Abstract
The postnatal mammalian heart is considered a terminally differentiated organ unable to efficiently regenerate after injury. In contrast, we have recently shown a remarkable regenerative capacity of the prenatal heart using myocardial tissue mosaicism for mitochondrial dysfunction in mice. This model is based on inactivation of the X-linked gene encoding holocytochrome c synthase (Hccs) specifically in the developing heart. Loss of HCCS activity results in respiratory chain dysfunction, disturbed cardiomyocyte differentiation and reduced cell cycle activity. The Hccs gene is subjected to X chromosome inactivation, such that in females heterozygous for the heart conditional Hccs knockout approximately 50% of cardiac cells keep the defective X chromosome active and develop mitochondrial dysfunction while the other 50% remain healthy. During heart development the contribution of HCCS deficient cells to the cardiac tissue decreases from 50% at mid-gestation to 10% at birth. This regeneration of the prenatal heart is mediated by increased proliferation of the healthy cardiac cell population, which compensates for the defective cells allowing the formation of a fully functional heart by birth. Here we performed microarray RNA expression analyses on 13.5 dpc control and heterozygous Hccs knockout hearts to identify molecular mechanisms that drive embryonic heart regeneration. Array data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE72054.
Keywords: Heart development, Cardiac regeneration, Cardiomyocyte proliferation, Mitochondrial dysfunction, Cellular stress response
| Specifications | |
|---|---|
| Organism/cell line/tissue | Mus musculus (129Sv/C57Bl6 mixed genetic background)/embryonic hearts (13.5 days post coitum) |
| Sex | female |
| Sequencer or array type | Affymetrix GeneChip Mouse Genome 430 2.0 arrays |
| Data format | Raw and processed |
| Experimental factors | Heterozygous heart conditional Hccs (Holocytochrome c synthase) knockout (cHccs+/−) versus littermate control female embryos |
| Experimental features | Total RNA was isolated from whole 13.5 dpc embryonic hearts and processed for hybridization to Affymetrix arrays. 5 biological replicates per genotype (i.e. cHccs+/− versus controls) each containing 4–5 pooled hearts were analyzed. |
| Consent | Not applicable |
| Sample source location | Münster, Germany |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Experimental design
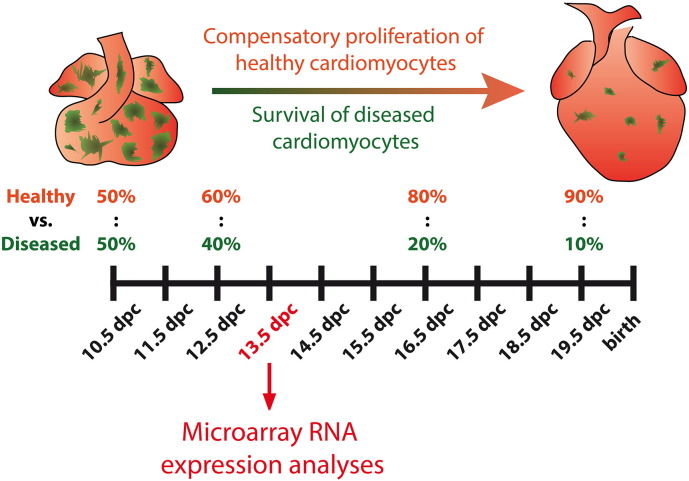
The prenatal mammalian heart has a remarkable regenerative capacity and can compensate for the presence of diseased cells or cardiomyocyte loss to build fully functional hearts by birth [1], [2]. In order to identify molecular mechanisms that drive embryonic heart regeneration, we utilized a genetic mouse model based on inactivation of the X-linked gene encoding holocytochrome c synthase (Hccs) in the developing heart [1]. Heterozygous heart conditional Hccs knockout female embryos (cHccs+/−) develop a tissue mosaic of mitochondrial dysfunction at 10.5 dpc, such that the myocardium is composed of 50% healthy and diseased cardiomyocytes, respectively. During heart development the contribution of HCCS deficient cells to the cardiac tissue decreases from 50% at mid-gestation to 10% at birth. This regeneration of the prenatal heart is mediated by increased proliferation of the healthy cardiac cell population, allowing the formation of a fully functional heart at birth. Nevertheless, HCCS deficient cardiomyocytes do not undergo cell death but survive embryonic and fetal development and are still detectable in the postnatal heart. Here we performed microarray RNA expression analyses on 13.5 dpc control and cHccs+/− hearts. At this developmental stage the most dramatic changes in tissue composition occur in cHccs+/− ventricular myocardium, as hyperproliferation of healthy cardiomyocytes is readily detectable while at the same time a substantial contribution of HCCS deficient cells is still present (Fig. 1). This should allow the identification of genes involved in compensatory growth of healthy cells as well as survival of defective cells.
