Abstract
Preservation of osteochondral allografts used for transplantation is critical to ensure favorable outcomes for patients after surgical treatment of cartilage defects. To study the biological effects of protocols currently used for cartilage storage, we investigated differences in gene expression between stored allograft cartilage and fresh cartilage from living donors using high throughput molecular screening strategies. We applied next generation RNA sequencing (RNA-seq) and real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR) to assess genome-wide differences in mRNA expression between stored allograft cartilage and fresh cartilage tissue from living donors. Gene ontology analysis was used to characterize biological pathways associated with differentially expressed genes. Our studies establish reduced levels of mRNAs encoding cartilage related extracellular matrix (ECM) proteins (i.e., COL1A1, COL2A1, COL10A1, ACAN, DCN, HAPLN1, TNC, and COMP) in stored cartilage. These changes occur concomitantly with increased expression of “early response genes” that encode transcription factors mediating stress/cytoprotective responses (i.e., EGR1, EGR2, EGR3, MYC, FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, and JUND). The elevated expression of “early response genes” and reduced levels of ECM-related mRNAs in stored cartilage allografts suggests that tissue viability may be maintained by a cytoprotective program that reduces cell metabolic activity. These findings have potential implications for future studies focused on quality assessment and clinical optimization of osteochondral allografts used for cartilage transplantation.
Keywords: cartilage, allograft, osteochondral, transplant, RNA sequencing
Focal cartilage lesions caused by disorders, such as osteochondritis dissecans, osteonecrosis, and traumatic injury, can lead to the early onset of osteoarthritis requiring early joint replacement. Surgical options for treating articular cartilage lesions depend on the location of the lesion (e.g., distal femoral, patella, talus, etc.), as well as size and thickness.1 Popular treatment options include bone marrow stimulation (BMS; i.e., microfracture), osteochondral autograft transfer, autologous chondrocyte implantation (ACI), particulated juvenile articular cartilage allograft (PJACA), and transplantation of freshly stored osteochondral allografts.2,3 Each method has advantages and disadvantages. BMS leads to healing with mature fibrocartilage, which lacks the durability of articular cartilage.4 Autograft harvests can result in donor site morbidity, and are limited in terms of amount and number of harvest locations.5 ACI is costly and labor intensive, because it requires multiple surgeries including an initial biopsy, followed by isolation, growth, and expansion of cells prior to surgical delivery. Osteochondral allograft transplantation is an effective technique for treating condylar lesions.6 Surgery involves the transfer of size-matched subchondral bone and cartilage from a cadaveric donor into a focal chondral or osteochondral defect. Optimal criteria for donor selection and graft preparation have not been established. Some studies advocate the use of grafts from donors between 15 and 40 years of age, with graft harvest within 24 h of donor death.7,8 Grafts are typically treated with pulse lavage of bone marrow, followed by storage in antibiotic solution for 24 h. The allografts are then refrigerated in either lactated ringer’s solution or proprietary physiologic culture medium at 4°C. Allograft transplantation is usually performed within 28 days of graft harvest at a time when the majority of allograft chondrocytes are thought to be viable7,9. Long-term refrigerated storage of grafts may affect graft performance and hence surgical outcome. Therefore, it would be desirable to have clinically informative biomarkers for measuring graft quality.
Although osteochondral allograft transplantation is a common technique with a high degree of clinical success,10 early failure can still occur.11,12 Quality assessment of allograft donor cartilage has focused on radiographic imaging, biomechanical assessment, as well as histological and metabolic assessment of cell viability.13 Several laboratory studies evaluating osteochondral allografts reveal relationships between allograft storage time and chondrocyte viability, function, and the integrity of the ECM.14,15 Although useful techniques for assessing graft viability, these measures are not fully able to predict the clinical efficacy of osteochondral allografts. To improve the accuracy of clinical graft assessment over current techniques, we have applied next generation RNA-seq to evaluate the expression of all known human genes (n > 23,000) in stored and fresh cartilage specimens.
The primary goal of this study is to evaluate the effects of conventional storage techniques on cartilage gene expression in clinical-grade osteochondral allografts at the highest level of molecular resolution currently achievable. We show that a selective subset of mRNAs encoding genes linked to stress/cytoprotective responses and ECM production exhibit prominent differences in expression. Our study suggests that chondrocytes embedded in stored but not fresh allografts mount a cytoprotective response that mitigates apoptosis and downregulates cartilage anabolic genes to preserve cell viability. The biomarkers identified in our study can facilitate molecular assessment of osteochondral allograft preservation to help optimize graft storage conditions. They may also have future potential to complement radiographic, biomechanical, histological and metabolic assessments for predicting clinical outcomes after cartilage transplantation.
MATERIALS AND METHODS
Collection of Cartilage Tissues
Allograft cartilage specimens were collected from osteochondral grafts leftover from surgical transplantation procedures. All allograft specimens were collected within 24 h of being used for cartilage transplantation procedures. Each allograft had been hypothermically stored at 4°C between 18 and 30 days in proprietary storage medium, prior to surgical use and research collection. Osteochondral allograft samples included: nine femoral hemi-condyles, three whole femoral condyles, two talus samples, and one humeral head. These clinical-grade grafts were obtained from either the Joint Restoration Foundation or the Musculoskeletal Transplant Foundation, and thus conform to highly stringent selection criteria for cartilage of surgical quality.
For molecular comparisons, fresh cartilage specimens were collected from either the distal femur or proximal tibia, as surgical waste tissue from living patients undergoing orthopedic procedures (e.g., amputations, joint replacement). We selected fresh cartilage samples from a diverse group of patients that are least likely to have to have undergone major therapies (e.g., chemotherapy) that would affect the cellular activity of chondrocytes embedded in cartilage. The clinical characteristics of the allograft and fresh cartilage specimens used in this investigation are summarized in Supplementary Table S1.
At the time of harvest, cartilage samples were separated from subchondral bone using a scalpel, snap frozen in liquid nitrogen, and subsequently stored at −80°C. Ten fresh cartilage samples were collected in total for this study. Written informed consent was obtained for the use of all fresh cartilage specimens. Allograft specimens were de-identified at an outside agency prior to clinical and research applications. All fresh cartilage specimens used in this investigation were collected under protocols approved by the Institutional Review Board.
