Abstract
Originally found in a Scottish family with diverse mental disorders, the DISC1 protein has been characterized as an intracellular scaffold protein that associates with diverse binding partners in neural development. To explore its functions in a genetically tractable system, we expressed the human DISC1 in fruit flies (Drosophila melanogaster). As in mammalian neurons, DISC1 is localized to diverse subcellular domains of developing fly neurons including the nuclei, axons and dendrites. Overexpression of DISC1 impairs associative memory. Experiments with deletion/mutation constructs have revealed the importance of amino terminal domain (46–290) for memory suppression whereas carboxyl domain (598–854) and the amino terminal residues (1–45) including the nuclear localization signal (NLS1) are dispensable. DISC1 overexpression also causes suppression of axonal and dendritic branching of mushroom body neurons, which mediate a variety of cognitive functions in the fly brain. Analyses with deletion constructs reveal that protein domains 598–854 and 349–402 are both required for the suppression of axonal branching while amino-terminal domains including NLS1 are dispensable. In contrast, NLS1 was required for the suppression of dendritic branching, suggesting a mechanism involving gene expression. Moreover, domain 403–596 is also required for the suppression of dendritic branching. We also show that overexpression of DISC1 suppresses glutamatergic synaptogenesis in developing neuromuscular junctions. Deletion/mutation experiments have revealed the importance of protein domains 403–596 and 349–402 for synaptic suppression, while amino terminal domains including NLS1 are dispensable. Finally, we show that DISC1 functionally interacts with the fly homolog of Dysbindin (DTNBP1) via direct protein-protein interaction in developing synapses.
Keywords: DISC1, memory, axons, dendrites, synapse, dysbindin
Introduction
Since the original discovery of the DISC1 gene from a Scottish pedigree with a wide range of mental disorders, biological evidence has accumulated that DISC1 is a multifunctional protein important for brain development and physiology.1–4 These studies have indicated that the cellular and biochemical processes involving DISC1 are relevant to the endophenotypes associated with psychiatric dysfunctions. On the other hand, except for the unique Scottish pedigree, how the DISC1 locus is implicated at the genetic level in major mental disorders is still elusive with little success in genome-wide association studies to identify the locus as a prominent hit for schizophrenia. 5, 6 This enigma in turn might imply an intricate underlying mechanism that involves complex genetic interactions with other loci that exert functional modifications of neuronal DISC1 functions in the brain.
The fruit fly (Drosophila melanogaster) has been used as a powerful model for understanding cellular and molecular mechanisms of diverse neurological disorders.7–9 In conjunction with the commonality of molecular genetic mechanisms in brain development,7–9 the vast array of transgenic techniques in fruit flies allows us to study human neurological disease genes either by loss-of-function or by gain-of-function approaches, involving overexpression of human disease genes in the fly brain. Whereas phenocopying human psychiatric symptoms is a major challenge for animal models, particularly in evolutionary distant species such as fruit flies, recent efforts toward a framework for basic research on mental disorders highlight the importance of elucidating the underlying mechanisms of mental dysfunction.10–12 In such a framework, mental disorders can be addressed as disorders of brain circuits, which can be studied at multiple biological and genetic levels using non-human models. In line with this concept, several studies have utilized the fly model in the past few years to reveal the underlying mechanisms of diverse mental disorders at the molecular and genetic levels.13–18
In this study, we expressed the DISC1 protein in developing mushroom bodies (MBs), the centers for cognitive functions of the fly brain.19–21 Although the DISC1 gene is not conserved in the fly genome,22 fruit flies exhibit significant conservation (93%) of the genes that encode the DISC1 interacting proteins23 (Table S1), providing a platform for molecular genetic analyses of its functions in brain circuit formation. We first demonstrate evolutionarily conserved mechanisms of subcellular localization of the DISC1 protein in developing fly neurons. We then show that overexpression of DISC1 in larval MB neurons impairs associative olfactory memory. Experiments with deletion/mutation constructs have revealed the importance of amino terminal domains (amino acid 46–290) for memory suppression while carboxyl domains (amino acid 598–854) and the amino terminal residues (amino acid 1–45) including the nuclear localization signal (NLS1) are dispensable. Using a mosaic technique with single-cell clones, we also show that DISC1 overexpression causes suppression of axonal and dendritic branching of MB neurons. Analyses with deletion constructs have revealed that protein domains 598–854 and 349–402 are both required for the suppression of axonal branching while amino-terminal domains including NLS1 are dispensable.. In contrast, NLS1 was required for the suppression of dendritic branching, suggesting a mechanism involving gene expression. Moreover, domain 403–596 is also required for the suppression of dendritic branching. In addition, we show that overexpression of DISC1 suppresses glutamatergic synaptogenesis in developing neuromuscular junctions (NMJs). Deletion/mutation experiments have revealed the importance of protein domains 403–596 and 349–402 for synaptic suppression, while amino terminal domains including NLS1 are dispensable. Finally, we show that DISC1 interacts with Dysbindin (DTNBP1) in synaptic development via direct protein-protein interaction.
Materials and Methods
Fly stocks
A white (w) stock outcrossed with Canton S ten times (w (CS10) ) was used as the standard stock. Flies carrying UAS-HA-DISC1 transgene14 were generated by P-element-mediated transformation. To ensure homogeneous genetic background, the transformed DISC1 stocks were outcrossed to the w (CS10) stock 5–10 times. All stocks were raised at 25 °C on a standard food consisting of 5.5 g/L agar, 40 g/L yeast extract, 90 g/L cornmeal, 100 g/L glucose. Propionic acid (3 ml/L) and n-butyl-p-hydroxybenzoate (0.7 g/L) were added as fungicides.
Immunohistochemistry
Immunological staining was performed as described previously.24 Rabbit anti-DISC1 antibody (Lys101-Arg260, AF6699, R&D Systems, Minneapolis, MN) or rabbit anti-DISC1 antibody (carboxyl terminal)25 were used to monitor DISC1 expression. Other antibodies used were: mouse anti-synaptotagmin diluted 1:2 (3H2 2D7, Developmental Studies Hybridoma Bank, University of Iowa, IA, USA), anti-horseradish peroxidase protein (HRP) conjugated with fluorescein-isothiocyanate diluted 1:50 (Jackson ImmunoResearch, West Grove, PA, USA), Rat anti-CD8α (Caltag, Burlingame, CA) diluted 1:50 and Alexa-conjugated secondary antibodies (Molecular Probes, Eugene, OR) diluted 1:1000. Confocal images were captured with Zeiss LSM510 or LSM710. Axonal and dendritic branching patterns were analyzed using Image J with 3D reconstruction. Expression levels of DISC1 proteins were determined based on the fluorescence intensities of confocal images using Image J.
Western blotting
Proteins were extracted from third instar larvae expressing the HA-tag DISC1 driven by tubP-GAL4. Third instar larvae were collected, rinsed with PBS and homogenized in Sarkosyl extraction buffer26 (50 mM HEPES (pH 7.5), 300 mM NaCl, 5mM EDTA, 5mM reduced glutathione, 1% NP-40, 0.2 % Sarkosyl, 1mM PMSF, 1× Protein Inhibitor Mix (Roche, Mannheim, Germany)). Animal carcass was removed by low speed centrifugation at 5,000 g (Eppendorf 5415R) for 3 min. The lysate was centrifuged at 16,100 g (Eppendorf 5415R) for 15 min to separate soluble and insoluble fractions. Precipitates were then taken up in the Sarkosyl extraction buffer. All steps were performed on ice or at 4 °C. Extracted proteins were then separated with 10 % SDS gel, transferred onto nylon membrane, and probed with anti-HA antibody (3F10, Roche, Mannheim, Germany).
Clonal analysis
Single-cell clones were generated using the mosaic analysis with a repressible cell marker (MARCM) technique,27, 28 which is based on the GAL4/GAL80 system coupled with FRT/FLT-mediated chromosomal recombination, which was induced by a brief heat shock. DISC1 constructs were driven under the UAS promoter sequence. The anatomy of the induced clones were visualized with a membrane-bound marker UAS-mCD8::GFP.
To induce MARCM clones, newly emerged larvae were collected at 24 hours after the end of a 4-hour egg collection, and heat shocked for 1 hour at 37°C. Larvae were then raised at 25°C and the brains of adult flies were dissected 2–3 days after hatching. Gal80 flies (hs-FLP UAS-mCD8::GFP; +/+; Gal80 FRT82B; OK107) were crossed with flies (1) w: FRT82B or (2) w; UAS-DISC1; FRT82B. Progenies of the following genotypes were examined for (1) wild-type clones: hs-FLP UAS-mCD8::GFP/w; +/+; Gal80 FRT82B/FRT82B; OK107/+ or (2) DISC1-expressing clones: hs-FLP UAS-mCD8::GFP/w; UAS-DISC1/+; Gal80 FRT82B/FRT82B/+; OK107/+.
