Abstract
Transcription activator-like effectors (TALEs) are sequence-specific DNA binding proteins found in a range of plant pathogenic bacteria, where they play important roles in host-pathogen interactions. However, it has been unclear how TALEs, after they have been injected into the host cells, activate transcription of host genes required for infection success. Here, we show that the basal transcription factor IIA gamma subunit TFIIAγ5 from rice is a key component for infection by the TALE-carrying bacterium Xanthomonas oryzae pv. oryzae, the causal agent for bacterial blight. Direct interaction of several TALEs with TFIIAγ5 is required for activation of disease susceptibility genes. Conversely, reduced expression of the TFIIAγ5 host gene limits the induction of susceptibility genes and thus decreases bacterial blight symptoms. Suppression or mutation of TFIIAγ5 can also reduce bacterial streak, another devastating disease of rice caused by TALE-carrying X. oryzae pv. oryzicola. These results have important implications for formulating a widely applicable strategy with which to improve resistance of plants to TALE-carrying pathogens.
DOI: http://dx.doi.org/10.7554/eLife.19605.001
Research Organism: Other
eLife digest
Around the world, bacterial infections reduce the yields of many important crops like rice, tomatoes, peppers and citrus fruits. Xanthomonas is a particularly widespread genus of bacteria; it consists of almost 30 species that cause diseases in more than 400 plant hosts, including bacterial blight and bacterial streak in rice plants.
Plants do have an immune system that is able to detect invading microbes and trigger a defensive response against them; however, many disease-causing bacteria have evolved ways to avoid or counteract this response. For example, at least five Xanthomonas species use proteins called transcription activator-like effectors (or TALEs for short) to infect their host plants. The bacterial proteins are essentially injected into the plant’s cells where they activate specific plant genes that make the host more susceptible to infection. Like other organisms, plants use proteins called transcription factors to switch genes on or off. However, it was not clear if the TALEs hijack the plant’s transcriptional machinery to activate these “susceptibility genes” or if they activate the genes via some other means.
Now, Yuan et al. show that TALE-carrying bacteria do make use of at least one of rice’s own transcription factors to cause bacterial blight and bacterial streak. The transcription factor in question is rice’s version of a general transcription factor, called TFIIAγ, which is essential for gene activation in plants, animals and fungi. Yuan et al. also identify the region of the TALE that binds to the transcription factor, and show that rice plants with lower levels of the transcription factor are protected against bacterial blight and bacterial streak.
Uncovering how disease-causing Xanthomonas bacteria use TALEs to infect plants will hopefully help researchers to develop crop plants that are more resistant to these harmful bacteria. Further work is now needed to see if the gene that encodes TFIIAγ in crop plants can be edited to achieve this goal, or whether genes encoding resistant variants of the protein already exist in other plant species.
Introduction
Transcription activator-like effectors (TALEs) are important effectors of plant pathogenic bacteria of the genus Xanthomonas (Boch et al., 2009). The bacteria inject TALEs via their Type III secretion system (T3SS) into host cells, where they translocate to the nucleus and bind host gene promoters in a sequence-specific manner. The DNA binding domain consists of variable repeats that together account for a predictable DNA recognition code (Boch et al., 2009; Moscou and Bogdanove, 2009). This property has been exploited for programmable DNA binding, and has allowed targeted genome editing by combining TALE DNA binding domains with nucleases (TALENs) (Maggio and Gonçalves, 2015). TALE-like proteins are not restricted to the genus Xanthomonas, and have also been found in the plant pathogen Ralstonia solanacearum (de Lange et al., 2014), and in the endosymbiont Burkholderia rhizoxinica (de Lange et al., 2014; Juillerat et al., 2014). TALE-like proteins thus may play not only antagonistic roles in host-microbe interactions.
Xanthomonas infect many important crops including barley, bean, brassica, cassava, citrus, cotton, mango, pepper, rice, rye, tomato, triticale, and wheat (Schornack et al., 2013; Boch et al., 2014). In rice, Xanthomonas oryzae pv. oryzae (Xoo) causes bacterial blight and X. oryzae pv. oryzicola (Xoc) causes bacterial streak, both of which are highly devastating diseases. The recessive resistance gene xa5 is widely used to improve rice resistance to Xoo (Kottapalli et al., 2007). xa5 is a natural allele of the gene for the transcription factor IIA gamma subunit 5 (TFIIAγ5), changing a valine to a glutamine (TFIIAγ5V39E thereafter) (Iyer and McCouch, 2004; Sugio et al., 2007). TFIIA is a basal transcription factor of eukaryotes and it is essential for polymerase II–dependent transcription (Høiby et al., 2007). It consists of two subunits, the large subunit TFIIAαβ and the small subunit TFIIAγ (Li et al., 1999).
Rice TFIIAγ5 has been suggested to be a cofactor that directly enables TALEs to induce host gene expression (Iyer-Pascuzzi and McCouch, 2007), either as a helper of TALE function (Boch et al., 2014), or as a TALE-targeted host gene (Gu et al., 2009). The latter scenario is supported by the finding that the TALE PthXo7 directly activates expression of another TFIIAγ encoding gene, TFIIAγ1 (Sugio et al., 2007).
In this paper, we reveal that TALEs from two Xanthomonas pathogens, Xoo and Xoc directly interact with TFIIAγ5 to activate host susceptibility genes, and that RNAi-mediated suppression or mutation of TFIIAγ5 confers disease resistance. Our results suggest that modifying host TFIIAγ genes by mutation or suppression may provide a widely applicable strategy to improve plant resistance to TALE-carrying pathogens.
Results
TFIIAγ5 is required for TALE-dependent induction of host genes
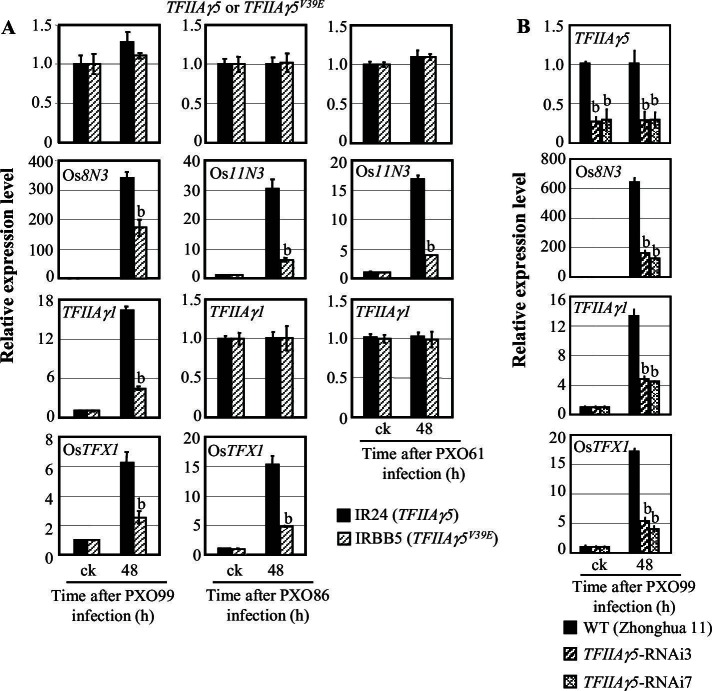
To assess whether host TFIIAγ is required for TALE-regulated transcriptional activation of rice susceptibility genes, we first assessed how pair of rice near-isogenic lines, IR24 carrying TFIIAγ5 and IRBB5 carrying mutant TFIIAγ5V39E in the IR24 background, responded to 15 different TALE-carrying Xoo strains (Yang and White, 2004). IRBB5 always showed fewer disease symptoms than IR24 (Figure 1—figure supplement 1A). Xoo infection did not induce RNA expression of TFIIAγ5 in IR24 or TFIIAγ5V39E in IRBB5 (Figure 1A), which correlates with the absence of predicted DNA binding motifs for known TALEs in the TFIIAγ5 promoter. In contrast, expression of known disease susceptibility genes Os8N3, TFIIAγ1, OsTFX1, and Os11N3, each of which is targeted by a different TALE (Römer et al., 2010; Sugio et al., 2007; Yang et al., 2006), was always lower in IRBB5 (p<0.01), although not necessarily completely abolished (Figure 1A). Together, these results point to TFIIAγ5 being a host co-factor for TALE-dependent induction of susceptibility genes.
Figure 1. Effects of TFIIAγ5 on the expression of disease susceptibility genes Os8N3, Os11N3, TFIIAγ1, or OsTFX1, after Xoo infection.
Plants were inoculated with Xoo strain PXO99 (harbouring TALEs PthXo1, PthXo7, and PthXo6), PXO86 (harbouring TALE PthXo3) or PXO61 (harbouring TALE AvrXa7) at the booting (panicle development) stage. It is known that PthXo1, PthXo7, and PthXo6 induce Os8N3, TFIIAγ1, and OsTFX1, respectively, and PthXo3 and AvrXa7 all induce Os11N3. Each bar represents mean (three replicates) ± standard deviation. (A) Mutation of TFIIAγ5 (rice line IRBB5). b, significant difference between IR24 and IRBB5 at p<0.01. (B) TFIIAγ5-RNAi lines. b, significant difference between wild-type (WT) and transgenic plants at p<0.01.
