Abstract
DNA polymerases synthesize DNA in only one direction, but large genomes require RNA priming and bi-directional replication from internal origins. Here we review the physical, chemical and evolutionary constraints underlying these requirements. We then consider the roles of the major eukaryotic replicases, DNA Polymerases α, δ and ε, in replicating the nuclear genome. Pol α has long been known to extend RNA primers at origins and on Okazaki fragments that give rise to the nascent lagging strand. Taken together, more recent results of mutation and ribonucleotide incorporation mapping, electron microscopy and immunoprecipitation of nascent DNA now lead to a model wherein Pol ε and Pol δ synthesize the majority of the nascent leading and lagging strands of undamaged DNA, respectively.
Keywords: DNA replication, polymerase, replication fork, genomic ribonucleotide, evolution
The asymmetry of DNA replication
Inheritance requires accurate duplication of genetic information. The genomes of cellular organisms encode genetic information in long chains of deoxyribonucleic acid (DNA) synthesized from deoxyribonucleoside triphosphate (dNTP) by DNA polymerases. These polymerases catalyze the formation of phosphodiester linkages between monomers via nucleophilic attack on the α-phosphate of an incoming dNTP by the 3´-hydroxyl of the deoxyribose at the end of a growing chain1 (Figure 1A, black arrow). Thus DNA strands are directional, with one 5´-terminus and one 3´-terminus. Two DNA strands can assemble into an antiparallel DNA double helix via complementary pairing of adenine (A) with thymine (T) and guanine (G) with cytosine (C)2. Thus, each serves as the template strand (see Glossary) for accurate replication of the other. By convention, the base sequence of a fully complementary double helix is reported as the sequence of one strand in the 5´-to-3´ direction. This strand is commonly called the top or Watson strand, with the sequence of the other (bottom or Crick) strand inferred as the reverse of the complementary sequence. DNA polymerases require a primer in order to initiate DNA synthesis. Cellular organisms initiate DNA synthesis with RNA priming (Figure 1A, orange bases) at origins of replication within double-stranded DNA genomes. The two strands of the double helix are replicated in concert, making a forked structure. Two replication forks usually proceed from each origin, one in each direction. One strand in each fork, called the leading strand, is replicated more or less continuously. The other strand is repeatedly primed and synthesized as discontinuous fragments, known as Okazaki fragments, which are eventually processed into a continuous lagging strand3.
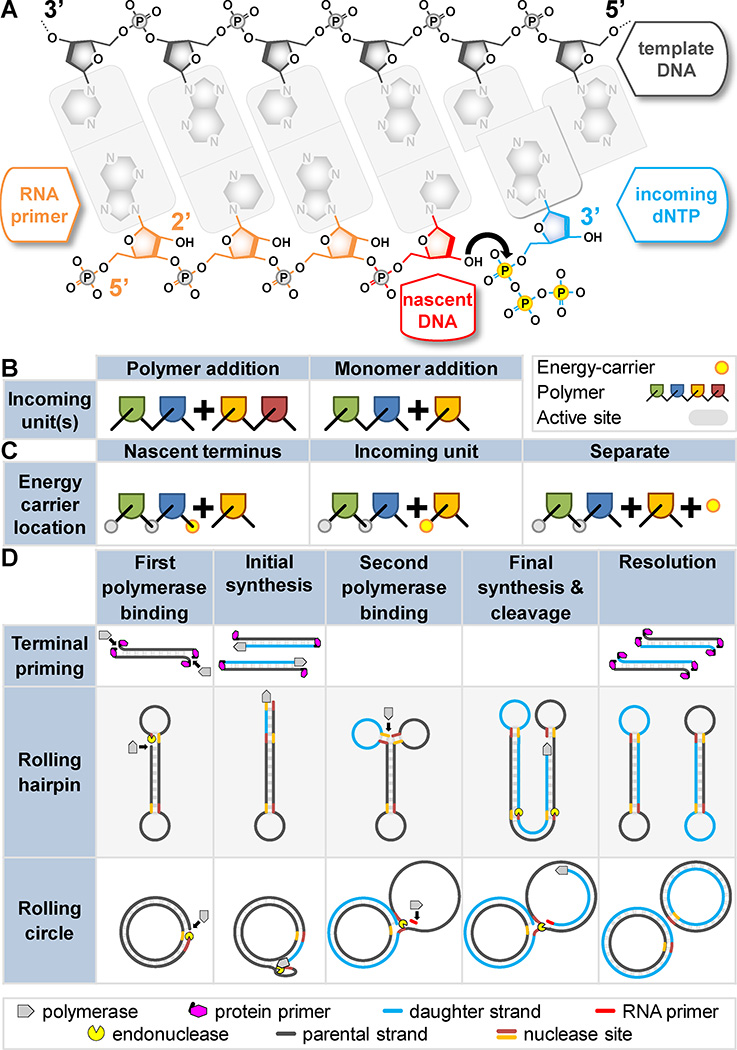
Figure 1. Why DNA replication is asymmetric and how simple replicators cope.
A) Termini of each DNA strand are named according to the hydroxyl groups on the deoxyribose sugar: 5’ and 3’ (labeled). DNA polymerases synthesize DNA via nucleophilic attack (black arrow) by the 3’ hydroxyl of the growing strand (the primer, red) on the α-phosphate of the incoming nucleotide triphosphate (blue). Chemical energy carried by the triphosphate (yellow) drives the reaction. RNA (orange) can also serve as a primer. RNA has a 2’ hydroxyl (labeled) that is absent in DNA. B–D) Genome synthesis could theoretically proceed by many modes. Arguments to which these panels refer hold for any hypothetical information-carrying polymer, thus polymer and terminal identities are herein deliberately ambiguous. Non-replicative polymerases and other enzymes show that alternate chemistries are possible, but replicases are limited: B) monomers (not polymers) are added in each synthesis step; C) the energy carrier is a moiety of the incoming monomer (not the growing strand or another reaction component); D) The leading/lagging asymmetry is avoidable. Terminal priming and rolling hairpin replication, sufficient for small linear genomes, require either a terminal-binding protein (magenta), or terminal hairpins (exposed single-strand), respectively. Rolling circle replication, sufficient for small circular genomes, requires RNA priming (red) on the second strand. Note: a stepwise nature is exaggerated for clarity; second strand synthesis is often simultaneous.
The rigors of replication asymmetry encourage specialization of those polymerases (replicases) tasked with genome replication and its precise regulation. This review begins by explaining the origins of replication asymmetry and how different organisms cope with this asymmetry. It then focuses on strategies practiced by eukaryotic cells. We explore the evolution of eukaryotic replicases and evidence of their division of labor in leading and lagging strand replication, and finish with a model of the normal eukaryotic replication fork.
Why DNA replication must be asymmetric
In theory, the outline of genome replication mentioned above need not be the only pathway. Why add one nucleotide at a time? Would polynucleotide addition be just as accurate and perhaps faster (Figure 1B)? Why are all known DNA polymerases incapable of 3´-to-5´ synthesis? After all, the 5´-OH should be just as capable of nucleophilic attack as the 3´-OH. Why not use 3´-triphosphate nucleotides? Why not use a triphosphate on the nascent chain rather than on the incoming nucleotide (Figure 1C)? Some of the answers to these questions have a sound chemical underpinning. Others are rooted in evolution and selection.
DNA polymerases always add one monomer (nucleotide) per synthesis step (Figure 1B). In contrast, DNA ligases and spliceosomes efficiently catalyze the joining of polynucleotides. However, splicing is not templated, and though many ligases prefer templates, in the form of complementary DNA overhangs (e.g. E. coli DNA ligase), ligation works with relatively rare ends. DNA polymerases, in contrast, require a monomer pool with four constituents (dATP, dCTP, dGTP, and dTTP) whose relative concentrations are carefully regulated, with mutagenic consequences when imbalances occur4, 5. If polymerases worked by stepwise polymer (polynucleotide triphosphate) addition, then both the number of reactant species and the number of balanced relationships would increase exponentially. More reactant species would also mean more potential mismatches in the polymerase active site, thus more unproductive binding events and less efficient synthesis.
