Abstract
Until recently, parasitic infections have been primarily studied as interactions between the parasite and the host, leaving out crucial players: microbes. The recent realization that microbes play key roles in the biology of all living organisms is not only challenging our understanding of host-parasite evolution, but it also provides new clues to develop new therapies and remediation strategies. In this paper we provide a review of promising and advanced experimental organismal systems to examine the dynamic of host-parasite-microbe interactions. We address the benefits of developing new experimental models appropriate to this new research area and identify systems that offer the best promises considering the nature of the interactions among hosts, parasites, and microbes. Based on these systems, we identify key criteria for selecting experimental models to elucidate the fundamental principles of these complex webs of interactions. It appears that no model is ideal and that complementary studies should be performed on different systems in order to understand the driving roles of microbes in host and parasite evolution.
Keywords: holobiont, symbioses, microbiome, microbe, mutualism, experimental models
Introduction
The term ‘symbiosis’ (from Greek “living together”) was first coined by Albert Bernhardt Frank in 1877, when he described the relationship between fungi and algae in lichens, and was then used to define “the living together of unlike organisms” by de Bary (1879) and Theis et al. (2016). The modern definition of symbiosis encompasses all beneficial, neutral, and harmful long-term and intimate relationships, which correspond to a continuum of mutualistic, communal, and parasitic symbiosis (Bronstein, 2009; Cheng, 2012). Symbionts can contribute to phenotypic variations and thus to evolutionary innovation in their hosts (Margulis, 1991). Recent developments in molecular techniques have propelled a vast expansion of our knowledge regarding the presence, diversity, and abundance of non-cultivable microbes. It has revealed that most often, organisms are not associated with one single symbiont, but with a myriad of microbial symbionts that can independently or synergistically participate in their physiology and evolution (Rosenberg and Zilber-Rosenberg, 2011). The recognition of this inherent complexity of organisms led to the concept of the holobiont that integrates the host organism and all its associated microbes (Margulis, 1991). Two divergent opinions have recently risen. Some argue that the term holobiont and the concepts associated to it can lead to misunderstanding of evolutionary concepts in host-associated microbes (Moran and Sloan, 2015; Douglas and Werren, 2016). Others maintain that this new concept is necessary to embrace the pluralistic attributes of any plants and animals, whose phenotype result from multilevel selection on the host genome, the microbe genomes, and the hologenome (Bordenstein and Theis, 2015; Theis et al., 2016). This complexity and its implications has renewed interest in symbionts and is revolutionizing many fields of biology including parasitology and immunology. Of interest in this review, host-parasite interactions are now being re-considered in light of the microbiome (Dheilly, 2014). This has led to a better understanding of the pluralistic role of microbes in host defense and to new theories of parasite virulence strategies driven by their interaction with microbes (Dheilly et al., 2015b). Host and parasite microbiomes contribute to host and parasite fitness and impose new selection pressures on parasites and hosts, respectively (Figure 1). Alteration of any partner may cause a chain of responses in those remaining and change the outcome of infection.
FIGURE 1.
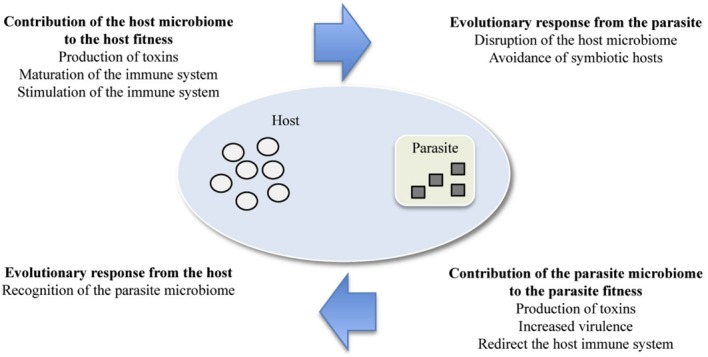
Contribution of the host and parasite microbiome to the host and parasite fitness and evolutionary responses expected from the parasite and host, respectively. The smaller symbols represent the microbes; the shape and color indicate whether it is associated with the host (white circle) or parasite (gray square).
Host Defensive Symbionts
Scientists have long recognized and accepted that mixed infection is the rule in natural environments and that microbes in the same host can directly and indirectly interfere with each other. Concomitant immunity was shown in Schistosomes (blood flukes) as early as 1969 and facilitation in 1971 (Kloetzel et al., 1971, 1977). We now know that resistance, competition, and facilitation can result from direct and indirect interactions among protozoa, helminthes, viruses, and bacteria (Cox, 2001). Thus, the net outcome of the host-parasite interaction can vary greatly in presence of other microbes, from harmful to neutral and beneficial. In particular, the concept of defensive mutualism led to the discovery of microbes acting in defense to parasites in many taxa, including fungi, mollusks, insects, plants, and vertebrates (Clay, 1988; White and Torres, 2009). Microbial symbionts are necessary for maturation of the immune system during early-life (Gollwitzer and Marsland, 2015) and later protect the host against pathogens via immune activation (Hall et al., 2008; Benson et al., 2009), production of antimicrobial factors, competition for resources or by limiting parasite adhesion to host cells (Cash, 2006; Kamada et al., 2012; Buffie and Pamer, 2013). Most importantly, microbe-mediated protection can evolve rapidly due to their large population size and short generation times, thus participating in host adaptation (King et al., 2016). In fact, symbionts may take over the evolutionary arms race against parasites, and alleviate the selection pressures on the host (Kwiatkowski et al., 2012), as demonstrated by the reduction in Drosophila melanogaster resistance to Drosophila C virus when associated with the defensive symbiont Wolbachia (Martinez et al., 2016a). The inter-regulation between microbiome and immune system has led to redefinition of this new unit that integrates host and microbe-encoded defenses into a single “holo-immunome” (Dheilly, 2014) or “microimmunosome” (Dietert, 2016).
