Abstract
A salt- and drought-responsive novel gene SbSDR1 is predominantly localised to the nucleus, up-regulated under abiotic stresses and is involved in the regulation of metabolic processes. SbSDR1 showed DNA-binding activity to genomic DNA, microarray analysis revealed the upregulation of host stress-responsive genes and the results suggest that SbSDR1 acts as a transcription factor. Overexpression of SbSDR1 did not affect the growth and yield of transgenic plants in non-stress conditions. Moreover, the overexpression of SbSDR1 stimulates the growth of plants and enhances their physiological status by modulating the physiology and inhibiting the accumulation of reactive oxygen species under salt and osmotic stress. Transgenic plants that overexpressed SbSDR1 had a higher relative water content, membrane integrity and concentration of proline and total soluble sugars, whereas they showed less electrolyte leakage and lipid peroxidation than wild type plants under stress conditions. In field conditions, SbSDR1 plants recovered from stress-induced injuries and could complete their life cycle. This study suggests that SbSDR1 functions as a molecular switch and contributes to salt and osmotic tolerance at different growth stages. Overall, SbSDR1 is a potential candidate to be used for engineering salt and drought tolerance in crops without adverse effects on growth and yield.
In nature, plants are often exposed to environmental stresses, which affect the bio-physiological activities of the plant. Salinity and drought stresses are major constraints for agricultural productivity worldwide1. Salinity costs agricultural crop production approximately $27 billion every year and therefore, particular attention is being focused on sustainable agriculture in salt-affected areas2,3. Salinity is predominant in arid or semi-arid areas, where water is limiting, and thus, agriculture is limited by drought and salinity under scarce rainfall3. In an economic cost model of salinity, it was estimated that farmers are often on the edge of profit or loss, and a small decrease in yield can have devastating economic consequences3.
The majority of agricultural crops grown nowadays are glycophytes (salt sensitive), and their productivity becomes commercially non-viable with an increase in salinity in the 4–8 dS/m range4, due to a decrease in yield of about 10%. Salt and drought stresses have similar effects on water potential, but salinity has additional cytotoxic effects. Salinity and drought both adversely affect photosynthesis, metabolic pathways and physiology. Consequently, this retards plant growth and can also lead to death5,6. Halophytes are naturally adapted plants that have the ability to complete their life cycle in a NaCl-rich environment; therefore, it is important to understand their abiotic stress tolerance mechanism. A signalling network is initiated under salt and drought stress conditions, which reprogrammes the expression of stress-responsive genes that are involved in different cellular, metabolic, biochemical and physiological processes7. Salt-tolerance genes from halophytes are considered potential key players with which to engineer plants to increase their salt tolerance8.
Abiotic stress causes a metabolic imbalance in cells, which promotes the production of reactive oxygen species (ROS) and results in damage to cell membranes, nucleic acids and cellular organelles, including the chloroplast9,10,11. Plants have developed a number of mechanisms to tolerate various stresses and stress-inducible genes play a key role in the regulation of stress tolerance and the maintenance of cellular homeostasis12,13. Previous studies have shown that the ectopic expression of abiotic stress-responsive gene(s) can positively regulate the stress tolerance of plants. However, bioengineering plants for stress tolerance requires knowledge concerning the key players of the stress-response network. However, it is difficult to distinguish critical genes that regulate the stress tolerance of the plant. Global transcript profiling has revealed that many genes are co-ordinately regulated by salt and drought stress14,15. A novel MsZEP gene was cloned from alfalfa for functional validation by heterologous expression in tobacco, and the gene confers drought and salt tolerance in transgenic plants16. The overexpression of a novel Zmhdz10 in rice and Arabidopsis led to a significantly improved tolerance to oxidative stress caused by drought and salt stresses17. Similarly, the IbZFP1 gene of sweet potato, which encodes a novel Cys2/His2 zinc finger protein, regulates ABA signalling, proline biosynthesis and ROS scavenging, and thus, improves salt and drought tolerance in transgenic Arabidopsis18.
