ABSTRACT
The Long Interspersed Element 1 (LINE1 or L1) ORF2 protein (ORF2p) can cause DNA damage through the activity of its endonuclease domain (EN). The DNA double-strand breaks (DSB) introduced by the ORF2p EN have the potential to be mutagenic. Previously, our lab has shown that ORF2p fragments containing the EN domain could be expressed in mammalian cells and have variable cytotoxicity. Inclusion of the ORF2p sequence C-terminal to the EN domain in these fragments both reduced the cytotoxicity of these fragments and increased their presence in the nucleus as detected by Western blot analysis. Here, we identify the amino acids (aa 270–274) in the newly-identified ORF2p Cryptic region (Cry) that may be important to the subcellular localization and cytotoxic potential of these EN-containing ORF2p fragments.
KEYWORDS: DNA damage, endonuclease, L1, ORF2, retrotransposition
Introduction
Long Interspersed Element 1 (LINE1 or L1) is the only currently active autonomous retroelement in the human genome.1 L1 is composed of a 5′ Untranslated Region (UTR) with internal PolII promoter, 2 Open Reading Frames (ORFs), a 3′ UTR, and PolyA signal. ORF1 codes for the ORF1 protein (ORF1p), the main structural protein of the retrotransposition intermediate L1 ribonucleoprotein particle (RNP).2,3 ORF2 codes for the ORF2p, which contains the enzymatic machinery necessary for successful L1 mobilization.1,4,5 ORF2p contains 2 enzymatic activities, an N-terminal endonuclease (EN) and a central reverse transcriptase (RT), both of which are required to drive successful L1 retrotransposition.
L1 via the activity of the ORF2p has been shown to cause DNA double-strand breaks (DSBs) in an EN-dependent manner.6-8 The expression of L1s with stop codons in the ORF2p can lead to the expression of ORF2p fragments containing the EN domain.7 These fragments have the potential to be cytotoxic to mammalian cells.7 We have recently demonstrated that the EN domain of the ORF2p is highly cytotoxic to mammalian cells when expressed alone, and that expression of EN-containing truncated ORF2 fragments is possible in vivo.7 Addition of ORF2p sequence C-terminal to the EN domain, recently termed the Cryptic region,9 reduced the cytotoxicity associated with these ORF2p fragments compared to the EN domain alone.7 Interestingly, this reduction in cytotoxicity was accompanied by an increase in fragment localization to the nucleus.7 In short, the more toxic, smaller EN-containing fragment was detected at similar levels in both the cytoplasm and nucleus, while the longer EN-containing fragment that possessed ORF2p Cryptic sequence were less cytotoxic and largely nuclear.7 The same phenomenon was observed with other EN-containing fragments of various lengths. The Cryptic area of the ORF2p contains amino acids important for retrotransposition.9 We hypothesized that this area of the ORF2p also contains the amino acid sequence that influences the subcellular localization of the ORF2p fragments and the different cytotoxic potential of these fragments.
Results
In addition to previously described ORF2 fragments (EN239Δ, EN264Δ, EN289Δ, EN314Δ, EN347Δ, and ENZ490Δ),9 we generated Gal4-tagged and VP16-tagged EN-containing ORF2 fragment expression constructs (Fig. 1: EN269Δ, EN274Δ, EN279Δ, and EN330Δ). The cytotoxic potential of these EN-containing ORF2 fragments was assessed using the previously described colonogenic cytotoxicity assay involving expression of EN-containing ORF2 fragments. In chronic toxicity assays, the antibiotic selection gene (either hygromycinr or neomycinr) is supplied in cis of the ORF2 fragment expression gene on the same transfected plasmid. Selection with antibiotic also selects for chronic expression of the ORF2p fragment with more toxic EN-containing ORF2 fragments, yielding fewer colonies. For acute toxicity assays, the antibiotic selection gene (neomycinr) is supplied in trans of the ORF2 fragment expression gene on a different plasmid cotransfected with the EN-containing ORF2p fragment expression plasmid. Selected for with antibiotics does not select for the maintenance of the EN fragment expression plasmid. Therefore, this approach measures toxicity resulting from transient expression of specific EN-containing ORF2 fragments. Following transfection into HeLa cells and chronic expression, EN239Δ, EN264Δ, EN269Δ, EN274Δ, and EN279Δ VP16-tagged ORF2 fragments were observed to be cytotoxic relative to empty vector control (Fig. 2A: Chronic Toxicity). Similarly, following transient transfection into HeLa cells, EN239Δ, EN264Δ, and EN269Δ Gal4-tagged ORF2p fragments were cytotoxic relative to empty vector control (Fig. 2A: Acute Toxicity). It is of note here that the chronic expression of EN-containing constructs is consistently more cytotoxic than acute expression of constructs,7 though in this case the influence of the different tags cannot be ruled out. Interestingly, EN-containing fragment chronic toxicity potential appeared group with one another in clusters rather than be reduced linearly with the inclusion of ORF2 Cryptic sequence C-terminal to the EN domain. In the chronic toxicity assay, EN264Δ and EN269Δ are not statistically different from one another, while EN274Δ and EN289Δ are not statistically different from one another. However, EN269Δ and EN274Δ are statistically different from one another, with EN269Δ being more cytotoxic (Fig. 2B). In the acute toxicity assay, EN239Δ, EN264Δ, and EN269Δ are not statistically different from one another, while EN274Δ to EN347Δ are not statistically different from one another (Fig. 2C) as they are not cytotoxic relative to control (Fig. 2A: Acute Toxicity). However, EN269Δ and EN274Δ are statistically different from one another, with EN269Δ being more cytotoxic (Fig. 2C). The inclusion of amino acids 270–274 appeared to be responsible for this decrease in cytotoxicity and grouping in both chronic and acute toxicity experiments.
