Abstract
BRCA2 is responsible for familial breast and ovarian cancer and has been linked to DNA repair and centrosome duplication. Here we analyzed the mechanism by which the centrosomal localization signal (CLS) of BRCA2 interacts with cytoplasmic dynein 1 to localize BRCA2 to the centrosome. In vitro pull-down assays demonstrated that BRCA2 directly binds to the cytoplasmic dynein 1 light intermediate chain 2. A dominant-negative HA-CLS-DsRed fusion protein, the depletion of dynein by siRNA, and the inactivation of dynein by EHNA, inhibited the localization of BRCA2 at centrosomes and caused the separation of centrosome pairs during the S-phase. The double depletion of BRCA2 and C-Nap1 caused a larger dispersion of centrosome distances than the silencing of C-Nap1. These results suggest that cytoplasmic dynein 1 binds to BRCA2 through the latter's CLS and BRCA2 mediates the cohesion between centrosomes during the S phase, potentially serving as a cell-cycle checkpoint.
Keywords: BRCA2, centrosome, CLS, C-Nap1, dynein, LIC2
Introduction
BRCA2 is known to play a role in the development of familial breast and ovarian cancer, and it plays a pivotal role during homologous recombination.1 BRCA2 is also important in centrosome positioning and amplification.2,3 BRCA2 possesses a functional centrosomal localization signal (CLS) that is essential for centrosomal targeting.4 The CLS was first identified in cyclin E as a modular domain that is necessary for both the centrosomal localization of cyclin E and its acceleration of S phase entry.5 Cyclin A also contains a CLS, the expression of which displaces endogenous cyclins A and E from centrosomes.6 However, the mechanism by which the CLS mediates the localization of these proteins to the centrosome is unclear.
Dynein is a microtubule minus end-directed molecular motor comprising 4 subunit classes as follows: heavy chains (DHCs), intermediate chains, light intermediate chains (LICs), and light chains (LCs).7 The heavy chains carry 4 ATPase domains to support motor activity, and the LIC subunits are involved in the binding of intracellular cargos and are believed to contribute to the transport of intercellular cargos.8 The centrosomal proteins pericentrin and Par3 bind to LIC1 and LIC2, respectively.9,10 Dynein is involved in the positioning of the mitotic spindle and microtubule organizing centers with respect to the cell cortex.11
The centrosome is the main microtubule-nucleating organelle in animal cells. During progression from G1 to the S phase, the centrosome duplicates itself, resulting in cells with 2 centrosomes held together by a linker composed of centrosomal NINA-related kinase 2-associated protein 1 (C-Nap1) and rootletin.12,13 Before the onset of mitosis, C-Nap1 and rootletin are phosphorylated by Nek2 kinase,14 followed by the separation of centrosome pairs and the formation of spindle poles.15-17 After mitosis, daughter cells inherit a single centrosome. In this study, we analyzed the mechanism of BRCA2 transport to the centrosome and determined that BRCA2 associates with dynein through its CLS. Furthermore, we investigated the possibility that BRCA2 mediates cohesion between centrosomes during S phase.
Results
Analysis of CLS-binding proteins using mass spectrometry
To identify new CLS-binding partners of BRCA2, we subjected lysates prepared from HA-CLS-DsRed- or H A-DsRed-expressing HeLa S3 cells to immunoprecipitation using an anti-HA antibody. To avoid contamination of the final eluate with immunoglobulins, we used the crosslinker 1-[8-(2,5-dioxo-3-sulfopyrrolidin-1-yl) oxy-8-oxooctanoyl] oxy-2,5-dioxopyrolidine-3-sulfonic acid (BS3) to couple the antibodies to magnetic protein G Dynabeads. Immunoprecipitated protein complexes were digested by trypsin in solution and analyzed by tandem mass spectroscopy LC/MS/MS (QTRAP 5500 system). Analysis of the tryptic peptides revealed 58 CLS-associated proteins, which are listed in Fig. S1 and grouped according to a gene ontology database search. Two subunits of dynein, cytoplasmic DHC1 and cytoplasmic LIC2, were identified as candidate CLS-associated proteins (Fig. 1A).
Figure 1.
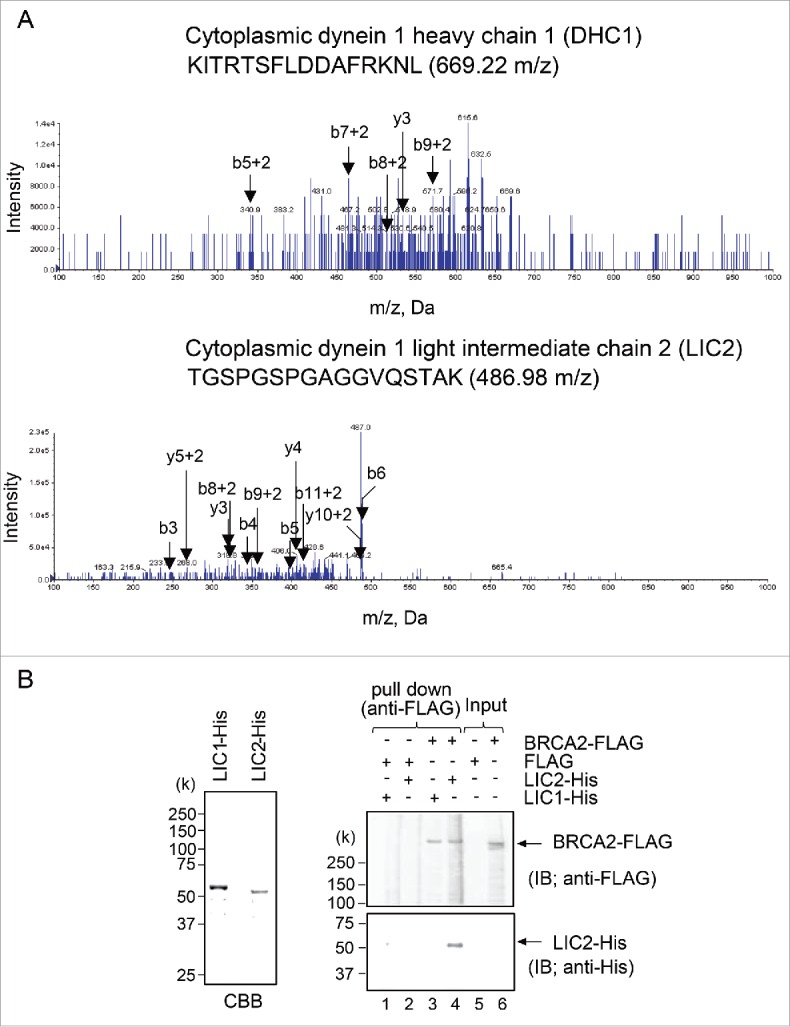
Identification of the BRCA2 centrosomal localization signal (CLS)-interacting protein. (A) MS/MS product ion spectra obtained by nano flow LC/MS/MS, showing cytoplasmic dynein 1 heavy chain 1 (DHC1) (upper) and cytoplasmic dynein 1 light intermediate chain 2 (LIC2) (lower), in immunoprecipitated protein complexes using anti-DsRed antibody from HA-CLS-DsRed-overexpressing HeLa S3 cells. The spectrum of the peptide clearly shows b- and y-ion fragments. (B) Pull-down assays of the ability of BRCA2-FLAG to bind the LIC1- and LIC2-His purified from E. coli. Bacterial lysate induced to express His fusions of LIC1 and LIC2 was resolved using SDS-PAGE and Coomassie brilliant blue-stained gels. FLAG-tagged full-length BRCA2 was bound to anti-FLAG M2 antibody agarose and incubated with His-tagged full-length LIC1 and LIC2.
