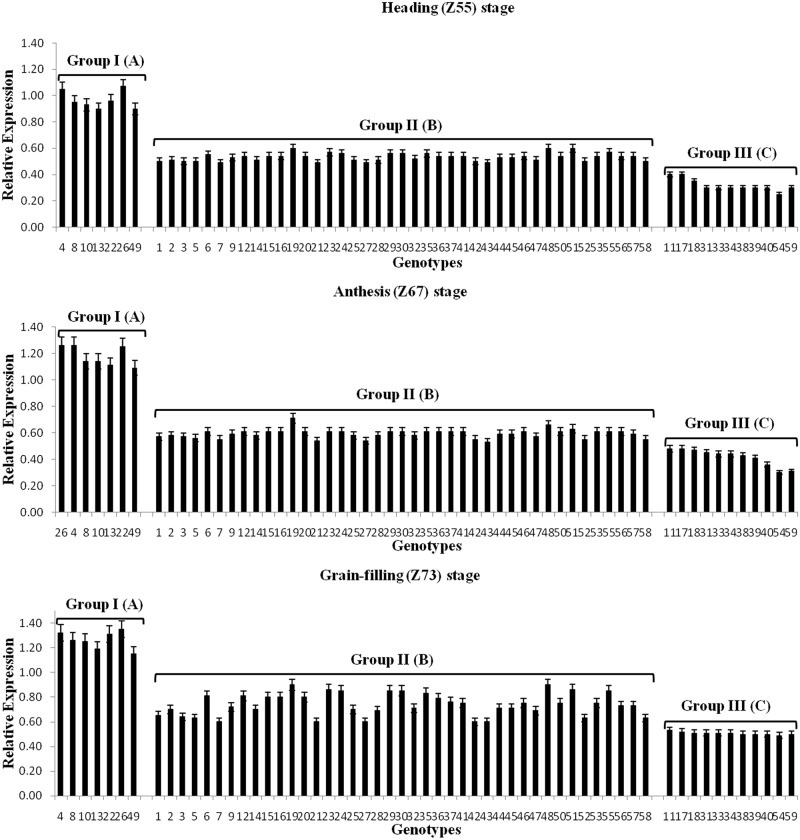
Significant differences were also observed among individual genotypes for the expressions of the three copies of TaRca2-α at the three growth stages (Fig 2, S2 Table). In general, the individual genotypes showed similar expression trends for the three copies at the three growth stages. For instance, the 7 genotypes (Zhoumai 16, Linhan 51329, Youmai 2, Zhonghan 110, Changwu 135, Linhan 536 and Luohan 6) in group I expressed TaRca2-α-4BS at higher level at all three stages (Fig 2, S2 Table), while the genotypes Jinan 18 and Aifeng 3 in group III expressed the lowest at all three stages.
Fig 2. TaRca2-α-4BS expression in flag leaves of the three groups of 59 bread wheat genotypes at Heading (Z55), Anthesis (Z67) and Grain-filling (Z73) stages.
Group I: high expression; Group II: intermediate expression; Group III: low expression. Uppercase letter represent significant differences among the three groups (P≤0.01). The numbers on X-axis correspond to the codes of individual genotypes in Table 1.