Figure 1.
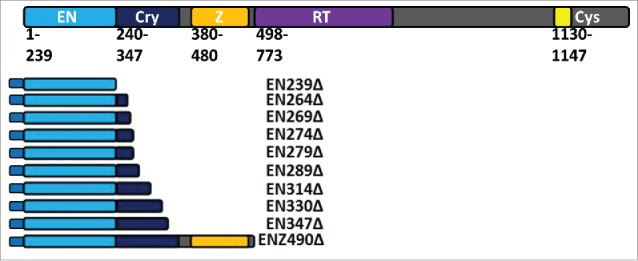
Schematic of ORF2 and tagged ORF2 fragments. The ORF2p molecule has multiple annotated domains important for retrotransposition. These include the enzymatically necessary endonuclease (EN: light blue) and reverse transcriptase (RT: purple) domains. Between the EN and RT domains is the Cryptic region (Cry: dark blue) and Z domain (Z: orange). Both contain amino acids essential to retrotransposition and ORF2p function. At the C-terminal end of the ORF2p is a cysteine-rich domain (Cys: yellow) that also contains amino acids essential to retrotransposition. EN-containing ORF2 fragments detailed here were generated in 2 formats: one with an N-terminal Gal4 tag and one with an N-terminal VP16 tag. VP16-tagged expression plasmids also contain a neomycin resistance gene. EN239Δ, EN269Δ, EN274Δ, EN289Δ, EN347Δ, and ENZ490Δ were also generated in an untagged format with a Hygromycin resistance gene in the expression plasmid. As described in materials and methods, reported domains are used as the body of the name, followed by the number corresponding to the terminal amino acid as it would be in the full length ORF2p, with the truncated ORF2p sequence denoted by a Δ.
Figure 2.
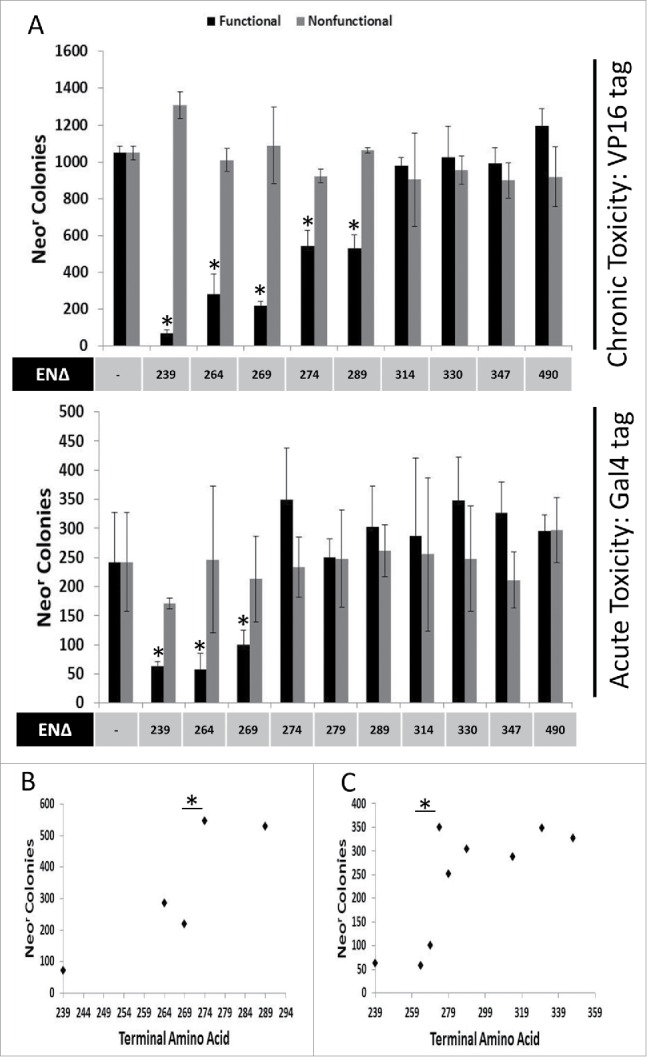
Chronic and Acute Toxicity of EN-containing ORF2 fragments. VP16-tagged and Gal4-tagged EN-containing ORF2 fragments are toxic to HeLa cells. (A) Chronic and Acute toxicity assays of EN-containing ORF2 fragments in HeLa cells. Amino acid that ORF2 fragment terminates in (ENΔ) denoted below graph bars for both functional (black) and nonfunctional (gray) EN-containing ORF2 fragments. Nonfunctional EN-containing fragments have catalytically necessary amino acids mutated in the EN domain (D205A, H230A). Error bars denote standard deviation (n = 3). Statistical significance was assessed using Student's t-test (*p < 0.05). (B) Grouping of chronic toxicity. Colony counts for EN264Δ and EN269Δ are not significantly different from one another. Colony counts for EN274Δ and EN279Δ are not significantly different from one another. Colony counts for EN264Δ and EN269Δ are significantly different from colony counts for EN274Δ and EN279Δ. Statistical significance was assessed using Student's t-test (*p < 0.05). (C) Grouping of acute toxicity. Colony counts for EN239Δ, EN264Δ and EN269Δ are not significantly different from one another. Colony counts for EN274Δ, EN279Δ, EN289Δ, EN314Δ, EN330Δ, EN347Δ, and ENZ490Δ are not significantly different from one another. Colony counts for EN239Δ, EN264Δ and EN269Δ significantly different from colony counts for EN274Δ, EN279Δ, EN289Δ, EN314Δ, EN330Δ, EN347Δ, and ENZ490Δ. Statistical significance was assessed using Student's t-test (*p < 0.05).