Interaction of BRCA2 with cytoplasmic LIC2 at the centrosome
Cytoplasmic dynein transports various cargos through LIC as a component of the dynein complex. To directly demonstrate the BRCA2-binding ability of the LIC2 protein identified by LC/MS/MS, we determined whether BRCA2 associates with LIC2 via pull-down assays. The immobilized BRCA2-FLAG to anti-FLAG antibody-affinity resin was challenged with purified recombinant His-fused LIC1 or LIC2 (Fig. 1B, left panel), and bound protein was revealed using anti-His antibody. These data revealed a direct association between BRCA2 and LIC2, but not with LIC1 (Fig. 1B, right panel), even though human LIC1 and LIC2 are highly homologous. As controls, we confirmed FLAG alone was not bound to LIC2.
Next, to determine the association between both endogenous BRCA2 and LIC2, we searched for BRCA2-associated LIC2 by mass spectrometry. First, we isolated the centrosome fraction from HeLa S3 cells by sucrose gradient centrifuge, in which we confirmed the presence of BRCA2 by immunoblotting using anti-BRCA2 antibody (Fig. 2A, lane 7). The centrosome fraction was subjected to LC/MS/MS following immunoprecipitation with anti-BRCA2 antibody, and LIC2, DHC1, and BRCA2 were detected in the centrosome fraction (Fig. 2B). However, these proteins were not detected in the centrosome fraction immunoprecipitated with normal mouse IgG. These results illustrated that BRCA2 interacts with LIC2 through the BRCA2 CLS in dynein complexes at the centrosome.
Figure 2.
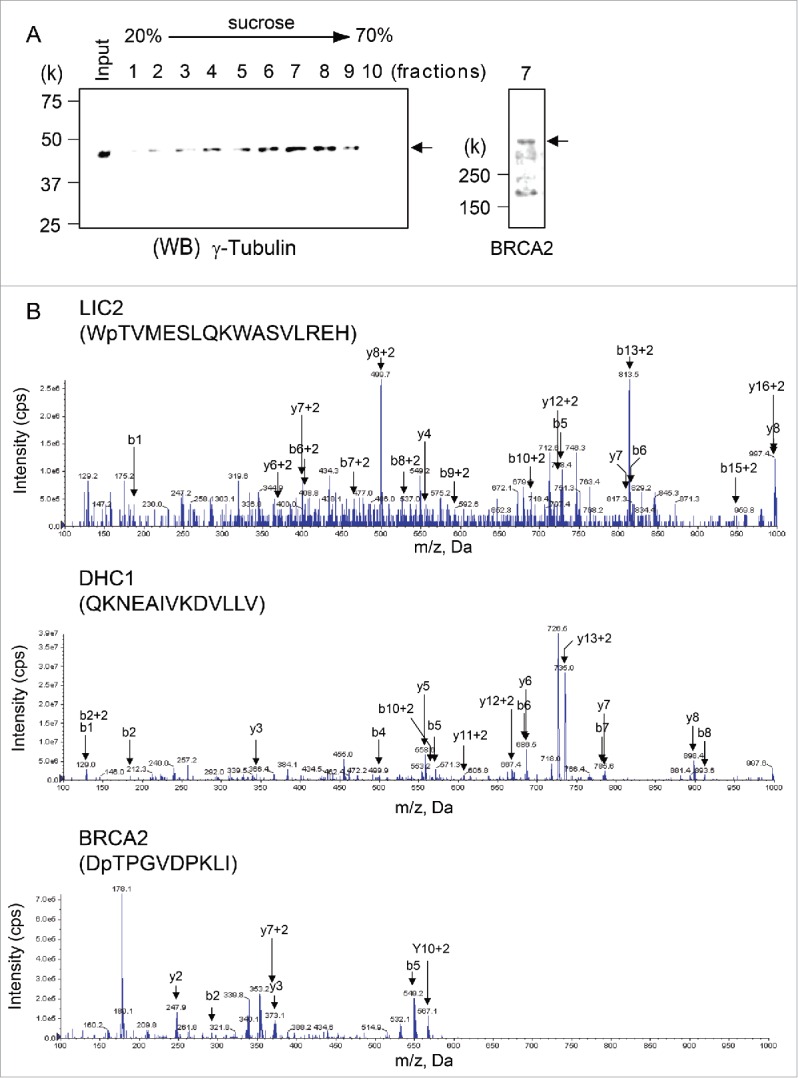
Dynein 1 heavy chain 1 (DHC1) and dynein 1 light intermediate chain 2 (LIC2) interacts with BRCA2 in centrosomes. (A) Centrosomes were isolated from HeLa S3 cells by sucrose density gradient centrifugation. The centrosome-containing fractions were analyzed by immunoblotting using the anti-γ-tubulin antibody. BRCA2 was detected in centrosome-containing fraction 7. (B) Anti-BRCA2 and anti-IgG immunoprecipitates were subjected to in-solution digestion and analyzed by liquid chromatography-tandem mass spectrometry. Mass spectra for the DHC1 peptide (AA 1440–1452), LIC2 peptide (AA 143–159), and BRCA2 peptide (AA 2606–2615) in anti-BRCA2 immunoprecipitates.
Expression of CLS dissociated the BRCA2-dynein complex
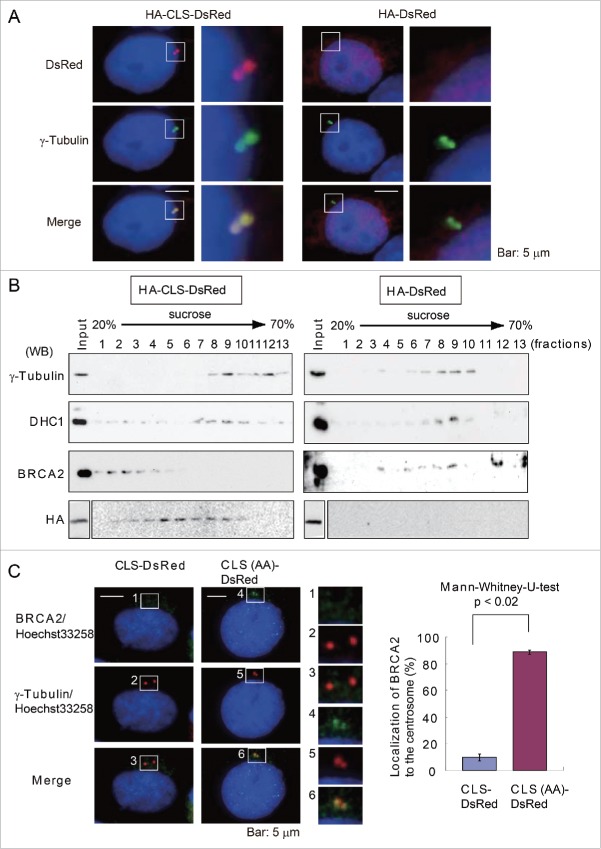
An analysis of HA-CLS-DsRed-transfected cells by immunofluorescence microscopy using anti-DsRed antibody revealed the HA-CLS-DsRed fusion protein localized to centrosomes (Fig. 3A). To validate our findings in HeLa cells, we also transfected human breast cancer MCF7 cells with HA-CLS-DsRed and observed protein localization to the centrosome (Fig. S3A). As described previously, we hypothesized that expression of the HA-CLS-DsRed protein would promote the release of endogenous BRCA2 from its interaction with dynein at centrosomes owing to competition for dynein binding by the CLS domain in the fusion protein. To test this hypothesis, we isolated centrosome-containing fractions from lysates prepared from HA-CLS-DsRed-transfected HeLa S3 cells by sucrose gradient centrifugation. Western blot analysis identified fractions containing HA-CLS-DsRed, γ-tubulin, and dynein, but failed to detect endogenous BRCA2 at the centrosome (Fig. 3B; left panel, fraction 9). However, we did detect BRCA2 in the tubulin-containing fractions from lysates prepared from the control HA-DsRed-transfected cells (Fig. 3B; right panel, fraction 9), suggesting the exogenous BRCA2 CLS-containing fusion protein prevented endogenous BRCA2 from binding to dynein, and BRCA2 failed to localize to the centrosome.