Associations between TaRca2-α expression and Pn
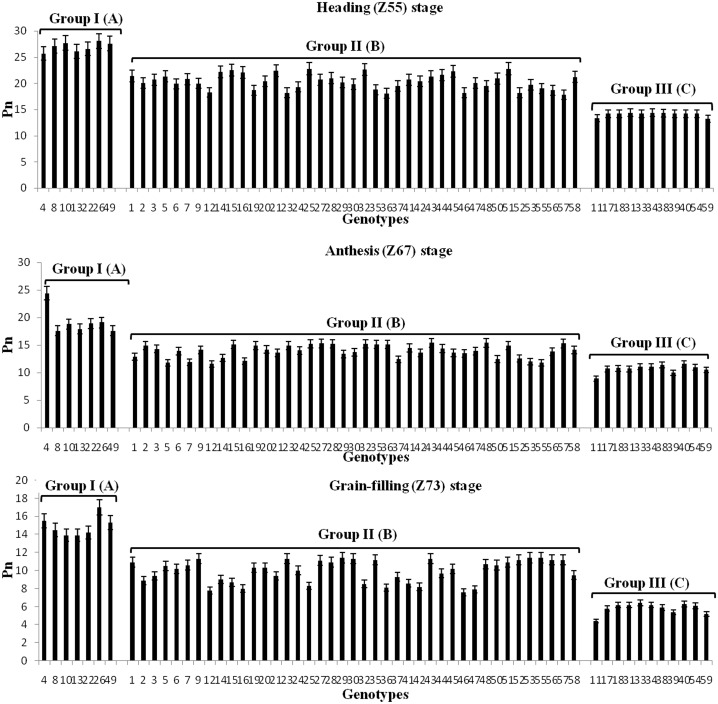
Significant differences for Pn among the wheat genotypes of the three groups were observed at all three growth stages, with the highest (20.4 μmol m-2 s-1) at heading and the lowest (10.2 μmol m-2 s-1) at grain-filling stage (Table 5; Fig 3). In general, the genotypes with high TaRca2-α expression at the three growth stages also showed higher Pn at the three growth stages. For instance, at heading stage, the highest Pn was recorded in all seven genotypes in group I (25.7 to 28.1 μmol m-2 s-1), whereas the lowest Pn was observed in Hanxuan 10 (13.2 μmol m-2 s-1), Linfen 10 (13.5 μmol m-2 s-1) and Jinan 18 (14.3 μmol m-2 s-1) from group III (S3 Table).
Table 5. Mean Pn (μmol m-2 s-1) of the three groups of 59 bread wheat genotypes at heading (Z55), anthesis (Z67) and heading (Z73) stages.
| Pn | Item | Group I | Group II | Group III | Average |
|---|---|---|---|---|---|
| Pn-Heading (Z55) | Mean | 26.9±0.336 A | 20.4±0.229 B | 14.0±0.120 C | 20.4±0.454 A |
| Range | 25.7–28.1 | 17.8–22.8 | 13.2–14.5 | 13.2–28.1 | |
| Pn-Anthesis (Z67) | Mean | 19.2±0.901 A | 13.8±0.186 B | 10.8±0.223 C | 14.6±0.342 B |
| Range | 17.6–24.4 | 11.6–15.4 | 8.9–11.6 | 8.9–24.4 | |
| Pn-Grain-filling (Z73) | Mean | 14.9±0.425 A | 9.9±0.195 B | 5.8±0.183 C | 10.2±0.354 C |
| Range | 13.9–17.0 | 7.6–11.4 | 4.4–6.4 | 4.4–17 |
Data are shown as the mean ± SE (standard error) of the genotypes in each group; Group I: High expression; Group II: medium expression; Group III: low expression. Uppercase letters represent significant differences between the three groups of (P<0.01).
Fig 3. Pn (μmol m-2 s-1) of 59 bread wheat genotypes in the three groups at heading (Z55), anthesis (Z67) and grain-filling (Z73) stages.
Group I: high expression; Group II: intermediate expression; Group III: low expression. Uppercase letter represent significant differences (P≤0.01) among the three groups. The numbers on X-axis correspond to the codes of individual genotypes in Table 1.
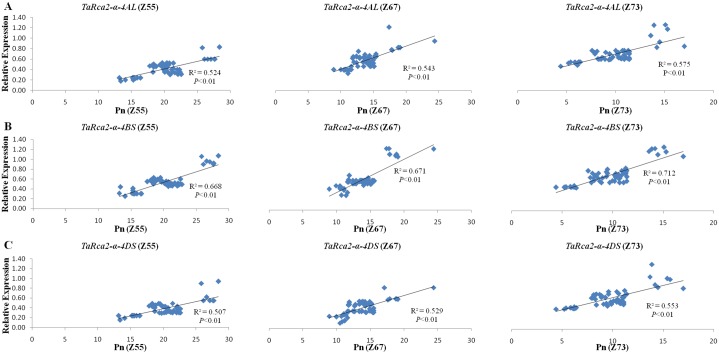
Regression analysis showed that expressions of the three copies of TaRca2-α at heading, anthesis and grain-filling stages were significantly and positively (P<0.01) associated with Pn at all the corresponding growth stages (Fig 4). The expressions of TaRca2-α-4BS were more strongly correlated with Pn than that of TaRca2-α-4AL and TaRca2-α-4DS, with regression coefficients of 0.678, 0.671 and 0.712 at heading, anthesis and grain-filling stages, respectively. The expressions of TaRca2-α-4AL, TaRca2-α-4BS and TaRca2-α-4DS were highly correlated with Pn at grain-filling stage, with regression coefficients of 0.575, 0.712 and 0.553, respectively, than at heading and grain-filling stages.