Previously, we had observed that longer, less-toxic EN-containing ORF2 fragments favored nuclear localization, while the minimal highly cytotoxic EN fragment was detected in both the cytoplasm and nucleus.7 We hypothesized that the same amino acids that are responsible for the transition from more cytotoxic to less cytotoxic EN-containing ORF2 fragments could also be responsible for this difference in subcellular localization between various fragments. Constructs designed to express Gal4-tagged EN-containing ORF2p fragment were transiently transfected into HeLa cells that were then subjected to Western blot analysis using commercially available Gal4 antibodies (Fig. 3). Gal4-tagged fragments were used because they generated a single, unambiguous protein band when analyzed via Western blot, unlike untagged ORF2p fragments detected with ORF2p antibodies.7 Both whole cell lysate and lysates from cytoplasm/nucleus-fractionated cells were analyzed. While Gal4-tagged EN-containing ORF2p fragments were expressed at similar levels in whole cell lysate, there was a shift in subcellular localization for EN274Δ, EN289Δ, EN314Δ, and EN330Δ when nuclear and cytoplasmic fractions were analyzed separately. Together, these results demonstrate that the inclusion of amino acids 270–274 promotes the nuclear localization of the Gal4-tagged EN-containing ORF2p fragments which also coincides with the change in cytotoxic potential of the VP16- and Gal4-tagged ORF2p fragments.
Figure 3.

Subcellular distribution of Gal4-tagged EN-containing ORF2 fragments. Western blot analysis of EN-containing ORF2 fragments. Indicated Gal4-tagged EN-containing constructs were transiently transfected into HeLa cells and subjected to Western blot analysis. Both whole cell lysate and cytoplasmic and nuclear cell fractions were analyzed using commercially available anti-Gal4 antibodies, yielding a single band in each sample lane corresponding to the expected size of each ORF2p fragment. Control is cells transfected with empty vector. Cell fractions are indicated on the right. Molecular weight markers are indicated on the left. GAPDH (total cell lysate and cytoplasmic fraction) and Lamin A/C (L A/C: nuclear fraction) were used as loading controls.