Figure 3.
Interaction between BRCA2 and dynein is disrupted by the overexpression of the BRCA2 centrosome localization signal (CLS) in the context of the HA-DsRed fusion protein. (A) Immunofluorescence microscopy of HA-CLS-DsRed and HA-DsRed overexpressing cells using anti-γ-tubulin (green) and anti-DsRed (red) antibody. The boxed areas are shown at higher magnification in the right panels. Nuclei were stained with Hoechst 33258. (B) Centrosome-containing fractions from a sucrose density gradient from the HA-CLS-DsRed-transfected (left panel) and HA-DsRed-transfected (right panel) HeLa S3 cell lysates were subjected to Western blot analysis using anti-γ-tubulin, anti-dynein, anti-BRCA2, and anti-HA antibodies. (C) Immunofluorescence microscopy of CLS-DsRed and CLS (AA: S2887A and R2888A)-DsRed-overexpressing cells using anti-γ-tubulin (red) and anti-BRCA2 (green) antibody. Magnifications of the areas indicated by the box are shown in the right panels. Quantification of BRCA2 localization to the centrosome in CLS-DsRed-transfected and CLS-DsRed- and CLS (AA)-DsRed-transfected cells. Endogenous BRCA2 in CLS-DsRed- and CLS (AA)-DsRed-transfected cells was immunostained with antibodies against BRCA2 and γ-tubulin. Nuclei were stained with Hoechst 33258. We then counted the number of cells in which endogenous BRCA2 was localized to the centrosome in the CLS-DsRed-transfected and CLS-DsRed- and CLS (AA)-DsRed-transfected cells.
We have previously reported that the localization of endogenous BRCA2 at the centrosome is disrupted in cells expressing a GFP-CLS fusion protein.4 In the current study, we have analyzed the effect of expressing the BRCA2 CLS in greater detail (Fig. 3C). The localization of endogenous BRCA2 to the centrosome in the CLS-DsRed- or CLS (AA: S2887A and R2888A)-DsRed-transfected cells, was assessed by immunofluorescence using antibodies against BRCA2 and γ-tubulin (Fig. 3C). Figure 3C shows the average number of cells in which the endogenous BRCA2 localized to the centrosome in 3 experiments, comparing CLS-DsRed (10.0 ± 2.6%, n = 293) and DsRed (88.7 ± 1.5%, n = 314). This analysis confirmed that the expression of the BRCA2 CLS in the context of an exogenous fusion protein inhibits the centrosomal localization of endogenous BRCA2.
Dynein is necessary for the centrosomal translocation of BRCA2
To test whether dynein transports BRCA2 to the centrosome, dynein expression was first depleted in HeLa S3 cells using 2 types of siRNA (DHC1-1, DHC1-2) (Fig. 4A), and the control and depleted cells were then subjected to immunofluorescence microscopy. In cells treated with the control siRNA, BRCA2 was observed to localize to the centrosomes, whereas in DHC1-depleted cells, BRCA2 was broadly distributed in the vicinity of the centrosome (Fig. 4B). The silencing of DHC1 reduced the localization of BRCA2 to the centrosomes (DHC1-1 siRNA, 14.3 ± 13.6%: n = 289 in 3 experiments; DHC1-2 siRNA, 47.1 ± 5.7%: n = 348 in 3 experiments), compared with control cells transfected with an siRNA of random sequence (control siRNA, 85.7 ± 10.7%: n = 353 in 3 experiments) or transfected without siRNA (non-transfected cells, 80.5 ± 3.5%: n = 365 in 3 experiments) (p < 0.05) (Fig. 4C). Furthermore, when HeLa S3 cells were treated with EHNA (an inhibitor of dynein) and labeled with anti-centrin 3 and anti-BRCA2 antibodies, BRCA2 was not localized to the centrosome (Fig. 4D). These findings suggest BRCA2 binds to dynein through their respective CLS domains and are thereby translocated to the centrosome. Dynein has also been implicated in cancer cell motility. However, a previous study has demonstrated that EHNA does not prevent the proliferation of cancer cells,18 therefore we did not examine cell motility in EHNA-treated cells in this study.
Figure 4.
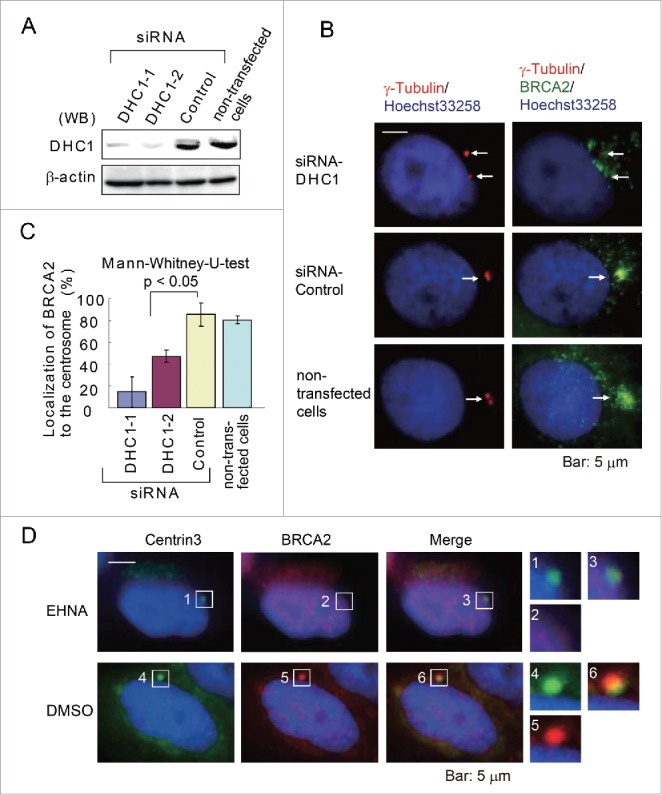
Dynein is necessary for the localization of BRCA2 to the centrosome. (A) Western blot analysis of lysates prepared from HeLa S3 cells transfected with siRNA targeting dynein 1 heavy chain 1 (DHC1) (DHC1-1 and DHC1-2 siRNA) or control siRNA or from non-transfected cells (siRNA-free) using anti-DHC1 and anti-β-actin antibodies. (B) Representative immunofluorescence microscopy of HeLa S3 cells treated as in (A) using anti-γ-tubulin (red) and anti-BRCA2 (green) antibodies. Nuclei were stained with Hoechst 33258. (C) Quantification of BRCA2 localization to the centrosome in DHC1 siRNA-transfected and transfection control cells. (D) Staining of control (DMSO-treated) and EHNA-treated HeLa S3 cells with antibodies against anti-centrin 3 (green) and anti-BRCA2 (red) antibodies. Nuclei were stained with Hoechst 33258. The boxed areas are shown at higher magnification in the right panels.