Figure 1—figure supplement 1. Effects of TFIIAγ5 on rice resistance to Xoo strains known to carry TALEs.
Figure 1—figure supplement 2. Effect of TFIIAγ5 on Xa23-mediated resistance to Xoo strain PXO99.
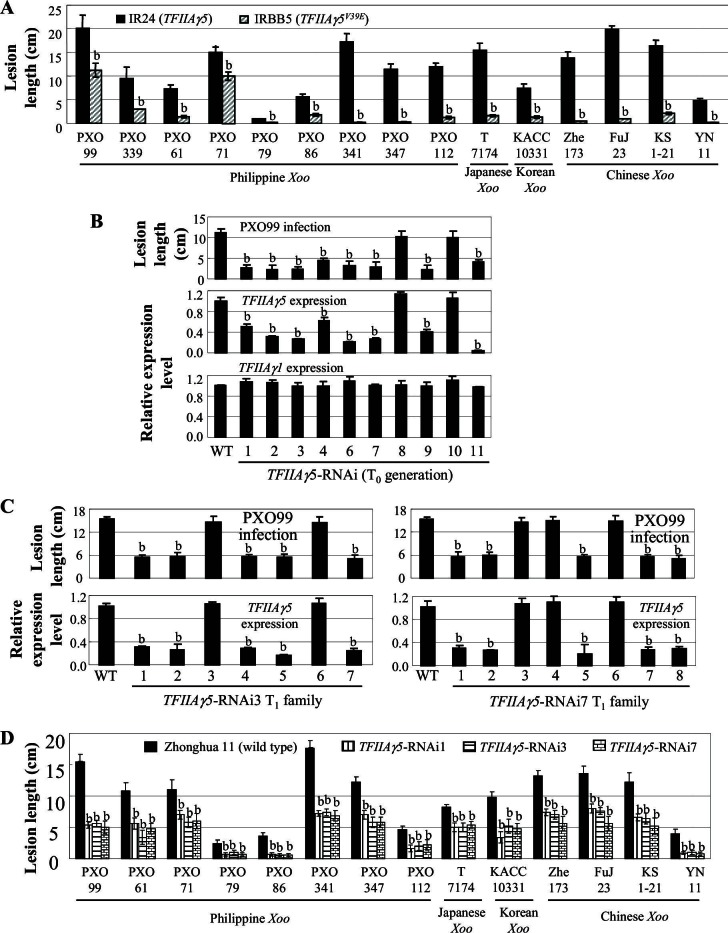
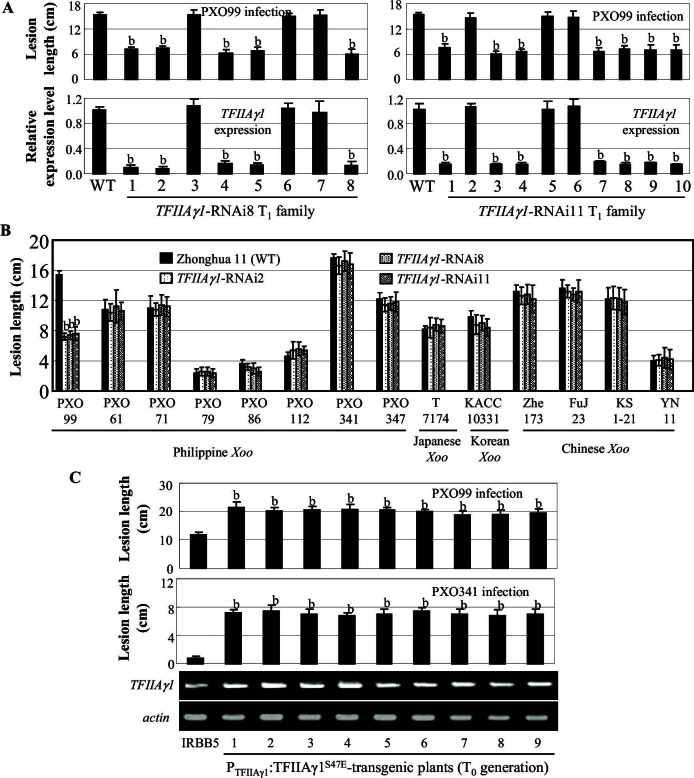
To determine directly the role of TFIIAγ5 in host gene expression, we suppressed its activity by RNA interference (RNAi). Only the expression of TFIIAγ5, but not of TFIIAγ1 was reduced in T0 transgenic plants, and the reduction in TFIIAγ5 expression correlated with enhanced resistance to Xoo PXO99 in T0 and T1 plants (Figure 1—figure supplement 1B,C). TFIIAγ5-RNAi plants also had enhanced resistance to a diverse collection of 13 additional Xoo strains (Figure 1—figure supplement 1D), and Xoo-induced expression of Os8N3 and OsTFX1 was reduced in TFIIAγ5-RNAi plants (Figure 1B). Suppressing TFIIAγ5 did not obviously influence growth and development of the transgenic plants.
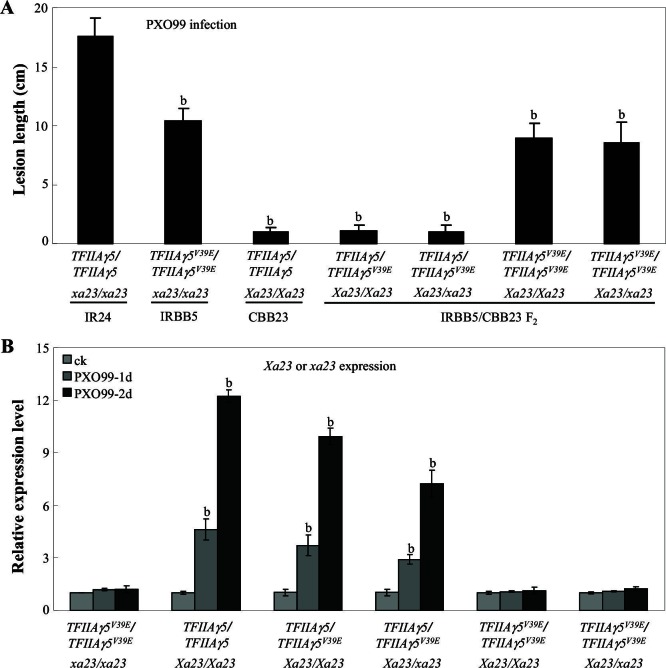
TALE DNA-binding motifs have been detected in the promoters of some disease resistance genes, an apparent evolutionary response against TALE-carrying bacteria (Gu et al., 2005; Römer et al., 2010; Wang et al., 2015). Xoo TALE AvrXa23 activates the Xa23 resistance gene, resulting in resistance to Xoo (Wang et al., 2015). To investigate the role of TFIIAγ5 in Xa23 resistance, we crossed rice lines IRBB5, with a xa23 susceptibility and a TFIIAγ5V39E resistance allele, and CBB23, with a Xa23 resistance and a TFIIAγ5 susceptibility allele. F2 plants of genotypes Xa23/Xa23 or Xa23/xa23 were completely resistant to PXO99 in the TFIIAγ5/TFIIAγ5 or TFIIAγ5/TFIIAγ5V39E background, but showed the reduced resistance in the TFIIAγ5V39E/TFIIAγ5V39E background (Figure 1—figure supplement 2A). Consistent with the resistance phenotype, Xa23 expression was rapidly induced by PXO99 in Xa23/Xa23 or Xa23/xa23 plants when they also were of genotype TFIIAγ5/TFIIAγ5 or TFIIAγ5/TFIIAγ5V39E (Figure 1—figure supplement 2B). Xa23 induction was completely lost in TFIIAγ5V39E/TFIIAγ5V39E plants. These results suggest that TFIIAγ5 plays dual roles in Xoo−rice interactions: it is required by TALE-containing Xoo to cause disease, but at the same time it can help to protect against disease in the presence of certain resistance genes that have TALE-binding motifs in their promoters.