The polarity of replication may be an accident of evolution. Alternative sugars, nucleobases, and backbone moieties are capable of information storage and templated replication, including replication by some present-day polymerases (reviewed6). Though many forms of genetic information storage and replication may once have existed, it is reasonable to suppose that RNA genomes preceded the DNA genomes that are now universal among cellular life (reviewed7). Replication machines in descendants of this RNA World must have evolved within the constraints of using ribonucleotides. Given triphosphates as the energy carriers, a hypothetical 3´-triphosphate (or 2´-triphosphate) would be subject to nucleophilic attack by the oxygen of the 2'-OH (or 3´-OH). Thus, ribonucleotide biosynthesis in all known organisms proceeds from an initial 5´-phosphoribose, and a 5'-to-3' replication polarity is fixed by the relative stability of the 5'-triphosphate.
Why do stable 5'-triphosphates set the directionality of synthesis? What if the 5´-triphosphate was on the nascent strand terminus rather than the incoming nucleotide? The answers deal with replication fidelity. Polymerases are selective for properly base-paired incoming nucleotides, but imperfect selections (mismatches) occur about once per 1–10 kbp in vitro8, 9. The first line of defense against these mistakes is a proofreading 3´-to-5´ exonuclease. The exonuclease trims mismatched nucleotides from the growing strand before they can leave the polymerase complex (unlike post-replication DNA mismatch repair (MMR)), improving fidelity by 100–1000 fold10. If the energy-carrying moiety (triphosphate) were on the nascent strand (Figure 1C), then exonucleolytic proofreading would require recharging before the next synthesis step. This would be difficult without first removing the polymerase, allowing access to mono- and di-phosphate kinases, slowing replication and negating one of the primary advantages of proofreading. Thus, replication with nascent strand terminal energy-carriers would be more mutagenic and/or less efficient than replication with the energy carrier on the incoming nucleotide.
Some non-replicative polymerases, like terminal deoxynucleotidyl transferase, poly(A) polymerases and terminal uridylyl transferases, and telomerase can add to a growing strand (DNA, RNA, and DNA, respectively) without an external template (reviewed11–13). This is because their purposes are to add either random diversity during antibody gene recombination, polyA or polyU tails during messenger and microRNA processing, or short repeats to 3’ chromosome ends, respectively. The last example uses a short RNA template that is part of the telomerase holoenzyme itself. These processes differ from inheritance, which requires templated synthesis. Taken together then, the requirements of efficient and high fidelity replication using polymerases descended from the RNA-world necessitate the antiparallel double helix and the directionality of replication. DNA replication is asymmetric because of inflexible chemistry and implacable natural selection.
How simple replicators cope with asymmetric replication
Simple replicators (i.e. some viruses, viroids, and plasmids) can manage asymmetric replication by treating the DNA strands as functionally independent. For instance, terminal protein priming (Figure 1D, top) has evolved independently among bacteriophages and vertebrate viruses1. In this mode, amino acid side chains of proteins bound to the termini of a linear genome take the place of primer 3´-hydroxyls of nucleic acid primers. The side chains used are serine (adenovirus14, phage Φ2915), threonine (phage Cp-1) and tyrosine (phage PRD11). All synthesis is continuous, so only one polymerase is needed. No primase is required and the terminal proteins even protect the genome from exonucleolytic attack.
Rolling hairpin replication (Figure 1D, middle) allows synthesis of linear genomes without protein primers; a nick is sufficient. These genomes (e.g. vaccinia and rabbit poxvirus; reviewed16) are essentially one strand each, thanks to terminal hairpins. As for terminal priming, all synthesis is continuous and multiple copies may be generated. Again, no primase is needed, and termini are protected from exonucleases. Many plasmids and viruses instead use rolling circle replication (Figure 1D, bottom)17, 18. This is similar to rolling hairpin replication, in that initial priming occurs at a nick and multiple copies of one strand may be generated by one continuous synthesis step. However, synthesis of the second strand requires RNA priming, usually at a defined site. In single-copy mode, this is followed by one continuous synthesis step. If multiple sequential copies of the first strand are generated, the second strand is re-primed at each priming site18 and second strand synthesis is discontinuous, with unit length fragments. Did lagging strand synthesis evolve from a similar process? Again, most synthesis is continuous, multiple copies may be generated, and there are no termini to protect from exonucleases.
Replicases specialize when replication is strand-coordinated
A simple way to increase the rate of genome replication is to increase the number of concurrently active replicases. For the large genomes of cellular organisms, the advantages of rapid replication outweigh the added complexity of discontinuous lagging strand synthesis. Thus the two DNA strands are replicated in concert at each replication fork, usually with two forks originating from each bidirectional origin (Figure 2A). The disparate needs of leading and lagging strand synthesis place varied demands on replicases (Figure 2B–C). Efficient extension from the RNA primer might favor active site flexibility over accuracy or processivity (the ability to synthesize long tracts before dissociation). Continuous leading strand synthesis may require processivity, but accuracy would also be emphasized, given synthesis of about half the genome and perhaps few entry points for repair proteins that require exposed termini. Although Okazaki fragment synthesis might also favor processivity, accuracy is perhaps more important, especially if the lagging replicase is expected to replace upstream Okazaki termini made by a less accurate primer extension specialist. Alternatively, yet another replicase could specialize in primer removal, perhaps forgoing processivity in favor of primer displacement or degradation capability (Figure 2C).
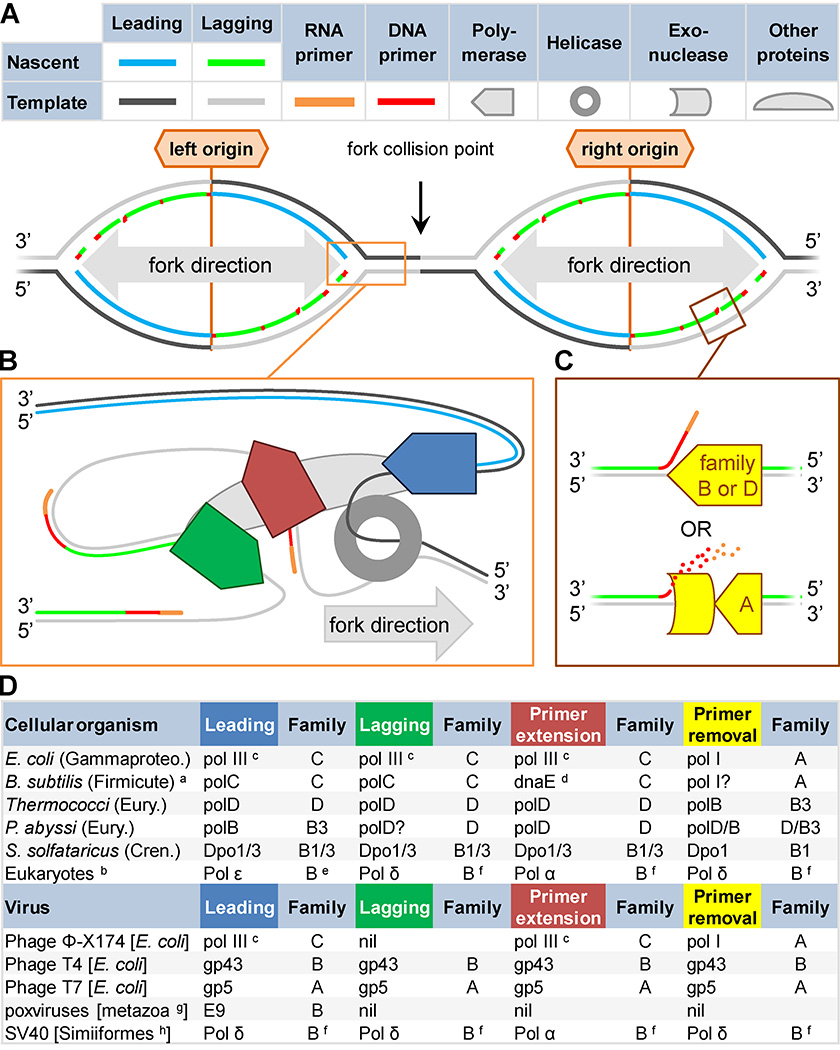
Figure 2. Organisms cope with the demands of simultaneous continuous and discontinuous replication in diverse ways.
