Abstract
Plant hormones cytokinins (CKs) are one of the major mediators of physiological responses throughout plant life span. Therefore, a proper homeostasis is maintained by regulation of their active levels. Besides degradation, CKs are deactivated by uridine diphosphate glycosyltransferases (UGTs). Physiologically, CKs active levels decline in senescing organs, providing a signal to nutrients that a shift to reproductive tissues has begun. In this work, we show CK glucosides distribution in Arabidopsis leaves during major developmental transition phases. Besides continuous accumulation of N-glucosides we detected sharp maximum of the glucosides in senescence. This is caused prevalently by N7-glucosides followed by N9-glucosides and specifically also by trans-zeatin-O-glucoside (tZOG). Interestingly, we observed a similar trend in response to exogenously applied CK. In Arabidopsis, only three UGTs deactivate CKs in vivo: UGT76C1, UGT76C2 and UGT85A1. We thereby show that UGT85A1 is specifically expressed in senescent leaves whereas UGT76C2 is activated rapidly in response to exogenously applied CK. To shed more light on the UGTs physiological roles, we performed a comparative study on UGTs loss-of-function mutants, characterizing a true ugt85a1-1 loss-of-function mutant for the first time. Although no altered phenotype was detected under standard condition we observed reduced chlorophyll degradation with increased anthocyanin accumulation in our experiment on detached leaves accompanied by senescence and stress related genes modulated expression. Among the mutants, ugt76c2 possessed extremely diminished CK N-glucosides levels whereas ugt76c1 showed some specificity toward cis-zeatin (cZ). Besides tZOG, a broader range of CK glucosides was decreased in ugt85a1-1. Performing CK metabolism gene expression profiling, we revealed that activation of CK degradation pathway serves as a general regulatory mechanism of disturbed CK homeostasis followed by decreased CK signaling in all UGT mutants. In contrast, a specific regulation of CKX7, CKX1 and CKX2 was observed for each individual UGT mutant isoform after exogenous CK uptake. Employing an in silico prediction we proposed cytosolic localization of UGT76C1 and UGT76C2, that we further confirmed by GFP tagging of UGT76C2. Integrating all the results, we therefore hypothesize that UGTs possess different physiological roles in Arabidopsis and serve as a fine-tuning mechanism of active CK levels in cytosol.
Keywords: cytokinin, glycosyltransferase, Arabidopsis, senescence, GFP subcellular localization
Introduction
Cytokinin (CK) homeostasis is tightly and precisely regulated in plant cells, enabling adequate response to distinct developmental and environmental requirements. CK metabolism is provided by a variety of enzymes that ensure proper hormone level maintenance. One of such regulatory enzymes is a uridine diphosphate glycosyltransferase (UGT; EC 2.4.1.) that deactivates the molecule of CK by its conjugation with a sugar moiety, mostly glucose; at O- and N- position (Entsch and Letham, 1979; Entsch et al., 1979; Martin et al., 1999; Mok et al., 2000a). Therefore, CKs can form various types of glycosides that can possess different roles. Whereas CK N-glucosides are thought to be terminal products of the irreversible deactivation or a detoxification pathway (McGaw et al., 1984; McGaw and Horgan, 1985; Blagoeva et al., 2004; Sakakibara, 2006; Bairu et al., 2011), CK O-glucosides were shown to be reversibly deglycosylated by β-glucosidase action (Brzobohatý et al., 1993; Falk and Rask, 1995; Kristoffersen et al., 2000) and therefore are thought to serve as inactive storage forms of CKs. UGTs belong to a large multi-gene family (Mackenzie et al., 1997) that deactivates a wide range of molecules. Within the last years, a molecular approach has been used to elucidate the function of the CK-specific glycosyltransferases. Mutant lines overexpressing genes for CK O-glycosyltransferases in maize (Pineda Rodó et al., 2008), tobacco (Mok et al., 2000a,b; Martin et al., 2001; Havlová et al., 2008), rice (Kudo et al., 2010) and Arabidopsis (Hou et al., 2004; Wang et al., 2011, 2013; Jin et al., 2013; Li et al., 2015) have been well characterized to date. Experiments utilizing a cZ O-glucosyltransferase gene form Phaseolus lunatus L. as the overexpressed transgene have confirmed increased accumulation of O-glucosides in different tissues. In certain conditions, these experiments have confirmed more or less severe phenotypic alteration (Mok et al., 2000a; Martin et al., 2001; Havlová et al., 2008; Pineda Rodó et al., 2008; Kudo et al., 2010).
Up to now, CK-specific UGTs have been shown to be expressed to various levels in different tissues such as developing seeds, roots and leaves of young seedling or maturing embryo (Martin et al., 1999; Veach et al., 2003; Wang et al., 2011, 2013; Jin et al., 2013) with low expression in vegetative tissues (Martin et al., 1999). On a subcellular level, no signal sequence or transmembrane domain was found in any of the plant UGTs (Li et al., 2001) investigated so far. However, it was suggested that the proteins may be associated as peripheral components of the endomembrane system (Winkel-Shirley, 1999). Glycosylation alters chemical properties of the inactivated molecules enabling their inter-compartment re-localization driven by newly gained increased hydrophilicity (Bowles et al., 2005). This raises a question whether inactive CK glucosides are destined to retargeting to cellular compartments or whether they serve as long-distance transport forms, which is more likely, since high levels of glucosides were found in xylem sap of cucumber (Kato et al., 2002) or extracellular space of Arabidopsis and barley leaves (Jiskrová et al., 2016). This hypothesis was further supported by Kato et al. (2002) whose research documented shoot greening mediated by CK glucosides uptake by roots. In contrast to speculative localization of CK-specific UGTs in vacuoles (Meek et al., 2008; Pineda Rodó et al., 2008; Bajguz and Piotrowska, 2009), the trans-zeatin O-glycosyltransferase enzyme from Zea mays and Phaseolus vulgaris was immunodetected in the nucleus, cytosol, and closely associated with the plasma membrane and in the cell wall of Z. mays root cells (Li et al., 2001). Further, GFP tagged UGT85A1 from Arabidopsis has been up to now detect in cytosol, and nucleus (Jin et al., 2013). Although dual subcellular localization was observed in plant UGTs (Hong et al., 2001), this was an exceptional case. Besides the cited works, only little is known about localization of the CK glucosides on the subcellular level. Although CK O-glucosides were observed to be present within vacuoles of Chenopodium rubrum (Fusseder and Ziegler, 1988; Mok et al., 1992), our recent work shows predominant localization of both types of CK glucosides in extracellular space (Jiskrová et al., 2016).
As previous reviewers summarized, the same UGT can recognizes multiple substrates in vitro and, conversely, different UGTs can glycosylate the same substrate (Lim and Bowles, 2004). However, since this does not reflect physiological functions of UGTs in planta, identification of their mutants were shown to be the most powerful tool to address their direct roles (Bowles et al., 2005). In Arabidopsis, five CK-specific UGTs (UGT73C1, UGT73C5, UGT76C1, UGT76C2, UGT85A1) were biochemically characterized in the past using recombinant proteins (Hou et al., 2004). Although three of them were further confirmed to be specific toward CKs in planta (Wang et al., 2011, 2013; Jin et al., 2013; Li et al., 2015), UGT73C5 was shown to be specific toward brassinosteroids (BR) (Poppenberger et al., 2005) and UGT73C1 was shown to be specific to trinitrotoluene compounds (Gandia-Herrero et al., 2008) with much higher affinity than to CKs. Amongst the three CK-specific UGTs, increased sensitivity to exogenously applied CK was detected in ugt76c2 and ugt76c1mutant (Wang et al., 2011, 2013) as a result of impaired CK glucosylation but only ugt76c2 resulted in a modified phenotype manifested by smaller seeds (Wang et al., 2011). Enhanced root elongation was observed in UGT85A1 overexpressing line (Jin et al., 2013) as a result of accelerated CK deactivation. Former studies showed that UGT76C1 and UGT76C2 are N-glucosylation specific (Hou et al., 2004; Wang et al., 2011, 2013) whilst UGT85A1 was proposed to be trans-zeatin-specific O-glucosyltransferase (Jin et al., 2013). Further experiments on UGT76C2 mutants demonstrated involvement of the CK deactivation pathway during drought and osmotic stress and. Moreover, these experiments also indicated temporal importance of CK glucosylation process under the stress conditions (Li et al., 2015).
The present study is aimed to extend the knowledge of the CK deactivation pathway. We take a multidisciplinary approach combining molecular biology and quantitative analysis to answer some intriguing questions regarding physiological relevance of CK glucosides and the significance of the glycosylation process in Arabidopsis. In this work, we describe CK glucosides formation during main developmental transition phases and further, we perform a comparative study of loss-of-function mutants of CK-specific UGTs to elucidate their roles in CK homeostasis maintenance during plant development and in response to exogenous stimuli. We also characterize ugt85a1 mutant in context to CK for the first time and discuss UGT85A1 ability to deactivate a broader range of substrates as well as its specificity in senescence process. Finally, our research also attempts to bring more light into CK metabolism compartmentation in this work.
Materials and Methods
Plant Materials
Arabidopsis thaliana ecotype Columbia-0 was used in this work. Seeds of ugt76c2 (SALK 135793C), ugt76c1 (SALK 144355C), ugt85A1-1 (SALK 085809C), and ugt85a1-2 (SALK 146306C) were obtained from the European Arabidopsis Stock Center (for the description of the lines see Supplementary Table S1). Surface sterilized seeds were sown on half strength MS medium (Murashige and Skoog, 1962) supplemented with 1% sucrose and stratified at 4°C for 4 days in the dark prior to germination. Seedlings were grown either on MS plates or in soil under standard Arabidopsis growth condition in an environmental chamber (16 h fluorescence light of 150 μmol photons∗m-2∗s-1 intensity/8 h dark, 22°C, 55% relative humidity). A green mature fully expanded leaf (sixth and seventh) from a 4-week-old rosette was detached for experiments with exogenously applied CK and further for gene expression profiling. The leaves were incubated in water containing 10 μM KIN, 6-benzylaminopurine (BAP), isopentenyladenine (iP) or trans-zeatin (tZ) with final concentration of dimethylsulfoxid (DMSO) 0.05% for 3 days under the same conditions as the donor plants. Further, intact whole 4-week-old rosettes were sprayed with 10 μM solution of tZ containing 0.005% surfactant Silweet L-77 every 24 h for 3 days. In both cases, mock treatment contained 0.05% DMSO. Solanum lycopersicum L., Peto 343 was used for overexpression of SU:UGT76C2-GFP for the subcellular localization study described below.
Identification of T-DNA Insertion Mutants
Although UGT85A1 mutant was described before, its expression was not abolished completely (Carviel et al., 2009). In this work, we characterize a new T-DNA insertion mutant of UGT85A1 (SALK_146306C), labeled as ugt85a1-1, and compare it with the previously characterized SALK_085809C (ugt85a1-2). Supplementary Figures S1A,C illustrates schematic of T-DNA positions within the UGT85A1 gene. The ugt85a1-1 and ugt85a1-2 lines were confirmed for their T-DNA insertion position, homozygosity (data not shown) and gene expression level in RT-PCR (Supplementary Figures S1B,D). Our results showed that UGT85A1 was expressed in WT but not in the ugt85a1-1 mutant, while a weak expression was detected in ugt85a1-2 mutant. Remaining loss-of-function mutants of CK-specific UGTs used in this work were analyzed for presence of their T-DNA insertion and its effect in previous publications: ugt76c2 (Wang et al., 2011), ugt76c1 (Wang et al., 2013). No redundancy effect of any of the CK-specific UGTs was detected in any of the mutants used in this work based on their gene expression quantification (Supplementary Figure S2).
Phenotyping and Root Growth Assay
Sterilized seeds of Arabidopsis wild-type (WT) and ugt85A1-1 were transferred to vertical square Petri dishes on a solid MS medium containing 0.5% MES and 1% sucrose as well as various concentrations of BAP, iP, dihydrozeatin (DHZ) and tZ, respectively. 0.05% DMSO was used in control (mock treatment) since that was the final concentration in all CK treatments. After stratification, the seeds were germinated and grown in an environmental chamber for 14 days. Within the growing period the length of the primary root was evaluated after seven and 14 days respectively, using Scion Image software (Scion Corporation, Frederick, MD, USA). The number of fully emerged lateral roots was scored under a SMZ800 Stereoscopic Microscope (Nikon, Japan). For a senescence induced experiment, sixth leaves from 5-week-old WT and ugt85A1-1 rosettes were detached from ten individual plants for each genotype. The experiment was performed in two replicates.