RNA Extraction From Cartilage
For RNA extraction, 50 mg of tissue frozen in liquid nitrogen was crushed into a fine powder using a hammer. Total RNA was then extracted using the Biochain RNA Isolation Kit (BioChain Institute, Inc., Newark, CA) according to the manufacturer’s protocol. Genomic RNA sample concentrations and purities were measured by NanoDrop spectrophotometer (ThermoFisher Scientific, Inc., Waltham, MA).
Next Generation mRNA Sequencing and Bioinformatics Analysis
RNA sequencing and bioinformatics analysis were performed as previously described,16 for three fresh and two allograft cartilage samples using the TruSeq RNA method (Illumina, San Diego, CA) for analysis of polyadenylated mRNAs that were selected using oligo dT magnetic beads. TruSeq Kits (12-Set A and 12-Set B) were used for indexing to permit multiplex sample loading on the flow cells of an Illumina HiSeq 2000 sequencer. Quality control for concentration and library size distribution was performed using an Agilent Bioanalyzer DNA 1000 chip and Qubit fluorometry (Invitrogen, Carlsbad, CA). Sequence alignment of reads and determination of normalized gene counts were performed using MAPRSeq (v.1.2.1), TopHat 2.0.6,17 HTSeq, and edgeR 2.6.218 work flows. Gene expression was normalized to one million reads and corrected for gene length (reads per kilobasepair per million mapped reads, RPKM). Functional gene annotation clustering analysis was performed using DAVID Bioinformatics Resources 6.7 database (DAVID 6.7)19 to obtain a ranking of primary gene ontology categories for genes preferentially expressed in stored versus fresh cartilage.
Real-Time Reverse Transcription Guantitative PCR (RT-qPCR) Analysis
Isolated RNA was transcribed into cDNA using the Super-Script III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). A gene panel was determined by screening the RNA sequencing results. Gene expression was quantified by using real-time qPCR reactions performed with 25 ng of cDNA per 10 μl using the QuantiTect SYBR Green PCR Kit (Qiagen, Hilden, Germany) and CFX384 Real-Time System (BioRad, Hercules, CA). Gene-specific primers are shown in Supplementary Table S2. Transcript quantity measurements were normalized to AKT1, and gene expression levels were quantified using the 2(−ΔCt) method. We note that AKT1 is among the most consistently expressed housekeeping genes (e.g., compared to GAPDH, HPRT, ACTB) in more than 400 mesenchymal cell types and musculoskeletal tissues analyzed by our investigative team (unpublished data).
Allograft Chondrocyte Viability Recovery Examination
Allograft cartilage was harvested as thin sections (approximately 1 mm) using a scalpel blade from osteochondral allografts immediately after use for surgical implantation. Cartilage tissues were stored at 4°C in the original preservation solution that specimens were shipped in. A subset of cartilage specimens was incubated at 37°C for either a 24 h or 10-day period. At the end of the incubation period, the cartilage specimens were fixed in 4% paraformaldehyde. Mitochondrial activity of the chondrocytes within the cartilage tissue was assessed using Mito Tracker staining (Life Technologies, Carlsbad, CA) with 1 mM MitoTracker Deep Red FM according to the manufacturer’s protocol at room temperature. Cartilage sections were counter-stained with 4′,6-diamidino-2-phenylindole (DAPI) 0.2 μg/ml solution. Stained cartilage sections were imaged using confocal microscopy. In addition, total RNA was extracted and quantified for each of the cartilage specimens harvested at the end of each incubation period.
Quantitative and Statistical Analyses
RNA-seq data were analyzed to assess relevant genes that differ between stored (n = 2) versus fresh allografts (n=3). Gene selection was initially achieved by filtering normalized reads for relative expression at a minimal arbitrary value (RPKM > 0.1) that reduces noise in the analysis. We also filtered genes for fold-changes greater than threefold to increase our focus on biologically interesting genes. Selected genes for representative pathways that are altered were subjected to RT-qPCR validation using gene-specific primers with mRNA samples from 24 cartilage specimens (i.e., n=15 allograft and n=9 fresh cartilage samples). Changes in gene expression obtained by RT-qPCR were statistically evaluated using a two-tailed Student’s t-test with statistical significance set at p-value <0.05. Error bars in all figures are given as ±1 standard deviation from the mean.
RESULTS
To examine the biological effects of long-term storage of cartilage grafts at 4°C before they are otherwise implanted into patients, we compared gene expression profiles of stored allograft cartilage with those of fresh cartilage from living donors using high throughput RNA-seq. RNA-seq analysis defines phenotype-specific genome-wide expression levels, as well as permits discovery and evaluation of new biomarkers that may inform about cartilage allograft viability and chondrocyte function. Osteochondral allografts used for cartilage transplantation in patients are stored for an extended period of time under hypothermic conditions (i.e., 4°C) and in storage solutions that differ markedly from normal synovial fluid. Therefore, we compared gene expression profiles of stored allografts to fresh cartilage that is snap frozen in liquid nitrogen immediately after surgical retrieval from living donors to preserve gene expression of chondrocytes in situ at the time of harvest.
One technical difficulty we encountered for RNA-seq analysis with all cartilage samples is the technical problem of low RNA yields and generally poor RNA quality. Based on statistical and economic considerations, the design of our experiments called for a two-way comparison of three independent patient samples (as biological replicates). Eventually, we obtained RNA from 25 cartilage specimens with variable RNA integrity (RIN) scores. We subjected three samples each for fresh or stored allografts with acceptable RIN scores. Because one of the stored allograft samples had a low read yield, our studies continued with the remaining five datasets (n=3 fresh versus n=2 stored allografts). Analysis of the RNA-seq data was, therefore, primarily used to obtain preliminary data for generating working hypotheses (Figs. 1–3). These hypotheses were subsequently validated by RT-qPCR with the entire sample cohort (n=24) to permit robust statistical comparisons (Figs. 4 and 6).
Figure 1.