A) Inherent DNA asymmetry and obligate 5’-to-3’ directional synthesis mean that one nascent strand (leading; blue) is synthesized continuously while the other (lagging; green) is synthesized in short discontinuous stretches (Okazaki fragments), creating a forked structure (orange box). Synthesis is initiated with an RNA primer (orange), followed by a short DNA primer (red), at replication origins (labeled). Efficiency dictates that two forks proceed in opposite directions from each origin (bidirectional), resulting in collisions (black arrow). B) Replicases (pentagons) can specialize in different replicative roles: extension from the RNA primer (red); discontinuous Okazaki fragment synthesis (green); or continuous leading strand synthesis (blue). C) Polymerases may specialize in synthesis during primer removal (yellow shapes), either through primer displacement or exonucleolysis. D) Replicases are diverse. Polymerase family (A–D), and subfamily (e.g. B3) are indicated. Viral hosts are listed in square brackets. Except in eukaryotes and Escherichia coli, most analyses of polymerase roles in cellular organisms have relied on inferences based on in vitro capabilities. Superscripts: a, not typical of all Firmicutes, Clostridia not shown; b, most studied in Saccharomyces cerevisiae; c, class dnaE1; d, class dnaE3; e, possibly B1 or B2; f, possibly B3; g, observed in vertebrates and arthropods; h, monkeys and apes. Abbreviations: Gammaproteo.→Gammaproteobacteria; B.→Bacillus; Eury.→Euryarchaeota; Chren.→Crenarchaeota; P.→Pyrococcus; S.→Sulfolobus.
Organisms fill replicase niches in a variety of ways (Figure 2D). Four of the seven DNA polymerase families, as defined by sequence similarity, contain known replicases: C and A in bacteria; B and D in archaea; and B in eukaryotes. Replicase specialties (Figure 2D) were inferred either from in vitro properties of purified replicases or from genetic or genomic activity mapping.
Bacteria fall into three categories based upon their Family C polymerase complement19. In the first category, most replication is accomplished by one highly processive Family C replicase (subclass DnaE1), exemplified by Escherichia coli DNA polymerase III (pol III)20. The holoenzyme is extremely accurate, thanks in part to a tethered proofreading exonuclease subunit21. Bacteria of the second category (i.e. Bacillus subtilis, Staphylococci, and Streptococci) have two C Family replicases (subclasses PolC and DnaE3). PolC is proofreading-proficient and does the bulk of genome replication while DnaE3 is proofreading-deficient but capable of RNA primer extension in vitro22, 23. Bacteria of the third category, typified by Clostridia, also have two C Family replicases, PolC and DnaE1, which are similar to the major replicases of B. subtilis and E. coli, respectively. A small subset of Clostridia lack either PolC or its proofreading domain, but the rare lack of an enzyme does not disprove its general importance (see pol2–16 below). Do Clostridia divide leading and lagging replication between distinct Family C polymerases? Primer removal (Figure 2C, bottom) is accomplished by the concurrent polymerase and 5’-to-3’ exonuclease activity of DNA polymerase I (pol I; Family A), which is ubiquitous and highly conserved among bacteria (reviewed24).
Archaea have three classes of replicases: one Family D (polD) and two Family B (subclasses B1 and B3)25, all of which have been implicated in each possible replicase specialty. In Thermococcus (Euryarchaea), polD is the major replicase but seems to lack strand displacement ability, so primer displacement during Okazaki fragment maturation (Figure 2C, top) is handled by polB (subclass B3)26, 27. In Pyrococcus abyssi (Euryarchaea), polB (subclass B3) appears to be the leading strand replicase, polD is responsible for primer extension, and the two share strand displacement duties depending on whether or not an RNaseH removed most of the RNA primer28, 29. The lagging strand replicase is unknown. In Sulfolobus solfataricus (Crenarchaea), Dpo1 and Dpo3 (subclasses B1 and B3, respectively) are both essential.While there is evidence that Dpo1 displaces primers, other replicase responsibilities are unknown30, 31.
Eukaryotic replicases are derived from archaeal enzymes
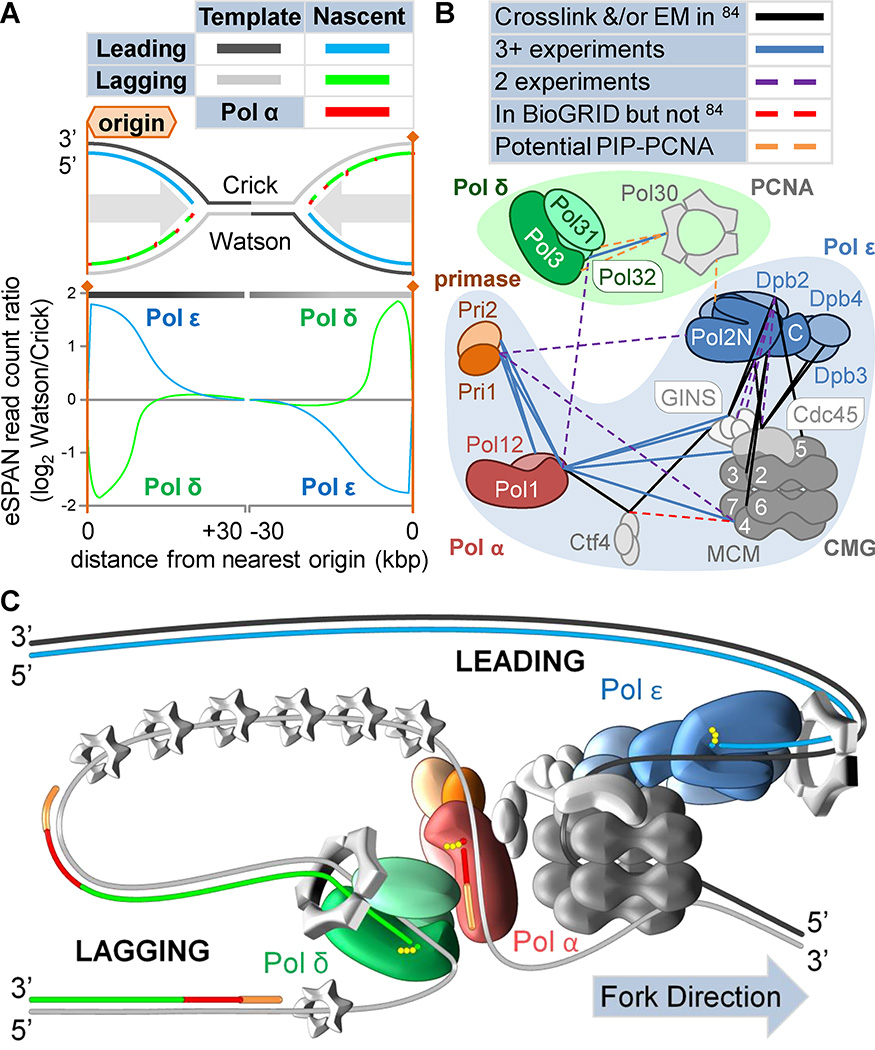
DNA polymerases α, δ and ε replicate undamaged eukaryotic nuclear genomes. That all are Family B may seem surprising after the riot of alternative sets among Bacteria and Archaea. However, bear in mind that the eukaryotic replication machinery originated in Archaea only between 1.6 and 2.1 billion years ago25, 32. Eukaryotes experienced a founder effect, left with whatever polymerases were bequeathed by an archaeal ancestor after the prokaryotes had already undergone at least 2 billion years of diversification33. Assuming that PolD (archaeal Family D) is derived from Family B (based on Zinc finger and accessory subunit homology), all eukaryotic and archaeal replicases may descend from one proto-Family B polymerase25 (Figure 3A). All Family B polymerases possess homologous exonuclease domains (active in eukaryotic Pols δ and ε and archaeal Pols B1 and B3), as do Family B/D accessory subunits (active in PolD). Figure 3A shows a speculative cladogram of eukaryotic and archaeal Families B and D based on25. Gene loss reduced the resulting replicase sets after the last common ancestors.
