Abstract
The Hippo signaling network integrates diverse upstream signals to control cell fate decisions and regulate organ growth. Recent studies have provided new insights into the cellular organization of Hippo signaling, its relationship to cell-cell junctions, and how the cytoskeleton modulates Hippo signaling. Cell-cell junctions serve as platforms for Hippo signaling by localizing scaffolding proteins that interact with core components of the pathway. Interactions of Hippo pathway components with cell-cell junctions and the cytoskeleton also suggest potential mechanisms for the regulation of the pathway by cell contact and cell polarity. As our understanding of the complexity of Hippo signaling increases, a future challenge will be to understand how the diverse inputs into the pathway are integrated, and to define their respective contributions in vivo.
Keywords: Hippo, signaling, cytoskeleton, mechanical force, cell junctions
The Hippo Signaling Network
The Hippo signaling network integrates diverse upstream signals to control cell fate decisions and regulate organ growth. It was first discovered in Drosophila through the identification and characterization of genes that, when mutated, cause severe over-growth phenotypes [1]. Hippo signaling is highly conserved amongst animals, and dysregulation of the pathway has been linked to many human cancers [2]. One remarkable feature of Hippo signaling is its role as an integrator of growth control signals. Indeed, Hippo signaling is influenced by, or cross-talks with, multiple pathways that respond to growth factors, that promote growth linked to positional information, or that influence growth in response to nutritional and metabolic status [3-5]. Hippo signaling is also affected by contacts with neighboring cells and the extracellular matrix, and by mechanical forces. In this review, we first briefly describe new insights into core components of the Hippo pathway, and then focus on recent discoveries that have enhanced our understanding of the cellular organization of the Hippo pathway and its regulation by cell junctions, the actin cytoskeleton, and mechanical force.
Expansion of the Hippo Core
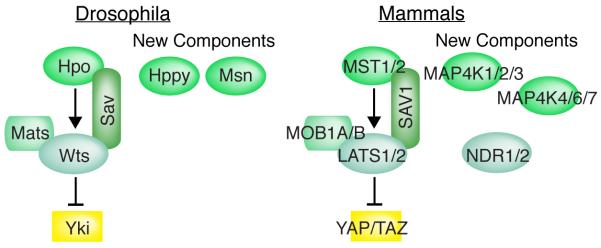
Hippo signaling regulates growth by controlling the localization of a transcriptional co-activator protein that in Drosophila is known as Yorkie (Yki) [6-8]. Transcriptional activation by Yki, which is achieved in part by recruiting chromatin and histone modifying complexes [9-11], leads to increased growth. Yki is down-regulated through phosphorylation by the kinase Warts (Wts), which promotes cytoplasmic localization of Yki [6, 7]. As most upstream inputs of Hippo signaling affect Wts, and Wts directly regulates Yki, Wts serves as a central regulatory node within the Hippo pathway. Wts is regulated in several ways, including phosphorylation by the kinase Hippo (Hpo) [12-16], and regulation of Wts abundance [17], Wts localization [18, 19], and Wts interaction with co-factors and inhibitors [20-24]. Activation of Wts is dependent upon two additional core components of the Hippo pathway: Mob-as-tumor suppressor (Mats), which is a Wts co-factor [23], and Salvador (Sav) [25, 26], which promotes Wts activation by acting as a scaffold that links Wts to Hpo [12-16]. The four proteins that regulate Yki, Hpo, Wts, Sav, and Mats, have been generally considered as the “core” of the Hippo network (Fig. 1A).
Figure 1. Core proteins of the Hippo network.
In Drosophila (left), the kinase Hpo phosphorylates and activates the kinase Wts; the kinase Wts phosphorylates and inhibits the transcriptional co-activator Yki. This requires the Wts co-factor Mats, and is facilitated by the scaffolding protein Sav. The Hppy and Msn kinases can also phosphorylate and activate Wts. In mammals (right), the kinases MST1 or MST2 phosphorylate and activate the kinases LATS1 and LATS2; the kinase LATS1 and LATS2 phosphorylate and inhibit the transcriptional co-activators YAP and TAZ. This requires the LATS co-factors MOB1A or MOB1B, and is facilitated by the scaffolding protein SAV1. MAP4K kinases can also phosphorylate and activate LATS kinases, and NDR kinases can also phosphorylate and inhibit YAP.
Mammals have an analogous Hippo network that includes the core components identified in Drosophila (though assigned different names). However, mammalian Hippo signaling has greater complexity and includes two Wts homologues, LATS1 and LATS2, two Hippo homologues MST1 and MST2, and two Yki homologues, YAP and TAZ (Fig 1B) [2, 3]. As in Drosophila, LATS proteins are the major, though not exclusive, regulators of YAP and TAZ. In mammals, YAP and TAZ localization as well as their stability are both regulated through LATS-dependent and LATS-independent processes [3]. Moreover, several ubiquitin ligases that influence LATS stability have been identified [27-32]. LATS kinases are members of a larger family of protein kinases, the Nuclear Dbf2-related kinases, and two other family members, NDR1 and NDR2, have recently been reported to phosphorylate Yap, and regulate Yap activity [33]. Recent studies have also led to the identification, in both Drosophila and in mammals, of multiple MAP4K-type kinases (which, like Hpo/Mst, are within the Ste20 family of protein kinases) that phosphorylate and activate Wts and LATS [34-37]. The identification of these additional “core” components of the pathway explains some instances of MST-independent regulation of LATS, and emphasizes that different inputs into the Hippo pathway could act through regulation of distinct kinases.