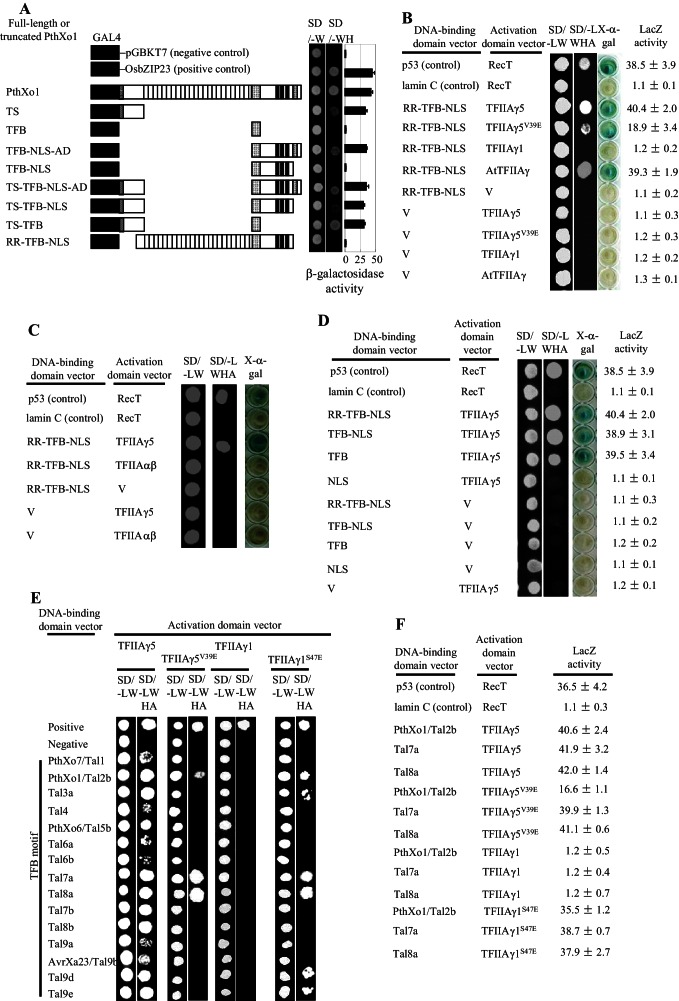
Xoo TALEs directly interact with TFIIAγ5
Xoo TALEs typically have an amino-terminal translocation signal (TS), a central repeat region (RR), a transcription factor binding (TFB) region, a nuclear localization signal (NLS), and a carboxyl-terminal transcription activation domain (AD) (Figure 2—figure supplement 1, Figure 2—source data 1) (Yang et al., 2006; Schreiber et al., 2015). When fused to the GAL4 DNA binding domain, Xoo TALE PthXo1 on its own could activate reporter gene expression in yeast. This was observed whenever the TS or AD were present, but not with the RR, TFB or NLS (Figure 2—figure supplement 1A). This is similar to what has been reported for Xoo TALE AvrXa10 and X. euvesicatoria TALE AvrBs3 (Szurek et al., 2001; Zhu et al., 1998).
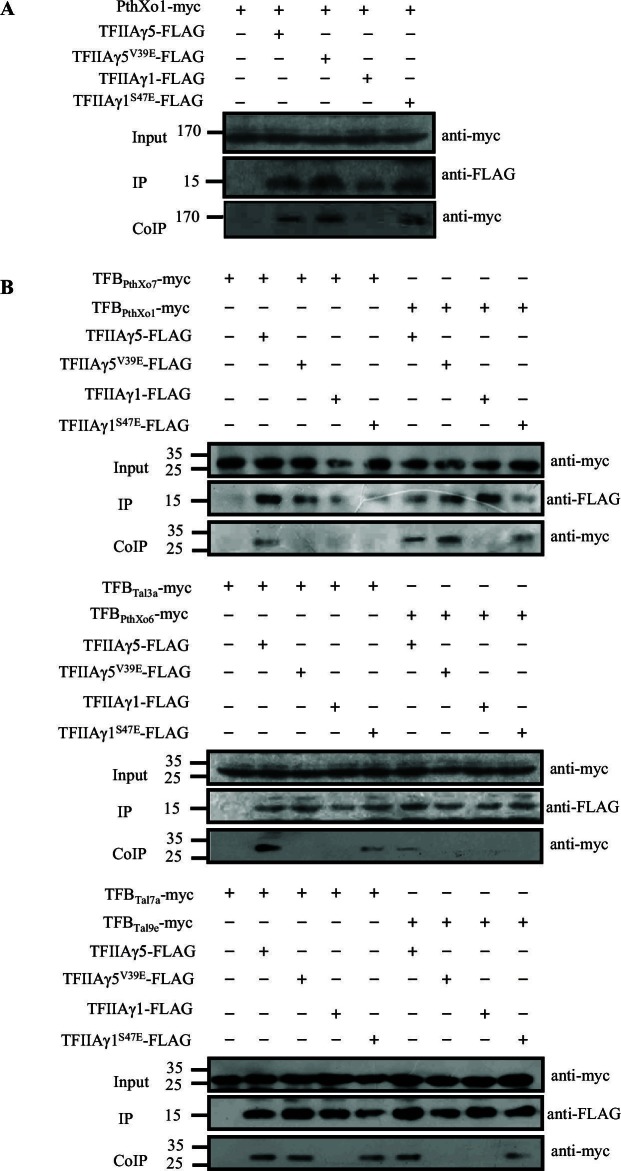
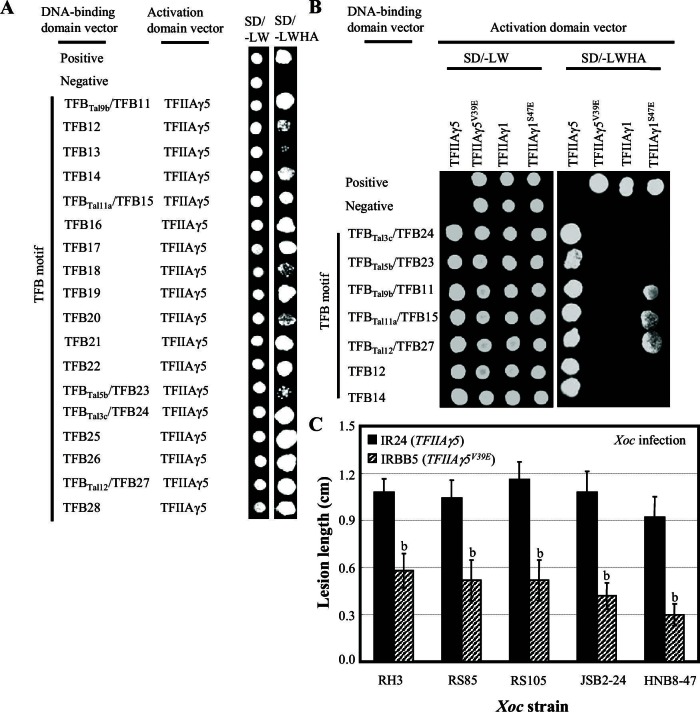
We hypothesized that TALEs use TFIIAγ5 directly as a co-factor to induce transcription of susceptibility genes. In yeast two-hybrid (Y2H) assays, truncated PthXo1, RR-TFB-NLS, lacking transcriptional activation ability, interacted strongly with TFIIAγ5, somewhat less so with the mutant TFIIAγ5V39E, and not at all with the large subunit of TFIIA, TFIIAαβ (Figure 2—figure supplement 1B,C). The interaction with TFIIAγ5 required the TFB (Figure 2—figure supplement 1D).
To determine whether this observation of interaction of a TALE TFB with TFIIAγ5, was general, we isolated the TFB encoding DNA fragments from 14 of the 18 other TALE genes in Xoo pv. PXO99 (Salzberg et al., 2008). These TFBs are 134 to 145 amino acids long, with the Tal7b and Tal8b TFBs predicted to be identical (Figure 2—source data 2). All 14 TFB fragments interacted with TFIIAγ5, but only two (Tal7a and Tal8a) with TFIIAγ5V39E (Figure 2—figure supplement 1E,F). Notably, different from PthXo1, Tal7a and Tal8a interacted equally well with TFIIAγ5 and TFIIAγ5V39E. The TFBs of Tal7a, Tal8a, and PthXo1 differed by 1 to 20 residues from the other 12 TFBs that interacted only with TFIIAγ5 (Figure 2—source data 2).
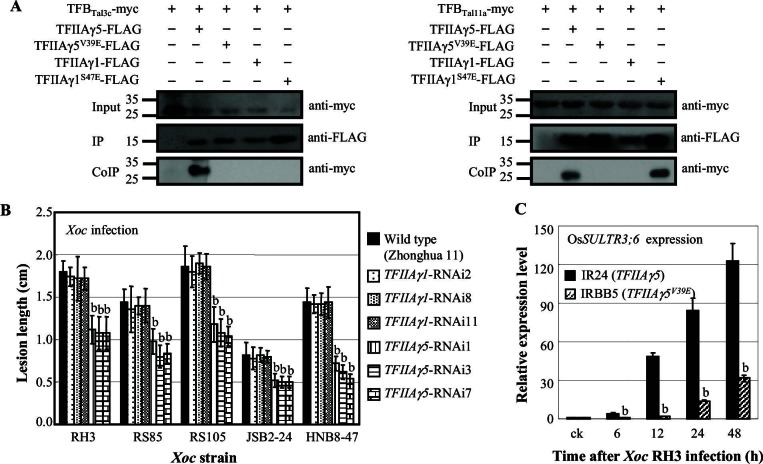
We confirmed the interactions observed in the Y2H system by transient expression of Myc- and FLAG-labeled proteins in Nicotiana benthamiana, followed by co-immunoprecipitation (CoIP) (Figure 2A). We found the interaction of full-length PthXo1 with TFIIAγ5 or TFIIAγ5V39E, of the TFBs of PthXo1, PthXo6, PthXo7, Tal3a, Tal7a, and Tal9e with TFIIAγ5, and of the TFBs of PthXo1 and Tal7a with TFIIAγ5V39E (Figure 2B).