Although several abiotic stress-responsive genes have been characterised, the quest for genes with novel functions continues, especially from halophytes. One extreme halophyte, Salicornia brachiata, which grows profusely on salt marshes, is a suitable candidate with which to study the salt stress-tolerance mechanism19. Several abiotic stress-responsive genes and promoters have been cloned from this halophyte to develop abiotic stress-tolerant transgenic plants20,21,22,23,24,25,26,27,28. Salicornia also possesses sulphur-rich seed-storage proteins29, unique oligosaccharides30 and metabolites31. The EST database of S. brachiata contains about 30% novel/unknown abiotic stress-responsive genes32 and we charactered a novel gene that is involved in salt and drought tolerance. A high transcript accumulation of the clone Sal-C-53.e1 (EB484704) was observed under salt and drought stress32, and was therefore selected for functional validation through a transgenic approach. In this study, we performed a detailed functional analysis of the SbSDR1 gene by overexpressing it in tobacco plants to explore its roles in abiotic stress tolerance. The functional characterisation of this gene will reveal a novel regulatory molecular mechanism that can also be used to engineer crop plants that are tolerant to environmental stresses.
Results
A nuclear-localised gene SbSDR1 differentially expressed under abiotic stress
The SbSDR1 cDNA sequence was 728 bp (gene accession no. KF015229) long and consisted of a 5′-untranslated leader sequence (5′-UTR; 1–93 bp), an open reading frame (ORF; 94–483 bp), a 3′-UTR (483–705 bp) and a poly(A) tail of 23 base pairs (Figure S1). The 390-bp ORF encodes a peptide (accession no. AGU69247) of 129 amino acids with a molecular mass of 13.14 kD, a pI of 5.22 and an instability index of 36.93, which confirms that protein is stable in nature. The gene showed no sequence homology with sequences in the existing database, whereas the deduced peptide showed 68% sequence identity (with 98% query coverage) with an unknown/hypothetical or uncharacterised protein from Beta vulgaris. In silico analyses revealed that SbSDR1 consists of helices and coils and contains an internal nuclear localisation signal (NLS; 41–53 amino-acid residues; IDKAKVAGAAEDV) without a cleavage site.
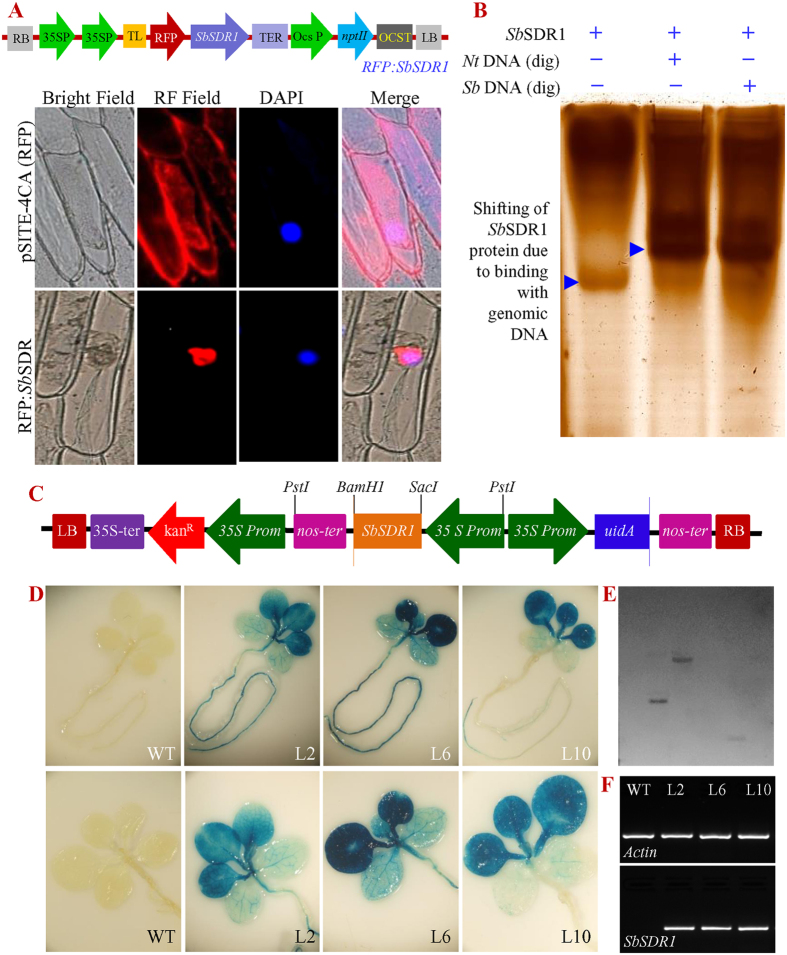
Furthermore, transient expression of the recombinant RFP:SbSDR1 protein confirmed that SbSDR1 is a nuclear protein; in contrast, the evenly distributed expression of RFP alone was detected in the entire cell region in a subcellular localisation study (Fig. 1A). The SbSDR1 ORF of 390 bp was amplified from cDNA and genomic DNA of S. brachiata. Comparative sequence and Southern blot analyses revealed a single exon structure and a single copy of the gene, respectively, and confirmed an intron-less genomic organisation of the gene (Figure S2).
