Abstract
Incorporation of chemical modifications into small interfering RNAs (siRNAs) increases metabolic stability and improves tissue distribution. However, how these modifications impact interactions with Argonaute-2 (Ago2), the molecular target of siRNAs, is not known. Herein, we present the crystal structure of human Ago2 bound to a metabolically stable siRNA containing extensive backbone modifications. Comparison to the structure of an equivalent unmodified-siRNA complex indicates that Ago2 structure is relatively unaffected by chemical modifications in the bound siRNA. In contrast, the modified siRNA appears to be much more plastic and shifts, relative to the unmodified siR-NA, to optimize contacts with Ago2. Structure/activity analysis reveals that even major conformational perturbations in the 3′ half of the siRNA seed region have a relatively modest effect on knock-down potency. These findings provide an explanation for a variety of modification patterns tolerated in siRNAs and a structural basis for advancing therapeutic siRNA design.
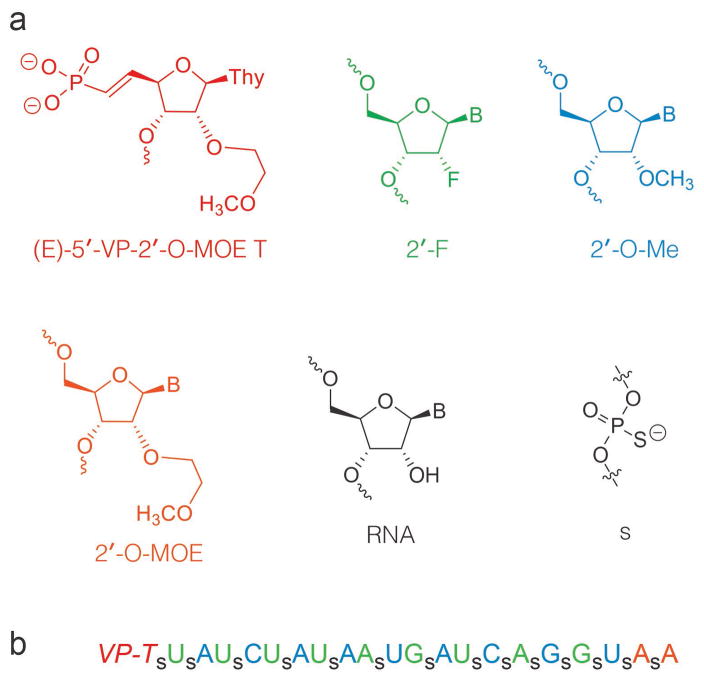
siRNA therapeutics hold tremendous therapeutic potential to treat unmet medical needs and several siRNA drugs are progressing in the clinic with excellent promise.1–5 Chemically modifed siRNA is necessary to improve pharmacokinetic properties and numerous nucleic acid modifications have been developed to improve properties of siRNA therpeutics.6–8 In general, chemical modifications led to reduced potency when compared with parent unmodified siRNAs.6,7 However, optimization of the placement of modifications in each strand can lead to derivatives with both improved potency and stability.9–11 Commonly used modifications include 2′-O-(2-methoxyethyl (2′-O-MOE), 2′-fluoro (2′-F), and 2′-O-methyl (2′-O-Me) sugar modifications and phosphorothioate (s) backbone modifications (Figure 1). Additionally, synthetic siRNA containing a metabolically stable (E)-5′-vinylphosphonate (Figure 1, 5′-VP) modification is 5–10 fold more potent than siRNA containing natural phosphate in mice.12
Figure 1.
(a) RNA modifications used in this study. (b) Sequence and structure of modified siRNA. Blue: 2′-O-Me, Green: 2′-F, Orange: 2′-O-MOE, VP-T = 2′-O-MOE-thymidine-(E)-5′-vinylphosphonate; s: phosphorothioate. All backbone linkages as phosphodiesters except those indicated with s.
Generally, siRNAs are administered as small RNA duplexes containing 2 nucleotide 3′ overhangs.2 Upon entering a target cell the duplex is loaded into the protein Ago2. One RNA strand (termed the “passenger”) is removed and degraded while the other strand (termed the “guide”) is retained in Ago2.13,14 Ago2 uses the guide strand to identify and cleave complementary target messenger RNAs.15 How commonly used guide RNA modifications impact interactions with Ago2 is not known.