Figure 2. Detection of interactions between rice TFIIAγs and TALEs from Xoo in planta by co-immunoprecipitation.
The protein–protein interaction assays were performed in N. benthamiana leaf cells. Proteins before (input) and after immunoprecipitation (IP) were detected with anti-myc and anti-FLAG antibodies. (A) Interaction of the myc-labelled full-length PthXo1 with FLAG-labelled TFIIAγ5, TFIIAγ5V39E, and mutated rice TFIIAγ1 (TFIIAγ1S47E). (B) Interactions of the myc-labelled TFB regions of six TALEs with FLAG-labelled rice TFIIAγs.
DOI: http://dx.doi.org/10.7554/eLife.19605.006
Figure 2—figure supplement 1. Interactions between Xoo TALEs and plant TFIIAγs in yeast cells.
TALE-dependent induction of host genes requires interaction with TFIIAγ5 but not TFIIAγ1
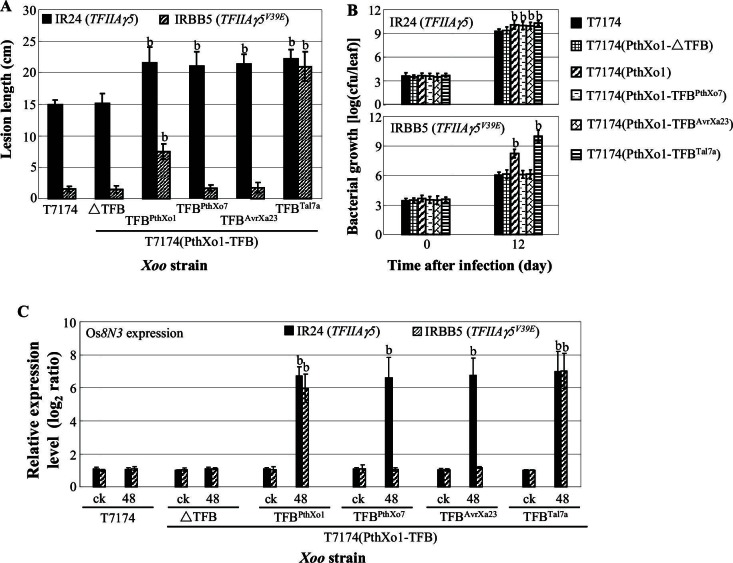
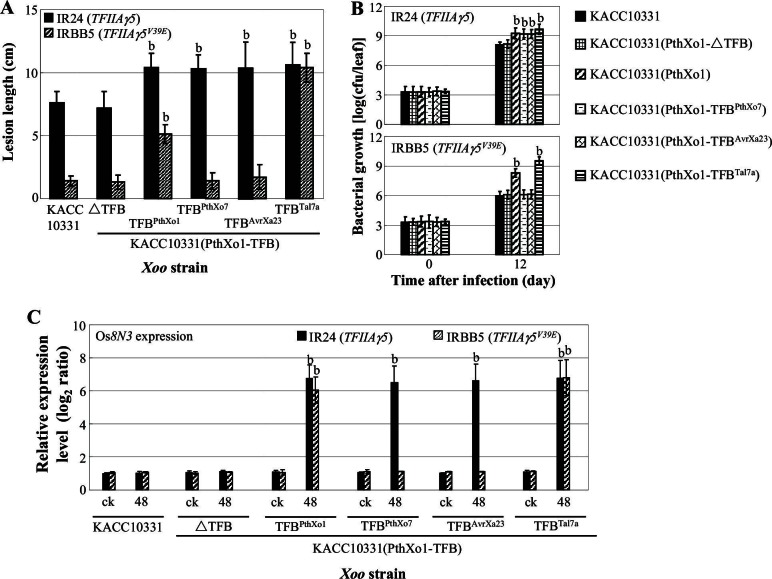
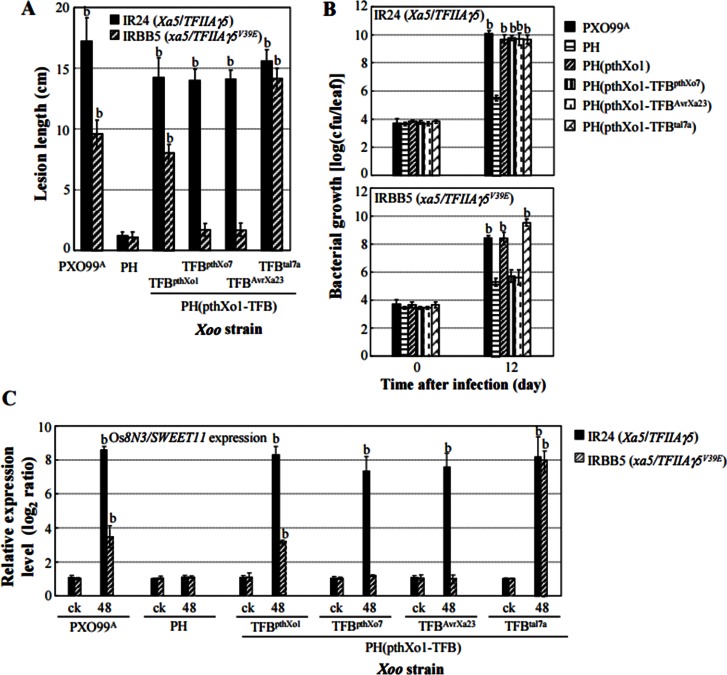
To learn whether the TFB region of TALEs is directly responsible for TALE-induced host gene expression, we generated recombinant Xoo strains in which the TFB of PthXo1 was replaced with different TFBs, chosen based on their differential interaction with TFIIAγ5 and TFIIAγ5V39E: PthXo1 (TFIIAγ5 > TFIIAγ5V39E), Tal7a (TFIIAγ5 = TFIIAγ5V39E), and PthXo7 and AvrXa23 (TFIIAγ5 but not TFIIAγ5V39E) (Figure 2—figure supplement 1B,E,F). In addition, we generated a TFB deletion in PthXo1. The constructs were introduced into Xoo pv. T7174 and KACC10331, both of which lack PthXo1. Rice strain IR24, which carries TFIIAγ5, is strongly susceptible to T7174 and moderately susceptible to KACC10331, while IRBB5, which carries TFIIAγ5V39E, is resistant to both Xoo strains (Figure 3A, Figure 1—figure supplement 1A, and Figure 3—figure supplement 1A).
Figure 3. Effects of the TFB region of TALE PthXo1 on the virulence of Xoo strains and on the expression of rice susceptibility gene in rice−Xoo interaction.
Each bar represents mean (total 30 to 35 leaves from five plants for lesion length; three replicates for gene expression and bacterial growth rate) ± standard deviation. (A) Virulence of wild-type strain T7174 and recombinant strains carrying PthXo1 and its derivatives in IR24 and IRBB5. b, significant difference between T7174 and recombinant strains in each rice line at p<0.01. (B) Growth of different Xoo strains in rice leaves. b, significant difference between 0 day (30 min after infection) and 12 days after infection of each strain at p<0.01. (C) Expression of susceptibility gene Os8N3 after infection of different strains. b, significant difference between non-inoculated (ck) and inoculated (at 48 hr after infection of a strain) plants in each rice line at p<0.01.
DOI: http://dx.doi.org/10.7554/eLife.19605.010
Figure 3—figure supplement 1. Effects of the TFB region of TALE PthXo1 on the virulence of Xoo strains and on the expression of rice susceptibility gene in rice−Xoo interaction. .
As expected, the deletion control PthXo1-ΔTFB did not change the success of infection by T7174 or KACC10331 (Figure 3A, and Figure 3—figure supplement 1A), while the TFBs from PthXo1 and Tal7a, which can interact with both TFIIAγ5 and TFIIAγ5V39E, enhanced infection success in both hosts, IR24 (TFIIAγ5) and IRBB5 (TFIIAγ5V39E). Consistent with Tal7a, but not PthXo1, interacting equally well with TFIIAγ5 and TFIIAγ5V39E, only the Tal7a TFB caused similar sized lesions in both IR24 and IRBB5 (Figure 3A, and Figure 3—figure supplement 1A). The TFBs of PthXo7 and AvrXa23, which can interact only with TFIIAγ5, accordingly increased disease symptoms only on IR24. Lesion size in these experiments was correlated with titer of bacterial growth (Figure 3B, and Figure 3—figure supplement 1B) and expression of Os8N3 (Figure 3C, and Figure 3—figure supplement 1C).
The TFB region of the TALEs harbours an imperfect leucine zipper motif, a known protein-protein interaction domain (Schreiber et al., 2015). We generated three TFB mutants of PthXo1, substituting leucine with alanine residues (Figure 3—source data 1A). The muations did, however, not compromise interaction with TFIIAγ5, nor infection success (Figure 3—source data 1B,C).