Fig 4. Regression analysis between the expressions of the three copies of TaRca2-αwith Pn (μmol m-2 s-1) at heading, anthesis and grain filling stages.
A: TaRca2-α-4AL; B: TaRca2-α-4BS; C: TaRca2-α-4DS. Pn (Z55), Pn (Z67), Pn (Z73), indicate the Pn at heading (Z55), anthesis (Z67) and grain filling (Z73) stages, respectively.
Correlations between TaRca2-α expression with BMPP and GYPP
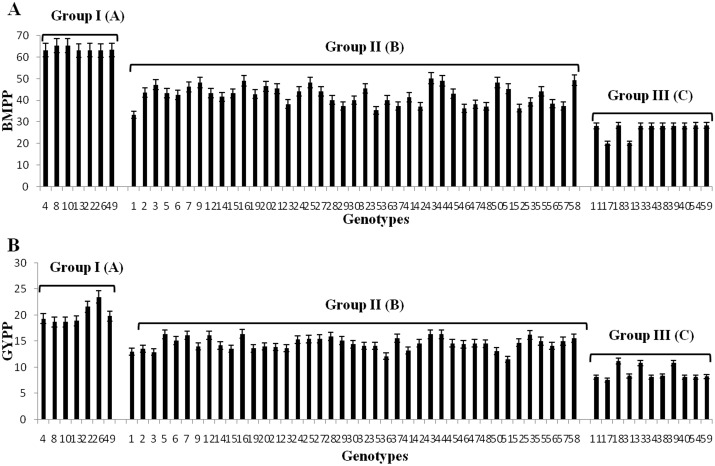
Significant differences were observed among the three groups of bread wheat genotypes for biomass plant-1 (BMPP) and grain yield plant-1 (GYPP) (Table 6, Fig 5). Generally, genotypes showing high TaRca2-α expression produced higher BMPP and GYPP. Group I genotypes with high expression produced the highest mean BMPP of 63.8 g plant-1and GYPP of 20.0 g plant-1, group II genotypes with intermediate expression showed medium mean BMPP of 42.3 g plant-1 and GYPP of 14.5 g plant-1, whereas group III genotypes with low expression produced the lowest average BMPP of 26.7 g plant-1 and GYPP of 8.8 g plant-1. The maximum BMPP was produced by Changwu 135 (65.4 g plant-1) and Zhonghan 110 (65.3 g plant-1) in group I, whereas the lowest BMPP was recorded in Xinyuan 958 (20.1g plant-1) and Aifeng 3 (20.2 g plant-1) in group III. The highest GYPP was recorded in genotype Zhoumai 16 (23.5 g plant-1) of group I, while the lowest GYPP was in genotype Xinyuan 958 (7.5g plant-1) of group III (S3 Table).
Table 6. Mean BMPP (g plant-1) and GYPP (g plant-1) of three groups of 59 bread wheat genotypes.
| Trait | Item | Group I | Group II | Group II |
|---|---|---|---|---|
| BMPP | Mean | 63.8±0.401 A | 42.3±0.702 B | 26.7±0.987 C |
| Minimum | 63.1 | 33.3 | 20.1 | |
| Maximum | 65.4 | 50.2 | 28.4 | |
| GYPP | Mean | 20.0±0.688 A | 14.5±0.187 B | 8.8±0.407 C |
| Minimum | 18.7 | 11.5 | 7.5 | |
| Maximum | 23.5 | 16.4 | 11.2 |
Data are shown as the mean ± SE (standard error) of the genotypes in each group; Group I: high expression; Group II: intermediate expression; Group III: low expression. Uppercase letters represent significant difference among the three groups (P<0.01).