Depletion of BRCA2 increased centrosome separation
The centrosomes are observed as 2 closely-linked punctate signals in normal S phase cells. However, the separation of centrosomes was observed by immunofluorescence microscopy using anti-γ-tubulin antibody following the siRNA-mediated depletion of BRCA2 (Figs. 5A and B), or dynein (Fig. 4B) in HeLa S3 cells. To quantify this effect, we measured the distance between the centrosomes during S phase. γ-Tubulin was co-visualized with the S phase marker PCNA (Fig. 5B-F). The BRCA2 CLS exerted a dominant-negative effect on BRCA2 function by competing for binding to dynein, and the native BRCA2 could not be translocated to the centrosome (Fig. 3). We hypothesized the CLS overexpression results in a greater effect on centrosome linkage. To test this hypothesis, we transfected CLS-DsRed into HeLa S3 cells and measured during S phase the distance between the centrosomes, which were visualized via the immunofluorescence of γ-tubulin. As expected, the transient expression of CLS-DsRed caused a greater variation in the distance separating the centrosomes than was observed in cells transiently expressing the control DsRed (F-test, p < 0.01; Fig. 5C). To validate our findings in HeLa cells, this result was also confirmed in MCF7 cells (Fig. S3A). This analysis revealed that in each test siRNA-treated group (DHC1 and BRCA2 siRNA), there was greater variation in the distance between the centrosomes than in the control siRNA group (F-test, p < 0.01; Fig. 5D, supplementary Fig. S2). Treatment with BRCA2 siRNA also increased centrosome separation in MCF7 cells, compared with control siRNA-treated cells (Fig. S3B). The depletion of DHC1 or BRCA2 has been found to cause centrosome splitting during interphase, suggesting the normal function of BRCA2 has been disrupted in the centrosome. The centrosomal protein C-Nap1 is considered to play an important role in centrosome cohesion during interphase.19 This study illustrated C-Nap1 depletion by siRNA caused a variation in the distance between the centrosomes that was similar to that observed following the depletion of BRCA2 by siRNA (Fig. 5E, supplementary Fig. S2). To further investigate the role of endogenous BRCA2 and C-Nap1 in centrosome cohesion, siRNA was used to silence both BRCA2 and C-Nap1 expression in HeLa S3 cells. There was greater variation in the inter-centrosome distance following the depletion of BRCA2 and C-Nap1 than was observed following the depletion of BRCA2 alone (F-test, p < 0.01, Fig. 5F). Based on these results, we suggest the interaction between endogenous BRCA2 and dynein is inhibited by the overexpression of HA-CLS-DsRed and that the dislocation of BRCA2 caused by this inhibition is responsible for the change in inter-centrosome distances.
Figure 5.
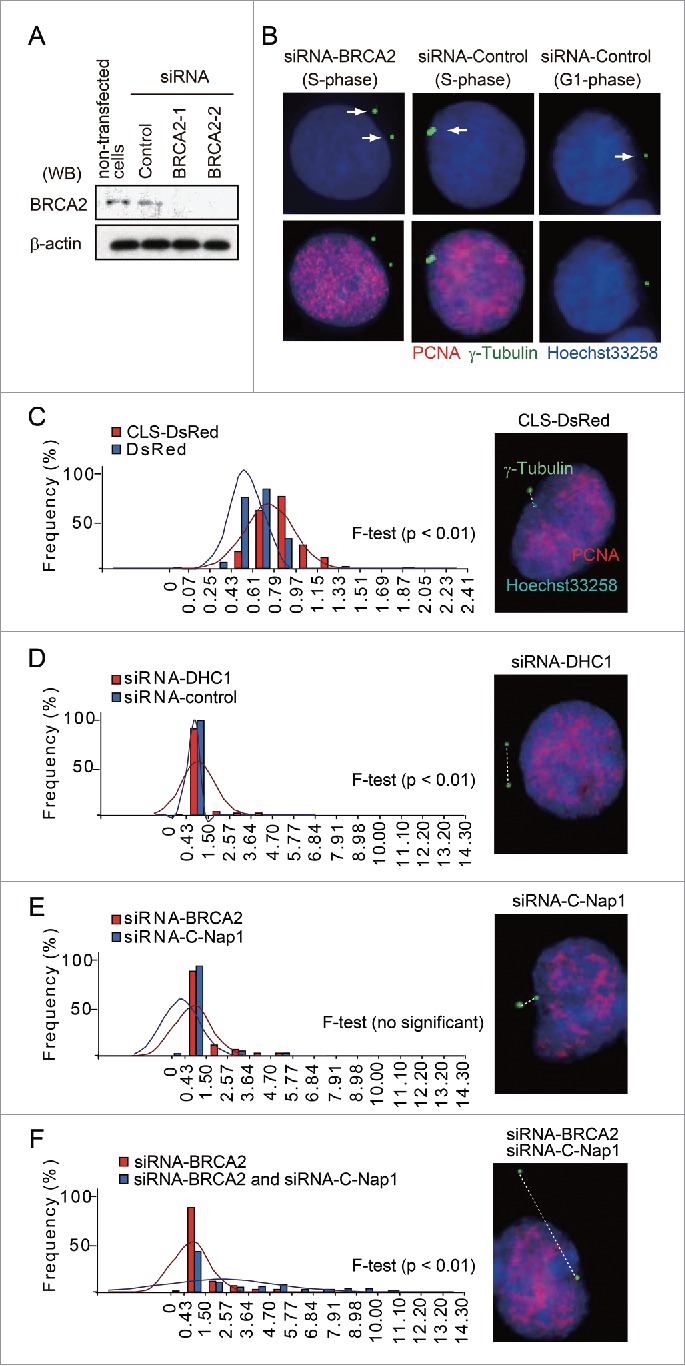
BRCA2 mediates centrosome cohesion during S phase in HeLa S3 cells. (A) Western blot analysis of lysates prepared from mock-transfected cells or cells transfected with control or BRCA2 siRNA using anti-BRCA2 and anti-β-actin antibodies. (B) Immunofluorescence microscopy of control siRNA- and BRCA2 siRNA-transfected HeLa S3 cells using anti-γ-tubulin (green) and anti-PCNA (red) antibodies. Nuclei were stained with Hoechst 33258. Centrosomes are indicated by arrows. (C) The minimum distance between the centers of the centrosomes was measured using Adobe Photoshop CS3 software. Distances were measured in CLS-DsRed- and DsRed-transfected cells (both n = 100; p = 0.01). (D) Distances were measured in DHC1 siRNA- and control siRNA-transfected cells (both n = 100; p = 0.01). (E) Distances were measured in BRCA2 siRNA- and C-Nap1 siRNA-transfected cells (both n = 100; no significant difference). (F) Distances were measured in cells following double depletion of BRCA2 and C-Nap1 by siRNA, or by depletion of BRCA2 alone (both n = 100; p = 0.01). (C-F) Centrosomes were stained with Hoechst 33258 (blue) and anti-γ-tubulin (green), PCNA (red) antibodies, and the measured distance between centrosomes is indicated by the white dotted line.
To investigate the possibility that the CLS-DsRed fragment disrupts interactions of other cellular proteins with dynein, thereby affecting centrosome function indirectly, we specifically mutated the S2887 and R2888 residues of the CLS, substituting them for alanine. We expressed the mutant BRCA2–FLAG protein (BRCA2-(CLS-AA)-FLAG) or a wild type BRCA2 FLAG protein following the siRNA-mediated depletion of endogenous BRCA2 in HeLa S3 and MCF7 cells. We confirmed expression of wild type and BRCA2-(CLS-AA)–FLAG by western blotting with FLAG antibody (Fig. 6A and Fig. S4A). The mutant protein did not localize to centrosomes and its expression increased the separation of the centrosomes during S phase. The transient expression of BRCA2-(CLS-AA)-FLAG increased the distance separating the centrosomes than was observed in cells expressing the wild type BRCA2-FLAG (F-test, p < 0.01; Fig. 6B and Fig. S4B). These findings suggest that the CLS fragment does not disrupt interactions of other cellular proteins with dynein.
Figure 6.
