Abstract
Biodiversity and ecosystem functioning research typically shows positive diversity- productivity relationships. However, local increases in species richness can increase competition within trophic levels, reducing the efficacy of intertrophic level population control. Pseudomonas spp. are a dominant group of soil bacteria that play key roles in plant growth promotion and control of crop fungal pathogens. Here we show that Pseudomonas spp. richness is positively correlated with take-all disease in wheat and with yield losses of ~3 t/ha in the field. We modeled the interactions between Pseudomonas and the take-all pathogen in abstract experimental microcosms, and show that increased bacterial genotypic richness escalates bacterial antagonism and decreases the ability of the bacterial community to inhibit growth of the take-all pathogen. Future work is required to determine the generality of these negative biodiversity effects on different media and directly at infection zones on root surfaces. However, the increase in competition between bacteria at high genotypic richness and the potential loss of fungal biocontrol activity highlights an important mechanism to explain the negative Pseudomonas diversity-wheat yield relationship we observed in the field. Together our results suggest that the effect of biodiversity on ecosystem functioning can depend on both the function and trophic level of interest.
Biodiversity has both positive and negative effects on ecosystem functioning1,2. These different effects are known to occur locally from competitive dominance of species that have a high or low functional output and from complementary or antagonistic interactions that increase with species richness3,4,5. Positive effects of biodiversity on productivity and resource capture within trophic levels are most commonly reported in the literature, but for practical management of ecosystems it is important we understand the range of conditions under which diversity causes negative effects on functioning. This is particularly relevant in conventional agriculture, where locally antagonistic interactions, such as between weeds and crop plants, have underpinned the rationale for the creation of species-poor production systems. However, lack of research, alongside potential mismatches between ecosystem functions and ecosystem services highlight the large gap in the practical application of current biodiversity and ecosystem functioning research to agriculture2,6.
Crop diseases pose some of the greatest challenges to food security of the 21st Century7,8,9,10. Root-associated soil bacteria might offer potential solutions to this problem by combating pathogens with anti-microbial compounds and improving plant growth11,12. However, a limiting factor to this solution is that rhizosphere bacteria can compete with each other with a range of antibacterial agents13,14,15. Whilst the spatial dynamics of antibiotic production in the rhizosphere are poorly understood, bacteria which possess the ability to control fungal pathogens can be rendered less effective at biocontrol if they are defeated by other bacteria16,17,18. This means that in agricultural crops where fungal pathogens are problematic, reduced crop yield may result from increased bacterial richness of potential biocontrol genotypes. An open question remains as to (1) the extent to which bacterial diversity patterns in soils conducive to fungal pathogens match this expectation, and (2) the degree that other aspects of bacterial diversity, such as evolutionary relatedness or resource use overlap, might alleviate the intensity of competition between bacteria19,20.
Take-all disease is caused by a pervasive fungal pathogen (Gaeumannomyces graminis var. tritici) in wheat (Tritium aestivum) and leads to devastating yield losses11,12. Populations of antifungal Pseudomonas spp. bacteria have been shown to naturally control take-all, but only after a period of yield losses lasting 4–6 cropping seasons11,12. However, a novel phenomenon was recently discovered in which particular wheat varieties are able to limit the build up of take-all in a single season and create disease suppressive soils prior to an epidemic taking place21. The putative role of some Pseudomonas spp. in take-all control, and their general dominance in the soil bacterial community, positions them as a useful focal group taxon to explore the phenomenon of take-all build up, and to test more general ideas of how bacterial diversity in wheat root system might be related to fungal pathogen suppression.
We used field trials to test if the genotypic richness Pseudomonas spp. in wheat root systems is correlated to severity of infection by the take-all pathogen and yield loss (e.g. Fig. 1). We pre-cultured field plots with different wheat varieties to generate treatments for high or low build up of the take-all inoculum21, and undertook our field assays in the following season on genetically identical clones of a commercial wheat variety (T. aestivum var. Xi19). We tested for generic changes in the background rhizosphere bacterial community structure based on relative abundance of 16S rRNA genes and then sub-sampled culturable isolates in both the endosphere and rhizosphere of wheat roots to describe the patterns of Pseudomonas spp. genotypic richness and phylogenetic diversity across pre-culture treatments at finer resolution.
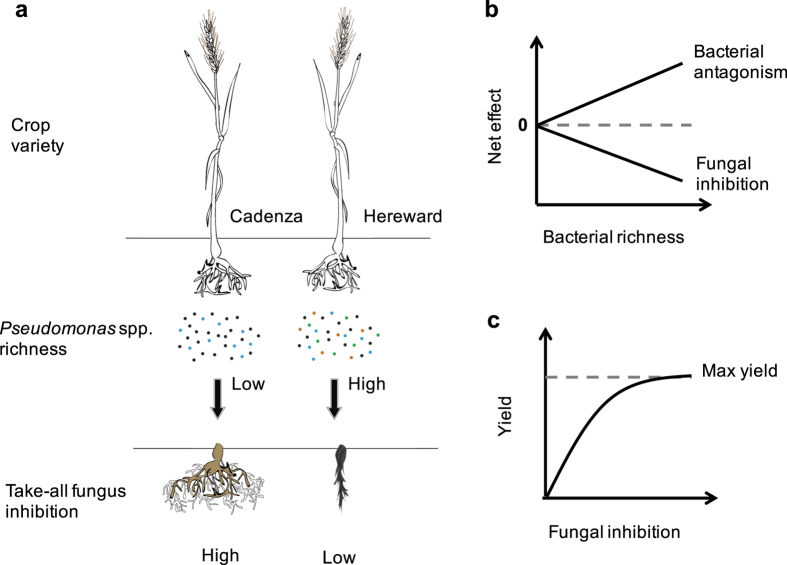
Figure 1. Hypothetical effects of bacterial richness on fungal pathogen inhibition at the plant-soil interface.