Fig 5. BMPP and GYPP of bread wheat genotypes in three groups.
Group I: high expression; Group II: intermediate expression; Group III: low expression. Uppercase letters represent significant differences (P<0.01) level. The numbers on X-axis correspond to the codes of individual genotypes in Table 1.
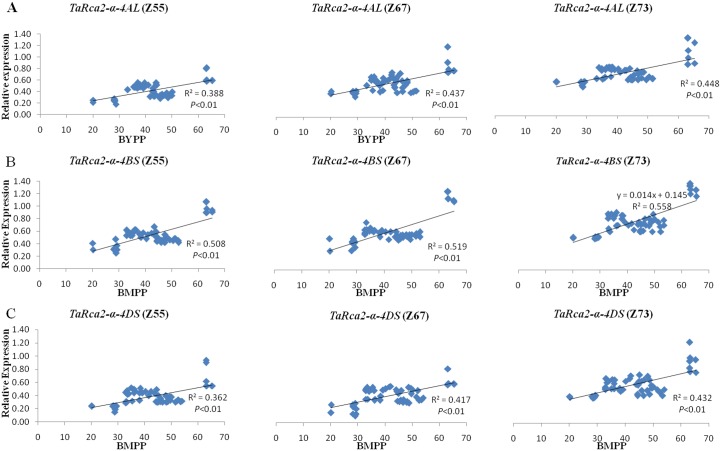
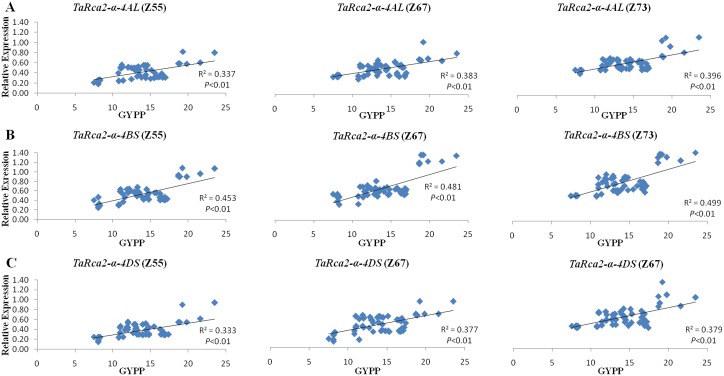
Significant and positive associations (P<0.01) were observed between the expression of all the three copies at the three growth stages with BMPP and GYPP as revealed by regression analysis (Figs 6 and 7). The expressions of all three copies of TaRca2-α at grain-filling were more strongly correlated with BMPP and GYPP with regression coefficients of 0.448, 0.558 and 0.432 for BMPP and of 0.396, 0.499 and 0.379 for GYPP with the three copies of TaRca2-α, respectively than that at heading and anthesis stages (Figs 6 and 7). In comparison with TaRca2-α-4AL and TaRca2-α-4DS, TaRca2-α-4BS expression showed stronger positive correlations with BMPP and GYPP at all three growth stages, with regression coefficient of 0.508, 0.519 and 0.588 for BMPP and of 0.453, 0.481 and 0.499 for GYPP with its expression at heading, anthesis and grain-filling stages, respectively (Figs 6 and 7).
Fig 6. Regression analysis between the expressions of the three copies of TaRca2-α at heading, anthesis and grain filling stages, with BMPP.
A: TaRca2-α-4AL; B: TaRca2-α-4BS; C: TaRca2-α-4DS. Z55, Z67 and Z73, indicate the expression measured at heading (Z55), anthesis (Z67) and grain filling (Z73) stages, respectively.
Fig 7. Regression analysis between the expressions of the three copies of TaRca2-α at heading, anthesis and grain filling stages, with GYPP.
A: TaRca2-α-4AL; B: TaRca2-α-4BS; C: TaRca2-α-4DS. Z55, Z67 and Z73, indicate the expression measured at heading (Z55), anthesis (Z67) and grain filling (Z73) stages, respectively.
