Abstract
ULK1 and ULK2 are thought to be essential for initiating autophagy, and Ulk1/2-deficient mice die perinatally of autophagy-related defects. Therefore, we used a conditional-knockout approach to investigate the roles of ULK1/2 in the brain. Although the mice showed neuronal degeneration, the neurons showed no accumulation of P62+/ubiquitin+ inclusions or abnormal membranous structures, which are observed in mice lacking other autophagy genes. Rather, neuronal death was associated with activation of the unfolded protein response (UPR) pathway. An unbiased proteomics approach identified SEC16A as an ULK1/2-interacting partner. ULK-mediated phosphorylation of SEC16A regulated the assembly of endoplasmic reticulum (ER) exit sites and ER-to-Golgi trafficking of specific cargo, which did not require other autophagy proteins (e.g. ATG13). The defect in ER-to-Golgi trafficking activated the UPR pathway in ULK-deficient cells; both processes were reversed upon expression of SEC16A with a phosphomimetic substitution. Thus, the regulation of ER-to-Golgi trafficking by ULK1/2 is essential for cellular homeostasis.
INTRODUCTION
Autophagy maintains cellular homeostasis by recycling intermediate metabolites sequestered within double-membrane–bound vesicles (i.e., autophagosomes) (Levine and Kroemer, 2008). The endoplasmic reticulum (ER) serves as both a platform for autophagosome formation and a source of membrane (Mizushima et al., 2011). Membrane nucleation for selective (i.e., mitophagy) and starvation-induced autophagy is regulated by the ULK1/ATG1 complex, comprising ULK1, ATG13, FIP200, and ATG101 (Mizushima et al., 2011). ULK1 interacts with the HSP90–CDC37 chaperone complex, which stabilizes a kinase-competent form of ULK1 (Joo et al., 2011). ULK1 is also regulated by multiple signaling kinases, including mTOR and AMPK, whose activities are directly influenced by nutrient/energy availability (Alers et al., 2012). In response to nutrient/energy depletion, ULK1 activates the ATG14-BECN1-VPS34–containing PI3-kinase complex by phosphorylating multiple subunits (Egan et al., 2015; Russell et al., 2013), thereby promoting the recruitment of PI3P-binding proteins to sites of autophagosome formation. However, even in the absence of metabolic stress, autophagy-inducing activity of the ULK1/ATG1 complex can be stimulated to eliminate intracellular pathogens and cargo via interaction with cargo receptors (Huang and Brumell, 2014).
There are single copies of the ATG1 gene in yeast (ATG1), flies (Atg1), and worms (unc-51). In mammals, the gene appears to be duplicated, and at least 2 genes, ULK1 and ULK2, have redundant roles in autophagy. We do not know how the functions of these genes overlap; but, mice with germline deletions of Ulk1 or Ulk2 are viable, while those lacking both die shortly after birth (Cheong et al., 2011; Cheong et al., 2014). Premature death is at least in part due to autophagy-related defects in glycogen metabolism, similar to those in mice lacking Atg5 or Atg7, two nonredundant autophagy genes (Komatsu et al., 2005; Kuma et al., 2004).
Disruption of autophagy is implicated in the pathogenesis of various neurodegenerative conditions (Hara et al., 2006; Komatsu et al., 2006; Liang et al., 2010; Nixon, 2013; Menzies et al., 2015). Given the importance of autophagy in neuronal homeostasis and the redundant roles of ULK1 and ULK2 in autophagy, we decided to investigate the consequence of deleting both kinases from the CNS of developing mice.
RESULTS
Mice Lacking Ulk1/Ulk2 Expression in the CNS Have a Distinct Pattern of Neuronal Loss
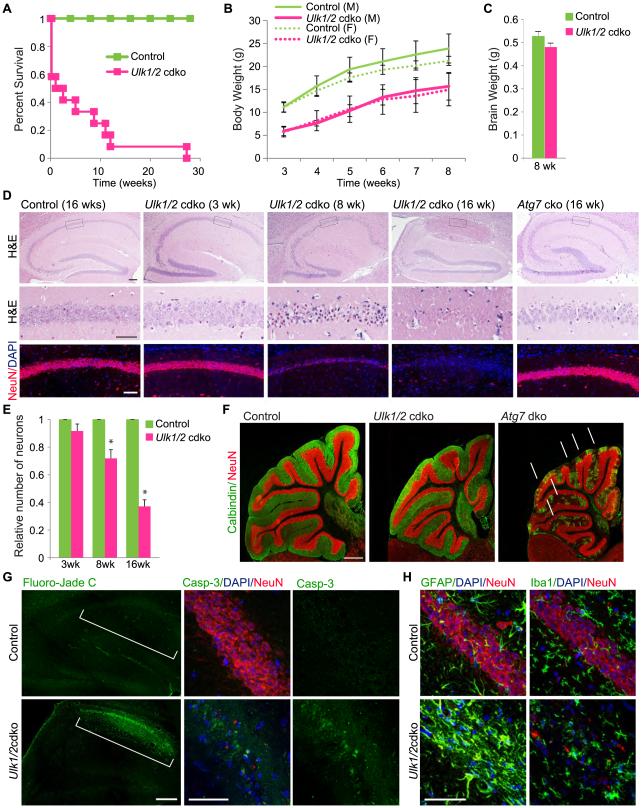
Ulk1 and Ulk2 mRNA are expressed throughout the CNS, with the highest levels in the cerebellum (Ulk1>Ulk2) and hippocampus (Ulk2>Ulk1) (Figure S1A). To investigate the role(s) of these kinases in the CNS, we generated mice lacking both Ulk1 and Ulk2 in the CNS [Ulk1−/−;Ulk2−/flox;Nestin-Cre; hereafter referred to as Ulk1/2-cdko (conditional–double-knockout) mice] and those expressing Ulk1 but not Ulk2 in the CNS (Ulk1+/−;Ulk2−/flox; Nestin-Cre and Ulk1+/+;Ulk2−/flox;Nestin-Cre; hereafter referred to as “controls”) (Figure S1C). We confirmed the loss of Ulk1 and Ulk2 mRNA expression in the brains (i.e., hippocampal region) of Ulk1/2-cdko mice by RT-qPCR analyses (Figure S1D). Although the Ulk1/2-cdko mice were born at the expected Mendelian ratio, approximately 40% died within 24 h (Figure 1A). Survival of the remaining Ulk1/2-cdko mice diminished over the ensuing 12 wk, and all were dead by 28 wk (Figure 1A). Newborn Ulk1/2-cdko mice were visibly indistinguishable from their littermates, but at 3 wk showed substantial, sex-independent weight loss (Figure 1B). The weight of the brains of 8-wk-old Ulk1/2-cdko mice did not differ from that of littermate controls (Figure 1C).
Figure 1. Degeneration of pyramidal neurons in the CA1 region of ULK1/2-deficient mice.
(A) Survival of control (n=20) and Ulk1/2-cdko (n=15) mice. (B) Average body weight ±SEM of male (M) and female (F) Ulk1/2-cdko (n=11) mice compared to control (n=26) mice. (C) The average brain weights±SEM of control (n=5) and Ulk1/2-cdko (n=5) mice did not significantly differ (Student’s t-test). (D) – Representative images of serial brain sections stained with hematoxylin and eosin (H&E), DAPI, and antibodies against the neuronal marker NeuN. Scale bars: 200 μm (E) Average number of pyramidal neurons (normalized to that in littermate controls) ±SEM in a 500-μm2 area of CA1 (n=3 mice/genotype for each age group).. *P <0.001 (Student’s t-test) when compared with control. (F) Representative images of serial brain sections stained with antibodies against NeuN and the cerebellar Purkinje cell marker, calbindin. Scale bar: 500 μm. (G) Representative images of Fluoro-Jade C stained and cleaved Caspase 3 (Casp-3) immunostained brain sections from the f 16-wk-old mice. The CA1 region is indicated by brackets. Scale bars: 200 μm (Fluoro-Jade C); 50 μm (cleaved Casp-3). (H) Representative images of serial brain sections from hippocampal region of16-wk-old mice stained with antibodies against GFAP or IBA1 and counterstained with anti-NeuN and DAPI. Scale bars: 50 μm. See also Figure S1.
Ulk1/2-cdko mice showed abnormal limb-clasping reflexes (Figure S1E) similar to that reported in Atg5–conditional knockout (cko) and Atg7-cko animals (Hara et al., 2006; Komatsu et al., 2006). However, unlike ATG5- or ATG7-deficient mice, which develop cerebellar ataxia (Hara et al., 2006; Komatsu et al., 2006), Ulk1/2-cdko mice had a normal gait (Figure S1F) and only a mild delay in motor skill learning compared to littermate controls (Figure S1G).