(a), different wheat varieties culture high or low richness of soil bacteria (Pseudomonas spp.) which leads to low or high suppression of fungal pathogens in the soil (take-all fungus), respectively. (b), as bacterial richness increases, antagonism between bacterial genotypes increases which causes a decrease in ability of the bacterial community to combat fungal pathogens. (c), an increase in fungal pathogen inhibition decreases disease incidence and leads to an increase in crop yield.
We then set up a series of abstract in vitro experimental model microcosms to test for a causal effect of Pseudomonas genotypic richness on growth inhibition of the take-all pathogen. We orthogonally partitioned the effect of Pseudomonas genotypic richness with Pseudomonas phylogenetic diversity (based on the gyrB housekeeping gene) and Pseudomonas functional trait diversity (based on carbon use profiles) to separate the effects of each component of diversity on the sign of bacterial interactions20,22. We then conducted a complementary set of competition experiments to test if the effects of Pseudomonas genotypic richness on inhibition of the take-all pathogen could be explained by the degree of antagonism between bacteria expressed in Pseudomonas mixtures (defined here as the compound effects of direct antagonistic toxin production and indirect exploitative resource competition).
Methods
Field trials
The field trial was undertaken on the Rothamsted Research experimental farm UK (N51°48′27″, W0°21′44″) during 2010–2012. A commercial winter wheat variety (Xi19) was sown in the autumn on soils in 2011, that in 2010, had been pre-cultured either high (Hereward) or low (Cadenza) take-all inoculum ‘build up’ wheat varieties21. Seeding was at 350 seeds m−2 on each of eight 10 m × 3 m plots (4 replicates × 2 soil culture treatments). Take-all disease intensity on wheat was characterized with the take-all intensity index (TAI)23. To determine the diversity of the dominant members of Pseudomonas spp. we randomly sampled 5 roots systems from each plot (plants were at late milk stage), pooled the roots, and extracted bacteria from both the rhizosphere soil and the root endosphere (after washing roots and surface sterilizing with hypochlorite followed by ethanol24). We pooled the roots (1 pool per plot, each containing 5 root systems), to obtain plot-level estimates of the dominant members. One caveat of this sampling procedure is that the details of alpha diversity at level of the root system may have been lost in place of plot level bacterial beta-diversity. Extracts were dilution plated onto Pseudomonas selective agar with CFC antibiotics (Oxoid) and then incubated for 96 h at 24 °C. Fifteen single colonies (or the maximum if less) from both the endosphere and rhizosphere of each plot were randomly sub-sampled and DNA from each colony was extracted by cell lysis with MicroLysis + (Microzone, UK) (Total isolates = 174, 91 from Hereward and 87 from Cadenza plots, see Table S1). This sub-sampling and selection procedure did not attempt to represent the full diversity of the endosphere and rhizosphere, but provide a feasible representation of the diversity of most dominant culturable isolates, with the caveat that more ‘rare’ strains may have been overlooked. Bacterial genotype was determined by genetic fingerprinting of enterobacterial repetitive intergenic consensus sequences (ERIC)25. For unique ERIC profile genotypes, we PCR amplified, sequenced and aligned a 940 bp fragment of the DNA Gyrase subunit B (gyrB) gene for each distinct genotype present in the study26 and perfomed maximum-likelihood based estimation using RAxML (v.7.0.4), optimized under the GTR-GAMMA substitution model27. We define phylogenetic diversity (PD) throughout the study as the total phylogenetic branch length of the gyrB sequence-determined genotypes present in a given assemblage28. Importantly, we used this gene only as a phylogenetic proxy, and did not perform a full multi-locus sequence analysis, as would be required for more in depth phylogenetic analysis. The sequence data generated in this project have been deposited in the GenBank Nucleotide database (accession numbers KF059880-KF059937). The field trial was combine-harvested and fresh grain weights of Xi19 recorded per plot at grain maturity in August 2012. Grain dry matter was determined by oven-drying 80 g sub-samples of the fresh grain for 16 h at 105 °C. Grain yields were then adjusted to 85% dry matter and scaled to tonnes ha−1.