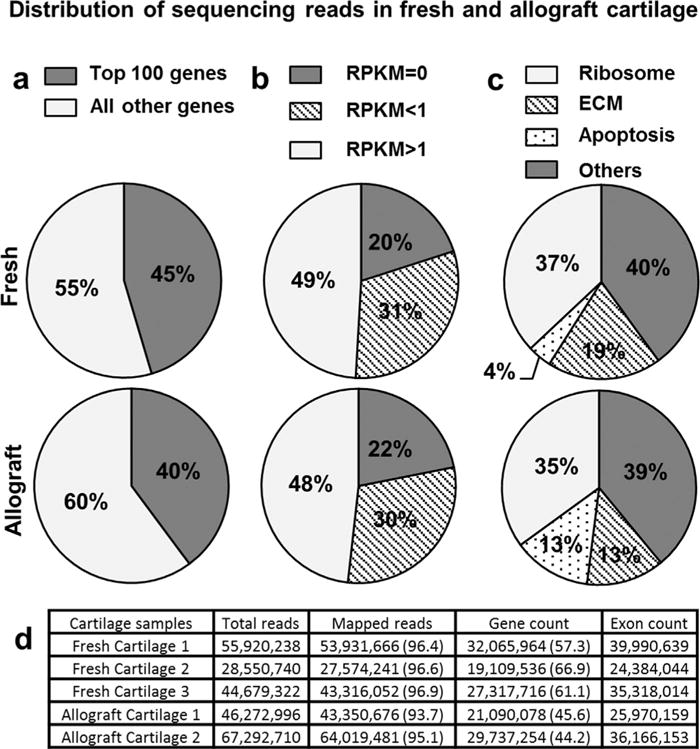
(a, b) The number and distribution of sequencing reads are similar between allograft and fresh cartilage specimens. (c) Among the top 100 expressed genes, there are a greater percentage of genes associated with apoptosis in allograft cartilage. (d) Total exon counts for allograft and fresh cartilage samples were similar, indicating that gene expression differences are unlikely to be due to overall abundance of transcripts.
Figure 3.
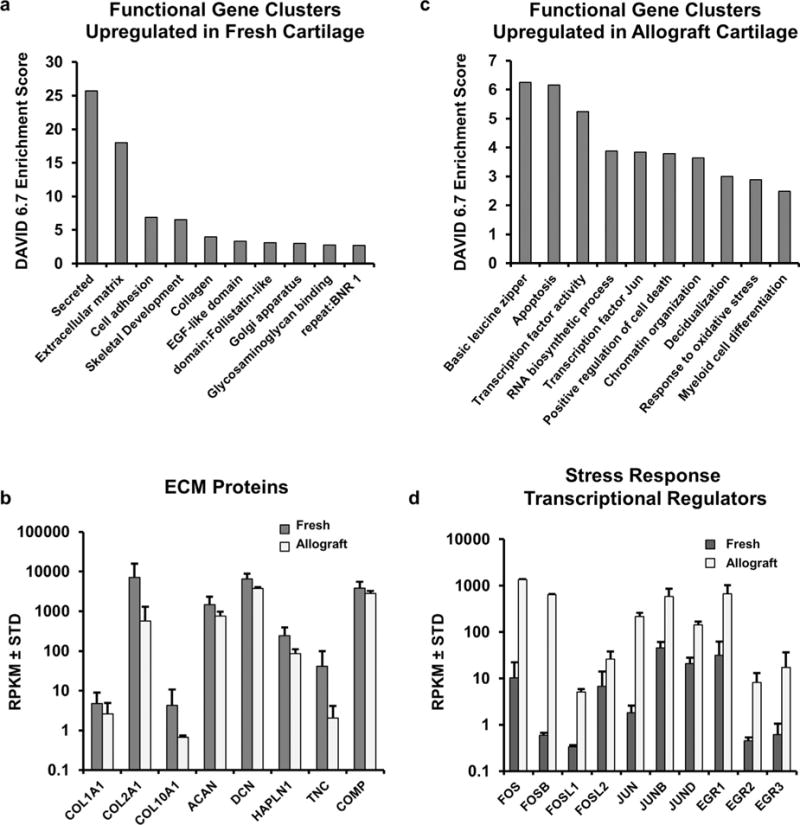
(a) Gene ontology analysis using DAVID 6.7 for genes with a fold change of threefold shows functional gene clusters linked to extracellular matrix production that are elevated in fresh cartilage compared to allograft cartilage. (b) Examination of the expression of mRNAs encoding ECM proteins linked to cartilage phenotype (COL2A1, ACAN, DCN) and general anabolism (COL1A1, COL10A1), from the RNA-seq data shows they are enriched in the fresh cartilage tissues. (c) Gene ontology analysis demonstrates upregulation of genes linked to apoptosis and stress response in allograft cartilage. (d) Specific genes linked to stress response from RNA-seq data are shown to be expressed at higher levels in allograft cartilage.
Figure 4.
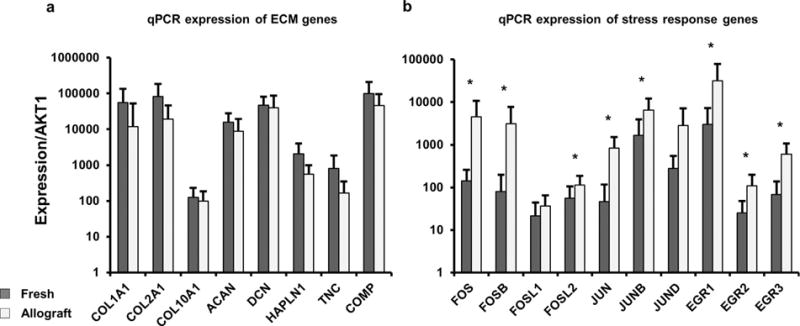
(a) RT-qPCR validation was performed using 25 cartilage specimens (15 allograft and 10 fresh cartilage samples), these data confirmed upregulation of ECM encoding mRNAs in fresh cartilage specimens. (b) RT-qPCR also confirmed differential expression of genes linked to stress response in accordance with RNA-seq results.
Figure 6.