Chlorophyll and Anthocyanin Content Assay
Chlorophyll (Chl) content was performed according to published protocol (Chory, 1991) with the following modification. After 4 days of incubation under continuous light, the detached leaves were frozen in liquid nitrogen and homogenized to extract Chl in 80% acetone. Chl a nad b portions of total Chl (expressed as relative amounts) were calculated using following equation: a = 12.7 (A663) – (A645) and b = 22.9 (A645) – 4.68 (A663). Further, the same samples were used for anthocyanin content measurement which was performed according to a published protocol (Neff and Chory, 1998) using acidified methanol and counting the relative content by subtracting A657 from A530.
In silico Prediction of Subcellular Localization
To predict the subcellular localization of the UGTs, the following prediction web based tools were used: TargetP (Emanuelsson et al., 2000), ProtComp v9.01, SignalP 4.0 (Petersen et al., 2011), ChloroP 1.1 (Emanuelsson et al., 1999), WoLF PSORT2, iPSORT (Bannai et al., 2002), Plant-mPLoc (Chou and Shen, 2010).
Cytokinin Content Determination
The procedure used for CKs purification was performed according to the described method (Svačinová et al., 2012) with subsequent modifications. The samples were extracted in modified Bieleski buffer (methanol/water/formic acid, 15/4/1, v/v/v) and then purified using two solid phase extraction columns, a C18 octadecylsilica-based column (500 mg of reversed-phase sorbent; Applied Separations) and, after that, an Oasis MCX column (30 mg of mixed-mode sorbent with reversed-phase/cation-exchange properties; Waters) (Dobrev and Kamínek, 2002). The samples were analyzed employing ultra-high performance liquid chromatography (Acquity UPLC system; Waters), coupled to a triple quadrupole mass spectrometer (Xevo TQ-S; Waters) equipped with an electrospray interface. Deuterium-labeled CK internal standards (OlChemIm) were used to validate the determination, each at 0.5 pmol per sample (Novák et al., 2008). Quantification was achieved by multiple reaction monitoring of [M + H]+ and the appropriate product ion. The quantification was performed by Masslynx software (v4.1; Waters) using a standard isotope dilution method. The ratio of endogenous CK to the appropriate labeled standard was determined and further used to quantify the level of endogenous compounds in the original extract according to the known quantity of the added internal standard.
RT-PCR and Quantitative RT-PCR (qPCR) Assay
Plant material was processed for reverse transcription according to the method described in our former publication (Vyroubalová et al., 2009). The primers used in our experiment amplified specifically for the following genes: glucosyltransferases UGT73C1, UGT73C5, UGT76C1, UGT76C2, UGT85A1; CK dehydrogenases CKX1-7; isopentenyltransferases IPT1-9; CK nucleoside 5′-monophosphate phosphoribohydrolase LOG2, LOG8; CK trans-hydroxylase CYP735A2; response regulators type A (ARR5, ARR15, ARR16), type B (ARR1, ARR2, ARR10, ARR14); CK receptors AHK4, AHK2, AHK3. All the listed primers were designed according to our previously published work (Mik et al., 2011; Motte et al., 2013; also see Supplementary material of these publications for details), Rubisco small chain RUBsc (Hensel et al., 1993), cysteine protease senescence-associated gene SAG12 (Gepstein et al., 2003; Wagstaff et al., 2009) and chlorophyll a/b binding protein CAB2 (Kim et al., 2006), for transcription factors MYB2 (Guo and Gan, 2011) and WRKY53 (Hinderhofer and Zentgraf, 2001; Balazadeh et al., 2008); for 1-aminocyklopropan-1-carboxylate synthase ACS8 as a key regulatory enzyme in the biosynthesis of the plant hormone ethylene (Tian et al., 2009); for chalcone flavanone isomerase CHI as anthocyanins biosynthetic enzyme (Catala et al., 2011); 9-cis-epoxycarotenoid dioxygenase NCED3, a key enzyme in the biosynthesis of abscisic acid (Tan et al., 2003). RNA from three biological replicates was transcribed in at least two independent reactions and each cDNA sample was run in at least three technical replications on Viia7TM Real-Time PCR System in a default program (Life Technologies). To evaluate a relative quantification of analyzed genes, cycle threshold (Ct) values were analyzed with the Formula method (Livak and Schmittgen, 2001) and normalized with respect to actin 2 (ACT2) and small nuclear ribonucleoprotein D1 (SnRNPD1) (Quilliam et al., 2006) that were used as internal standards. The expression data are relative quantities (RQ) calculated as 2-ΔCq extrapolated to controls that are given as 1.0. To confirm UGT85A1 expression level of ugt85a1 mutants, 85A1fw and 85A1rev primers were used (Carviel et al., 2009) in RT-PCR.
Cloning and Generation of GFP Translational Fusions
A Modular Binary Construct System with AKK1436 as a shuttle vector and AKK1472B binary vector (Christopher Taylor Lab, Donald Danforth Plant Science Center, St. Louis, MO, USA) was used for GFP gene fusion and subsequent overexpression of the GFP tagged UGT76C2 gene in tomato hairy roots under a super ubiquitin (SU) promoter as described in our previous work (Šmehilová et al., 2009). The gene for UGT76C2 was synthetized using GeneArt® commercial service (Thermo Fisher Scientific, USA) with At5g05860 CDS as a template sequence without any modifications. BamHI restriction sites were added to the sequence to enable further sub-cloning of the synthetic gene from the source pMK-RQ vector. Use of BamHI digestion and further ligation to AKK1436 vector resulted in in-frame UGT76C2-GFP fusion further incorporated into AKK1472B through PacI digest/ligation.
Transgenic Tissue Preparation and Analysis
Root transformation was performed as described previously (Collier et al., 2005). The binary constructs containing SU:UGT76C2-GFP and SU:GFP were introduced into Agrobacterium rhizogenes strain 15834, verified for transgenes presence and further used for transgenic tomato hairy roots production following the procedure used in our previous work (Šmehilová et al., 2009).
Subcellular Localization of GFP-Fused Proteins
The transgenic roots were first evaluated for GFP fluorescence with a SMZ800 Stereoscopic Microscope (Nikon) and analyzed for transgene expression in RT-PCR. Three to five individual roots were selected from the confirmed lines and mounted in 100 mM phosphate buffered saline (PBS) pH 6.5 prior to observation in confocal microscopy. Zeiss LSM710 laser-scanning confocal microscope (Zeiss) was used to detect the GFP signal of the SU:UGT76C2-GFP and SU:GFP. GFP was excited at 488nm and detected between 500 and 535 nm. Images were processed using Zeiss ZEN software and Adobe Photoshop software.
Results
Cytokinin N7-Glucosides Accumulate during Whole Plant Development, Whereas O-Glucosides Fluctuate with Sharp Maximum of tZOG in Senescent Leaf
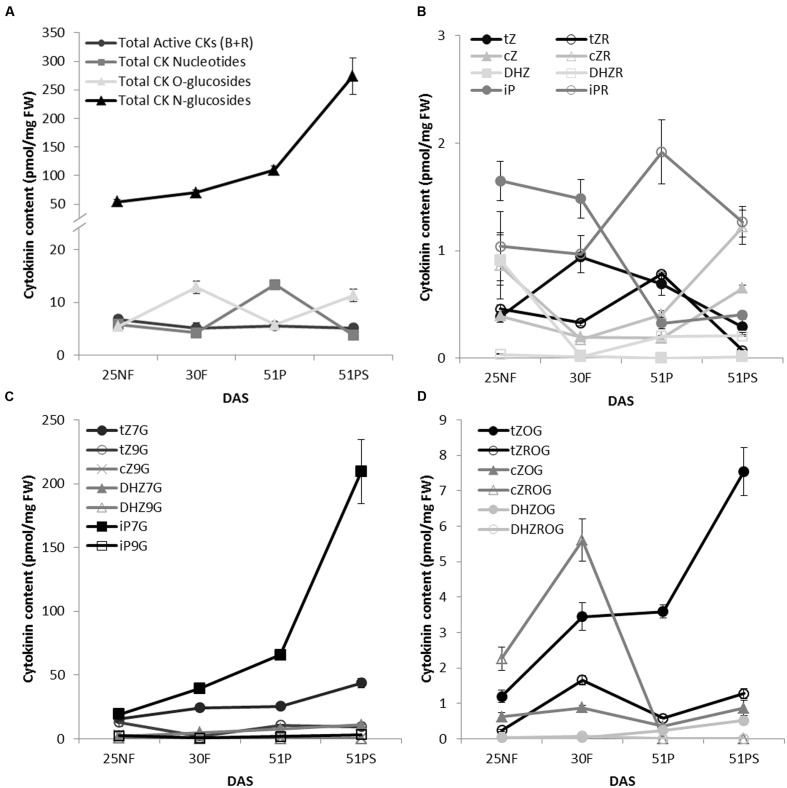
To determine CK glucosylation status during Arabidopsis leaf senescence, levels of isoprenoid CKs and their respective ribosides, ribotides and glucosides were determined under standard growth conditions in four time points covering the main developmental transition phases: green mature (fully expanded) leaves of non-flowering and flowering rosettes, and green and senescent leaves from pod forming plants. Generally, N-glucosides accumulated during leaf aging (Figure 1A) with sharp maximum of N7-glucosides in yellowing stage of the leaf aging (Figure 1C). It might seem that CK O-glucosides fluctuated in an opposite way to CK nucleotides when summed up (Figure 1A); however, Figure 1D illustrates flowering time-related increase of all O-glucosides except DHZOG and tZOG that accumulated in aging-dependent manner. Our results point out that tZOG is the main O-glucoside present in Arabidopsis senescent leaves since its level arose almost seven times in comparison to other CK O-glucosides in this time point. Indeed, we observed a decline in active CK forms (Figure 1A), with fluctuation characteristic for each form (Figure 1B). Interestingly, cZ and particularly cZR levels increased in senescent leaf which is in accordance with previous work (Gajdošová et al., 2011). It should be noted that the content of CK glucosides was more than one order of magnitude higher than active forms.
FIGURE 1.
Cytokinin content of WT A. thaliana leaves during transitions between vegetative, reproductive and senescence stages. The time points were: green fully developed leaves from a non-flowering (NF) rosette, a flowering (F) rosette, from a plant with maturing pods (P); and senescent leaves from the same plant (PS); days after sowing (DAS). The overall distribution of cytokinins grouped by types of their forms (A) and with a focus on individual forms: active cytokinins (B), cytokinin N-glucosides (C) and cytokinin O-glucosides (D). Values are the means of three biological replicates ± SD.
Gene Expression Profile of Cytokinin-Specific UGTs
Up to date, the UGTs expression was assessed only in young developing tissues (Wang et al., 2011, 2013). According to our data, Arabidopsis accumulated large quantities of CK glucosides in leaves in age-dependent manner so that we employed gene expression profiling to determine which UGTs are specific in CK deactivation process during senescence. We show that the most upregulated CK-specific UGT was UGT85A1 whose expression is specifically boosted in senescent leaves (Figure 2A) followed by BR-specific UGT73C5 (Poppenberger et al., 2005). We detected significantly downregulated expression of UGT76C1 with age-dependent manner but without specificity in yellowing leaf. Steady expression was detected for UGT76C2 throughout the developmental stages, whereas varying expression was detected for UGT73C1. CAB2 gene was used as a control of senescence progress that reflects photosynthesis status. Consistently, downregulated expression of CAB2 in senescent leaves correlates with previously published data (Kim et al., 2006).
FIGURE 2.
Gene expression profile in leaves of Arabidopsis cytokinin-specific UGTs during plant development (A) and after cytokinin treatment (B). YGL, Young green leaf 30 DAS; GL, green fully developed leaf; SL, senescent leaf. In the experiment where transcript level in response to exogenously applied cytokinin was analyzed, green mature fully expanded leaves (sixth and seventh) from 4 weeks old rosette were used and treated by 10 μM cytokinins for 3 days. The gene expressions are expressed as relative quantities and extrapolated relative to the GL 30 DAS (A) and mock treatment (B), respectively, given as 1.0. The values represent means of three biological replicates (three technical replicates) with standard deviations. Asterisks indicate significant differences between the controls and the samples according to unpaired Student’s t-test, ∗P < 0.05; <LOD, below limit of detection.