Recent studies have also identified additional proteins that could act as scaffolds and promote the interaction of core Hippo pathway components. APC, which is best known as a key component of the β-catenin destruction complex, was observed in mammalian cells to have an additional function as a scaffold that promotes association of LATS and SAV [38]. In Drosophila, βPix and Git form a scaffold that promotes activation of Hpo [39]. In mammalian cells, βPix was identified as interacting with LATS and YAP/TAZ to stimulate YAP/TAZ phosphorylation [40]. Schip1 was recently identified in Drosophila as a protein that activates Hpo by binding to both Expanded and Tao-1 [41], a kinase that induces Hpo [42, 43].
Cellular organization of the Hippo network and regulation at cell junctions
The first decade of Hippo pathway research was characterized by tremendous progress in genetic and biochemical characterization of the pathway. More recently, our understanding of the cell biology of Hippo signaling has advanced significantly – where pathway components localize, where key events happen inside the cell, and how changes in protein localization modulate pathway activity. Many Hippo pathway components localize to cell-cell junctions, such that in addition to their role in maintaining tissue integrity and polarity, these junctions effectively provide a platform for regulation of Hippo signaling. The links between cell junctions and Hippo pathway components can help explain how cell contacts and cell polarity modulate Hippo pathway activity. In both Drosophila and mammalian cells, cadherin-mediated cell-cell adhesion occurs at adherens junctions, and these sites of cell attachment are connected to the actin-myosin cytoskeleton through catenins and associated proteins. Apical to the adherens junctions, mammalian epithelial cells have tight junctions, which form a paracellular diffusion barrier. In Drosophila, the paracellular diffusion barrier is formed by septate junctions, which are basal to the adherens junctions. Nonetheless, many proteins that are found at tight junctions in mammals, such as Crumbs, are conserved in Drosophila, and as in mammalian cells they localize to cell junctions just apical to adherens junctions [44]; in Drosophila epithelia this region is referred to as the marginal zone, or sub-apical region. Several of the proteins first identified as upstream activators of Hippo signaling, including Dachsous (Ds), Fat, Expanded (Ex), and Merlin (Mer), localize near the marginal zone [18, 45, 46], suggesting that this could be a site of pathway activation. The concept that activation of core components happens at the membrane was further supported by observations that forced membrane localization of over-expressed Hpo could increase Hpo activity [47], and forced membrane localization of over-expressed Mats or Wts, or their mammalian homologues, could increase Wts/LATS activity [19, 48, 49].
More recently, progress has been made in visualizing the endogenous localization of components in Drosophila, and in characterizing mechanisms that contribute to their localization (Fig. 2A). Sav localizes to cell membranes through interaction with the transmembrane immunoglobulin-domain protein echinoid (Ed) [50], which localizes to the membrane overlapping adherens junctions and the marginal zone. Ed participates in homophilic binding to Ed in neighboring cells [51], which serves as a mechanism linking cell-cell contacts to Hippo pathway regulation. Sav, in turn, can recruit Hpo to apical cell-cell junctions, although Hpo is normally found predominantly in the cytoplasm [18, 19, 52]. Crumbs localizes to the marginal zone, and this depends upon homophilic binding between Crumbs proteins in neighboring cells, which serves as another mechanism linking cell contact and polarity to Hippo signaling [53]. Cell contact-dependent regulation of Hippo signaling through Ed and Crb has recently been implicated in maintaining quiescence of neural stem cells in the larval brain [54]. Crumbs, in turn, is required for membrane localization of Ex [53, 55, 56]. By contrast, Wts localizes to adherens junctions, where it is recruited by the Drosophila Ajuba LIM family protein, Jub [20]. Jub is an inhibitor of Wts [21, 22], implying that in this context Wts localization to cell-cell junctions is associated with Wts inhibition, rather than activation. Consistent with this role, disruption of adherens junctions in Drosophila epithelia can be associated with increased Hippo pathway activity [57]. Wts can also be down-regulated by a signaling pathway initiated by the large cadherin family proteins Ds and Fat, which localize to the marginal zone, and regulate Hippo signaling through the Myosin family protein Dachs [1]. Earlier studies identified an influence of Dachs on Wts protein levels [17], and a more recent study identified an ability of Dachs to inhibit Wts association with Mats [24].
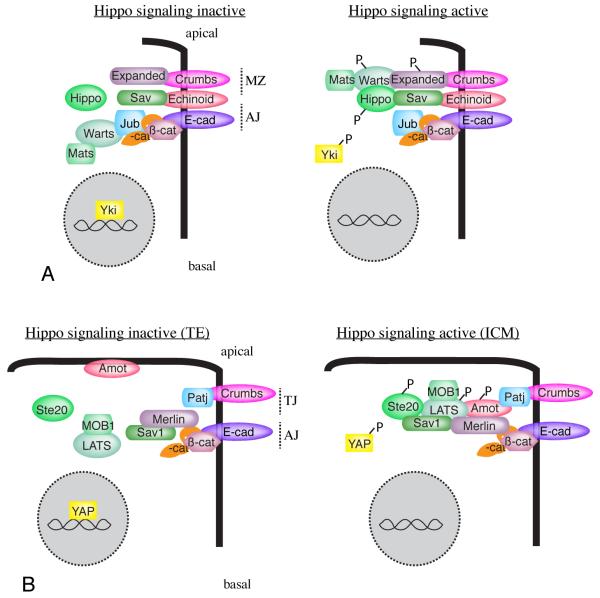
Figure 2. Localization and re-localization of core Hippo pathway components.