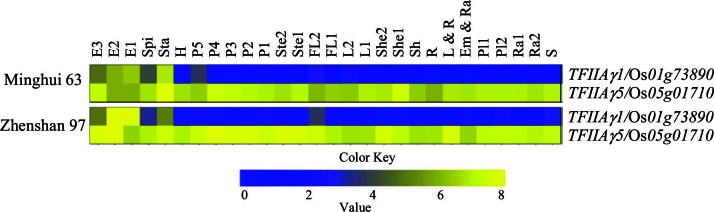
The other TFIIAγ encoded in the rice genome, TFIIAγ1, shares 86% sequence identity with TFIIAγ5 (Figure 4—source data 1), but has a very restricted expression profile, with highest expression in endosperm and stamens (Figure 4—figure supplement 1). TFIIAγ1 did not interact with full-length or truncated PthXo1 or other Xoo TALE TFBs in yeast or in planta (Figure 2, and Figure 2—figure supplement 1B,E,F).
We produced eight TFIIAγ1 derivatives with TFIIAγ5 substitutions at six positions (Figure 4—figure supplement 2). Of 15 TFBs tested, those of PthXo1, Tal3a, Tal7a, Tal8a, Tal9d and Tal9e could interact in yeast with TFIIAγ1S47E, but not with other TFIIAγ1 mutants (Figure 4—figure supplement 2, and Figure 2—figure supplement 1E). Four of these interactions could be confirmed in planta (Figure 2B).
We then generated TFIIAγ1-RNAi plants as well as transgenic plants expressing the TFIIAγ1S47E mutant from TFIIAγ1 regulatory sequence. Both types of plants were morphologically normal. Some T0 TFIIAγ1-RNAi plants showed enhanced resistance to Xoo pv. PXO99 (Figure 4—figure supplement 3). Increased resistance was associated with reduced TFIIAγ1 expression, whereas TFIIAγ5 expression was unaffected (Figure 4—figure supplement 3), which was confirmed in two T1 families (Figure 4A). However, these plants did not show enhanced resistance to other 13 Xoo strains (Figure 4B). This is in agreement with previous suggestions that the TFIIAγ1 promoter is a target of the TALE PthXo7 from PXO99 (Boch et al., 2009; Sugio et al., 2007). PthXo7-induced TFIIAγ1 expression is dependent on TFIIAγ5 (Figure 1).
Figure 4. Effects of TFIIAγ1 on response to infections by different Xoo strains.
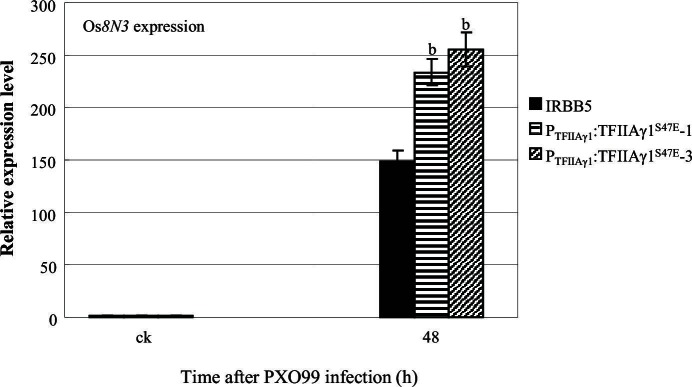
Plants were inoculated with Xoo at the booting stage. Each bar represents mean (three replicates for gene expression and total 35 to 40 leaves from five plants for lesion length) ± standard deviation. (A) Suppressing TFIIAγ1 enhanced rice resistance to strain PXO99. b, significant difference between wild-type (WT) Zhonghua 11 and transgenic plants at p<0.01. (B) Suppressing TFIIAγ1 did not change rice response to other strains. b, significant difference between WT and transgenic plants at p<0.01. (C) PTFIIAγ1:TFIIAγ1S47E-transgenic plants showed susceptibility to PXO99 and PXO341 compared to IRBB5. b, significant difference between IRBB5 and transgenic plants at p<0.01.
DOI: http://dx.doi.org/10.7554/eLife.19605.013
Figure 4—figure supplement 1. Expression profiles of TFIIAγ5 and TFIIAγ1 in 28 tissues covering the entire life cycle of rice varieties Minghui 63 and Zhenshan 97.
Figure 4—figure supplement 2. Interactions between the TFB region of TALE PthXo1 and mutated TFIIAγ1s in yeast cells.
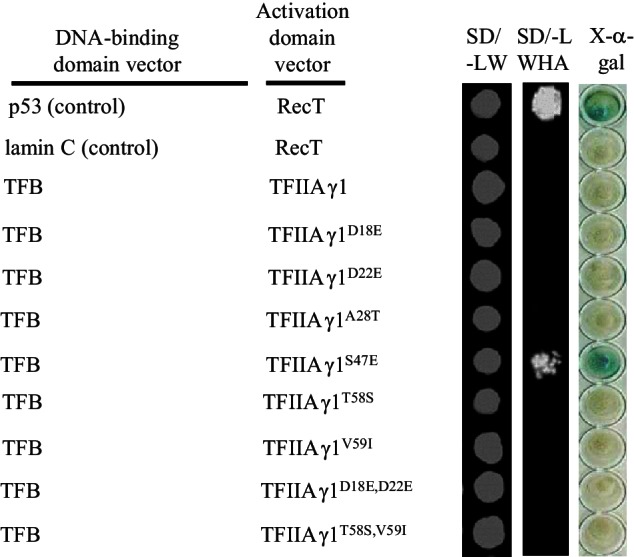
Figure 4—figure supplement 3. Effect of suppressing TFIIAγ1 on rice resistance to Xoo strain PXO99.
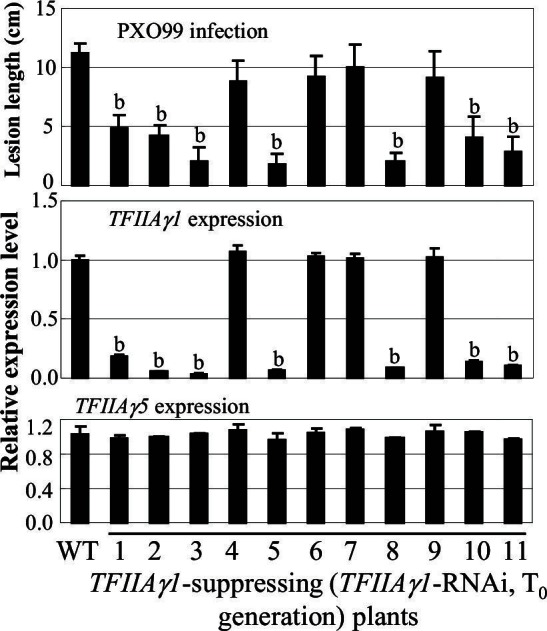
Figure 4—figure supplement 4. Effect of mutation of TFIIAγ1 on the expression of disease susceptibility gene Os8N3 after Xoo infection.
In the background of TFIIAγ5V39E, the TFIIAγ1S47E-transgenic plants showed increased susceptibility to Xoo pv. PXO99 and PXO341 (Figure 4C). The increased susceptibility to PXO99 might be due to an interaction between TFIIAγ1S47E and PthXo1 (Figure 2B) to induce the susceptibility gene Os8N3 (Figure 4—figure supplement 4), while the susceptibility to PXO341 may be explained by another TALE (see the TFBs tested in Figure 2—figure supplement 1E,F) that can interact with TFIIAγ1S47E.
Genetic variation in TFIIAγ5 and TFIIAγ1 genes
We searched the RiceVarMap database of 1419 rice accessions (http://ricevarmap.ncpgr.cn; Zhao et al., 2015) for allelic variation at TFIIAγ1 and TFIIAγ5. There were no non-synonymous single nucleotide polymorphisms (SNPs) in TFIIAγ1 (Figure 4—source data 2). Thirty-three rice accessions shared the same two non-synonymous SNPs diagnostic for the TFIIAγ5V39E allele (Figure 4—source data 3). Twenty-nine of these belong to the Aus group, which is mainly from South Asia, and the other four accessions belong to the Indica II group, mainly from Southeast Asia (Xie et al., 2015) (Figure 4—source data 3). The regional distribution of the TFIIAγ5V39E resistance allele likely reflects the high disease pressure in these regions.
Xoc TALEs hijack TFIIAγ5 to cause bacterial streak
To learn whether TALEs of other pathogenic bacteria also exploit TFIIAγ5 to cause disease, we investigated the interaction of TFIIAγ5 with TALEs from Xoc, which causes bacterial streak. Xoc pv. RH3 has at least 11 TALE genes based on DNA blot analysis (Figure 5—figure supplement 1). All TFBs of RH3 TALEs (GenBank accession numbers KU163014 to KU163031) interacted with TFIIAγ5 in yeast, and two were confirmed in planta (Figure 5A, and Figure 5—figure supplement 2A). Seven randomly chosen TFBs did not interact with TFIIAγ5V39E or TFIIAγ1, but three interacted with TFIIAγ1S47E in yeast, and at least one in planta (Figure 5A, and Figure 5—figure supplement 2B). Consistent with these results, rice accession IRBB5 (TFIIAγ5V39E) was more resistant to infection by different Xoc strains than IR24 (TFIIAγ5) (Figure 5—figure supplement 2C). TFIIAγ5-RNAi plants also showed enhanced resistance to Xoc, whereas suppressing TFIIAγ1 had no effect on resistance to Xoc (Figure 5B).