To visualize how siRNA modifications impact interactions with Ago2, we determined crystal structures of Ago2 bound to a modified siRNA, and of Ago2 bound to an unmodified siRNA of the same length and nucleotide sequence. The modified siRNA contained a 5′-VP, extensive phosphorothioate linkages, with 2′-MOE, 2′-F and 2′-O-Me sugar modifications (Figure 1) and was synthesized according to a previously reported procedure.13
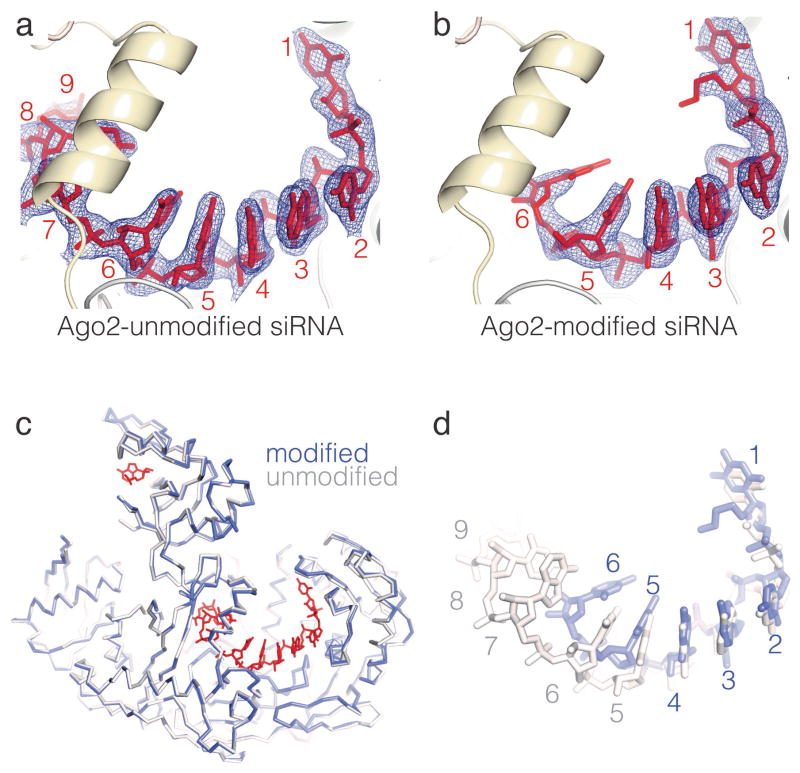
Ago2 samples were loaded with either a modified or unmodified siRNA targeting the PTEN gene, and the resulting complexes were purified and crystallized as described previously16,17. Diffraction data from both crystal forms were refined against protein atoms in the original Ago2 structure (PDB ID 4OLA) (Table S1). The conformation of Ago2 in complex with the modified siRNA was essentially identical to the unmodified complex (RMSD of 0.303 Å for 735 equivalent Cα atoms), indicating that the modifications did not substantially alter the structure of Ago2 (Figure 2).
Figure 2.
Structures of Ago2-siRNA complexes. (a) unmodified-siRNA, and (b) modified-siRNA guides bound to Ago2. 2Fo-Fc electron density maps surrounding the siRNAs are displayed as wire mesh. (c) Superposition of Ago2 Cα backbones from unmodified and modified structures. Unmodified siRNA in red. (d) Superposition of unmodified and modified siRNAs. Nucleotides numbered from 5′ ends.
Inspection of electron density maps revealed unambiguous density for nucleotides 1–9 and 21 of the unmodified siRNA, and nucleotides 1–6 of the modified siRNA (Figure 2). Discontinuous electron density corresponding to the central and 3′ regions of the modified siRNA was also observed, but could not be modeled with confidence. The well-ordered regions of both siRNAs follow a similar trajectory relative to Ago2, with 5′ ends extending from the MID domain into the Ago2 central cleft. However, in contrast to the protein atoms in the two structures, there are pronounced differences between the modified and unmodified siRNA conformations (Figure S1).