A) Localization of core components in Drosophila epithelia. Under conditions of low Hippo pathway activity (left), Wts is associated with its inhibitor, Jub, at adherens junctions (AJ), and Hpo is predominantly cytoplasmic, while Yki is nuclear. Under conditions of high Hippo pathway activity, Wts localizes with Ex at the marginal zone (MZ), and Hpo is recruited to Sav, causing Yki to be cytoplasmic. B) Localization of core components in 32 cell mouse blastocysts. In outer TE cells, Hippo signaling is low, YAP is nuclear, and Amot is localized to the apical membrane. This is presumed to prevent the formation of a LATS activation complex, although the localization of LATS proteins has not been determined in this tissue. In inner ICM cells, Hippo signaling is high, YAP is cytoplasmic, and Motins (Amot) are localized to cell-cell junctions, where, together with Merlin/NF2, they promote phosphorylation and activation of LATS [118].
Dynamic localization of Hippo pathway components
The observation that Wts normally localizes with an inhibitor, Jub, raised the question of how and where Wts normally gets activated. Under conditions of pathway activation in Drosophila imaginal discs, Wts re-localized from Jub to Ex, where it is activated by Hpo , as revealed by phospho-Wts staining [18] (Fig. 2A). This re-localization requires both Ex, which physically interacts with Wts, and Hpo, which promotes Ex-Wts binding. Ex also physically interacts with Hpo [58], and thus could act as a scaffold to link Hpo to Wts (Fig. 3A). Interestingly, examination of Merlin suggests that it plays a similar role, both in Drosophila and mammalian cells [19]. While it was initially thought that Merlin functions as an activator of Hpo, Mer was found instead to promote Wts/LATS activation by bringing Wts/LATS and Hpo/MST together at cell membranes [19]. This scaffolding occurs because, under some conditions, Mer binds to Wts/LATS, and Mer also binds Sav, therefore, linking Mer to Hpo (Fig. 3B). In addition, both APC (in mammals) [38] and Kibra, which in Drosophila localizes near Mer and Ex and acts genetically at a similar point in the Hippo pathway, can bind to both Sav and Wts/LATS [58-61]. Together, these observations imply that assembly of an activation complex, in which Hpo and Wts are linked through scaffolding by Ex, Mer, Kibra, or APC, is a key step in Hippo signaling. There has, however, been controversy in mammalian cells over how Merlin influences Hippo signaling, as it has also been reported that Merlin regulates LATS through the ubiquitin ligase, CRL4/DCAF1, and that CRL4/DCAF1 regulates LATS in the nucleus [62, 63]. Merlin might regulate LATS by multiple mechanisms, but immunolocalization studies of LATS proteins are discordant, and further studies are needed to determine whether this reflects differences in experimental conditions, or in the reagents employed.
Figure 3. Hippo activation complexes.
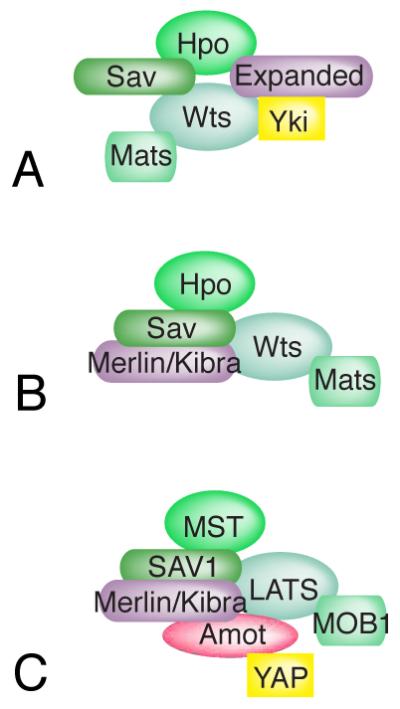
Hippo activation appears to require the participation of multiple scaffolds that assemble multi-protein complexes. A) In Drosophila, both Sav and Ex can interact with both Hpo and Wts. B) Ex is partially redundant with Merlin and Kibra (depending upon the tissue), which can interact with Wts and Sav. C) In mammals, Merlin (NF2) and Motins play key roles in Hippo pathway activation. Merlin and Kibra can each interact with SAV1 and LATS, and Motins can interact with LATS, Merlin, and Kibra.
There are also differences between upstream regulation of the pathway in Drosophila and that in mammals. Loss of E-cadherin or α-catenin in mammalian cells has been associated with increased YAP activity [64, 65], rather than decreased Yki activity as in Drosophila [57], which suggests that instead of, or at least in addition to, a role in promoting Wts inactivation, adherens junctions in mammalian cells have a role in promoting Wts activation. One possible mechanism for this observation could be physical interactions between α-catenin and Merlin [66], which could localize Merlin to adherens junctions, although Merlin also associates with tight junction proteins [67]. It was also recently reported that α-catenin inhibits a direct activation of YAP by Src [68]. Whether mammalian homologues of Ds and Fat (Dchs1 and Fat4) regulate the Hippo pathway in mammalian cells is also controversial, as there have been conflicting reports about whether they influence Yap activity [69-71]. Moreover, how they might influence Hippo signaling in mammals is unclear, as Dachs, which is essential for Wts regulation by Ds-Fat signaling in Drosophila [17], is not conserved in vertebrates [72]. Crumbs is an upstream regulator of Hippo signaling in both Drosophila and mammalian cells [53, 55, 56, 73, 74], but may act through distinct mechanisms: in Drosophila it influences Hippo signaling by localizing Ex, which is not fully conserved in mammals, although it has some similarity to mammalian Willin [75]. In mammals, a Crumbs homologue (Crb3) promotes Lats phosphorylation [74], possibly by recruiting tight junction proteins that regulate Lats (Fig. 2B).