To compare UGTs in their response to various exogenously applied CK we analyzed modulation of UGTs expression in detached leaves in response to 10 μM KIN, BAP, iP and tZ, respectively. Generally, no significant difference in transcript level was detected in all but UGT76C2 whose transcript level was increased enormously in response to all the tested CKs, with the highest increase after KIN uptake (Figure 2B) in comparison to mock treatment. UGT85A1 was sensitive only to tZ treatment which increased its expression only mildly, 1.59-times. Surprisingly, the second CK N-specific UGT76C1 glycosyltransferase showed no response to any of the CKs on transcript level.
Cytokinin Content after Exogenous Application of Cytokinins
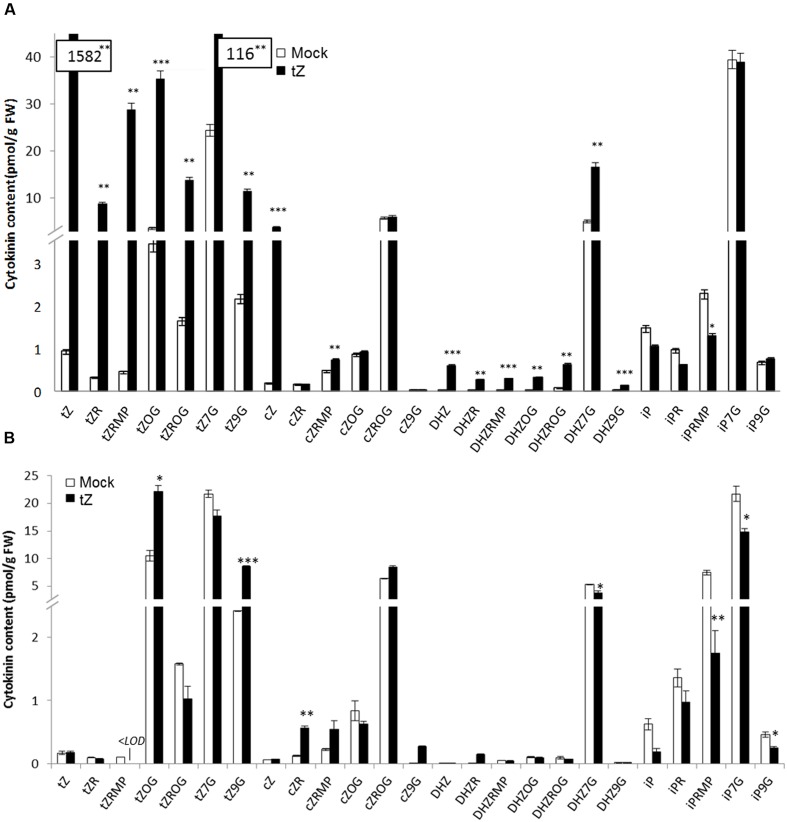
In order to correlate increased transcript level of UGT76C2 after exogenously applied CK, we determined CK content in detached leaves after KIN treatment (as a non-native Arabidopsis CK not interfering with natural CK content) and in whole 4-week-old rosettes in response to tZ. Subsequently, the most prevalent CK types (Mok and Mok, 2001) and forms were analyzed. After tZ uptake, a strong interfering effect in terms of extremely elevated tZ-type CKs was observed (Figure 3A). Further, all DHZ-type CKs arose as well. Consistently with previous studies (Cowley et al., 1978; Letham et al., 1978; Hou et al., 2004), N7-glucosides were the predominant forms of CK deactivation pathway, since strong increase of tZ7G and DHZ7G were detected after tZ uptake, followed by its O-glucosides. tZ9G and tZOG were the most significantly elevated CKs when leaves were treated by KIN (Figure 3B). Both treatments resulted in abolished CK biosynthesis as manifested by general decline of all iP-type CKs and particularly of tZRMP in case of KIN treatment. Interestingly, cZ level rose after tZ uptake, whereas no change was detected in KIN experiment. Since KIN does not interfere with native Arabidopsis CK content but triggers the same CK response in Arabidopsis (Mik et al., 2011; Motte et al., 2013), we used KIN to study CK-mediated response in UGT mutants.
FIGURE 3.
Cytokinin content in whole plants of WT A. thaliana after tZ treatment (A) and in detached leaves after KIN treatment (B). Values are the means of three biological replicates with ± error bars that represent standard deviations. Asterisks over bars indicate significant differences between KIN treated and mock control plants according to two-tailed unpaired Student’s t-test, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
ugt85a1-1 Loss-of-Function Mutant Shows Delayed Senescence Phenotype under Stress Condition
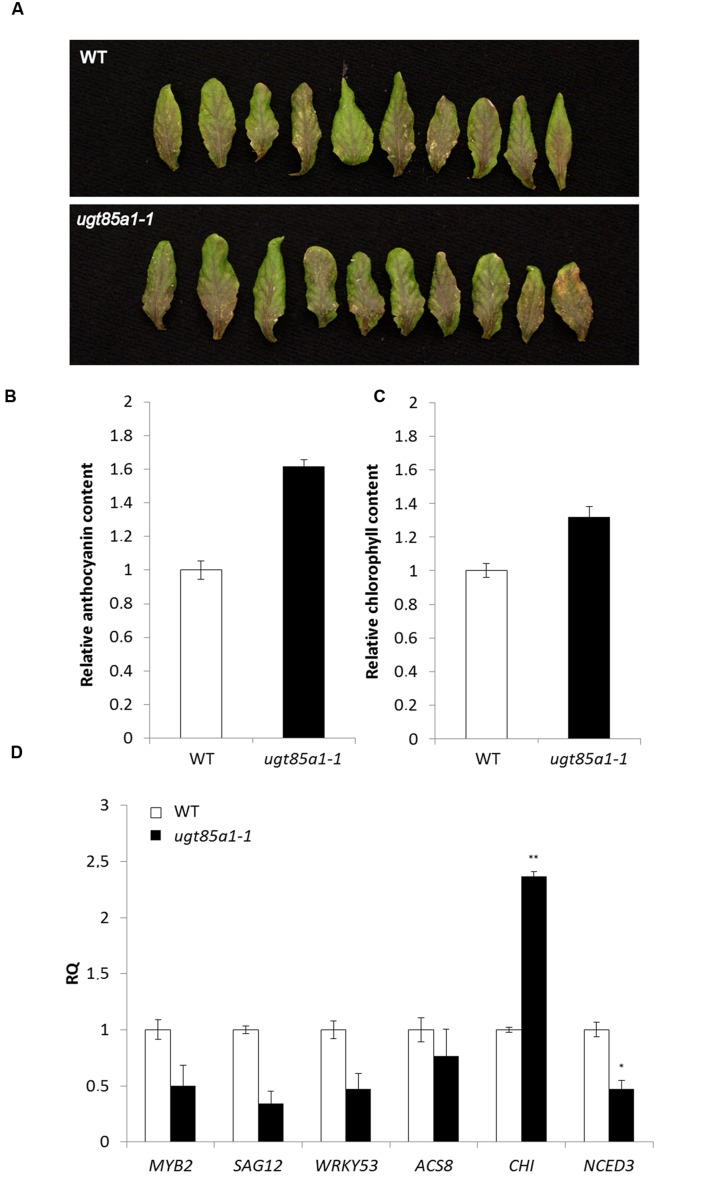
In this study, UGT85A1 was shown on transcript level to be the major form responsible for CK inactivation during senescence. However, the ugt85a1-1 mutant did not show apparent phenotypic alteration under normal growth conditions. In order to investigate expected enhanced sensitivity of ugt85a1-1 mutant to exogenously applied CK, a rooting test was performed using BAP, iP and tZ in 0.1, 0.5, and 1.0 μM concentrations. No significant difference in main root length or in number of lateral roots was observed after any of the treatments. Since no phenotype difference was observed on whole plant level in any of developmental stages analyzed, we performed an experiment where senescence was induced by leaves detachment. After 3 days of incubation, the leaves of ugt85a1-1 mutant showed apparent enhanced accumulation of anthocyanins (Figure 4A). This was further verified by the anthocyanin assay that confirmed 1.6-fold increase of anthocyanins in ugt85a1-1 (Figure 4B). Similarly, chlorophyll content was measured showing reduced chlorophyll degradation in ugt85a1-1 mutant (Figure 4C). In order to show senescence status of the mutant in this experiment, we determined gene expression of senescence and stress associated genes together with genes responsible for anthocyanin biosynthesis CHI (Figure 4D). Relative expression of all the senescence and stress marker genes was lower in ugt85a1-1 mutant in comparison to the WT control with significant reduction of gene transcript of the key enzyme responsible for abscisic acid biosynthesis NCED3. Higher relative expression of CHI gene coding for one of the core anthocyanin biosynthetic enzymes further supports observed enhanced anthocyanin accumulation. Taken together, our data suggest important role of UGT85A1 in senescence process.
FIGURE 4.
Characterization of ugt85a1-1 mutant. Detached leaves of WT and the mutant after 3 days of continuous light (A), chlorophyll content (B), anthocyanin content (C) and gene expression profile of senescence and stress related markers (D) including anthocyanin biosynthetic gene CHI. Values are the means of two independent experiments where leaves of ten individual plants were pooled and expressed as relative quantities extrapolated to WT control (white) that is given as 1.0 with error bars representing standard deviations. Asterisks indicate significant difference between WT and transgenic tissue according to two-tailed unpaired Student’s t-test, ∗P < 0.05, ∗∗P < 0.01.
UGT85A1 Can Be Specific toward a Broader Range of Substrates
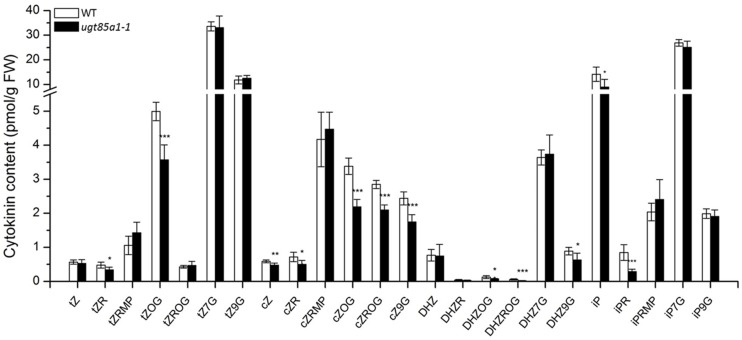
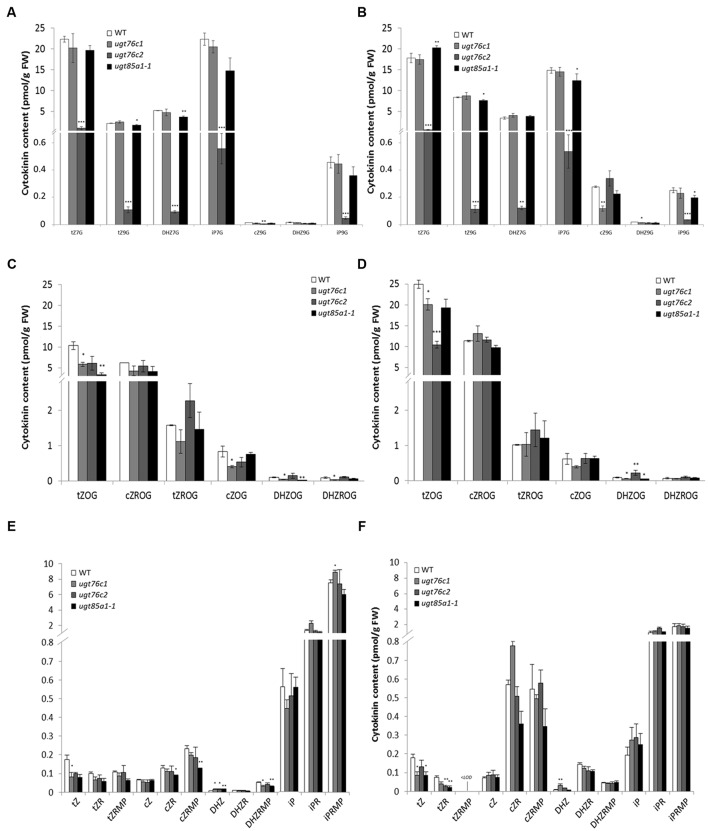
We determined CK profile of ugt85a1-1 in 25-day-old seedlings. As indicated in Figure 5, our data show significant decrease in tZOG level that is in accordance with previously published data where UGT85A1 was proposed to specifically O-glucosylate tZ based on UGT85A1 overexpressor characterization (Jin et al., 2013). Our data further point out that this UGT might possibly glucosylate also other CKs since significantly decreased levels of almost all O-glucosides as well as cZ9G and DHZ9G were detected. However, this could be due to other uncharacterized regulatory processes that occur in the mutant. Although some of the active CKs decreased, particularly iP, iPR, cZ, cZR, and tZR, CK nucleotides somewhat increased. No change in any of N7-glucosides was observed in our data.