A number of organismal systems were developed to study the role of defensive symbionts in the ecology and evolution of hosts and parasites. Many insects harbor microbial mutualists and studies of their role in defense to parasites have long been successful. Aphids have provided the most remarkable example of insect-associated defensive symbionts (see Oliver et al., 2014 for detailed review). In Acyrthosiphon pisum, the facultative symbionts Hamiltonella and Serratia provide defense against the parasitic wasp Aphidius ervi (Oliver et al., 2003, 2005) whereas Regiella provides resistance to the fungus Pandora neoaphidis (Scarborough et al., 2005). Because aphids have a parthenogenetic reproduction, clonal lines of aphids can be maintained indefinitely and the microbiome is manipulated by a combination of antibiotic treatments and micro-injection of symbionts. Aphids are thus ideal experimental models to understand the impact of defensive symbionts on the evolution of their hosts and parasites. For instance, it is employed to study the cost of defensive symbionts (Vorburger et al., 2013; Polin et al., 2014; Cayetano et al., 2015), their population dynamics (Oliver et al., 2008), their role in parasitoid community composition (Rothacher et al., 2016), the specificity of symbiont-associated defense (Martinez et al., 2016b), and to demonstrate that host symbiont’s role in defense imposes new selection pressure on parasites (Oliver et al., 2012). Another biological system worth mentioning is the role of Wolbachia against viruses. Wolbachia is a reproductive parasite of the fruit fly Drosophila melanogaster (Fry et al., 2004) but it also imparts resistance to Drosophila C viruses, Cricket Paralysis Virus, Flock house virus, Nora virus, and West Nile Virus by reducing the viral load (Hedges et al., 2008; Teixeira et al., 2008; Osborne et al., 2009; Glaser and Meola, 2010). Knowledge gained from studying this organismal system has led to the discovery of a Wolbachia-induced resistance to many human parasites including Dengue virus, Chikungunya virus, Yellow fever virus, West Nile Virus, and malaria parasites in mosquitoes (Moreira et al., 2009; Bian et al., 2010; Van den Hurk et al., 2012). Clearly, integrating the role of defensive symbionts into our understanding of vector competence has implications for the control of disease transmission. For example, Wolbachia-infected mosquitoes have been released in natural environments as a preventative measure to limit the prevalence of Dengue virus (Hoffmann et al., 2011; Iturbe-Ormaetxe et al., 2011). The diversity of insects carrying defensive symbionts provides opportunities to answer many including the extent of host system reliance on defensive symbionts, the role of competition among symbionts, or the potential co-diversification of defensive symbionts with parasites.
In vertebrates, emerging evidence shows that microbes inhabiting many sites in the body have a role in maintaining health even though few studies examine the direct role of defensive symbionts in resistance against parasites. Promising experimental systems include the red-back salamander, Plethodon cinereus that hosts cutaneous, antifungal metabolite-producing bacteria, which prevent infection by pathogenic fungi Batrachochytrium dendrobatidis (Brucker et al., 2007, 2008). In birds, Upupa epops and Phoeniculus purpureus, the uropygial glands house symbiotic bacteria that secrete protective antimicrobial substances providing protection to the feathers and eggs (Martín-Platero et al., 2006; Ruiz-Rodriguez et al., 2009; Martín-Vivaldi et al., 2010, 2014; Ruiz-Rodríguez et al., 2013). Further studies of these tractable vertebrate biological systems are necessary to characterize the significance of the increased resistance and test ecological and evolutionary theories that are mostly developed on invertebrate systems.
Parasite Disruptive Strategies
Many parasites share the same environment as host-associated microbes, including defensive symbionts. Interestingly, parasites from all major phyla, including helminthes, viruses, bacteria, and fungi modify their host microbiome. For example, parasitic helminth infection results in significant modifications of gut microbiome in humans, pigs, cattle, hamsters, goats, and sheep (Nicholls, 1987; Li et al., 2011, 2012, 2016; Wu et al., 2012; Plieskatt et al., 2013; Cantacessi et al., 2014; Lee et al., 2014). But it is unknown if the modifications in the composition of the host microbiome are beneficial for the parasite, a defense mechanism of the host, or by-products of the pathology. It was proposed that parasites can directly modify their microbial environment to make it more favorable, or to exercise immune-modulatory activity and reduce the host resistance to infection, a process called the disruptive strategy (Dheilly et al., 2015b). In helminths, evidence suggests that alteration of the host microbiome indeed participates in parasite strategies to modulate the host immune system (Plieskatt et al., 2013). This theory is also supported by the observation that hatching by the nematode Trichuris muris is dependent on an abundant microflora (Hayes et al., 2010; Kane et al., 2011; Kuss et al., 2011) but the beneficial role of parasite-associated disruption of the host microbiome remains to be experimentally tested in a tractable system. Trophically transmitted parasites such as cestodes, are of interest because they have the potential to interact with microbes associated with both definitive and intermediate hosts. Triaenophorus nodulosus and Eubortium rugosum have been found to physically interact with many gut bacteria of their intermediate hosts, Esox Lucius (pike) and Lota lota (burbot), respectively, (Izvekova and Lapteva, 2004). In the rat, Hymenolepis diminuta infection results in a significant modification of the gut microbiome composition and a stronger stability in response to a mild inflammatory challenge (McKenney et al., 2015). The cestode parasite, Schistocephalus solidus, appears as a model of choice to investigate cestode parasite interactions with host gut microbes because all parasite stages can be cultured in vitro by mimicking either the avian intestinal environment, or the copepod or the stickleback fish body cavity (Smyth, 1990; Jakobsen et al., 2012), which allows rigorous functional experiments. Of interest, the highly specific second intermediate host of S. solidus, the threespine stickleback has a broad geographical variation (Hagen and Gilbertson, 1972) and has become a model species in biology for both ecological and evolutionary studies of the interaction (Barber and Nettleship, 2010).