Mammals also have a distinct family of proteins, the Motins, which are upstream regulators of Hippo signaling that can localize to tight junctions, and that have some functional similarities to Ex [76]. The Motins include Angiomotin, which exists in distinct p80 and p130 isoforms (created by alternative splicing), Angiomotin-like 1 (AmotL1), and Angiomotin-like 2 (AmotL2). Motins can act as scaffolding proteins that bring together multiple components of the Hippo pathway: Amot-p130, AmotL1 and AmotL2 can bind LATS, YAP, Merlin, and Kibra (Fig. 3C). Their ability to bind YAP could enable them to modulate YAP activity both by promoting its phosphorylation, and by directly sequestering it in the cytoplasm [77-80]; Ex has a similar ability to sequester Yki [81, 82]. Motins have also been implicated in a feed-forward loop that promotes Hippo pathway activation: Motins are substrates of LATS [83-86], and phosphorylation by LATS both stabilizes Motins, and promotes their binding to Merlin. Binding of Motins to Merlin appears to influence Merlin conformation, such that its binding to LATS is enhanced, which presumably promotes further LATS activation [87].
Studies of early cell fate specification in the mouse embryo have identified a role for differential Motin localization in controlling Hippo signaling [88]. At the 32-cell stage, mouse blastomeres are subdivided into inner cells, which form the inner cell mass (ICM), and outer cells, which form the trophectoderm (TE). This subdivision requires Hippo signaling, which is high in the ICM, leading to cytoplasmic YAP, and low in the TE, leading to nuclear YAP [89]. Both cell contacts and cell polarity influence Hippo signaling at this stage. In the ICM, Motins localizes to cell-cell junctions and Hippo signaling is active. However, the outer cells become polarized, causing Motins to localize to the apical domain rather than to cell-cell junctions [86, 90]. Evidently, this loss of Motins from cell junctions prevents formation of the Hippo pathway activation complex needed to promote LATS activation (Fig. 2B). Thus, while there are some differences in the specific proteins involved between Drosophila and mammals, a common theme has emerged regarding the existence and importance of platforms for Hippo signal transduction at cell junctions.
Regulation of Hippo signaling by the extracellular matrix
Hippo signaling is also regulated by attachment to the extracellular matrix (ECM). For example, the extent of cell-ECM contacts influences Hippo signaling, and detachment of cells can result in cell death through activation of the Hippo pathway [91-93]. The requirement for cell-substrate attachment has both biochemical and biomechanical components. One biochemical mechanism involves the modulation of Hippo signaling by Integrin-linked Kinase (ILK), which could inhibit Merlin activation by inhibiting the phosphatase MYPT1 [94]. More recently, a link between integrin and Hippo signaling that depends upon Focal adhesion kinase (FAK) was identified [95]. Integrins bound to fibronectin stimulate FAK, thereby activating Src, which activates PI3K. The PI3K downstream kinase PDK1 then disrupts the core kinase cassette, resulting in the inhibition of Hippo signaling [96]. As activation of FAK by integrins can be modulated by substrate stiffness [97], regulation of Hippo signaling through FAK could also contribute to influences of the mechanical environment on Hippo signaling, . An alternative mechanism by which Src can promote YAP activity, involving direct phosphorylation of tyrosines residues on YAP by Src, has also recently been described [68, 98].
Regulation of Hippo signaling by F-actin levels
Indications of the key influence of the cytoskeleton on Hippo signaling first came from observations that mutations in Drosophila that increase F-actin accumulation could be associated with increased Yki activity [99, 100]; this also occurs in mammalian cells [101]. Demonstration of the influence of mechanical force on YAP and TAZ activity then came from observations that cell shape and rigidity of the extracellular matrix could influence YAP/TAZ activity [93], and that this influence requires myosin, which generates tension in the actin cytoskeleton. It was also reported that regulation of YAP/TAZ by cell attachment and cell shape occurs independently of LATS [93], but others have reported LATS-dependent effects [91, 92]. Modulation of the actin cytoskeleton is also correlated with cell density-dependent effects on Yap activity [101, 102].
The influences of cell shape, cell attachment, cell density, and matrix rigidity on Hippo signaling are suppressed by inhibiting the key cytoskeletal regulator Rho [91-93, 101]. This is also true for other upstream inputs of Hippo signaling, such as GPCR pathways, which may influence Hippo signaling through cytoskeleton regulation [103]. In Drosophila, Zyxin may also regulate Hippo signaling in part through modulation of the cytoskeleton [21, 104]. Initially, how the actin cytoskeleton influences Hippo signaling remained unknown, but a series of recent studies have begun to make progress on identifying molecular mechanisms that link the cytoskeleton to regulation of Hippo signaling.
In mammalian cells, Motins have been identified as a key link between F-actin and Hippo pathway regulation, as knockdown of all three Motins increased Yap activity, even in the presence of cytoskeletal disruption [105]. Motins can physically associate with F-actin, but this association is blocked by phosphorylation of Motins by Lats kinases [83, 84, 105]. Moreover, F-actin competes with YAP for binding to Motins. Thus, when LATS phosphorylates Amot-p130 to inhibit its binding to F-actin, it increases Amot-p130 binding to YAP, and hence inhibition of YAP [105]. Notably, the influence of F-actin on Motin-YAP binding, together with potential sequestration of YAP through direct binding to Motins, also provides a possible explanation for observations of LATS-independent regulation of YAP by the cytoskeleton. Down-regulation of YAP induced by disruption of the actin cytoskeleton also requires Protein kinase A in mammalian cells, which can directly phosphorylate LATS and enhance LATS activity [106].