Figure 1. Sub-cellular localisation, DNA binding activity and confirmation of transgenic tobacco plants.
(A) Gene construct RFP:SbSDR1, transient expression of RFP alone and RFP:SbSDR1 translational fusion protein in transformed onion epidermal cells; (B) DNA binding activity of SbSDR1 protein with genomic DNA of Salicornia brachiata and tobacco; (C) Schematic representation of SbSDR1-pCAMBIA2301 plant transformation vector construct; (D) Histochemical GUS assay; (E) Southern hybridization to determine transgene copy number, and (F) Semi-quantitative RT-PCR of transgenic lines along with control plants.
Differential expression of SbSDR1 was observed in S. brachiata under varying stress conditions; expression was up-regulated at each time-point compared to the control treatment (Figure S3). The expression level was rapidly induced by NaCl (two-fold at 6 h), then decreased after 12 h (compared to at 6 h) and again increased at 24 h. The expression of the gene increased concomitantly with increasing drought stress and reached a maximum (3.28-fold) at 24 h. Similarly, the transcript level increased gradually up to 12 h (4.03-fold) following heat stress and subsequently decreased slightly (2.78-fold). The transcript was also up-regulated (1.7- to 2.1-fold) by cold.
The SbSDR1 acts as transcription factor
In silico analysis predicted that the biological function of SbSDR1 is to regulate metabolic processes and gene expression. Subcellular localisation analysis confirmed that the protein is localised to the nucleus. Furthermore, DNA binding property was assayed by a electrophoretic mobility shift assay. For this, SbSDR1 protein was expressed in E. coli and the 13-kDa protein was extracted (Figure S4). The DNA/protein interaction showed a clear shift of the protein band, which confirmed the binding specificity of SbSDR1 to genomic DNA (Fig. 1B).
Molecular analyses of transgenic tobacco plants
Healthy leaves from T1 transgenic tobacco lines (T1 to T16 lines) overexpressing the SbSDR1 gene (Fig. 1C), and wild type (WT) plants were harvested and screened for the presence of transgenes by PCR and histochemical GUS assays (Figure S5). The three lines L2, L6 and L10 that showed the highest GUS expression were selected (Fig. 1D) for further functional analysis using morphological, physio-biochemical and molecular analyses. Southern blot and semi-quantitative RT-PCR analysis confirmed the presence of a single copy of the transgene and a high expression of the SbSDR1 gene, respectively in all selected lines (Fig. 1E,F).