We next wanted to confirm our results concerning the tagged ORF2 fragments using corresponding untagged ORF2 fragments. Previously, our lab had shown that endogenous expression of truncated ORF2 fragments is possible.7 In addition to previously characterized ORF2 fragment expression plasmids (EN239Δ and ENZ490Δ), we generated expression plasmids corresponding to the key ORF2 fragments from Figs. 2 and 3 (EN269Δ, EN274Δ, EN289Δ, and EN347Δ) (Fig. 4A). All EN-containing ORF2 fragments were cytotoxic in HeLa cells compared to their nonfunctional controls (Fig. 4B). The same grouping of the level of cytotoxicity observed with Gal4-tagged fragments in Figure 2B and 2C was also detected with the untagged ORF2 fragments (Fig. 4C). Western blot analysis using monoclonal ORF2p antibodies10 confirmed that the inclusion of amino acids 270 through 274 in the EN-containing fragments led to a shift in their relative distribution (Fig. 4D), as demonstrated by quantitation of relative protein amounts of EN269Δ and EN274Δ in the cytoplasmic and nuclear fractions (Fig. 4E). These data show that amino acids 270 and 274 are important for EN-containing ORF2 fragment cytotoxicity and subcellular distribution, albeit through an unknown mechanism.
Figure 4.
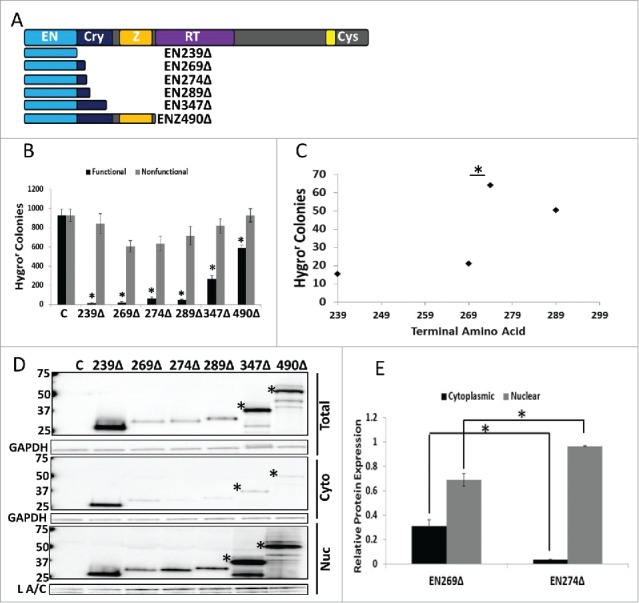
Analysis of Untagged EN-containing ORF2 Fragments: Selected untagged EN-containing ORF2 fragments behaved similarly to the corresponding VP16- and Gal4-tagged EN-containing ORF2 fragments in regards to cytotoxicity and subcellular distribution. (A) Schematic of untagged ORF2 fragments used in this experiment with ORF2 displayed above for reference. (B) Chronic toxicity of EN-containing ORF2 fragments. All ORF2 fragments were cytotoxic as compared to their nonfunctional controls. Error bars denote standard deviation (n = 3). Statistical significance assessed using Student's t-test (*p < 0.05). (C) Analysis of toxicity grouping of EN-containing ORF2 fragments. Analysis is identical to Figure 2B and 2C (*p < 0.05). (D) Western blot analysis of HeLa cells transiently transfected with indicated constructs using anti-ORF2p monoclonal antibodies. Both whole cell lysate and cytoplasmic and nuclear cell fractions were analyzed. Highest molecular weight band in each sample corresponds to expected size of each construct, with larger EN-containing fragments undergoing processing to yield smaller products (*: EN347Δ and ENZ490Δ). Control (C) is cells transfected with empty vector. Cellular fraction is indicated on the right. Molecular weights are indicated on the left. GAPDH (total and cytoplasmic (cyto)) and Lamin A/C (L A/C: nuclear (nuc)) used as loading control. Relative distribution in subcellular compartments of EN269Δ and EN274Δ is quantified in (E). (E) Quantitation of relative distribution of EN-containing ORF2 fragments. Signal intensity of EN-containing ORF2p fragments in each compartment was normalized to loading control. Normalized signal was summed for each cellular compartment and proportion protein expression quantitated per compartment (Example for Cytoplasmic: Normalized Cytoplasmic Signal/(Normalized Cytoplasmic Signal+Normalized Nuclear Signal)). Error bars denote standard deviation (n = 3). Statistical significance was assessed using Student's t-test (*p < 0.05).