In cultured Drosophila cells, cytoskeletal disruption increased Merlin-Wts binding, suggesting that F-actin accumulation could potentially modulate Hippo signaling by influencing interaction between Merlin and Wts [19]. Regulation of Wts activity by F-actin in Drosophila was also partially dependent upon JNK activity [99]. JNK also contributes to influences of cyclic stretch on YAP activity in mammalian cells [107], which occurs over a time scale that correlates with reorganization of F-actin. JNK has a complex relationship to Hippo signaling, as in Drosophila, depending upon the context, it can activate or inhibit Yki [108-110]. A mechanism by which Yki gets activated by JNK involves phosphorylation of Jub, or one of its mammalian homologues, LIMD1; this phosphorylation promotes its ability to bind to, and hence inhibit, LATS [111].
Regulation of Hippo signaling by cytoskeletal tension
In addition to mechanisms that appear to depend upon accumulation of F-actin, mechanisms that could provide a basis for influences of tension within the actin cytoskeleton on Hippo signaling have been identified. As noted above, one such mechanism is the influence of cytoskeletal tension on integrin-dependent signaling. The actin cytoskeleton also forms attachments to the nuclear envelope. Intriguingly, a recent study reported that Nesprin 1 Giant, a protein required for attachment of the actin cytoskeleton to the nuclear membrane, is required for the activation of YAP in response to dynamic stretch in mesenchymal stem cells [112]. How this attachment is able to influence YAP activity remains to be determined.
Epithelial cells are also mechanically coupled to each other at adherens junctions, which are attached to the actin cytoskeleton. In growing Drosophila epithelia, these cell-cell junctions are under tension, and this tension promotes Yki activity [20]. Activation of YAP that is promoted by stretching cells, and dependent upon adherens junctions, has also been observed in cultured mammalian cells [113]. A mechanism for how tension at adherens junctions promotes Yki activity has been identified in Drosophila, where the localization of Jub to adherens junctions is regulated by myosin activity [20]. This recruitment is mediated through α-catenin, which can act as a mechanotransducer: studies of the association between α -catenin and Vinculin have indicated that α -catenin, which links adherens junctions to the actin cytoskeleton, can undergo a tension-dependent conformational change that exposes, under high tension, a Vinculin binding site [114]. This same conformational change might also influence binding between Jub and α -catenin. The Jub recruited to adherens junctions then recruits Wts to adherens junctions, which, leads to increased Yki activity because Jub is a Wts inhibitor. Conversely, when tension is lowered by reducing myosin activity, Jub and Wts recruitment to junctions is decreased, as is Yki activity [20].
The Spectrin cytoskeleton also appears to provide a link between tension and Hippo signaling, but the nature of this link remains unclear. Spectrins were found to influence Hippo signaling, both in Drosophila and in cultured mammalian cells, in three independent studies [115-117]. Two of these suggested that Spectrins might be regulated by cytoskeletal tension, and help transduce the effects of tension onto Hippo signaling, possibly through upstream regulators of Hippo signaling including Crumbs, Merlin, and Kibra [115, 116]. The other study, by contrast, reported that Spectrins influence myosin phosphorylation, and suggested that Spectrins might influence Hippo signaling by affecting actomyosin contractility [117]. Thus, while Spectrins clearly have an influence on Hippo signaling, defining the mechanism by which they regulate the Hippo network requires further study.
Concluding Remarks
From its simple beginnings as the Salvador-Warts-Hippo pathway, our conception of Hippo signaling has expanded to a complex network including dozens of interacting proteins that cross-talk with numerous other cellular pathways. A key aspect of Hippo signal transduction emphasized by recent studies is the fundamental role of protein scaffolds, like Ex and the Motins, which assemble activation complexes through their ability to bind multiple pathway components. The localization of these scaffolds to cell junctions provides a basis for the sensitivity of Hippo signaling to cell-cell contact and cell polarity. Moreover, modulation of the localization of these scaffolds, or of the ability of other pathway components to bind to them, has emerged as a key mechanism for modulating Hippo signaling.
One of the remarkable features of Hippo signaling is its sensitivity to the cytoskeleton and mechanical forces. Progress has been made recently in identifying molecular mechanisms by which the cytoskeleton can influence Hippo signaling, but among the many outstanding questions that remain to be answered (see Outstanding Questions), a key challenge for the future will be to define the respective contributions of these different mechanisms in vivo, and understand how these contributions vary in different contexts to control cell fate decisions and regulate organ growth.
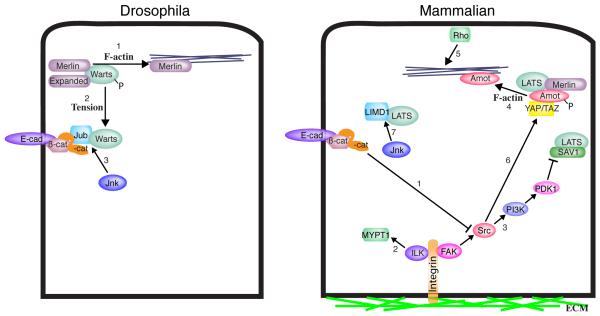
Figure 4. Regulation of Hippo signaling by F-actin and cytoskeletal tension.