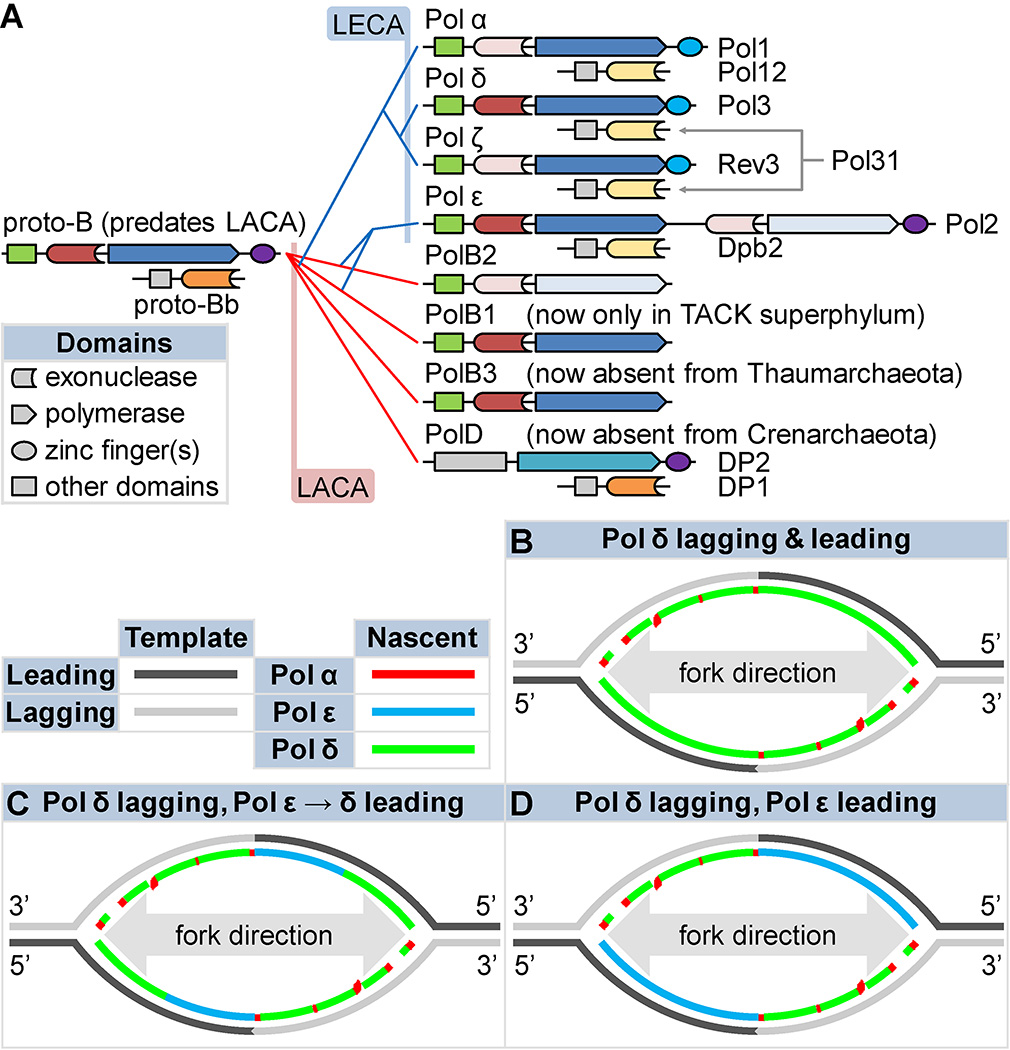
Figure 3. Origins of eukaryotic replicases and models for their division of labor.
A) A speculative cladogram of eukaryotic (blue branches) and archaeal (red branches) B and D family polymerases based on25. See the text for notes on the topology and rationale. Presumably, a proto-B family polymerase preceded the last common ancestors of Archaea and Eukaryota (LACA and LECA, respectively) and begat the archaeal and eukaryotic subclasses. Gene losses reduced the resulting set in both clades. LACA and LECA each possessed at least four subfamilies (B1–3 and D; α, δ, ε, and ζ, respectively). Family D is highly derived. Potential temporal positions of LACA and LECA are indicated (pink and blue areas, respectively). Schematics indicate domain architecture and activities. Catalytic subunits (right of schematics), associated B-subunits (right of schematics), and holoenzymes (above schematics) are indicated (Saccharomyces cerevisiae names for eukaryotes). Colors denote homology, with lighter shades indicating inactivation. B–D) Three models of eukaryotic replication origins, assuming that Pol α extends from RNA primers before passing synthesis to Pols δ and ε. B) Pol δ replicates the bulk of both DNA strands47, as in SV40 viruses41. C) Pols δ and ε replicate the lagging and leading strands, respectively. D) As in C, but Pol ε can cede the leading strand to Pol δ under special circumstances52, 53.
Though diverse specialization in Archaea shows that Family B polymerases can adopt niche roles, direct comparisons with eukaryotes are complicated by the fact that no known archaeon possesses eukaryote-like replicases. Figure 3A assumes that Eukaryota is either a sister group to or a descendant of the TACK superphylum, the latest version of the Eocyte Hypothesis34 (reviewed35). The presence of subclass B1 (from which Pol ε may descend) and the occasional absence of PolD (as in eukaryotes), are unique to TACK25. The recent discovery of the surprisingly eukaryote-like and TACK-like Lokiarchaeota36 suggests that our inventory of archaeal diversity is incomplete. Perhaps an archaeon with a eukaryotic replicase complement awaits discovery. Meanwhile, clues to eukaryotic replicase division of labor have arisen from biochemical, genetic and genomic data.
Three models for eukaryotic nuclear replicase usage
Pol δ was discovered in 197637, as a polymerase similar to the original eukaryotic replicase Pol α, but containing an intrinsic proofreading exonuclease. More than a decade later, the discovery of mammalian Pol ε (originally called Pol δII38) and its yeast homolog39, 40, and the realization that it too has proofreading activity and is highly accurate, led to the suggestion40 that Pol δ and Pol ε might synthesize opposite strands during DNA replication. Since then, three predominant models of polymerase usage during eukaryotic replication have emerged. They all propose that Pol α initiates replicative synthesis and that Pol δ is the major lagging strand replicase involved in Okazaki fragment synthesis and maturation. However, they differ in the fraction of leading strand synthesis performed by Pol ε (Figure 3B–D).
The first model emerged when Pols α and δ were demonstrated to replicate the mammalian SV40 viral genome in vitro without the assistance of Pol ε41. Though viral replicase usage often differs from the host (Figure 2D, bottom), this observation is consistent with the fact that yeast cells with an in-frame deletion of the active polymerase and exonuclease domains of Pol ε (pol2–16) survive, albeit poorly42–46. Those studies, and a recent study of the pattern of replication errors observed in a Pol δ mutator strain47, have led to the proposal that Pol δ is the primary replicase for both the leading and lagging strands of the nuclear genome (Figure 3B). In this model, Pol ε only proofreads errors made by Pol δ during leading strand replication, and not during lagging strand replication. We do not favor this model for several reasons48, including recent evidence suggesting that Pol ε does not proofread errors made by Pol δ49. However, biochemical50 and genetic data49, 51 suggest that Pol δ can proofread some errors made by Pol α and Pol ε, so it remains theoretically possible that Pol ε may also proofread errors made by other polymerases, perhaps including mismatches made during DNA synthesis under stress. A second model52, 53 is that after Pol ε-dependent leading strand replication begins, a switch to Pol δ occurs (Figure 3C), e.g., following stress. Evidence for such a switch comes from a recent study indicating that yeast Pol δ replicates both the leading and lagging DNA strands after encountering a replication fork barrier54. This idea that Pol δ catalyzes a minority of leading strand replication under certain circumstances is further supported by ribonucleotide incorporation data (see below). A third model suggests that Pols ε and δ are the primary leading strand and lagging strand replicases, respectively, during normal replication of undamaged nuclear DNA (Figure 3D).