FIGURE 5.
Cytokinin content of 4-week-old seedlings of ugt85a1-1 mutant. Values are the means of three biological replicates with error bars representing standard deviations. Asterisks over bars indicate significant difference between wild-type and the mutant according to two-tailed unpaired Student’s t-test, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
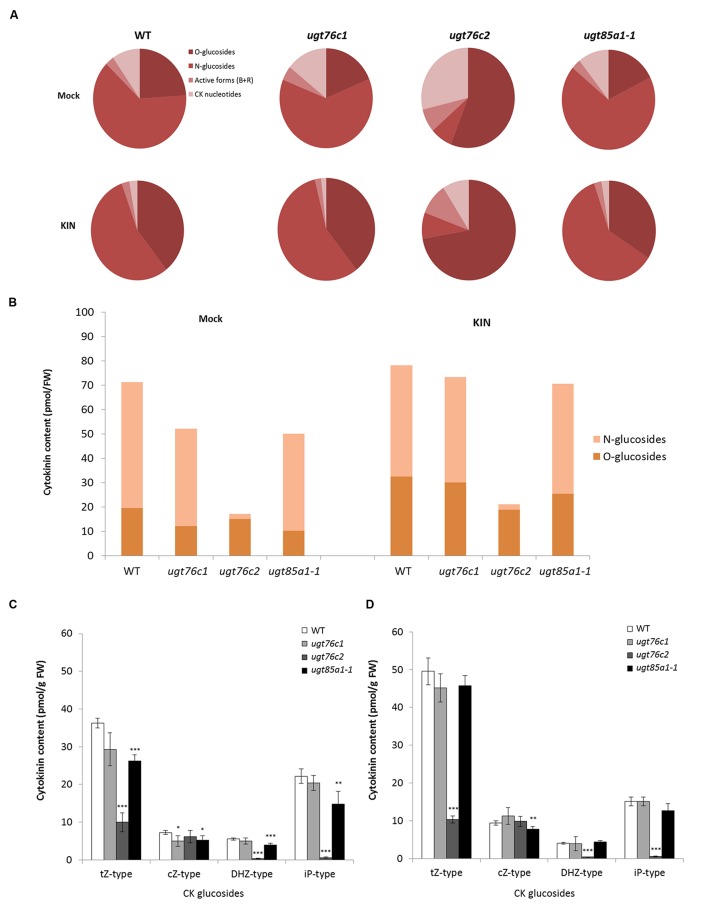
Comparison of Cytokinin Content in Detached Leaves of UGT Loss-of-Function Mutants after KIN Treatment
Based on our knowledge of plant ability to restore impaired CK homeostasis by modulation of gene expression (Mrízová et al., 2013; Pospíšilová et al., 2016) and of the fact that only minor regulation effect was detected in ugt76c1 mutant (Wang et al., 2013), we performed an experiment on detached leaves to uncouple the leaf response from whole plant level. We further treated the leaves with exogenously applied KIN to cease CK biosynthesis, thereby boosting visibility of plant’s response to applied CK with focus on glucosylation. Subsequently, we analyzed CK content of all the CK-specific UGT loss-of-function mutants in response to exogenously applied KIN. In order to compare the CK content of WT and ugts in general, we grouped CK by forms (active, nucleotides, O- and N-glucosides) and compared their overall ratios as illustrates Figure 6A. Here it can be noticed that ugt76c1 and ugt85a1-1 shared a similar pattern of CK forms distribution in general, whereas ugt76c2 showed significantly altered ratio caused mainly by decreased N-glucosides portion. N-glucosides were in fact almost depleted in ugt76c2 as shown in Figure 6B (comparing total glucosides amounts) whereas they were just moderately decreased in ugt76c1 and ugt85a1-1. Figure 7B also shows a portion of O-glucosides that are decreased in all the mutants and particularly in ugt76c1 and ugt85a1-1 under non-treated condition. Interestingly, higher amounts of O-glucosides were detected after KIN uptake in contrast to non-treated leaves in all genotypes, with the lowest amounts in ugt76c2. When CK glucosides are presented as individual CK types in absolute numbers (Figures 6C,D) it can be concluded that the UGTs are not specific for any particular CK type since glucosides of all CK types were decreased in all the three analyzed mutants. Figure 7 describes the contents of all individual CK in detail. Neither ugt76c2 nor ugt85a1-1 showed any specificity for N7- or N9- glucosylation since both groups of glucosides were decreased (Figures 7A,B) which is in accordance with previous data (Wang et al., 2011, 2013). However, KIN treatment of ugt76c1 revealed significantly lower level of cZ9G (Figure 7B) that was not determined in previous studies. Major contributors of total O-glucosides levels are tZOG and cZROG. We detected significantly decreased tZOG levels generally in all the mutants under both conditions, whereas the levels of cZROG stayed steady (Figures 7C,D). Interestingly, lower levels of cZOG were measured in ugt76c1 under both conditions. The levels of active CK and their biosynthetic forms are in accordance with our previous experiment with KIN treatment since lower levels of iP and iPRMP were detected after KIN uptake in general as a result of proposed abolished CK biosynthesis (Figure 7F). Lower levels of tZ under non-treated condition together with lower levels of tZR and depleted tZRMP levels after KIN uptake were assessed in all the mutants, pointing to the plant’s need to restore the homeostasis caused by impaired glucosylation, especially when more stress caused by excessive supply of exogenous CK was applied.
FIGURE 6.
Cytokinin content in detached leaves of ugt76c1, ugt76c2 and ugt85a1-1 mutants after 10 μM kinetin treatment. Ratio among CK forms in mock treatment and KIN treated leaves (A). Distribution of total amounts of CK O- and N-glucosides in the genotypes (B). Cytokinin content by CK types; each individual CK-type group sums up free base and riboside with their corresponding O-glucosides and N-glucosides plus ribotides; values are the means of three biological replicates with error bars representing standard deviations. Asterisks over bars indicate significant difference according to two-tailed unpaired Student’s t-test, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 where for each replicate sixth and seventh green fully developed leaves from ten independent 28-day-old rosettes were pooled for control mock treatment (C) and KIN treatment (D).
FIGURE 7.
Detailed cytokinin content in detached leaves of WT, ugt76c1, ugt76c2 and ugt85a1-1 mutants. N-glucosides content in control mock treatment (A) and after KIN treatment (B), O-glucosides content in control mock treatment (C) and after KIN treatment (D), and CK free bases together with their ribosides and ribotides in control mock treatment (E) and after KIN treatment (F). Values are the means of three biological replicates with error bars representing standard deviations. Asterisks over bars indicate significant differences between wild-type and the mutants according to two-tailed unpaired Student’s t-test, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
Cytokinin Metabolism Gene Expression Profile in ugt Mutants
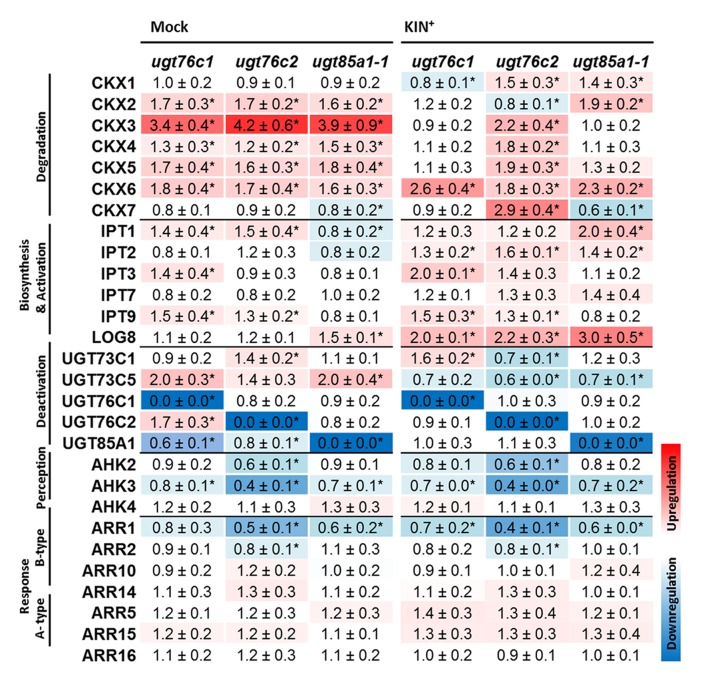
Gene expression of major CK metabolic pathways, perception and response was analyzed in detached leaves of ugt mutants after KIN treatment as well as in control mock treatment. Figure 8 illustrates RQ of the gene transcripts in a heat map. Here, expression of majority of degradation pathway genes was increased in all the mutants, particularly in non-treated group and specifically also in ugt76c2 after KIN uptake. The most upregulated form was CKX3 followed by CKX5 and CKX6. A different pattern was detected for CKX1 since this isoform was downregulated in ugt76c1 mutant after KIN uptake, whereas upregulated in the remaining two mutants. Similarly, CKX2 transcript level was specifically increased in ugt85a1-1 mutant, but slightly suppressed in ugt76c2 and not modulated in ugt76c1. CKX7 form was slightly downregulated in ugt85a1-1 in both conditions but upregulated in ugt76c2 after KIN uptake. CK biosynthesis was generally only modestly decreased when not treated by CK with mutant dependent exceptions. IPT1 and IPT9 transcripts were more abundant in ugt76c1 and ugt76c2, whereas transcript level of IPT3 was increased specifically in ugt76c1 mutant. Majority of IPT genes were slightly elevated after KIN treatment when compared to WT. Although IPT3 and IPT9 shared a somewhat similar trend as when not treated, IPT1 behaved in an opposite way in ugt85a1-1 mutant. Expression of LOG8 increased in order WT < ugt76c1 < ugt76c2 < ugt85a1-1 both, in plants not treated and treated with KIN, but the increase was more pronounced after the treatment. UGT85A1 was downregulated in ugt76c1 and ugt76c2 mutants when not treated, and UGT76C2 increased in ugt76c1. AHK3 transcript decreased in all the mutants in both conditions as well as ARR1with significance for ugt76c2. AHK2 was specifically decreased in both ugt76c mutants after KIN uptake. Two out of three examined A-type ARR were upregulated in response to KIN.
FIGURE 8.
Expression profiles of cytokinin metabolism, perception and response genes under standard conditions (Mock treatment) and after kinetin treatment (KIN+) in detached leaves of A. thaliana ugt76c1, ugt76c2 or ugt85a1-1 mutants. The gene expressions of the mutant plants are expressed as relative quantities and extrapolated relative to the mock treatment and KIN treated WT, respectively, given as 1.0. The values represent means of three biological replicates (three technical replicates) with standard deviations. Asterisks indicate significant differences between controls and samples according to two-tailed unpaired Student’s t-test, ∗P < 0.05; < LOD, below limit of detection. No expression was detected for LOG2, LOG6 and LOG7.