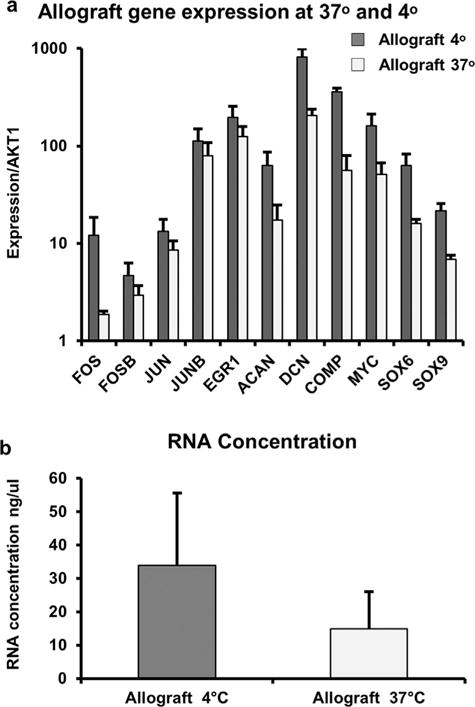
To assess the potential of pre-warming of allograft cartilage to decrease stress response and enhance the expression of ECM proteins observed in fresh cartilage, we incubated cartilage specimens from allografts at 37°C and 4°C for 10 days. (a) Evaluation of gene expression using RT-qPCR shows a downregulation in stress response, as well as a decrease in ECM encoding mRNAs. (b) Changes in gene expression are also associated with a decrease in total RNA yields from tissue samples. These findings could be seen with a loss of chondrocyte viability in warmed allograft cartilage.
Our initial analysis of the RNA-seq data using simple sorting algorithms revealed that all specimens express relatively high levels (>100 RPKM) of well-established cartilage ECM biomarkers, including collagen type II (COL2A1), aggrecan (ACAN), and decorin (DCN; data not shown; further discussed below). This finding indicates that allografts from both deceased and living donors contain detectable mRNAs derived from chondrocytes functionally producing cartilage-related ECM proteins.
From a genomic perspective, both the number and the distribution of sequencing reads are similar between fresh cartilage and allograft cartilage specimens (Fig. 1). The top 100 of genes that are most highly expressed in any specimen account for 40–45% of all mapped reads (Fig. 1a), consistent with our previous observations for RNA-seq data sets using cultured human cells.16 About half of all mRNAs are robustly expressed (RPKM>1), whereas less than a quarter of all genes are not detected at all (RPKM=0) in all specimens (Fig. 1b). As expected, the most abundant mRNAs encode proteins involved in translation (e.g., cytoplasmic and mitochondrial ribosomal proteins) and ECM formation (Fig. 1c). However, there appear to be relative differences in the expression of ECM proteins (lower mRNA levels) and apoptosis-related genes (higher mRNA levels) in stored versus fresh tissues (Fig. 1c). Thus, allograft tissues derived from deceased versus living donors may be biologically distinct.
To provide an unbiased assessment of whether fresh and stored cartilage allograft specimens are different, we compared the expression values for all genes among all samples using unsupervised hierarchical clustering with the Pearson correlation method. The three fresh cartilage specimens we analyzed cluster independently from the two stored allograft cartilage tissues (Fig. 2). This initial analysis suggests that, first, gene expression patterns for fresh and allograft cartilage are different, and second, that there may be quantifiable and potentially biologically relevant differences in gene expression between the two groups.
Figure 2.
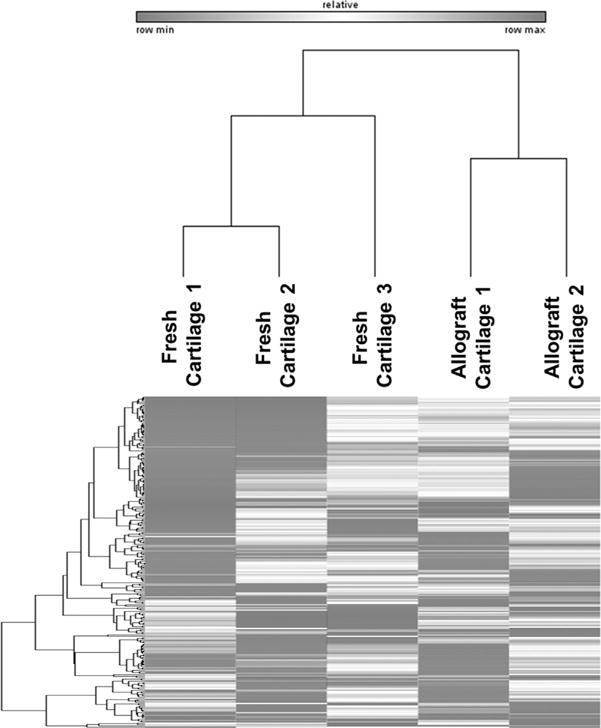
Unsupervised hierarchical clustering using the Pearson correlation method shows independent clustering of allograft and fresh cartilage specimens. This unbiased analysis suggests that changes in gene expression are due to biological differences in stored versus fresh cartilage rather than patient to patient variation.
To determine cellular pathways and biological roles of genes that are differentially expressed between fresh and stored cartilage, we filtered our datasets initially for genes with expression levels of RPKM > 0.1, and biologically interesting changes in expression (fold-change > 3; Table 1). Functional gene annotation clustering of strongly modulated genes was performed using the bioinformatics tool DAVID 6.7.19 Genes enriched in fresh cartilage in comparison with allograft cartilage, show enrichment in genes linked to ECM synthesis, collagen organization, and glycosaminoglycan binding (e.g., COL1A1, COL2A1, COL10A1, ACAN, DCN, HAPLN1, TNC, COMP; Fig. 3a and b). In contrast, allograft cartilage specimens show increased expression of early response genes linked to apoptosis/cell survival and stress/cytoprotective responses, including the key transcriptional regulators FOS, FOSB, FOSL1, FOSL2, JUN, JUNB, JUND, EGR1, EGR2, and EGR3 (Fig. 3c and d). In addition to showing strong enrichment among differentially expressed genes, stress response regulatory mRNAs were also represented among the top most differentially expressed transcripts (Table 1). This increase in stress response genes correlates with a loss of mRNAs encoding chondrocyte-specific ECM proteins. Hence, the RNA-seq data suggest that chondrocytes in stored allografts compared to fresh cartilage may undergo a stress/cytoprotective response and reduce ECM anabolic activity to maintain cell viability within the articular cartilage matrix.