Larval memory assay
Larval memory assay was performed as described previously.29, 30 Larvae were raised with the standard food described above without propionic acid. Behavioral experiments were performed at 25°C using early third instar larvae (72–76 h after egg laying). Larvae were harvested from vials with 15% glucose solution, and rinsed three times with DW. For training, larvae were placed on the surface of a 2.5% agar plate (diameter 85 mm), on which 1 ml of 1M sucrose (Nacalai, Kyoto, Japan) or DW was spread shortly before experiments. Undiluted odor (10 μl), linalool (Nacalai, Kyoto, Japan) was spotted on a filter disk (55 mm in diameter) placed on the inside of the lid, and larvae were exposed to the odor for 30 min. Typically, several hundreds animals are placed on a plate and conditioned en masse. Larval olfactory and gustatory responses were determined as described previously. 29, 30
Genetic interaction analyses
Motoneuron termini of the muscle 6/7 of the second abdominal segment were examined using confocal microscopy as described previously.31 Mutant flies were balanced with a Cantonized double balancer stock (w/w; Sp/CyO Act-GFP; Pr Dr/TM6B ubi-GFP), and the resulting mutant progeny were then crossed either with control (w; +; tubP-GAL4/TM6B ubi-GFP) or with DISC1 (w; UAS-DISC1(CS10)6-6(II); tubP-GAL4/TM6B ubi-GFP) flies. The following genotypes were examined. Control; (1) w; +/+; +/tubP-GAL and (2) w; +/UAS-DISC1; +/tubP-GAL4. dysbe01028 background; (3) w; +/+; dysbe01028/tubP-GAL and (4) w; +/UAS-DISC1; dysbe01028/tubP-GAL4. Larvae were raised at 25 °C and fixed at 116–120 hours after egg laying. Total synaptic area was determined using Image J based on the confocal images of anti-Synaptotagmin staining.
Proximity Ligation Assay
To detect protein-protein interaction in the larval NMJ, UAS-Venus::Drosophila-Dysbindin (gift from Graeme Davis)32 and UAS-DISC1 were co-expressed with tubP-GAL4. Proximity ligation assay (PLA)33 was performed as described previously.34 Briefly, NMJ of third instar larvae were dissected in ice-cold phosphate saline, fixed for 30 min with 4% paraformaldehyde, and incubated with 1% bovine serum albumin in PBT (phosphate buffered saline containing 0.1 % Triton X-100). Tissues were then incubated with 1:50 dilution of sheep anti-DISC1 (AF6699, R&D systems, Minneapolis, MN, USA) and 1:500 dilution of rabbit anti-GFP (Molecular Probes, Eugene, OR) overnight at 4°C. After washing with PBT, tissues were further incubated with anti-horseradish peroxidase (HRP) conjugated with FITC for three hours. After washing with PBT, tissues were incubated with 1:15 dilution of anti-rabbit PLUS (DUO092002, Sigma-Aldrich, St. Louis, MO) and anti-goat MINUS (DUO092006, Sigma-Aldrich, St. Louis, MO) PLA probes for two hours at 37°C. After washing with wash buffer A (Duolink, Sigma-Aldrich, St. Louis, MO), Ligation reaction was done for one hour at 37 °C followed by amplification for two hours at 37°C. Tissues were then washed with wash buffer B (Duolink, Sigma-Aldrich, St. Louis, MO) and mounted on a slide. PLA signals were captured with confocal microscopy LSM510 or LSM700 and analyzed with Image J.
Statistics
Experimental data of anatomical alterations including the analyses of MB clones and NMJ synapses were analyzed using parametric tests (Students’ t-test and one-way ANOVA) based on the previous studies28, 31 without randomization and blinding. For multiple comparisons among relevant groups, Dunnett’s post hoc test was used in conjunction with one-way ANOVA. For simplicity, larval memory data are also presented based on parametric tests (multiple comparison with one way ANOVA followed by Dunnett’s test). Considering the small number of samples, the data were also examined with nonparametric tests (Kruskal-Wallis) to further examine statistical significance. The conclusions were unaltered between the parametric and nonparametric tests.29, 30
Results
Expression of the human DISC1 protein in fruit fly larval neurons
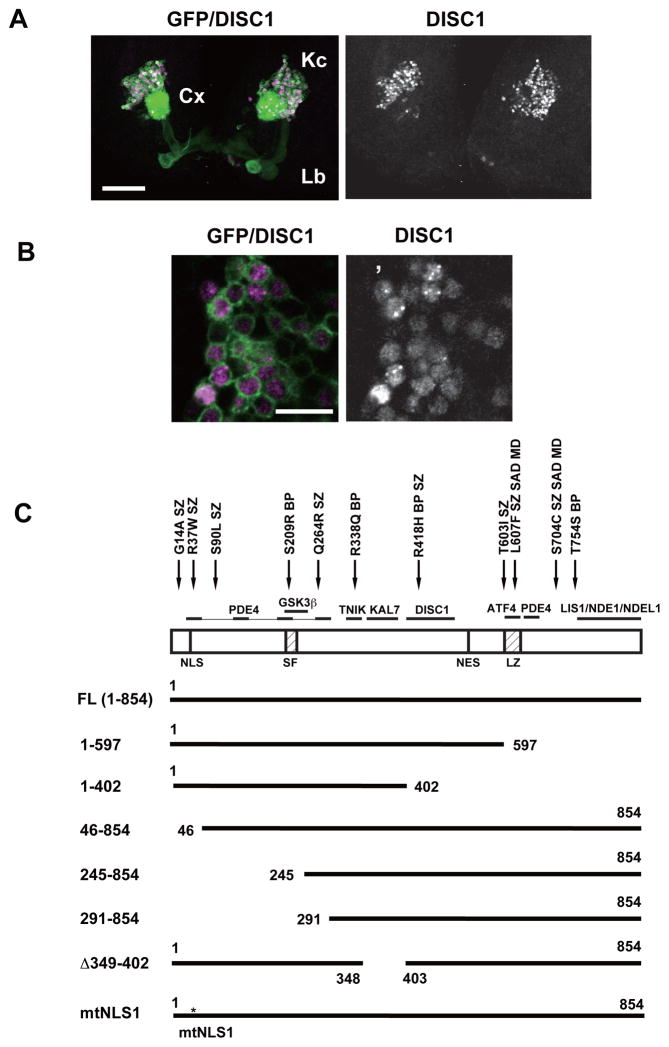
To express the human DISC1 gene in the fly brain, we utilized the GAL4-UAS system, which enables targeted expression in defined sets of neurons in the fly brain (Fig. 1A, 1B). Sawamura et al.14 has shown that DISC1 is localized exclusively to the nuclei in the adult MB neurons (Fig. S2J). but expression and functional analysis in developing neurons has yet to be done. In this study, we overexpressed DISC1 in larval MB neurons, which mediate cognitive functions such as learning and memory and are thus pertinent targets of DISC1 overexpression in the developing fly brain. The nervous system of fruit fly larvae consists of only 10,000 neurons, enabling in depth analyses of neurocircuitry and synapses at high resolution35. Intriguingly, we found that DISC1 showed a diffuse pattern in both the nucleus and the cytoplasm with nuclear puncta in a subset of cells. (Fig. S1A, S2A, S2I). We also examined subcellular localization of DISC1 (1–597), which corresponds to the Scottish family mutation. In the adult MBs, DISC1 (1–597) shows diffuse appearance in the nuclei but is not detected in the dendrites and axons.14 By contrast, we observed weak expression of DISC1 (1–597) in dendrites and axons of larval neurons (Fig. S1A and Table S2). These results demonstrate stage-dependent subcellular dynamics of DISC1 proteins in fly neurons, reminiscent of the subcellular localization dynamics observed in vertebrate neurons.36
Figure 1. Expression of the human DISC1 protein in larval MB neurons.
A. Full-length DISC1 (magenta) was expressed using a MB-GAL4 driver, 201Y, which drives gene expression in MB neurons. Cells were labeled with a membrane-bound marker, UAS-mCD8::GFP (green). Third instar larval stage. Kc (Kenyon cells): the cell bodies of MB neurons. Cx (calyx): the dendritic structure. Lb (lobe): the axonal extensions. Scale bar, 50 μm.
B. Subcellular localization patterns of DISC1 in larval MB neurons. The DISC1 protein FL (1-854) exhibited punctate dots in many of the nuclei. DISC1 also showed diffuse pattern in the nucleus and the perinuclear cytoplasm. Magenta, DISC1. Green, mCD8::GFP. Scale bar, 10 μm.
C. DISC protein domains and the deletion/mutant constructs. NLS, nuclear localization signal; SF, Ser-Phe rich domain; NES, nuclear exclusion signal; LZ, leucine-zipper domain. Representative interacting proteins and DISC1 genetic variants associated with mental disorders1–4 are shown above the structure. Q264R, L607F and S704C are common variants associated with schizophrenia. L607F and S704C are also associated with schizoaffective disorder (SAD) and major depression (MD). G14A, R37W, S90L and T603I are rare variants associated with schizophrenia (SZ). S209R, R338Q and T754S are rare variants associated with bipolar disorder (BP).
Protein domains controlling subcellular localization in fruit fly larval neurons
Subcellular localization of the DISC1 protein is controlled by other neuronally expressed proteins that bind to distinctive domains on DISC1.1–4 To dissect the functional protein domains in the larval neurons, we overexpressed a series of mutant DISC1 proteins (Fig. 1C) and analyzed their localization (Fig. S1, S2 and Table S2).
Nuclear targeting was abolished by mutation of the amino-terminal region that included the nuclear localization signal (NLS1) (Fig. S2D–F, 2H) while punctate nuclear localization required the carboxyl-terminal region that included the leucine zipper (LZ) domain (Fig. S2B, 2C). We also noted that mutant Δ349–402 lacked punctate localization in the nuclei (Fig. S2G). The nuclear export signal (NES) at 546–555 also seemed functional in the larval neurons; except for DISC1 (1–402), which lacked NES, all DISC1 constructs were detected in the perinuclear cytoplasm (Fig. S1, S2 and Table S2).