Subcellular Localization of UGTs
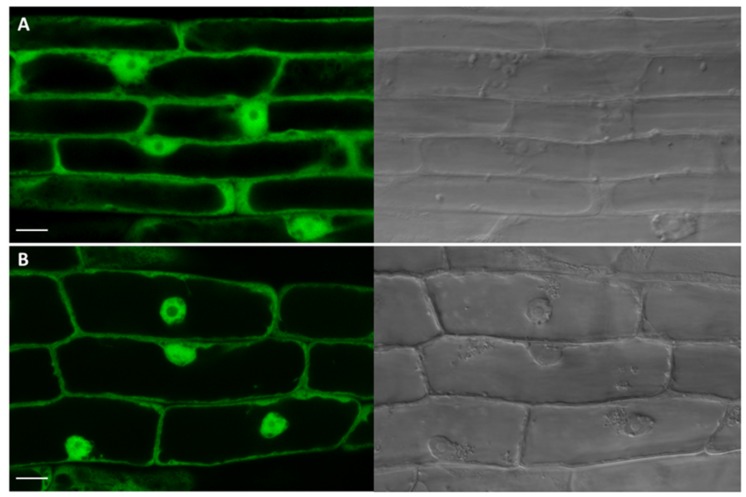
The only CK-specific UGT with confirmed subcellular localization is UGT85A1 detected in cytosol (Jin et al., 2013). That was in strong contrast with its in silico prediction by WoLF PSORT prediction tool3 (Woo et al., 2007). In that study, UGT85A1 was predicted as part of the AtUGT85A-(sub)class to be a membrane-associated enzyme, with targeting to chloroplasts and endoplasmic reticulum (ER). The authors further showed that the AtUGT85A1 protein contains the xKQxxEF motif for microsome retention and ER membrane retention signal QKSQ at the C terminus as well as several posttranslational modification sites suggesting high level of posttranslational modification (Woo et al., 2007). However, confirmed localization to cytosol accords with generally accepted presumption that plant UGTs are cytosolic enzymes (Bowles et al., 2005; Sun et al., 2013). Here, we performed a signal peptide prediction of the remaining CK-specific UGTs using available prediction software (Table 1). Similar to UGT85A1, UGT76C1and UGT73C1 localization is predicted to chloroplast according to signal peptide prediction analysis. Since UGT76C2 is one of the core CK homeostasis genes, we examined its subcellular localization using confocal laser scanning microscopy using GFP tagging. As shown in Figure 9B, the fluorescent pattern of UGT76C2-GFP displayed diffuse fluorescence in cytosol, accumulating along the plasma membrane and in nuclei (Grebenok et al., 1997a,b; Köhler et al., 1997; Figure 9A), thus proving UGT76C2 to be a cytosolic enzyme. We suggest the same localization for UGT76C1 based on our signal peptide prediction and comparison (see Supplementary Figure S3).
Table 1.
Prediction of a signal peptide in protein sequences of UGTs and model proteins.
| Protein | TargetP | Prot Comp Plant | SignalP | ChloroP | WoLF PSORT | iPSORT | Plant-mPLoc | Confirmed |
|---|---|---|---|---|---|---|---|---|
| UGT73C1 | O5 | Cl1 | SP3 | Cl | Cl3 | Cl3 | TM | C (unpublished) |
| UGT73C5 | O3 | Cl TM2 | SP3 | – | N M3 | Cl1 | TM | C (Husar et al., 2011) |
| UGT76C1 | O3 | Cl TM3 | – | – | Cl N4 | M3 | TM Cl | C (unpublished) |
| UGT76C2 | O4 | C1 | – | – | N1 | SP1 | TM | C (this work) |
| UGT85A1 | O3 | Cl TM1 | – | Cl | Cl1 | Cl1 | TM | C (Jin et al., 2013) |
| CKX1 | M4 | V3 | SP3 | Cl | V5 | M4 | V | V (Werner et al., 2003) |
| CKX2 | SP2 | EC1 | SP1 | – | C2 | – | ER | EC∗ (Werner et al., 2003) |
| CKX7 | O3 | EC2 | SP1 | – | C3 | – | EC | C (Köllmer et al., 2014) |
| RubSC† | Cl3 | EC1 | SP1 | Cl | Cl1 | Cl1 | Cl | Cl |
Subcellular compartments: mitochondria (M), chloroplast (Cl), membrane bound protein (TM), unspecified signal peptide (SP), nucleus (N), vacuole (V), cytosol (C), extracellular protein (EC) and other location (O), no signal peptide in the sequence (–). Prediction reliability class (from most reliable to the lowest reliable): 1.000–0.800 (1), 0.800–0.600 (2), 0.600–0.400 (3), 0.400–0.200 (4), 0.200–0.000 (5). ∗AtCKX2 is proposed to function in cytosol based on structure and function analogy to ZmCKX1 (Šmehilová et al., 2009). †RubSC, Ribulose bisphosphate carboxylase small chain, functions in ribulose bisphosphate carboxylase complex in CO2 fixation localized in chloroplast.
FIGURE 9.
Subcellular localization analysis of UGT76C2-GFP fusion protein. Green channel and transmission light images captured by confocal microscopy. (A) A control root overexpressing SU:GFP with typical GFP pattern in cytosol and nuclei. (B) The root cells overexpressing SU:UGT76C2-GFP with signal indicating the cytosolic localization of UGT76C2. Scale bars 10 μm.
Discussion
It is widely accepted that CK levels decrease during leaf senescence; however, this is based on former studies where the information was assumed based only on restricted CK types and forms of only limited time points and plant species analyzed. Specifically, a decreased zeatin level was observed in senescent tobacco leaves (Singh et al., 1992), followed by finding that higher amount of glucosides are present in mature than young tobacco leaves (Benková et al., 1999). In recent studies, CK glucosides were shown to be generally prevalent in the species analyzed so far (Gajdošová et al., 2011). Furthermore, particularly N-glucosides were shown to be predominant CK in tobacco (Havlová et al., 2008; Mýtinová et al., 2011; Uzelac et al., 2015). In our experiment, we assessed distribution of CK forms during Arabidopsis development with focus on leaf senescence, demonstrating that particularly N7- are the most predominant glucosides in Arabidopsis under physiological conditions (Figure 1C) with enormous accumulation in senescence leaf. Since no particular UGT is responsible for the dramatic increase according to our data, we can therefore assume that the accumulation is caused by fact that the N-glucosides are terminal metabolites. Besides N-glucosides, tZOG increased specifically in senescent leaf (Figure 1D) which agrees with strongly upregulated expression of UGT85A1 (Figure 2A). In senescent leaf, active CK level drops down to approx. 60% of their initial content, which is in strong contrast with more than fivefold increase of CK glucosides. In fact, free bases and ribosides are active in nanomolar concentrations and are maintained within this physiological range ensured by their precise regulation. Thus, this observation raises a question whether CK glucosides, particularly their terminal N- forms, possess any physiological role when they accumulate in such enormous concentrations in comparison to active CK. Here it has to be noted that CK O-glucosides are not substrates for any maize CKX enzymes (Zalabák et al., 2014), which suggests that glucosylation presumably protects CKs from their rapid degradation. Neither N- nor O-glucosides were found to be able to trigger known CK receptors (Spíchal et al., 2004; Romanov et al., 2006; Stolz et al., 2011), and no inhibition effect on the receptors was observed for tZOG (Romanov et al., 2005). The question was, whether N-glucosides could possibly possess an inhibitory effect on the receptors. In order to extend the current knowledge, we analyzed the most accumulated CK glucosides (iP7G, iP9G, tZ7G and tZ9G) for their inhibition potency on AHK3 and AHK4 receptors in a competition test (Romanov et al., 2005, 2006) in concentration ratios similar to those detected in senescence stage. However, no inhibition was detected for any of the tested glucosides (data not shown). Therefore we confirmed that glucosylation of CKs has a physiological significance as storage or terminal deactivation mechanism rather than interference of glucosides with the CK signalization pathway.
Up to date, the knowledge of in vivo function of UGT85A1 as a CK deactivation enzyme was based on gain-of-function mutants that showed a potential to O-glucosylate preferentially tZ. Our results showed that UGT85A1 is specifically expressed in senescence leaves. Since characterization of a loss-of-function mutant is one of the most powerful approaches commonly used to directly unravel the studied protein’s function we used this approach to investigate possible significance of UGT85A1 in senescence. We characterized a true loss-of-function mutant of UGT85A1 in the present work and extended the knowledge of its proposed specific O-glucosylation of tZ (Jin et al., 2013) by observation that this isoform could partially contribute also to other CK-glucosides formation (Figure 5) if this fact is not caused by other regulatory processes in the mutant. The suggested broader range of CK acceptors of UGT85A1 could be supported by the knowledge that UGTs can glycosylate a rather wider range of substrates (Bowles et al., 2005) and further by the fact that a recombinant UGT85A1 showed comparable specificity to tZ and cZ substrate as well as to DHZ (Hou et al., 2004). Since detected lower CK-glucosides concentrations of ugt85a1-1 did not manifest in phenotype changes in comparison to ugt76c1 (Wang et al., 2013), we performed experiment with induced senescence in detached leaves and observed decrease in chlorophyll degradation together with enhanced anthocyanin accumulation, further supported by modulated expression of stress and senescence related genes (Figure 4). Taken together, our results suggest that UGT85A1 plays important role in proper senescence process progress in Arabidopsis leaf.
Former studies pointed out that UGT76C2 could possess a significant role in CK regulation. UGT76C2 transcript was shown to be upregulated in response to exogenously applied iP (Motte et al., 2013), KIN (Mik et al., 2011) or BAP (Bhargava et al., 2013; Vylíčilová et al., 2016), whereas no regulation of UGT76C1 or UGT85A1 was detected. This led us to hypothesis that UGT76C2 might play a prominent role in CK homeostasis maintenance. No difference in expression level of UGT76C1 and UGT85A1 can be explained by the fact that BAP, KIN or iP are not the best substrates for the above two enzymes (Hou et al., 2004). Therefore, we performed a comparative study where we assessed an expression profile of individual UGTs with all the above substrates implementing tZ. Our results showed only slight response of UGT85A1 to tZ uptake, but more importantly, they indicated enormous upregulation of UGT76C2 expression in response to each of the tested CK (Figure 2B). The slight increase in UGT85A1 transcript after tZ uptake was further confronted with publicly available transcriptomic data (not shown here), but no significant response was observed in analyzed experiments. Brenner et al. (2012) performed a comparative meta-analysis of the above transcriptomic experiments and proposed UGT76C2 to be part of CK response core genes. That is in good agreement with data published by research group of Professor Hou who demonstrated that CK homeostasis is maintained by regulation on CK perception and response level in ugt76c2 mutant (Wang et al., 2011). Although the same pattern of regulation was shown in ugt76c1 mutant under standard conditions, the differences in relative quantity in comparison to WT are rather moderate (Wang et al., 2013). Moreover, when comparing CK glucosides content of all the UGTs mutants under standard conditions and after KIN uptake (Figures 7A–F), ugt76c2 possesses the lowest concentrations. Thus, taking into account all our findings together with previously published data, we conclude that UGT76C2 possesses an exclusive role in overall CK homeostasis maintenance.
Besides UGT76C2, fast response to CK treatment is ensured by CKX whose transcripts rise as early as 15 min after the treatment in our previous studies (Mik et al., 2011; Motte et al., 2013). In our experiment with exogenously applied CK, we compared CK content after tZ and KIN treatment and found out that, surprisingly, except expected N-glucosides formation, tZOG accumulation was also detected in both treatments. Although UGT85A1 was shown to be the main contributor to tZOG pool (Jin et al., 2013), we did not detect any activation on transcript level. It could be hypothesized that the enzyme is activated on protein level or can simply produce more tZOG when needed. In addition, it can be hypothesized that UGT76C2 contributes to increased tZOG level, since we detected decreased levels of tZOG in ugt76c2 after KIN uptake by detached leaves. Further, it has to be noted that even though KIN was not detected to be a suitable substrate for recombinant UGT85A1 (Hou et al., 2004), upregulated expression of UGT85A1 was detected after KIN treatment in vivo when incubated in continuous dark (Mik et al., 2011). The ratio of tZ and tZOG in ugt76c2 suggests importance of maintenance of adequate tZOG level when UGT76C2 capability of CK glucosylation is impaired.
Our results on CK content determination revealed a significantly different regulation in ugt76c2 mutant, whereas ugt76c1 and ugt85a1-1 shared a similar CK profile with minor differences. The data suggest UGT76C1 to be more cZ specific, which is not in contrast with previous study (Wang et al., 2013). Although we did not conclude any specific physiological function of UGT76C1 in our study in contrast to UGT76C2 and UGT85A1, we detected slightly higher expression of UGT76C2 in ugt76c1mutant. This could eventually explain only slightly decreased glucosides in this mutant in our experimental condition and thus point to speculative co-expression of these closely related isoforms. Yet, since we did not detect any redundancy of these two isoforms in our experiment, the role of UGT76C1 remains unclear. Gene profiling of CK metabolism brought more light to regulatory mechanism of the plant’s need to balance the impaired glucosylation. We confirmed the mechanism of enhanced CK degradation and decreased CK perception (ensured particularly by CKX3, CKX6, AHK2, AHK3 and ARR1) as general when CK glucosylation is impaired, since we observed this pattern in ugt85a1-1 mutant as well as in ugt76c1 and ugt76c2, as was described before (Wang et al., 2011, 2013). The plants’ enhanced CKX expression agrees with observed decreased levels of tZ (Figures 7E,F) since it is one of the best substrates of these isoforms (Galuszka et al., 2007). CKX3 was also shown to be the main isoform responsible for CK excesses degradation after KIN uptake in WT Arabidopsis (Mik et al., 2011). We detected a specific regulation in individual UGT mutants mediated by CKX1 (in case of ugt76c2) and further by CKX2 and CKX7 (in case of ugt85a1-1 and ugt76c2). A specific regulation was also detected in ugt85a1-1, since this was the only case when downregulation of CK biosynthesis was observed under standard condition (Figure 8). The specific regulation on transcript level goes well with CK content determined based on the enzymes substrate preferences (Miyawaki et al., 2006; Galuszka et al., 2007; Kuroha et al., 2009) and likewise correlates well with previous studies on CKX overexpressing lines (Köllmer et al., 2014). Interestingly, we reported increased levels of cZ(R) in ugt76c2 after KIN uptake (Figure 7F) and, reversely, lowered cZs in ugt85a1-1 in whole plants (Figure 5) as well as in detached leaves (Figure 7F). Increased levels of cZ(R)s were found in many plant species in response to both, abiotic and biotic stress (Schäfer et al., 2015), as well as after exogenous application of CK (Vyroubalová et al., 2009). This phenomenon can support the importance of UGT76C2 action and thus increased cZ(R)s accumulation when the action of the protein is impaired. Reversely, lowered cZs could be an accompanying effect of possibly better stress performance of ugt85a1-1, especially when taking into account that that lowered level of ABA biosynthetic gene was detected in detached leaves in the UGT85A1 loss-of-function mutant in our stress experiment.