As predicted from modeling studies13, the 5′-VP group of the modified siRNA bound the 5′-phosphate binding pocket at the interface of the Ago2 MID and PIWI domains16,18–21 (Figure 3). The double bond character of the 5′-VP group restricts rotation around the C5′–C6′ bond, and thus the 5′-VP cannot perfectly mimic the unmodified 5′ phosphate (in which the ε dihedral angle is ~140°). This difference appears to be accommodated by a minor shift (~1 Å) in the position of nucleotide-1, which allows the 5′-VP to maintain the major interactions observed between Ago2 and the unmodified 5′-phosphate. Specifically, the 5′-VP oxygen atoms are within hydrogen bond/salt linkage distance (≤ 3.2 Å) of the Y529 hydroxyl and the K566, K533, and K570 ε-amines. Repositioning of nucleotide-1 is subtle enough to preserve stacking interactions between the modified nucleotide-1 base and the Y529 phenolic ring. Therefore, although the 5′-VP is not identical to the unmodified siRNA subtle repositioning of nucleotide-1 allows the majority of contacts to Ago2 to be retained.
Figure 3.
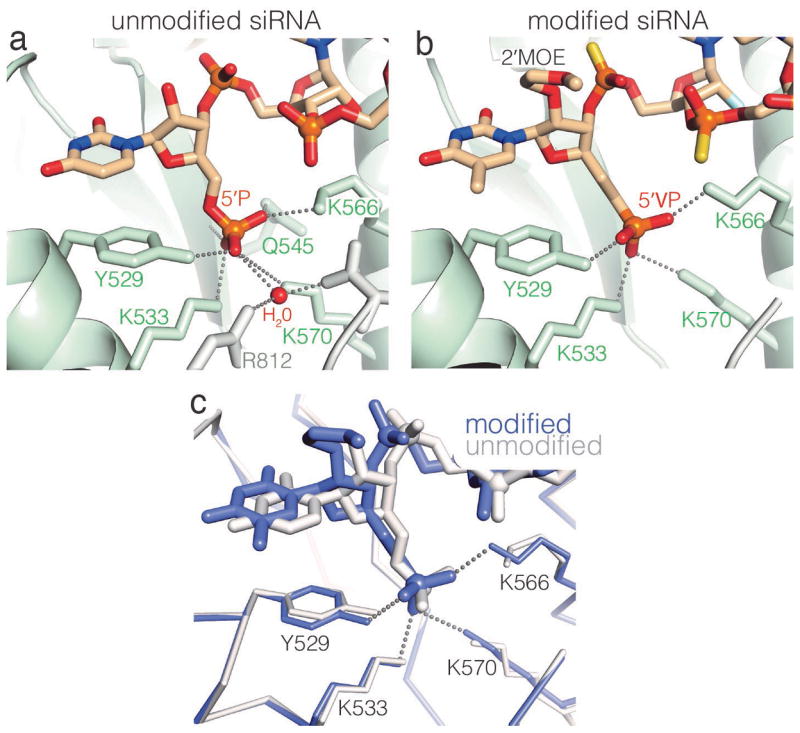
5′-VP is accommodated by subtle repositioning of the 5′ nucleotide. Close-up views of (a) unmodified and (b) modified 5′ siRNA ends. The refined 5′-VP model lacks hydrogen bonds to the side chain of Q545, the C546 main chain amine, and to an ordered water molecule. (c) Superposition of modified and unmodified 5′ nucleotides with surrounding protein atoms displayed.