To identify the neurologic deficits in Ulk1/2-cdko animals, we analyzed their brain histology at various time points. Hematoxylin and eosin staining showed progressive degeneration of pyramidal neurons in the hippocampal CA1 region after 3 wk (Figure 1D-E). The dystrophic hippocampal neurons also showed loss of expression of the neuronal marker NeuN (Figure 1D). The hippocampal degeneration in these mice was more severe than that in age-matched Atg7-cko mice (Figure 1D). In contrast, neuronal loss from other brain regions (e.g., cerebellum) was more severe in Atg7-cko mice than in Ulk1/2-cdko mice (Figures 1F and S1H-I). The hippocampal degeneration in Ulk1/2-cdko mice was accompanied by increased fluoro-Jade C+ staining, which labels degenerating neurons, and activated caspase-3 staining in the CA1 region (Figure 1G). Immunostaining for the glial marker GFAP and microglial marker IBA1 was increased in the hippocampal CA1 region (Figure 1H), which was consistent with glial activation in response to neuronal damage. These histopathologic changes were not seen in littermate controls. These results highlight the functional redundancy of ULK1 and ULK2 in the CNS and the differential sensitivity of neurons to their loss.
ULK1/2 Deficiency Activates the Unfolded Protein Response in Hippocampal Neurons
To determine whether the hippocampal degeneration in Ulk1/2-cdko mice is associated with defective autophagy, we examined steady-state levels of the ubiquitin-binding protein P62/SQSTM1 (hereafter referred to as P62), an autophagy substrate whose levels correlate inversely with autophagy flux (Ichimura and Komatsu, 2010). Immunoblot analyses revealed minimal increase in P62 in extracts from hippocampal regions of Ulk1/2-cdko mice compared to that in littermate controls (Figure S2A-B). The level of P62 in the brains of Ulk1/2-cdko mice (<2-fold increase) was much lower than that in corresponding regions of Atg7-cko mice (>40-fold increase) and was not accompanied by a change in lipidated LC3 (LC3-II) levels (Figure S2A-B). Moreover, we did not detect any P62+ or ubiquitin+ inclusions in the hippocampal neurons of Ulk1/2-cdko mice (Figure 2A). These results indicate that in neurons, the autophagy-mediated turnover of ubiquitinated proteins that occurs under basal physiologic conditions proceeds in the absence of ULK1 and ULK2, and that defective autophagy is probably not the primary cause of neuronal degeneration in Ulk1/2-cdko mice. Given the surprising absence of P62 accumulation in the brains of the Ulk1/2-cdko mice, we examined autophagy flux in SV40 immortalized murine embryonic fibroblasts (MEFs) generated from wild-type (WT) and Ulk1/2-dko mice. Although the levels of LC3-lipidation and turnover and p62 were comparable between WT and ULK1/2-deficient MEFs grown in complete medium, ULK1/2-deficient MEFs showed a marked reduction in LC3 lipidation and turnover and increase in P62 levels compared to those in WT MEFs upon amino acid deprivation (Figure S1C). These results are consistent with the expected defect in starvation-induced autophagy in ULK1/2-deficient MEFs (Cheong et al., 2011).
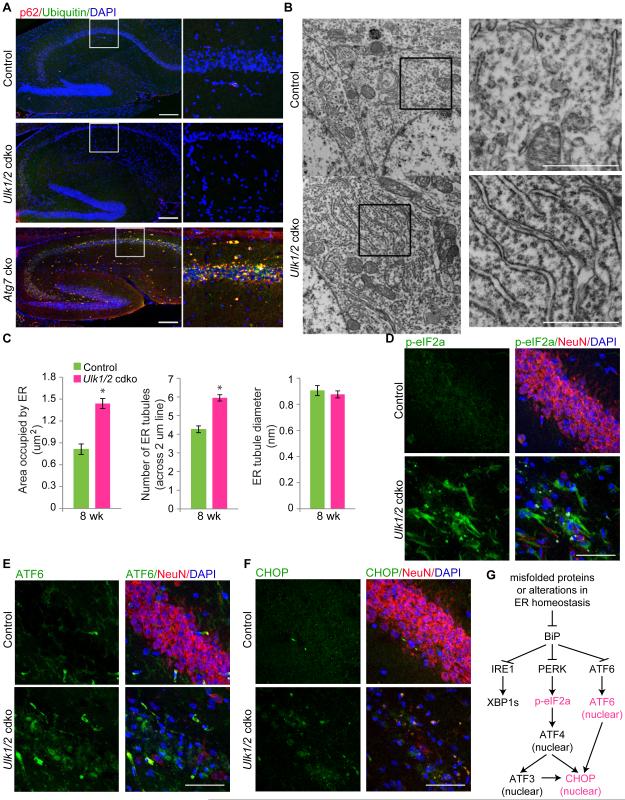
Figure 2. Ulk1/2 deficiency in hippocampal neurons is associated with activation of the UPR pathway.
(A) Representative images of brain sections from 4-wk-old Atg7-cko, 8-wk-old Ulk1/2-cdko and 8-wk-old control mice stained with antibodies against ubiquitin and P62, and counterstained with DAPI. Scale bars: 200 μm. (B) Representative electron micrographs of neurons in CA1 region from 8-wk-old mice. Scale bars: 1 μm. (C) Morphometric analyses of electron micrographs of hippocampal CA1 neurons from 8-wk-old control (n=2) and Ulk1/2-cdko (n=2) mice. *P <0.001 (Student's t-test). (D-F) Representative images of brain sections from the hippocampal region of16-wk-old mice stained with antibodies against p-eIF2α, ATF6 or CHOP, and counterstained with anti-NeuN. Sections were counterstained with anti-NeuN and DAPI. Scale bars: 50 μm. (G) Diagram of the UPR pathway, highlighting the activation of the PERK and ATF6 arms in the Ulk1/2-cdko mice. See also Figure S2.
To learn the cause of the neuronal degeneration in Ulk1/2-cdko mice, we examined the hippocampal region by transmission electron microscopy. Although we did not detect any accumulation of the atypical membranous structures or abnormal mitochondria that are seen with deficiency of other autophagy-related genes (Hara et al., 2006; Komatsu et al., 2006; Liang et al., 2010), the ER compartment was expanded (Figure 2B-C). The phosphorylated ribosomal protein S6 levels were also lower in hippocampal neurons of Ulk1/2-cdko (Figure S2D), suggesting suppression of mTOR activity in those cells.
Because ER expansion (Schuck et al., 2009) and mTOR inhibition in neurons (Di Nardo et al., 2009) can result from ER stress and activation of the unfolded protein response (UPR) pathway, which may ultimately lead to cell death, we examined the expression of markers associated with UPR activation in hippocampal pyramidal neurons. Three major sensors of ER stress initiate the UPR: PERK, ATF6, and IRE1. These sensors are activated upon release from the ER chaperone BiP, which is competed away by accumulation of misfolded proteins in the ER (Walter and Ron, 2011). We did not detect spliced XBP1, an alternative product that is produced by activating IRE1 in mRNA samples prepared from the CA1 region (data not shown). However, the levels of phosphorylated eIF2α, a substrate of PERK, and nuclear localization of ATF6 were both increased in the CA1 region of Ulk1/2-cdko mice with degenerating neurons (Figure 2D-E). Consistent with activation of the PERK–eIF2α and ATF6 pathways in the CA1 region of Ulk1/2-cdko mice, the levels and nuclear localization of CHOP, a transcription factor that promotes ER stress–mediated apoptosis, were increased (Figure 2F-G). These results indicate that the degeneration of Ulk1/2-deficient pyramidal neurons in the CA1 region disrupts a process that leads to ER stress and UPR activation but not the accumulation of p62+/ubiquitin+ inclusions or membranous structures seen in mice lacking other autophagy genes (e.g., FIP200, Atg5, or Atg7).
ULK1 and ULK2 Mediate the Phosphorylation of SEC16A
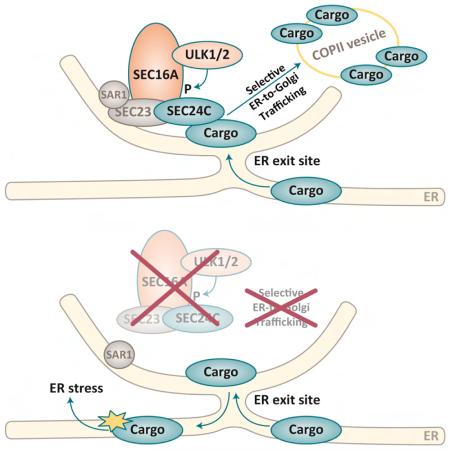
We used an unbiased proteomics-based approach to identify ULK/ATG1-interacting proteins and gain insight into ULK/ATG1 function. We analyzed immunoprecipitates (IPs) of endogenous ULK1 from WT MEFs by liquid chromatography/mass spectrometry. SEC16A was among proteins identified only in ULK1-containing samples (Figure S3A). The ULK1–SEC16 interaction was confirmed by immunoblot analyses of ULK1 IPs in WT MEFs (Figure 3A) and hippocampal extracts from WT mice (Figure 3B). SEC16A is a large protein that localizes to ER-exit sites (ERES) and facilitates the recruitment of soluble subunits of the coatomer protein complex II (COPII), including SEC23, SEC24, SEC13, and SEC31 (Miller and Barlowe, 2010). The COPII machinery distinguishes transmembrane and luminal secretory cargo from resident ER proteins and packages them into transport vesicles destined for Golgi. Defects in ER-to-Golgi trafficking can lead to the accumulation of protein in the ER and activation of the UPR (Fang et al., 2015; Preston et al., 2009); therefore, we further characterized the interaction between the ULKs and SEC16A.