Discussion
Expression patterns of TaRca2-α in wheat
In bread wheat, TaRca2-α is produced as a result of splicing event at the end of exon-5 of TaRca2-β [8]. The α and β isoforms are capable of supporting photosynthesis with increased expression and contribution of α isoform to plant productivity under different growth conditions [25, 26, 27]. It suggests the important role of α isoform of Rca as a molecular chaperon in protecting other functional proteins from damage [16]. The present research was conducted to study the expression of TaRca2-α at heading (Z55), anthesis (Z67) and grain-filling (Z73) stages under natural field conditions where macro/micro climate is not steady and plants face certain limiting factors. Overall, our results confirmed the higher variation in the expression of TaRca2-α in the panel of 59 bread wheat genotypes grown under field conditions (Table 3). Most of the previous studies were conducted under controlled growth conditions, whereas little information is available regarding investigation of Rca genes under natural conditions especially in wheat. However, results of the current study are in agreement with earlier studies in other crops [16, 17] in which the investigators reported higher expression of α isoform with positive effects on plant phenotypes under controlled as well as natural field conditions. Being allopolyploid, wheat has three homoeologous copies of a gene in general and study of the three copies helps to know the contribution of a specific copy [18]. In the current study, the three copies of TaRca2-α were identified in bread wheat, and then genome-specific primers were designed to investigate their expression levels in flag leaves at the three growth stages under field condition. The expressions of all three copies of TaRca2-α were higher at grain-filling (Z73) stage than at heading (Z55) and anthesis (Z67) stages, as similar expression patterns for Rca-α were also reported in other crops [28, 29], which suggests positive contribution of expression at the grain-filling stage to the plant productivity and grain yield under prevailing environmental conditions. TaRca2-α-4BS was more highly expressed than TaRca2-α-4AL and TaRca2-α-4DS, which were in consistency with those by Carmo-Silva et al. [8], which suggests that TaRca2-α-4BS might be more important and may contribute to enhanced plant performance. In a research on wheat, Edae et al [30] studied the association of homoeologous copies of DREB1, ERA1 and 1-FEH, and reported relatively strong association of one copy with traits compared to other copies indicating the important role of a specific homoeologue in controlling agronomic traits.
Diversity in gene expression is one of the mechanisms underlying phenotypic variation among genotypes [17] and aids in identification of genotypes with better traits [16, 20, 31]. In the present work, significant variations on the expression levels of the three copies of TaRca2-α at the three growth stages were also observed among the bread wheat genotypes in the panel, therefore the genotypes were clustered into groups based on the relative expression of TaRca2-α to facilitate the analysis for determining whether those variations were associated with their photosynthetic capability and performance regarding biomass and grain yield. As shown in the cluster results, 41 bread wheat genotypes (69.5%) were with intermediate expressions of TaRca2-α, and only 7 genotypes with higher expressions, similar results were found in our previous work with TaER genes in bread wheat [20].
Association of TaRca2-α expression with Pn
Rubisco activase is the key enzyme for net CO2 assimilation in C3 crops through activating Rubisco [9, 32, 33] and the α isoform of Rca plays an important role in maintaining Rubisco’s activity and hence plant efficiency under various conditions [15, 16]. In the present study, we observed that the genotypes showing high TaRca2-α expression levels at the three growth stages also expressed high Pn values at the corresponding stages, and the expression levels of the three copies of TaRca2-α at the three stages were all significantly and positively correlated with Pn at the corresponding stages with comparatively stronger association for TaRca2-α-4BS and at grain-filling stage. This possibly suggested the great contribution of TaRca2-α to Pn, with a more significant contribution by TaRca2-α-4BS. These are in agreement with the previous findings in soybean, maize and other crops [16, 20, 28, 29, 34]. Grain-filling is a crucial stage for achieving optimum grain yield, the relatively stronger association of the expression levels of TaRca2-α with Pn at this stage suggested that higher expression may contribute more to grain yield, although Pn reduced greatly at this stage as shown in other studies [5, 17]. These results support the hypothesis that TaRca2-α contributed to photosynthesis substantially, detection of its expression levels could be utilized as a selection tool for the improvement of photosynthetic efficiency in bread wheat The genomic variations resulting in this expression difference should be further clarified for execution in marker assisted selection.