Dendritic localization was found for the mutant proteins that showed cytoplasmic localization. On the other hand, axonal localization was found for the mutants 1–597, 46–854, 245–854, 291–854, and mtNLS1 but not for the mutants 1–402 and Δ349–402, indicating the requirement of the dual domains of amino acids 403–597 and 349–402 for efficient axonal targeting in the developing neurons. Notably, the amino acids 403–504 are known to be required for DISC1 self-interaction (Fig. 1C).1, 2, 23
Subcellular targeting of DISC1 has been studied previously with cultured mammalian cells and primary mouse cortical neurons.14, 37, 38 Importantly, our results with the developing fruit fly neurons are consistent with the domain studies with the mammalian cells, suggesting evolutionally conserved mechanisms of subcellular targeting of the DISC1 protein in developing fly neurons. The mutant proteins exhibited expression levels in the MB neurons comparable with that of the intact protein, except for DISC1 (245–854), which showed reduced levels (Fig. S1B), suggesting that the different localization patterns were caused by the lack of the functional domains rather than reduced expression levels.
A pool of DISC1 protein is recovered in the Sarkosyl insoluble fraction but does not elicit neuronal degeneration in fruit fly neurons
Recent studies have suggested that DISC1 has a tendency to form aggregates.26, 39–44 To investigate potential aggregate formation in fruit flies, we extracted DISC1 proteins from third instar larvae to perform Western blotting. We detected full-length and mutant DISC1 proteins in the Sarkosyl insoluble fraction (Fig. S3A–D). In particular, the mNLS1 protein exhibited marked accumulation in the precipitate fraction, consistent with the formation of prominent puncta in the cytoplasm of the MB neurons (Fig. S2H). The levels of the DISC1 proteins in the Sarkosyl insoluble fraction were comparable to or higher than that of GAPDH, which can form aggregates in cells.45, 46
To investigate the effects of DISC1 aggregates on cellular differentiation, we overexpressed the full-length (1–854) and a truncated (1–597) DISC1 proteins in larval eye primordia (Fig. S3E). Owing to the crystal-like arrangement of a large number of ommatidia, the fruit fly compound eye is a highly sensitive system to detect neuronal degeneration, and has been used in functional studies of human neurodegeneration genes7, 47, 48. The localization patterns of the full-length (1–854) and the truncated (1–597) proteins in eye primordia were similar to those observed in the developing MB neurons. We also coexpressed the full-length and the truncated proteins in developing ommatidia (Fig. S3E). Intriguingly, although the number of the nuclear puncta was not altered (Fig. S3F), coexpression of the full-length and truncated form (FL + TR) caused robust nuclear puncta formation and higher nuclear DISC1 levels (Fig. S3G), suggesting the participation of the truncated protein in normal nuclear structure formation. To further investigate the effects of DISC1 overexpression, we examined the morphology of compound eyes of aged flies (35 days) using scanning microscopy; again no alterations were detected with both the full-length and the truncated proteins (Fig. S4). These results thus suggest that, although a pool of the DISC1 protein is recovered in the Sarkosyl insoluble fraction, it may not elicit robust neurodegeneration in the fruit fly neurons.
Overexpression of DISC1 impairs associative memory in fruit fly larvae
The above results argue for the relevance of the larval stage to the analysis of diverse DISC1 functions both in the cytoplasm and nuclei. Among cognitive deficits found in psychiatric patients, memory deficit is an important endophenotype, and has been shown to correlate with genetical susceptibility associated with DISC1 haplotypes.49–51 Studies using murine models have shown that suppression of DISC1 with either RNAi techniques or dominant-negative expression result in frontal lobe deficits that are characterized by working memory impairment.1–4, 49–51 To investigate neural functions of DISC1 in memory formation, we analyzed olfactory associative memory using fruit fly larvae, which provide an attractive model to study learning and memory in the developing brain29, 30, 35, 52–54. Despite its simple neural network, memory in the fruit fly shares a number of molecular features with human, including the critical requirement of intracellular cAMP signaling.19–21, 29, 55, 56 The fruit fly PDE (encoded by dunce), which exhibits significant sequence conservation with the human PDE, is preferentially expressed in MB neurons to play critical roles in learning and memory.19, 21
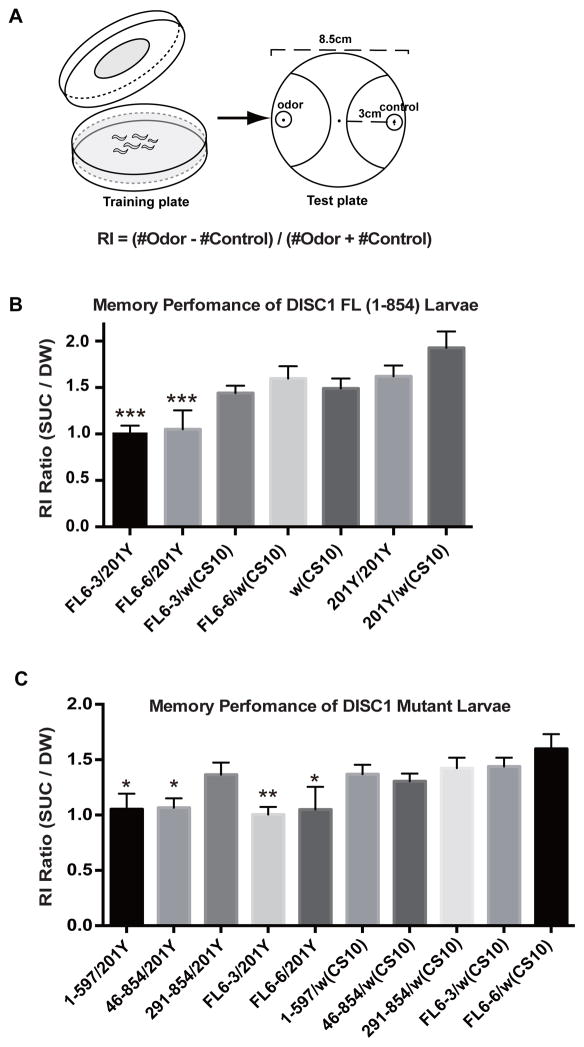
The paradigm utilized in this study involves associative training using a specific odorant (linalool) in conjunction with sucrose, which serves as an appetitive reinforcer (Fig. 2A). Memory performance is determined as enhancement of olfactory responses to the test odorant after appetitive training. We expressed DISC1 in the larval MB neurons using the GAL4-UAS system. Previous studies have shown that neuronal signaling of olfactory information in MB neurons is critical for memory formation.19–21, 29, 30, 52 While control larvae (w (CS10)) exhibited higher olfactory response after appetitive training with sucrose than with DW (Fig. 2B and Table S3), larvae expressing DISC1 in MB neurons (FL-6-3/201Y and FL-6-6/201Y) failed to exhibit olfactory memory. On the other hand, larvae carrying the DISC1 construct but not the GAL4 driver (FL-6-3/w (CS10) and FL-6-6/w (CS10)) exhibited normal memory performance, indicating that memory suppression was caused by DISC1 expression in the larval MB neurons. Normal memory was observed in larvae having only the GAL4 driver (201Y/201Y and 201Y/w (CS10)).
Figure 2. Overexpression of DISC1 impairs olfactory associative memory in fruit fly larvae.
A. Olfactory associative training and memory performance test of fruit fly larvae. For associative olfactory training, larvae were placed on 2.5% agar plate spread either with 1M sucrose or with distilled water (control), and exposed for 30 min to an odorant (linalool) spotted on a filter disk attached to the lid. Larvae were then transferred to the center of an olfactory test plate. A small filter disc spotted with the test odorant was placed on one side of the plate, and a control disc was placed on the opposite side. The numbers of animals which had moved into the semicircular areas were counted after 3 min. Response index (RI) was calculated as indicated. Typically, 50–100 larvae were used for a single olfactory response test.
B. Olfactory memory performance of DISC1 and control larvae. Memory performance was measured as increment in the olfactory response to the test odor. RI ratio (SUC/DW) = 1.0 corresponds to no learning. DISC1 was expressed in the larval MB neurons by a GAL4 driver, 201Y. Associative memory was suppressed in FL-6-3/201Y and FL-6-6/201Y, two independent DISC1-expressing stocks. ***p < 0.001 with one-way ANOVA and Dunnett’s post hoc test, n = 14 ~25.
C. Olfactory memory performance of mutant DISC1 larvae. Associative memory was suppressed in larvae expressing 1-597/201Y and 46-854/201Y but not 291-854/201Y. *p < 0.05 and **p < 0.01 with one-way ANOVA and Dunnett’s post hoc test, n = 11 ~32.
DISC1 amino-terminal domain is required for memory suppression in fruit fly larvae
To gain insight into the mechanism of memory suppression, we analyzed memory performance in transgenic larvae expressing DISC1 deletion constructs. Intriguingly, larvae expressing the DISC1 (1-597) protein failed to form memory (1-597/201Y), as did the larvae expressing the intact DISC1 protein (FL6-3/201Y and FL6-6/201Y) (Fig. 2C). Likewise, larvae expressing the DISC1 construct 46-854, which removed the nuclear localization signal NLS1, also failed to form memory, consistent with the notion that cytoplasmic signaling plays essential roles in memory formation.19, 21, 29 On the other hand, larvae expressing the DISC1 construct with a large amino terminal deletion exhibited normal memory performance (291-854/201Y). As anticipated, larvae carrying the mutated DISC1 construct but not the GAL4 driver exhibited normal memory performance (1-597/w (CS10), 46-854/w (CS10), and 291-854/w (CS10)).