Hippo signaling is regulated by levels of F-actin, tension within the actin cytoskeleton, and cell attachments. Several processes that could contribute to these effects have been identified, some of which are illustrated here. In Drosophila, these include 1) inhibition of Merlin-Wts interactions by F-actin, 2) cytoskeletal tension dependent recruitment of Wts into a complex with Jub at adherens junctions, and 3) promotion of Jub-Wts binding by Jnk phosphorylation. In mammals, these include 1) α-catenin-mediated inhibition of Src activation by integrins, 2) activation of MYPT1 by ILK, 3) inhibition of Sav-Lats association through a FAK-Src-PI3K-PDK1 pathway, 4) association of Amot with F-actin, which prevents Amot from associating with YAP/TAZ, 5) increases in F-actin promoted by Rho, 6) activation of YAP through phosphorylation of YAP by Src, and 7) promotion of LATS-LIMD1 binding by Jnk phosphorylation. In most cases potential conservation of these processes between Drosophila and mammals has not yet been investigated.
Outstanding questions.
Are different core components of the Hippo signaling network regulated by different upstream inputs?
Where do LATS proteins localize in vivo and what does regulation of their localization contribute to Hippo pathway regulation in mammals?
How are different cytoskeletal-dependent forms of regulation integrated and coordinated?
What do each of the distinct mechanisms for cytoskeletal regulation of Hippo signaling contribute to modulation of Hippo signaling in vivo during different developmental, physiological or pathological processes?
What additional cellular sites of Hippo/Mst and Warts/Lats activation remain to be discovered?
Which of the regulatory mechanisms so far identified only in Drosophila or only in mammals are evolutionarily conserved?
Trends box.
Hippo signaling is a complex network that integrates multiple growth control signals through an expanding set of core kinases.
Recent studies have provided insights into the cellular organization of Hippo signaling, including where pathway components localize, where key events happen inside the cell, and how changes in protein localization modulate pathway activity.
Scaffolds play an essential role in Hippo signal transduction by assembling kinase activation complexes through their ability to bind multiple pathway components. Localization of these scaffolds to cell junctions provides a basis for the sensitivity of Hippo signaling to cell contact and cell polarity.
Progress has been made in identifying mechanisms by which the cytoskeleton and mechanical forces can regulate Hippo signaling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- 2.Harvey KF, et al. The Hippo pathway and human cancer. Nature reviews. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 3.Yu F-X, et al. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santinon G, et al. Control of YAP/TAZ Activity by Metabolic and Nutrient-Sensing Pathways. Trends in cell biology. 2015 doi: 10.1016/j.tcb.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Irvine KD, Harvey KF. Control of organ growth by patterning and hippo signaling in Drosophila. Cold Spring Harbor perspectives in biology. 2015;7 doi: 10.1101/cshperspect.a019224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, et al. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 9.Oh H, et al. Yorkie promotes transcription by recruiting a histone methyltransferase complex. Cell Reports. 2014;8:449–459. doi: 10.1016/j.celrep.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oh H, et al. Genome-wide Association of Yorkie with Chromatin and Chromatin-Remodeling Complexes. Cell Reports. 2013;3:309–318. doi: 10.1016/j.celrep.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qing Y, et al. The Hippo effector Yorkie activates transcription by interacting with a histone methyltransferase complex through Ncoa6. eLife. 2014:e02564. doi: 10.7554/eLife.02564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu S, et al. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 13.Udan RS, et al. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 14.Pantalacci S, et al. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 15.Jia J, et al. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey KF, et al. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 17.Cho E, et al. Delineation of a Fat tumor suppressor pathway. Nat Genet. 2006;38:1142–1150. doi: 10.1038/ng1887. [DOI] [PubMed] [Google Scholar]
- 18.Sun S, et al. Localization of Hippo Signaling complexes and Warts activation in vivo. Nature Communications. 2015;6:8402. doi: 10.1038/ncomms9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin F, et al. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rauskolb C, et al. Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell. 2014;158:143–156. doi: 10.1016/j.cell.2014.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauskolb C, et al. Zyxin links fat signaling to the hippo pathway. PLoS biology. 2011;9:e1000624. doi: 10.1371/journal.pbio.1000624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das Thakur M, et al. Ajuba LIM proteins are negative regulators of the Hippo signaling pathway. Curr Biol. 2010;20:657–662. doi: 10.1016/j.cub.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai ZC, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 24.Vrabioiu AM, Struhl G. Fat/Dachsous Signaling Promotes Drosophila Wing Growth by Regulating the Conformational State of the NDR Kinase Warts. Developmental Cell. 2015;35:737–749. doi: 10.1016/j.devcel.2015.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tapon N, et al. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 26.Kango-Singh M, et al. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- 27.Salah Z, et al. NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell cycle. 2013;12:3817–3823. doi: 10.4161/cc.26672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeung B, et al. WWP1 E3 ligase targets LATS1 for ubiquitin-mediated degradation in breast cancer cells. PLoS ONE. 2013;8:e61027. doi: 10.1371/journal.pone.0061027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho KC, et al. Itch E3 ubiquitin ligase regulates large tumor suppressor 1 stability [corrected] Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4870–4875. doi: 10.1073/pnas.1101273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salah Z, et al. Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Research. 2011;71:2010–2020. doi: 10.1158/0008-5472.CAN-10-3516. [DOI] [PubMed] [Google Scholar]
- 31.Bae SJ, et al. NEDD4 controls intestinal stem cell homeostasis by regulating the Hippo signalling pathway. Nature communications. 2015;6:6314. doi: 10.1038/ncomms7314. [DOI] [PubMed] [Google Scholar]