Association of TaRca2-α expression with BMPP and GYPP
Studies in wheat and other C3 crops have shown that there exists a positive association between Pn and biomass [35] and grain yield [5], improvement in plant biomass can translate in reasonable gains in crop yield. A significant and positive correlation between Pn at heading, anthesis and grain-filling stages with BMPP and GYPP (P<0.01) were also observed in this study, which was more significant (r = 0.647 for BMPP and r = 0.511 for GYPP) at the grain-filling stage (Table 7). This indicates that there is a potential for an increase in grain yield through improvement in photosynthetic efficiency and biomass production. In wheat, Rubisco can potentially enhance CO2 assimilation resulting in biomass gain, the endogenous levels of Rca expression can be of importance to plant photosynthesis and biomass production [27, 28]. Investigation in this study revealed that, the expression levels of the three copies of TaRca2-α at the three growth stages were strongly and positively correlated with BMPP and GYPP with stronger effect of TaRca2-α-4BS, and also at grain-filling stage. In general, the genotypes showing high expression also produced higher BMPP and GYPP. These results are in concurrence with those reported in other crops under variable environmental conditions [15, 16, 17, 36, 37]. This suggests that regulation of TaRca-α expression may efficiently improve BMPP and GYPP in wheat and the variations in the expression levels of TaRca-α may be utilized for selection of biomass and grain yield among wheat genotypes under natural field conditions.
Table 7. Correlation coefficients between Pn at heading (Z55), anthesis (Z67) and grain-filling (Z73) stages with BMPP (g plant-1) and GYPP (g plant-1) in 59 bread wheat genotypes.
| Pn-Heading (Z55) | Pn-Anthesis (Z67) | Pn-Grain-filling (Z73) | BMPP | GYPP | |
| Pn-Heading (Z55) | 1 | ||||
| Pn-Anthesis (Z67) | 0.575** | 1 | |||
| Pn-Grain-filling (Z73) | 0.594** | 0.611** | 1 | ||
| BMPP | 0.558** | 0.574** | 0.647** | 1 | |
| GYPP | 0.476** | 0.486** | 0.511** | 0.455** | 1 |
**. Correlation is significant at the 0.01 level
Pn: net photosynthesis rate (μmol m-2 s-1); BMPP: biomass plant-1; GYPP: grain yield plant-1
The associations between the expression levels of TaRca2-α copies with Pn, BMPP and GYPP were studied in a panel of 59 bread wheat genotypes under field conditions. TaRca2-α-4BS was highly expressed as compared to TaRc2-α-4AL and TaRca2-4DS. The expression of the three copies of TaRca2-α was more profound at grain-filling (Z73) stage than at heading (Z55) and anthesis (Z67) stages, and were significantly and positively correlated with Pn, BMPP, and GYPP, which were stronger at grain-filling stage than at heading and anthesis stages, with comparatively stronger association with Pn. These results suggested that the expression of TaRca2-α contribute greatly to Pn, BMPP and GYPP, and regulation of TaRca-α expression may efficiently improve Pn, BMPP and GYPP in wheat, and the variations detected in TaRca2-α expression levels with special emphasis on TaRca2-α-4BS may be utilized for selection in improving photosynthetic efficiency and grain yield of bread wheat.
Supporting Information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work is supported by sub-project of the 863 Program (2013AA102902) of the Ministry of Science and Technology, and the China 111 Project (B12007), P. R. China. YGH received the funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Reynolds M, Bonnett D, Chapman SC, Furbank RT, Manes Y, Mather DE, et al. Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. J Exp Bot. 2011; 62(2): 439–452. 10.1093/jxb/erq311 [DOI] [PubMed] [Google Scholar]
- 2.FAO. 2015. http://faostat3.fao.org/home/E.