Figure 5. Effect of TFIIAγ on rice-Xoc interaction.
(A) Interactions of myc-labelled TFB regions of TALEs from Xoc RH3 and FLAG-labelled rice TFIIAγs in N. benthamiana leaf cells analysed by CoIP assays. Proteins before (input) and after immunoprecipitation (IP) were detected with anti-myc and anti-FLAG antibodies. (B) TFIIAγ5-RNAi but not TFIIAγ1-RNAi plants showed enhanced resistance to Xoc strains. Each bar represents mean (total 30 to 35 leaves from five plants) ± standard deviation. b, significant difference between wild-type and transgenic plants after infection of a strain at p<0.01. (C) Mutation of TFIIAγ5 (rice line IRBB5) reduced expression of disease susceptibility gene OsSULTR3;6 after infection. Each bar represents mean (three replicates) ± standard deviation. b, significant difference between IR24 and IRBB5 at p<0.01.
Figure 5—figure supplement 1. Southern hybridization analysis of TALE genes in different Xanthomonas species.
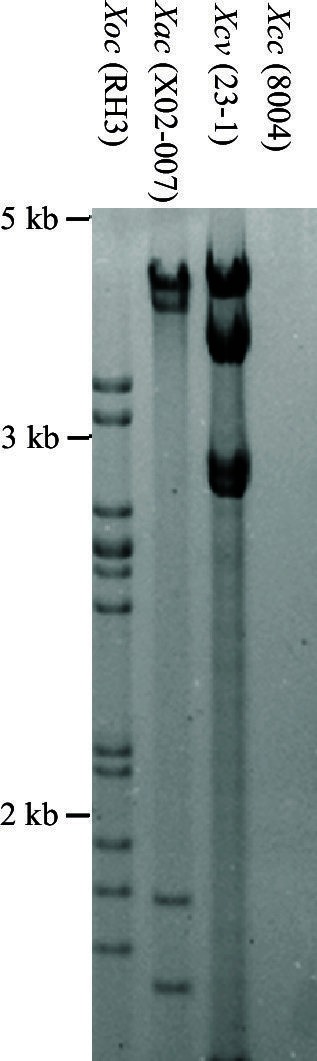
Figure 5—figure supplement 2. Analysis of interactions between Xoc TALEs and rice TFIIAγs.
A recent study has shown that a major quantitative trait locus for resistance to Xoc col-localizes with TFIIAγ5 (Xie et al., 2014). Two additional studies have revealed that a TALE that occurs in at least 10 sequenced Xoc strains transcriptionally activates the gene for the sulphate transporter OsSULTR3;6, a major susceptibility gene in rice−Xoc interactions (Cernadas et al., 2014; Wilkins et al., 2015). Xoc-induced expression of OsSULTR3;6 was significantly reduced (p<0.01) in IRBB5 relative to IR24 (Figure 5C), suggesting that TALE-containing Xoc also requires TFIIAγ5 to infect rice via TALE-induced expression of host susceptibility genes.
Discussion
TFIIAγ is indispensable for polymerase II–dependent transcription (Li et al., 1999). We have shown here how TALE-carrying Xoo and Xoc exploit rice TFIIAγ5 for activating transcription of downstream susceptibility genes (Figure 6). TALEs from Xoo and Xoc bind to TFIIAγ5 through their TFB regions, and the binding and binding strength are associated with the induction of susceptibility genes. Thus, TFIIAγ5 functions as a key component for TALE-induced host gene expression.
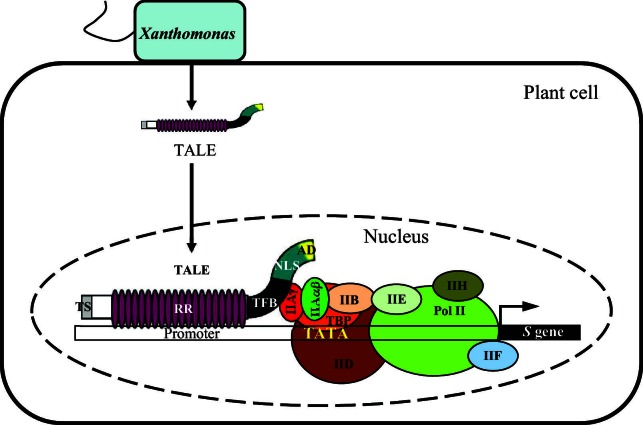
Figure 6. A model showing TFIIAγ5 as a key component of rice infection by Xanthomonas bacteria.
The bacteria hijack rice basal transcription factor TFIIAγ5 (IIAγ) by the transcription factor binding (TFB) region of their TALEs to induce host susceptibility (S) genes for infection. TS, amino-terminal translocation signal; RR, central repeat region; NLS, nuclear localization signal; AD, carboxyl-terminal transcription activation domain. The IIAγ belongs to the transcription pre-initiation complex. This complex consists of transcription factors IIA, which is composed of IIAβα subunit and IIAγ subunit, IIB, IID, IIE, IIF, and IIH, RNA polymerase II (Pol II), and TATA-binding protein (TBP). The binding of transcription pre-initiation complex to the TATA box of promoter was adopted and modified based on Yudkovsky et al. (2000).
It is striking that the only TFIIAγ5 paralog in rice, TFIIAγ1, apparently functions as a downstream susceptibility gene for Xoo PXO99, such that the TALE PthXo7 directly activates TFIIAγ1 transcription (Sugio et al., 2007), which differs from the protein-protein interaction of several Xoo TALEs with TFIIAγ5.
The recessive disease resistance allele TFIIAγ5V39E confers markedly reduced TALE-dependent induction of downstream susceptibility genes, apparently without compromising the overall activity of TFIIA. The rice accession IRBB5 carrying TFIIAγ5V39E is indistinguishable from the near-isogenic line IR24 in plant morphology and agronomic performance, including heading date, flag leaf length, number of panicles per plant, panicle length, grains per panicle, 1000-grain weight, yield per plant, seed setting rate, grain length, width, and thickness, with only slightly reduced plant height (Supplementary file 1). Here, we have shown that not only the specific point mutant TFIIAγ5V39E has increased Xoo resistance, but also that this can also be achieved by RNAi mediated knockdown of TFIIAγ5. In addition, we have shown that TALEs from other Xanthomonas pathogens, such as Xoc, exploit TFIIAγ5. Alteration of TFIIAγ5 activity, either through the introduction of the TFIIAγ5V39E allele, or through other reduction-of-function mutations, can provide a general strategy for improving rice resistance to TALE-carrying pathogens. TALE-carrying bacteria cause diseases in many other crops (Schornack et al., 2013; Boch et al., 2014). If these bacteria also exploit the host TFIIAγ for infection, modification of TFIIAγ may provide a road to improving disease resistance in other crops as well.
Materials and methods
Plant and bacterial materials
A pair of near-isogenic lines, IR24 (TFIIAγ5) and IRBB5 (TFIIAγ5V39E), and the variety Zhonghua 11 were used in this study. Plants were grown during the normal rice growing season under natural field conditions in the Experimental Stations of Huazhong Agricultural University, Wuhan, China.
Chinese, Japanese, Korean, and Pilipino Xoo strains were used to study rice resistance to bacterial blight disease (Gao et al. 2010; Li et al., 2012). Resistance to Xoc was tested using Chinese strains (Ke et al., 2014). X. campestris pv. campestris strain was used for Southern blot analysis of TALE genes (He et al., 2007). All Xanthomonas strains were grown at 28°C on nutrient agar medium. Antibiotics were used at the following final concentrations as required: ampicillin at 100 μg ml−1, rifampicin at 75 μg ml−1, kanamycin at 25 μg ml−1, and spectinomycin at 50 μg ml−1 when genetic manipulation of bacteria.
Transformation
To construct RNA interference vector, the 3′ untranslated regions of TFIIAγ5 and TFIIAγ1 were amplified with primers listed in Supplementary file 2 and inserted into vector pDS1301 (Yuan et al., 2010). Agrobacterium-mediated transformation of rice was performed (Lin and Zhang, 2005; Ge et al., 2006).
Pathogen inoculation
Xoo inoculation described in more detail at Bio-protocol (Ke et al., 2017). To evaluate reaction of rice plants to Xoo, plants were inoculated with the Xoo strains by the leaf-clipping method at the booting (panicle development) stage (Chen et al., 2002). The disease was scored by measuring the lesion length at 14 days after inoculation. Each bacterial inoculation assay was repeated at least twice. The disease of some plants was also evaluated by analysing bacterial growth based on a count of the colony-forming units as described previously (Sun et al., 2004). For measuring bacterial growth, one Xoo-infected leaf from each plant was examined as one replicate, and a total of three plants for each sample were analysed.
To evaluate Xoc resistance, rice plants were inoculated with Xoc strains by the penetration method using a needleless syringe at the booting stage (Ke et al., 2014). Disease was scored by lesion length at 14 days after inoculation. Each bacterial inoculation assay was repeated twice.