Fig. 1.
Regeneration of the embryonic heart in cHccs+/− female mice. The prenatal cHccs+/− heart is composed of healthy (depicted in red) and diseased (i.e. HCCS deficient) cardiomyocytes harboring a defect in the mitochondrial respiratory chain (depicted in green). At mid-gestation (i.e. 10.5 dpc) a 50:50 ratio of healthy versus diseased cells is observed in the myocardium, whereas the contribution of diseased tissue progressively declines to just 10% prior to birth. This embryonic heart regeneration is mediated by increased proliferation of healthy cardiomyocytes, which compensate for the defective cells to build a fully functional heart by birth. Nevertheless, HCCS deficient cells survive fetal development and are still detectable in the postnatal heart. Transcriptional profiling of regenerating cHccs+/− hearts has been performed at 13.5 dpc.
2.2. Mice
The generation and characterization of heart conditional Hccs knockout (KO) mice has been described previously [1]. Briefly, “floxed” (fl) Hccs mice were bred to mice expressing Cre recombinase under the control of the Nkx2.5 promoter. All mice were maintained on a mixed 129Sv/C57Bl6 genetic background and all embryo experiments were performed on heterozygous Hccs KO females (Hccsfl/+/Nkx2.5Cre, referred to as cHccs+/−) and their respective Cre positive female littermate controls (Hccs+/+/Nkx2.5Cre, referred to as Hccs+/+).
2.3. Preparation of embryonic hearts
For embryo generation females were mated to the respective males and checked for vaginal plug the next morning. The day of an observed plug was defined as 0.5 dpc and embryos were prepared at 13.5 dpc. Dams were sacrificed by cervical dislocation and embryos were quickly dissected from the uterus into cold PBS. Whole hearts (including ventricles, atria and parts of the outflow tract) were removed from the thoracic cavity, rinsed in cold PBS to wash out blood and snap frozen in liquid nitrogen. Tail tissue was used for DNA preparation and PCR genotyping [1].
2.4. RNA isolation
Given the small size of 13.5 dpc mouse hearts, to purify sufficient RNA amounts suitable for microarray analyses an average of 4 to 5 hearts of the same genotype were pooled. This procedure furthermore accounts for interindividual as well as environmental or maternal differences during pregnancy. RNA was purified using the RNeasy Mini Kit (Qiagen). The tissue was homogenized in RLT buffer supplemented with β-mercaptoethanol using a micropestle. Total RNA was isolated via spin columns according to the manufacturer's instructions, including digestion of genomic DNA on the column using the RNase-Free DNase Set (Qiagen). RNA purity and quality was tested using spectrophotometric parameters and a Bioanalyzer (Agilent), respectively. All samples used for subsequent microarray analyses had RNA integrity numbers (RIN) of ≥ 8.8.
2.5. Microarray RNA expression analyses
For microarray expression analyses a total of 10 Affymetrix GeneChip Mouse Genome 430 2.0 arrays were used. Five pooled samples per genotype (i.e. cHccs+/− and control female embryos) were analyzed. cDNA synthesis was performed using Live Technologies SuperScript® One-Cycle cDNA Kit followed by in vitro transcription using the MEGAScript T7 Kit. After fragmenting of the cRNA for target preparation using the standard Affymetrix protocol, 15 μg fragmented cRNA was hybridized for 16 h at 45 °C to the Mouse Genome 430 array. Arrays were washed and stained with streptavidin-phycoerythrin in the Affymetrix Fluidics Station 450 following standard procedures and further scanned using the Affymetrix GeneChip Scanner 3000 7G.