Table 1.
Top 20 Genes Differently Expressed Between Allograft and Fresh Cartilage
| Genes Enriched in Fresh Cartilage
|
Genes Enriched in Allograft Cartilage
|
||||||
|---|---|---|---|---|---|---|---|
| Genes | Allograft | Fresh | Fold Change | Genes | Allograft | Fresh | Fold Change |
| EPYC | 0.01 | 2.41 | 237.18 | XIST | 0.02 | 24.05 | 1421.10 |
| LOC391322 | 0.03 | 4.10 | 147.80 | FOSB | 0.60 | 642.45 | 1075.69 |
| EFEMP1 | 0.14 | 17.36 | 127.82 | CDC42BPG | 0.01 | 3.41 | 546.64 |
| IGFBP2 | 0.31 | 29.30 | 95.74 | LDHC | 0.01 | 4.98 | 517.09 |
| ZNF285 | 0.01 | 0.46 | 78.93 | STC1 | 0.00 | 1.52 | 381.80 |
| GALNT14 | 0.00 | 0.30 | 66.71 | PRKCG | 0.01 | 3.00 | 236.78 |
| VCAM1 | 0.66 | 42.77 | 64.62 | LIF | 0.01 | 1.37 | 222.42 |
| C17orf104 | 0.00 | 0.20 | 58.81 | TMEM72 | 0.02 | 3.80 | 170.56 |
| ODZ2 | 0.00 | 0.27 | 55.58 | PLK2 | 0.08 | 13.39 | 158.58 |
| APOB | 0.00 | 0.12 | 52.20 | RHO | 0.00 | 0.62 | 137.74 |
| ANKRD20A8P | 0.00 | 0.21 | 50.32 | LOC729966 | 0.18 | 24.30 | 135.03 |
| ZIC5 | 0.00 | 0.17 | 48.77 | FOS | 10.40 | 1364.11 | 131.22 |
| LTF | 0.01 | 0.38 | 46.08 | ATP2B3 | 0.01 | 0.97 | 122.70 |
| MRPL42P5 | 0.02 | 1.06 | 45.40 | JUN | 1.82 | 216.48 | 118.74 |
| MUC15 | 0.00 | 0.21 | 45.17 | ATF3 | 0.43 | 47.08 | 110.06 |
| PAK3 | 0.00 | 0.21 | 43.82 | RNU12 | 0.61 | 66.49 | 108.28 |
| GREM1 | 1.31 | 55.07 | 41.89 | MIR4505 | 0.21 | 22.52 | 106.81 |
| LOC100130417 | 0.19 | 7.41 | 39.20 | DUSP2 | 0.10 | 9.55 | 93.29 |
| PAQR4 | 0.03 | 1.12 | 38.59 | PPM1N | 0.29 | 25.54 | 89.00 |
| CYP1B1-AS1 | 0.02 | 0.68 | 38.37 | DNAH2 | 0.01 | 0.44 | 80.87 |
RT-qPCR was used to validate the working hypotheses derived from RNA-seq data by establishing which differences in gene expression between allografts and fresh cartilage tissues are statistically significant in our larger set of 15 allograft and 10 fresh cartilage samples (Fig. 4). These data clearly corroborate differences in expression for all early response genes (i.e., FOS/JUN, MYC, and EGR genes) that were detected by RNA-seq. For comparison, reduced expression of ECM proteins showed a clear trend in reduced expression for all ECM genes we examined, although these differences did not reach significance due to sample variation. The observation that these two gene classes (i.e., stress response versus ECM) differ in statistical significance favors the interpretation that increased expression of stress/cytoprotective early response genes is a primary event, and that reduced expression of ECM proteins may be a secondary consequence.
The statistically significant increase in expression of stress-related early response transcripts in our cohort of 25 samples indicates that random patient-to-patient variation cannot account for the observed differences in mRNA levels. Hence, we postulate that our results are associated with procedural parameters related to allograft storage, including cold storage, poor nutrient supply, and/or cellular reactions to preservation solution. Discriminating among these possibilities may inform clinical procedures for cartilage handling prior to surgical implantation. To begin addressing this question, we assessed whether short-term incubation of specimens at 37°C would restore gene expression patterns and cell viability observed in fresh cartilage. Strikingly, there is no evidence of mitochondrial activity in chondrocytes stored at 4°C. However, samples that were warmed during storage at 37°C for 24 h exhibited positive staining for mitochondrial activity, indicative of chondrocyte viability and metabolic activity (Fig. 5). From a clinical perspective, our finding establishes that stored allografts are capable of regaining cellular functions characteristic of live tissues, consistent with their surgical use in joint repair.
Figure 5.
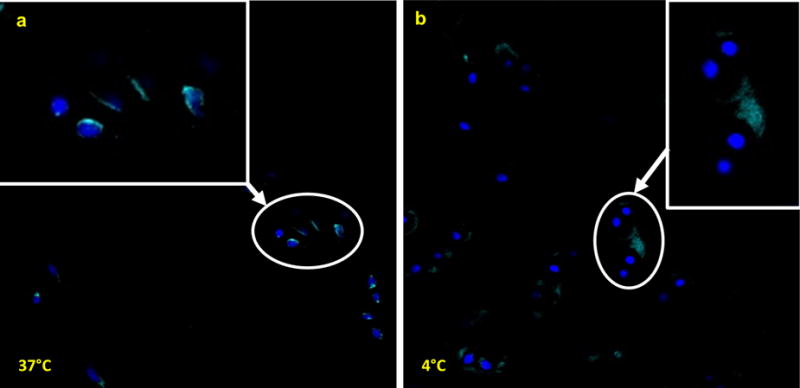
(a) Allograft chondrocytes incubated at 37°C show positive staining using MitoTracker to evaluate mitochondrial activity as a surrogate for cell viability. (b) In contrast chondrocytes from the same viable allograft incubated at 4°C do not exhibit positive MitoTracker staining. Thus pre-warming of allograft cartilage should be performed prior to the use of metabolic assays to assess chondrocyte metabolic activity.