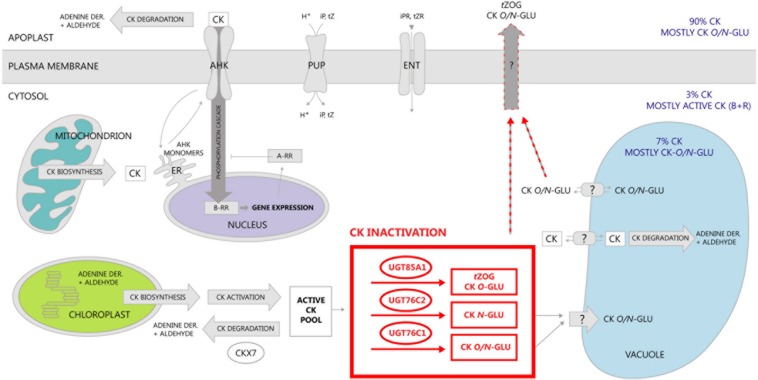
In this study, we further investigated compartmentation of CK glucosylation. The fact that UGT76C2 possess an exclusive role in CK homeostasis maintenance raised a question of its subcellular localization that could elucidate compartmentation of CK action. We localized this enzyme using GFP-tagging in cytosol. Analogically to the results obtained for UGT76C2, our results on signal peptide homology and in silico prediction for UGT76C1 suggest cytosolic localization of this isoform. UGT85A1 was confirmed to be a cytosolic enzyme before (Jin et al., 2013). As discussed in an outstanding review by Lim and Bowels, glycosylation and deglycosylation can regulate levels of metabolites in pathways through controlling their exit and re-entry from cytosol into their reaction environments (Lim and Bowles, 2004). Based on our newly gained knowledge of CK glucosylation taking place in cytosol, along with the current finding of CK forms compartmentation in Arabidopsis (Jiskrová et al., 2016), we present a new hypothesis of CK metabolism compartmentation on subcellular level schematically illustrated in Figure 10. We employed former schemes of exquisite reviews (Kasahara et al., 2004; Frébort et al., 2011; Ha et al., 2012; Spíchal, 2012; Zalabák et al., 2013) and supplemented them with all currently known information on CK metabolism compartmentation. Among all Arabidopsis CKX enzymes, CKX7 possesses a prominent role in CK homeostasis maintenance inside the cell, since it is the only cytosolic CKX (Köllmer et al., 2014). Interestingly, this is the only CKX isoform with a rather constitutive expression pattern (Mik et al., 2011; Motte et al., 2013). Further, when taking into account kinetic parameters of CKX and UGT enzymes in general (Bilyeu et al., 2001; Hou et al., 2004), we propose that CK glucosylation within the cytosol might serve as an immediate fine-tuning mechanism of inner CK homeostasis that subsequently contributes to overall CK metabolism status once the formed CK-glucosides are accumulated in extracellular space (Jiskrová et al., 2016). Since glycosylation makes molecules more hydrophilic and better accessible to membrane-bound transporters (Lim and Bowles, 2004), we further hypothesize a transport mechanism of CK-glucosides out of the cytosol with specificity to tZOG. In fact, glucosides can enter other more hydrophilic compartments such vacuoles, which was proved for CK-glucosides except tZOG (Jiskrová et al., 2016). Apart from the CK non-specific transport mediated by PUP and ENT transporters across plasma membrane (Gillissen et al., 2000; Bürkle et al., 2003; Hirose et al., 2005, 2007; Sun et al., 2005), not much is known about the CK transport mechanism in general; thus, we emphasize importance of elucidation of CK inter-compartmental transport to overall understanding of CK signalization.
FIGURE 10.
Schematic distribution of CK and compartmentation of CK metabolism in Arabidosis. CK biosynthesis was localized mainly to plastids and further to mitochondrion and cytosol (Kasahara et al., 2004) followed by their activation that occurs within cytosol (Chen and Kristopeit, 1981a,b; Kuroha et al., 2009). Distribution of distinct CK forms within cellular compartments was elucidated in our recent publication (Jiskrová et al., 2016). Almost no active CK content in apoplast in comparison to CK glucosides is given by the fact that CK degradation takes place prevalently there (Werner et al., 2003). Besides CK degradation in apoplast, two of its enzymes were localized in vacuoles (Werner et al., 2003), whereas only one enzyme (CKX7) was revealed to play its role in cytoplasm (Köllmer et al., 2014). In this study, we confirmed localization of CK glucosylation in cytosol, thereby suggesting a transport mechanism of CK glucosides across plasma membrane out of the cytosol. The only known transporters of CK up to date are purine permeases (PUP) transporting iP and tZ (Gillissen et al., 2000; Bürkle et al., 2003; Cedzich et al., 2008), and equilibrative nucleoside transporters (ENT) (Sun et al., 2005; Hirose et al., 2007). Perception and signal transduction of CK was described in many works up to now (for review, see Kakimoto, 2003; or Spíchal, 2012). In this scheme, we adopt a hypothesis from our review (Zalabák et al., 2013) where recycling of AHK monomers between endosomes and plasma membrane (Dortay et al., 2008; Lomin et al., 2011) is proposed as a possible compromise scenario.
Conclusion
In this study, we present results of our comprehensive targeted analysis of CK metabolites and related transcript levels of mutants of three CK-specific UGTs. We confronted our results on loss-of-function mutants of cytokinin-specific glycosyltransferases with previous studies and propose physiological significance rather of cytokinin glycosylation process than of CK-glucosides themselves. We hypothesize a quick fine-tuning effect of cytokinin-specific glycosyltransferases on active CK levels in cytoplasm alongside their rapid degradation by CKX enzymes that take place predominantly in extracellular space in Arabidopsis.
Author Contributions
MŠ served as principal investigator, designed and performed the experiments, conceived the project, drafted and finalized the manuscript. JD assisted with the experiments and data analysis, ON with cytokinin content analysis and TT with confocal microscopy. PG and ON assisted with manuscript preparation and revision.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Christopher G. Taylor for providing AKK 1436, AKK 1472B vectors and Agrobacterium rhizogenes 15834 strain. We wish to thank Michaela Mrvková and Hana Martinková for CK analysis, Věra Chytilová and Kateřina Střelcová for assistance with plant material, Zuzana Mičková for the AHK receptors inhibition test, Veronika Kořínková for assistance with graphics and Tomáš Hluska for his critical reading of the manuscript. Special thanks belong to Julie Škrabišová.
Abbreviations
- BAP
N6-benzylaminopurine
- cZ
cis-zeatin
- cZ9G
cZ N9-glucoside
- cZOG
cZ O-glucoside
- cZROG
cZR O-glucoside
- cZR
cZ riboside
- cZRMP
cZR monophosphate
- DHZ
dihydrozeatin
- DHZR
DHZ riboside
- DHZOG
DHZ O-glucoside
- DHZR
O-glucoside
- DHZ7G
DHZ N7-glucoside
- DHZ9G
DHZ N9-glucoside
- DHZRMP
DHZR monophosphate
- iP
N6-(Δ2-isopentenyl)adenine
- iPR
iP riboside
- iP7G
iP N7-glucoside
- iP9G
iPN9-glucoside
- iPRMP
iPR monophosphate
- Kin
kinetin
- tZ
trans-zeatin
- tZ7G
tZ N7-glucoside
- tZ9G
tZ N9-glucoside
- tZr
tZ riboside
- tZOG
tZ O-glucoside
- tZROG
tZR O-glucoside
- tZRMP
tZR monophosphate
- UGT
uridine diphosphate glucose glycosyltransferase
Funding. This work was solely supported by the grant P501/12/P160 from the Czech Science Foundation, Czech Republic.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01264
References
- Bairu M. W., Novák O., Doležal K., van Staden J. (2011). Changes in endogenous cytokinin profiles in micropropagated Harpagophytum procumbens in relation to shoot-tip necrosis and cytokinin treatments. Plant Growth Regul. 63 105–114. 10.1007/s10725-010-9558-6 [DOI] [Google Scholar]
- Bajguz A., Piotrowska A. (2009). Conjugates of auxin and cytokinin. Phytochemistry 70 957–969. 10.1016/j.phytochem.2009.05.006 [DOI] [PubMed] [Google Scholar]
- Balazadeh S., Riaño-Pachón D. M., Mueller-Roeber B. (2008). Transcription factors regulating leaf senescence in Arabidopsis thaliana. Plant Biol. 10 63–75. 10.1111/j.1438-8677.2008.00088.x [DOI] [PubMed] [Google Scholar]
- Bannai H., Tamada Y., Maruyama O., Nakai K., Miyano S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18 298–305. 10.1093/bioinformatics/18.2.298 [DOI] [PubMed] [Google Scholar]
- Benková E., Witters E., Van Dongen W., Kolár J., Motyka V., Brzobohatý B., et al. (1999). Cytokinins in tobacco and wheat chloroplasts. Occurrence and changes due to light/dark treatment. Plant Physiol. 121 245–252. 10.1104/pp.121.1.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava A., Clabaugh I., To J. P., Maxwell B. B., Chiang Y.-H., Schaller G. E., et al. (2013). Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol. 162 272–294. 10.1104/pp.113.217026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilyeu K. D., Cole J. L., Laskey J. G., Riekhof W. R., Esparza T. J., Kramer M. D., et al. (2001). Molecular and biochemical characterization of a cytokinin oxidase from maize. Plant Physiol. 125 378–386. 10.1104/pp.125.1.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoeva E., Dobrev P. I., Malbeck J., Motyka V., Strnad M., Hanuš J., et al. (2004). Cytokinin N-glucosylation inhibitors suppress deactivation of exogenous cytokinins in radish, but their effect on active endogenous cytokinins is counteracted by other regulatory mechanisms. Physiol. Plant. 121 215–222. 10.1111/j.1399-3054.2004.00320.x [DOI] [PubMed] [Google Scholar]
- Bowles D., Isayenkova J., Lim E.-K., Poppenberger B. (2005). Glycosyltransferases: managers of small molecules. Curr. Opin. Plant Biol. 8 254–263. 10.1016/j.pbi.2005.03.007 [DOI] [PubMed] [Google Scholar]
- Brenner W. G., Ramireddy E., Heyl A., Schmülling T. (2012). Gene regulation by cytokinin in Arabidopsis. Front. Plant Sci. 3:8 10.3389/fpls.2012.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzobohatý B., Moore I., Kristoffersen P., Bako L., Campos N., Schell J., et al. (1993). Release of active cytokinin by a beta-glucosidase localized to the maize root meristem. Science 262 1051–1054. 10.1126/science.8235622 [DOI] [PubMed] [Google Scholar]
- Bürkle L., Cedzich A., Döpke C., Stransky H., Okumoto S., Gillissen B., et al. (2003). Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. Plant J. 34 13–26. 10.1046/j.1365-313X.2003.01700.x [DOI] [PubMed] [Google Scholar]
- Carviel J. L., Al-Daoud F., Neumann M., Mohammad A., Provart N. J., Moeder W., et al. (2009). Forward and reverse genetics to identify genes involved in the age-related resistance response in Arabidopsis thaliana. Mol. Plant Pathol. 10 621–634. 10.1111/j.1364-3703.2009.00557.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R., Medina J., Salinas J. (2011). Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108 16475–16480. 10.1073/pnas.1107161108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedzich A., Stransky H., Schulz B., Frommer W. B. (2008). Characterization of cytokinin and adenine transport in Arabidopsis cell cultures. Plant Physiol. 148 1857–1867. 10.1104/pp.108.128454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-M., Kristopeit S. M. (1981a). Metabolism of cytokinin: dephosphorylation of cytokinin ribonucleotide by 5′-nucleotidases from wheat germ cytosol. Plant Physiol. 67 494–498. 10.1104/pp.67.3.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-M., Kristopeit S. M. (1981b). Metabolism of cytokinin: deribosylation of cytokinin ribonucleoside by adenosine nucleosidase from wheat germ cells. Plant Physiol. 68 1020–1023. 10.1104/pp.68.5.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J. (1991). Light signals in leaf and chloroplast development – photoreceptors and downstream responses in search of a transduction pathway. New Biol. 3 538–548. [PubMed] [Google Scholar]