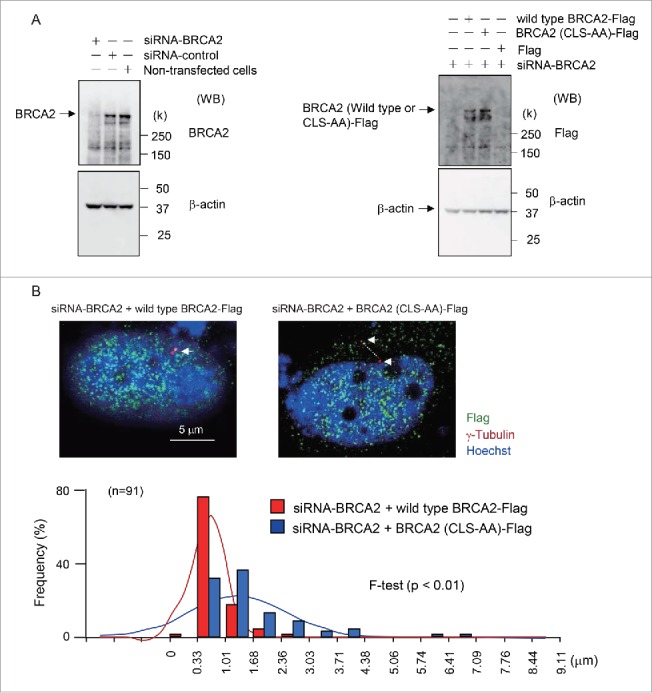
Expression of wild type BRCA2-FLAG and mutant BRCA2-(CLS-AA)-FLAG protein in endogenous BRCA2-knockdown HeLa S3 cells. (A) Western blot analysis showing BRAC2 and β-actin expression in cells transfected with control or BRCA2 siRNA (left panel). The expression of wild type BRCA2-FLAG and a mutant BRCA2–FLAG in which the S2887 and R2888 residues of the CLS were substituted with alanine (BRCA2-(CLS-AA)-FLAG) following siRNA-mediated depletion of endogenous BRCA2 in HeLa S3 cells (right panel). (B) Immunofluorescence microscopy of wild type BRCA2-FLAG- and BRCA2-(CLS-AA)-FLAG-transfected cells following BRCA2-knockdown using anti-γ-tubulin (red) and anti-FLAG (green) antibodies. Nuclei were stained with Hoechst 33258. Centrosomes are indicated by arrows. The minimum distance between the centers of the centrosomes was measured using Adobe Photoshop CS3 software. Distances were measured in wild type BRCA2-FLAG- and BRCA2-(CLS-AA)-FLAG-transfected cells (both n = 91; p < 0.01).
Effects of CLS on HeLa S3 cell-cycle regulation
To determine the physiological role of centrosome cohesion during S phase, we transfected HA-CLS-DsRed into HeLa S3 cells and measured cell-cycle progression using BrdU, a modified nucleoside incorporated during DNA synthesis. DNA content in nuclei was determined on the basis of the fluorescence intensity of 7-aminoactinomycin D (7-AAD), which intercalates into dsDNA. In total, 37.1% of cells expressing HA-DsRed exhibited BrdU incorporation (S phase) (G1: 48.1%, G2/M: 14.8%) (Fig. 6). By contrast, the expression of HA-CLS-DsRed induced G1 arrest (G1: 75.6%, S: 17.3, G2/M: 7.1%) (Fig. 7A). This result illustrates that the depletion of BRCA2 from the centrosome causes cell-cycle arrest during G1.
Figure 7.
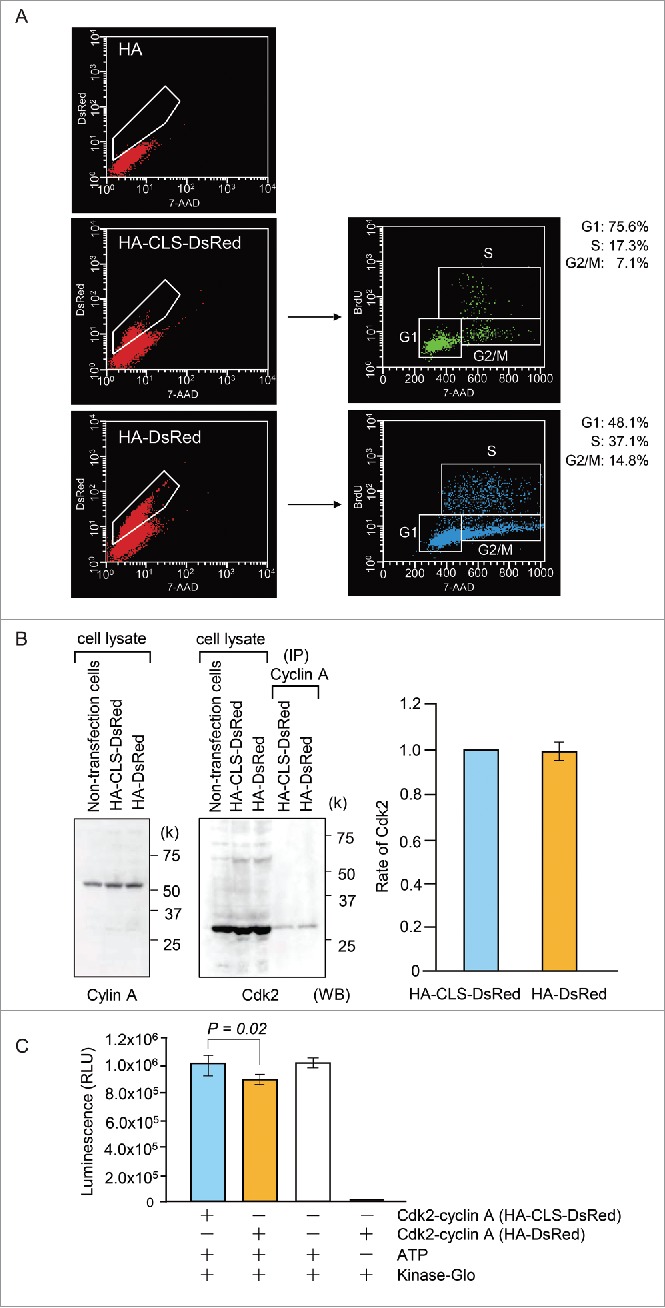
The effects of HA-CLS-DsRed on the cell cycle. (A) The cell-cycle phases (right panels) of HA-CLS-DsRed- and HA-DsRed-positive HeLa S3 cells (boxed areas: Left panels) were measured by BrdU incorporation into DNA using flow cytometry. (B) Western blot analysis showing cyclin A and Cdk2 expression in HA-CLS-DsRed- and HA-DsRed-transfected cells. Cell extracts were used for anti-cyclin A immunoprecipitations. The immunoprecipitates were immunoblotted and probed with anti-Cdk2 antibody to detect Cdk2 expression in Cdk2-cyclin A complexes from HA-CLS-DsRed- and HA-DsRed-transfected cells. Presented results are representative of 3 experiments. The histograms show quantification of Cdk2 expression. (C) Kinase assays of Cyclin A immunoprecipitations from cells transfected with HA-CLS-DsRed and HA-DsRed. The Cdk2-cyclin A activity was measured by Kinase-Glo Plus Luminescent Kinase Assay (Promega). Presented results are representative of 3 experiments. The histograms present quantified luminescence, which is related to kinase activity.
Reduced Cdk2-cyclin A activity was previously implicated in G1 arrest. Therefore, to further define the molecular mechanism of the G1 arrest, we examined the expression and activity of Cdk2-cyclinA complexes. These complexes were prepared by immunoprecipitation from HeLa S3 cells expressing HA-CLS-DsRed or HA-DsRed using an anti-cyclin A antibody. The level of Cdk2 in anti-cyclin A immunoprecipitates was measured by immunoblot analysis using an anti-Cdk2 antibody (Fig. 7B). Cdk2-cyclin A activity was measured using a Kinase-Glo Plus Luminescent Kinase Assay (Promega) (Fig. 7C). The activity but not the expression of Cdk2-cyclin A was diminished in HA-CLS-DsRed-expressing cells compared with HA-DsRed-expressing cells.