While most microbiome disruptions by parasites result in subtle changes with unknown consequences for the outcome of infection, some have spectacular consequences. Vibrio shiloi, a bacterium infecting the coral host Oculina patagonica, produces toxins specifically targeting the coral’s symbiotic zooxanthellae (Kushmaro et al., 1997, 1998, 2001). Following the temperature-dependent attachment to the coral surface, V. shiloi infiltrates the tissue and multiplies rapidly (Kushmaro et al., 1998; Toren et al., 1998; Banin et al., 2000). The bacterium then releases photosynthesis-inhibiting toxins and lyses zooxanthellae, eventually leading to coral bleaching and death (Ben-Haim et al., 1999). In another key example, the microsporidian parasite Paranosema locustae relies on gut acidification of its locust host (Locusta migratoria manilensis) and increased production of reactive oxygen species to modify the gut microbial community and insure its own development (Shi et al., 2014). Because the locust microbiome is involved in producing pheromones responsible for the locust swarming behavior, infection by P. locustae prevents swarming (Dillon et al., 2000; Shi et al., 2014). The adaptive value and specificity of V. shiloi and P. locustae for their host-associated microbes remain to be demonstrated but the obvious phenotypic changes associated with the microbiome disruption increases the value of these organismal systems. In addition, they provide the opportunity to study evolutionary trade-offs of parasite induced disruption of the host microbiome.
Parasite Biological Weapon Strategies
The presence of microbes associated with hosts has been demonstrated in many systems, but the presence of microbes associated with parasites has been tested less frequently. For example, Wolbachia is an obligate bacterial symbiont of parasitic filarial nematodes (Kozek, 1977; Hoerauf et al., 1999). Wolbachia incite inflammatory reactions in the human host and it has been implicated in many debilitating diseases (Taylor and Hoerauf, 1999). For instance, Wolbachia-induced inflammation is responsible for the loss of vision and blindness following O. volvulus infection (Francis et al., 1985; Hall and Pearlman, 1999; Saint André et al., 2002) and for the inflammation of lymphatic vessels following W. bancrofti infection (Dreyer et al., 2000; Hoerauf et al., 2003). Similarly, the trematode Opisthorchis viverrini, also known as liver fluke, often leads to cholangiocarcinoma (Braconi and Patel, 2010). Recent studies demonstrated that O. viverrini is associated with a complex microbiome (Plieskatt et al., 2013) and that some of these bacteria are transmitted to the host and present in abundance in tumors, calling for more basic experiments to determine their contribution in cancer development (Chng et al., 2016; Osborne and WegenerParfrey, 2016). Other parasites are associated with viruses, such as Trichomonas vaginalis, a protozoan parasite that infects the human vagina (Goodman et al., 2011). Approximately half of all clinical samples carry a dsRNA virus called Trichomonavirus that is recognized by the human immune system and causes an inflammatory response (Fichorova et al., 2012) responsible for increased susceptibility to sexually transmitted diseases (Van Der Pol et al., 2008), infertility (Grys, 1972; Grodstein et al., 1993), miscarriage (Minkoff et al., 1984), pre-term delivery (Hardy et al., 1984; Minkoff et al., 1984), low birth weight (Hardy et al., 1984), and cervical cancer (Gram et al., 1992; Kharsany et al., 1993). The presence of the Trichomonavirus also explains why symptoms can worsen upon successful antibiotic treatment: the dying parasite releases its viral load within the human vagina (Fichorova et al., 2012). Notably, parasites from various phyla are associated with viruses, including trematodes and parasitic protozoans but their impact on the host has been rarely investigated (Ip and Desser, 1984; Wang and Wang, 1991; Ives et al., 2011). Clearly, the significance of parasite-virus and parasite-bacteria associations for treatment of the diseases is evident. But, what is the significance of the phenomenon for the parasites themselves? It has been proposed that parasites can utilize symbionts to increase their virulence, a process called the biological weapon strategy (Dheilly et al., 2015b). Clearly, further experimental studies are needed to determine if symbiotic bacteria or viruses and associated inflammation are beneficial for the parasite.
A promising tractable experimental system involves the parasitoid wasp, Dinocampus coccinellae that infects ladybeetles and carries a RNA virus, D. coccinellae paralysis virus (DcPV; Dheilly et al., 2015a). In adult wasps, DcPV is stored in the oviduct. The wasp injects its eggs into the body cavity of the ladybeetle and the growing larvae feed off the ladybeetle’s fat body. During larval development, the virus replicates actively and is transmitted to the ladybeetle. DcPV then targets the ladybeetle nervous tissue. When later a single mature larva egresses from the host, DcPV replication in the ladybeetle nervous system results in the induction of antiviral immune responses and damage to the nervous tissue associated with ladybeetle paralysis. The larva stills between the legs of the immobilized ladybeetle and spins a cocoon within which it pupates into an adult wasp. The ladybeetle appears crouched protectively over the cocoon of the wasp and makes erratic movements efficiently deterring predators (Maure et al., 2011). Upon larva pupation, clearance of DcPV and nervous system regeneration results in the ladybeetle recovery of normal behavior (Dheilly et al., 2015a). Therefore, it appears that D. coccinellae uses DcPV as a biological weapon to manipulate its ladybeetle host and protect its progeny from predation (Maure et al., 2011; Dheilly et al., 2015a). This organismal system appears ideal to investigate the role of associated viruses in the evolution of parasites because of the short generation time of host and parasite, the broad geographic range of the parasite that infect ladybeetle species on all continents, and because of the possibility to track variations in virus genome in experimental evolution experiments. Most interestingly, ladybeetles are also associated with a diverse array of bacteria including multiple members of the genus Flavobacteria, Rickettsia, Spiroplasma, and Wolbachia, that may be involved in ladybeetle defense against the parasitoid (Hurst et al., 1994, 1997; Weinert et al., 2007; Elnagdy et al., 2013). Using techniques developed previously to study the defensive role of aphid symbionts, the microbiome of ladybeetles could be manipulated using a combination of antibiotic treatments and microinjections. Thus, this organismal system provides an opportunity to investigate the possibility for direct interactions between host-associated microbes and parasite-associated microbes within the body cavity of the host.