- Chou K.-C., Shen H.-B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5:e11335 10.1371/journal.pone.0011335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier R., Fuchs B., Walter N., Lutke W. K., Taylor C. G. (2005). Ex vitro composite plants: an inexpensive, rapid method for root biology. Plant J. 43 449–457. 10.1111/j.1365-313X.2005.02454.x [DOI] [PubMed] [Google Scholar]
- Cowley D. E., Duke C. C., Liepa A. J., Macleod J. K., Letham D. S. (1978). Structure and synthesis of cytokinin metabolites. 1. The 7- and 9- β-D-glucofuranosides and pyranosides of zeatin and 6-benzylaminopurine. Aust. J. Chem. 31 1095–1111. 10.1071/CH9781095 [DOI] [Google Scholar]
- Dobrev P. I., Kamínek M. (2002). Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A 950 21–29. 10.1016/S0021-9673(02)00024-9 [DOI] [PubMed] [Google Scholar]
- Dortay H., Gruhn N., Pfeifer A., Schwerdtner M., Schmülling T., Heyl A. (2008). Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. J. Proteome Res. 7 3649–3660. 10.1021/pr0703831 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300 1005–1016. 10.1006/jmbi.2000.3903 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., Von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. 10.1110/ps.8.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entsch B., Letham D. S. (1979). Enzymic glucosylation of the cytokinin, 6-benzylaminopurine. Plant Sci. Lett. 14 205–212. 10.1016/0304-4211(79)90061-0 [DOI] [Google Scholar]
- Entsch B., Parker C. W., Letham D. S., Summons R. E. (1979). Preparation and characterization, using high-performance liquid chromatography, of an enzyme forming glucosides of cytokinins. Biochim. Biophys. Acta 570 124–139. 10.1016/0005-2744(79)90207-9 [DOI] [PubMed] [Google Scholar]
- Falk A., Rask L. (1995). Expression of a zeatin-O-glucoside-degrading β-glucosidase in Brassica napus. Plant Physiol. 108 1369–1377. 10.1104/pp.108.4.1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frébort I., Kowalska M., Hluska T., Frébortová J., Galuszka P. (2011). Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 62 2431–2452. 10.1093/jxb/err004 [DOI] [PubMed] [Google Scholar]
- Fusseder A., Ziegler P. (1988). Metabolism and compartmentation of dihydrozeatin exogenously supplied to photoautotrophic suspension cultures of Chenopodium rubrum. Planta 173 104–109. 10.1007/bf00394494 [DOI] [PubMed] [Google Scholar]
- Gajdošová S., Spíchal L., Kamínek M., Hoyerová K., Novák O., Dobrev P. I., et al. (2011). Distribution, biological activities, metabolism, and the conceivable function of cis-zeatin-type cytokinins in plants. J. Exp. Bot. 62 2827–2840. 10.1093/jxb/erq457 [DOI] [PubMed] [Google Scholar]
- Galuszka P., Popelková H., Werner T., Frébortová J., Pospíšilová H., Mik V., et al. (2007). Biochemical characterization of cytokinin oxidases/dehydrogenases from Arabidopsis thaliana expressed in Nicotiana tabacum L. J. Plant Growth Regul. 26 255–267. 10.1007/s00344-007-9008-5 [DOI] [Google Scholar]
- Gandia-Herrero F., Lorenz A., Larson T., Graham I. A., Bowles D. J., Rylott E. L., et al. (2008). Detoxification of the explosive 2,4,6-trinitrotoluene in Arabidopsis: discovery of bifunctional O- and C-glucosyltransferases. Plant J. 56 963–974. 10.1111/j.1365-313X.2008.03653.x [DOI] [PubMed] [Google Scholar]
- Gepstein S., Sabehi G., Carp M. J., Hajouj T., Nesher M. F. O., Yariv I., et al. (2003). Large-scale identification of leaf senescence-associated genes. Plant J. 36 629–642. 10.1046/j.1365-313X.2003.01908.x [DOI] [PubMed] [Google Scholar]
- Gillissen B., Burkle L., Andre B., Kuhn C., Rentsch D., Brandl B., et al. (2000). A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12 291–300. 10.1105/tpc.12.2.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebenok R. J., Lambert G. M., Galbraith D. W. (1997a). Characterization of targeted nuclear accumulation of gfp in cells of transgenic plants. Plant J. 12 685–696. 10.1046/j.1365-313X.1997.00685.x [DOI] [Google Scholar]
- Grebenok R. J., Pierson E., Lambert G. M., Gong F.-C., Afonso C. L., Haldeman-Cahill R., et al. (1997b). Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11 573–586. 10.1046/j.1365-313X.1997.11030573.x [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 156 1612–1619. 10.1104/pp.111.177022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S., Vankova R., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.-S. P. (2012). Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci. 17 172–179. 10.1016/j.tplants.2011.12.005 [DOI] [PubMed] [Google Scholar]
- Havlová M., Dobrev P. I., Motyka V., Štorchová H., Libus J., Dobrá J., et al. (2008). The role of cytokinins in responses to water deficit in tobacco plants over-expressing trans-zeatin O-glucosyltransferase gene under 35S or SAG12 promoters. Plant Cell Environ. 31 341–353. 10.1111/j.1365-3040.2007.01766.x [DOI] [PubMed] [Google Scholar]
- Hensel L. L., Grbiæ V., Baumgarten D. A., Bleecker A. B. (1993). Developmental and age-related processes that influence the longevity and senescence of photosynthetic tissues in Arabidopsis. Plant Cell 5 553–564. 10.1105/tpc.5.5.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinderhofer K., Zentgraf U. (2001). Identification of a transcription factor specifically expressed at the onset of leaf senescence. Planta 213 469–473. 10.1007/s004250000512 [DOI] [PubMed] [Google Scholar]
- Hirose N., Makita N., Yamaya T., Sakakibara H. (2005). Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 138 196–206. 10.1104/pp.105.060137.syltransferase [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Takei K., Kuroha T., Kamada-Nobusada T., Hayashi H., Sakakibara H. (2007). Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59 75–83. 10.1093/jxb/erm157 [DOI] [PubMed] [Google Scholar]
- Hong Z. L., Zhang Z. M., Olson J. M., Verma D. P. S. (2001). A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 13 769–779. 10.1105/tpc.13.4.769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou B., Lim E.-K., Higgins G. S., Bowles D. J. (2004). N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J. Biol. Chem. 279 47822–47832. 10.1074/jbc.M409569200 [DOI] [PubMed] [Google Scholar]
- Husar S., Berthiller F., Fujioka S., Rozhon W., Khan M., Kalaivanan F., et al. (2011). Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol. 11:51 10.1186/1471-2229-11-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. H., Ma X. M., Kojima M., Sakakibara H., Wang Y. W., Hou B. K. (2013). Overexpression of glucosyltransferase UGT85A1 influences trans-zeatin homeostasis and trans-zeatin responses likely through O-glucosylation. Planta 237 991–999. 10.1007/s00425-012-1818-4 [DOI] [PubMed] [Google Scholar]
- Jiskrová E., Novák O., Pospíšilová H., Holubová K., Karády M., Galuszka P., et al. (2016). Extra- and intracellular distribution of cytokinins in the leaves of monocots and dicots. N. Biotechnol. 33(5 Pt B), 735–742. 10.1016/j.nbt.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Kakimoto T. (2003). Perception and signal transduction of cytokinins. Annu. Rev. Plant Biol. 54 605–627. 10.1146/annurev.arplant.54.031902.134802 [DOI] [PubMed] [Google Scholar]
- Kasahara H., Takei K., Ueda N., Hishiyama S., Yamaya T., Kamiya Y., et al. (2004). Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 279 14049–14054. 10.1074/jbc.M314195200 [DOI] [PubMed] [Google Scholar]
- Kato C., Kato H., Asami T., Yoshida S., Noda H., Kamada H., et al. (2002). Involvement of xylem sap zeatin-O-glucoside in cucumber shoot greening. Plant Physiol. Biochem. 40 949–954. 10.1016/S0981-9428(02)01458-4 [DOI] [Google Scholar]
- Kim H. J., Ryu H., Hong S. H., Woo H. R., Lim P. O., Lee I. C., et al. (2006). Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103 814–819. 10.1073/pnas.0505150103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler R. H., Zipfel W. R., Webb W. W., Hanson M. R. (1997). The green fluorescent protein as a marker to visualize plant mitochondria in vivo. Plant J. 11 613–621. 10.1046/j.1365-313X.1997.11030613.x [DOI] [PubMed] [Google Scholar]
- Köllmer I., Novák O., Strnad M., Schmülling T., Werner T. (2014). Overexpression of the cytosolic cytokinin oxidase/dehydrogenase (CKX7) from Arabidopsis causes specific changes in root growth and xylem differentiation. Plant J. 78 359–371. 10.1111/tpj.12477 [DOI] [PubMed] [Google Scholar]
- Kristoffersen P., Brzobohatý B., Höhfeld I., Bako L., Melkonian M., Palme K. (2000). Developmental regulation of the maize Zm-p60.1 gene encoding a β-glucosidase located to plastids. Planta 210 407–415. 10.1007/PL00008149 [DOI] [PubMed] [Google Scholar]
- Kudo T., Kiba T., Sakakibara H. (2010). Metabolism and long-distance translocation of cytokinins. J. Integr. Plant Biol. 52 53–60. 10.1111/j.1744-7909.2010.00898.x [DOI] [PubMed] [Google Scholar]
- Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., et al. (2009). Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21 3152–3169. 10.1105/tpc.109.068676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letham D. S., Summons R. E., Entsch B., Gollnow B. I., Parker C. W., MacLeod J. K. (1978). Glucosylation of cytokinin analogues. Phytochemistry 17 2053–2057. 10.1016/S0031-9422(00)89279-1 [DOI] [Google Scholar]
- Li Y., Baldauf S., Lim E.-K., Bowles D. J. (2001). Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J. Biol. Chem. 276 4338–4343. 10.1074/jbc.M007447200 [DOI] [PubMed] [Google Scholar]
- Li Y., Wang B., Dong R., Hou B. (2015). AtUGT76C2, an Arabidopsis cytokinin glycosyltransferase is involved in drought stress adaptation. Plant Sci. 236 157–167. 10.1016/j.plantsci.2015.04.002 [DOI] [PubMed] [Google Scholar]
- Lim E.-K., Bowles D. J. (2004). A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 23 2915–2922. 10.1038/sj.emboj.7600295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lomin S. N., Yonekura-Sakakibara K., Romanov G. A., Sakakibara H. (2011). Ligand-binding properties and subcellular localization of maize cytokinin receptors. J. Exp. Bot. 62 5149–5159. 10.1093/jxb/err220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie P. I., Owens I. S., Burchell B., Bock K. W., Bairoch A., Belanger A., et al. (1997). The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence. Pharmacogenetics 7 255–269. 10.1097/00008571-199708000-00001 [DOI] [PubMed] [Google Scholar]
- Martin R. C., Mok M. C., Habben J. E., Mok D. W. S. (2001). A maize cytokinin gene encoding an O-glucosyltransferase specific to cis-zeatin. Proc. Natl. Acad. Sci. U.S.A. 98 5922–5926. 10.1073/pnas.101128798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R. C., Mok M. C., Mok D. W. S. (1999). A gene encoding the cytokinin enzyme zeatin O-xylosyltransferase of Phaseolus vulgaris. Plant Physiol. 120 553–558. 10.1104/pp.120.2.553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaw B. A., Heald J. K., Horgan R. (1984). Dihydrozeatin metabolism in radish seedlings. Phytochemistry 23 1373–1377. 10.1016/S0031-9422(00)80468-9 [DOI] [Google Scholar]
- McGaw B. A., Horgan R. (1985). Cytokinin metabolism and the control of cytokinin activity. Phytochemistry 27 180–187. 10.1007/BF02902158 [DOI] [Google Scholar]