Discussion
Using several approaches to examine centrosome protein assembly in HeLa S3 cells, we demonstrated that BRCA2 is recruited to centrosomes via an interaction of its CLS domain with dynein (Fig. 1A). In this study, we uncovered the modular domain of BRCA2 associates with cytoplasmic DHC1 and cytoplasmic LIC2, both of which are components of cytoplasmic dynein. In this study, we showed that the centrosomal accumulation of BRCA2 was inhibited by interfering with the interaction between BRCA2 and dynein via the expression of HA-CLS-DsRed, depletion of dynein using siRNA, or via inactivation of dynein by EHNA.
We propose a model in which dynein binds to the CLS of BRCA2 and directly transports the latter to centrosomes (Fig. 8A). The current centrosome cohesion model proposes C-Nap1 and rootletin are a part of a linker structure that plays a role in the cohesion of duplicated centrioles during interphase.12 Thus, our study suggests BRCA2 also participates in centrosome cohesion during the cell cycle (Fig. 8B). We present several lines of evidence in support of this conclusion. First, expression of the dominant-negative HA-CLS-DsRed and CLS-DsRed inhibits the localization of BRCA2 at centrosomes and leads to separation of the centrosome pairs during interphase (Figs. 3B, C, 5C, and Fig. S3A). Second, the depletion of BRCA2 or dynein by siRNA induces centrosome splitting similar to that caused by C-Nap1 siRNA (Fig. 5D, E, and Fig. S3B). Thirdly, the double depletion of BRCA2 and C-Nap1 causes a larger dispersion between centrosomes than the silencing of C-Nap1 alone (Fig. 5F). Upon the initiation of mitosis, C-Nap1 is phosphorylated by Nek2A, which induces dissociation of the former from the centrioles, which in turn results in the separation of centrosomes.14 Previous co-immunoprecipitation studies using anti-BRCA2 and anti-γ-tubulin antibodies suggested BRCA2 interacts with γ-tubulin.4 BRCA2 localizes to the centrosomes during S phase and early M phase, but this centrosomal localization gradually decreases from prophase. We recently reported BRCA2 is cleaved into 2 fragments by membrane type-1 matrix metalloproteinase (MT1-MMP) during M-phase, and the silencing of MT1-MMP prevents the removal of BRCA2 from centrosomes.20 These results strongly suggest BRCA2 may be involved in centrosome cohesion and further suggest it mediates this effect by a mechanism distinct from that of C-Nap1 and rootletin.
Figure 8.
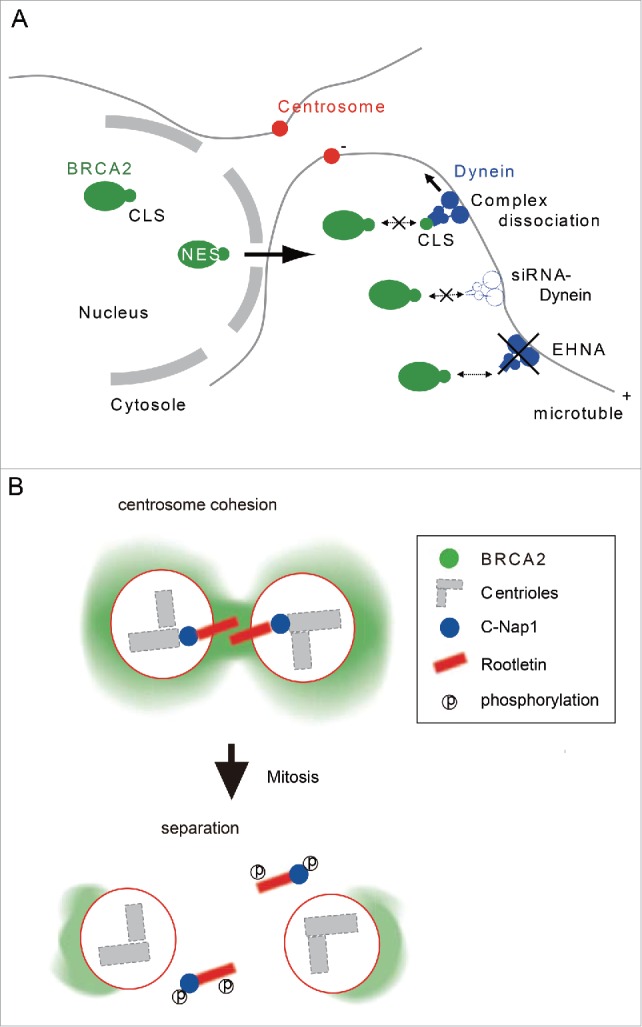
Model for BRCA2 localization to the centrosome through its centrosome localization signal. (A) BRCA2 interacts with dynein through its CLS and localizes to the centrosome. (B) BRCA2 is required for centrosome cohesion with C-Nap1 and rootletin during S phase.
In this study, we demonstrated dynein mediates the translocation of BRCA2 to the centrosomes. The regulation of centrosome proteins by dynein is likely to be important in the control of cell-cycle progression from G1 to S phase. The depletion of some centrosome proteins causes cell-cycle arrest during G1.21-23 In this regulatory pathway, it is reported that p53 activation in turn activates p21 and inhibits Cdk2.21 The transient expression of HA-CLS-DsRed caused cell-cycle arrest during G1 (Fig. 7). The centrosomal localization of endogenous BRCA2 was disrupted in cells expressing HA-CLS-DsRed, suggesting exogenous BRCA2 CLS can act in a dominant-negative manner (Fig. 3). Although the mechanism of inhibition of cell-cycle progression following the expression of CLS-DsRed remains unclear, centrosome separation during S phase might underlie cell-cycle arrest during G1. We observed that the activity but not the expression of Cdk2-cyclin A complexes was diminished by HA-CLS-DsRed. These findings suggested a regulatory mechanism involving Cdk inhibitory activity. We propose the existence of a cell-cycle checkpoint that monitors centrosome integrity during G1-S phase.
These results suggest BRCA2 may block the separation of centrosomes during S phase and further indicate the onset of mitosis may be caused by the separation of centrosomes. This study reveals new functions of BRCA2 at centrosomes, which should help elucidate the complex mechanism underlying the molecular basis for understanding centrosome function.
Materials and methods
Cell culture and transfection
HeLa S3 cells were purchased from RIKEN Cell Bank (Tsukuba, Japan), and MCF7 (human breast cancer cell line) cells were purchased from ATCC. All transfections were performed using Lipofectamine™ LTX with PLUS™ Reagent (Invitrogen), according to the manufacturer's instructions.
Antibodies and reagents
The following antibodies were used in this study: BRCA2 (H-300) rabbit polyclonal antibody (Santa Cruz); γ-tubulin (C-11) mouse mAb (Santa Cruz); centrin 3 (E-6) mouse mAb (Santa Cruz); γ-tubulin rabbit polyclonal antibody (Sigma-Aldrich); β-actin (MAB1501R) mouse mAb (Chemicon); PCNA mouse mAb (MBL); dynein HC (sc-9115) rabbit polyclonal antibody (Santa Cruz); DsRed rabbit polyclonal antibody (Clontech); cyclin E (ab2094) rabbit polyclonal antibody (Abcam); HA Tag (6E2) mouse mAb (Cell Signaling Technology); anti-HA tag (clone 3F10) rat mAb (Roche); DYNC1LI2 (D01) rabbit polyclonal antibody (Abnova); and centrin 3 (E-6) mouse mAb (Santa Cruz). Anti-FLAG M2 monoclonal antibody-affinity resin and EHNA (dynein inhibitor) was purchased from Sigma, and BS3 [bis (sulfosuccinimidyl) suberate] was purchased from Thermo-Fisher Scientific.