Encouraged by this result, we investigated whether allograft tissue can be maintained at 37°C for a longer period by monitoring expression of stress response and ECM-related genes using RT-qPCR. We selected a 10-day time point because this period would have practical advantages for allograft use considering constraints in surgical scheduling. We note that a longer period is not desirable because we observe an approximately twofold decrease in total RNA yields following extraction from cartilage tissue warmed at 37°C for 10 days most likely reflecting increased cell death (Fig. 6a). Consistent with diminished total RNA yields and cell viability, RT-qPCR analysis reveals a global decrease in stress response and ECM-related genes (Fig. 6b). Because these mRNA expression data are normalized, the decreased expression of stress/cytoprotective genes is an inconclusive finding (either reflecting less cellular stress or a global down regulation in gene expression), reduced expression of ECM-related genes may reflect reduced ECM anabolic activity in chondrocytes that occurs concomitant with compromised cell viability. These preliminary studies suggest that while long-term warming (day 10) may not be desirable, short-term warming of allografts (24 h) increases mitochondrial activity in chondrocytes without compromising cell viability and may warrant further investigation. In addition, these studies suggest that global down regulation of both early response and ECM genes may act as biomarkers to indicate loss or retention of chondrocyte viability and recovery. Furthemore, loss of early response gene expression together with maintenance or increased expression of ECM mRNAs may indicate a favorable response to storage conditions, as well as supports allograft rejuvenation and clinical optimization.
DISCUSSION
Osteochondral allograft transplantation has been widely used for clinical restoration of damaged articular cartilage and subchondral bone. Previous studies have demonstrated time-dependent loss of chondrocytes when grafts are stored at low temperatures and suggest that the number of viable chondrocytes in osteochondral allografts at the time of transplantation determines long-term clinical outcomes.20–23 Our present work complements these previous studies by showing that stored cartilage allografts mount a cytoprotective response that may prevent unscheduled cell death, as well as retain viability and resume mitochondrial functions required for ECM production when refrigerated tissues are warmed to body temperature.
To improve clinical outcomes, all aspects of allografts, including donor selection, allograft procurement, processing, storage, transportation, and surgical implantation, need to be optimized. Prior studies have examined methods to prolong the viability of allograft chondrocytes by modifying the preservation methods or the formulation of preservation solutions. Using a canine model, Cook et al.13 found 90% chondrocyte viability at day 60 in the “Missouri Osteochondral Allograft Preservation System” compared to 53% viability with standard media. Linn et al.24 showed that the cytokine inhibitor etanercept improves cell viability of osteochondral allografts after 28 days of storage. As an anti-tumor necrosis factor (TNF) antibody, etanercept has the potential to inhibit the stress-induced activation of early response genes, thus studies examining changes in gene expression in allografts treated with etanercept are potentially interesting. Other studies25,26 suggest that articular cartilage allograft storage at a physiologic temperature of 37°C improves chondrocyte viability compared to storage at 4°C.
Although previous studies have investigated the effects of storage on cell viability, our study pioneers molecular phenotyping of allograft quality at high resolution using RNA-seq. We have identified changes in allograft gene expression with the potential to assess graft quality and guide the development of more effective storage protocols. Our data show major distinctions in gene expression that provide new insights into the biological status of stored versus fresh cartilage specimens, which differ by increased expression of stress/cytoprotective response pathways and as well as decreased expression of ECM proteins. It is important to note that cold storage can attenuate RNA degradation, and thus the RNA signature we are detecting is partially representative of the state of the chondrocytes before and during their transition to cold storage conditions. A chondrocyte-specific pattern of gene expression (e.g., collagen type II and ACAN) clearly exists, with a super-imposed gene signature that we can clearly identify as related to classical early response factors (e.g., EGR proteins and FOS/JUN proteins).
Allografts from deceased donors remain a primary choice in joint restorative surgery, especially in view of inherent limitations in the availability and size of fresh living donor allografts. Yet, because storage conditions and time from explantation affect allograft viability, assessment of allograft quality is highly important. The differentially expressed early response and ECM-related genes identified in this investigation, which are readily quantifiable using RT-qPCR, may be able to provide additional information regarding chondrocyte activity and clinical viability of cartilage grafts.
Our observation that expression of ECM-related mRNAs exhibit differential expression between allograft and fresh cartilage implicates these genes as potential biomarkers for future studies aimed at graft storage and optimization. Previous studies have shown that some ECM components, such as collagens and aggrecan, play a key role in chondrocyte survival by specifically binding to integrins on the surface of cells, which can mediate cell survival through downstream kinase-mediated signaling pathways.27 Therefore, ECM disruption and concomitant loss of integrin-mediated pro-survival signals may promote apoptosis.28,29 Although many factors can initiate chondrocyte apoptosis,28–32 activation of cysteinyl aspartate-specific proteases (Caspases)33 is a major downstream enzymatic pathway that mediates programmed cell death.34–36 Indeed, broad-spectrum caspase inhibitors block apoptosis and enhance the survival of bovine osteochondral allograft chondrocytes during storage.37 Our study reveals the mechanistic underpinning of this latter finding by showing upregulation of genes associated with apoptosis in stored allografts.
Early response genes are often upregulated under a variety of stress associated factors as a cytoprotective mechanism to maintain cellular homeostasis. However, prolonged activation due to sustained environmental stresses can ultimately induce inflammation and apoptosis.38 Thus, examination of stress response genes using RT-qPCR biomarkers may facilitate the development of environmental conditions and preservation methods that mitigate stress responses (e.g., temperature, storage solutions) to improve the clinical efficacy of allograft cartilage.
In conclusion, this study provides a framework for evaluating the quality of cartilage materials used for surgical transplantation. The genes identified in this investigation represent potential diagnostic biomarkers for evaluating and tracking allograft response to different types of storage medium and environmental conditions. These markers are thus potentially useful for future experimental work focusing on graft optimization.
Supplementary Material
Acknowledgments
This work was supported, in whole or in part, by National Institutes of Health grants R01 AR049069 (to Andre J. van Wijnen), R03 AR066342-02 (to Annalise N. Larson), T32 AR56950 (to Eric A. Lewallen), and F32 AR066508 (to Amel Dudakovic). We also appreciate the generous philanthropic support of William and Karen Eby, and the charitable foundation in their names. We also thank Susan Puffer and Renae Olson for assistance with handling surgical specimens and the members of our laboratory, as well as our colleagues David Deyle and David Lewallen for sharing reagents and/or stimulating discussions.