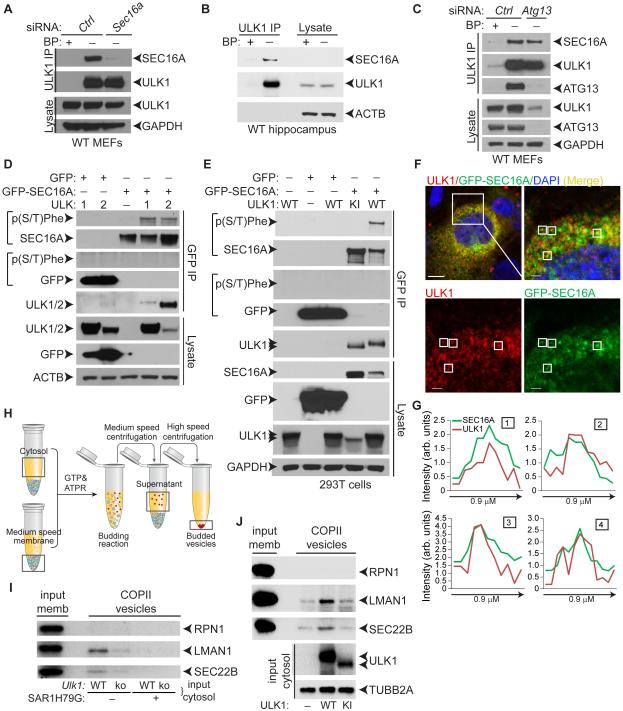
Figure 3. ULKs mediate the phosphorylation of SEC16A and facilitate in vitro budding of COPII vesicles.
(A) Representative immunoblot analyses of IPs from WT MEFs transfected with either control nontargeting siRNA (Ctrl) or Sec16a siRNA using anti-ULK1 antibodies in the presence or absence of epitope-specific blocking peptides (BP). (B) Representative immunoblot analyses of ULK1 IPs from the hippocampus of 4-wk-old WT mice. (C) Representative immunoblot analyses of ULK1 IPs from WT MEFs transfected with either control nontargeting siRNA (Ctrl) or Atg13 siRNA. (D) Representative immunoblot analyses of 293T cells transfected with GFP–SEC16A and either C-terminal–MYC-DDK–tagged ULK1 or ULK2 expression constructs. Phosphorylated SEC16A was detected using the anti-p(S/T)Phe antibody. (E) Representative immunoblot analyses of GFP IPs from 293T cells transfected with GFP or GFP–SEC16A and untagged WT or KI ULK1 expression constructs. (F) Representative low- and high-magnification pseudocolored images of Ulk1-ko MEFs cotransfected with ULK1 and GFP–SEC16A and stained with antibodies against ULK1 and DAPI. (G) Line scans showing the degree of colocalization between ULK1 (red) and SEC16A (green) in lines (not shown) drawn within the boxes labeled 1-4 in the high magnification images from panel F. Scale bars: 10 μm (low magnification) and 5 μm (high magnification). (H) Diagram of the in vitro COPII-budding reaction. (I) Representative immunoblot analyses of budded vesicles from in vitro COPII-budding reactions using cytosolic fractions from WT or Ulk1-ko MEFs and membrane fractions from Ulk1-ko MEFs. RPN1 is an ER-resident protein; LMAN1/ERGIC53 and SEC22B are COPII cargo whose incorporation into budded vesicles is inhibited by SAR1H79G, a dominant-negative form of the COPII-specific GTPase. (J) Representative immunoblot analyses of budded vesicles from in vitro COPII-budding reactions using cytosolic fractions from 293T cells transfected with empty vector (—), WT, or KI ULK1 combined with membrane fractions from untransfected 293T cells. See also Figure S3.
ULK1 exists in a complex with ATG13, FIP200, and ATG101; the stability of the complex and ULK1 levels are reduced in ATG13-deficient cells (Hosokawa et al., 2009). RNAi-mediated silencing of Atg13 expression reduced ULK1 steady-state levels but did not diminish the ULK1–SEC16A interaction (Figure 3C). Next, we identified the ULK1 domain that interacts with SEC16A. Immunoblot analyses of cells expressing FLAG-tagged ULK1-deletion constructs revealed that the N-terminal domain of ULK1 is necessary and sufficient to bind SEC16A (Figure S3B-C). Given the high degree of homology between ULK1 and ULK2 in their N-terminal domains (Mizushima et al., 2011), it was not surprising that we detected MYC-DDK–tagged ULK2 in GFP–SEC16A+ IPs (Figure 3D). ULK1 was also detected in GFP IPs containing GFP-tagged SEC16B (Figure S3D). The region of homology between SEC16A and SEC16B is restricted to a 469-residue central domain that is conserved among all SEC16 isoforms (Bhattacharyya and Glick, 2007). These results suggest that the interaction between the ULKs and SEC16A is mediated by the evolutionarily conserved domains of each protein (Figure S3E) and does not require ATG13.
Many ULK-interacting proteins are phosphorylated by ULK1 and ULK2. Therefore, we wondered if SEC16A is an ULK substrate. To test this hypothesis, we used a phospho-serine/threonine–specific antibody that recognizes phosphorylated serine/threonine residues with tyrosine, tryptophan, or phenylalanine at the −1 position or phenylalanine at the +1 position [hereafter referred to as p(S/T)Phe] (Kalabis et al., 2006). This antibody is predicted to recognize the S15 (MQVSFVCQ) and S30 (LDTSFKILD) residues of Beclin-1 and S249 (ILKSFELVK) of VPS34 that are phosphorylated by ULK1 (Egan et al., 2015; Russell et al., 2013). GFP–SEC16A coexpressed with ULK1 or ULK2 but not with the kinase-inactive (KI) ULK1 mutant (K46A) showed robust serine phosphorylation (Figure 3D-E), which was abolished by treating IPs with calf alkaline intestinal phosphatase prior to SDS-PAGE (Figure S3F). These results indicate that SEC16A is phosphorylated in an ULK1/2-dependent manner.
ULK1 Activity Regulates the Budding of COPII Vesicles in Vitro
The assembly and budding of COPII vesicles is initiated upon activation of the small COPII GTPase SAR1. Sar1 promotes the assembly of soluble SEC23–SEC24 heterodimers, which form the inner coat of the transport vesicles, and soluble SEC13–SEC31 heterodimers, which form an outer lattice (Zanetti et al., 2012). SEC16A has a punctate-distribution pattern in cells in culture, consistent with its localization to ERES, where it facilitates the assembly of the soluble COPII components (Bhattacharyya and Glick, 2007; Watson et al., 2006). ULK1 localized to a subset of GFP–SEC16A+ puncta (Figure 3F-G), suggesting that it is recruited to ERES, where it may regulate COPII transport.
We examined the contribution of ULK1 activity in the budding of COPII vesicles in vitro. The assay combines the cargo-containing membrane fraction from 1 cell source with cytosolic extracts, which supply soluble COPII proteins and putative regulatory proteins, from another cell source (Figure 3H). Cytosolic extracts derived from autophagy-deficient Atg5-ko MEFs support COPII budding in vitro; and the incorporation of 2 endogenous cargo, SEC22B and LMAN1, into budded COPII is not inhibited by adding the type-III PI3 kinase inhibitor 3-methyladenine (Ge et al., 2014), indicating that essential components of the autophagy machinery are not involved in the budding reaction. In contrast, budded COPII (i.e., SAR1-dependent) vesicles containing SEC22B and LMAN1 were reduced in reactions containing cytosolic extracts derived from Ulk1-ko MEFs (Figures 3I and S3G). Moreover, SEC22B and LMAN1 signals in budded vesicles were increased in reactions containing cytosolic extracts from 293T cells overexpressing WT ULK1, but not KI ULK1 (Figures 3J and S3H). These results suggest a role for ULK/ATG1 activity in the budding of COPII vesicles and/or incorporation of cargo into COPII vesicles.