Fungal pathogen inhibition
We conducted a biodiversity ecosystem functioning experiment to determine how increasing genotypic, phylogenetic and functional diversity of Pseudomonas spp. influenced inhibition of take-all fungus (Gaeumannomyces graminis var. tritici). Eight isolates (Fig. 2) representing the phylogenetic breadth of genotypes in our Pseudomonas spp. library were grown up in Lysogeny broth (LB) at 25 °C for 12 h, shaken at 200 rpm. Late exponential phase bacteria were then pelleted by centrifugation, washed twice in 1× phosphate buffered saline (PBS), incubated for 6 h at room temperature to allow terminating division cycles and adjusted to an OD600 of 1.0, with 1 × PBS. We assembled the genotypes into 49 mixtures in a substitutive design (e.g at constant density), randomly constructing 20 assemblages representing a gradient of phylogenetic diversity within each of the 2 and 4 genotype richness treatments (8 × 1 genotype; 20 × 2; 20 × 4 genotypes; 1 × 8 genotypes = Total 49). To do this first the phylogenetic diversity of all possible combinations of 2, or 4 genotypes was computed, and then 20 assemblages with unique phylogenetic diversity values were randomly sampled from each pool. This design allowed us to orthogonally test for the role of phylogenetic diversity vs. genotypic richness in inhibition of the fungus. Eight microliters of each community (at OD600 1.0) was spotted 2 cm from the edge of Petri dishes containing potato dextrose agar (PDA, OxoidTM, pH 5.6), and incubated at 27 °C for 48 hours. We then placed a 5 mm plug from the leading edge of a culture of the take-all pathogen (grown for 7 days at 24 °C) in the center of the dish, and plates were incubated for a further 7 days at 24 °C before photographing at a standardized height. All plates were spotted with a negative control of PBS, which had no effect on fungal growth. Photographs were analyzed using Fiji29, and inhibition measured as the total pixels free from fungal growth. Functional traits relevant to resource use were proxied by measuring the differential ability of each isolate to utilize 95 different carbon substrates. Assays were completed using GN2 MicroPlates (Biolog, Inc.), with 100 μL of each OD600 adjusted population per well, incubation at 25 °C for 72 h and measurement at OD590. We define functional diversity (FD) as: 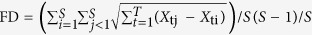 where T is the total number of traits, Xti and Xtj are the values of the trait “t” of genotypes “i” and “j” and S is the total number of genotypes in a given assemblage30. The experiment and functional trait assays were repeated in triplicate.
where T is the total number of traits, Xti and Xtj are the values of the trait “t” of genotypes “i” and “j” and S is the total number of genotypes in a given assemblage30. The experiment and functional trait assays were repeated in triplicate.
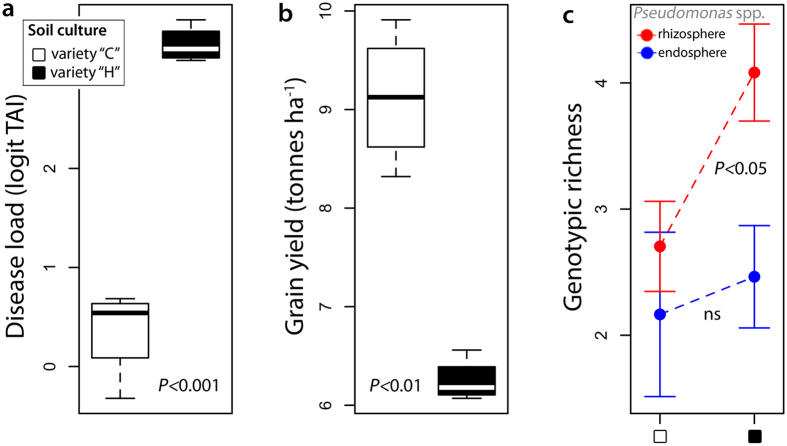
Figure 2. Increased bacterial richness is associated with increased fungal disease load (TAI) and wheat yield losses in the field.
(a), wheat variety choice in the first year of cropping directly influences the intensity of take-all disease in the following isogenic wheat crop (cv. Xi19). (b), this has a marked effect on grain yield. (c), these negative effects of the take-all fungal pathogen are associated with an increased rarefied genotypic richness of Pseudomonas spp. in the rhizospheres of wheat plants (data show means ± s.e.m). (174 isolates in total; Cadenza plots = 87, Hereward plots = 91). All comparisons are made with Welch’s t-test.“ns” = non-significant. “C” = Cadenza; “H” = Hereward; TAI = take all intensity index (see METHODS for details). In all cases n = 4.
Bacterial antagonism
To assess how different levels of genotypic, phylogenetic or functional diversity influenced antagonistic interactions between bacteria, we conducted a second biodiversity ecosystem functioning experiment. Here we used 45 of the same assemblages used in the take-all pathogen inhibition experiment (8 × 1 genotype; 19 × 2 genotype; 19 × 4 genotypes; 1 × 8 genotypes = Total 45; with a reduction to 19 for 2–4 genotypes due to practicalities of 48 wells per replicate) and cultured (from an initial density of ∼107 cells ml−1) them in LB broth at 27 °C for 18 h, shaken at 200 rpm (Note: LB is a rich medium which we used to match initial culture conditions and accelerate growth, future work might also repeat this with 1/10th Tryptone Soya Broth or root exudates). We centrifuged each mixture and deposited 150 μl of the cell free supernatant into separate wells of a microtitre plate. We then overlaid 50 μl of OD600 adjusted populations of each of the 8 different genotypes, subjecting each genotype to the compound legacy effects of antagonistic toxin production and nutrient depletion by each community. Productivity differences were proxied as simply OD600 after 6 h incubation at 25 °C, shaken at 200 rpm. OD scales well to dry weight biomass and is a useful proxy for integrated productivity (i.e. the product of cell sizes and counts)31,32. The whole experiment was repeated in triplicate.