To confirm sensory integrity and locomotor activity of DISC1-expressing larvae, we examined the naïve olfactory response to a test odor using the same agar plate assay used for the memory tests. Normal olfactory response was observed for all the DISC1-expressing larvae (Table S4). Gustatory response for sucrose was also unaffected in all the DISC1-expressing larvae (Table S4). These results imply that the reduced response to odorant in DISC1-expressing larvae was caused neither by sensory nor locomotor defects, but was in fact due to deficits in associative memory formation.
DISC1 regulates axonal branching in MB neurons
In order to investigate the regulatory functions of DISC1 in neuronal connectivity, we examined anatomical integrity of MB neurons using a mosaic technique. Our previous study has shown that overexpression of DISC1 induces no gross anatomical change of the adult MBs,14 but whether developmental expression of DISC1 affects the branching patterns of individual neurons has not been addressed. The fruit fly MB neurons are grouped into three classes, α/β, α′/β′ and γ, with distinctive birth order and axonal trajectory.19, 21 Among them, γ neurons are generated during the embryonic and larval periods, and undergo complex remodeling including pruning and re-extension of neurites during metamorphosis to achieve the characteristic branching structure in the mature brain (Fig. 3A).
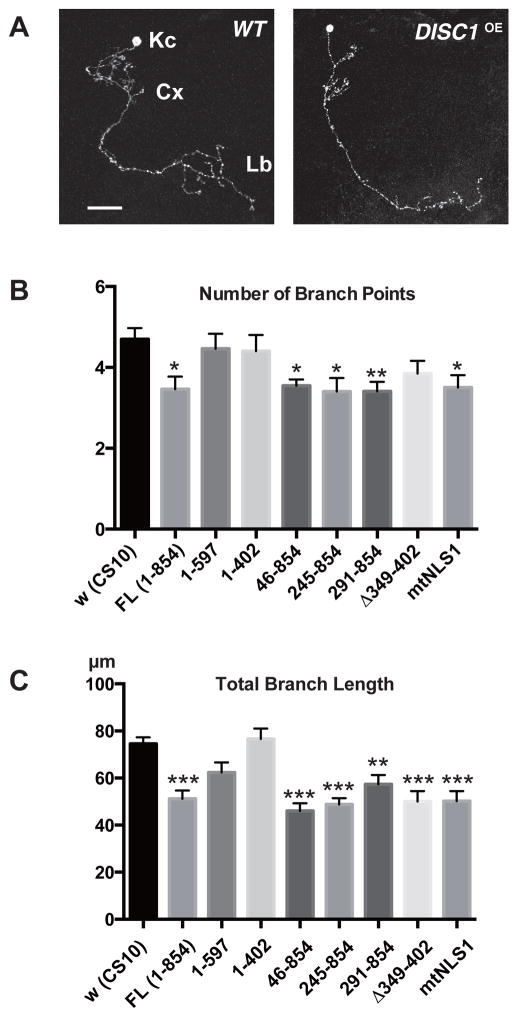
Figure 3. Overexpression of DISC1 suppresses axonal branching of MB neurons.
A. Branching patterns of wild-type and DISC1-expressing (DISC1OE) γ-neurons. Kc (Kenyon cells). Cx (calyx). Lb (lobe). Cells were labeled with a membrane-bound GFP (UAS-mCD8::GFP) driven by a MB GAL4-driver, OK107. Scale bar, 20 μm.
B, C. Quantification of axonal branching phenotypes of DISC1 single-cell clones. B. Number of branch points. C. Total branch length. Single-cell clones were induced at an early larval stage and analyzed at young adult stage. *p < 0.05, **p < 0.01 and ***p < 0.001 with one-way ANOVA and Dunnett’s post hoc test, n = 10 ~20.
To investigate the developmental changes caused by DISC1 at high resolution, we generated γ-neuron clones using a mosaic technique (MARCM),27 which is based on the GAL4/GAL80 expression control coupled with the FRT/FLT chromosomal recombination. This technique enables us to specifically express the target gene in the induced clones under the UAS promoter sequence (UAS-DISC1). The anatomy of the induced clones could be visualized at the single-cell resolution with simultaneous expression of UAS-mCD8::GFP, which encodes a membrane bound GFP. We induced the DISC1 clones at the onset of the larval stage and analyzed anatomical integrity of individual neurons at the adult stage after neural remodeling. We found that, whereas the overall axonal extension and guidance of γ-neurons was unaffected, overexpression of DISC1 suppressed the elaborate terminal branching (Fig. 3A–C). Quantification of the branching patterns revealed that not only the number of branch points but also the total branch length was suppressed in γ-neuron clones expressing the full-length DISC1 (FL (1-854)).
To elucidate the protein domains that control axonal branching, we then analyzed single-cell clones that expressed various deletion constructs. Whereas amino terminal deletion constructs, including 291–854, caused axonal branch suppression, carboxyl truncation constructs (1–597 and 1–402) failed to do so. In addition, a small deletion construct (Δ349–402) exhibited an intermediate phenotype with significant suppression of branch length but not branching points (Fig. 2B, C). Finally, mtNLS1 also caused axonal branch suppression, indicating that nuclear localization was not required for axonal terminal modulation.
DISC1 regulates dendritic branching in MB neurons
Abnormalities in dendritic formations such as over-pruning or neurite maintenance failures are among the critical developmental alterations underlying mental disorders.1–4 DISC1 regulates dendritic development, including spines of glutamate synapses in the murine brain.1–4 In order to further analyze DISC1 functions in neuronal connectivity, we extended our analysis to dendritic development. As with many invertebrate neurons, the MB γ-neurons are unipolar and exhibit elaborate dendritic branches near the cell body (Fig. 4A). The characteristic branching patterns are established through pruning and re-extension during metamorphosis. Many of the dendritic branches have claw-like endings that are post-synaptic to olfactory projection neurons.57
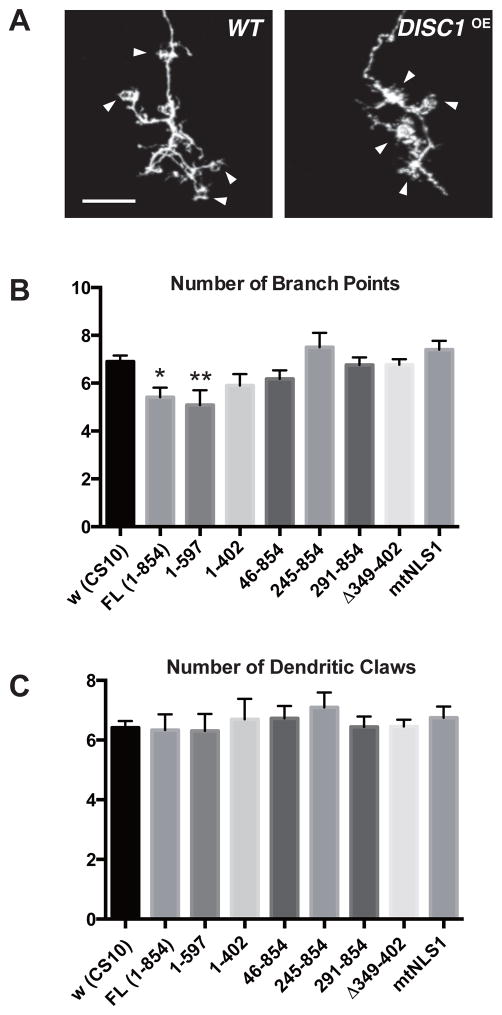
Figure 4. Overexpression of DISC1 suppresses dendritic branching of MB neurons.
A. Dendrite branching patterns of wild-type and DISC1-expressing (DISC1OE) γ-neurons. Arrowheads indicate dendritic termini (claws), postsynaptic structures receiving inputs from olfactory projection neurons. Cells were labeled with a membrane-bound GFP (UAS-mCD8::GFP) driven by a MB GAL4-driver, OK107. Scale bar, 10 μm.
B, C. Quantification of dendritic phenotypes of DISC1 single-cell clones. B. Number of dendritic branch points. C. Number of dendritic claws. Partial claws were counted as 0.5. Single-cell clones were induced at an early larval stage and analyzed at young adult stage. *p < 0.05 and **p < 0.01 with one-way ANOVA and Dunnett’s post hoc test, n = 10 ~22.
Intriguingly, DISC1 overexpression caused moderate suppression of dendritic branching of γ-neurons (Fig. 4B) whereas formation of terminal claws was not affected (Fig. 4C). Analysis with deletion constructs revealed that, in contrast to its requirement for axonal branch suppression, the carboxyl region was dispensable for the suppression of dendritic branching (construct 1–597) while further removal of the carboxyl sequences (construct 1–402) abolished it. In addition, the deletion construct (Δ349–402) failed to suppress dendritic branching. On the other hand, the amino-terminal deletion constructs, including the small deletion (46–854), did not suppress dendritic branching, suggesting a critical role of the amino terminal region, which harbors a nuclear localization signal. Indeed, the nuclear localization mutant (mtNLS1) failed to suppress dendritic branching, suggesting critical involvement of nuclear DISC1 in dendritic modulation as opposed to axonal modulation.