- 3.RE Evenson, Gollin D. Assessing the impact of the green revolution, 1960 to 2000. Science. 2003; 300(5620): 758–762. [DOI] [PubMed] [Google Scholar]
- 4.Rosegrant MW, Agcaoili M. Global food demand, supply, and price prospects to 2010. Washington, DC: International Food Policy Research Institute; 2010. [Google Scholar]
- 5.Fischer RA, Ress D, Sayre KD, Lu ZM, Condon AG, Saavedra AL. Wheat yield progress associated with higher stomatal conductance and photosynthetic rate, and cooler canopies. Crop Sci. 1998; 38: 1467–1475. [Google Scholar]
- 6.Raven JA. Rubisco: still the most abundant protein on Earth? New Phytologist. 2013; 198, 1–3. 10.1111/nph.12197 [DOI] [PubMed] [Google Scholar]
- 7.McNevin D, von Caemmerer S, Farquhar GD. Determining Rubisco activation kinetics and other rate and equilibrium constants by simultaneous multiple non-linear regression of a kinetic model. J Exp Bot. 2006; 57(14): 3883–3900. [DOI] [PubMed] [Google Scholar]
- 8.Carmo-Silva E, Scales JC, Madjwick PJ, Parry MA. Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ. 2014; 38(9): 1817–32. 10.1111/pce.12425 [DOI] [PubMed] [Google Scholar]
- 9.Portis AR Jr. Rubisco activase: Rubisco’s catalytic chaperone. Photosynth Res. 2003; 75(1):11–27. [DOI] [PubMed] [Google Scholar]
- 10.Spreitzer RJ, Salvucci ME. Rubisco: Structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol. 2002; 53: 449–475. 10.1146/annurev.arplant.53.100301.135233 [DOI] [PubMed] [Google Scholar]
- 11.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation and disassembly of protein complexes. Genome Res. 1999; 9(1): 27–43. [PubMed] [Google Scholar]
- 12.Portis AR Jr. The regulation of Rubisco by Rubisco activase. J Exp Bot. 1995; 46 (Special Issue):1285–1291. 10.1093/jxb/46.special_issue.1285 [DOI] [Google Scholar]
- 13.Salvucci MM, Ogren WL. The mechanism of Rubisco activase: insights from studies of the properties and structure of the enzyme. Photosynth Res. 1996; 47(1)1–11. 10.1007/BF00017748 [DOI] [PubMed] [Google Scholar]
- 14.Shen JB, Orozco EM Jr, Ogren WL. Expression of the isoforms of spinach ribulose 1,5-bisphosphate carboxylase acitvase and essentially of the conserved lysine in the consensus nucleotide-binding domain. J Biol Chem. 1991; 266(14): 8963–8968. [PubMed] [Google Scholar]
- 15.Wang D, Li XF, Zhou ZJ, Feng XP, Yang WJ, Jiang DA. Two Rubisco activase isoforms may play different roles in photosynthetic heat acclimation in the rice plant. Physiol Plant. 2010; 139(1):55–67. 10.1111/j.1399-3054.2009.01344.x [DOI] [PubMed] [Google Scholar]
- 16.Chao M, Yin Z, Hao D, Zhang J, Song H, Ning A et al. Variation in Rubisco activase (RCAα) gene promoters and expression soybean [Glycine max (L) Merr.]. J Exp Bot 2013; 65(1): 47–59. 10.1093/jxb/ert346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin Z, Zhenliang Z, Dexiang D, Maoni C, Qingsong G, Yijun W et al. Characterization of Rubisco Activase Genes in Maize: An α-Isoform Gene Functions alongside a β-Isofrom Gene. Plant Physiol. 2014; 164(4): 2096–2106. 10.1104/pp.113.230854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X-Q, Anita B-B. Development of genome-specific primers for homoeologous genes in allopolyploid species: the waxy and starch synthase II genes in allohexaploid wheat (Triticum aestivum L.) as example. BMC Research Notes. 2010; 3:140 10.1186/1756-0500-3-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999; 41: 95–98. [Google Scholar]