Statistical analysis
To determine the differences between take-all disease intensity (logit transformed), log total grain yield and bacterial diversity (genotypic richness/phylogenetic diversity) in our field plots we used Welch’s t-test. We rarefied phylogenetic diversity measures to account for species differences between plots, using the standalone phylorare() function33 in R 3.034. We also used individual-based rarefaction on genotypic richness in each field plot to account for sampling effort biases that resulted from plots that did not yield 15 single separate colonies35. Analysis of Similarity36 based on presence-absence data and Bray-Curtis distances were used to test for differences in community composition between plots. We used linear models, to orthogonally test for the effect of genotypic richness and phylogenetic diversity on inhibition of take-all and bacterial antagonism in the 2–4 genotypic richness assemblages. Genotypic richness was fitted as a categorical variable first in all the models and phylogenetic (or functional) diversity fitted after as a continuous variable, with first order interactions. As no significant interactions were present in any of the models, we removed the interaction terms and assumed fixed slope estimates for the models presented in the main text. The biodiversity metrics we used as our response were characterized using the net biodiversity effect37: log(Φobs/Φexp), where, for the fungal inhibition experiment, Φobs is the overall inhibition of a given assemblage, and Φexp is the mean inhibition from each genotype present in the assemblage in monoculture.; and where, for the bacterial inhibition experiment, Φobs is the overall productivity of a genotype on the spent medium from a given assemblage, and Φexp is the mean productivity of that same genotype on the spent medium from each genotype present in the assemblage. All models were run and assumptions validated in R38.
Results
Field trials
In our field trials we found that, the take-all severity on wheat roots of isogenic T. aestivum var. Xi19 was dependent on the wheat variety used in soil pre-culture treatments (Welch’s t-test, t3.91 = 11.64, P < 0.001; Fig. 2a). This difference in disease load was related to large differences in overall grain yields of wheat (31% ± 4% SE [~3t ha−1] higher yields in plots with low take-all disease) (Welch’s t-test, t4.24 = 9.26, P = 0.0006; Fig. 2b). To investigate whether this difference in disease load and yield was associated with Pseudomonas spp. we sampled Pseudomonas communities in the endosphere (inside) and rhizosphere (outside) of the roots of the isogenic wheat plants, and then compared genotypic richness and phylogenetic diversity across plots. We found that the plots with highest take-all disease loads, and lowest wheat grain yields, contained the highest genotypic richness of Pseudomonas in the rhizosphere (Welch’s t-test, t5.98 = 2.63, P = 0.039; Fig. 2c). We did not detect clear differences in rhizosphere Pseudomonas community composition per se between plots with high or low disease load (Analysis of Similarity, Bray-Curtis, R = 0.19, P = 0.08). We found no differences in phylogenetic diversity of Pseudomonas in the rhizosphere of wheat roots between low or high take-all plots (Welch’s t-test, t4.96 = 0.40, P = 0.70; Fig. S5). Endosphere Pseudomonas genotypic richness or phylogenetic diversity also showed no significant relationships with take-all intensity or yield (Fig. 2, Fig. S5). On average 1.7% of 16S rRNA reads were identified as Pseudomonas across the study plots. Pseudomonas relative abundances and generic microbial community composition based on 16S rRNA sequences did not show obvious differences between treatments (Fig. S6. Supplementary Notes 1).
In vitro experimental isolates
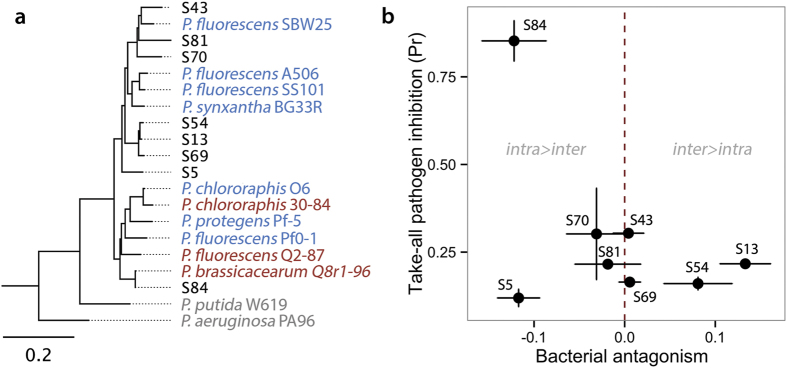
The relationship between the genotypes we used in our in vitro experiments, and other well-known biocontrol Pseudomonas genotypes is given in Fig. 3a. Our genotypes expressed differences in in vitro take-all pathogen inhibitory activity and intragenotypic vs. intergenotypic bacterial antagonism. However, we did not observe any genotype level trade-off between take-all pathogen inhibitory ability and bacterial antagonism. The most potent inhibitor of the take-all pathogen (S84: 85% ± 6% SE inhibition of mycelial growth), nevertheless expressed the classic self-limiting trait where intragenotypic antagonism > intergenotypic antagonism (Fig. 3b). Strain S84 also clustered with other antibiotic producing strains P. brassicacearum Q8r1-96 and P. fluorescens Q2-87, previously implicated in control of the take-all fungal pathogen39. We found no correlation between pairwise distances in carbon resource use profiles and phylogenetic distances based on gyrB (Mantel test, P = 0.74; Fig. S2).
Figure 3. Pseudomonas spp. used in the experiments.