Ectopic expression of the SbSDR1 gene improves plant growth under salt and osmotic stress
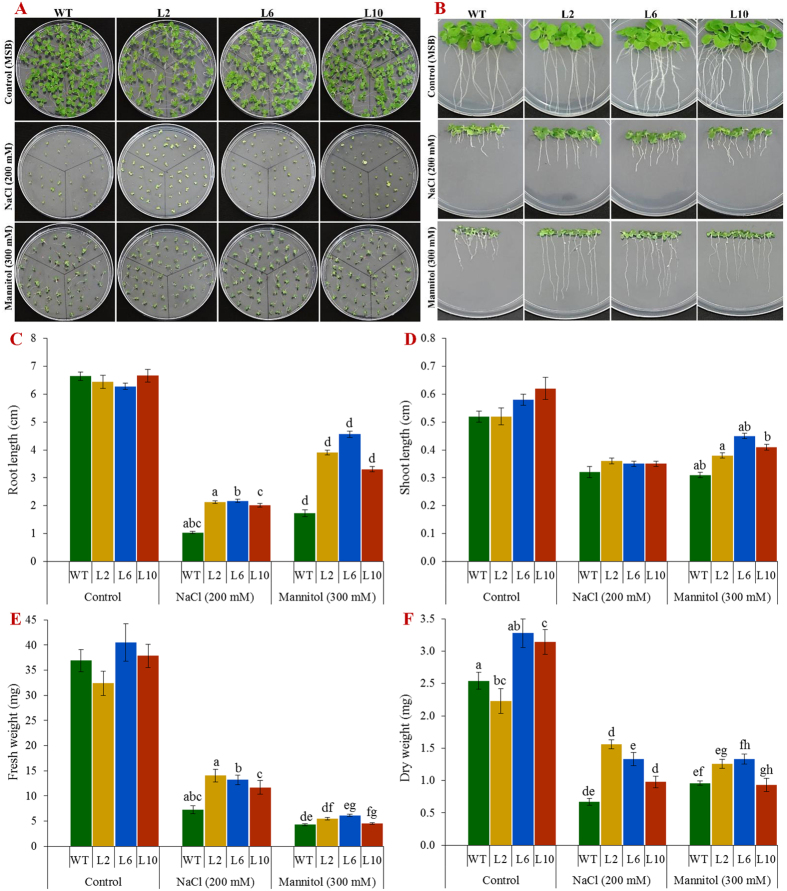
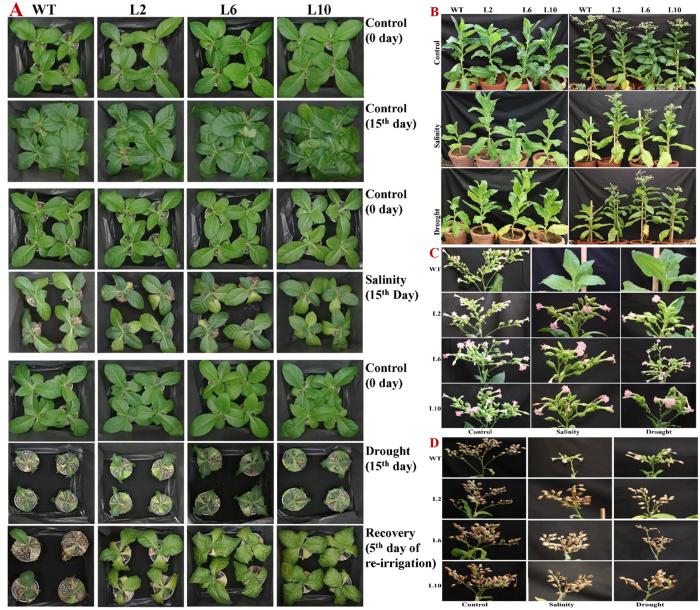
Transgenic lines (L2, L6 and L10) showed a significantly (p < 0.05) higher percentage seed germination than control plants (WT) in stress conditions (Figs 2 and S6). The percentage seed germination of WT was about 58 and 82%, whereas that of the transgenic lines was 73–95 and 93–97% under salinity and osmotic stress, respectively. Although the growth of all plants under stress was lower than that of control plants, the reduction in growth was smaller in transgenic lines compared to in WT plants (Figs 2 and S7). Growth parameters, including fresh weight, dry weight, shoot and root length were significantly (p < 0.05) higher in transgenic lines (L2, L6 and L10) compared to those in WT plants under stress conditions (Fig. 2).
Figure 2. Plant growth analyses under salt and osmotic stress.
(A) Seed germination of transgenic tobacco plants under different abiotic stress; (B) Comparative growth analysis of WT and SbSDR1 plants under control and stress conditions. Comparative study of (C) root length; (D) shoot length; (E) fresh weight, and (F) dry weight of transgenic (L2, L6 and L10) and WT plants under salt and osmotic stress. Bars represent means ± SE and values with similar letters are significant at P < 0.05.
Ectopic expression of the SbSDR1 gene enhances physiological status of plant under stress
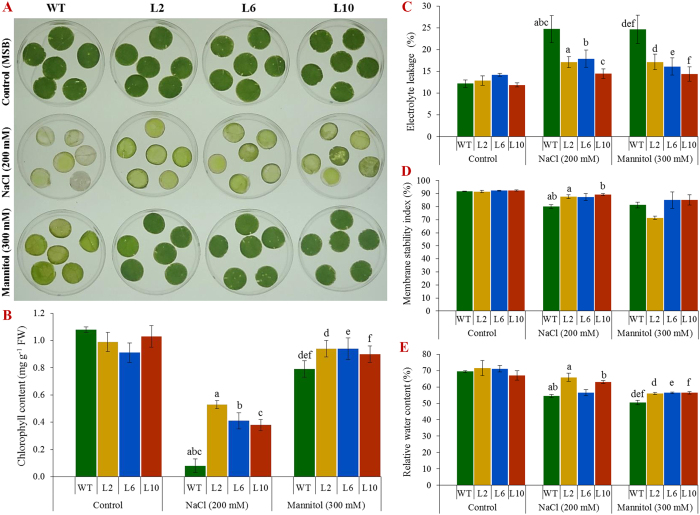
Salt and osmotic stress induced less damage in transgenic lines compared to in control plants (Figs 3 and 4). Leaf segments of wild type plants showed a lower viability and senesced, whereas transgenic lines exhibited less necrosis under salt and osmotic stress (Fig. 3A). Furthermore, a reduction in cellular chlorophyll content was observed under stress conditions in all plants, but a significantly (p < 0.05) a higher chlorophyll content was observed in transgenic plants than in WT plants (Fig. 3B).