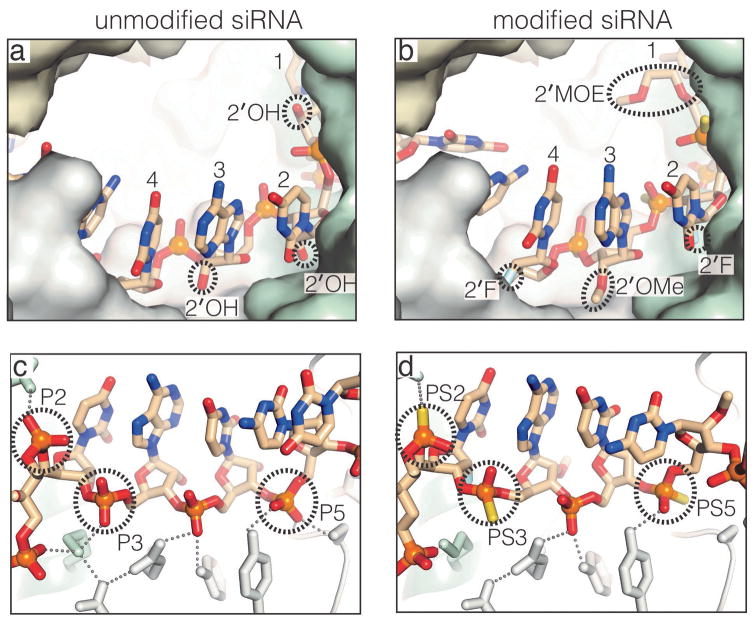
Nucleotides 2–4 of the modified-siRNA bound Ago2 in a conformation similar to their unmodified counterparts, indicating that the modifications incorporated into these nucleotides have a negligible impact on interactions with Ago2 (Figure 4). Specifically, phosphorothioate groups in the siRNA backbone (on nucleotides 2, 3 and 5) reside in the same positions as the equivalent phosphates in the unmodified siR-NA. This may explain why phosphorothioates in the siRNA seed region (nt. 2–8) are not detrimental to silencing.6,22 Sulfur atoms in the phosphorothioates could be modeled at either unesterfied position, indicating that Ago2 binds both enantiomeric forms at each PS position. The 2′-O-MOE modification on nucleotide-1 extends into the Ago2 central cleft without steric clash with the protein. Similarly, the 2′-O-Me group on nucleotide-3 fits into a narrow surface cleft between the Ago2 MID and PIWI domains. The 2′-F atoms on nucleotides-2 and -4 occupy positions equivalent to the 2′-OH groups in the unmodified siRNA and do not directly contact Ago2. These findings may explain why an alternating pattern of 2′-O-Me/2′-F, with 2′-O-Me on the odd numbered nucleotides, is well tolerated in siRNAs.23
Figure 4.
Modifications in nucleotides 2–4 do not impact interactions with Ago2. Views highlighting the sugars (a) and (b), and phosphate backbones (c) and (d) in unmodified and modified siRNA structures.
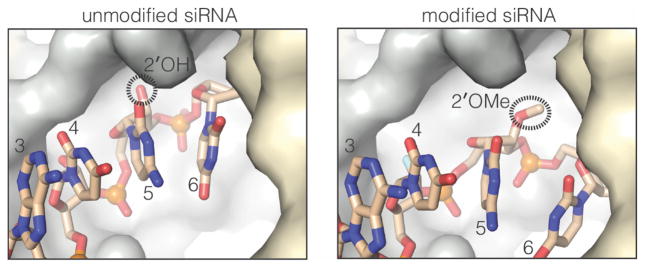
In contrast to nucleotides 2–4, the positions of modified nucleotides 5 and 6 deviate substantially from the unmodified siRNA (Figure 5). In the unmodified structure, the nucleotide-5 2′-OH inserts into a small pocket on the surface of Ago2 and makes a hydrogen bond with the main chain amide of I756.18 In the modified siRNA, the nucleotide-5 2′-O-Me is instead inserted into a larger adjacent surface pocket, which is normally occupied by the ribose of nucleotide-6 in unmodified guide RNA structures18,20,21,24. Thus, the nucleotide-5 2′-O-Me appears to displace the nucleotide-6 ribose, leading to a major shift (~6 Å) in the position of the modified-siRNA compared to the unmodified. The positional shift of modified nucleotide-6 also likely propagates to nucleotides 7–9, which are disordered in the modified siRNA structure.
Figure 5.
Repositioning of nucleotides 5–9 in the modified siRNA. The 2′-O-Me on nucleotide-5 shifts into the binding pocket of the nucleotide-6 ribose.