ULK1 and Related Kinases Have a Conserved Role in Assembling COPII Components at ERES
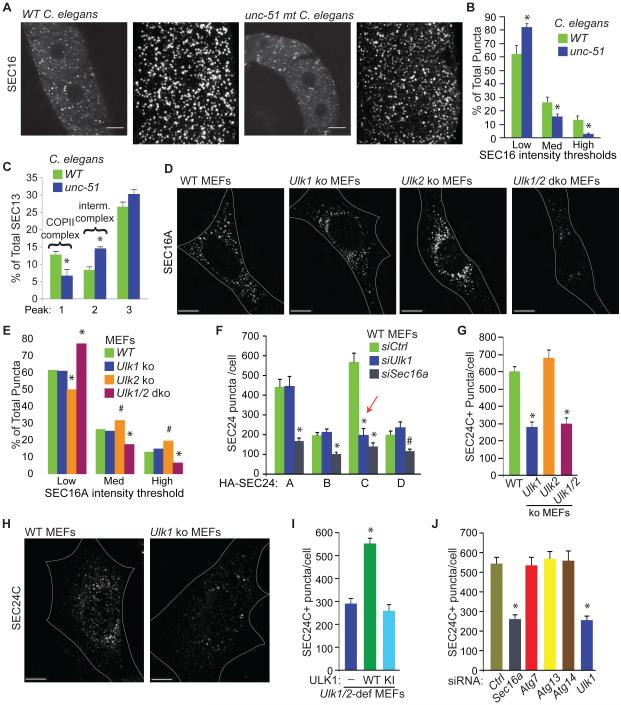
Because SEC16A promotes the assembly of soluble COPII components at ERES, and ULK1 localized to a subset of SEC16A+ puncta in MEFs and was required for efficient formation of LMAN1 and SEC22B-containing vesicles in vitro, we wondered if ULK1 and homologs regulate the assembly of COPII components at ERES. First, we examined the staining pattern of endogenous SEC16 in oocytes from loss-of-function mutant unc-51(e369) Caenorhabditis elegans. The proportion of high-intensity SEC16+ ERES in unc-51 mutant oocytes was significantly lower (P <0.05) than that in WT controls (Figure 4A-B). The amount of SEC13 in high molecular–weight gel-filtration fractions, which contain fully assembled COPII complexes comprising SEC13-containing heterotetramers and SEC23-containing heterodimers, was also lower (P <0.05) (Figures 4C and S4A-B). Intensity profiles of SEC23 and SEC13 further revealed shifts in the intermediate–molecular weight peaks (Figures 4C and S4A-B), suggesting the presence of abnormal partially assembled COPII complexes in animals expressing the mutant unc-51 allele. These results indicate that UNC-51/ATG1 is involved in the proper assembly of ERES in C. elegans.
Figure 4. ULKs regulate the assembly of COPII complexes at ERES.
(A) Representative low- and high-magnification images of distal gonads from WT and unc-51–mutant C. elegans stained using a fluorescently labelled anti-SEC16 antibody. (B) Mean percentages ±SEM of ERES that fall within specified intensity thresholds of SEC16 staining. For each condition, at least 1000 unique ERES from C. elegans gonads were examined. Scale bars: 10 μm. *P <0.05 (Student’s t-test) when compared with WT. (C) Mean percentages (±SEM) of total SEC13 in peaks 1-3 from Superose 6 gel filtration experiments similar to the one shown in Figure S4A. *P <0.05 (Student’s t-test) when compared with WT. (D) Representative images of endogenous SEC16A immunostaining in WT, Ulk1-ko, Ulk2-ko, and Ulk1/2-dko MEFs. Scale bars: 10 μm. (E) Mean percentages (±SEM) of all ERES that fall within specified intensity thresholds of SEC16A staining. *P <0.001 and #P <0.05 (ANOVA) when compared with WT. (F) Mean number (±SEM) of SEC24+ puncta per cell in WT MEFs transfected with the indicated HA-tagged SEC24 isoform (A, B, C, or D) and control nontargeting (Ctrl), Ulk1, or Sec16a siRNA. Ten HA+ cells per population were scored. Red arrow highlights the decrease in SEC24C puncta number in Ulk1-depleted cells. *P <0.001 and #P <0.05 (ANOVA) when compared with siCtrl. (G) Mean number (±SEM) of SEC24C+ puncta per cell in WT, Ulk1-ko, Ulk2-ko, and Ulk1/2-dko MEFs. Ten cells per population were scored. *P <0.001 (ANOVA) when compared with WT. (H) Representative images of endogenous SEC24C staining in WT and Ulk1-ko MEFs. Scale bars: 10 μm. (I) Mean number (±SEM) of SEC24C+puncta per cell in Ulk1/2-deficient (i.e., Ulk1 ko and Ulk2 shRNA) MEFs stably transduced with the indicated viral vector [i.e., (—) empty vector; WT ULK; or KI ULK1 mutant]. Ten cells per population were scored. *P <0.001 (ANOVA) when compared with empty vector-transduced cells. (J) Mean number (±SEM) of SEC24C+ puncta per cell in WT MEFs transfected with the indicated siRNA. Ten cells per population were scored. *P <0.001 (ANOVA) when compared with Ctrl siRNA-transfected cells. See also Figure S4.
The proportion of high-intensity SEC16A+ ERES in Ulk1/2-dko MEFs (Figure 4D-E) was significantly reduced (P <0.05), similar to that in unc-51–mutant oocytes (Figure 4A-B). Silencing Sec16a expression in mammalian cells decreases the accumulation of soluble COPII components at ERES (Watson et al., 2006). Given the reduced number of high-intensity SEC16A+ puncta in Ulk1/2-deficient MEFs, we compared the effects of silencing Sec16a and Ulk1 in WT and Ulk2-ko MEFs on the accumulation of cargo adapters, SEC24 A-D, at ERES (Figures 4F and S4C-F). RNAi-mediated knockdown of Sec16a significantly reduced (P <0.001) the number of puncta formed by each of the overexpressed SEC24 proteins in MEFs (Figures 4F and S4C-F). To our surprise, RNAi-mediated knockdown of Ulk1 in WT or Ulk2-ko MEFs selectively reduced the number of SEC24C+ puncta (Figures 4F and S4C-F) without affecting the overall levels of SEC24C (Figure S4C, F). The number of endogenous SEC24C+ puncta was also significantly reduced (P <0.001) in Ulk1-ko and Ulk1/2-dko MEFs (Figure 4G-H), as was the colocalization of SEC24C with SEC16A (Figure S4G-H), which is consistent with impaired assembly of SEC24C-containing COPII complexes at ERES in cells lacking ULK1. The defect in ERES assembly in either Ulk1- or Ulk1/2-deficient MEFs was rescued by ectopic expression of WT ULK1 but not the KI mutant (Figure 4I and Figure S4I), indicating that ULK1 catalytic activity is required for recruiting SEC24C to ERES. Whereas transient RNAi-mediated knockdown of Ulk1 or Sec16a in WT MEFs caused significant decreases (P<0.001) in the number of SEC24C+ puncta, knockdown or knockout of Atg7, Atg13, or Atg14 did not (Figure 4J and Figure S4J-L).
These results indicate that ULK/ATG1 has an evolutionarily conserved role in regulating ERES assembly, which is not shared by other autophagy-related proteins. ULK1 and ULK2 activities appear to converge on multiple aspects of SEC16A function, including the normal accumulation of SEC16A at ERES, and the assembly of SEC24C-containing ERES. These functions, however, appear to have different thresholds of sensitivity to loss of ULK/ATG1 function. SEC24C expression was relatively increased in hippocampal neurons, compared to the other SEC24 isoforms (Figure S4M) and neurons in other brain regions (data not shown). Therefore, these neurons may be particularly sensitive to the disrupted assembly of SEC24C-containing ERES, which occurs in the absence of Ulk1/2 expression.
ULK1 and Related Kinases Regulate ER-to-Golgi Trafficking
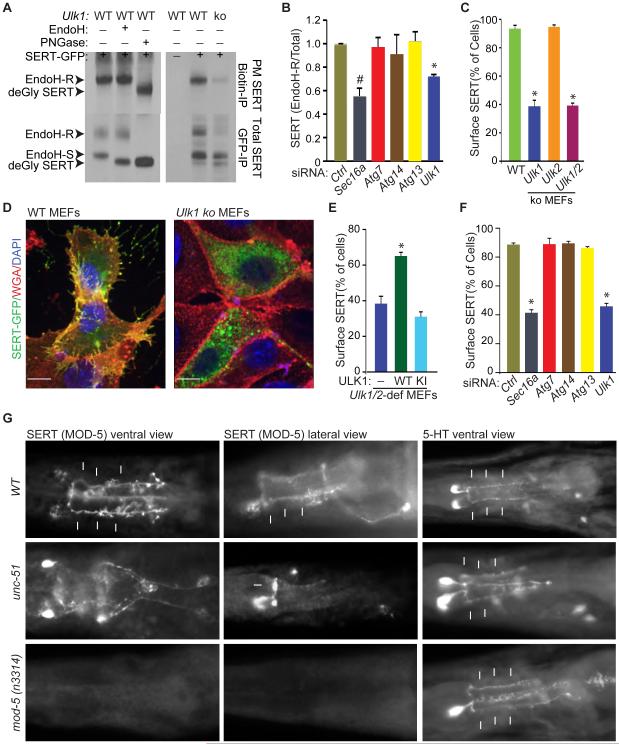
Sec16a depletion profoundly affects ER-to-Golgi trafficking in mammalian cells (Bhattacharyya and Glick, 2007; Iinuma et al., 2007; Watson et al., 2006), one manifestation of which is reduced delivery of the ts045-mutant strain of vesicular stomatitis virus envelope glycoprotein G (VSVG) to plasma membranes (PMs) (Presley et al., 1997). VSVG–GFP localizes exclusively to the ER in cells incubated at 40°C due to protein misfolding. After temperature reduction, VSVG–GFP in WT MEFs is rapidly transported from the ER to the PM via the Golgi apparatus, where it acquires a complex glycan structure that is resistant to Endoglycosidase H (Endo H) treatment (Presley et al., 1997). As expected, VSVG–GFP localization to the PMs of Sec16a-depleted cells was reduced (Figure S5A-B). PM localization of VSVG–GFP and acquisition of Endo H-resistant posttranslational modifications appeared normal in Ulk1-ko MEFs (Figure S5A-D1), and only a mild defect in VSVG trafficking was detected in Ulk1/2-dko MEFs (Figure S5C-D).