Figure 3. Leaf disc assay and physiological analyses of transgenic lines.
(A) Leaf disc assay; (B) total chlorophyll content; (C) electrolyte leakage; (D) membrane stability index, and (E) relative water content from leaves of WT and transgenic (L2, L6 and L10) plant under control, salinity and osmotic stress conditions. Bars represent means ± SE and values with similar letters are significant at P < 0.05.
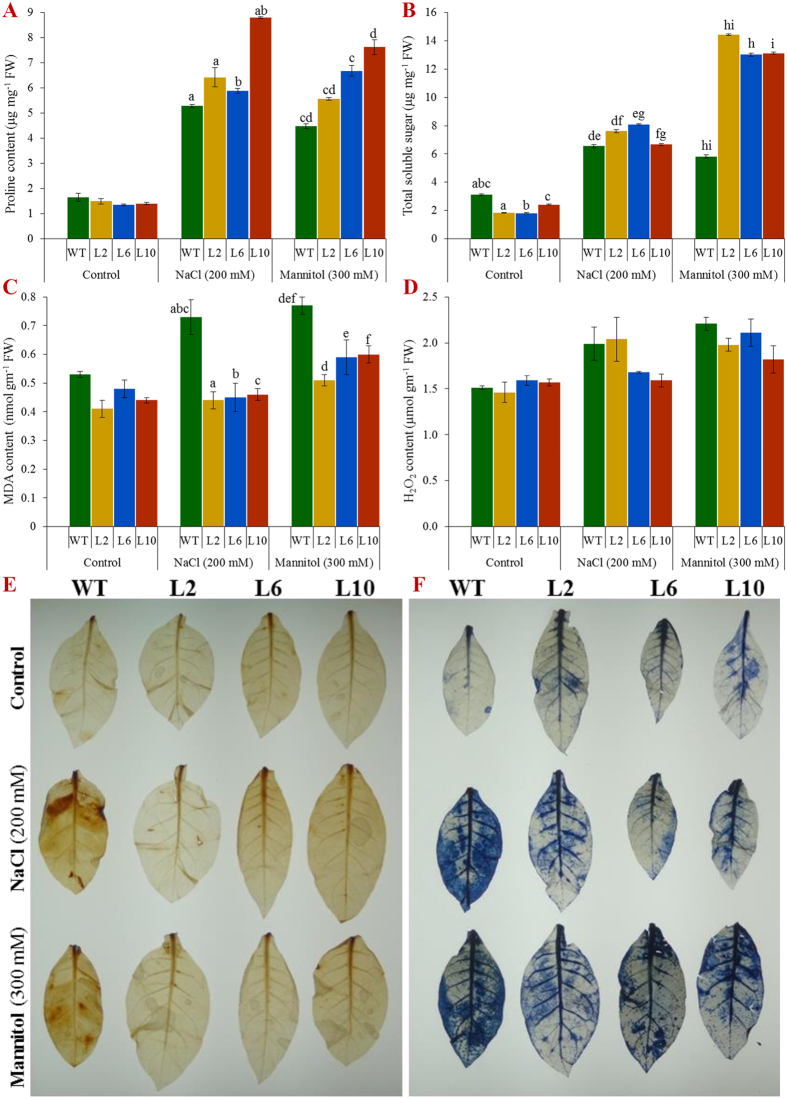
Figure 4. Biochemical analyses and in-vivo localisation of peroxide and superoxide free radicals of transgenic tobacco plants under abiotic stress.