Conclusion
Understanding the role of microbes in host and parasite ecology and evolution is important for both basic and applied biology, as so many disease-causing organisms carry their own microbes or interact with host-associated microbes. Empirical studies of host-parasite-microbes interactions at a mechanistic and evolutionary level can help us understand the ecological circumstances where symbiosis benefits host or parasite fitness. The current review of experimental systems enabling studies of host-parasite-microbe interactions has allowed us to identify key criteria to improve upon and further study these complex interactions (Box 1, Supplementary Table S1). We would like to emphasize that it is now crucial to investigate the role of community of microbes as well as individual microbial symbionts. Indeed, microbes may independently, synergistically, or antagonistically impact and participate in the physiology and evolution of their hosts Therefore, it is of particular importance to develop models where it is possible to intimately manipulate the host and parasite microbiomes to alter their composition. We expect that future research project will couple such manipulation with new sequencing technologies, proteomics or metabolomics to elucidate the molecular dialog between the different partners, as demonstrated in studies of host-symbiont and host-microbiome interactions (Chaston and Douglas, 2012; Rooks and Garrett, 2016). Development of these systems can provide a basis for developing new therapeutic and prevention strategies: vector hosts could be inoculated with defensive symbionts to reduce the transmission rate of a parasite; Parasite-associated microbes could be targeted to limit parasite development and allow its elimination by the host immune system; New parasite-specific probiotics could be selected to increase resistance or tolerance to parasitic infection. Developing these experimental systems for host-parasite-microbes’ interactions is now critical to determine which partner in the interaction should be targeted for effective therapy. Finally, investigating the role of microbes and microbiomes in host-parasite interactions may provide opportunities to confirm or infirm the hologenome theory of evolution by demonstrating multi-level selection.
BOX 1 | Key Criteria for experimental model evaluation.
-
(1)
The host and parasite could be bred and maintained in the laboratory. It is a necessary step to perform controlled experimental infections and understand the transmission mechanisms of symbionts of interest. It is also crucial for testing the environmental and host genetic factors that regulate the abundance, diversity and stability of the microbiome.
-
(2)
A core microbiome would have been identified in the host suggesting that it selects for specific microbes. This in itself suggests that the host should be considered as a holobiont.
-
(3)
The parasite shares the same environment as microbes in the host body, which would suggest potential direct parasite-host-associated microbe interaction that remain to be tested.
-
(4)
The parasite would be associated with microbes for which the role in virulence will be assessed. The presence of a core microbiome in parasites and the role of parasite-associated microbes in driving the evolution of host defenses remain to be demonstrated.
-
(5)
Techniques have been developed to manipulate the composition of the microbiome of the host and/or parasite. The role of microbes in the result of infection can only be tested through manipulation of the microbiome and injection of isolated microbes. It is necessary to characterize the role of microbes individually and as a community in susceptibility, resistance, and infectivity.
-
(6)
The host and the parasite would have a wide geographical species range and ecological diversity, which would allow comprehensive fields studies to understand how environmental factors influence host-parasite-microbe interaction. It would allow to examine the role of microbes in local adaptation of parasites through cross-infection experiments.
-
(7)
The host and/or parasite would have short generation time, which would facilitate the use experimental evolution to test the role of microbes in host or parasite adaptation. For instance, the hologenome theory would predict that associated microbes participate in the arms race through shorter generation time and higher mutation rate but this hypothesis remains to be tested.
-
(8)
Closely related species of hosts could be parasitized with closely related species of parasites, thus allowing comparative studies of the role of microbes in the interaction and testing their role during parasite evolution, host switch, and host or parasite speciation.
-
(9)
The parasite would have various effects on the phenotype of its host, so that the role of microbes in any aspect of the ’normal’ behavior or appearance of infected organism may be more readily detected and more reliably assessed for fitness consequences.
-
(10)
The genome of the host and parasite would have been sequenced, facilitating the use of high throughput comparative genomics approaches to characterize the genes involved in
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. The authors acknowledge support from Stony Brook University.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01300
References
- Banin E., Israely T., Kushmaro A., Loya Y., Orr E., Rosenberg E. (2000). Penetration of the coral-bleaching bacterium Vibrio shiloi into Oculina patagonica. Appl. Environ. Microbiol. 66 3031–3036. 10.1128/AEM.66.7.3031-3036.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber I., Nettleship S. (2010). From ‘trash fish’to supermodel: the rise and rise of the three-spined stickleback in evolution and ecology. Biologist 57 15–21. [Google Scholar]
- Benson A., Pifer R., Behrendt C. L., Hooper L. V., Yarovinsky F. (2009). Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe 6 187–196. 10.1016/j.chom.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Haim Y., Banim E., Kushmaro A., Loya Y., Rosenberg E. (1999). Inhibition of photosynthesis and bleaching of zooxanthellae by the coral pathogen Vibrio shiloi. Appl. Environ. Microbiol. 1 223–229. 10.1046/j.1462-2920.1999.00027.x [DOI] [PubMed] [Google Scholar]