Because Ulk1/2-deficiency impaired the accumulation of SEC24C at ERES and only minimally affected VSVG trafficking, which relies to a greater extent on the A and B isoforms of SEC24 than on the C and D isoforms for ER export (Bonnon et al., 2010), we examined the role of the ULKs in trafficking SEC24C-specific cargo. The serotonin transporter SERT interacts specifically with SEC24C, and its PM localization depends on SEC24C-dependent export from the ER (Montgomery et al., 2014; Sucic et al., 2011). SERT is expressed on the surface of platelets and specific types of neurons. It regulates the concentration of serotonin (5-HT) at synapses and in blood plasma via uptake of 5-HT into neurons and platelets, respectively (Mercado and Kilic, 2010). Ulk1-ko MEFs showed reduced levels of the Endo H–resistant forms of SERT in whole-cell extracts (Figure 5A). Similarly, transient RNAi-mediated knockdown of Sec16a or Ulk1 in WT MEFs significantly reduced (P <0.05 and P <0.01, respectively) the ratio of Endo H–resistant to Endo H–sensitive SERT (Figures 5B and S5E-F). However, knockdown of other autophagy-related genes (e.g., Atg7, Atg13, or Atg14) had no effect on SERT trafficking (Figures 5B and S5E).
Figure 5. ULKs regulate ER-to-Golgi trafficking.
(A) Representative immunoblots of Biotin IPs (top panels) and GFP IPs (bottom panels) from SERT-GFP—transfected WT and Ulk1-ko MEFs. WT MEFs were incubated in the presence or absence of Endo H or peptide-N-glycosidase F (PNGase) to establish the migration pattern of the different glycosylated forms of SERT. (B) Mean ratios (±SD) of Endo H–R SERT to total SERT in RNAi-treated samples from 2 independent experiments. *P <0.01 and #P <0.05 and (ANOVA) when compared with Ctrl. (C) Mean percentages (±SEM) of cells showing colocalization of AlexaFluor 647–conjugated wheat germ agglutinin (WGA) and SERT-GFP. Data were acquired from 3 independent experiments, and more than 100 cells per population were scored. *P <0.001 (ANOVA) when compared with WT. (D) Representative merged pseudocolored images of SERT-GFP—transfected WT and Ulk1-ko MEFs stained with AlexaFluor 647–conjugated wheat germ agglutinin (WGA) and DAPI. Scale bar: 10 μm. (E) Mean percentages (±SEM) of cells with colocalized WGA and SERT-GFP. Data were acquired from 3 independent experiments, and more than 100 cells per population were scored in each experiment. *P <0.002 (ANOVA) when compared with empty vector–transduced cells. (F) Mean percentages (±SEM) of siRNA transfected cells with colocalized WGA and SERT-GFP. Data were acquired from 3 independent experiments, and more than 100 cells per population were scored. *P <0.001 (ANOVA) when compared with siCtrl-transfected cells. (G) Representative images of WT, unc-51 mutant and mod-5 mutant C. elegans stained with antibodies against MOD-5/SERT and 5-HT. The arrows highlight the 5-HT or MOD-5/SERT staining of NSM processes. See also Figure S5.
Treating cells with NHS-biotin prior to lysis allows selective biotinylation of surface proteins and provides a sensitive biochemical method of detecting defects in trafficking of proteins to the PM. Consistent with defective ER-to-Golgi trafficking of SERT, the levels of biotinylated surface SERT were reduced in anti-biotin IPs prepared from Ulk1-ko MEFs (Figure 5A). Ulk1-ko and Ulk1/2-dko MEFs also showed significant reduction (P <0.001) in PM localization of SERT–GFP (Figure 5C-D). The defect in cell surface localization of SERT in Ulk1/2-deficient MEFs was rescued by expressing WT but not KI ULK1 (Figure 5E), indicating that ULK1 catalytic activity is required for SERT trafficking. Transient RNAi-mediated knockdown of Sec16a or Ulk1 significantly reduced (P <0.001) SERT localization to the PM of WT MEFs, but knockdown of other autophagy-related genes (e.g., Atg7, Atg13, or Atg14) did not (Figures 5F and S5G).
We next examined the role of ULKs in endogenous SERT trafficking in vivo. Platelet SERT can be used as a peripheral biomarker for predicting serotonergic neurotransmission (Yubero-Lahoz et al., 2013). Therefore, we examined the steady-state levels of SERT on the surface of platelets from Ulk1-ko mice and their WT littermates by using 2 independent approaches: surface biotinylation followed by immunoblot analysis of SERT (Figure S5H-I) and flow cytometry with an anti-SERT antibody (Figure S5J). Consistent with a role for ULK1 in ER-to-Golgi trafficking of SERT, platelets isolated from Ulk1-ko mice showed a relative reduction in the proportion of SERT on their PMs (Figure S5H-J). The rate of 5-HT uptake into Ulk1-ko platelets was also significantly decreased (P <0.001; Figure S5K). These findings are consistent with ULKs functioning in SERT trafficking under physiologic conditions.
In C. elegans, SERT (MOD-5) is expressed on the PM, along the length of axons of 5-HT–producing pharyngeal neurosecretory motor (NSM) neurons, with minimal localization of SERT/MOD-5 in cell bodies. Thus, C. elegans is a sensitive system for detecting defects in SERT trafficking in neurons in vivo. In contrast to its pattern of expression in WT NSM neurons, SERT/MOD-5 accumulated in the cell bodies of unc-51–mutant NSM neurons and failed to localize to 5-HT–stained NSM axons (Figure 5G). These findings are consistent with a defect in ER-to-Golgi trafficking associated with disruption of UNC-51/ATG1 function. In addition to NSM neurons, AIM and RIH interneurons express SERT/MOD-5. These interneurons do not synthesize 5-HT; instead, they regulate 5-HT levels at extrasynaptic 5-HT targets via uptake mediated by SERT/MOD-5 (Jafari et al., 2011). As reported previously, SERT/MOD-5 is essential for accumulating 5-HT in AIM and RIH interneurons (Jafari et al., 2011), and these neurons in loss of function mod-5–mutant C. elegans showed no 5-HT accumulation (Figure 5G). However, the unc-51–mutant C. elegans showed normal levels of 5-HT accumulation (Figure 5G), suggesting that SERT trafficking and SERT-mediated 5-HT uptake is normal in AIM and RIH neurons. The basis for the cell-type specificity of ULK/ATG1-mediated SERT trafficking in C. elegans remains unknown. These findings indicate that ULK1 and homologs have an evolutionarily conserved and cell-type specific regulatory role in the trafficking of cargo such as SERT, which is not shared by other autophagy-related proteins, including ATG13.