Grant sponsor: National Institutes of Health; Grant numbers: R01 AR049069, R03 AR066342-02, T32 AR56950, F32 AR066508; Grant sponsor: Mayo Graduate School, Clinical and Translational Sciences Track; Grant sponsor: Center for Regenerative Medicine at Mayo Clinic.
Footnotes
Conflict of interest: Dr. Bruce Levy receives royalties from Arthrex and VOT Solutions. Dr. Aaron Krych is a paid consultant for Arthrex and Vericel. Dr. Michael Stuart is a paid consultant for Arthrex and receives royalties from Arthrex. The remaining authors have no conflicts relevant to this publication.
AUTHORS’ CONTRIBUTIONS
Yang Lin—participated in the design of the study, carried out experiments, interpreted data, prepared and approved final manuscript. Eric A. Lewallen—participated in the design of the study, carried out experiments, interpreted data, prepared and approved final manuscript. Emily T. Camilleri—participated in the design of the study, carried out experiments, interpreted data, helped revise and approve final manuscript. Carolina A. Bonin—participated in the design of the study, carried out experiments, interpreted data, helped revise and approve final manuscript. Dakota Jones—participated in the design of the study, carried out experiments, interpreted data, helped revise and approve final manuscript. Amel Dudakovic—participated in the design of the study, carried out experiments, interpreted data, helped revise and approve final manuscript. Catalina, Galeano-Garces—participated in the design of the study, carried out experiments, interpreted data, helped revise and approve final manuscript. Wei Wang—participated in the design of the study, carried out experiments, interpreted data, helped revise and approve final manuscript. Marcel J. Karperien—analysis and interpretation of data, critical revision of the draft and approval of the final manuscript. A. Noelle Larson—assisted with design of the study, helped obtain funding, helped revise and approve final manuscript. Diane L. Dahm—assisted with design of the study, provided research/study specimens, helped revise and approve final manuscript. Michael J. Stuart—assisted with design of the study, provided research/study specimens, helped revise and approve final manuscript. Bruce A. Levy—assisted with design of the study, provided research/study specimens, helped revise and approve final manuscript. Daniel B. Ryssman—assisted with design of the study, provided research/study specimens, helped revise and approve final manuscript. Jay Smith—assisted with design of the study, helped draft and revise final manuscript. Jennifer J. Westendorf—assisted with design of the study, helped revise and approve final manuscript. Hee-Jeong Im—assisted with design of the study, helped revise and approve final manuscript. Andre J. van Wijnen—assisted with design of the study, provided funding for this project, helped revise and approve final manuscript. Scott M. Riester—assisted with design of the study, carried out experiments, coordinate collection of surgical specimens, helped draft and prepare final manuscript. Aaron J. Krych—assisted with design of the study, provided funding for this project, provided surgical specimens for the study, helped revise and approve final manuscript.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
References
- 1.Alford JW, Cole BJ. Cartilage restoration, part 1: basic science, historical perspective, patient evaluation, and treatment options. Am J Sports Med. 2005;33:295–306. doi: 10.1177/0363546504273510. [DOI] [PubMed] [Google Scholar]
- 2.LaPrade RF, Botker J, Herzog M, et al. Refrigerated osteoarticular allografts to treat articular cartilage defects of the femoral condyles. A prospective outcomes study. J Bone Joint Surg. 2015;91:805–811. doi: 10.2106/JBJS.H.00703. [DOI] [PubMed] [Google Scholar]
- 3.Alford JW, Cole BJ. Cartilage restoration, part 2: techniques, outcomes, and future directions. Am J Sports Med. 2005;33:443–460. doi: 10.1177/0363546505274578. [DOI] [PubMed] [Google Scholar]
- 4.Krych AJ, Robertson CM, Williams RJ., 3rd Return to athletic activity after osteochondral allograft transplantation in the knee. Am J Sports Med. 2012;40:1053–1059. doi: 10.1177/0363546511435780. [DOI] [PubMed] [Google Scholar]
- 5.LaPrade RF, Botker JC. Donor-site morbidity after osteochondral autograft transfer procedures. Arthroscopy. 2004;20:e69–e73. doi: 10.1016/j.arthro.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 6.Versier G, Dubrana F. Treatment of knee cartilage defect in 2010. Orthop Traumatol Surg Res. 2011;97:S140–S153. doi: 10.1016/j.otsr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Vangsness CT, Jr, Triffon MJ, Joyce MJ. Soft tissue for allograft reconstruction of the human knee: a survey of the American Association of Tissue Banks. Am J Sports Med. 1996;24:230–234. doi: 10.1177/036354659602400221. [DOI] [PubMed] [Google Scholar]
- 8.Gortz S, Bugbee WD. Fresh osteochondral allografts: graft processing and clinical applications. J Knee Surg. 2006;19:231–240. doi: 10.1055/s-0030-1248112. [DOI] [PubMed] [Google Scholar]
- 9.Gross AE, Silverstein EA, Falk J, et al. The allotransplantation of partial joints in the treatment of osteoarthritis of the knee. Clin Orthop Relat Res. 1975;108:7–14. doi: 10.1097/00003086-197505000-00003. [DOI] [PubMed] [Google Scholar]
- 10.Sherman SL, Garrity J, Bauer K, et al. Fresh osteochondral allograft transplantation for the knee: current concepts. J Am Acad Orthop Surg. 2014;22:121–133. doi: 10.5435/JAAOS-22-02-121. [DOI] [PubMed] [Google Scholar]
- 11.Emmerson BC, Gortz S, Jamali AA, et al. Fresh osteochondral allografting in the treatment of osteochondritis dissecans of the femoral condyle. Am J Sports Med. 2007;35:907–914. doi: 10.1177/0363546507299932. [DOI] [PubMed] [Google Scholar]