- Bian G., Xu Y., Lu P., Xie Y., Xi Z. (2010). The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS Pathog. 6:e1000833 10.1371/journal.ppat.1000833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein S. R., Theis K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226 10.1371/journal.pbio.1002226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braconi C., Patel T. (2010). Cholangiocarcinoma: new insights into disease pathogenesis and biology. Infect. Dis. Clin. North Am. 24 871–884. 10.1016/j.idc.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein J. L. (2009). The evolution of facilitation and mutualism. J. Ecol. 97 1160–1170. 10.1111/j.1365-2745.2009.01566.x [DOI] [Google Scholar]
- Brucker R. M., Baylor C. M., Walters R. L., Lauer A., Harris R. N., Minbiole K. P. C. (2007). The identification of 2,4-diacetylphloroglucinol as an antifungal metabolite produced by cutaneous bacteria of the salamander Plethodon cinereus. J. Chem. Ecol. 34 39–43. 10.1007/s10886-007-9352-8 [DOI] [PubMed] [Google Scholar]
- Brucker R. M., Harris R. N., Schwantes C. R., Gallaher T. N., Flaherty D. C., Lam B. A., et al. (2008). Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J. Chem. Ecol. 34 1422–1429. 10.1007/s10886-008-9555-7 [DOI] [PubMed] [Google Scholar]
- Buffie C. G., Pamer E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13 790–801. 10.1038/nri3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantacessi C., Giacomin P., Croese J., Zakrzewski M., Sotillo J., McCann L., et al. (2014). Impact of experimental hookworm infection on the human gut microbiota. J. Infect. Dis. 210 1431–1434. 10.1093/infdis/jiu256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash H. L. (2006). The Ligand and Function of the RegIII Family of Bactericidal C-Type Lectins. Master’s thesis, University of Texas Southwestern Medical Center, Dallas, TX. [Google Scholar]
- Cayetano L., Rothacher L., Simon J.-C., Vorburger C. (2015). Cheaper is not always worse: strongly protective isolates of a defensive symbiont are less costly to the aphid host. Proc. R. Soc.B 282 20142333 10.1098/rspb.2014.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaston J., Douglas A. (2012). Making the most of “omics” for symbiosis research. Biol. Bull. 223 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T. C. (2012). General Parasitology. Amsterdam: Elsevier. [Google Scholar]
- Chng K. R., Chan S. H., Ng A. H. Q., Li C., Jusakul A., Bertrand D., et al. (2016). Tissue microbiome profiling identifies an enrichment of specific enteric bacteria in Opisthorchis viverrini associated cholangiocarcinoma. EBioMedicine 8 195–202. 10.1016/j.ebiom.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clay K. (1988). Fungal endophytes of grasses: a defensive mutualism between plants and fungi. Ecology 69 10–16. 10.2307/1943155 [DOI] [Google Scholar]
- Cox F. E. G. (2001). Concomitant infections, parasites and immune responses. Parasitology 122 S23–S38. 10.1017/S003118200001698X [DOI] [PubMed] [Google Scholar]
- de Bary A. (1879). Die Erscheinung der Symbiose: Vortrag gehalten auf der Versammlung Deutscher Naturforscher und Aerzte zu Cassel. Strassburg: Verlag von Karl J. Trübner. [Google Scholar]
- Dheilly N. M. (2014). Holobiont–Holobiont interactions: redefining host–parasite interactions. PLoS Pathog. 10:e1004093 10.1371/journal.ppat.1004093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheilly N. M., Maure F., Ravallec M., Galinier R., Doyon J., Duval D., et al. (2015a). Who is the puppet master? Replication of a parasitic wasp-associated virus correlates with host behaviour manipulation. Proc. R. Soc. B 282 20142773 10.1098/rspb.2014.2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheilly N. M., Poulin R., Thomas F. (2015b). Biological warfare: microorganisms as drivers of host–parasite interactions. Infect. Genet. Evol. 34 251–259. 10.1016/j.meegid.2015.05.027 [DOI] [PubMed] [Google Scholar]
- Dietert R. R. (2016). The microbiome-immune-host defense barrier complex (microimmunosome) and developmental programming of noncommunicable diseases. Reprod. Toxicol. (in press) 10.1016/j.reprotox.2016.04.026 [DOI] [PubMed] [Google Scholar]
- Dillon R. J., Vennard C. T., Charnley A. K. (2000). Pheromones: exploitation of gut bacteria in the locust. Nature 403 851–851. 10.1038/35002669 [DOI] [PubMed] [Google Scholar]
- Douglas A. E., Werren J. H. (2016). Holes in the hologenome: why host-microbe symbioses are not holobionts. mBio 7 e02099 10.1128/mBio.02099-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer G., Noroes J., Figueredo-Silva J., Piessens W. (2000). Pathogenesis of lymphatic disease in bancroftian filariasis: a clinical perspective. Parasitol. Today. 16 544–548. 10.1016/S0169-4758(00)01778-6 [DOI] [PubMed] [Google Scholar]
- Elnagdy S., Messing S., Majerus M. E. N. (2013). Two strains of male-killing Wolbachia in a ladybird, Coccinella undecimpunctata, from a hot climate. PLoS ONE 8:e54218 10.1371/journal.pone.0054218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichorova R. N., Lee Y., Yamamoto H. S., Takagi Y., Hayes G. R., Goodman R. P., et al. (2012). Endobiont viruses sensed by the human host–beyond conventional antiparasitic therapy. PLoS ONE 7:e48418 10.1371/journal.pone.0048418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis H., Awadzi K., Ottesen E. (1985). The Mazzotti reaction following treatment of onchocerciasis with diethylcarbamazine: clinical severity as a function of infection intensity. Am. J. Trop. Med. Hyg. 34 529–536. [DOI] [PubMed] [Google Scholar]
- Fry A. J., Palmer M. R., Rand D. M. (2004). Variable fitness effects of Wolbachia infection in Drosophila melanogaster. Heredity (Edinb) 93 379–389. 10.1038/sj.hdy.6800514 [DOI] [PubMed] [Google Scholar]
- Glaser R. L., Meola M. A. (2010). The native Wolbachia endosymbionts of Drosophila melanogaster and Culex quinquefasciatus increase host resistance to West Nile virus infection. PLoS ONE 5:e11977 10.1371/journal.pone.0011977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollwitzer E. S., Marsland B. J. (2015). Impact of early-life exposures on immune maturation and susceptibility to disease. Trends Immunol. 36 684–696. 10.1016/j.it.2015.09.009 [DOI] [PubMed] [Google Scholar]