ULK-Mediated Phosphorylation of SEC16A at S846 is Required for ER-to-Golgi Trafficking and ERES Assembly
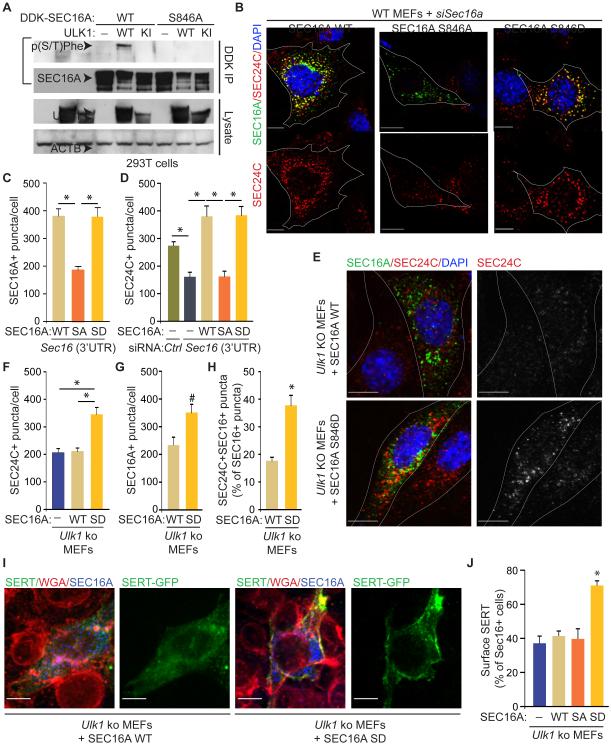
ULKs mediate SEC16A phosphorylation, and both proteins function in ER-to-Golgi trafficking; thus, we investigated the role of ULK-mediated phosphorylation of SEC16 in this process. As mentioned above, we detected ULK1/2-dependent phosphorylation of SEC16A by using a p(S/T)Phe antibody. Site-directed mutagenesis was used to substitute serine and threonine residues in SEC16A that matched the antibody-recognition motif with alanines. The resulting GFP–SEC16A mutants were screened for their ability to disrupt ULK1-dependent phosphorylation (Figure S6A and Table S1). We observed a reproducible decrease in the ULK1-dependent phosphorylation signal on the GFP–SEC16A S846A–mutant protein compared to WT (Figure S6A and Table S1). Independently, each Ser/Thr residue in the SEC16A protein sequence (NP_055681.1) was given a score (Tables S2 and S3) calculated based on the ULK1 position–specific scoring matrix (PSSM) (Egan et al., 2015); the conservation of each site among vertebrates and invertebrates was also evaluated (Table S4). S846, which was highly conserved among vertebrates (Table S4), had a PSSM score of 4.76 and was ranked 9th among all putative SEC16A phosphorylation sites (Table S3). To confirm that S846 was phosphorylated in an ULK1-dependent manner, we reintroduced the S846A mutation into a MYC-DDK–tagged SEC16A-expression construct. Use of this construct demonstrated that the S846A substitution was sufficient to abolish the ULK1-dependent phosphorylation signal on SEC16A that was recognized by the p(S/T)Phe antibody (Figure 6A). Moreover, phospho-specific rabbit polyclonal antibodies generated against immunogenic peptides containing the phosphorylated S846 residue detected ULK1- and ULK2-dependent phosphorylation of WT SEC16A (Figure S6B and data not shown); this signal was not detected in cells coexpressing the S846A SEC16A-mutant and WT ULK1 or ULK2 (Figure S6B and data not shown).
Figure 6. ULK-mediated phosphorylation of SEC16A at S846 is required for assembly of SEC24C+ ERES and ER-to-Golgi trafficking.
(A) Representative immunoblots of DDK IPs prepared from 293T cells transfected with either MYC-DDK–tagged WT SEC16A or a mutant form of MYC-DDK–tagged SEC16A harboring the S846A and the specified ULK1-expression construct. (B) Representative pseudocolored images of MEFs stained with DAPI and antibodies against MYC-DDK-tagged SEC16A and SEC24C. SA: SEC16A S846A mutant; SD: SEC16A S846D mutant. (C) Mean numbers (±SEM) of SEC16A+ puncta per cell. (D) Mean numbers (±SEM) of SEC24C+ puncta per cell . Ten cells (DDK+, if appropriate) per population were scored in C and D. *P <0.001 (ANOVA). (E) Representative pseudocolored images of MEFs stained with DAPI and antibodies against SEC24C and MYC-DDK-tagged SEC16A. (F) Mean numbers (±SEM) of SEC24C+ puncta per cell. (G) Mean numbers (±SEM) of SEC16A+ puncta per cell. (H) Mean percentages (±SEM) of SEC16A+ puncta that were also SEC24C+. Ten cells (DDK+, if appropriate) per population were scored in F-H. Scale bar = 10 μm. *P <0.001 and #P <0.05 when compared with WT (ANOVA). (I) Representative merged pseudocolored images of MEFs stained with AlexaFluor 647–conjugated WGA and antibodies against MYC-DDK-tagged SEC16A. White arrowheads indicate PM. (J) Mean percentages (±SEM) of cells from each population with colocalized WGA and GFP. Data were acquired from 3 independent experiments, and more than 100 cells per population were scored in each experiment. Scale bar: 10 μm. *P <0.001 (ANOVA) when compared with empty vector. See also Figure S6.
To determine the consequence of ULK-mediated phosphorylation of SEC16A at S846, we expressed WT SEC16A, the phospho-defective S846A mutant, and the phosphomimetic S846D mutant in Sec16a-depleted cells and assessed their ability to restore the defects in ERES assembly and ER-to-Golgi trafficking common to Ulk1/2-deficient and Sec16a-depleted MEFs. First, we detected fewer SEC16A+ puncta in Sec16a-depleted cells upon overexpression of the S846A mutant than in cells expressing WT SEC16A or the S846D mutant, despite comparable levels of protein (Figures 6B-C and S6C-F). The number of SEC24C+ puncta was also fewer in Sec16a-depleted cells expressing the S846A mutant than in those expressing WT SEC16A or the S846D mutant (Figure 6B, D). These results are consistent with the notion that ULK-mediated phosphorylation of SEC16A is required for efficient recruitment and/or retention of SEC16A and SEC24C at ERES. WT SEC16A did not increase the number of SEC24C+ puncta in Ulk1-deficient MEFs (Figure 6E-F), suggesting that the residual Ulk2 expression in these MEFs was insufficient to yield high enough levels of phosphorylated SEC16A to promote accumulation of SEC24C at ERES. In contrast, expressing the S846D mutant in Ulk1-deficient MEFs significantly increased (P <0.001) the number of SEC16A+ puncta and SEC24C+ puncta and the proportion of puncta showing colocalization of SEC16A and SEC24C (Figure 6E-H). Similarly, the defect in SERT trafficking in Ulk1-ko MEFs was rescued by overexpressing the S846D SEC16A mutant but not the WT SEC16A or the S846A mutant (Figure 6I-J and Figure S6G). The S846A mutant decreased SERT trafficking to the PM when overexpressed in WT MEFs (Figure S6H-I), suggesting a dominant-negative function of the phospho-defective mutant. These data indicate that ULK1/2-mediated phosphorylation of SEC16A at S846 drives the assembly of COPII components at ERES and trafficking of associated secretory cargo.
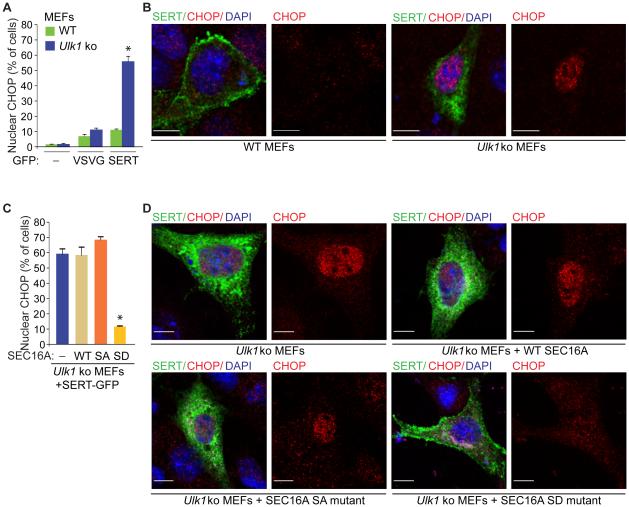
The Defect in ER-to-Golgi Trafficking in ULK-Deficient Cells Leads to an ER–Stress Response that Is Reversed by the S846D SEC16A Mutant
Defects in ER-to-Golgi trafficking can cause proteins to accumulate in the ER and activate the UPR system, but we did not detect any evidence of UPR activation in Ulk1- and/or Ulk2-ko MEFs under basal growth conditions (data not shown) or in Ulk1-ko MEFs expressing VSVG–GFP (Figure 7A). By contrast, Ulk1-ko MEFs expressing SERT–GFP showed a significant increase (P <0.001) in nuclear localization of CHOP (Figure 7A-B). Similarly, expression of SERT–GFP in 293T cells increased steady-state levels of BiP, ATF3, and CHOP (Figure S7A). The increase in CHOP expression was reversed upon overexpression of WT ULK1, but not the KI ULK1 mutant, and either WT SEC16A or the S846D SEC16A mutant in 293T cells (Figure S7B-C). Moreover, expression of the S846D SEC16A mutant in Ulk1-ko MEFs, which rescued the defect in ER-to-Golgi trafficking of SERT, prevented nuclear localization of CHOP (Figure 7C-D). These findings indicate that the defect in ULK- and SEC16A-dependent ER-to-Golgi trafficking of certain cargo activates the UPR pathway, similar to that observed in Ulk1/2-deficient hippocampal neurons.
Figure 7. SEC16A mutant with the phosphomimetic S846D overcomes the defect in ER-to-Golgi trafficking and prevents activation of the UPR in ULK-deficient cells.
(A) Mean percentages (±SEM) of cells with nuclear staining of CHOP. Data were acquired from 3 independent experiments, and more than 100 GFP+ cells per population were scored in each experiment. *P <0.001 (ANOVA) when compared with empty vector-transduced cells. (B) Representative pseudocolored images of WT and Ulk1-ko MEFs transfected with indicated GFP-tagged cargo [i.e, (−), empty vector; VSVG-GFP or SERT-GFP] stained with an antibody against CHOP and DAPI. Scale bar: 10 μm. (C) Mean percentages (±SEM) of cells with nuclear staining of CHOP. Data were acquired from 3 independent experiments, and more than 100 GFP+DDK+ cells per population were scored in each experiment. *P <0.001 (ANOVA) when compared with empty vector- transduced cells. SA: S846A SEC16A mutant; SD, S846D SEC16A mutant. (D) Representative pseudocolored images of MEFs stained with antibodies against MYC-DDK-tagged SEC16A (channel not shown) and CHOP and DAPI. Scale bar: 10 μm. See also Figure S7.