Mutator evidence for specialized replicase action
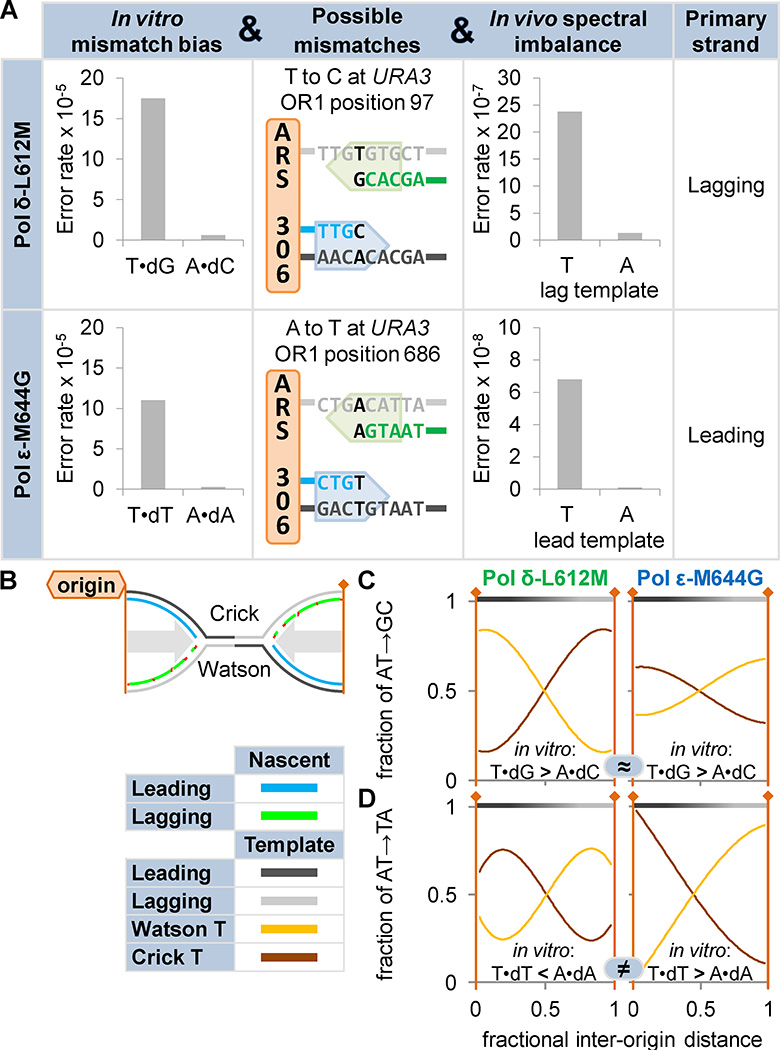
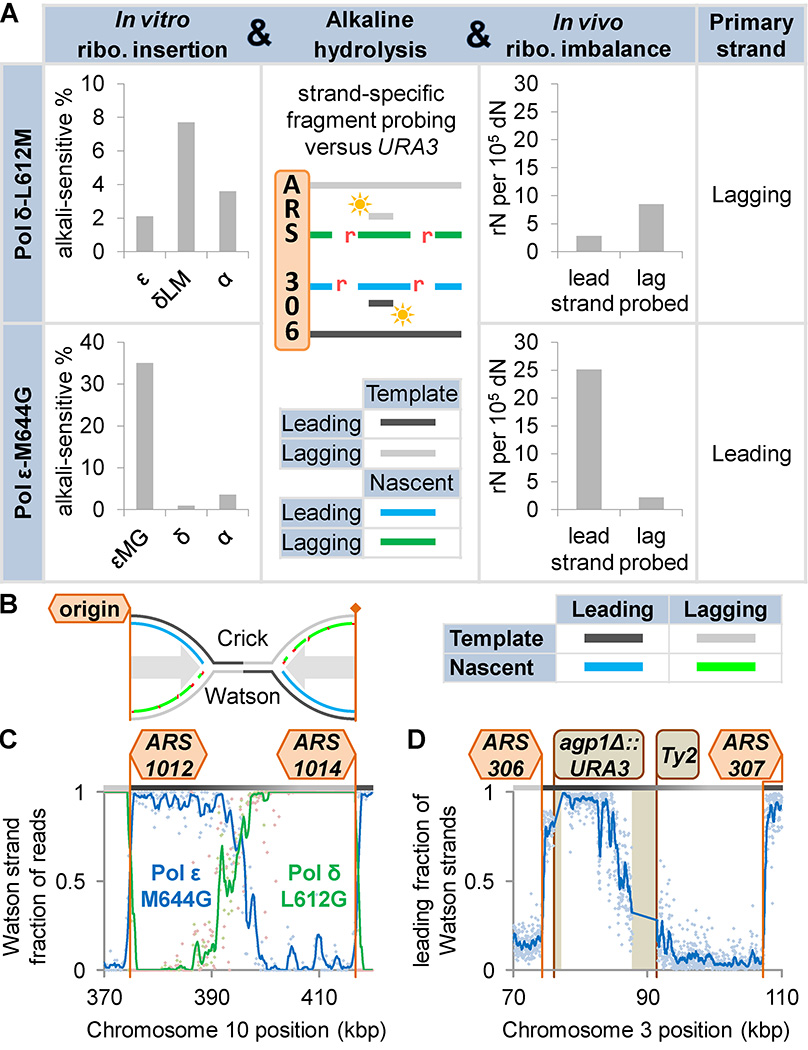
The idea that Pols δ and ε might synthesize different DNA strands emerged from observations that the mutation spectra of their exonuclease-deficient mutants vary significantly by reporter gene orientation55 and that they proofread base analog-induced DNA replication errors on opposite DNA strands56. More recently, several studies57 used derivatives of Pols α, δ and ε with amino acid replacements in the polymerase active site that render these polymerases error-prone. These enzymes retain robust catalytic activity and the resulting cells show little or no growth defects but have strongly biased mutation rates58, 59. For example, a yeast Pol ε-M644G mutant generates T•dT mismatches at a much higher rate than the complementary A•dA mispair58 and a yeast Pol δ-L612M mutant generates T•dG mismatches in vitro at a higher rate than the complementary A•dC mismatch60 (Figure 4A, left). These biased mutational specificities are seen in vivo at the URA3 reporter gene placed in opposite orientations at the AGP1 locus adjacent an early, highly-efficient replication origin (ARS306; Figure 4A, second column). The mutation results are most easily explained if Pol ε as the primary leading strand replicase and Pol δ is modeled as the primary lagging strand replicase (Figure 4A, right). Pol δ results were later recapitulated in Schizosaccharomyces pombe61. Strand-specific mutational patterns between adjacent replication origins (Figure 4B) have been reported in whole genome sequencing studies of mutagenesis in S. cerevisiae62, 63. Where Pols δ and ε variants share an in vitro mutation bias, they show reciprocal patterns between replication origins (Figure 4C). Where their in vitro preferences are opposite, mutation biases between origins match (Figure 4D). Together, these findings indicate that Pols δ and ε work primarily on opposite strands. Further, the directionality of their inter-origin patterns, again given their in vitro biases, places them on the nascent lagging and leading strands, respectively. Human tumor cells with proofreading-defective Pol ε alleles show similar inter-origin mutation patterns, suggesting leading strand-specific Pol ε activity64, 65.
Figure 4. DNA polymerase errors reveal the division of labor between eukaryotic replicases.
A) Pols δ and ε variants Pol δ-L612M and Pol ε-M644G have wild type replication reduced and biased fidelity. For example, Pol δ-L612M makes T•dG errors more frequently than A•dC mispairs59, 85, 86 and Pol ε-M644G makes T•dT more frequently than A•dA58. Mutation rates at the URA3 reporter gene, inserted adjacent to the ARS306 replication origin on S. cerevisiae chromosome 3. Example mutation hotspots are shown. In all cases, substitution rates indicated a division of labor between Pols δ and ε58, 60. In a Pol δ-L612M strain, the AT→GC mutation rate was higher with a lagging strand template T and Pol ε-M644G AT→TA rate was higher with a leading strand template T. B) A schematic of converging replication forks. C–D) Thousands of mutations were accumulated in mismatch repair-deficient S. cerevisiae bearing either Pol δ-L612M or Pol ε-M644G63. C) Where these variants possess similar in vitro mutation biases (denoted “≈”; e.g. T•dG>A•dC), opposite bias patterns exist between adjacent origins. D) Where in vitro mutation biases are opposite (denoted “≠”; e.g. T•dT>A•dA versus T•dT<A•dA), similar bias patterns exist. Thus Pols δ and ε must operate on different strands. The logic used at the URA3 locus, applied across the genome, indicates that Pols δ and ε primarily replicate the lagging and leading strands, respectively.