- 20.Zheng J, Yang Z, Madgwick PJ, Carmo-Silva E, Parry MAJ, Hu Y-G. TaER Expression is Associated with Transpiration Efficiency Traits and Yield in Bread Wheat. PLoS ONE. 2015; 10(6): e0128415 10.1371/journal.pone.0128415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinmachi F, Buchner P, Stroud JL, Parmar S, Zhao FJ, McGrath SP et al. Influence of sulfur deficiency on the expression of specific sulfate transporters and the distribution of sulfur, selenium and molybdenum in wheat. Plant Physiol. 2010; 153(1): 327–336. 10.1104/pp.110.153759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verwoerd TC, Dekker BM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 1989; 17(6): 2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuro Sci Lett. 2003; 339(1): 62–66. . [DOI] [PubMed] [Google Scholar]
- 24.Rieu I, Powers SJ. Real-time quantitative RT-PCR: design, calculations and statistics. Plant Cell. 2009; 21(4): 1031–1033. 10.1105/tpc.109.066001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, Xiao-Man W, Li Z, Yi H, Dun W, Yan-Hua Q et al. Rubisco Activase Is Also a Multiple Responder to Abiotic Stresses in Rice. PLoSONE. 2015; 10(10): e0140934 10.1371/journal.pone.0140934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurek I, Thom KC, Sean BM, Alfredo M, Lu L, Michael WL, Z. et al. Enhanced thermostability of Arabidopsis rubisco activase improves photosynthesis and growth rates under moderate heat stress. The Plant Cell. 2007; 19: 3230–3241. 10.1105/tpc.107.054171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ristic Z, Momcilovic I, Bukovnik U, Prasad PV, Fu J, Deridder BP, et al. Rubisco activase and wheat productivity under heat-stress conditions. J Exp Bot. 2009; 60(14): 4003–4014. 10.1093/jxb/erp241 [DOI] [PubMed] [Google Scholar]
- 28.Martinez-Barajas E, Molina-Galan J, Sanchez de Jimenez ES. Regulation of Rubisco activity during grain-fill in maize: Possible role of Rubisco activase. J Agri Sci. 1997; 128(2): 155–161. [Google Scholar]
- 29.Morales A, Ortega-Delgado ML, Molina-Galan J, de Jimenez ES. Importance of Rubisco activase in maize productivity based on mass selection procedures. J Exp Bot. 1999; 50(335): 823–829. 10.1093/jxb/50.335.823 [DOI] [Google Scholar]
- 30.Edae EA, Patrick FB, Harish M, Scott DH, Marc M, Marta SL et al. Association mapping and nucleotide sequence variation in five drought tolerance candidate genes in spring wheat. The Plant Genome. 2013; 6(2). 10.3835/plantgenome2013.04.0010 [DOI] [Google Scholar]
- 31.Whitney SM, Houtz RL, Alonso H. Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011a; 155(1): 27–35. 10.1104/pp.110.164814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmo-Silva E, Salvucci ME. The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transition. Plant Physiol. 2013; 161: 1645–1655. 10.1104/pp.112.213348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang N, Kallis RP, Ewy RG, Portis AR Jr. Light modulation of Rubisco in Arabidopsis requires a capacity for redox regulation of the larger Rubisco activase isoform. Proceedings of the National Academy of Sciences of the United States of America. 2002; 99: 3330–3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portis AR Jr, Li C, Wang D, Salvucci ME. Regulation of Rubisco activase and its interaction with Rubisco. J Exp Bot. 2008; 59(7):1597–1604. 10.1093/jxb/erm240 [DOI] [PubMed] [Google Scholar]
- 35.Kruger EL, Volin JC. Reexamining the empirical relation between plant growth and leaf photosynthesis. Functional Plant Biology. 2006; 33(5): 421–429. 10.1071/FP05310. [DOI] [PubMed] [Google Scholar]
- 36.Kumar A, Li C, Portis AR Jr. Arabidopsis thaliana expressing a thermostable chimeric Rubisco activase exhibits enhanced growth and higher rates of photosynthesis at moderately high temperatures. Photosynth Res. 2009; 100(3): 143–153. 10.1007/s11120-009-9438-y [DOI] [PubMed] [Google Scholar]
- 37.Yin Z, Meng F, Song H, Wang X, Xu X, Yu D. Expression quantitative trait loci analysis of two genes encoding Rubisco activase in soybean. Plant Physiol. 2010b; 152(3): 1625–1637. 10.1104/pp.109.148312 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.