(a), phylogenetic relationships between the genotypes used in the in vitro experiments and other well known biocontrol genotypes based on maximum likelihood tree estimated on a 940 kb fragment of the universal houskeeping gene gyrB, using the RAxML algorithm (with GTR-GAMMA substitution). Scale bar represents nucleotide substitutions per site. Blue strains are known biocontrol agents, red strains were previously isolated from wheat root systems and have been implicated in take-all control, grey strains are outgroups38. (b), Take-all pathogen inhibition and bacterial antagonistic ability of the strains. Take-all pathogen inhibition is the area of fungal mycelial growth inhibited by each Pseudomonas genotype in Petri dish assays (1 = no fungal growth, and 0 = complete fungal growth). Bacterial antagonism is the log ratio of strain productivity (OD 600) on spent liquid medium cultured by its own genotype versus its mean productivity on spent medium cultured by all other genotypes. Data show means ± s.e.m. n = 3.
Fungal pathogen inhibition
To model the effect of Pseudomonas spp. diversity on take-all pathogen growth, we created experimental microcosms and presented the take-all fungal pathogen in vitro to 49 different synthetic Pseudomonas communities of varying phylogenetic diversity, orthogonally nested within different genotypic richness levels. We characterized the response with the net biodiversity effect37: log(Φobs/Φexp), where Φobs is the overall inhibition by a given assemblage, and Φexp is the mean inhibition from each genotype present in the assemblage in monoculture. We tested if the ratio of log(Φobs/Φexp) differed between levels of richness and phylogenetic diversity (the null hypothesis being that the ratio remained constant).We found that increasing Pseudomonas genotypic richness was associated with a reduced inhibition of the fungal pathogen, beyond that expected from the additive inhibitory effects of individual Pseudomonas genotypes (F1, 37 = 10.45, P = 0.003; Fig. 4a). We fitted the presence of each genotype before the genotypic richness term in separate linear models, but none were able to remove the signal of genotypic richness, giving no evidence to suggest a single genotype was completely dominating the communities’ ability to inhibit take-all. We found no influence of Pseudomonas phylogenetic diversity on the fungal inhibition net biodiversity effect (R2 = 0.03; F1, 37 = 1.4, P = 0.23; Fig. S3a). We fitted functional diversity in carbon resource use traits in place of phylogenetic diversity, and found it was also a poor predictor of the fungal inhibition net biodiversity effect (R2 = 0.02; F1, 37 = 0.7, P = 0.38; Fig. S4a).
Figure 4. Negative effects of bacterial genotypic richness on fungal pathogen inhibition result from increasing intergenotypic antagonism between soil bacteria.
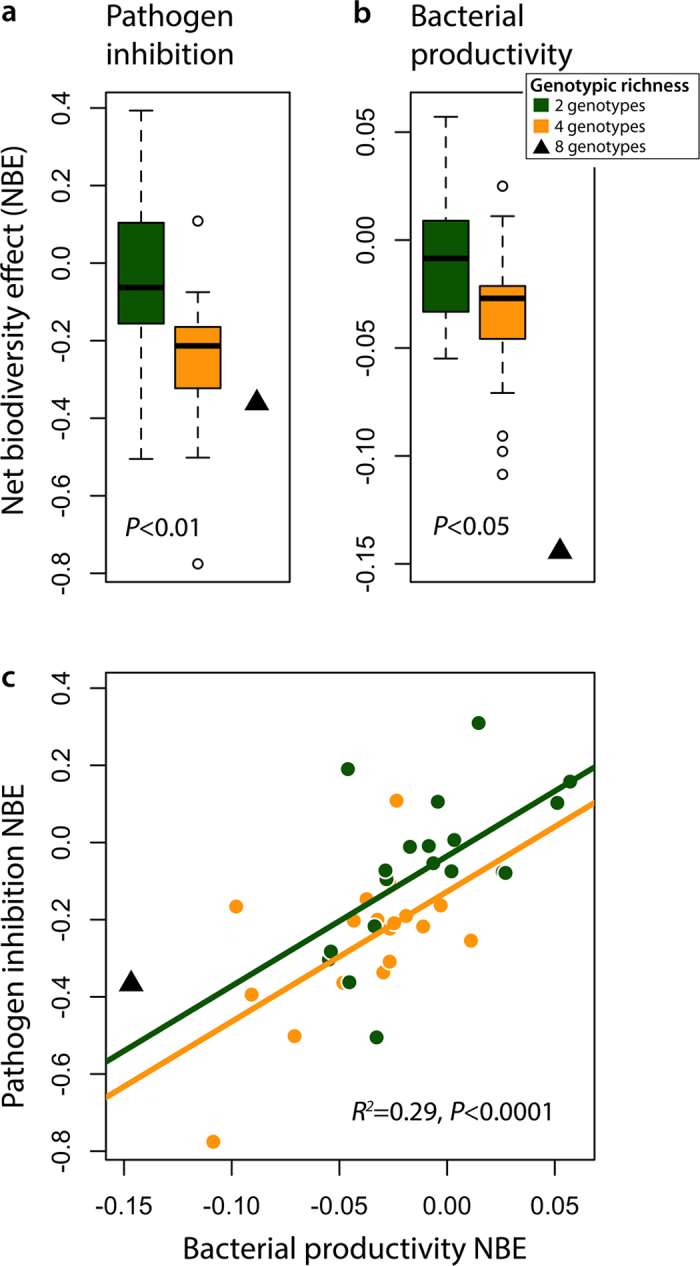
(a), net negative effects of increasing Pseudomonas genotypic richness on inhibition of wheat take-all fungal pathogen (n = 20). (b), net negative effects of increasing Pseudomonas genotypic richness on Pseudomonas productivity (OD600) via intergenotypic bacterial antagonism (n = 19). (c), relationship between net effects of Pseudomonas intergenotypic antagonism and fungal pathogen inhibition at 2 and 4 genotypic richness treatments (n = 19). R2 calculated from the variance in pathogen inhibition NBE explained by bacterial productivity NBE, independent of species richness effects (Total R2 = 0.48, P < 0.001). All comparisons are made with Analysis of Covariance (see METHODS for details). All points are the mean of three experimental replicates. The values of net biodiversity effects of 8 genotypes are included for visual comparison. See main text for details on how Net Biodiversity Effects (NBE) responses are calculated.