Discussion
While much study has been given to the impact of retrotransposons and retrotransposition as a gross process on human health (Reviewed in refs.11,12), many of the specifics of the L1 replication cycle and ORF2p biology remain poorly understood. Our lab has previously demonstrated that EN-containing ORF2 fragments can be cytotoxic and that fragments containing different amounts of the ORF2 Cryptic sequence display different patterns of subcellular localization.7 Somewhat counter-intuitively, longer EN-containing fragments that are predominately nuclear have a reduced capacity to induce DNA DSBs and cause cytotoxicity, while the smallest protein fragment containing the minimal EN domain is detected in both cytoplasmic and nuclear fractions and is highly cytotoxic.7 The area of the ORF2p that may be involved in this phenomenon (between the EN and Z domains) was previously unannotated and served no known function. However, our lab has recently discovered that this region (called Cryptic) of the ORF2p has several amino acids and putative motifs important to retrotransposition.9 Our data presented here show that there are amino acids in the Cryptic region of the ORF2p molecule that may be modulating EN function. It may be the case that Cryptic, in addition to harboring amino acids necessary for RT function, possesses amino acids that affect EN activity. Interestingly, the amino acids identified here (aa 270–274) within the Cryptic region of the ORF2p overlap with a putative PCNA binding site that has been shown to be important for Alu retrotransposition (FF 273–274).9 Our data suggest that it is possible that PCNA may play a role in the localization or nuclear retention of these longer fragments. We speculate that fragments containing the putative PCNA binding site may remain anchored to chromatin, which could reduce their cytotoxic potential by tethering them to the chromatin in a single location. This scenario would also explain their increased presence in the nucleus: the fragments bound to PCNA would be retained in the nucleus on the chromatin. Additionally, PCNA is known to be involved in DNA repair processes,13-17 and L1 can be inhibited by DNA repair proteins.18-20 We speculate that the EN-containing fragment could be brought into close proximity to DNA repair proteins via PCNA interaction, thereby limiting EN-containing ORF2p fragment-mediated cytotoxicity. However, the inverse is also logically possible, as PCNA could theoretically make the EN fragments in question more toxic by keeping them in close proximity to the host DNA. Alternatively, it is possible that the inclusion of these amino acids facilitates ORF2p fragment binding to another regulatory factor, or that these amino acids themselves are somehow directly responsible for modulating the EN activity within the fragments. All of these possibilities are highly speculative and require further experimental interrogation.
How the L1 RNP and the ORF2p molecule localize to the nucleus has been a subject of intense debate within the field, often centering on whether this process is active or a result of the dissolution of the nuclear envelope in actively cycling cells.21-24 Previously, our lab mutated predicted nuclear localization signals within the Z domain with no effect on toxicity or subcellular localization of longer EN-containing ORF2p fragments.7 Here, we empirically identify one area that may be involved in nuclear localization of said fragments. However, the importance of these amino acids to the full-length ORF2p may be different than their importance in the context of the EN-containing ORF2 fragments. As the ORF2p is a large and enzymatically complex molecule, it is possible that it has redundant mechanisms for nuclear localization. Indeed, prior to the identification of the Cryptic region, over 50% of the 1275 amino acids that comprise the human ORF2p have no known function. Of the 1275 amino acids that comprise the ORF2p molecule, only 631 are contained within the annotated domains (EN, Z, RT, and Cys). It is possible that some of this sequence has multiple, redundant nuclear localization signals. Further studies using the full-length ORF2 and L1 will be needed to fully interrogate the importance of these amino acids to ORF2p function and retrotransposition.
Materials and methods
Naming conventions
ORF2 fragments are named as described.9 For C-terminally truncated fragments, the previously reported domains are used as the body of the name, followed by the number corresponding to the terminal amino acid as it would be in the full length ORF2p, with the truncated ORF2p sequence denoted by a Δ. For example, the ORF2p fragment that contains the EN and Z domains that ends at ORF2p amino acid 490 is written as ENZ490Δ.