- Goodman R. P., Ghabrial S. A., Fichorova R. N., Nibert M. L. (2011). Trichomonasvirus: a new genus of protozoan viruses in the family Totiviridae. Arch. Virol. 156 171–179. 10.1007/s00705-010-0832-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gram I. T., Macaluso M., Churchill J., Stalsberg H. (1992). Trichomonas vaginalis (TV) and human papillomavirus (HPV) infection and the incidence of cervical intraepithelial neoplasia (CIN) grade III. Cancer Causes Control 3 231–236. 10.1007/BF00124256 [DOI] [PubMed] [Google Scholar]
- Grodstein F., Goldman M. B., Cramer D. W. (1993). Relation of tubal infertility to history of sexually transmitted diseases. Am. J. Epidemiol. 137 577–584. [DOI] [PubMed] [Google Scholar]
- Grys E. (1972). Diagnosis of trichomonadosis in the urinary tract of women (preliminary report). Ann. Parasitol. 19 461–462. [PubMed] [Google Scholar]
- Hagen D., Gilbertson L. (1972). Geographic variation and environmental selection in Gasterosteus aculeatus L. in the Pacific Northwest, America. Evolution 26 32–51. 10.2307/2406981 [DOI] [PubMed] [Google Scholar]
- Hall J. A., Bouladoux N., Sun C. M., Wohlfert E. A., Blank R. B., Zhu Q., et al. (2008). Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity 29 637–649. 10.1016/j.immuni.2008.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. R., Pearlman E. (1999). Pathogenesis of onchocercal keratitis (river blindness). Clin. Microbiol. Rev. 12 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy P., Nell E. E., Spence M., Hardy J., Graham D., Rosenbaum R. (1984). Prevalence of six sexually transmitted disease agents among pregnant inner-city adolescents and pregnancy outcome. Lancet. 324 333–337. 10.1016/S0140-6736(84)92698-9 [DOI] [PubMed] [Google Scholar]
- Hayes K., Bancroft A., Goldrick M., Portsmouth C., Roberts I., Grencis R. (2010). Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science 328 1391–1394. 10.1126/science.1187703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges L. M., Brownlie J. C., O’Neill S. L., Johnson K. N. (2008). Wolbachia and virus protection in insects. Science 322 702–702. 10.1126/science.1162418 [DOI] [PubMed] [Google Scholar]
- Hoerauf A., Mand S., Volkmann L., Büttner M., Marfo-Debrekyei Y., Taylor M., et al. (2003). Doxycycline in the treatment of human onchocerciasis: kinetics of Wolbachia endobacteria reduction and of inhibition of embryogenesis in female Onchocerca worms. Microbes Infect. 5 261–273. 10.1016/S1286-4579(03)00026-1 [DOI] [PubMed] [Google Scholar]
- Hoerauf A., Nissen-Pähle K., Schmetz C., Henkle-Dührsen K., Blaxter M. L., Büttner D. W., et al. (1999). Tetracycline therapy targets intracellular bacteria in the filarial nematode Litomosoides sigmodontis and results in filarial infertility. J. Clin. Invest. 103 11–18. 10.1172/JCI4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Montgomery B., Popovici J., Iturbe-Ormaetxe I., Johnson P., Muzzi F., et al. (2011). Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476 454–457. 10.1038/nature10356 [DOI] [PubMed] [Google Scholar]
- Hurst G. D. D., Hammarton T. C., Bandi C., Majerus T. M. O., Bertrand D., Majerus M. E. N. (1997). The diversity of inherited parasites of insects: the male-killing agent of the ladybird beetle Coleomegilla maculata is a member of the Flavobacteria. Genet. Res. 70 1–6. 10.1017/S0016672397002838 [DOI] [Google Scholar]
- Hurst G. D. D., Purvis E. L., Sloggett J. J., Majerus M. E. N. (1994). The effect of infection with male-killing Rickettsia on the demography of female Adalia bipunctata L. (two spot ladybird). Heredity 73 309–316. 10.1038/hdy.1994.138 [DOI] [Google Scholar]
- Ip H. S., Desser S. S. (1984). A picornavirus-like pathogen of Cotylogaster occidentalis (Trematoda: Aspidogastrea), an intestinal parasite of freshwater mollusks. J. Invertebr. Pathol. 43 197–206. 10.1016/0022-2011(84)90138-1 [DOI] [Google Scholar]
- Iturbe-Ormaetxe I., Walker T., O’ Neill S. L. (2011). Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 12 508–518. 10.1038/embor.2011.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives A., Ronet C., Prevel F., Ruzzante G., Fuertes-Marraco S., Schutz F., et al. (2011). Leishmania RNA virus controls the severity of Mucocutaneous leishmaniasis. Science 331 775–778. 10.1126/science.1199326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izvekova G., Lapteva N. (2004). Microflora associated with the digestive-transport surfaces of fish and their parasitic cestodes. Russ. J. Ecol. 35 176–180. 10.1023/B:RUSE.0000025968.97632.30 [DOI] [Google Scholar]
- Jakobsen P., Scharsack J., Hammerschmidt K., Deines P., Kalbe M., Milinski M. (2012). In vitro transition of Schistocephalus solidus (Cestoda) from coracidium to procercoid and from procercoid to plerocercoid. Exp. Parasitol. 130 267–273. 10.1016/j.exppara.2011.09.009 [DOI] [PubMed] [Google Scholar]
- Kamada N., Kim Y.-G., Sham H. P., Vallance B. A., Puente J. L., Martens E. C., et al. (2012). Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336 1325–1329. 10.1126/science.1222195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M., Case L. K., Kopaskie K., Kozlova A., MacDearmid C., Chervonsky A. V., et al. (2011). Successful transmission of a retrovirus depends on the commensal microbiota. Science 334 245–249. 10.1126/science.1210718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharsany A., Hoosen A., Moodley J., Bagaratee J., Gouws E. (1993). The association between sexually transmitted pathogens and cervical intra-epithelial neoplasia in a developing community. Genitourin. Med. 69 357–360. 10.1136/sti.69.5.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King K. C., Brockhurst M. A., Vasieva O., Paterson S., Betts A., Ford S. A., et al. (2016). Rapid evolution of microbe-mediated protection against pathogens in a worm host. ISME J. 10 1915–1924. 10.1038/ismej.2015.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloetzel K., Chieffi P., Faleiros J. (1977). Mortality and other parameters of concomitant infections in albino mice: the Schistosoma-Toxoplasma model. Trop. Geogr. Med. 29 407–410. [PubMed] [Google Scholar]