DISC1 interacts with dysbindin in glutamatergic synaptogenesis
To further investigate the neurodevelopmental functions of DISC1, we examined synapse development using the fruit fly larval NMJs, which exhibit several key features in common with the excitatory synapses in the vertebrate brain.58–61 In particular, the fly NMJs utilize glutamate as the major neurotransmitter, and have been used as a powerful model to study the molecular mechanisms of synaptic development and functions.58–61
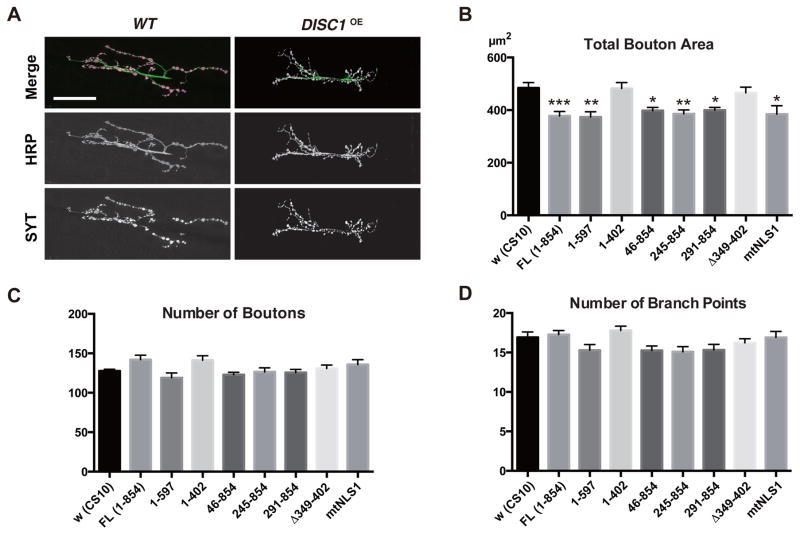
We found that overexpression of DISC1 with a ubiquitous driver caused overall shrinkage of the synaptic termini of the motoneurons (Fig. 5A) with significant reduction in total bouton area (Fig. 5B) but not the numbers of boutons (Fig. 5C) and the axonal branch points (Fig. 5D). Analysis with deletion constructs revealed that the carboxyl deletion construct (1–597) suppressed the synaptic bouton area, as did the intact protein. However, construct 1–402 failed to do so. Moreover, the small deletion construct (Δ349–402) also failed to suppress the synaptic bouton area. On the other hand, deletions of the amino terminal regions had no effect (constructs 46–854, 245–854 and 291–854). Notably, mtNLS1 also suppressed the synaptic bouton area as did the intact protein, indicating that nuclear localization was not required for synaptic suppression.
Figure 5. Overexpression of DISC1 suppresses synaptic development in larval NMJ.
A. Branching patterns of wild-type and DISC1 overexpression (DISC1OE) NMJs. Green, motor neuron termini labeled with an anti-neuronal antibody (anti-HRP). Magenta, synaptic boutons labeled with anti-Synaptotagmin (SYT). Muscle 6/7 of the second abdominal segment in late third instar larvae (116–120 hours after egg collection). UAS-DISC1 was driven with a ubiquitous driver, tubP-GAL4. Scale bar, 40 μm.
B–D. Quantification of NMJ synaptogenesis in DISC1 overexpression NMJ. B. Total synaptic bouton area. C. Number of synaptic boutons. D. Number of axonal branching points. *p < 0.05 and **p < 0.01 with one-way ANOVA and Dunnett’s post hoc test, n = 10 ~26.
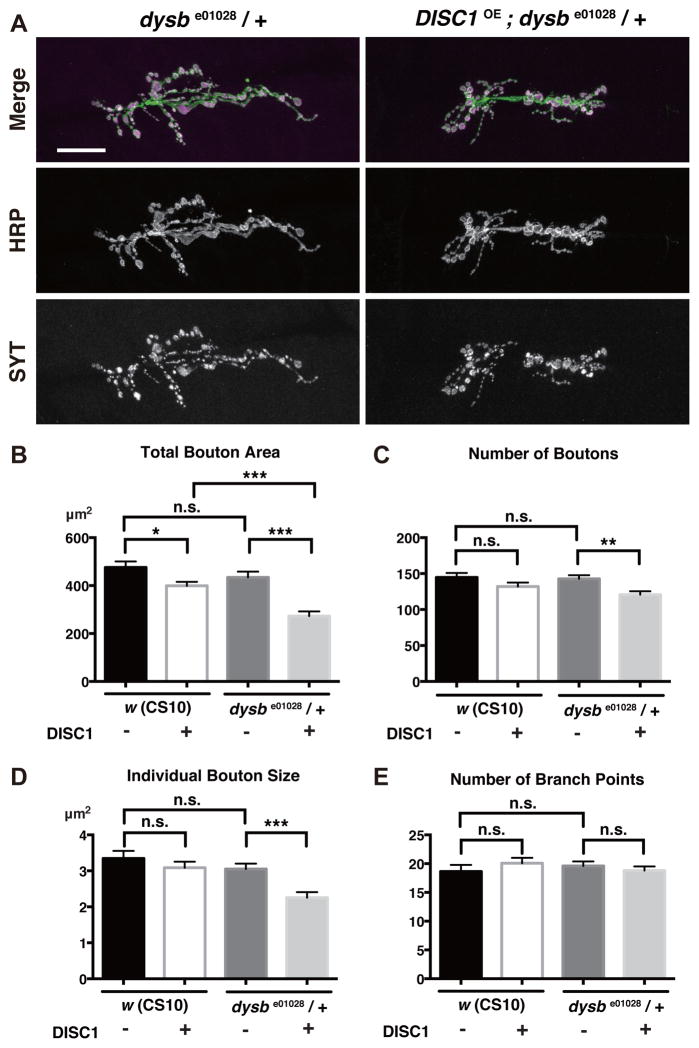
To reveal underlying mechanisms, we conducted a screening for genes that cooperatively function with DISC1 in synaptic development. We expressed DISC1 in the heterozygous background of fly mutations implicated in neural circuit formation and functions (details will be published elsewhere), and compared their NMJ phenotypes with the phenotype caused by DISC1 expression in the wild type background. Among the genes identified through this screening, mutation of dysbindin (dysb), the fruit fly homolog of the human DTNBP1 gene, caused intriguing enhancement of the DISC1 phenotype (Fig. 6A). The dysbe01028 mutation is recessive per se, and did not alter total bouton area on its own in the heterozygous background (Fig. 6B, comparisons of DISC1 minus data between w (CS10) and dysbe01028/+). However, DISC1 suppressed the synaptic area more profoundly in dysbe01028/+ heterozygous background (Fig. 6B, comparisons of DISC1 plus data between w (CS10) and dysbe01028/+). Moreover, DISC1 caused significant reductions in the number of boutons and the individual bouton size in the dysbe01028/+ heterozygous background (Fig. 6C and 6D). On the other hand, the number of axonal branch points of the motor neurons were not changed, arguing for specific suppression of synaptic bouton formation (Fig. 6E).
Figure 6. DISC1 genetically interacts with dysbindin in the development of glutamatergic synapses.
A. Synaptic terminal branches of dysbe01028 heterozygous NMJs with DISC1 overexpression (DISC1OE). Green, motor neuron termini labeled with an anti-neuronal antibody (anti-HRP). Magenta, synaptic boutons labeled with anti-Synaptotagmin (SYT). Scale bar, 40 μm.
B–D. Quantification of NMJ synaptogenesis in w (CS10) control and dysb/+ backgrounds. B. Total synaptic bouton area. C. Number of synaptic boutons. D. Individual synaptic bouton size. E. Number of axonal branch points. *p < 0.05, **p < 0.01 and ***p<0.001 with t-test, n = 10.
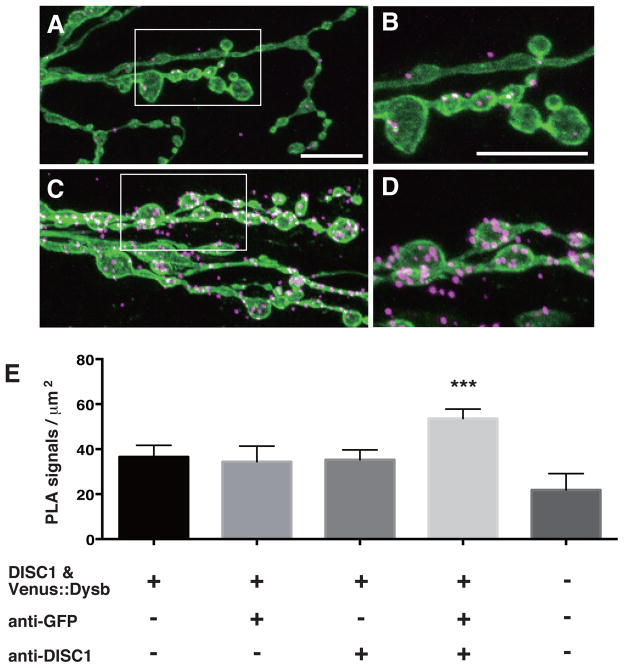
In order to determine whether DISC1 directly interacts with the fruit fly Dysbindin in synaptogenesis, we applied the PLA technique33 in the larval NMJs. In this assay, the target proteins are localized with specific primary antibodies, which are then detected with secondary antibodies conjugated to PLA-PLUS or MINUS oligonucleotide probes. The attached probes can be bridged through hybridization of connector oligonucleotides only when the two proteins are in close proximity to make direct contact. The annealed oligonucleotides are then closed by ligation into circular DNA molecules, which serve as templates for subsequent rolling circle amplification to be visualized by in situ hybridization with a fluorescently labeled probe.
Consistent with the genetic results, we detected significant PLA signals between DISC1 and the fruit fly Dysbindin in the developing synapses (Fig. 7). These results as a whole suggest an intriguing functional interaction between DISC1 and Dysbindin in the development of glutamatergic synapses.