Estimation of (A) proline; (B) total soluble sugar; (C) MDA, and (D) H2O2 contents in WT and transgenic (L2, L6 and L10) plants under salinity and osmotic stress condition. In-vivo localisation of (E) peroxide by DAB, and (F) superoxide free radicals by NBT staining of transgenic (L2, L6 and L10) and WT leaves. Bars represent means ± SE and values with similar letters are significant at P < 0.05.
In control conditions, physiological parameters such as electrolyte leakage (EL), the membrane stability index (MSI), relative water content (RWC) and the concentration of proline and total soluble sugars were comparable in wild type and transgenic lines (Figs 3 and 4). Transgenic lines showed a significantly (p < 0.05) higher RWC and MSI, but a reduced EL under stress conditions compared to control plants (Fig. 3C–E). Furthermore, transgenic plants exhibited a considerably (p < 0.05) higher concentration of the osmoprotectants proline and total soluble sugars in transgenic lines compared to in wild type under salinity and osmotic stress (Fig. 4A,B). Thus, transgenic lines exhibited a better physiological status under stress conditions.
The SbSDR1 regulates ROS buildup under salinity and oxidative stress
The MDA and H2O2 contents were similar in all plants under control conditions, but showed a significantly (p < 0.05) lower accumulation in transgenic plants than in wild type under stress conditions (Fig. 4C,D). Furthermore, in an in vivo localisation study, WT plant leaves accumulated more O2− and H2O2 than transgenic lines exposed to salt and osmotic stress (Fig. 4E,F).
Ectopic expression of the SbSDR1 gene provides salt and osmotic endurance
Thirty-day-old wild type and transgenic tobacco plants (L2, L6 and L10) with uniform growth were subjected to salt and drought stress in greenhouse conditions (Fig. 5A). The growth of all plants was comparable in control conditions, whereas that of WT plants was significantly affected by salt and drought stress. Although the growth of transgenic plants was suppressed, they showed better growth than WT. Furthermore, a higher degree of chlorosis was observed in wild type plants than in transgenic plants under salt stress. Retarded growth and wilting were observed in wild type plants compared to transgenic lines under drought stress and after re-irrigation, transgenic lines recovered rapidly compared to wild type plants.
Figure 5. Comparative plant growth studies under salt and osmotic stress.
(A) Morphological studies; (B) life cycle study; (C) flowering stage, and (D) mature (seed) stage of WT and SbSDR1 transgenic plants at control and stress conditions. Plant images were documented as per stages of plants grown under control condition.
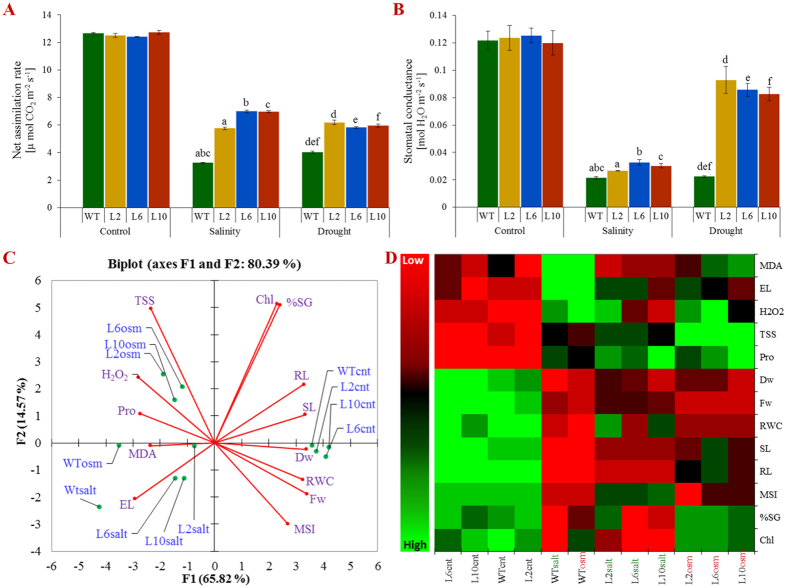
Similarly, the growth of WT plants in the field study was severely inhibited by salinity and drought treatment, whereas the growth of transgenic tobacco plants was less affected. Initially, all plants showed retarded growth, but subsequently, the growth of the transgenic lines gradually improved and the plants completed their life cycle under the stress treatment, whereas the WT plants did not (Fig. 5B). The transgenic plants were significantly taller and healthier than the WT plants and eventually flowered (Fig. 5C,D). In unstressed soil conditions, the number of inflorescences, flowers, pods and pod weight were the same in the transgenic and control plants (Figs 5 and S8). Furthermore, in the field, net photosynthesis and stomatal conductance were also similar under the normal (control) conditions for wild type and transgenic plants, but decreased with increasing salinity and drought stress (Fig. 6A,B). However, in comparison to WT, transgenic lines performed much better and showed a higher net photosynthesis and stomatal conductance.