Gene expression analysis
The 2-cm leaf segments next to the bacterial infection sites in the rice plants were collected for RNA isolation. Quantitative reverse transcription-PCR (qRT-PCR) was conducted using gene-specific primers (Supplementary file 3) as described previously (Qiu et al., 2007). The expression level of the rice actin gene was used to normalize the measurement of the expression. Each rice sample was a mixture of leaf tissue from at least five plants, with 8 to 10 leaves per plant. For transgenic plants, segments from three to five leaves of the plant were sampled. Each qRT-PCR assay was repeated at least twice, with each repetition having three technical replicates.
Vector construction
The TALE PthXo1 was cloned into pHM1 vector to produce pHM1pthXo1, and transferred into Xoo strains T7174 and KACC10331 following published method (Yang and White, 2004). The TFB region of PthXo1 was replaced with TFB regions of other TALEs by Gibson assembly (Gibson et al., 2009). The recombinant strains were confirmed by PCR amplification of TALE fragments.
Southern hybridization analysis
A standard procedure for Southern hybridization of the bacterial DNA was performed (Gu et al., 2009). Genomic DNA from different Xanthomonas strains was digested with SphI, separated by electrophoresis on 1.2% agarose gel in TAE buffer, blotted onto a nylon membrane, and hybridized using a 32P-labeled 2.9-kb SphI fragment of PthXo1.
Transactivation activity assay
The transactivation activity of PthXo1 was analysed in yeast cells as described previously (Deng et al., 2012). The open reading frame of pthXo1 was ligated into pGBKT7 vector and fused in frame with the yeast GAL4 DNA binding domain. The recombinant vector was transformed into yeast strain AH109. The transformed yeast cells were plated on SD/−Trp or SD/−Trp-His medium and cultured for 3 days as described previously (Yuan et al., 2010).
Protein–protein interaction assay
The interaction between bacterial TALE proteins and host proteins in yeast cells was assayed using MATCHMAKER GAL4 Two-Hybrid System 3 (Clontech, Mountain View, CA) according the manufacturer’s instructions. To construct the interaction vectors, full-length and truncated TALEs and the TFB regions of TALEs and plant genes were amplified using the PCR primers listed in Supplementary file 2. The amplified DNA fragments were first inserted into vector pBluescript (Agilent Technologies, Santa Clara, CA) for sequencing confirmation. The confirmed bacterial DNA fragments were then ligated into pGBKT7 vector, and the confirmed plant DNA fragments were then ligated into pGADT7 Rec vector. The recombinant pGBKT7 and pGADT7 plasmids were co-transformed into yeast strain AH109 for yeast two-hybrid assays following the lithium acetate method (Yuan et al., 2010). The yeast clones were first scribed on the synthetic defined premixes (SD) medium lacking leucine (L) and tryptophan (W) (−LW). The growth of yeast cells on SD/−LW medium indicated that they carried both pGBKT7 and pGADT7 plasmids. An aliquot (10 μl) of 1:10 diluted stationary phase cultured yeast clones grown on the SD/−LW medium was then scribed on the selective SD medium lacking L, W, histidine (H), and adenine (A) (−LWHA). The growth of yeast cells on SD/−LWHA medium indicated that the examined proteins interacted with each other. The interactions of these proteins were also assessed by examination of β-D-galactopyranoside (X-α-gal) activity and β-galactosidase (LacZ) activity as described previously (Yuan et al., 2010). Each yeast two-hybrid assay was repeated at least twice.
CoIP assays were performed to study the interaction between TALE proteins and plant proteins in planta. The 9×myc DNA fragment was amplified from pN-TAPa vector (Rubio et al., 2005) by using myc-specific primers (Supplementary file 2) and inserted into the SmaI- and BamHI-digested pU1301 vector (Cao et al., 2007), resulting in a vector that we named pU1301-9myc. The DNA fragments of full-length, truncated, or TFB region of TALEs were ligated into the pU1031-9myc vector. The DNA fragments of plant genes were ligated into the pU1301-3FLAG vector (Yuan et al., 2010). The recombinant vectors were introduced into Agrobacterium tumefaciens strain GV3101. Agrobacterium-mediated transformation was performed by infiltration into N. benthamiana leaves using a needleless syringe (Yuan et al., 2010). CoIP assays were conducted using anti-FLAG antibody (RRID:AB_259529, Sigma-Aldrich, St. Louis, MO) and anti-myc antibody (Tiangen, Beijing, China) as described previously (Yuan et al., 2010). Each CoIP assay was repeated at least twice.
Site-directed mutation
Mutations of plant genes and the Xoo TALE genes were made using the GeneTailor Site-Directed Mutagenesis System (Invitrogen Life Technologies, Carlsbad, CA) as described previously (Yuan et al., 2011). The mutagenic primers are listed in Supplementary file 2.
Statistical analysis
Differences between samples were analysed for statistical significance by t-test in Microsoft Excel (Microsoft, Redmond, WA). Correlations between gene transcript level and disease level were calculated using CORREL analysis in the Microsoft Office Excel program.
Acknowledgements
We thank Professor Gongyou Chen of Shanghai Jiao Tong University for kindly providing Xanthomonas bacterial strains and Professor Qifa Zhang of Huazhong Agricultural University and the eLife editorial team for their help in editing the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31330062, 31100875, and 31371926), the National Program on the Development of Basic Research in China (2012CB114005), the Special Key Project by the Ministry of Science and Technology of China (2016YFD0100903), and the Fundamental Research Funds for the Central Universities (2014PY039).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Funding Information
This paper was supported by the following grants:
National Natural Science Foundation of China 31100875 to Meng Yuan.
National Natural Science Foundation of China 31371926 to Meng Yuan.
Ministry of Science and Technology of the People's Republic of China 2012CB114005 to Meng Yuan, Shiping Wang.
Ministry of Science and Technology of the People's Republic of China 2014PY039 to Meng Yuan.
Ministry of Science and Technology of the People's Republic of China 2016YFD0100903 to Shiping Wang.
National Natural Science Foundation of China 31330062 to Shiping Wang.
Additional information
Competing interests
The authors declare that no competing interests exist.
Author contributions
MY, Designed and performed most of the experiments, Analysed the data, Drafted the manuscript.
YK, Helped to generate transgenic rice plants and Xoo mutant, analyse protein-protein interactions and amplify the transcription activator-like effector.
RH, Helped to generate transgenic rice plants and Xoo mutant, analyse protein-protein interactions, and amplify the transcription activator-like effector.
LM, Helped to generate transgenic rice plants and Xoo mutant, analyse protein-protein interactions, and amplify the transcription activator-like effector.
ZY, Helped to generate transgenic rice plants and Xoo mutant, analyse protein-protein interactions, and amplify the transcription activator-like effector.
ZC, Helped to generate transgenic rice plants and Xoo mutant, analyse protein-protein interactions, and amplify the transcription activator-like effector.
JX, Provided biochemical and molecular analysis support and management and final approval of the manuscript.
XL, Provided biochemical and molecular analysis support and management and final approval of the manuscript.
SW, Supervised the project, Designed some of the experiments, Interpreted data, Revised the manuscript.