- 12.Ghazavi MT, Pritzker KP, Davis AM, et al. Fresh osteochondral allografts for post-traumatic osteochondral defects of the knee. J Bone Joint Surg Br. 1997;79:1008–1013. doi: 10.1302/0301-620x.79b6.7534. [DOI] [PubMed] [Google Scholar]
- 13.Cook JL, Stoker AM, Stannard JP, et al. A novel system improves preservation of osteochondral allografts. Clin Orthop Relat Res. 2014;472:3404–3414. doi: 10.1007/s11999-014-3773-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LaPrade RF, Botker J, Herzog M, et al. Refrigerated osteoarticular allografts to treat articular cartilage defects of the femoral condyles. A prospective outcomes study. J Bone Joint Surg Am. 2009;91:805–811. doi: 10.2106/JBJS.H.00703. [DOI] [PubMed] [Google Scholar]
- 15.Pennock AT, Robertson CM, Wagner F, et al. Does subchondral bone affect the fate of osteochondral allografts during storage? Am J Sports Med. 2006;34:586–591. doi: 10.1177/0363546505281815. [DOI] [PubMed] [Google Scholar]
- 16.Dudakovic A, Camilleri E, Riester SM, et al. High-resolution molecular validation of self-renewal and spontaneous differentiation in clinical-grade adipose-tissue derived human mesenchymal stem cells. J Cell Biochem. 2014;115:1816–1828. doi: 10.1002/jcb.24852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D, Pertea G, Trapnell C, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Allen RT, Robertson CM, Pennock AT, et al. Analysis of stored osteochondral allografts at the time of surgical implantation. Am J Sports Med. 2005;33:1479–1484. doi: 10.1177/0363546505275010. [DOI] [PubMed] [Google Scholar]
- 21.Malinin T, Temple HT, Buck BE. Transplantation of osteochondral allografts after cold storage. J Bone Joint Surg Am. 2006;88:762–770. doi: 10.2106/JBJS.D.02991. [DOI] [PubMed] [Google Scholar]
- 22.Williams SK, Amiel D, Ball ST, et al. Analysis of cartilage tissue on a cellular level in fresh osteochondral allograft retrievals. Am J Sports Med. 2007;35:2022–2032. doi: 10.1177/0363546507305017. [DOI] [PubMed] [Google Scholar]
- 23.Pallante AL, Chen AC, Ball ST, et al. The in vivo performance of osteochondral allografts in the goat is diminished with extended storage and decreased cartilage cellularity. Am J Sports Med. 2012;40:1814–1823. doi: 10.1177/0363546512449321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linn MS, Chase DC, Healey RM, et al. Etanercept enhances preservation of osteochondral allograft viability. Am J Sports Med. 2011;39:1494–1499. doi: 10.1177/0363546511398645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallante AL, Bae WC, Chen AC, et al. Chondrocyte viability is higher after prolonged storage at 37 degrees C than at 4 degrees C for osteochondral grafts. Am J Sports Med. 2009;37:24S–32S. doi: 10.1177/0363546509351496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrity JT, Stoker AM, Sims HJ. Improved osteochondral allograft preservation using serum-free media at body temperature. Am J Sports Med. 2012;40:2542–2548. doi: 10.1177/0363546512458575. [DOI] [PubMed] [Google Scholar]
- 27.Dimicco MA, Kisiday JD, Gong H. Structure of pericellular matrix around agarose-embedded chondrocytes. Osteoarthritis Cartilage. 2007;15:1207–1216. doi: 10.1016/j.joca.2007.03.023. Epub 2007 May 23. [DOI] [PubMed] [Google Scholar]
- 28.Cao L, Lee V, Adams ME, et al. Beta-integrin-collagen interaction reduces chondrocyte apoptosis. Matrix Biol. 1999;18:343–355. doi: 10.1016/s0945-053x(99)00027-x. [DOI] [PubMed] [Google Scholar]
- 29.Cao L, Yang BB. Chondrocyte apoptosis induced by aggrecan G1 domain as a result of decreased cell adhesion. Exp Cell Res. 1999;246:527–537. doi: 10.1006/excr.1998.4335. [DOI] [PubMed] [Google Scholar]
- 30.Aizawa T, Kon T, Einhorn TA, et al. Induction of apoptosis in chondrocytes by tumor necrosis factor-alpha. J Orthop Res. 2001;19:785–796. doi: 10.1016/S0736-0266(00)00078-4. [DOI] [PubMed] [Google Scholar]
- 31.Asada S, Fukuda K, Nishisaka F, et al. Hydrogen peroxide induces apoptosis of chondrocytes; involvement of calcium ion and extracellular signal-regulated protein kinase. Inflamm Res. 2001;50:19–23. doi: 10.1007/s000110050719. [DOI] [PubMed] [Google Scholar]
- 32.Islam N, Haqqi TM, Jepsen KJ, et al. Hydrostatic pressure induces apoptosis in human chondrocytes from osteoarthritic cartilage through up-regulation of tumor necrosis factor-alpha, inducible nitric oxide synthase, p53, c-myc, and bax-alpha, and suppression of bcl-2. J Cell Biochem. 2002;87:266–278. doi: 10.1002/jcb.10317. [DOI] [PubMed] [Google Scholar]
- 33.Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 34.D’Lima D, Hermida J, Hashimoto S, et al. Caspase inhibitors reduce severity of cartilage lesions in experimental osteoarthritis. Arthritis Rheum. 2006;54:1814–1821. doi: 10.1002/art.21874. [DOI] [PubMed] [Google Scholar]
- 35.Lo MY, Kim HT. Chondrocyte apoptosis induced by collagen degradation: inhibition by caspase inhibitors and IGF-1. J Orthop Res. 2004;22:140–144. doi: 10.1016/S0736-0266(03)00117-7. [DOI] [PubMed] [Google Scholar]
- 36.Nuttall ME, Nadeau DP, Fisher PW, et al. Inhibition of caspase-3-like activity prevents apoptosis while retaining functionality of human chondrocytes in vitro. J Orthop Res. 2000;18:356–363. doi: 10.1002/jor.1100180306. [DOI] [PubMed] [Google Scholar]
- 37.Kim HT, Teng MS, Dang AC. Chondrocyte apoptosis: implications for osteochondral allograft transplantation. Clin Orthop Relat Res. 2008;466:1819–1825. doi: 10.1007/s11999-008-0304-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Molecular Cell. 2014;54:281–288. doi: 10.1016/j.molcel.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