Cloning
Plasmids containing codon optimized L1 sequence (L1PA1 Chang)6 were used as templates to generate truncated ORF2 PCR fragments for subcloning into pcDNA 3.1/Hygro+ (Life Technologies), pAct (Promega), and pBind (Promega). ORF2 fragments containing inactivating D205A and H230A mutations within the EN sequence were cloned using previously reported expression plasmid containing codon optimized ORF2 sequence to generate ENn expression plasmids designed to produce protein fragments with non-functional endonucleases.7 To generate untagged ORF2p fragment expression constructs, an NheI restriction site-Kozac -ATG and TGA-HindIII restriction site were added 5′ and 3′ of the ORF2 DNA sequence of interest. PCR products were digested with NheI and HindIII and cloned into pcDNA3.1/Hygro+ (Life Technologies). To generate Gal4 tagged ORF2p fragment expression constructs, PCR primers were designed using the Flexi Vector primer design tool (Promega). These PCR primers added a 5′ SgfI restriction site and a 3′ PmeI restriction site containing a valine (V) codon (GTT) and a stop codon (TAA) to the ORF2 sequence of interest. PCR products were digested with SgfI/PmeI blend (Promega: catalog number R1852) and cloned into both the pAct and pBind vector (Promega Checkmate Mammalian Two Hybrid System: catalog number C934A).
Chronic toxicity assay: Figure 2 VP16 tagged and figure 4
HeLa cells were maintained in MEM supplemented with 1% sodium pyruvate, L-glutamate, NEAA, and 10% FBS as previously described.25 ORF2p EN chronic toxicity assay was performed in HeLa cells as previously described.7 Five hundred thousand cells were seeded 16–18 hours prior to transfection in T75 flasks. 1 μg EN or ENn plasmid containing a Neomycin resistance gene (VP16-tagged: Fig. 2) or Hygromycin resistance gene (untagged: Fig. 4) were transfected with 8 μL Lipofectamine reagent (Life Technologies) and 4 μL Plus reagent (Life Technologies). Cell culture media was supplemented with 0.45 mg/mL neomycin (Fig. 2) or hygromycin (Fig. 4) ∼24 hours post transfection. Colonies were stained after 2 weeks with of culture with appropriate antibiotic selection with crystal violet solution (0.2% crystal violet, 5% acetic acid, 2.5% isopropanol) and counted with Oxford Optronics ColCount. Statistical significance assessed using Student's t-test for paired samples (n = 3), with error bars denoting standard deviation.
Acute toxicity assay: Figure 2 Gal4 tagged
HeLa cells were maintained as described above. ORF2p EN acute toxicity assay was performed in HeLa cells as previously described.7 Five hundred thousand cells were seeded 16–18 hours prior to transfection in T75 flasks. 1 μg EN or ENn plasmid and 1 μg previously described pIRES2-GFP plasmid (source of neomycin resistance) cotransfected with 8 μL Lipofectamine reagent (Life Technologies) and 4 μL Plus reagent (Life Technologies). Colonies were selected, stained, and counted as above. Statistical significance assessed as above.
Immunoblot analysis
Performed as previously described.7,9,26 2 million cells seeded 16–18 hours prior to transfection in T75 flasks. Six μg of appropriate expression construct or appropriate empty vector (control) were transfected with 24 μL Lipofectamine reagent (Life Technologies) and 12 μL Plus reagent (Life Technologies). For Cytoplasmic/Nuclear fractionation, approximately 24 hours post transfection, cells were washed 1x with phosphate buffered saline (PBS) and then harvested in 500 μL Lysis Buffer (LB: 50 mM Tris, 150 mM NaCl, 10 mM EDTA, 0.5% Triton-X, pH 7.2) supplemented with 10 μL/mL of Halt protease inhibitor cocktail, phosphate inhibitor cocktail 2, and phosphate inhibitor cocktail 3 (Sigma). Cell lysates were then centrifuged at 4C at 14 thousand rpm for 15 minutes. This cytoplasmic fraction was removed. Remaining nuclear fraction pellet was suspended in 250 μL Total Lysis Buffer with SDS (TLB SDS: 50 mM Tris, 150 mM NaCl, 10 mM EDTA, 0.5% SDS, 0.5% Triton-X, pH 7.2) supplemented with 10 μL/mL of Halt protease inhibitor cocktail, phosphate inhibitor cocktail 2, and phosphate inhibitor cocktail 3 (Sigma). Cells were sonicated with a Microson XL-2000 sonicator (Misonix) 3x (10 seconds sonication/10 seconds rest on ice), and cell lysates were then centrifuged at 4C at 14 