Bacterial antagonism
We tested the idea that antagonistic interactions between bacterial genotypes increase with richness by conducting a second microcosm experiment. We measured the productivity (OD600,) of each genotype when grown on medium that had been cultured by the assemblages we used in the in vitro take-all fungal inhibition experiment (Methods). This assay accounts for the compound antagonistic effects of toxin production and nutrient depletion on bacterial growth, which both potentially operate when increasing richness17,20. Here we define Φobs as the overall productivity of a genotype on the spent medium from a given assemblage, and Φexp as the mean productivity of that genotype on the spent medium from each genotype present in that assemblage. As before, we tested if the ratio of log(Φobs/Φexp) differed between levels richness, phylogenetic or functional diversity. We found that Pseudomonas community productivity decreased with increasing genotypic richness beyond what would be expected if antagonism between bacterial genotypes did not increase with richness (F1, 35 = 5.6, P = 0.024; Fig. 4b). Again, we fitted the presence of each genotype before the genotypic richness term in separate linear models, but as before dominant selection effects by any one genotype were not observed. We were unable to detect any clear signal on the effect of phylogenetic (R 2= 0.00; F1, 35 = 0.03, P = 0.85; Fig. S3b), or functional diversity based on carbon utilization traits (R2 = 0.01; F1, 35 = 0.33, P = 0.57, Fig. S4b) on the increases in antagonism between bacteria in mixtures within richness levels. If antagonism between Pseudomonas genotypes was an important factor driving fungal inhibition, and if our results were robust across the different media we used on our microcosms, we would have the expectation that the net biodiversity effects observed in our fungal inhibition experiment could be predicted by the variation in the net biodiversity effects in the Pseudomonas inhibition experiment. And indeed, we found that the increase in bacterial antagonism in bacterial mixtures was a positively related to the reduction in inhibition of the take-all pathogen by those same bacterial communities (R 2= 0.29; F1, 35 = 19.56, P < 0.0001; Fig. 4c).
Discussion
Our field trials show that take-all disease severity, and crop grain losses are positively associated with increased Pseudomonas genotypic richness in wheat rhizospheres. Our in vitro experiments show that antagonistic interactions between Pseudomonas at high genotypic richness can reduce the ability of those bacteria to inhibit growth of the take-all pathogen. This mechanism offers a plausible explanation for our field observations of high take-all disease and crop yield loss in high Pseudomonas spp. richness plots. Future work is required to determine the generality of these effects on different media, directly at infection zones on root surfaces, and in different study systems. Our findings support the view that the sign of the biodiversity and ecosystem functioning relationship can be highly context and function specific.
The negative effects of genotypic richness we observed in our model experiments do run contrary to the commonly reported phenomenon that diversity has a positive effect on ecosystem functioning1,40. Other laboratory experiments have previously shown bacterial antagonism to cause ecosystem functioning to collapse at high richness17,20, but see Jousset et al.16. Whilst, (1) the presence of competitively dominant but ‘functionally poor’ species (‘poor’ in a subjective sense, e.g. for a desired function), or (2) the non-additive effects of exploitative and interference competition, are likely to occur in other taxonomic groups, we suggest it may be of limited use to look for general statements of positive vs. negative biodiversity effects. Cautioning against a dichotomy seems important because the direction of the biodiversity ecosystem functioning relationship can depend on both the function and the trophic level of interest. Simply replacing the fungal pathogen in our study with a bacterial pathogen would have likely yielded the opposite results: with bacterial diversity having a positive effect on pathogen control.
Our in vitro experiments were designed to orthogonally test for the effects of bacterial phylogenetic (or functional) diversity or genotypic richness on fungal pathogen suppression. We found no effects of phylogenetic functional trait diversity use, but consistent negative effects of genotypic richness, on bacterial antagonism and fungal pathogen suppression. Previous studies have suggested the inability to detect an effect of phylogenetic or functional trait diversity, might result from resource scarcity and limited possibilities for niche partitioning in experimental settings20,22,41. However, it is also possible that (1) core genomes are poor predictors of functional traits due to convergent evolution and horizontal gene transfer42; that (2) characterizing functional traits in species grown in isolation are inadequate because trait values can be plastic and change in the presence of different species in mixtures; or (3) simply that trait dissimilarity itself is a poor predictor of niche partitioning43.
Our study did not account for local ecological history effects (e.g. specific root location or individual plant clone) which are also likely to influence the biodiversity-ecosystem functioning relationship19. Whilst there is evidence to suggest that sympatrically coevolved bacteria can be more antagonistic44 there is also evidence to suggest that some background environments enhance positive interactions between species over time32. More fine scaled sampling, or real-time imaging of bacterial interactions on the root system, in future could be useful for identifying the impact of isolate sampling location on our results.