DISCUSSION
This study revealed an unexpected, evolutionarily conserved role for ULK/ATG1 in ER-to-Golgi trafficking under physiologic conditions, which is essential for maintaining cellular homeostasis. ULK1 and ULK2 play important roles in selective and nonselective autophagy induced in response to mitochondrial damage, infection, proteotoxic stress, or various forms of metabolic stress (Joo et al., 2011; Lim et al., 2015; Noda and Fujioka, 2015). Given the presumed importance of the ULKs in both forms of autophagy, the lack of ubiquitin+ and P62+ inclusions or accumulation of abnormal mitochondria in the neurons of Ulk1/2-cdko animals was surprising. These results suggest that unlike ATG5, ATG7, and FIP200, all of which mediate the degradation of ubiquitinated proteins in neurons under basal physiologic conditions (Hara et al., 2006; Komatsu et al., 2006; Liang et al., 2010), ULK1 and ULK2 are not required for “basal” autophagy. Although we cannot exclude the possibility that another ULK/Atg1 homolog (e.g., ULK3) compensates for the lack of ULK1 and ULK2 in CNS neurons, our findings raise the possibility that an ATG13/FIP200 complex regulates the autophagy-mediated turnover of ubiquitinated proteins in an ULK/ATG1-independent manner. Recent studies in isolated cells have shown that ATG13 and FIP200 support starvation-induced autophagy in the absence of ULK1/2, albeit to a lesser extent than in their presence (Alers et al., 2011; Hieke et al., 2015). Such a pathway may support sufficient levels of autophagy in neurons under basal physiologic conditions to prevent the accumulation of ubiquitinated protein deposits.
Although ULK1 and ULK2 are not required for basal autophagy in neurons, our data indicate that a primary function of the ULKs in the absence of metabolic stress is regulating ER-to-Golgi trafficking. The CNS is spared in response to most metabolic stresses, perhaps making it one of a few environments in which the ULKs’ role in ER-to-Golgi trafficking can be readily separated from that in autophagy. ULK/ATG1-mediated COPII transport does not require the presence of other autophagy-related proteins (e.g., ATG13, ATG14, and ATG7), confirming that ULK/ATG1-regulated ER-to-Golgi trafficking is not an indirect consequence of ULKs affecting autophagy. Moreover, the absence of ATG13 does not disrupt the ULK1–SEC16A complex, as it does the canonical ULK/ATG1 complex composed of ULK1, ATG13, FIP200, and ATG101. These results indicate that regulating ER-to-Golgi trafficking under basal physiologic conditions is a noncanonical function (i.e., one that does not require ATG13) of the ULKs.
The uncoordinated phenotype of C. elegans unc-51 mutants is associated with defective axon guidance and is not recapitulated in autophagy-deficient mutants, such as epg-1 (Atg13) and epg-9 (Atg101) mutants (Liang et al., 2012; Tian et al., 2009). RNAi-mediated silencing of other autophagy-related genes, e.g., bec-1 (Atg6), M7.5 (Atg7), lgg-1 (Atg8), or F41E6.5 (Atg18), also does not replicate the neuronal defects observed in unc-51 mutants (Ogura and Goshima, 2006). Our data suggest that the N-terminal domain of ULK/ATG1 and the central domain of SEC16A, which are the most highly evolutionarily conserved regions of both proteins, mediate the interaction between ULK/ATG1 and SEC16A. Indeed, the role of ULK/ATG1 in ER-to-Golgi trafficking appears to be conserved in C. elegans, as unc-51 mutants show evidence of abnormal COPII assembly and trafficking of MOD-5 (SERT). Therefore, it is tempting to speculate that the defect in ER-to-Golgi trafficking in unc-51 mutants contributes to their uncoordinated phenotype.
Although the noncanonical function of the ULKs in ER-to-Golgi trafficking does not require other autophagy-related proteins, recent studies have implicated the COPII pathway in autophagosome biogenesis (Ishihara et al., 2001; Zoppino et al., 2010; Graef et al., 2013; Suzuki et al., 2013; Ge et al., 2013; Ge et al., 2014). Given that ULK/ATG1 activity is regulated by energy/nutrient availability, we speculate that the ULK/ATG1–SEC16A interaction represents a specific point of convergence among the metabolism, autophagy, and COPII-transport pathways.
EXPERIMENTAL PROCEDURES
Mice
The strategy for targeting the Ulk2 locus was described previously (Cheong et al., 2011). Mice harboring the targeted Ulk2 allele (Ulk2flox-neo) were crossed with EIIaCre transgenic mice (Jackson Laboratory, stock: 003724), and progeny with Ulk2ko or Ulk2flox alleles were crossed with WT mice to eliminate the expression of the Cre-recombinase. Ulk1−/− mice have been described previously (Kundu et al., 2008). The Nestin-Cre line was obtained from the Jackson Laboratory (stock: 003771). Ulk1+/−;Ulk2flox/flox females were bred with Ulk1+/−;Ulk2−/−;Nestin-Cre males to generate Ulk1/2-cdko (Ulk1−/−;Ulk2−/flox;Nestin-Cre) mice. Atg7flox/flox females (a generous gift from Dr. Masaaki Komatsu, Tokyo Metropolitan Institute of Medical Science) (Komatsu et al., 2005) were crossed with Nestin-Cre males to generate Atg7-cko mice. All animals were maintained in a mixed background between C57BL/6 and 129. All animal experiments were approved by and performed in accordance with guidelines provided by the Institutional Animal Care and Use Committee at St. Jude Children’s Research Hospital.
Immunoprecipitation
Endogenous ULK1 was extracted from the hippocampi of 4-wk-old WT mice or from MEFs by using a Triton-based cell lysis buffer (40 mM HEPES, 120 mM NaCl, 1 mM EDTA, 1.5 mM Na3VO4, 50 mM NaF, 10 mM β-glycerophosphate, 20 mM MoO4, 0.5% Triton X-100, protease inhibitor, phosphatase inhibitor). The lysates were incubated with anti-ULK1 antibody (Santa Cruz Biotechnology, sc10900) overnight at 4°C and precipitated with Protein G agarose beads (Thermo Scientific). For immunoprecipitation of GFP–SEC16A, whole-cell extracts were prepared from 293T cells by using the described Triton-based buffer and precipitated with anti-GFP antibody–conjugated sepharose beads (Abcam, ab69314) after overnight incubation at 4°C. The beads were washed 5 times with cold Triton-based buffer and incubated at 95°C for 5 min in SDS sample buffer (Sigma Aldrich). Phosphatase treatment was performed on GFP IPs by using calf alkaline intestinal phosphatase (Sigma Aldrich) per the manufacturer’s protocol, before elution in SDS sample buffer. Anti–FLAG M2-agarose beads (Sigma Aldrich, A2220) were used for immunoprecipitation of FLAG-tagged proteins.
COPII Vesicle–Formation Assay
The preparation of cytosol (source of soluble COPII proteins) and medium-speed membrane-pellet (source of membrane and cargo) fractions has been described before (Ge et al., 2013; Ge et al., 2014). Each vesicle-budding reaction consists of 3 μL membrane from the medium-speed pellet (OD600 = 10 for total membrane), 25 μL cytosol (5 mg/mL), 0.75 μL GTP, and 5 μL 10× ATP regeneration (Ge et al., 2013; Ge et al., 2014). B88 buffer (20 mM HEPES-KOH, pH 7.2, 250 mM sorbitol, 150 mM potassium acetate, and 5 mM magnesium acetate) was added last to adjust the reaction mixture to a final volume of 50 μL. The reaction was performed at 30°C for 1 h followed by centrifugation at 20,000 xg for 20 min. Supernatant aliquots (35 μL) were transferred to an ultracentrifuge tube for sedimentation at 100,000 xg in a Beckman TLA100.3 rotor for 30 min. The supernatant fractions were removed, and the small membranes were analyzed by immunoblotting with the antibodies indicated in the figures.
Statistical Analyses
Statistical analyses were performed using SigmaPlot; significance was assessed by 2-tailed Student’s t-test or by 1- or 2-factor ANOVA analysis followed by Holm-Sidak post-hoc analysis.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Terrence Geiger [St. Jude Children’s Research Hospital (St. Jude), Memphis, TN], Toshifumi Tomoda (Beckman Research Institute of City of Hope, Duarte, CA), Masaaki Komatsu (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) and Do-Hyung Kim (University of Minnesota, Twin Cities, MN) for reagents. We are grateful to Angela McArthur and Keith Laycock (St. Jude) for editing the manuscript. The mass spectrometry analysis was performed by staff in the St. Jude Proteomics Facility; histological analyses were performed by Peter Vogel in the St. Jude Veterinary Pathology Core; and most of the imaging data was acquired in the St. Jude Cell & Tissue Imaging Center. The St. Jude core facility resources are supported by Cancer Center Support Grant (P30 CA021765) from the National Cancer Institute and by ALSAC. The research was partially supported by grants from the National Heart, Lung and Blood Institute (R01 HL114697 to M.K. and R01 HL091196 to F.K.), the National Institute of Child Health and Human Development (R01 HD058697 and R01 HD053477 to F.K.), the National Institute of General Medical Sciences (R01 GM110567 to A.A. and K99 GM114397 to L.G.), the National Institute of Ageing (AG047928 to J. P.), the National Institute of Mental Health (MH105389 to J.S.) the Burroughs Welcome Fund (1006062.05 to M.K.), the American Society of Hematology (to M.K.), the American Heart Association (GRNT17240014 to F.K.) and ALSAC (to M.K., J.P, D.R.G., L.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Data Items (7 figures and 4 tables), Supplemental Experimental Procedures, and Supplemental References.