- Kloetzel K., Faleiros J. J., Mendes S. R. (1971). Concurrent infection of white mice with T. cruzi and S. mansoni. Trans. R. Soc. Trop. Med. Hyg. 65 530–531. 10.1016/0035-9203(71)90167-2 [DOI] [PubMed] [Google Scholar]
- Kozek W. J. (1977). Transovarially-transmitted intracellular microorganisms in adult and larval stages of Brugia malayi. J. Parasitol. 63 992–1000. 10.2307/3279832 [DOI] [PubMed] [Google Scholar]
- Kushmaro A., Banin E., Loya Y., Stackebrandt E., Rosenberg E. (2001). Vibrio shiloi sp. nov., the causative agent of bleaching of the coral Oculina patagonica. Int. J. Syst. Evol. Microbiol. 51 1383–1388. 10.1099/00207713-51-4-1383 [DOI] [PubMed] [Google Scholar]
- Kushmaro A., Rosenberg E., Fine M., Ben Haim Y., Loya Y. (1998). Effect of temperature on bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar. Ecol. Prog. Ser. 171 131–137. 10.3354/meps171131 [DOI] [Google Scholar]
- Kushmaro A., Rosenberg E., Fine M., Loya Y. (1997). Bleaching of the coral Oculina patagonica by Vibrio AK-1. Mar. Ecol. Prog. Ser. 147 159–165. 10.3354/meps147159 [DOI] [Google Scholar]
- Kuss S. K., Best G. T., Etheredge C. A., Pruijssers A. J., Frierson J. M., Hooper L. V., et al. (2011). Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334 249–252. 10.1126/science.1211057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski M., Engelstädter J., Vorburger C. (2012). On genetic specificity in symbiont-mediated host-parasite coevolution. PLoS Comput. Biol. 8:e1002633 10.1371/journal.pcbi.1002633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. C., San Tang M., Lim Y. A., Choy S. H., Kurtz Z. D., Cox L. M., et al. (2014). Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl. Trop. Dis. 8:e2880 10.1371/journal.pntd.0002880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. W., Li W., Sun J., Yu P., Baldwin R. L., Urban J. F. (2016). The effect of helminth infection on the microbial composition and structure of the caprine abomasal microbiome. Sci. Rep. 6:20606 10.1038/srep20606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. W., Wu S., Li W., Huang Y., Gasbarre L. C. (2011). Metagenome plasticity of the bovine abomasal microbiota in immune animals in response to Ostertagia ostertagi infection. PLoS ONE 6:e24417 10.1371/journal.pone.0024417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. W., Wu S., Li W., Navarro K., Couch R. D., Hill D., et al. (2012). Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect. Immun. 80 2150–2157. 10.1128/IAI.00141-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis L. (1991). “Symbiogenesis and Symbionticism,” in Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis, eds Margulis L., Fester R. (Cambridge, MA: MIT Press; ), 1–13. [PubMed] [Google Scholar]
- Martinez J., Cogni R., Cao C., Smith S., Illingworth C. J., Jiggins F. M. (2016a). Addicted? Reduced host resistance in populations with defensive symbionts. Proc. R. Soc. B 283 20160778 10.1098/rspb.2016.0778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A. J., Kim K. L., Harmon J. P., Oliver K. M. (2016b). Specificity of multi-modal aphid defenses against two rival parasitoids. PLoS ONE 11:e0154670 10.1371/journal.pone.0154670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Platero A. M., Valdivia E., Ruíz-Rodríguez M., Soler J. J., Martín-Vivaldi M., Maqueda M., et al. (2006). Characterization of antimicrobial substances produced by Enterococcus faecalis MRR 10-3, isolated from the uropygial gland of the hoopoe (Upupa epops). Appl. Environ. Microbiol. 72 4245–4249. 10.1128/AEM.02940-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Vivaldi M., Peña A., Peralta-Sánchez J. M., Sánchez L., Ananou S., Ruiz-Rodríguez M., et al. (2010). Antimicrobial chemicals in hoopoe preen secretions are produced by symbiotic bacteria. Proc. R. Soc. B 277 123–130. 10.1098/rspb.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Vivaldi M., Soler J. J., Peralta-Sánchez J. M., Arco L., Martín-Platero A. M., Martínez-Bueno M., et al. (2014). Special structures of hoopoe eggshells enhance the adhesion of symbiont-carrying uropygial secretion that increase hatching success. J. Anim. Ecol. 83 1289–1301. 10.1111/1365-2656.12243 [DOI] [PubMed] [Google Scholar]
- Maure F., Brodeur J., Ponlet N., Doyon J., Firlej A., Elguero É, et al. (2011). The cost of a bodyguard. Biol. Lett. 7 843–846. 10.1098/rsbl.2011.0415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney E. A., Williamson L., Yoder A. D., Rawls J. F., Bilbo S. D., Parker W. (2015). Alteration of the rat cecal microbiome during colonization with the helminth Hymenolepis diminuta. Gut Microbes 6 182–193. 10.1080/19490976.2015.1047128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkoff H., Grunebaum A. N., Schwarz R. H., Feldman J., Cummings M., Crombleholme W., et al. (1984). Risk factors for prematurity and premature rupture of membranes: a prospective study of the vaginal flora in pregnancy. Am. J. Obstet. Gynecol. 150 965–972. 10.1016/0002-9378(84)90392-2 [DOI] [PubMed] [Google Scholar]
- Moran N. A., Sloan D. B. (2015). The hologenome concept: helpful or hollow? PLoS Biol. 13:e1002311 10.1371/journal.pbio.1002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira L. A., Iturbe-Ormaetxe I., Jeffery J. A., Lu G., Pyke A. T., Hedges L. M., et al. (2009). A Wolbachia symbiont in Aedes aegypti limits infection with dengue, Chikungunya, and Plasmodium. Cell 139 1268–1278. 10.1016/j.cell.2009.11.042 [DOI] [PubMed] [Google Scholar]
- Nicholls C. (1987). Endoscopy, Physiology and Bacterial Flora of Sheep Infected with Abomasal Nematodes. Ph.D. thesis, University of Leeds, Leeds. [Google Scholar]