- 32.Ma B, et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nature Cell Biology. 2015;17:95–103. doi: 10.1038/ncb3073. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, et al. NDR Functions as a Physiological YAP1 Kinase in the Intestinal Epithelium. Current biology : CB. 2015;25:296–305. doi: 10.1016/j.cub.2014.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S, et al. Overlapping functions of the MAP4K family kinases Hppy and Msn in Hippo signaling. Cell Discovery. 2015;1:15038. doi: 10.1038/celldisc.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, et al. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Developmental Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, et al. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Developmental Cell. 2015;34:642–655. doi: 10.1016/j.devcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng Z, et al. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nature communications. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai J, et al. β-Catenin destruction complex-independent regulation of Hippo-YAP signaling by APC in intestinal tumorigenesis. Genes & Development. 2015;29:1493–1506. doi: 10.1101/gad.264515.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dent LG, et al. The GTPase regulatory proteins Pix and Git control tissue growth via the Hippo pathway. Current biology. 2015;25:124–130. doi: 10.1016/j.cub.2014.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arash EH, et al. Arhgef7 promotes activation of the Hippo pathway core kinase Lats. The EMBO Journal. 2014:e201490230. doi: 10.15252/embj.201490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung H-L, et al. Drosophila Schip1 Links Expanded and Tao-1 to Regulate Hippo Signaling. Developmental Cell. 2016;36:511–524. doi: 10.1016/j.devcel.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Poon CLC, et al. The Sterile 20-like Kinase Tao-1 Controls Tissue Growth by Regulating the Salvador-Warts-Hippo Pathway. Developmental Cell. 2011;21:896–906. doi: 10.1016/j.devcel.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 43.Boggiano JC, et al. Tao-1 Phosphorylates Hippo/MST Kinases to Regulate the Hippo-Salvador-Warts Tumor Suppressor Pathway. Developmental Cell. 2011;21:888–895. doi: 10.1016/j.devcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tepass U, et al. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell. 1990;61:787–799. doi: 10.1016/0092-8674(90)90189-l. [DOI] [PubMed] [Google Scholar]
- 45.Ma D, et al. Fidelity in planar cell polarity signalling. Nature. 2003;421:543–547. doi: 10.1038/nature01366. [DOI] [PubMed] [Google Scholar]
- 46.McCartney BM, et al. The neurofibromatosis-2 homologue, Merlin, and the tumor suppressor expanded function together in Drosophila to regulate cell proliferation and differentiation. Development. 2000;127:1315–1324. doi: 10.1242/dev.127.6.1315. [DOI] [PubMed] [Google Scholar]
- 47.Deng Y, et al. Hippo activation through homodimerization and membrane association for growth inhibition and organ size control. Developmental Biology. 2013;375:152–159. doi: 10.1016/j.ydbio.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 48.Hergovich A, et al. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochemical and biophysical research communications. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 49.Ho LL, et al. Mob as tumor suppressor is activated at the cell membrane to control tissue growth and organ size in Drosophila. Dev Biol. 2010;337:274–283. doi: 10.1016/j.ydbio.2009.10.042. [DOI] [PubMed] [Google Scholar]
- 50.Yue T, et al. The cell adhesion molecule echinoid functions as a tumor suppressor and upstream regulator of the Hippo signaling pathway. Dev Cell. 2012;22:255–267. doi: 10.1016/j.devcel.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Islam R, et al. Neuroglian activates Echinoid to antagonize the Drosophila EGF receptor signaling pathway. Development. 2003;130:2051–2059. doi: 10.1242/dev.00415. [DOI] [PubMed] [Google Scholar]
- 52.Grzeschik NA, et al. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 53.Chen CL, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci U S A. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding R, et al. The Hippo signalling pathway maintains quiescence in Drosophila neural stem cells. Nature communications. 2016;7:10510. doi: 10.1038/ncomms10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Robinson BS, et al. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ling C, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci U S A. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang C-C, et al. Differential regulation of the Hippo pathway by adherens junctions and apical-basal cell polarity modules. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:1785–1790. doi: 10.1073/pnas.1420850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu J, et al. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Genevet A, et al. Kibra is a regulator of the Salvador/Warts/Hippo signalling network. Dev Cell. 2010;18:300–308. doi: 10.1016/j.devcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baumgartner R, et al. The WW domain protein Kibra acts upstream of Hippo in Drosophila. Dev Cell. 2010;18:309–316. doi: 10.1016/j.devcel.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 61.Xiao L, et al. KIBRA regulates Hippo signaling activity via interactions with large tumor suppressor kinases. Journal of Biological Chemistry. 2011;286:7788–7796. doi: 10.1074/jbc.M110.173468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, et al. Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer cell. 2014;26:48–60. doi: 10.1016/j.ccr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schlegelmilch K, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim NG, et al. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gladden AB, et al. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Developmental Cell. 2010;19:727–739. doi: 10.1016/j.devcel.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi C, et al. A Tight Junction-Associated Merlin-Angiomotin Complex Mediates Merlin's Regulation of Mitogenic Signaling and Tumor Suppressive Functions. Cancer cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li P, et al. αE-catenin inhibits a Src-YAP1 oncogenic module that couples tyrosine kinases and the effector of Hippo signaling pathway. Genes & Development. 2016 doi: 10.1101/gad.274951.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mao Y, et al. Fat4/Dchs1 signaling between stromal and cap mesenchyme cells influences nephrogenesis and ureteric bud branching. Development. 2015;142:2474–2585. doi: 10.1242/dev.122630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bagherie-Lachidan M, et al. Stromal Fat4 acts non-autonomously with Dachsous1/2 to restrict the nephron progenitor pool. Development. 2015;142:2564–2573. doi: 10.1242/dev.122648. [DOI] [PubMed] [Google Scholar]
- 71.Das A, et al. Stromal-epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol. 2013;15:1035–1044. doi: 10.1038/ncb2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bossuyt W, et al. An evolutionary shift in the regulation of the Hippo pathway between mice and flies. Oncogene. 2014;33:1218–1228. doi: 10.1038/onc.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Varelas X, et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Developmental cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 74.Szymaniak AD, et al. Crumbs3-Mediated Polarity Directs Airway Epithelial Cell Fate through the Hippo Pathway Effector Yap. Developmental Cell. 2015;34:283–296. doi: 10.1016/j.devcel.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Angus L, et al. Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene. 2012;31:238–250. doi: 10.1038/onc.2011.224. [DOI] [PubMed] [Google Scholar]