It is important to emphasize our field trials were unable to conclude if the negative relationship between Pseudomonas spp. genotypic richness and take-all in the field was causal. Sequential sampling of bacterial isolates to map the dynamic interactions between the microbiota of wheat varieties and take-all disease, alongside microbiome transplant experiments and further in planta manipulations would help validate this idea. Similarly, although we assayed the most virulent strain of a field isolated take-all pathogen in our model microcosms; we did not assay the effect of different take-all pathogen strains or consider the intraspecific population diversity of the take-all pathogen in our study.
In summary, our findings argue for an integrated view that appreciates the importance of both the negative and positive effects of biodiversity in driving specific ecosystem functions within and across trophic levels. Notably we have focused on the diversity of a single bacterial group, containing putative biocontrol strains highly relevant to our focal fungal pathogen. An analysis of generic groupings of 16S rRNA sequences in the soil at each of our trial plots showed that other bacterial groups are also highly relatively abundant, some of which may play important roles in plant fitness. Testing how co-cultures of larger numbers of isolates from different bacterial genera respond to diversity manipulations, and perform one or more ecosystem functions, remains an important and exciting avenue of research. In particular, future work could identify the applicability of our findings to functions involved in both concurrent disease control and nutrient supply, across multiple bacterial indicator groups and trophic levels. A sharpened focus on these services may help to significantly reduce environmentally and economically costly inputs into global food systems.
Additional Information
How to cite this article: Mehrabi, Z. et al. Pseudomonas spp. diversity is negatively associated with suppression of the wheat take-all pathogen. Sci. Rep. 6, 29905; doi: 10.1038/srep29905 (2016).
Supplementary Material
Acknowledgments
We thank Josh Roworth and Julia Burnell for research assistance, Dominique Gravel for code, Tom Bell, Alex Jousset and Rebekah Robinson for many helpful discussions, and four anonymous reviewers whose comments helped to improve the manuscript. The field trial was conducted as part of the core project of the Wheat Genetic Improvement Network supported by DEFRA (Defra, IF0146). Rothamsted Research receives strategic funding from the BBSRC and we acknowledge funding from the BBSRC ISPG, Optimization of nutrients in soil-plant systems (BBS/E/C/0005196). V.M. was supported by a BBSRC-CASE quota studentship awarded to Rothamsted and additional supported by HGCA (3480). In addition, V.M. and K.H-K. were supported by the Biotechnology and Biological Sciences Research Council of the UK (BBSRC) through the Institute Strategic Programme Grant 20:20 Wheat® (BB/J/00426X/1). G.C. was supported by the BBSRC Wheat Improvement Strategic Programme Grant (WISP) BB/J004596/1. Z.M. was supported by the BBSRC [grant number BB/J014427/1].
Footnotes
Author Contributions Z.M., T.H.M., V.E.M., K.E.H.-K. and P.H. designed the research. Z.M., T.M., G.C. and V.E.M. did the experiments and collected the data. I.M.C and T.M. processed the 16S amplicon data. Z.M. analysed the data. Z.M. wrote the manuscript. Z.M., T.H.M., V.E.M., K.E.H.-K., G.P. and P.R.H. commented on the manuscript.
References
- Hooper D. U. et al. Effects of biodiversity on ecosystem functioning. Ecological Monographs 75, 3–35 (2005). [Google Scholar]
- Cardinale B. J. et al. Biodiversity loss and its impact on humanity. Nature 486, 59–67 (2012). [DOI] [PubMed] [Google Scholar]
- Flynn D. F. B., Mirotchnick N., Jain M., Palmer M. I. & Naeem S. Functional and phylogenetic diversity as predictors of biodiversity–ecosystem-function relationships. Ecology 92, 1573–81 (2011). [DOI] [PubMed] [Google Scholar]
- Huston, M. A. Hidden treatments in ecological experiments: Re-evaluating the ecosystem function of biodiversity. Oecologia 110, 449–460 (1997). [DOI] [PubMed] [Google Scholar]
- Gravel D. et al. Phylogenetic constraints on ecosystem functioning. Nature Communications 3, 1117 (2012). [DOI] [PubMed] [Google Scholar]
- Naeem S., Duffy J. E. & Zavaleta E. The Functions of Biological Diversity in an Age of Extinction. Science 336, 1401–1406 (2012). [DOI] [PubMed] [Google Scholar]
- Oerke E. C. Crop losses to pests. The Journal of Agricultural Science 144, 31 (2006). [Google Scholar]
- Crowder D. W., Northfield T. D., Strand M. R. & Snyder W. E. Organic agriculture promotes evenness and natural pest control. Nature 466, 109–112 (2010). [DOI] [PubMed] [Google Scholar]
- Tscharntke T. et al. Global food security, biodiversity conservation and the future of agricultural intensification. Biological Conservation 151, 53–59 (2012). [Google Scholar]
- Fisher M. C. et al. Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–94 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak Y. S. & Weller D. M. Take-all of wheat and natural disease suppression: A review. Plant Pathology Journal 29, 125–135 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers J. M., Paulitz T. C., Steinberg C., Alabouvette C. & Moënne-Loccoz Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant and Soil 321, 341–361 (2009). [Google Scholar]
- Riley, M. A. & Wertz J. E. Bacteriocins: evolution, ecology, and application. Annual review of microbiology 56, 117–137 (2002). [DOI] [PubMed] [Google Scholar]