Sirna treatment
The siRNA oligonucleotides used for human BRCA2, DHC1, and C-Nap1, and the non-specific negative control, were purchased from Dharmacon.
Plasmids and constructs
Wild type and mutant CLS-DsRed constructs were created by annealing double-stranded oligonucleotides into the expression vector pDsRed-Monomer (Clontech). For the construction of wild type CLS-DsRed, the oligonucleotides
Five′-TATTTACCAGCACGTGCACTAACAAGACAGCAAGTTCGTGCTTTGCAAGATGGTGCAGAGA-3′ and
Five′-CTCTGCACCATCTTGCAAAGCACGAACTTGCTGTCTTGTTAGTGCACGTGCTGGTAAATAA-3′, were used. To construct mutant CLS (AA: S2887A and R2888A)-DsRed, the oligonucleotides
Five′-TATTTACCAGCAGCTGCACTAACAAGACAGCAAGTTCGTGCTTTGCAAGAT-GGTGCAGAGA-3′ and
Five′-CTCTGCACCATCTTGCAAAGCACGAACTTGCTGTCTTGTTAGTGCAGCTGCTGGTAAATAA-3′ were annealed and ligated into pDsRed-Monome. For the construction of HA-CLS-DsRed, the oligonucleotides
Five′-CCGGAATTCCTATTTACCATCACGTGCACTAACAAGACAGCAAGTTCGTGCTTTGCAAGATGGTGCAGAGTAACTCGAGTAT-3′ and
Five′-ATACTCGAGTTACTCTGCACCATCTTGCAAAGCACGAACTTGCTGTCTTGTTAGTGCACGTGATGGTAAATAGGAATTCCGG-3′, were annealed and ligated into pDsRed-Monome. All plasmids were verified by DNA sequencing.
DNA encoding LIC1 was amplified by PCR and cloned into the bacterial expression vector pET16b (Novagen). LIC2 amplified by PCR was cloned into pET16b at the NdeI-XhoI sites in which the NdeI site was converted to a blunt end following cleavage, and then the EcoRV site was substituted with the modified NdeI site by site-directed mutagenesis. To construct LIC1, the oligonucleotides
Five′- ACCCATATGGCGGCCGTGGGGCGAG-3′ and
Five′-TTGCTCGAGTCAAGAAGCTTCTCCTTCCG-3′, were used.
To construct LIC2, the oligonucleotides
Five′-TATCGATGGCGCCGGTGGGGGT-3′ and
5′-TAACTCGAGTCAGGCTTCATTTTCTGTTGAAG-3′, were used. The PCR products cloned into the pET16b vector were transformed into Escherichia coli BL21 (DE3) pLysS competent cells (Thermo-Fisher Scientific). The final PCR products were subjected to DNA sequencing to verify the desired sequence.
Immunoprecipitation crosslinking
To eliminate antibody contamination in precipitated proteins, HA antibody was crosslinked to Dynabeads protein G using the crosslinking agent BS3, per the manufacturer's instructions. Antibody-coated beads were incubated with the lysates under rotation for 2 h at 4 °C. Dynabeads coated with the antigen-antibody complex were washed extensively (4×) with the wash buffer supplied by the manufacturer. The proteins on Dynabeads were eluted using 50 mM glycine (pH 2.8).
Immunoprecipitation and protein gel blot analysis
For immunoprecipitation (IP), whole cell lysates were obtained by harvesting the cells in lysis buffer [20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 0.5 M EDTA, 0.1% NP40] containing protease inhibitors (completely EDTA-free; Roche). The extract was centrifuged at 15,000 rpm for 30 min to remove cell debris and BRCA2, dynein, and HA antibodies were added to the cell extract and incubated for 30 min at 4°C with rotation. Protein G-sepharose (20 µl of a 50% suspension) was added and incubated for 1 h at 4°C with rotation. The immunoprecipitates were then washed 5 times with lysis buffer and subjected to SDS-PAGE. For western blot analysis, the proteins were transferred electrophoretically onto a PVDF membrane (Millipore, Billerica, MD). The blots were probed with primary antibodies for 2 h at room temperature and then with secondary antibodies coupled with horseradish peroxidase (ECL anti-mouse or anti-rabbit IgG) for 1 h at room temperature. The blots were developed using the enhanced chemiluminescent reagent SuperSignal (Pierce, Rockford, IL) and exposed to Kodak X-OMAT film, according to the manufacturer's instructions.
Immunofluorescence microscopy
The cells were fixed with 3.7% formaldehyde in PBS for 10 min on ice and permeabilized sequentially with 50, 75, and 95% ethanol on ice for 5 min each. The slides were blocked with PBS-containing blocking solution for 30 min at room temperature and then incubated with primary antibody (BRCA2, DsRed, γ-tubulin, or centrin 3) for 1 h at room temperature. Slides were then washed 3 times with PBS for 5 min each and incubated with Alexa 488- or 594-conjugated secondary antibody (Molecular Probes) for 30 min at 37°C. After washing twice with PBS, DNA was stained with 1 μg/ml bis-benzimide (Hoechst 33258) in the final PBS wash. The samples were examined using an Olympus power BX51 fluorescence microscope.
Centrosome isolation
Centrosome isolation was performed as described previously (Cell Biology: A Laboratory Handbook) with the following modifications. Cells were washed with 50 ml of TBS, with this step repeated once with TBS (0.1%)-sucrose (8%) buffer (using half the volume used in the previous step). Cells were then resuspended in 1 ml of TBS (0.1%)-sucrose (8%) buffer. Lysis buffer (1 M HEPES, 1% NP40, 1 M MgCl2, 2-mercaptoethanol) containing protease inhibitors (completely EDTA-free), and the samples were centrifuged at 3500 rpm for 10 min. The lysate supernatant was filtered through a nylon mesh (100 µm nylon cell strainer; BD Falcon). Next, HEPES buffer (1 M) and DNase (1 mg/ml) were added to final concentrations of 10 mM and 2 units/ml (1 µg/ml), respectively, and the suspension was allowed to sit for 30 min. The lysate supernatant was overlaid onto 500 µl of 60% sucrose solution in a centrifuge tube and then ultracentrifuged at 7600 rpm for 30 min. A discontinuous sucrose gradient was prepared in a centrifuge tube containing, from the bottom 5, 3, and 3 ml of 70, 50, and 40% sucrose solutions, respectively. The obtained centrosomal suspension was suspended, and the centrifuge tube was filled with this sample. The gradient was then ultracentrifuged at 17,000 rpm for 1 h.
Bacterial expression of recombinant proteins
Recombinant overexpression of His-tagged LIC1 and LIC2 was performed in E. coli BL21 (DE3) physS-competent cells using the same protocol. Cells were grown in LB medium supplemented with 100 μg/ml ampicillin at 37°C up to an OD600 of 0.6–0.8. Protein expression was induced with 0.5 mM isopropyl β-d-thiogalactoside, and cultures were grown further at 37°C for 2 h. Harvesting of the cells was performed by centrifugation. For purification, cell pellets were resuspended in a lysis buffer containing 50 mM Tris-HCl pH 7.4, 400 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, 5% glycerol, and 1 mM DTT supplemented with 1 mg/ml lysozyme and 0.5 mM PMSF. Cells were lysed by sonication. Lysates were clarified by centrifugation, and supernatants were applied to a Dynabeads His-Tag Isolation & Pulldown (Veritas), equilibrated in lysis buffer. Elution was performed using 200 mM imidazole. Eluted fractions were dialyzed against sample buffer (3.25 mM sodium phosphate buffer pH 7.4, 70 mM NaCl, and 0.01% Tween 20) for imidazole removal.