Figure 7. DISC1 directly interacts with the Drosophila Dysbindin in the larval NMJ.
A–D. Confocal images of PLA signals in the larval NMJ. A, B. PLA in w (CS10) control NMJs. C, D. PLA in NMJs expressing Drosophila-Dysbindin and DISC1. Third instar larval NMJ. UAS-Venus::Drosophila-Dysbindin and UAS-DISC1 were co-expressed with tubP-GAL4. B, D. Higher magnification of the area indicated in A and C. Green, motor neuron termini labeled with anti-HRP. Magenta, PLA signals. Scale bars, 10 μm.
E. Quantification of PLA signals. Venus::Dysbindin was detected with anti-GFP. ***p < 0.001 by one-way ANOVA followed by Dunnett’s post hoc test. n = 9–19.
Discussion
Molecular cellular studies in the past decade have suggested that DISC1 is a multifunctional intracellular scaffold protein that participates in diverse physiological and developmental processes.1–4 In this study, we expressed human DISC1 in developing fruit fly neurons, and performed behavioral and neurodevelopmental studies in living organisms (summarized in Table 1). We have shown that overexpression of DISC1 causes associative memory defects in fruit fly larvae. Mosaic analyses with single-cell clones revealed that overexpression of DISC1 suppresses axonal and dendritic branching of MB neurons. Moreover, we have shown that DISC1 functionally interacts with the fly homolog of DTNBP1 in synaptic development via direct protein-protein interaction.
Table 1.
Suppression of memory and neural development by DISC1 proteins
| DISC1 Construct | Associative Memory | Axonal Development | Dendritic Development | Synaptic Development | |||
|---|---|---|---|---|---|---|---|
| Axonal Branching | Total Branch Length | Dendritic Branching | Claw Formation | Bouton Area | Number of Boutons | ||
| FL (1-854) | ++ | ++ | ++ | ++ | − | ++ | − |
| 1-597 | ++ | − | − | ++ | − | ++ | − |
| 1-402 | n. t. | − | − | − | − | − | − |
| 46–854 | ++ | ++ | ++ | − | − | ++ | − |
| 245–854 | n. t. | ++ | ++ | − | − | ++ | − |
| 291–854 | − | ++ | ++ | − | − | ++ | − |
| Δ349-402 | n. t. | − | ++ | − | − | − | − |
| mtNLS1 | n. t. | ++ | ++ | − | − | ++ | − |
Recent studies have reported that DISC1 has a tendency to form aggregates in mammalian cells.26, 39–44 Here, we obtained the data that a pool of DISC1 can be recovered in the detergent insoluble precipitates in the fly cells too. In contrast to aggregate-prone proteins for neurodegenerative diseases7, 47, 48, insoluble DISC1 did not elicit robust neuronal cell death in the compound eyes of the fly. It is an important future question whether insoluble pools of DISC1 mediate the developmental and functional phenotypes that we observed in the fly neurons.
DISC1 exhibits diverse neurodevelopmental functions in fruit fly neurons via different mechanisms
We have shown that overexpression of DISC1 suppresses axonal branching of MB neurons. Experiments with deletion constructs have revealed the importance of the carboxyl and the middle domains for axonal branch suppression while the amino-terminal domains including NLS1 are dispensable. Several studies have shown that DISC1 regulates axonal development in the murine brain,1–4 and that cytoplasmic factors such as the NDEL1/LIS1 complex interact with the carboxyl regions of the DISC1 protein to regulate microtubule dynamics and axonal transport.1–4 Homologs of the dynein complex are found in flies and mediate cellular morphology and dynamics such as migration.62, 63 The fruit fly LIS1 is expressed in developing MB neurons to control neural progenitor proliferation and axodendritic development.64, 65 Furthermore, Hayashi-Takagi et al66 have shown that the middle domain of DISC1 interacts with Kalirin-7 to control Rac1 activity that is critical for actin cytoskeleton organization. Notably, the fly Kalirin homolog (Trio) is expressed in fruit fly MB neurons to regulate axodendritic development.67, 68 In addition, our results have suggested another domain (amino acid 349–402) in the middle of the DISC1 protein for axonal branch suppression. Intriguingly, the DISC1 amino acids 335–347, nearby the region deleted in Δ349–402, are bound by TNIK,23 a psychiatric risk factor and an activator of Wnt target genes.69 The fly TNIK homolog (misshapen) is known to control axon targeting and synaptogenesis in fruit flies.70
Our analyses with deletion constructs have shown that, as opposed to suppression of axonal branching, suppression of dendritic branching requires nuclear localization of the DISC1 protein. Molecular studies on the associated proteins have indicated that DISC1 interacts with diverse nuclear proteins as well.1–4, 14, 23 Sawamura et al14 have shown that DISC1 binds ATF4/CREB2 and the fly homolog Cryptocephal to regulate CRE-mediated gene transcription and sleep homeostasis in fruit flies. However, experiments with deletion constructs argue against the involvement of carboxyl binding factors, including ATF4/CREB2, in the suppression of MB dendrites. Instead, our data suggest other factors that target the domain 403–596, which harbors the DISC1 dimerization sequence (amino acids 403–504) along with the binding sites for other proteins.23 Further studies are required to elucidate the exact interacting proteins as well as the molecular mechanism through which nuclear DISC1 controls dendritic development.
Converging control of glutamatergic synaptogenesis by DISC1 and Dysbindin
It has been hypothesized that altered glutamate neurotransmission might be a critical cause for cognitive deficits in schizophrenia and other mental disorders.71, 72 Through a genetic interaction study in the fly NMJ, we have shown that DISC1 interacts with Dysbindin in synaptic development. Initially found as a component of the dystrophin-dystroglycan complex,73 Dysbindin was subsequently identified as a component of biogenesis of lysosome-related organelles complex (BLOC-1) that controls organelle biogenesis and intracellular membrane trafficking.74, 75 Dysbindin is expressed both pre-and postsynaptic cells to control dendritic spine formation through the association with WAVE-2 and Abi-1, key regulators of Rac1 that control actin cytoskeletal dynamics.76 Consistently, knockdown of dysbindin results in disorganization of actin cytoskeletons that accompanies neurite shortening and growth cone abnormality. 74, 77
As noted above, DISC1 controls Rac1 through association with Karilin-7.66 In addition, studies of DISC1 interactome23 suggested that Dysbindin and DISC1 bind several proteins in common, such as microtubule crosslinking factors Dystonin (DST/BPAG1) and microtubule-actin crosslinking factor 1 (MACF1). Both DISC1 and Dysbindin interact with members of the exocyst complex23, which regulate protein trafficking to synaptic terminals in both vertebrates and fruit flies.78–80 It is also noteworthy that DISC1 interacts directly with TNIK, which in turn interacts with DST.23 Interestingly, it has been shown that Dysbindin controls synaptic development and functions in fruit flies.32, 81 Moreover, the fly homolog (short stop) of MACF1 and DST plays critical roles in axonal, dendritic and synaptic development.82, 83
Recently, Ottis et al40 have suggested intriguing convergence of DISC1 and Dysbindin by showing that DISC1 recruits Dysbindin into protein aggresomes in mouse neuroblastoma cells via direct protein-protein interaction mediated by amino acids 316–597, which is consistent with the domains involved for synaptic modulation in fruit fly NMJs. Physical interaction between Dysbindin and DISC1 is also critical for the stability of Dysbindin and for neurite out growth in cultured neuronal cells.84 Taken together, our results as a whole suggest a complex but intriguing converging mechanism controlled by DISC1 and Dysbindin in the developing glutamatergic synapses.
In this study, we have presented a dissection of the functional DISC1 domains in fruit flies. Given the unparalleled power of the Drosophila genetics, it is feasible to systematically identify interacting genetic loci that collaboratively function in vivo through shared pathways. Combined with the recent advancement in human psychiatric genetics, the fruit fly provides insights relevant to the understanding of the etiology of mental disorders at the brain circuit level.
Supplementary Material
Acknowledgments
We are grateful to Daisuke Tanaka, Yuya Kawanaka, Yasushi Maruyama, Shinichiro Horigane, and Takato Honda for their help in diverse aspects of this study. We also thank Ryusuke Niwa, Satory Kobayashi, Fuminori Tsuruta, Yoshiki Hayashi and Ryoma Ohta for their help in biochemical analyses. We thank Developmental Studies Hybridoma Bank for antibodies and the Bloomington Stock Center for fly stocks. This study was supported by Grants-in-Aid for Scientific Research, MEXT, Japan (K. F. T.) and NIH grants of MH-084018 (A.S.), MH-094268 Silvo O. Conte center (A.S., A.K.), MH-069853 (A.S.), MH-085226 (A.S.), MH-088753 (A.S.), MH-092443 (A.S.), MH-091230 (A.K), as well as grants from Stanley (A.S.), S-R (A.S.), RUSK (A.S.), NARSAD (A.S., A.K.), JHU-BSI (A.S.), MSCRF (A.S.).