AUTHOR CONTRIBUTIONS
J.H.J, B.W., F.K., J.Y.S., A.A., and M.K. designed experiments. J.H.J., B.W., E.F., L.G., L.X., T.I.S., R.I., X.L.-H., C.W., and F.K. performed experiments and analyzed the data. T.L., D.R.G., and L.M.H. provided critical reagents. M.K., J.H.J. and B.W. wrote the manuscript. L.G., J.P., F.K., J.Y.S., A.A., D.R.G. and L.H. edited the manuscript.
REFERENCES
- Alers S, Loffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, Campbell DG, Fehrenbacher B, Schaller M, Wesselborg S, et al. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy. 2011;7:1423–1433. doi: 10.4161/auto.7.12.18027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol. Cell. Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Glick BS. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol. Biol. Cell. 2007;18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnon C, Wendeler MW, Paccaud JP, Hauri HP. Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J. Cell Sci. 2010;123:1705–1715. doi: 10.1242/jcs.062950. [DOI] [PubMed] [Google Scholar]
- Brenner B, Harney JT, Ahmed BA, Jeffus BC, Unal R, Mehta JL, Kilic F. Plasma serotonin levels and the platelet serotonin transporter. J. Neurochem. 2007;102:206–215. doi: 10.1111/j.1471-4159.2007.04542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Lindsten T, Wu J, Lu C, Thompson CB. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11121–11126. doi: 10.1073/pnas.1107969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H, Wu J, Gonzales LK, Guttentag SH, Thompson CB, Lindsten T. Analysis of a lung defect in autophagy-deficient mouse strains. Autophagy. 2014;10:45–56. doi: 10.4161/auto.26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Kramvis I, Cho N, Sadowski A, Meikle L, Kwiatkowski DJ, Sahin M. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J. Neurosci. 2009;29:5926–5937. doi: 10.1523/JNEUROSCI.0778-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol. Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Liu M, Zhang X, Sakamoto T, Taatjes DJ, Jena BP, Sun F, Woods J, Bryson T, Kowluru A, et al. COPII-Dependent ER Export: A Critical Component of Insulin Biogenesis and beta-Cell ER Homeostasis. Mol. Endocrinol. 2015;29:1156–1169. doi: 10.1210/me.2015-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley IG, Lam du H, Wang J, Ding X, Chen S, Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell. 2013;24:2918–2931. doi: 10.1091/mbc.E13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hieke N, Loffler AS, Kaizuka T, Berleth N, Bohler P, Driessen S, Stuhldreier F, Friesen O, Assani K, Schmitz K, et al. Expression of a ULK1/2 binding-deficient ATG13 variant can partially restore autophagic activity in ATG13-deficient cells. Autophagy. 2015;11:1471–1483. doi: 10.1080/15548627.2015.1068488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brumell JH. Bacteria-autophagy interplay: a battle for survival. Nat. Rev. Microbiol. 2014;12:101–114. doi: 10.1038/nrmicro3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y, Komatsu M. Selective degradation of p62 by autophagy. Semin. Immunopathol. 2010;32:431–436. doi: 10.1007/s00281-010-0220-1. [DOI] [PubMed] [Google Scholar]
- Iinuma T, Shiga A, Nakamoto K, O'Brien MB, Aridor M, Arimitsu N, Tagaya M, Tani K. Mammalian Sec16/p250 plays a role in membrane traffic from the endoplasmic reticulum. J. Biol. Chem. 2007;282:17632–17639. doi: 10.1074/jbc.M611237200. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari G, Xie Y, Kullyev A, Liang B, Sze JY. Regulation of extrasynaptic 5-HT by serotonin reuptake transporter function in 5-HT-absorbing neurons underscores adaptation behavior in Caenorhabditis elegans. J. Neurosci. 2011;31:8948–8957. doi: 10.1523/JNEUROSCI.1692-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol. Cell. 2011;43:572–585. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabis J, Rosenberg I, Podolsky DK. Vangl1 protein acts as a downstream effector of intestinal trefoil factor (ITF)/TFF3 signaling and regulates wound healing of intestinal epithelium. J. Biol. Chem. 2006;281:6434–6441. doi: 10.1074/jbc.M512905200. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 2010;285:3499–3509. doi: 10.1074/jbc.M109.072389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Yang P, Tian E, Han J, Zhang H. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy. 2012;8:1426–1433. doi: 10.4161/auto.21163. [DOI] [PubMed] [Google Scholar]
- Lim J, Lachenmayer ML, Wu S, Liu W, Kundu M, Wang R, Komatsu M, Oh YJ, Zhao Y, Yue Z. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS genetics. 2015;11:e1004987. doi: 10.1371/journal.pgen.1004987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 2015;16:345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- Mercado CP, Kilic F. Molecular mechanisms of SERT in platelets: regulation of plasma serotonin levels. Mol. Interv. 2010;10:231–241. doi: 10.1124/mi.10.4.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Barlowe C. Regulation of coat assembly--sorting things out at the ER. Curr. Opin. Cell Biol. 2010;22:447–453. doi: 10.1016/j.ceb.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Montgomery TR, Steinkellner T, Sucic S, Koban F, Schuchner S, Ogris E, Sitte HH, Freissmuth M. Axonal targeting of the serotonin transporter in cultured rat dorsal raphe neurons is specified by SEC24C-dependent export from the endoplasmic reticulum. J. Neurosci. 2014;34:6344–6351. doi: 10.1523/JNEUROSCI.2991-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat. Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Noda NN, Fujioka Y. Atg1 family kinases in autophagy initiation. Cell. Mol. Life Sci. 2015;72:3083–3096. doi: 10.1007/s00018-015-1917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura K, Goshima Y. The autophagy-related kinase UNC-51 and its binding partner UNC-14 regulate the subcellular localization of the Netrin receptor UNC-5 in Caenorhabditis elegans. Development. 2006;133:3441–3450. doi: 10.1242/dev.02503. [DOI] [PubMed] [Google Scholar]
- Petrova K, Oyadomari S, Hendershot LM, Ron D. Regulated association of misfolded endoplasmic reticulum lumenal proteins with P58/DNAJc3. EMBO J. 2008;27:2862–2872. doi: 10.1038/emboj.2008.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Hirschberg K, Zaal KJ, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Preston AM, Gurisik E, Bartley C, Laybutt DR, Biden TJ. Reduced endoplasmic reticulum (ER)-to-Golgi protein trafficking contributes to ER stress in lipotoxic mouse beta cells by promoting protein overload. Diabetologia. 2009;52:2369–2373. doi: 10.1007/s00125-009-1506-5. [DOI] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P. Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J. Cell Biol. 2009;187:525–536. doi: 10.1083/jcb.200907074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S, El-Kasaby A, Kudlacek O, Sarker S, Sitte HH, Marin P, Freissmuth M. The serotonin transporter is an exclusive client of the coat protein complex II (COPII) component SEC24C. J. Biol. Chem. 2011;286:16482–16490. doi: 10.1074/jbc.M111.230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013 doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- Tian E, Wang F, Han J, Zhang H. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy. 2009;5:608–615. doi: 10.4161/auto.5.5.8624. [DOI] [PubMed] [Google Scholar]
- Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Watson P, Townley AK, Koka P, Palmer KJ, Stephens DJ. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubero-Lahoz S, Robledo P, Farre M, de laTorre R. Platelet SERT as a peripheral biomarker of serotonergic neurotransmission in the central nervous system. Curr. Med. Chem. 2013;20:1382–1396. doi: 10.2174/0929867311320110003. [DOI] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R. COPII and the regulation of protein sorting in mammals. Nat. Cell Biol. 2012;14:20–28. doi: 10.1038/ncb2390. [DOI] [PubMed] [Google Scholar]
- Ziu E, Mercado CP, Li Y, Singh P, Ahmed BA, Freyaldenhoven S, Lensing S, Ware J, Kilic F. Down-regulation of the serotonin transporter in hyperreactive platelets counteracts the pro-thrombotic effect of serotonin. J. Mol. Cell. Cardiol. 2012;52:1112–1121. doi: 10.1016/j.yjmcc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic. 2010;11:1246–1261. doi: 10.1111/j.1600-0854.2010.01086.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.