GST pull-down assay
BRCA2-FLAG and FLAG were independently applied to 100 µl of anti-FLAG M2 affinity gel (Sigma). The resin was then washed 4 times with sample buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl). His-tagged LIC1 and LIC2 were applied to the anti-FLAG M2 affinity gel binding of BRCA2-FLAG or FLAG. The resin was then washed 4 times with sample buffer, and bound protein was eluted with 1 mM FLAG peptide (Sigma). Eluted fractions were dialyzed against the sample buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl) for FLAG peptide removal, and visualized via Coomassie blue staining.
Cell cycle analysis
A BrdU-APC kit (Becton Dickinson) was used for cell-cycle analysis. HA-CLS-DsRed- and HA-DsRed-transfected and non-transfected HeLa S3 cells were treated with 100 mg/ml BrdU (Sigma) for 2 h, and cells were fixed and permeabilized. Cells were stained with anti-BrdU-APC (Becton Dickinson) and FITC-HA (MBL) antibodies, and 7-AAD (Becton Dickinson). Flow cytometric detection was performed using a FACSCalibur cytometer (Becton Dickinson).
Kinase assay
Affinity-purified cyclin A antibody was linked to Dynabeads (life technologies) and incubated with cell lysates (5 mg/ml) for 10 min before being washed 3 times in buffer. Precipitated proteins were resuspended in 20 μl elution buffer (50 mM Glycine at pH 2.8) for immunoblot analysis and kinase assay. Immunoprecipitates were dissolved in 10 μl kinase buffer (25 mM MOPS, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, phosphate inhibitor Cocktail Set III (Calbiochem) at pH 7.2) with 10 mM DTT and 100 μM ATP. Each sample was incubated with 5 μg histone H1 (Signalchem) in a final volume of 50 μl for 15 min at 30°C. 50 μl kinase reaction was incubated with 50 μl Kinase-Glo Plus reagent (Promega) for a total volume of 100 μl for 10 min at room temperature, and then luminescence was recorded using an EnSpire (PerkinElmer).
Mass spectrometric identification
MS analysis was performed via LC-MS/MS (QTRAP 5500 system). Peak lists for protein database searches (ProteinPilot; Applied Biosystems, CA, USA) were extracted from the resulting data, and the peptide enrichment ratio was calculated to evaluate the certainty in peptide identification.
Supplementary Material
Abbreviations
- BRCA2
breast cancer susceptibility gene 2
- EHNA
erythro-9-[3-(2-Hydroxynonyl) Jadenine
- PCNA
proliferating cell nuclear antigen
- BrdU
bromodeoxyuridine.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all members of the Department of Molecular Genetics of the Tokyo Medical and Dental University, which provided helpful discussion.
Funding
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology in Japan.
References
- [1].Marmorstein LY, Ouchi T, Aaronson SA. The BRCA2 gene product functionally interacts with p53 and RAD51. Proc Natl Acad Sci U S A 1998; 95:13869-74; PMID:9811893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Niwa T, Saito H, Imajoh-ohmi S, Kaminishi M, Seto Y, Miki Y, Nakanishi A. BRCA2 interacts with the cytoskeletal linker protein plectin to form a complex controlling centrosome localization. Cancer Sci 2009; 100:2115-25; PMID:19709076; http://dx.doi.org/ 10.1111/j.1349-7006.2009.01282.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang HF, Takenaka K, Nakanishi A, Miki Y. BRCA2 and nucleophosmin coregulate centrosome amplification and form a complex with the rho effector kinase ROCK2. Cancer Res 2011; 71:68-77; PMID:21084279; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-0030 [DOI] [PubMed] [Google Scholar]
- [4].Nakanishi A, Han X, Saito H, Taguchi K, Ohta Y, Imajoh-Ohmi S, Miki Y. Interference with BRCA2, which localizes to the centrosome during S and early M phase, leads to abnormal nuclear division. Biochem Biophys Res Commun 2007; 355:34-40; PMID:17286961; http://dx.doi.org/ 10.1016/j.bbrc.2007.01.100 [DOI] [PubMed] [Google Scholar]
- [5].Matsumoto Y, Maller JL. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science 2004; 306:885-8; PMID:15514162; http://dx.doi.org/306/5697/885. [DOI] [PubMed] [Google Scholar]
- [6].Pascreau G, Eckerdt F, Churchill ME, Maller JL. Discovery of a distinct domain in cyclin A sufficient for centrosomal localization independently of cdk binding. Proc Natl Acad Sci U S A 2010; 107:2932-7; PMID:20133761; http://dx.doi.org/ 10.1073/pnas.0914874107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tynan SH, Gee MA, Vallee RB. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem 2000; 275:32769-74; PMID:10893223; http://dx.doi.org/ 10.1074/jbc.M001537200 [DOI] [PubMed] [Google Scholar]
- [8].Schroeder CM, Ostrem JM, Hertz NT, Vale RD. A ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. Elife 2014; 3:e03351; PMID:25272277; http://dx.doi.org/ 10.7554/eLife.03351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tynan SH, Purohit A, Doxsey SJ, Vallee RB. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J Biol Chem 2000; 275:32763-8; PMID:10893222; http://dx.doi.org/ 10.1074/jbc.M001536200 [DOI] [PubMed] [Google Scholar]
- [10].Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol 2009; 19:1065-74; PMID:19540120; http://dx.doi.org/ 10.1016/j.cub.2009.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Karki S, Holzbaur EL. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol 1999; 11:45-53; PMID:10047518; http://dx.doi.org/S0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- [12].Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol 2000; 151:837-46; PMID:11076968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell 2006; 17:1033-40; PMID:16339073; http://dx.doi.org/ 10.1091/mbc.E05-10-0943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol 1998; 141:1563-74; PMID:9647649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mayor T, Hacker U, Stierhof YD, Nigg EA. The mechanism regulating the dissociation of the centrosomal protein C-Nap1 from mitotic spindle poles. J Cell Sci 2002; 115:3275-84; PMID:12140259 [DOI] [PubMed] [Google Scholar]
- [16].Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol 2005; 171:27-33; PMID:16203858; http://dx.doi.org/ 10.1083/jcb.200504107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. Components of the hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol 2010; 12:1166-76; PMID:21076410; http://dx.doi.org/ 10.1038/ncb2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kim D, You E, Rhee S. Dynein regulates cell migration depending on substrate rigidity. Int J Mol Med 2012; 29(3):440-6; PMID:22200735; http://dx.doi.org/ 10.3892/ijmm.2011.867 [DOI] [PubMed] [Google Scholar]
- [19].Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell 2006; 17:1033-40; PMID:16339073; http://dx.doi.org/ 10.1091/mbc.E05-10-0943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wali N, Hosokawa K, Malik S, Saito H, Miyaguchi K, Imajoh-Ohmi S, Miki Y, Nakanishi A. Centrosomal BRCA2 is a target protein of membrane type-1 matrix metalloproteinase (MT1-MMP). Biochem Biophys Res Commun 2014; 443:1148-54; PMID:24384087; http://dx.doi.org/ 10.1016/j.bbrc.2013.12.103 [DOI] [PubMed] [Google Scholar]
- [21].Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol 2007; 9:160-70; PMID:17330329 [DOI] [PubMed] [Google Scholar]
- [22].Song L, Dai T, Xiong H, Lin C, Lin H, Shi T, Li J. Inhibition of centriole duplication by centrobin depletion leads to p38-p53 mediated cell-cycle arrest. Cell Signal 2010; 22:857-64; PMID:20085806; http://dx.doi.org/ 10.1016/j.cellsig.2010.01.009 [DOI] [PubMed] [Google Scholar]
- [23].Srsen V, Gnadt N, Dammermann A, Merdes A. Inhibition of centrosome protein assembly leads to p53-dependent exit from the cell cycle. J Cell Biol 2006; 174:625-30; PMID:16943179; http://dx.doi.org/ 10.1083/jcb.200606051 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.