Footnotes
Supplementary information is available at Molecular Psychiatry’s web site.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Brandon NJ, Sawa A. Linking neurodevelopmental and synaptic theories of mental illness through DISC1. Nature reviews Neuroscience. 2011;12(12):707–722. doi: 10.1038/nrn3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porteous DJ, Millar JK, Brandon NJ, Sawa A. DISC1 at 10: connecting psychiatric genetics and neuroscience. Trends in molecular medicine. 2011;17(12):699–706. doi: 10.1016/j.molmed.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Narayan S, Nakajima K, Sawa A. DISC1: a key lead in studying cortical development and associated brain disorders. Neuroscientist. 2013;19(5):451–464. doi: 10.1177/1073858412470168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hikida T, Gamo NJ, Sawa A. DISC1 as a therapeutic target for mental illnesses. Expert opinion on therapeutic targets. 2012;16(12):1151–1160. doi: 10.1517/14728222.2012.719879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porteous DJ, Thomson PA, Millar JK, Evans KL, Hennah W, Soares DC, et al. DISC1 as a genetic risk factor for schizophrenia and related major mental illness: response to Sullivan. Mol Psychiatry. 2014;19(2):141–143. doi: 10.1038/mp.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sullivan PF. Questions about DISC1 as a genetic risk factor for schizophrenia. Mol Psychiatry. 2013;18(10):1050–1052. doi: 10.1038/mp.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lessing D, Bonini NM. Maintaining the brain: insight into human neurodegeneration from Drosophila melanogaster mutants. Nat Rev Genet. 2009 doi: 10.1038/nrg2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellen HJ, Tong C, Tsuda H. 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat Rev Neurosci. 2010;11(7):514–522. doi: 10.1038/nrn2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wangler MF, Yamamoto S, Bellen HJ. Fruit flies in biomedical research. Genetics. 2015;199(3):639–653. doi: 10.1534/genetics.114.171785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuthbert BN. Research domain criteria: toward future psychiatric nosology. Asian journal of psychiatry. 2014;7(1):4–5. doi: 10.1016/j.ajp.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am J Psychiatry. 2010;167(7):748–751. doi: 10.1176/appi.ajp.2010.09091379. [DOI] [PubMed] [Google Scholar]
- 12.Morris SE, Cuthbert BN. Research Domain Criteria: cognitive systems, neural circuits, and dimensions of behavior. Dialogues Clin Neurosci. 2012;14(1):29–37. doi: 10.31887/DCNS.2012.14.1/smorris. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doll CA, Broadie K. Impaired activity-dependent neural circuit assembly and refinement in autism spectrum disorder genetic models. Front Cell Neurosci. 2014;8:30. doi: 10.3389/fncel.2014.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawamura N, Ando T, Maruyama Y, Fujimuro M, Mochizuki H, Honjo K, et al. Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol Psychiatry. 2008;13(12):1138–1148. 1069. doi: 10.1038/mp.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Alphen B, van Swinderen B. Drosophila strategies to study psychiatric disorders. Brain research bulletin. 2013;92:1–11. doi: 10.1016/j.brainresbull.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 16.Furukubo-Tokunaga K. Modeling schizophrenia in flies. Progress in brain research. 2009;179:107–115. doi: 10.1016/S0079-6123(09)17912-8. [DOI] [PubMed] [Google Scholar]
- 17.van der Voet M, Nijhof B, Oortveld MA, Schenck A. Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci Biobehav Rev. 2014;46(Pt 2):326–342. doi: 10.1016/j.neubiorev.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 18.Androschuk A, Al-Jabri B, Bolduc FV. From Learning to Memory: What Flies Can Tell Us about Intellectual Disability Treatment. Front Psychiatry. 2015;6:85. doi: 10.3389/fpsyt.2015.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heisenberg M. Mushroom body memoir: from maps to models. Nature reviews Neuroscience. 2003;4(4):266–275. doi: 10.1038/nrn1074. [DOI] [PubMed] [Google Scholar]
- 20.Furukubo-Tokunaga K, Ludlow ZN, Hirth F. Memory circuits in Drosophila. In: Giese PG, editor. The Memory Mechanisms in Health and Disease. World Scientific; London, UK: 2012. [Google Scholar]
- 21.Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annual review of neuroscience. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 22.Bord L, Wheeler J, Paek M, Saleh M, Lyons-Warren A, Ross CA, et al. Primate disrupted-in-schizophrenia-1 (DISC1): high divergence of a gene for major mental illnesses in recent evolutionary history. Neuroscience research. 2006;56(3):286–293. doi: 10.1016/j.neures.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Camargo LM, Collura V, Rain JC, Mizuguchi K, Hermjakob H, Kerrien S, et al. Disrupted in Schizophrenia 1 Interactome: evidence for the close connectivity of risk genes and a potential synaptic basis for schizophrenia. Mol Psychiatry. 2007;12(1):74–86. doi: 10.1038/sj.mp.4001880. [DOI] [PubMed] [Google Scholar]
- 24.Kurusu M, Awasaki T, Masuda-Nakagawa LM, Kawauchi H, Ito K, Furukubo-Tokunaga K. Embryonic and larval development of the Drosophila mushroom bodies: concentric layer subdivisions and the role of fasciclin II. Development. 2002;129(2):409–419. doi: 10.1242/dev.129.2.409. [DOI] [PubMed] [Google Scholar]
- 25.Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, Fujii K, et al. Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. P Natl Acad Sci USA. 2003;100(1):289–294. doi: 10.1073/pnas.0136913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leliveld SR, Bader V, Hendriks P, Prikulis I, Sajnani G, Requena JR, et al. Insolubility of disrupted-in-schizophrenia 1 disrupts oligomer-dependent interactions with nuclear distribution element 1 and is associated with sporadic mental disease. J Neurosci. 2008;28(15):3839–3845. doi: 10.1523/JNEUROSCI.5389-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 28.Kao CFLT. Genetic mosaic analyses of Drosophila brain by MARCM. In: Zhang BF, MR, Waddell S, editors. Drosophila neurobiology. Cold Spring Harbor Laboratory Press; New York: 2010. pp. 125–139. [Google Scholar]
- 29.Honjo K, Furukubo-Tokunaga K. Induction of cAMP response element-binding protein-dependent medium-term memory by appetitive gustatory reinforcement in Drosophila larvae. J Neurosci. 2005;25(35):7905–7913. doi: 10.1523/JNEUROSCI.2135-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honjo K, Furukubo-Tokunaga K. Distinctive neuronal networks and biochemical pathways for appetitive and aversive memory in Drosophila larvae. J Neurosci. 2009;29(3):852–862. doi: 10.1523/JNEUROSCI.1315-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramachandran PBV. Embryonic and larval neuromuscular junction: an overview with selected methods and protocols. In: Zhang BF, MR, Waddell S, editors. Drosophila neurobiology. Cold Spring Harbor Laboratory Press; New York: 2010. pp. 93–123. [Google Scholar]
- 32.Dickman DK, Davis GW. The schizophrenia susceptibility gene dysbindin controls synaptic homeostasis. Science. 2009;326(5956):1127–1130. doi: 10.1126/science.1179685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 34.Wang S, Yoo S, Kim HY, Wang M, Zheng C, Parkhouse W, et al. Detection of in situ protein-protein complexes at the Drosophila larval neuromuscular junction using proximity ligation assay. J Vis Exp. 2015;(95):52139. doi: 10.3791/52139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogelstein JT, Park Y, Ohyama T, Kerr RA, Truman JW, Priebe CE, et al. Discovery of brainwide neural-behavioral maps via multiscale unsupervised structure learning. Science. 2014;344(6182):386–392. doi: 10.1126/science.1250298. [DOI] [PubMed] [Google Scholar]
- 36.Schurov IL, Handford EJ, Brandon NJ, Whiting PJ. Expression of disrupted in schizophrenia 1 (DISC1) protein in the adult and developing mouse brain indicates its role in neurodevelopment. Mol Psychiatry. 2004;9(12):1100–1110. doi: 10.1038/sj.mp.4001574. [DOI] [PubMed] [Google Scholar]
- 37.Millar JK, James R, Christie S, Porteous DJ. Disrupted in schizophrenia 1 (DISC1): subcellular targeting and induction of ring mitochondria. Molecular and cellular neurosciences. 2005;30(4):477–484. doi: 10.1016/j.mcn.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Brandon NJ, Schurov I, Camargo LM, Handford EJ, Duran-Jimeniz B, Hunt P, et al. Subcellular targeting of DISC1 is dependent on a domain independent from the Nudel binding site. Molecular and cellular neurosciences. 2005;28(4):613–624. doi: 10.1016/j.mcn.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 39.Korth C. Aggregated proteins in schizophrenia and other chronic mental diseases: DISC1opathies. Prion. 2012;6(2):134–141. doi: 10.4161/pri.18989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ottis P, Bader V, Trossbach SV, Kretzschmar H, Michel M, Leliveld SR, et al. Convergence of two independent mental disease genes on the protein level: recruitment of dysbindin to cell-invasive disrupted-in-schizophrenia 1 aggresomes. Biol Psychiatry. 2011;70(7):604–610. doi: 10.1016/j.biopsych.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 41.Leliveld SR, Hendriks P, Michel M, Sajnani G, Bader V, Trossbach S, et al. Oligomer assembly of the C-terminal DISC1 domain (640–854) is controlled by self-association motifs and disease-associated polymorphism S704C. Biochemistry. 2009;48(32):7746–7755. doi: 10.1021/bi900901e. [DOI] [PubMed] [Google Scholar]