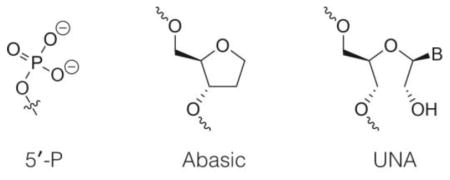
Upon observing major conformational differences between modified and unmodified siRNAs in the region surrounding nucleotides 5 and 6, we wondered how these differences impact siRNA potency, and what effects other modifications in this region might have on the efficiency of silencing by RNAi. To explore these questions we measured knock-down of the PTEN gene in HeLa cells using variants of the modified-siRNA with 2′ modifications at positions 5 and 6 (Table 1).12–13 Strikingly, all possible combinations of 2′-F and 2′-O-Me at positions 5 and 6 led to similar levels of PTEN knockdown (Table 2). Moreover, siRNAs with unmodified nucleotides at position 5, 6, or both were equivalently effective. In contrast, siRNAs including abasic or unlocked nucleotides at either position 5 or 6 were compromised.
Table 1.
Sequence and structure of siRNA guide strands. Blue: 2′-O-Me, Green: 2′-F, Orange: 2′-O-MOE, VP-T = 2′-O-MOE-thymidine-(E)-5′-vinylphosphonate; P: phosphate, s: phosphorothioate; all backbone linkages as phosphodiesters except those indicated with s; X1: abasic; X2: UNA.
| siRNA No. | Guide Sequence |
|---|---|
| 1 |
|
| 2 |
|
| 3 |
|
| 4 |
|
| 5 |
|
| 6 |
|
| 7 |
|
| 8 |
|
| 9 |
|
| 10 |
|
| 11 |
|
Table 2. IC50 values of siRNAs tested.
siRNA duplexes were generated by paring guide strand with a complementary unmodified passenger strand (5′-ACCUGAUCAU UAUAGAUAA-3′).
| siRNA No. | Position 5 chemistry | Position 6 chemistry | siRNA IC50 (nM) |
|---|---|---|---|
| 1 | 2′-O-Me | 2′-F |
|
| 2 | 2′-O-Me | 2′-F | 1.2 ± 0.9 |
| 3 | 2′-F | 2′-O-Me | 2.4 ± 0.8 |
| 4 | 2′-F | 2′-F | 3.2 ± 0.7 |
| 5 | RNA | 2′-F | 1.8 ± 0.9 |
| 6 | 2′-O-Me | RNA | 2.2 ± 0.8 |
| 7 | RNA | RNA |
|
| 8 | Abasic | 2′-F |
|
| 9 | 2′-O-Me | Abasic |
|
| 10 | UNA | 2′-F |
|
| 11 | 2′-O-Me | UNA |
|
Our results show how Ago2 binds a pharmacologically stable and potent siRNA containing extensive chemical modifications. Notably, modified nucleotides 1–4 bind in a conformation closely matching the unmodified siRNA. We suggest that this may be a critical feature of effective modified siRNAs because 1) the 5′ nucleotide serves as an anchor for siRNA binding25, and 2) nucleotides 2–4 play a major role in initiating pairing to target RNAs.26,27 Indeed, ss-siRNAs with (Z)-5′-vinylphosphonate, which cannot mimic the ε dihedral angle of the unmodified 5′-P, are significantly less active than ss-siRNA containing (E)-5′-vinylphosphonate in mammalian cells.28 Moreover, bulky 2′-O-MOE modifications in nucleotides 2–4 dramatically reduce siRNA activity.10 In contrast, the conformation of modified nucleotides 5–8 deviate substantially from the unmodified siRNA, revealing that strict structuring of the 3′ half of the seed region prior to target binding is not necessary for Ago2 function. Thus, these nucleotides are likely to be less sensitive to a wider range of chemical modifications. Consistent with this idea, 2′-O-MOE modifications are relatively well tolerated in siRNA nucleotides 6–8.10 The combined results provide mechanistic insights into modified siRNA function and a structural basis for advancing design of therapeutic siRNAs.
Supplementary Material
Acknowledgments
Funding Sources
This work was funded by NIH grants GM104475 and CA201861 to IJM.