Additional files
Major datasets
The following previously published dataset was used:
Wang L,Xie W,Chen Y,Tang W,Yang J,Ye R,Liu L,Xu C,Lin Y,Xiao J,Zhang Q,2010,Dissecting the developmental transcriptomes of rice,http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE19024,Publicly available at the NCBI Gene Expression Omnibus (accession no. GSE19024)
References
- Boch J, Bonas U, Lahaye T. TAL effectors--pathogen strategies and plant resistance engineering. New Phytologist. 2014;204:823–832. doi: 10.1111/nph.13015. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S. The expression pattern of a rice disease resistance gene xa3/xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics. 2007;177:523–533. doi: 10.1534/genetics.107.075176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas RA, Doyle EL, Niño-Liu DO, Wilkins KE, Bancroft T, Wang L, Schmidt CL, Caldo R, Yang B, White FF, Nettleton D, Wise RP, Bogdanove AJ. Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathogens. 2014;10:e1003972. doi: 10.1371/journal.ppat.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wang S, Zhang Q. New gene for bacterial blight resistance in rice located on chromosome 12 identified from minghui 63, an elite restorer line. Phytopathology. 2002;92:750–754. doi: 10.1094/PHYTO.2002.92.7.750. [DOI] [PubMed] [Google Scholar]
- de Lange O, Wolf C, Dietze J, Elsaesser J, Morbitzer R, Lahaye T. Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain. Nucleic Acids Research. 2014;42:7436–7449. doi: 10.1093/nar/gku329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Liu H, Li X, Xiao J, Wang S. A CCCH-type zinc finger nucleic acid-binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiology. 2012;158:876–889. doi: 10.1104/pp.111.191379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhao J, Xu C, Li X, Wang S. Development of rice germplasms conferring high-level and broad-spectrum resistance to Xanthomonas oryzae pv. oryzae at both seedling and adult stages. Molecular Plant Breeding. 2010;8:420–425. doi: 10.3969/mpb.008.000420. [DOI] [Google Scholar]
- Ge X, Chu Z, Lin Y, Wang S. A tissue culture system for different germplasms of indica rice. Plant Cell Reports. 2006;25:392–402. doi: 10.1007/s00299-005-0100-7. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gu K, Tian D, Qiu C, Yin Z. Transcription activator-like type III effector AvrXa27 depends on OsTFIIAgamma5 for the activation of Xa27 transcription in rice that triggers disease resistance to Xanthomonas oryzae pv. oryzae. Molecular Plant Pathology. 2009;10:829–835. doi: 10.1111/j.1364-3703.2009.00567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- He YQ, Zhang L, Jiang BL, Zhang ZC, Xu RQ, Tang DJ, Qin J, Jiang W, Zhang X, Liao J, Cao JR, Zhang SS, Wei ML, Liang XX, Lu GT, Feng JX, Chen B, Cheng J, Tang JL. Comparative and functional genomics reveals genetic diversity and determinants of host specificity among reference strains and a large collection of Chinese isolates of the phytopathogen Xanthomonas campestris pv. campestris. Genome Biology. 2007;8:R218. doi: 10.1186/gb-2007-8-10-r218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høiby T, Zhou H, Mitsiou DJ, Stunnenberg HG. A facelift for the general transcription factor TFIIA. Biochimica Et Biophysica Acta. 2007;1769:429–436. doi: 10.1016/j.bbaexp.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Iyer AS, McCouch SR. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Molecular Plant-Microbe Interactions. 2004;17:1348–1354. doi: 10.1094/MPMI.2004.17.12.1348. [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi AS, McCouch SR. Recessive resistance genes and the Oryza sativa-Xanthomonas oryzae pv. oryzae pathosystem. Molecular Plant-Microbe Interactions. 2007;20:731–739. doi: 10.1094/MPMI-20-7-0731. [DOI] [PubMed] [Google Scholar]
- Juillerat A, Bertonati C, Dubois G, Guyot V, Thomas S, Valton J, Beurdeley M, Silva GH, Daboussi F, Duchateau P. BurrH: a new modular DNA binding protein for genome engineering. Scientific Reports. 2014;4:3831. doi: 10.1038/srep03831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Hui S, Yuan M. Xanthomonas oryzae pv. oryzae Inoculation and Growth Rate on Rice by Leaf Clipping Method. BIO-PROTOCOL. 2017;7:e2568. doi: 10.21769/BioProtoc.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Liu H, Li X, Xiao J, Wang S. Rice OsPAD4 functions differently from Arabidopsis AtPAD4 in host-pathogen interactions. The Plant Journal. 2014;78:619–631. doi: 10.1111/tpj.12500. [DOI] [PubMed] [Google Scholar]
- Kottapalli KR, Kottapalli P, Agrawal GK, Kikuchi S, Rakwal R. Recessive bacterial leaf blight resistance in rice: complexity, challenges and strategy. Biochemical and Biophysical Research Communications. 2007;355:295–301. doi: 10.1016/j.bbrc.2007.01.134. [DOI] [PubMed] [Google Scholar]
- Li HJ, Li XH, Xiao JH, Wing RA, Wang SP. Ortholog alleles at Xa3/Xa26 locus confer conserved race-specific resistance against Xanthomonas oryzae in rice. Molecular Plant. 2012;5:281–290. doi: 10.1093/mp/ssr079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Le Gourierrec J, Torki M, Kim YJ, Guerineau F, Zhou DX. Characterization and functional analysis of Arabidopsis TFIIA reveal that the evolutionarily unconserved region of the large subunit has a transcription activation domain. Plant Molecular Biology. 1999;39:515–525. doi: 10.1023/A:1006139724849. [DOI] [PubMed] [Google Scholar]
- Lin YJ, Zhang Q. Optimising the tissue culture conditions for high efficiency transformation of indica rice. Plant Cell Reports. 2005;23:540–547. doi: 10.1007/s00299-004-0843-6. [DOI] [PubMed] [Google Scholar]
- Maggio I, Gonçalves MA. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends in Biotechnology. 2015;33:280–291. doi: 10.1016/j.tibtech.2015.02.011. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Qiu D, Xiao J, Ding X, Xiong M, Cai M, Cao Y, Li X, Xu C, Wang S. OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Molecular Plant-Microbe Interactions. 2007;20:492–499. doi: 10.1094/MPMI-20-5-0492. [DOI] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. The Plant Journal. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- Römer P, Recht S, Strauss T, Elsaesser J, Schornack S, Boch J, Wang S, Lahaye T. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytologist. 2010;187:1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, Furutani A, Ochiai H, Delcher AL, Kelley D, Madupu R, Puiu D, Radune D, Shumway M, Trapnell C, Aparna G, Jha G, Pandey A, Patil PB, Ishihara H, Meyer DF, Szurek B, Verdier V, Koebnik R, Dow JM, Ryan RP, Hirata H, Tsuyumu S, Won Lee S, Seo YS, Sriariyanum M, Ronald PC, Sonti RV, Van Sluys MA, Leach JE, White FF, Bogdanove AJ. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204. doi: 10.1186/1471-2164-9-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, Moscou MJ, Ward ER, Horvath DM. Engineering plant disease resistance based on TAL effectors. Annual Review of Phytopathology. 2013;51:383–406. doi: 10.1146/annurev-phyto-082712-102255. [DOI] [PubMed] [Google Scholar]
- Schreiber T, Sorgatz A, List F, Blüher D, Thieme S, Wilmanns M, Bonas U. Refined requirements for protein regions important for activity of the TALE AvrBs3. PLoS One. 2015;10:e0120214. doi: 10.1371/journal.pone.0120214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugio A, Yang B, Zhu T, White FF. Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIA 1 and OsTFX1 during bacterial blight of rice. PNAS. 2007;104:10720–10725. doi: 10.1073/pnas.0701742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Cao Y, Yang Z, Xu C, Li X, Wang S, Zhang Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. The Plant Journal. 2004;37:517–527. doi: 10.1046/j.1365-313X.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Szurek B, Marois E, Bonas U, Van den Ackerveken G. Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. The Plant Journal. 2001;26:523–534. doi: 10.1046/j.0960-7412.2001.01046.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhang X, Fan Y, Gao Y, Zhu Q, Zheng C, Qin T, Li Y, Che J, Zhang M, Yang B, Liu Y, Zhao K. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Molecular Plant. 2015;8:290–302. doi: 10.1016/j.molp.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Wilkins KE, Booher NJ, Wang L, Bogdanove AJ. TAL effectors and activation of predicted host targets distinguish Asian from African strains of the rice pathogen Xanthomonas oryzae pv. oryzicola while strict conservation suggests universal importance of five TAL effectors. Frontiers in Plant Science. 2015;6:536. doi: 10.3389/fpls.2015.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Wang G, Yuan M, Yao W, Lyu K, Zhao H, Yang M, Li P, Zhang X, Yuan J, Wang Q, Liu F, Dong H, Zhang L, Li X, Meng X, Zhang W, Xiong L, He Y, Wang S, Yu S, Xu C, Luo J, Li X, Xiao J, Lian X, Zhang Q. Breeding signatures of rice improvement revealed by a genomic variation map from a large germplasm collection. PNAS. 2015;112:E5411–E5419. doi: 10.1073/pnas.1515919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Chen Z, Cao J, Guan H, Lin D, Li C, Lan T, Duan Y, Mao D, Wu W. Toward the positional cloning of qBlsr5a, a QTL underlying resistance to bacterial leaf streak, using overlapping sub-CSSLs in rice. PLoS One. 2014;9:e95751. doi: 10.1371/journal.pone.0095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Sugio A, White FF. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. PNAS. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, White FF. Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Molecular Plant-Microbe Interactions. 2004;17:1192–1200. doi: 10.1094/MPMI.2004.17.11.1192. [DOI] [PubMed] [Google Scholar]
- Yuan M, Chu Z, Li X, Xu C, Wang S. The bacterial pathogen Xanthomonas oryzae overcomes rice defenses by regulating host copper redistribution. The Plant Cell. 2010;22:3164–3176. doi: 10.1105/tpc.110.078022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Li X, Xiao J, Wang S. Characterization of Xanthomonas oryzae-responsive cis-acting element in the promoter of rice race-specific susceptibility gene Xa13. Molecular Plant. 2011;4:300–309. doi: 10.1093/mp/ssq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate that is stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- Zhao H, Yao W, Ouyang Y, Yang W, Wang G, Lian X, Xing Y, Chen L, Xie W. RiceVarMap: a comprehensive database of rice genomic variations. Nucleic Acids Research. 2015;43:D1018–D1022. doi: 10.1093/nar/gku894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Yang B, Chittoor JM, Johnson LB, White FF. AvrXa10 contains an acidic transcriptional activation domain in the functionally conserved C terminus. Molecular Plant-Microbe Interactions. 1998;11:824–832. doi: 10.1094/MPMI.1998.11.8.824. [DOI] [PubMed] [Google Scholar]