Ribonucleotide incorporation data strongly support specialized replicase action
In 2010, wild-type S. cerevisiae Pols α, δ and ε from budding yeast were shown to incorporate ribonucleotides into DNA in vitro at rates that are much higher than the rates at which they generate mismatches66. Shortly thereafter, the mutator variants of S. cerevisiae Pols α, δ and ε and S. pombe Pol ε were examined for the ability to incorporate ribonucleotides into DNA during replication, especially in the absence of ribonucleotide excision repair (RER; reviewed67). The first studies involved alkaline hydrolysis of genomic DNA from S. cerevisiae that lack the ability to repair newly incorporated ribonucleotides, followed by strand-specific probing of the URA3 gene (Figure 5A)68. These studies indicated that the Pol ε variant preferentially incorporates ribonucleotides into the nascent leading strand, whereas Pol α and δ variants preferentially incorporate ribonucleotides into the nascent lagging strand61, 68. A more global view was obtained when ribonucleotides that incorporated across whole genomes were mapped at ~1000× higher resolution than previous replication error maps69–72 (reviewed73). A representative example for S. cerevisiae chromosome 1069 shows that the fraction of DNA ends resulting from hydrolysis of ribonucleotides incorporated during nuclear DNA replication and mapping to the Watson strand (Figure 5B) is maximal to the right of ARS1012 in Pol ε-M644G cells and to the left of ARS1014 in Pol δ-L612G variant cells (Figure 5C). In these regions, the Watson strand corresponds to the nascent leading and lagging strands, respectively. The same pattern is evident at the locus used for most single-locus studies mentioned above (Figure 5D)74. The vast majority of the data derived from ribonucleotide-mapping studies in yeast suggest that during replication in normal cells, Pol ε is the major leading strand replicase. This of course does not exclude the likely participation of Pols α and δ in synthesis of a smaller fraction of the leading strand, including at replication origins and under aberrant conditions (discussed in52, 53).
Figure 5. Incorrect sugar incorporation reveals the division of labor between eukaryotic replicases.
A) Pol δ-L612M and Pol ε-M644G also have higher in vitro ribonucleotide incorporation rates than wild type Pols α, δ, and ε66, 68, 74, 87. Strand-specific probing of alkaline hydrolyzed genomic DNA (nicked at ribonucleotides; red) revealed increased fragmentation (increased ribonucleotide frequency) on the nascent lagging and leading strands for ribonuclease H2 (RNase H2)-deficient cells expressing Pol δ-L612M and Pol ε-M644G, respectively. RNase H2 nicks at ribonucleotides, initiating their excision. B) A schematic of converging replication forks. C) Pol δ-L612G (green) and Pol ε-M644G (blue) incorporate ribonucleotides into genomic DNA much more frequently than wild type Pols α, δ, or ε. Millions of ribonucleotides were mapped across S. cerevisiae and Schizosaccharomyces pombe genomes using four similar techniques (reviewed73). For example, in RNase H2-deficient S. cerevisiae69, Pol ε-M644G drives a Watson strand bias in genomic ribonucleotides to the right of origins, indicating Pol ε activity on the nascent leading strand. Pol δ-L612G biases indicate lagging strand activity. Data shown are background-subtracted. Lines represent 1 kb moving averages. D) Assuming this division of labor, more sophisticated noise removal74 allows the confirmation of replication strand assignments at the AGP1 locus, reaffirming conclusions from mutagenesis studies (see Figure 4).
Physical interactions at the replication fork
Maps of replication biomarkers and direct measures of physical interaction between replication proteins and DNA strands have also helped to build a picture of the normal eukaryotic replication fork (Figure 6, Key Figure). For example, eSPAN has been used to measure physical associations between replication proteins and nascent DNA strands75. This approach involves immunoprecipitation of BrdU-labeled nascent DNA following chromatin immunoprecipitation of proteins associated with the replication fork. Pol α, Rfa1 (a component of the single-stranded DNA-binding protein RPA), and Rfc1 (a component of the PCNA loading complex, RFC) are enriched on the nascent lagging strand, confirming previous predictions76. Components of the CMG replicative helicase complex, Cdc45 and Mcm6, are enriched on the nascent leading strand, reinforcing predictions that the helicase traverses the leading template strand77. Moreover, Pols δ and ε were primarily associated with the lagging and leading strands, respectively (Figure 6A).
Figure 6. Key Figure. Physical interactions, combined with previous evidence, suggest a model for the normal eukaryotic replication fork.
A) A schematic of replication between adjacent origins and approximate traces of eSPAN data (enrichment and sequencing of protein-associated nascent DNA derived from75). Bromodeoxyuridine incorporation, crosslinking, immunoprecipitation, and high throughput DNA sequencing together map replication protein interactions with nascent DNA. To the right of the average S. cerevisiae Group I origin, Pol ε is associated primarily with the Watson strand and Pol δ primarily with the Crick strand. Left of origins, associations are reversed. Comparison with the schematic in panel B suggests that Pols δ and ε are enriched on the nascent lagging and leading strands, respectively. B) A schematic representation of physical interactions detected between proteins at the replication fork (outlined shapes; S. cerevisiae nomenclature). Obligatory interactions are represented by shape overlaps. Lines indicate inter-complex interactions (bold names). Solid lines denote interactions observed via electron microscopy and/or crosslinking (black;84) or by ≥3 experiments in the Biological General Repository for Interaction Datasets (BioGRID; blue;83). Dashed lines indicate interactions in two BioGRID experiments (purple), in BioGRID but not in84 (red), or assumed via interacting motifs but not in BioGRID (orange;78, 88). C) A model of the normal eukaryotic replication fork, influenced heavily by84. Small grey trimeric rings represent RPA. Small spheres represent deoxyribonucleotide triphosphates. Compare with models in Figure 3 C–D. Strands are differentiated by color (RNA primer orange; otherwise as per key in A).
The division of replicase labor can be also be inferred from divergent macromolecular interactions at origins. Protein interactions at the replication fork have been extensively mapped via a variety of techniques (Figure 6B, colored lines). While Pols δ and ε have comparable processivity in the presence of PCNA, they appear to load onto primer termini via separate mechanisms78, 79. Pol ε interacts with CMG, and lacks efficiency in the absence of either CMG or bridging subunits80. Conversely, CMG excludes Pol δ from the leading strand81, 82. The Biological General Repository for Interaction Datasets (BioGRID;83) maintains a comprehensive list of protein-protein interactions, and many components of the S. cerevisiae replication complex have been visualized directly through electron microscopy84. Extensive interactions between CMG, Ctf4, Pol α, a C-terminal fragment of Pol ε, and associated subunits were visualized and mapped via crosslinking (Figure 6B, black lines), and Pols ε and α appear to bind separate faces of the helicase complex.
Concluding remarks
Strand biases in eSPAN, ribonucleotide density, and mutations rates suggest lower bounds for Pol ε activity on the leading strand of roughly 77%, 90%, and nearly 100%, respectively63, 69, 72, 75. Similar comparisons place Pol δ on the lagging strand 77%, 90%, and 80–90% of the time, respectively. The highest boundaries suggest that Pol ε may be restricted to the leading strand and that Pol δ works primarily on the lagging strand. The less than 10% of Pol δ activity on the leading strand may be during replication restart, recombination and repair. The combination of these strand assignments with protein-protein interaction patterns and electron microscopy visualizations suggests a model for the normal eukaryotic replication fork (Figure 6C).
Does this division of labor transcend fungi? Mutation biases of human exonuclease-defective Pol ε (exo*) have been determined both biochemically and within POLE-exo* tumors64. Mutational strand biases near replication origins indicate that human Pol ε also works on the leading strand. Looking across origin clusters in many human tumor types, Pol ε-defective tumor cells have mutation patterns that imply leading strand mismatches while MMR-defective tumor cells have mutation patterns that imply lagging strand mismatches65, as seen in MMR-defective yeast63. This links the division of replicase labor with the outcomes of human disease and also supports the model across the opisthokonts (fungi, animals and their close relatives). Given conservation of replicase sequences, a conservation of the division of labor among the three replicases is implied across eukaryotes, but remains to be tested in a broader sample, including plants and amoebae (see Outstanding Questions).
Outstanding Questions.
Did lagging strand synthesis evolve from multi-copy rolling circle second strand synthesis?
Are there eukaryotes with vestigial archaeal replicases, or vice versa? What could this tell us about the origins of eukaryotes?
Can new methods for mapping polymerase responsibilities and interactions across genomes clarify the division of replicase labor in archaea (specifically the Lokiarchaea), bacteria (specifically Clostridia and B. subtilis), and in non-opisthokont eukaryotes (specifically plants and basal taxa)? What would the results suggest about the lifestyle and evolution of the Last Universal Common Ancestor (LUCA)?
With what frequency does Pol ε dissociate from the nascent leading strand and cede synthesis to Pol δ?
Is there a way to map replicase responsibilities in wild type organisms, perhaps allowing extension of new mapping methods to uncultured or genetically less-tractable organisms?
Can deviations from the normal division of replicase labor be mapped and characterized? If so, what are their implications for human development and health?
Trends.
In the presence of mutator polymerases variants and/or the absence of DNA mismatch repair, mutational analysis using both reporter genes and whole genome mutation mapping in yeasts and in human tumors reveal biases that suggest that eukaryotic replicases, DNA Polymerases (Pols) δ and ε, specialize in lagging and leading strand replication, respectively.