- 76.Moleirinho S, et al. The Angiomotins--from discovery to function. FEBS letters. 2014;588:2693–2703. doi: 10.1016/j.febslet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao B, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes & Development. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Paramasivam M, et al. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Molecular biology of the cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan SW, et al. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. The Journal of biological chemistry. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, et al. Angiomotin-like proteins associate with and negatively regulate YAP1. Journal of Biological Chemistry. 2011;286:4364–4370. doi: 10.1074/jbc.C110.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oh H, et al. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Badouel C, et al. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev Cell. 2009;16:411–420. doi: 10.1016/j.devcel.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 83.Dai X, et al. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. Journal of Biological Chemistry. 2013;288:34041–34051. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chan SW, et al. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. Journal of Biological Chemistry. 2013;288:37296–37307. doi: 10.1074/jbc.M113.527598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adler JJ, et al. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17368–17373. doi: 10.1073/pnas.1308236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hirate Y, et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Current biology. 2013;23:1181–1194. doi: 10.1016/j.cub.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y, et al. Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway. Cell research. 2015;25:801–817. doi: 10.1038/cr.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sasaki H. Position- and polarity-dependent Hippo signaling regulates cell fates in preimplantation mouse embryos. Seminars in cell & developmental biology. 2015;47-48:80–87. doi: 10.1016/j.semcdb.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 89.Nishioka N, et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 90.Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nature communications. 2013;4:2251. doi: 10.1038/ncomms3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao B, et al. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes & Development. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wada K-I, et al. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- 93.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 94.Serrano I, et al. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nature communications. 2013;4:2976. doi: 10.1038/ncomms3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim N-G, Gumbiner BM. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. The Journal of Cell Biology. 2015;210:503–515. doi: 10.1083/jcb.201501025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fan R, et al. Regulation of Hippo pathway by mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1216462110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Seong J, et al. Distinct biophysical mechanisms of focal adhesion kinase mechanoactivation by different extracellular matrix proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19372–19377. doi: 10.1073/pnas.1307405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Taniguchi K, et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sansores-Garcia L, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. The EMBO Journal. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fernández BG, et al. Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development. 2011;138:2337–2346. doi: 10.1242/dev.063545. [DOI] [PubMed] [Google Scholar]
- 101.Aragona M, et al. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 102.Mateus R, et al. Control of tissue growth by Yap relies on cell density and F-actin in zebrafish fin regeneration. Development. 2015;142:2752–2763. doi: 10.1242/dev.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu F-X, et al. Regulation of the Hippo-YAP Pathway by G-Protein-Coupled Receptor Signaling. Cell. 2012 doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gaspar P, et al. Zyxin Antagonizes the FERM Protein Expanded to Couple F-Actin and Yorkie-Dependent Organ Growth. Current biology. 2015;25:679–689. doi: 10.1016/j.cub.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 105.Mana-Capelli S, et al. Angiomotins link F-actin architecture to Hippo pathway signaling. Molecular biology of the cell. 2014;25:1676–1685. doi: 10.1091/mbc.E13-11-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim M, et al. cAMP/PKA signalling reinforces the LATS-YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. The EMBO Journal. 2013;32:1543–1555. doi: 10.1038/emboj.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Codelia VA, et al. Regulation of YAP by Mechanical Strain through Jnk and Hippo Signaling. Current biology. 2014;24:2012–2017. doi: 10.1016/j.cub.2014.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Enomoto M, et al. JNK signaling is converted from anti- to pro-tumor pathway by Ras-mediated switch of Warts activity. Developmental Biology. 2015;403:162–171. doi: 10.1016/j.ydbio.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 109.Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Developmental biology. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen C-L, et al. Tumor suppression by cell competition through regulation of the Hippo pathway. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1113882109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sun G, Irvine KD. Ajuba Family Proteins Link JNK to Hippo Signaling. Science signaling. 2013;6:ra81. doi: 10.1126/scisignal.2004324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Driscoll TP, et al. Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells. Biophysical journal. 2015;108:2783–2793. doi: 10.1016/j.bpj.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Benham-Pyle BW, et al. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science. 2015;348:1024–1027. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yonemura S, et al. alpha-Catenin as a tension transducer that induces adherens junction development. Nature Cell Biology. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 115.Wong KKL, et al. β-Spectrin regulates the hippo signaling pathway and modulates the basal actin network. Journal of Biological Chemistry. 2015;290:6397–6407. doi: 10.1074/jbc.M114.629493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fletcher GC, et al. The Spectrin cytoskeleton regulates the Hippo signalling pathway. The EMBO Journal. 2015;34:940–954. doi: 10.15252/embj.201489642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deng H, et al. Spectrin regulates Hippo signaling by modulating cortical actomyosin activity. eLife. 2015;4 doi: 10.7554/eLife.06567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cockburn K, et al. The Hippo pathway member Nf2 is required for inner cell mass specification. Current biology. 2013;23:1195–1201. doi: 10.1016/j.cub.2013.05.044. [DOI] [PubMed] [Google Scholar]