thousand rpm for 15 minutes. Protein concentrations of cleared cell lysates were determined using BioRad protein assay (Bradford method). For total cell lysis, approximately 24 hours post transfection, cells were washed 1x with phosphate buffered saline (PBS) and then harvested in 500 μL TLB SDS supplemented with 10 μL/mL of Halt protease inhibitor cocktail, phosphate inhibitor cocktail 2, and phosphate inhibitor cocktail 3 (Sigma). After one round of freeze (−80C) /thaw on ice, cells were sonicated with a Microson XL-2000 sonicator (Misonix) 3x (10 seconds sonication/10 seconds rest on ice), and cell lysates were then centrifuged at 4C at 14 thousand rpm for 15 minutes. Protein concentrations of cleared cell lysates were determined using BioRad protein assay (Bradford method). 30 μg of total cell lysate were boiled in equal volume of Laemmli buffer with 3.4% β-mercaptoethanol (Fig. 4) or heated at 85C for 5 minutes in Laemmli buffer without β-mercaptoethanol supplementation (Fig. 3). Samples were fractionated on 3–8% Tris-Acetate gels (Life Technologies) and transferred to nitrocellulose membranes using the iBlot system (Life Technologies). Membranes were blocked in PBS-Tween (PBS, 0.1% Tween) with 5% blotting-grade blocker (BioRad) and incubated with primary antibodies overnight at 4C. Primary antibodies for Gal4-tagged constructs were commercially available anti-Gal4 antibodies (Santa Cruz: sc-577 1:1000 dilution). Primary antibodies for untagged EN-containing fragments were previously reported custom monoclonal ORF2p antibodies.10 As appropriate, either HRP-goat anti-mouse (Fig. 4) or HRP-goat anti-rabbit (Fig. 3) were used. Western blots were developed using the Immun-Star WesternC kit (BioRad). Images were captured using a BioRad Gel Doc XR+ imager. GAPDH (Santa Cruz: sc-25778) was used as loading control (1:3000 in 3% blotting grade blocker/PBS-Tween). Statistical significance of ORF2p or protein fragment band signal intensity differences relative to GAPDH loading control (cytoplasmic) or Lamin A/C (nuclear) assessed using Student's t-test for paired samples (n = 3), with error bars representing standard deviation.
Abbreviations
- (LINE1 or L1)
Long Interspersed Element 1
- (ORF)
Open Reading Frame
- (UTR)
Untranslated Region
- (ORFp)
Open Reading Frame protein
- (EN)
endonuclease
- (RT)
reverse transcriptase
- (Cry)
Cryptic
- (DSB)
Double-Strand Break
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Dawn deHaro, Mark Sokolowski, and the members of COMET (Consortium of Mobile Elements at Tulane) for critical discussion.
Funding
This work was funded by Life Extension Foundation to VPB; National Institutes of Health [P20GM103518] to VPB; Kay Yow Cancer Fund to VPB.
References
- [1].Mathias SL, Scott AF, Kazazian HH, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science 1991; 254:1808-10; PMID:1722352; http://dx.doi.org/ 10.1126/science.1722352 [DOI] [PubMed] [Google Scholar]
- [2].Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG. Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci U S A 1990; 87:6990-4; PMID:1698287; http://dx.doi.org/ 10.1073/pnas.87.18.6990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kolosha VO, Martin SL. In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci U S A 1997; 94:10155-60; PMID:9294179; http://dx.doi.org/ 10.1073/pnas.94.19.10155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Feng Q, Moran JV, Kazazian HH, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996; 87:905-16; PMID:8945517; http://dx.doi.org/ 10.1016/S0092-8674(00)81997-2 [DOI] [PubMed] [Google Scholar]
- [5].Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. High frequency retrotransposition in cultured mammalian cells. Cell 1996; 87:917-27; PMID:8945518; http://dx.doi.org/ 10.1016/S0092-8674(00)81998-4 [DOI] [PubMed] [Google Scholar]