Ribonucleotides incorporated by DNA polymerase variants with increased incorporation rates serve as biomarkers of polymerase activity across the genome that likewise suggest that Pols δ and ε specialize in lagging and leading strand replication, respectively.
Electron microscopy of reconstituted replisome components, immunoprecipitation of nascent DNA and associated proteins, and other interaction studies have confirmed that Pols δ and ε localize to the nascent lagging and leading strands, respectively, and reveal the interactions that initiate and maintain this division.
Acknowledgments
We thank Roel Schaaper, Daniel Menendez and Rachel Krasich for critical reading of the manuscript. This work was supported by Project Z01 ES065070 to T.A.K. from the Division of Intramural Research of the National Institute of Environmental Health Sciences, which is a member of the National Institutes of Health. The authors apologize to the many colleagues whose primary research articles are not cited due to space limitations.
Glossary
- ARS####
autonomous replicating sequence; S. cerevisiae origin of replication; examples in text: ARS306, ARS1012, ARS1014.
- BrdU
bromodeoxyuridine; structural analog of deoxythymidine; incorporated opposite deoxyadenosine by DNA polymerases.
- Cdc45
component of the CMG replicative helicase complex; a replication initiation factor.
- chromatin immunoprecipitation
ChIP; technique used to investigate protein-DNA interactions
- CMG
replicative helicase complex; composed of Cdc45, MCM, and GINS.
- Crick strand
here, the "bottom" or 3'-to-5' reference genomic strand.
- Ctf4
Chromosome Transmission Fidelity; chromatin-associated protein; tethers Pol α to CMG.
- DNA polymerase families
polymerase sets defined by sequence homology; examples in text: Families A, B, C and D.
- DnaE1/DnaE3
bacterial Family C subclasses; proofreading proficient (via subunit)/deficient-.
- Dpo1/Dpo3
S. solfataricus DNA polymerases (Family B subclass B1/B3); also called PolB1/PolB3.
- Eocyte Hypothesis
suggests that eukaryotes descend from an archaeal ancestor, originally the Crenarchaeota; more recently interpreted as descent from within the TACK superphylum.
- eSPAN
enrichment and Sequencing of Protein-Associated Nascent DNA, a technique used to investigate protein-nascent DNA interactions.
- GINS
go-ichi-ni-san; in S. cerevisiae, composed of Sld5, Psf1, Psf2 and Psf3; involved in the assembly of the replication machinery.
- LACA/LECA
Last Archaea/Eukaryota Common Ancestors; the last organism from which all extant archaea/eukaryotes descend.
- leading/lagging strand
continuously/discontinuously synthesized nascent strands.
- MCM
MiniChromosome Maintenance; hexameric helicase component of CMG; in S. cerevisiae, composed of Mcm2–7.
- MMR
DNA Mismatch Repair; system for removing DNA mismatches after replication; analogous to the spellcheck function in word processing.
- nascent/template strand
growing/parental DNA strands.
- PCNA
Proliferating Cell Nuclear Antigen; eukaryotic DNA clamp; replication processivity factor.
- pol I
bacterial Family A polymerase; 5'-to-3' exonuclease activity.
- pol III
E. coli DNA polymerase (Family C subclass DnaE1); the holoenzyme has many modular subunits allowing optimization for specialized tasks.
- DNA polymerases α/δ/ε
eukaryotic Pols α/δ/ε (Family B); specialized in primer extension/lagging/leading strand synthesis; low/high/high processivity (latter two with PCNA/CMG); middling/high/high accuracy; proofreading deficient/proficient/proficient.
- PolC
bacterial Family C polymerases; usually proofreading-proficient.
- polD
archaeal Family D polymerase; composed of a polymerase subunit and a proofreading accessory subunit.
- proofreading
exonucleolytic removal of DNA mismatches before the replication fork has passed; analogous to the backspace function in word processing.
- RER
ribonucleotide excision repair; removes genomic ribonucleotides after replication; analogous to format- or font-specific find-and-replace functions in word processing.
- RFC
Replication Factor C; the PCNA clamp loading complex; in S. cerevisiae, composed of Rfc1–5.
- RNA World
hypothesized precursor to known life; RNA was responsible for genetic information storage and biological catalysis.
- TACK superphylum
archaeal superphylum composed of Thaum-, Aig-, Cren- and Kor-archaeota; provisionally designated as kingdom Proteoarchaeota.
- URA3
forward mutation reporter gene; encodes orotidine-5'-phosphate (OMP) decarboxylase, which converts 5-FOA into toxic 5-fluorouracil; mutations in URA3 can confer resistance to 5-FOA.
- Watson strand
here, the "top" or 5'-to-3' reference genomic strand.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kornberg A, Baker TA. DNA replication. W.H. Freeman; 1992. [Google Scholar]
- 2.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 3.Kainuma-Kuroda R, Okazaki R. Mechanism of DNA chain growth. XII. Asymmetry of replication of P2 phage DNA. J Mol Biol. 1975;94:213–228. doi: 10.1016/0022-2836(75)90079-0. [DOI] [PubMed] [Google Scholar]
- 4.Kumar D, et al. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38:3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar D, et al. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39:1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinheiro VB, Holliger P. Towards XNA nanotechnology: new materials from synthetic genetic polymers. Trends Biotechnol. 2014;32:321–328. doi: 10.1016/j.tibtech.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgs PG, Lehman N. The RNA World: molecular cooperation at the origins of life. Nat Rev Genet. 2015;16:7–17. doi: 10.1038/nrg3841. [DOI] [PubMed] [Google Scholar]
- 8.Fortune JM, et al. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 9.Shcherbakova PV, et al. Unique error signature of the four-subunit yeast DNA polymerase ε. J Biol Chem. 2003;278:43770–43780. doi: 10.1074/jbc.M306893200. [DOI] [PubMed] [Google Scholar]
- 10.St Charles JA, et al. Quantifying the contributions of base selectivity, proofreading and mismatch repair to nuclear DNA replication in Saccharomyces cerevisiae. DNA Repair (Amst) 2015;31:41–51. doi: 10.1016/j.dnarep.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motea EA, Berdis AJ. Terminal deoxynucleotidyl transferase: the story of a misguided DNA polymerase. Biochim Biophys Acta. 2010;1804:1151–1166. doi: 10.1016/j.bbapap.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norbury CJ. Cytoplasmic RNA: a case of the tail wagging the dog. Nat Rev Mol Cell Biol. 2013;14:643–653. doi: 10.1038/nrm3645. [DOI] [PubMed] [Google Scholar]
- 13.Berman AJ, Cech TR. SnapShot: telomeres and telomerase. Cell. 2012;151:1138–1138. e1131. doi: 10.1016/j.cell.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Nagata K, et al. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc Natl Acad Sci U S A. 1983;80:6177–6181. doi: 10.1073/pnas.80.20.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermoso JM, Salas M. Protein p3 is linked to the DNA of phage phi 29 through a phosphoester bond between serine and 5'-dAMP. Proc Natl Acad Sci U S A. 1980;77:6425–6428. doi: 10.1073/pnas.77.11.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moss B. Poxvirus DNA replication. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol Rev. 2006;30:321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 19.Timinskas K, et al. Comprehensive analysis of DNA polymerase III alpha subunits and their homologs in bacterial genomes. Nucleic Acids Res. 2014;42:1393–1413. doi: 10.1093/nar/gkt900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reyes-Lamothe R, et al. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozawa K, et al. Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, epsilon, theta and beta reveals a highly flexible arrangement of the proofreading domain. Nucleic Acids Res. 2013;41:5354–5367. doi: 10.1093/nar/gkt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanders GM, et al. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Beattie TR, Reyes-Lamothe R. A Replisome's journey through the bacterial chromosome. Front Microbiol. 2015;6:562. doi: 10.3389/fmicb.2015.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel PH, et al. Prokaryotic DNA polymerase I: evolution, structure, and "base flipping" mechanism for nucleotide selection. J Mol Biol. 2001;308:823–837. doi: 10.1006/jmbi.2001.4619. [DOI] [PubMed] [Google Scholar]
- 25.Makarova KS, et al. Evolution of replicative DNA polymerases in archaea and their contributions to the eukaryotic replication machinery. Front Microbiol. 2014;5:354. doi: 10.3389/fmicb.2014.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cubonová L, et al. Archaeal DNA polymerase D but not DNA polymerase B is required for genome replication in Thermococcus kodakarensis. J Bacteriol. 2013;195:2322–2328. doi: 10.1128/JB.02037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenough L, et al. The roles of family B and D DNA polymerases in Thermococcus species 9 degrees N Okazaki fragment maturation. J Biol Chem. 2015;290:12514–12522. doi: 10.1074/jbc.M115.638130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouillon C, et al. DNA polymerase switching on homotrimeric PCNA at the replication fork of the euryarchaea Pyrococcus abyssi. J Mol Biol. 2007;369:343–355. doi: 10.1016/j.jmb.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 29.Henneke G. In vitro reconstitution of RNA primer removal in Archaea reveals the existence of two pathways. Biochem J. 2012;447:271–280. doi: 10.1042/BJ20120959. [DOI] [PubMed] [Google Scholar]