- Meek L., Martin R. C., Shan X., Karplus P. A., Mok D. W. S., Mok M. C. (2008). Isolation of legume glycosyltransferases and active site mapping of the Phaseolus lunatus zeatin O-glucosyltransferase ZOG1. J. Plant Growth Regul. 27 192–201. 10.1007/s00344-008-9045-8 [DOI] [Google Scholar]
- Mik V., Szüčová L., Šmehilová M., Zatloukal M., Doležal K., Nisler J., et al. (2011). N9-substituted derivatives of kinetin: effective anti-senescence agents. Phytochemistry 72 821–831. 10.1016/j.phytochem.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Miyawaki K., Tarkowski P., Matsumoto-Kitano M., Kato T., Sato S., Tarkowska D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103 16598–16603. 10.1073/pnas.0603522103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok D. W. S., Martin R. C., Shan X., Mok M. C. (2000a). Genes encoding zeatin O-glycosyltransferases. Plant Growth Regul. 32 285–287. 10.1023/A:1010712102890 [DOI] [Google Scholar]
- Mok D. W. S., Mok M. C. (2001). Cytokinin Metabolism and Action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 89–118. 10.1146/annurev.arplant.52.1.89 [DOI] [PubMed] [Google Scholar]
- Mok M. C., Martin R. C., Mok D. W. S. (2000b). Cytokinins: biosynthesis metabolism and perception. Vitro. Cell. Dev. Biol. Plant 36 102–107. 10.1007/s11627-000-0021-7 [DOI] [Google Scholar]
- Mok M. C., Martin R. C., Mok D. W. S., Shaw G. (1992). “Cytokinin activity metabolism and function in Phaseolus,” in Physiology and Biochemistry of Cytokinins in Plants, eds Kamínek M., Mok D. W. S., Zažímalová E. (The Hague: SPB Academic Publishing; ), 41–46. [Google Scholar]
- Motte H., Galuszka P., Spíchal L., Tarkowski P., Plíhal O., Šmehilová M., et al. (2013). Phenyl-adenine, identified in a LIGHT-DEPENDENT SHORT HYPOCOTYLS4-assisted chemical screen, is a potent compound for shoot regeneration through the inhibition of CYTOKININ OXIDASE/DEHYDROGENASE activity. Plant Physiol. 161 1229–1241. 10.1104/pp.112.210716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrízová K., Jiskrová E., Vyroubalová Š., Novák O., Ohnoutková L., Pospíšilová H., et al. (2013). Overexpression of cytokinin dehydrogenase genes in barley (Hordeum vulgare cv. Golden promise) fundamentally affects morphology and fertility. PLoS ONE 8:79029 10.1371/journal.pone.0079029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assay with tobacco tissue cultures. Physiol. Plant. 15 473–497. 10.1111/j.1399-3054.1962.tb08052.x [DOI] [Google Scholar]
- Mýtinová Z., Motyka V., Haisel D., Lubovská Z., Trávníèková A., Dobrev P., et al. (2011). Antioxidant enzymatic protection during tobacco leaf ageing is affected by cytokinin depletion. Plant Growth Regul. 65 23–34. 10.1007/s10725-011-9571-4 [DOI] [Google Scholar]
- Neff M. M., Chory J. (1998). Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 118 27–36. 10.1104/pp.118.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O., Hauserová E., Amakorová P., Doležal K., Strnad M. (2008). Cytokinin profiling in plant tissues using ultra-performance liquid chromatography–electrospray tandem mass spectrometry. Phytochemistry 69 2214–2224. 10.1016/j.phytochem.2008.04.022 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Pineda Rodó A., Brugière N., Vankova R., Malbeck J., Olson J. M., Haines S. C., et al. (2008). Over-expression of a zeatin O-glucosylation gene in maize leads to growth retardation and tasselseed formation. J. Exp. Bot. 59 2673–2686. 10.1093/jxb/ern137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G. L., Vaistij F. E., Hiranuma S., et al. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. U.S.A. 102 15253–15258. 10.1073/pnas.0504279102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospíšilová H., Jiskrová E., Vojta P., Mrízová K., Kokáš F., Èudejková M. M., et al. (2016). Transgenic barley overexpressing a cytokinin dehydrogenase gene shows greater tolerance to drought stress. Natt. Biotechnol. 33 692–705. 10.1016/j.nbt.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Quilliam R. S., Swarbrick P. J., Scholes J. D., Rolfe S. A. (2006). Imaging photosynthesis in wounded leaves of Arabidopsis thaliana. J. Exp. Bot. 57 55–69. 10.1093/jxb/erj039 [DOI] [PubMed] [Google Scholar]
- Romanov G. A., Lomin S. N., Schmülling T. (2006). Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. J. Exp. Bot. 57 4051–4058. 10.1093/jxb/erl179 [DOI] [PubMed] [Google Scholar]
- Romanov G. A., Spíchal L., Lomin S. N., Strnad M., Schmülling T. (2005). A live cell hormone-binding assay on transgenic bacteria expressing a eukaryotic receptor protein. Anal. Biochem. 347 129–134. 10.1016/j.ab.2005.09.012 [DOI] [PubMed] [Google Scholar]
- Sakakibara H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57 431–449. 10.1146/annurev.arplant.57.032905.105231 [DOI] [PubMed] [Google Scholar]
- Schäfer M., Meza-Canales I. D., Brütting C., Baldwin I. T., Meldau S. (2015). Cytokinin concentrations and CHASE-DOMAIN CONTAINING HIS KINASE 2 (NaCHK2)- and NaCHK3-mediated perception modulate herbivory-induced defense signaling and defenses in Nicotiana attenuata. New Phytol. 207 645–658. 10.1111/nph.13404 [DOI] [PubMed] [Google Scholar]
- Singh S., Letham D. S., Palni L. M. S. (1992). Cytokinin biochemistry in relation to leaf senescence. VIII. Translocation, metabolism and biosynthesis of cytokinins in relation to sequential leaf senescence of tobacco. Physiol. Plant. 86 398–406. 10.1034/j.1399-3054.1992.860308.x [DOI] [Google Scholar]
- Šmehilová M., Galuszka P., Bilyeu K. D., Jaworek P., Kowalska M., Šebela M., et al. (2009). Subcellular localization and biochemical comparison of cytosolic and secreted cytokinin dehydrogenase enzymes from maize. J. Exp. Bot. 60 2701–2712. 10.1093/jxb/erp126 [DOI] [PubMed] [Google Scholar]
- Spíchal L. (2012). Cytokinins - Recent news and views of evolutionally old molecules. Funct. Plant Biol. 39 267–284. 10.1071/FP11276 [DOI] [PubMed] [Google Scholar]
- Spíchal L., Rakova N. Y., Riefler M., Mizuno T., Romanov G. A., Strnad M., et al. (2004). Two cytokinin receptors of Arabidopsis thaliana, CRE1/AHK4 and AHK3, differ in their ligand specificity in a bacterial assay. Plant Cell Physiol. 45 1299–1305. 10.1093/pcp/pch132 [DOI] [PubMed] [Google Scholar]
- Stolz A., Riefler M., Lomin S. N., Achazi K., Romanov G. A., Schmülling T. (2011). The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 67 157–168. 10.1111/j.1365-313X.2011.04584.x [DOI] [PubMed] [Google Scholar]
- Sun J., Hirose N., Wang X., Wen P., Xue L., Sakakibara H., et al. (2005). Arabidopsis SOI33/AtENT8 gene encodes a putative equilibrative nucleoside transporter that is involved in cytokinin transport in planta. J. Integr. Plant Biol. 47 588–603. 10.1111/j.1744-7909.2005.00104.x [DOI] [Google Scholar]
- Sun Y.-G., Wang B., Jin S.-H., Qu X.-X., Li Y.-J., Hou B.-K. (2013). Ectopic expression of Arabidopsis glycosyltransferase UGT85A5 enhances salt stress tolerance in tobacco. PLoS ONE 8:e59924 10.1371/journal.pone.0059924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svačinová J., Novák O., Plačková L., Lenobel R., Holík J., Strnad M., et al. (2012). A new approach for cytokinin isolation from Arabidopsis tissues using miniaturized purification: pipette tip solid-phase extraction. Plant Methods 8 17 10.1186/1746-4811-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B. C., Joseph L. M., Deng W. T., Liu L. J., Li Q. B., Cline K., et al. (2003). Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 35 44–56. 10.1046/j.1365-313X.2003.01786.x [DOI] [PubMed] [Google Scholar]
- Tian Q.-Y., Sun P., Zhang W.-H. (2009). Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytol. 184 918–931. 10.1111/j.1469-8137.2009.03004.x [DOI] [PubMed] [Google Scholar]
- Uzelac B., Janoševiæ D., Simonović A., Motyka V., Dobrev P. I., Budimir S. (2015). Characterization of natural leaf senescence in tobacco (Nicotiana tabacum) plants grown in vitro. Protoplasma 253 259–275. 10.1007/s00709-015-0802-9 [DOI] [PubMed] [Google Scholar]
- Veach Y. K., Martin R. C., Mok D. W. S., Malbeck J., Vankova R., Mok M. C. (2003). O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 131 1374–1380. 10.1104/pp.017210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vylíčilová H., Husičková A., Spíchal L., Srovnal J., Doležal K., Plíhal O., et al. (2016). C2-substituted aromatic cytokinin sugar conjugates delay the onset of senescence by maintaining the activity of the photosynthetic apparatus. Phytochemistry 122 22–33. 10.1016/j.phytochem.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Vyroubalová Š., Václavíková K., Turečková V., Novák O., Šmehilová M., Hluska T., et al. (2009). Characterization of new maize genes putatively involved in cytokinin metabolism and their expression during osmotic stress in relation to cytokinin levels. Plant Physiol 151 433–447. 10.1104/pp.109.142489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff C., Yang T. J. W., Stead A. D., Buchanan-Wollaston V., Roberts J. A. (2009). A molecular and structural characterization of senescing Arabidopsis siliques and comparison of transcriptional profiles with senescing petals and leaves. Plant J. 57 690–705. 10.1111/j.1365-313X.2008.03722.x [DOI] [PubMed] [Google Scholar]
- Wang J., Ma X.-M., Kojima M., Sakakibara H., Hou B.-K. (2013). Glucosyltransferase UGT76C1 finely modulates cytokinin responses via cytokinin N-glucosylation in Arabidopsis thaliana. Plant Physiol. Biochem. 65 9–16. 10.1016/j.plaphy.2013.01.012 [DOI] [PubMed] [Google Scholar]
- Wang J., Ma X.-M. X.-M., Kojima M., Sakakibara H., Hou B.-K. (2011). N-Glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol. 52 2200–2213. 10.1093/pcp/pcr152 [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H., Schmülling T. (2003). Cytokinin-deficient transgenic Arabidopsis plants show functions of cytokinins in the regulation of shoot and root meristem Activity. Plant Cell 15 2532–2550. 10.1105/tpc.014928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (1999). Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107 142–149. 10.1034/j.1399-3054.1999.100119.x [DOI] [Google Scholar]
- Woo H.-H. H., Jeong B. R., Hirsch A. M., Hawes M. C. (2007). Characterization of Arabidopsis AtUGT85A and AtGUS gene families and their expression in rapidly dividing tissues. Genomics 90 143–153. 10.1016/j.ygeno.2007.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalabák D., Galuszka P., Mrízová K., Podlešáková K., Gu R., Frébortová J. (2014). Biochemical characterization of the maize cytokinin dehydrogenase family and cytokinin profiling in developing maize plantlets in relation to the expression of cytokinin dehydrogenase genes. Plant Physiol. Biochem. 74 283–293. 10.1016/j.plaphy.2013.11.020 [DOI] [PubMed] [Google Scholar]
- Zalabák D., Pospíšilová H., Šmehilová M., Mrízová K., Frébort I., Galuszka P. (2013). Genetic engineering of cytokinin metabolism: prospective way to improve agricultural traits of crop plants. Biotechnol. Adv. 31 97–117. 10.1016/j.biotechadv.2011.12.003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.