- 42.Ji B, Higa KK, Kim M, Zhou L, Young JW, Geyer MA, et al. Inhibition of protein translation by the DISC1-Boymaw fusion gene from a Scottish family with major psychiatric disorders. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eykelenboom JE, Briggs GJ, Bradshaw NJ, Soares DC, Ogawa F, Christie S, et al. A t(1;11) translocation linked to schizophrenia and affective disorders gives rise to aberrant chimeric DISC1 transcripts that encode structurally altered, deleterious mitochondrial proteins. Hum Mol Genet. 2012;21(15):3374–3386. doi: 10.1093/hmg/dds169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Chen Q, Schaukowitch K, Kelsoe JR, Geyer MA. Insoluble DISC1-Boymaw fusion proteins generated by DISC1 translocation. Mol Psychiatry. 2010;15(7):669–672. doi: 10.1038/mp.2009.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itakura M, Nakajima H, Kubo T, Semi Y, Kume S, Higashida S, et al. Glyceraldehyde-3-phosphate Dehydrogenase Aggregates Accelerate Amyloid-beta Amyloidogenesis in Alzheimer Disease. J Biol Chem. 2015;290(43):26072–26087. doi: 10.1074/jbc.M115.669291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakajima H, Amano W, Kubo T, Fukuhara A, Ihara H, Azuma YT, et al. Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J Biol Chem. 2009;284(49):34331–34341. doi: 10.1074/jbc.M109.027698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annual review of genetics. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 48.Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nature reviews Genetics. 2005;6(1):9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- 49.Cannon TD, Hennah W, van Erp TG, Thompson PM, Lonnqvist J, Huttunen M, et al. Association of DISC1/TRAX haplotypes with schizophrenia, reduced prefrontal gray matter, and impaired short- and long-term memory. Archives of general psychiatry. 2005;62(11):1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- 50.Hennah W, Tuulio-Henriksson A, Paunio T, Ekelund J, Varilo T, Partonen T, et al. A haplotype within the DISC1 gene is associated with visual memory functions in families with a high density of schizophrenia. Mol Psychiatry. 2005;10(12):1097–1103. doi: 10.1038/sj.mp.4001731. [DOI] [PubMed] [Google Scholar]
- 51.Li W, Zhou Y, Jentsch JD, Brown RA, Tian X, Ehninger D, et al. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. P Natl Acad Sci USA. 2007;104(46):18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Honda T, Lee CY, Yoshida-Kasikawa M, Honjo K, Furukubo-Tokunaga K. Induction of associative olfactory memory by targeted activation of single olfactory neurons in Drosophila larvae. Scientific reports. 2014;4:4798. doi: 10.1038/srep04798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diegelmann S, Klagges B, Michels B, Schleyer M, Gerber B. Maggot learning and Synapsin function. J Exp Biol. 2013;216(Pt 6):939–951. doi: 10.1242/jeb.076208. [DOI] [PubMed] [Google Scholar]
- 54.Schleyer M, Saumweber T, Nahrendorf W, Fischer B, von Alpen D, Pauls D, et al. A behavior-based circuit model of how outcome expectations organize learned behavior in larval Drosophila. Learn Mem. 2011;18(10):639–653. doi: 10.1101/lm.2163411. [DOI] [PubMed] [Google Scholar]
- 55.Guven-Ozkan T, Davis RL. Functional neuroanatomy of Drosophila olfactory memory formation. Learn Mem. 2014;21(10):519–526. doi: 10.1101/lm.034363.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davis RL. Traces of Drosophila memory. Neuron. 2011;70(1):8–19. doi: 10.1016/j.neuron.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yasuyama K, Meinertzhagen IA, Schurmann FW. Synaptic organization of the mushroom body calyx in Drosophila melanogaster. The Journal of comparative neurology. 2002;445(3):211–226. doi: 10.1002/cne.10155. [DOI] [PubMed] [Google Scholar]
- 58.Collins CA, DiAntonio A. Synaptic development: insights from Drosophila. Curr Opin Neurobiol. 2007;17(1):35–42. doi: 10.1016/j.conb.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 59.Menon KP, Carrillo RA, Zinn K. Development and plasticity of the Drosophila larval neuromuscular junction. Wiley interdisciplinary reviews Developmental biology. 2013;2(5):647–670. doi: 10.1002/wdev.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Charng WL, Yamamoto S, Bellen HJ. Shared mechanisms between Drosophila peripheral nervous system development and human neurodegenerative diseases. Curr Opin Neurobiol. 2014;27:158–164. doi: 10.1016/j.conb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koles K, Budnik V. Wnt signaling in neuromuscular junction development. Cold Spring Harbor perspectives in biology. 2012;4(6) doi: 10.1101/cshperspect.a008045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wainman A, Creque J, Williams B, Williams EV, Bonaccorsi S, Gatti M, et al. Roles of the Drosophila NudE protein in kinetochore function and centrosome migration. Journal of cell science. 2009;122(Pt 11):1747–1758. doi: 10.1242/jcs.041798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang N, Inaki M, Cliffe A, Rorth P. Microtubules and Lis-1/NudE/dynein regulate invasive cell-on-cell migration in Drosophila. PLoS ONE. 2012;7(7):e40632. doi: 10.1371/journal.pone.0040632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Siller KH, Serr M, Steward R, Hays TS, Doe CQ. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Molecular biology of the cell. 2005;16(11):5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Z, Steward R, Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nature cell biology. 2000;2(11):776–783. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- 66.Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, Dunlop AJ, et al. Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nature neuroscience. 2010;13(3):327–332. doi: 10.1038/nn.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, et al. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;26(1):119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- 68.Iyer SC, Wang D, Iyer EP, Trunnell SA, Meduri R, Shinwari R, et al. The RhoGEF trio functions in sculpting class specific dendrite morphogenesis in Drosophila sensory neurons. PLoS ONE. 2012;7(3):e33634. doi: 10.1371/journal.pone.0033634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, et al. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatry. 2011;16(10):1006–1023. doi: 10.1038/mp.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kraut R, Menon K, Zinn K. A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Current biology : CB. 2001;11(6):417–430. doi: 10.1016/s0960-9822(01)00124-5. [DOI] [PubMed] [Google Scholar]
- 71.Laruelle M. Schizophrenia: from dopaminergic to glutamatergic interventions. Curr Opin Pharmacol. 2014;14:97–102. doi: 10.1016/j.coph.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Kantrowitz J, Javitt DC. Glutamatergic transmission in schizophrenia: from basic research to clinical practice. Curr Opin Psychiatry. 2012;25(2):96–102. doi: 10.1097/YCO.0b013e32835035b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benson MA, Newey SE, Martin-Rendon E, Hawkes R, Blake DJ. Dysbindin, a novel coiled-coil-containing protein that interacts with the dystrobrevins in muscle and brain. J Biol Chem. 2001;276(26):24232–24241. doi: 10.1074/jbc.M010418200. [DOI] [PubMed] [Google Scholar]
- 74.Ghiani CA, Starcevic M, Rodriguez-Fernandez IA, Nazarian R, Cheli VT, Chan LN, et al. The dysbindin-containing complex (BLOC-1) in brain: developmental regulation, interaction with SNARE proteins and role in neurite outgrowth. Mol Psychiatry. 2010;15(2):115, 204–115. doi: 10.1038/mp.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iizuka Y, Sei Y, Weinberger DR, Straub RE. Evidence that the BLOC-1 protein dysbindin modulates dopamine D2 receptor internalization and signaling but not D1 internalization. J Neurosci. 2007;27(45):12390–12395. doi: 10.1523/JNEUROSCI.1689-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ito H, Morishita R, Shinoda T, Iwamoto I, Sudo K, Okamoto K, et al. Dysbindin-1, WAVE2 and Abi-1 form a complex that regulates dendritic spine formation. Mol Psychiatry. 2010;15(10):976–986. doi: 10.1038/mp.2010.69. [DOI] [PubMed] [Google Scholar]
- 77.Ma X, Fei E, Fu C, Ren H, Wang G. Dysbindin-1, a schizophrenia-related protein, facilitates neurite outgrowth by promoting the transcriptional activity of p53. Mol Psychiatry. 2011;16(11):1105–1116. doi: 10.1038/mp.2011.43. [DOI] [PubMed] [Google Scholar]
- 78.Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13(7):898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mehta SQ, Hiesinger PR, Beronja S, Zhai RG, Schulze KL, Verstreken P, et al. Mutations in Drosophila sec15 reveal a function in neuronal targeting for a subset of exocyst components. Neuron. 2005;46(2):219–232. doi: 10.1016/j.neuron.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 80.Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, et al. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev Cell. 2005;9(3):351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 81.Shao L, Shuai Y, Wang J, Feng S, Lu B, Li Z, et al. Schizophrenia susceptibility gene dysbindin regulates glutamatergic and dopaminergic functions via distinctive mechanisms in Drosophila. Proc Natl Acad Sci U S A. 2011;108(46):18831–18836. doi: 10.1073/pnas.1114569108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alves-Silva J, Sanchez-Soriano N, Beaven R, Klein M, Parkin J, Millard TH, et al. Spectraplakins promote microtubule-mediated axonal growth by functioning as structural microtubule-associated proteins and EB1-dependent +TIPs (tip interacting proteins) J Neurosci. 2012;32(27):9143–9158. doi: 10.1523/JNEUROSCI.0416-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee S, Nahm M, Lee M, Kwon M, Kim E, Zadeh AD, et al. The F-actin-microtubule crosslinker Shot is a platform for Krasavietz-mediated translational regulation of midline axon repulsion. Development. 2007;134(9):1767–1777. doi: 10.1242/dev.02842. [DOI] [PubMed] [Google Scholar]
- 84.Lee SA, Kim SM, Suh BK, Sun HY, Park YU, Hong JH, et al. Disrupted-in-schizophrenia 1 (DISC1) regulates dysbindin function by enhancing its stability. J Biol Chem. 2015;290(11):7087–7096. doi: 10.1074/jbc.M114.614750. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.