- Little A. E. F., Robinson C. J., Peterson S. B., Raffa K. F. & Handelsman J. Rules of engagement: interspecies interactions that regulate microbial communities. Annual review of microbiology 62, 375–401 (2008). [DOI] [PubMed] [Google Scholar]
- Hibbing M. E., Fuqua C., Parsek M. R. & Peterson S. B. Bacterial competition: surviving and thriving in the microbial jungle. Nature reviews Microbiology 8, 15–25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousset A. et al. Biodiversity and species identity shape the antifungal activity of bacterial communities. Ecology 95, 1184–1190 (2014). [DOI] [PubMed] [Google Scholar]
- Becker J., Eisenhauer N., Scheu S. & Jousset A. Increasing antagonistic interactions cause bacterial communities to collapse at high diversity. Ecology Letters 15, 468–474 (2012). [DOI] [PubMed] [Google Scholar]
- Xu X. M., Jeffries P., Pautasso M. & Jeger M. J. Combined use of biocontrol agents to manage plant diseases in theory and practice. Phytopathology 101, 1024–1031 (2011). [DOI] [PubMed] [Google Scholar]
- Gravel D. et al. Experimental niche evolution alters the strength of the diversity–productivity relationship. Nature 469, 89–92 (2011). [DOI] [PubMed] [Google Scholar]
- Jousset A., Schmid B., Scheu S. & Eisenhauer N. Genotypic richness and dissimilarity opposingly affect ecosystem functioning. Ecology Letters 14, 537–545 (2011). [DOI] [PubMed] [Google Scholar]
- McMillan V. E., Hammond-Kosack K. E. & Gutteridge R. J. Evidence that wheat cultivars differ in their ability to build up inoculum of the take-all fungus, Gaeumannomyces graminis var. tritici, under a first wheat crop. Plant Pathology 60, 200–206 (2011). [Google Scholar]
- Jousset A., Schulz W., Scheu S. & Eisenhauer N. Intraspecific genotypic richness and relatedness predict the invasibility of microbial communities. The ISME journal 5, 1108–1114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman G. L., Gutteridge R. J. & Jenkyn J. F. Take-all and grain yields in sequences of winter wheat crops testing fluquinconazole seed treatment applied in different combinations of years. Annals of Applied Biology 145, 317–330 (2004). [Google Scholar]
- Robinson R. J. et al. Endophytic bacterial composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant and Soil 10.1007/s11104-015-2495-4 (2015). [Google Scholar]
- Versalovic J., Koeuth T. & Lupski J. R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic acids research 19, 6823–6831 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S. et al. Phylogeny of the genus Pseudomonas: Intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiology 146, 2385–2394 (2000). [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith D. P. Conservation evaluation and phylogenetic diversity. Biological Conservation 61, 1–10 (1992). [Google Scholar]
- Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B., Kinzig A. & Langridge J. Plant attribute diversity, resilience, and ecosystem function: The nature and significance of dominant and minor species. Ecosystems 2, 95–113 (1999). [Google Scholar]
- Myers J. A., Curtis B. S. & Curtis W. R. Improving accuracy of cell and chromophore concentration measurements using optical density. BMC biophysics 6, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegna F., Moreno-Letelier A., Bell T. & Barraclough T. G. Evolution of species interactions determines microbial community productivity in new environments. The ISME Journal 9, 1235–1245 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nipperess, D. A. & Matsen F. A. The mean and variance of phylogenetic diversity under rarefaction. Methods in Ecology and Evolution 4, 566–572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing ISBN 3-900051-07-0, 2013). [Google Scholar]
- Gotelli N. J. & Colwell R. K. Quantifying biodiversity: procedures and pitfalls in the measurment and comparison of species richness. Ecology Letters 4, 379–391 (2001). [Google Scholar]
- Clarke K. R. Non-parametric multivariate analyses of changes in community structure. Austr. J. Ecol. 18, 117–143 (1993). [Google Scholar]
- Loreau M. & Hector A. Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76 (2001). [DOI] [PubMed] [Google Scholar]
- Pena E. & Slate E. Global validation of linear modelling assumptions. Journal of the American statistics Association 101, 341–354 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loper J. E. et al. Comparative genomics of plant-associated pseudomonas spp.: Insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genetics 8, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. N., Byrnes J. E. K. & Cardinale B. J. Effects of predator richness on prey suppression: A meta-analysis. Ecology 94, 2180–2187 (2013). [DOI] [PubMed] [Google Scholar]
- Eisenhauer N., Schulz W., Scheu S. & Jousset A. Niche dimensionality links biodiversity and invasibility of microbial communities. Functional Ecology 27, 282–288 (2013). [Google Scholar]
- Mouquet N. et al. Ecophylogenetics: Advances and perspectives. Biological Reviews 87, 769–785 (2012). [DOI] [PubMed] [Google Scholar]
- Hillerislambers J., Adler P. B., Harpole W. S., Levine J. M. & Mayfield M. M. Rethinking Community Assembly through the Lens of Coexistence Theory. Annu. Rev. Ecol. Evol. Syst 43, 227–48 (2012). [Google Scholar]
- Kinkel L. L., Schlatter D. C., Xiao K. & Baines A. D. Sympatric inhibition and niche differentiation suggest alternative coevolutionary trajectories among Streptomycetes. The ISME journal 8, 249–56 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.