- [6].Wallace NA, Belancio VP, Deininger PL. L1 mobile element expression causes multiple types of toxicity. Gene 2008; 419:75-81; PMID:18555620; http://dx.doi.org/ 10.1016/j.gene.2008.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kines KJ, Sokolowski M, deHaro DL, Christian CM, Belancio VP. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res 2014; 42:10488-502; PMID:25143528; http://dx.doi.org/ 10.1093/nar/gku687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol 2006; 357:1383-93; PMID:16490214; http://dx.doi.org/ 10.1016/j.jmb.2006.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Christian CM, deHaro D, Kines KJ, Sokolowski M, Belancio VP. Identification of L1 ORF2p sequence important to retrotransposition using Bipartile Alu retrotransposition (BAR). Nucleic Acids Res 2016; 44(10):4818-34; PMID:27095191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sokolowski M, DeFreece CB, Servant G, Kines KJ, deHaro DL, Belancio VP. Development of a monoclonal antibody specific to the endonuclease domain of the human LINE-1 ORF2 protein. Mob DNA 2014; 5:29; PMID:25606060; http://dx.doi.org/ 10.1186/s13100-014-0029-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Belancio VP, Roy-Engel AM, Deininger PL. All y'all need to know'bout retroelements in cancer. Semin Cancer Biol 2010; 20:200-10; PMID:20600922; http://dx.doi.org/ 10.1016/j.semcancer.2010.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Belancio VP, Deininger PL, Roy-Engel AM. LINE dancing in the human genome: transposable elements and disease. Genome Med 2009; 1:97; PMID:19863772; http://dx.doi.org/ 10.1186/gm97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kelman Z. PCNA: structure, functions and interactions. Oncogene 1997; 14:629-40; PMID:9038370; http://dx.doi.org/ 10.1038/sj.onc.1200886 [DOI] [PubMed] [Google Scholar]
- [14].Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res 2008; 18:162-73; PMID:18157158; http://dx.doi.org/ 10.1038/cr.2007.114 [DOI] [PubMed] [Google Scholar]
- [15].Mailand N, Gibbs-Seymour I, Bekker-Jensen S. Regulation of PCNA-protein interactions for genome stability. Nat Rev Mol Cell Biol 2013; 14:269-82; PMID:23594953; http://dx.doi.org/ 10.1038/nrm3562 [DOI] [PubMed] [Google Scholar]
- [16].Zhu Q, Chang Y, Yang J, Wei Q. Post-translational modifications of proliferating cell nuclear antigen: A key signal integrator for DNA damage response (Review). Oncol Lett 2014; 7:1363-9; PMID:24765138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karimian A, Ahmadi Y, Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016; 42:63-71; PMID:27156098; http://dx.doi.org/ 10.1016/j.dnarep.2016.04.008 [DOI] [PubMed] [Google Scholar]
- [18].Gasior SL, Roy-Engel AM, Deininger PL. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 2008; 7:983-9; PMID:18396111; http://dx.doi.org/ 10.1016/j.dnarep.2008.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wallace NA, Gasior SL, Faber ZJ, Howie HL, Deininger PL, Galloway DA. HPV 5 and 8 E6 expression reduces ATM protein levels and attenuates LINE-1 retrotransposition. Virology 2013; 443(1):69-79; PMID: 23706308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Alderton GK. Tumour suppression: p53 suppresses retrotransposition. Nat Rev Cancer 2016; 16:70; PMID:2674303126743031 [Google Scholar]
- [21].Goodier JL, Ostertag EM, Engleka KA, Seleme MC, Kazazian HH. A potential role for the nucleolus in L1 retrotransposition. Hum Mol Genet 2004; 13:1041-8; PMID:15028673; http://dx.doi.org/ 10.1093/hmg/ddh118 [DOI] [PubMed] [Google Scholar]
- [22].Shi X, Seluanov A, Gorbunova V. Cell divisions are required for L1 retrotransposition. Mol Cell Biol 2007; 27:1264-70; PMID:17145770; http://dx.doi.org/ 10.1128/MCB.01888-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kubo S, Seleme MC, Soifer HS, Perez JL, Moran JV, Kazazian HH, Kasahara N. L1 retrotransposition in nondividing and primary human somatic cells. Proc Natl Acad Sci U S A 2006; 103:8036-41; PMID:16698926; http://dx.doi.org/ 10.1073/pnas.0601954103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xie Y, Mates L, Ivics Z, Izsvák Z, Martin SL, An W. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob DNA 2013; 4:10; PMID:23497436; http://dx.doi.org/ 10.1186/1759-8753-4-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet 2003; 35:363-6; PMID:14625551; http://dx.doi.org/ 10.1038/ng1269 [DOI] [PubMed] [Google Scholar]
- [26].Sokolowski M, Deharo D, Christian CM, Kines KJ, Belancio VP. Characterization of L1 ORF1p Self-Interaction and Cellular Localization Using a Mammalian Two-Hybrid System. PLoS One 2013; 8:e82021; PMID:24324740; http://dx.doi.org/ 10.1371/journal.pone.0082021 [DOI] [PMC free article] [PubMed] [Google Scholar]