2.6. Data processing and analysis
Arrays have been quantile-normalize using the RMA algorithm. RMA normalization was performed using Partek Genomic Suite version 6.3, RMA background correction, quantile normalization and median polish probeset summarization. Not or low expressed transcripts were removed by a (log2) maximum expression cutoff < 6. The data filtering resulted in 18,571 of 45,101 probe sets. After normalization the arrays were checked for outlier using the principal component analysis (PCA), a correlation dispersion matrix and normalized Eigenvector scaling. No outlier has been detected. Differential expression was ascertained using t-statistic followed by a FDR multiple testing correction [3]. Probes which undergo 5% FDR were further investigated by functional enrichment using g:Profiler [4], with a simulation based analytical threshold for significance estimation.
3. Discussion
The microarray RNA expression analyses described above revealed 437 genes differentially expressed in 13.5 dpc cHccs+/− compared to control hearts, the majority of which are involved in protein and amino acid metabolism, unfolded protein response (UPR), translational control, cellular stress response and cell death regulation (for details see (5)). Most of the genes and pathways regulating cell stress, UPR and apoptosis could be assigned to HCCS deficient cardiomyocytes, which likely contributes to their survival within the myocardium [5]. In contrast, the transcriptional signature of the healthy cardiomyocyte population is less clear. In this regard it is important to note that we used whole 13.5 dpc cHccs+/− hearts containing a tissue mosaic of healthy and HCCS deficient cardiomyocytes for microarray analyses. Subtle changes in one of the two cell populations are easily missed if not sufficient to alter expression levels in RNA samples isolated from whole hearts. Although the cellular response to mitochondrial dysfunction in the diseased cardiomyocyte population is the dominating outcome of our analyses, the microarray data is likely to also contain genes specifically regulated in the healthy cardiomyocyte population. Identification of the latter would be most relevant for cardiomyocyte cell cycle regulation and cardiac regeneration. Consequently, we do not claim to provide a full list of differentially expressed genes in the regenerating cHccs+/− heart [5], but future studies using laser microdissection or fluorescence activated cell sorting (FACS) will be required to separate the two cell populations. This would allow to more thoroughly define gene expression changes specific for healthy and HCCS deficient cardiomyocytes, respectively.
Acknowledgments
This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, grant number DR 446/3-1). We thank Sabine Schmidt and Gabriele Born for technical assistance with microarray hybridization procedures.
References
- 1.Drenckhahn J.D., Schwarz Q.P., Gray S., Laskowski A., Kiriazis H., Ming Z., Harvey R.P., Du X.J., Thorburn D.R., Cox T.C. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev. Cell. 2008;15:521–533. doi: 10.1016/j.devcel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Sturzu A.C., Rajarajan K., Passer D., Plonowska K., Riley A., Tan T.C., Sharma A., Xu A.F., Engels M.C., Feistritzer R., Li G., Selig M.K., Geissler R., Robertson K.D., Scherrer-Crosbie M., Domian I.J., Wu S.M. Fetal mammalian heart generates a robust compensatory response to cell loss. Circulation. 2015;132:109–121. doi: 10.1161/CIRCULATIONAHA.114.011490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 4.Reimand J., Kull M., Peterson H., Hansen J., Vilo J. g:profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Magarin M., Pohl T., Lill A., Schulz H., Blaschke F., Heuser A., Thierfelder L., Donath S., Drenckhahn J.D. Embryonic cardiomyocytes can orchestrate various cell protective mechanisms to survive mitochondrial stress. J. Mol. Cell. Cardiol. 2016;97:1–14. doi: 10.1016/j.yjmcc.2016.04.007. [DOI] [PubMed] [Google Scholar]