- Oliver K. M., Campos J., Moran N. A., Hunter M. S. (2008). Population dynamics of defensive symbionts in aphids. Proc. R. Soc. B 275 293–299. 10.1098/rspb.2007.1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K. M., Moran N. A., Hunter M. S. (2005). Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl. Acad. Sci. 102 12795–12800. 10.1073/pnas.0506131102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K. M., Noge K., Huang E. M., Campos J. M., Becerra J. X., Hunter M. S. (2012). Parasitic wasp responses to symbiont-based defense in aphids. BMC Biol. 10:11 10.1186/1741-7007-10-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K. M., Russell J. A., Moran N. A., Hunter M. S. (2003). Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc. Natl. Acad. Sci. 100 1803–1807. 10.1073/pnas.0335320100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver K. M., Smith A. H., Russell J. A. (2014). Defensive symbiosis in the real world–advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 28 341–355. 10.1111/1365-2435.12133 [DOI] [Google Scholar]
- Osborne L. C., WegenerParfrey L. (2016). Liver flukes and the microbiota in cancer. EBioMedicine 8 12–13. 10.1016/j.ebiom.2016.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne S. E., Leong Y. S., O’Neill S. L., Johnson K. N. (2009). Variation in antiviral protection mediated by different Wolbachia strains in Drosophila simulans. PLoS Pathog. 5:e1000656 10.1371/journal.ppat.1000656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plieskatt J. L., Deenonpoe R., Mulvenna J. P., Krause L., Sripa B., Bethony J. M., et al. (2013). Infection with the carcinogenic liver fluke Opisthorchis viverrini modifies intestinal and biliary microbiome. FASEB J. 27 4572–4584. 10.1096/fj.13-232751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polin S., Simon J. C., Outreman Y. (2014). An ecological cost associated with protective symbionts of aphids. Ecol. evol. 4 836–840. 10.1002/ece3.991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks M. G., Garrett W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16 341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Zilber-Rosenberg I. (2011). “The Hologenome Concept,” in Beneficial Microorganisms in Multicellular Life Forms, eds Rosenberg E., Gophna U. (Berlin: Springer; ), 323–340. [Google Scholar]
- Rothacher L., Ferrer-Suay M., Vorburger C. (2016). Bacterial endosymbionts protect aphids in the field and alter parasitoid community composition. Ecology 97 1712–1723. 10.1890/15-2022.1 [DOI] [PubMed] [Google Scholar]
- Ruiz-Rodríguez M., Martínez-Bueno M., Martín-Vivaldi M., Valdivia E., Soler J. J. (2013). Bacteriocins with a broader antimicrobial spectrum prevail in enterococcal symbionts isolated from the hoopoe’s uropygial gland. FEMS Microbiol. Ecol. 85 495–502. 10.1111/1574-6941.12138 [DOI] [PubMed] [Google Scholar]
- Ruiz-Rodriguez M., Valdivia E., Soler J. J., Martin-Vivaldi M., Martin-Platero A., Martinez-Bueno M. (2009). Symbiotic bacteria living in the hoopoe’s uropygial gland prevent feather degradation. J. Exp. Biol. 212 3621–3626. 10.1242/jeb.031336 [DOI] [PubMed] [Google Scholar]
- Saint André A. V., Blackwell N. M., Hall L. R., Hoerauf A., Brattig N. W., Volkmann L., et al. (2002). The role of endosymbiotic Wolbachia bacteria in the pathogenesis of river blindness. Science 295 1892–1895. 10.1126/science.1068732 [DOI] [PubMed] [Google Scholar]
- Scarborough C. L., Ferrari J., Godfray H. (2005). Aphid protected from pathogen by endosymbiont. Science 310 1781–1781. 10.1126/science.1120180 [DOI] [PubMed] [Google Scholar]
- Shi W., Guo Y., Xu C., Tan S., Miao J., Feng Y., et al. (2014). Unveiling the mechanism by which microsporidian parasites prevent locust swarm behavior. Proc. Nat. Acad. Sci. U.S.A 111 1343–1348. 10.1073/pnas.1314009111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth J. D. (1990). In Vitro Cultivation Of Parasitic Helminths. Boca Raton, FL: CRC press. [Google Scholar]
- Taylor M., Hoerauf A. (1999). Wolbachia bacteria of filarial nematodes. Parasitol. Today 15 437–442. 10.1016/S0169-4758(99)01533-1 [DOI] [PubMed] [Google Scholar]
- Teixeira L., Ferreira Á., Ashburner M. (2008). The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 6:e1000002 10.1371/journal.pbio.1000002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis K., Dheilly N., Klassen J., Brucker R. M., Bosch T. C. G., Cryan J. F., et al. (2016). Getting the hologenome concept right: an eco-evolutoinary framework for hosts and their microbiome. mSystems 1 10.1128/mSystems.00028-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toren A., Landau L., Kushmaro A., Loya Y., Rosenberg E. (1998). Effect of temperature on adhesion of Vibrio strain AK-1 to Oculina patagonica and on coral bleaching. Appl. Environ. Microbiol. 64 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Hurk A. F., Hall-Mendelin S., Pyke A. T., Frentiu F. D., McElroy K., Day A., et al. (2012). Impact of Wolbachia on infection with chikungunya and yellow fever viruses in the mosquito vector Aedes aegypti. PLoS Negl. Trop. Dis. 6:e1892 10.1371/journal.pntd.0001892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Pol B., Kwok C., Pierre-Louis B., Rinaldi A., Salata R. A., Chen P.-L., et al. (2008). Trichomonas vaginalis infection and human immunodeficiency virus acquisition in African women. J. Infect. Dis. 197 548–554. 10.1086/526496 [DOI] [PubMed] [Google Scholar]
- Vorburger C., Ganesanandamoorthy P., Kwiatkowski M. (2013). Comparing constitutive and induced costs of symbiont-conferred resistance to parasitoids in aphids. Ecol. evol. 3 706–713. 10.1002/ece3.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Wang C. (1991). Viruses of the protozoa. Annu. Rev. Microbiol. 45 251–263. 10.1146/annurev.mi.45.100191.001343 [DOI] [PubMed] [Google Scholar]
- Weinert L. A., Tinsley M. C., Temperley M., Jiggins F. M. (2007). Are we underestimating the diversity and incidence of insect bacterial symbionts? A case study in ladybird beetles. Biol. Lett. 3 678–681. 10.1098/rsbl.2007.0373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. F., Jr., Torres M. S. (2009). Defensive Mutualism in Microbial Symbiosis. Boca Raton, FL: CRC Press. [Google Scholar]
- Wu S., Li R. W., Li W., Beshah E., Dawson H. D., Urban J. F., Jr. (2012). Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PLoS ONE 7:e35470 10.1371/journal.pone.0035470 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