Figure 6. Physiological and multivariate data analyses of transgenic lines.
(A) Net photosynthesis; (B) stomatal conductance; (D) an integrated comparative Bi-plot based principal component analysis with first two principal components, and (E) Heat map showing the differential response of transgenic lines (L2, L6 and L10) and WT plants under un-stress and stress (NaCl and osmotic) conditions. Bars represent means ± SE and values with similar letters are significant at P < 0.05.
Differential morpho-physio-biochemical response of transgenic plants under stress condition
Principal component analysis (PCA) was used to distinguish the morphological, biochemical and physiological responses of transgenic and control plants under normal and various stress conditions (Fig. 6C,D). All plants (transgenic lines and control) showed comparable responses in the unstressed conditions, as revealed by the bi-plot analysis, in which transgenic and WT plants clustered together (cnt). Control plants showed similar responses under stress conditions, whereas transgenic lines exhibited a differential response, and clustered at an axis, and thus, revealed a similar response to environmental stress. The integrated heat-map shows the differential plant responses to the variables of the stress conditions. Individually, the morphological, physiological and biochemical responses of plants explained 96.15, 89.68 and 88.32% of the variation, respectively (data not shown), whereas together, they explained 80.39% of the variation, because of the varying response, as shown by the heat map.
The SbSDR1 gene involves in transcriptional regulation of host stress responsive genes and transcription factors
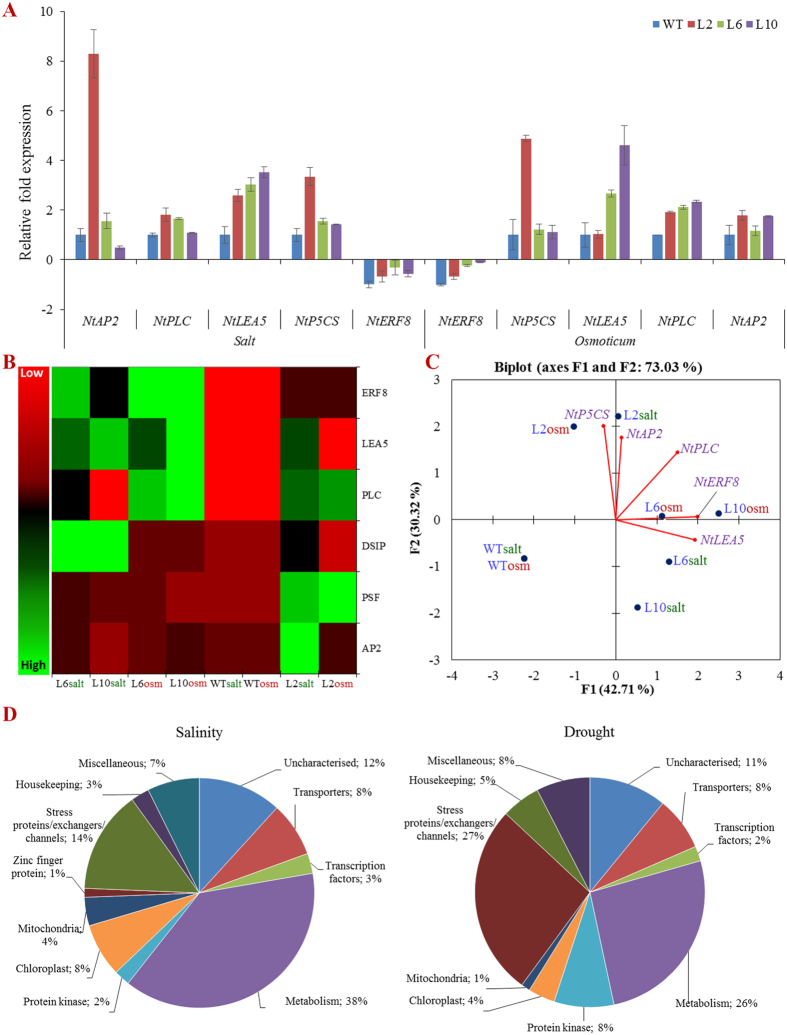
The expression of ten genes that encoded different transcription factors and other gene(s) of the host plant (i.e. tobacco) were studied. Five out of these ten genes were differentially expressed in transgenic plants under stress conditions (Fig. 7A). The expression of a gene encoding an AP2 domain-containing transcription factor (NtAP2) was highly upregulated (eight-fold) under salt stress in transgenic plants. The phosphoinositide-specific phospholipase C (NtPLC), late embryogenesis abundant protein 5 (NtLEA5) and delta 1-pyrroline-5-carboxylate synthetase (NtP5CS) genes were upregulated in transgenic lines upon salinity and osmotic stress. In contrast, the gene encoding ethylene response factor 8 (NtERF8) was downregulated in transgenic lines, but was more down-regulated in WT plants in stress conditions. The heat map and PCA analysis further confirmed that genes encoding transcription factors were differentially expressed in stress conditions (Fig. 7B,C).