- 30.Beattie TR, Bell SD. Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 2012;31:1556–1567. doi: 10.1038/emboj.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauer RJ, et al. Kinetics and fidelity of polymerization by DNA polymerase III from Sulfolobus solfataricus. Biochemistry. 2012;51:1996–2007. doi: 10.1021/bi201799a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knoll AH, et al. Eukaryotic organisms in Proterozoic oceans. Philos Trans R Soc Lond B Biol Sci. 2006;361:1023–1038. doi: 10.1098/rstb.2006.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohtomo Y, et al. Evidence for biogenic graphite in early Archaean Isua metasedimentary rocks. Nature Geoscience. 2014;7:25–28. [Google Scholar]
- 34.Lake JA, et al. Eocytes: a new ribosome structure indicates a kingdom with a close relationship to eukaryotes. Proc Natl Acad Sci U S A. 1984;81:3786–3790. doi: 10.1073/pnas.81.12.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koonin EV. Archaeal ancestors of eukaryotes: not so elusive any more. BMC Biol. 2015;13:84. doi: 10.1186/s12915-015-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spang A, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Byrnes JJ, et al. A new mammalian DNA polymerase with 3' to 5' exonuclease activity: DNA polymerase delta. Biochemistry. 1976;15:2817–2823. doi: 10.1021/bi00658a018. [DOI] [PubMed] [Google Scholar]
- 38.Kunkel TA, et al. Exonucleolytic proofreading by calf thymus DNA polymerase delta. Proc Natl Acad Sci U S A. 1987;84:4865–4869. doi: 10.1073/pnas.84.14.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syvaoja J, et al. DNA polymerases alpha, delta, and epsilon: three distinct enzymes from HeLa cells. Proc Natl Acad Sci U S A. 1990;87:6664–6668. doi: 10.1073/pnas.87.17.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrison A, et al. A third essential DNA polymerase in S. cerevisiae. Cell. 1990;62:1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- 41.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 42.Kesti T, et al. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 43.Dua R, et al. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol ε and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–22288. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- 44.Feng W, D'Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21:4495–4504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pavlov YI, et al. Mutator effects of overproducing DNA polymerase eta (Rad30) and its catalytically inactive variant in yeast. Mutat Res. 2001;478:129–139. doi: 10.1016/s0027-5107(01)00131-2. [DOI] [PubMed] [Google Scholar]
- 46.Ohya T, et al. The DNA polymerase domain of pol ε is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J Biol Chem. 2002;277:28099–28108. doi: 10.1074/jbc.M111573200. [DOI] [PubMed] [Google Scholar]
- 47.Johnson RE, et al. A Major Role of DNA Polymerase δ in Replication of Both the Leading and Lagging DNA Strands. Mol Cell. 2015;59:163–175. doi: 10.1016/j.molcel.2015.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burgers PM, et al. Who Is Leading the Replication Fork, Pol epsilon or Pol delta? Mol Cell. 2016;61:492–493. doi: 10.1016/j.molcel.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flood CL, et al. Replicative DNA polymerase δ but not ε proofreads errors in Cis and in Trans. PLoS Genet. 2015;11:e1005049. doi: 10.1371/journal.pgen.1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrino FW, Loeb LA. Hydrolysis of 3'-terminal mispairs in vitro by the 3'----5' exonuclease of DNA polymerase delta permits subsequent extension by DNA polymerase alpha. Biochemistry. 1990;29:5226–5231. doi: 10.1021/bi00474a002. [DOI] [PubMed] [Google Scholar]
- 51.Pavlov YI, et al. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr Biol. 2006;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 52.Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pavlov YI, Shcherbakova PV. DNA polymerases at the eukaryotic fork-20 years later. Mutat Res. 2010;685:45–53. doi: 10.1016/j.mrfmmm.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miyabe I, et al. Polymerase delta replicates both strands after homologous recombination-dependent fork restart. Nat Struct Mol Biol. 2015;22:932–938. doi: 10.1038/nsmb.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morrison A, Sugino A. The 3'-->5' exonucleases of both DNA polymerases delta and epsilon participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- 56.Shcherbakova PV, Pavlov YI. 3' –>5' exonucleases of DNA polymerases ε and δ correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niimi A, et al. Palm mutants in DNA polymerases α and η alter DNA replication fidelity and translesion activity. Mol Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pursell ZF, et al. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nick McElhinny SA, et al. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J Biol Chem. 2007;282:2324–2332. doi: 10.1074/jbc.M609591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nick McElhinny SA, et al. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyabe I, et al. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi: 10.1371/journal.pgen.1002407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larrea AA, et al. Genome-wide model for the normal eukaryotic DNA replication fork. Proc Natl Acad Sci U S A. 2010;107:17674–17679. doi: 10.1073/pnas.1010178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lujan SA, et al. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014;24:1751–1764. doi: 10.1101/gr.178335.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shinbrot E, et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014;24:1740–1750. doi: 10.1101/gr.174789.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haradhvala NJ, et al. Mutational Strand Asymmetries in Cancer Genomes Reveal Mechanisms of DNA Damage and Repair. Cell. 2016;164:538–549. doi: 10.1016/j.cell.2015.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nick McElhinny SA, et al. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc Natl Acad Sci U S A. 2010;107:4949–4954. doi: 10.1073/pnas.0914857107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams JS, et al. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nick McElhinny SA, et al. Genome instability due to ribonucleotide incorporation into DNA. Nat Chem Biol. 2010;6:774–781. doi: 10.1038/nchembio.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Clausen AR, et al. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat Struct Mol Biol. 2015;22:185–191. doi: 10.1038/nsmb.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reijns MA, et al. Lagging-strand replication shapes the mutational landscape of the genome. Nature. 2015;518:502–506. doi: 10.1038/nature14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koh KD, et al. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat Methods. 2015;12:251–257. doi: 10.1038/nmeth.3259. 253 p following 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Daigaku Y, et al. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 2015;22:192–198. doi: 10.1038/nsmb.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jinks-Robertson S, Klein HL. Ribonucleotides in DNA: hidden in plain sight. Nat Struct Mol Biol. 2015;22:176–178. doi: 10.1038/nsmb.2981. [DOI] [PubMed] [Google Scholar]
- 74.Williams JS, et al. Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nat Struct Mol Biol. 2015 doi: 10.1038/nsmb.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu C, et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 77.Fu YV, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chilkova O, et al. The eukaryotic leading and lagging strand DNA polymerases are loaded onto primer-ends via separate mechanisms but have comparable processivity in the presence of PCNA. Nucleic Acids Res. 2007;35:6588–6597. doi: 10.1093/nar/gkm741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johansson E, Macneill SA. The eukaryotic replicative DNA polymerases take shape. Trends Biochem Sci. 2010;35:339–347. doi: 10.1016/j.tibs.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 80.Langston LD, et al. CMG helicase and DNA polymerase ε form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc Natl Acad Sci U S A. 2014;111:15390–15395. doi: 10.1073/pnas.1418334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Georgescu RE, et al. Mechanism of asymmetric polymerase assembly at the eukaryotic replication fork. Nat Struct Mol Biol. 2014;21:664–670. doi: 10.1038/nsmb.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Georgescu RE, et al. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife. 2015;4:e04988. doi: 10.7554/eLife.04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oughtred R, et al. Use of the BioGRID Database for Analysis of Yeast Protein and Genetic Interactions. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.prot088880. pdb prot088880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sun J, et al. The architecture of a eukaryotic replisome. Nat Struct Mol Biol. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]