We are grateful to Punit P. Seth, Eric E. Swayze and Stanley T. Crooke for support and insights.
Footnotes
Supporting information includes crystallographic and refinement statistics, analytical data for guide strands, PTEN knockdown data, detailed methods, and supplemental references. This material is available free of charge via the Internet at http://pubs.acs.org.
Structures of the Ago2-unmodified-siRNA complex and Ago2-modified-siRNA complex have been deposited in the Protein Data Base (PDB IDs, 5JS1 and 5JS2, respectively).
References
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Nature. 1998;391:806. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir SM, Lendeckel W, Tuschl T. Genes & development. 2001;15:188. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sehgal A, Barros S, Ivanciu L, Cooley B, Qin J, Racie T, Hettinger J, Carioto M, Jiang Y, Brodsky J, Prabhala H, Zhang X, Attarwala H, Hutabarat R, Foster D, Milstein S, Charisse K, Kuchimanchi S, Maier MA, Nechev L, Kandasamy P, Kel’in AV, Nair JK, Rajeev KG, Manoharan M, Meyers R, Sorensen B, Simon AR, Dargaud Y, Negrier C, Camire RM, Akinc A. Nat Med. 2015;21:492. doi: 10.1038/nm.3847. [DOI] [PubMed] [Google Scholar]
- 5.Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, Chiesa J, Warrington S, Tranter E, Munisamy M, Falzone R, Harrop J, Cehelsky J, Bettencourt BR, Geissler M, Butler JS, Sehgal A, Meyers RE, Chen Q, Borland T, Hutabarat RM, Clausen VA, Alvarez R, Fitzgerald K, Gamba-Vitalo C, Nochur SV, Vaishnaw AK, Sah DW, Gollob JA, Suhr OB. The New England journal of medicine. 2013;369:819. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 6.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. Biochemistry. 2003;42:7967. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 7.Corey DR. J Clin Invest. 2007;117:3615. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deleavey GF, Damha MJ. Chem Biol. 2012;19:937. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Journal of medicinal chemistry. 2005;48:901. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 10.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. Journal of medicinal chemistry. 2005;48:4247. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 11.Chiu YL, Rana TM. RNA. 2003;9:1034. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prakash TP, Kinberger GA, Murray HM, Chappell A, Riney S, Graham MJ, Lima WF, Swayze EE, Seth PP. Bioorg Med Chem Lett. 2016;26:2817–2820. doi: 10.1016/j.bmcl.2016.04.063. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Cell. 2003;115:199. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 14.Khvorova A, Reynolds A, Jayasena SD. Cell. 2003;115:209. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Science. 2004;305:1437. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 16.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Science. 2014;346:608. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flores-Jasso CF, Salomon WE, Zamore PD. RNA. 2013;19:271. doi: 10.1261/rna.036921.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schirle NT, MacRae IJ. Science. 2012;336:1037. doi: 10.1126/science.1221551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakanishi K, Weinberg DE, Bartel DP, Patel DJ. Nature. 2012;486:368. doi: 10.1038/nature11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Cell Rep. 2013;3:1893. doi: 10.1016/j.celrep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. Cell Rep. 2013;3:1901. doi: 10.1016/j.celrep.2013.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Antisense Nucleic Acid Drug Dev. 2003;13:83. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 23.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Nucleic acids research. 2003;31:2705. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schirle NT, Sheu-Gruttadauria J, Chandradoss SD, Joo C, MacRae IJ. Elife. 2015;4 doi: 10.7554/eLife.07646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Nature structural & molecular biology. 2005;12:340. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 26.Chandradoss SD, Schirle NT, Szczepaniak M, MacRae IJ, Joo C. Cell. 2015;162:96. doi: 10.1016/j.cell.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salomon WE, Jolly SM, Moore MJ, Zamore PD, Serebrov V. Cell. 2015;162:84. doi: 10.1016/j.cell.2015.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prakash TP, Lima WF, Murray HM, Li W, Kinberger GA, Chappell AE, Gaus H, Seth PP, Bhat B, Crooke ST, Swayze EE. Nucleic acids research. 2015;43:2993. doi: 10.1093/nar/gkv162. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.