Figure 7. Transcript expression analysis and microarray-based functional classification of host stress responsive genes.
Comparative (A) transcript expression profile; (B) Heat map, and (C) principal component (Bi-plot) analysis of AP2 domain-containing transcription factor (NtAP2), phosphoinositide-specific phospholipase C (NtPLC), late embryogenesis abundant protein 5 (NtLEA5), delta 1-pyrroline-5-carboxylate synthetase (NtP5CS) and ethylene-responsive transcription factor 8 (NtERF8) genes under un-stress and stress (NaCl and osmotic) conditions. (D) Functional classification of differentially expressed genes of SbSDR1 overexpressing transgenic tobacco plant under abiotic stress conditions. Genes differentially expressed in SbSDR1 plant under stress condition were normalised with transcript of WT plants treated with same stress.
A whole-transcript expression array of tobacco, containing 272,410 EST gene-probe sets, was used to study the differential expression of host genes under high salinity and drought-stress conditions. A hierarchical cluster analysis and scattered plot confirmed the differential expression of host genes under abiotic stress conditions (Figures S9 and S10). A total of 12,490 and 5,142 genes showed at least a two-fold up- (>2) or down-(<−2) regulation under high salinity and drought stress, respectively, at p < 0.05 using ANOVA. In total, 1,998 genes were differentially expressed by more than eight-fold (<−8 or >8) under salinity stress, whereas 236 genes were differentially expressed more than five-fold (<−5 or >5) under drought stress (Figure S11). Out of the 1,198 genes with an eight-fold change in expression under high salinity, 1,169 genes were up-regulated, whereas 29 genes were down-regulated. Similarly, under drought stress, 167 genes were up-regulated, whereas the remaining 69 were down-regulated. In total, 31 genes were commonly expressed under salt and drought stress. The microarray analysis revealed the up- and down-regulation of many genes encoding transporters, ion exchangers, stress-related proteins, metabolic enzymes and transcription factors; some of the important genes/proteins are listed in Table 1. The differentially expressed genes/proteins were further categorised into different groups according to their biological functions (Fig. 7D). Approximately 38% and 26% genes/proteins that were differentially expressed under salt and drought stress, respectively, were associated with metabolism, whereas 14% and 27% of genes/proteins were stress-responsive or ion exchangers, respectively. Transporters represented 8% of all differentially expressed genes/proteins; however, about 12% of the genes/proteins were categorised as uncharacterised genes/proteins. About 2–3% of the transcription factors were upregulated under stress conditions.
Table 1. Selected transcripts that differentially expressed (up- or down-regulated) in SbSDR1 overexpressing transgenic tobacco plant compared with the wild type under stress (salt or osmotic) conditions.
| S. No. | Probe Id | Gene name | Gene accession | Fold Change (log2) |
|---|---|---|---|---|
| Transcripts significantly differentially expressed under salt (200 mM NaCl) stress | ||||
| Transporters | ||||
| 1 | NtPMIa1g79974e2_st | ABC transporter | FH327345 | 6.88 |
| 2 | NtPMIa1g179413e1_s_st | ABC transporter 1 (NtPDR1 gene for PDR-type) | FH465185 | 5.91 |
| 3 | NtPMIa1g58619e1_st | Sulphate transporter | FG143420 | 5.78 |
| 4 | NtPMIa1g52967e1_st | Sugar transporter ERD6 | ET768583 | 4.88 |
| 5 | NtPMIa1g150213e1_s_st | ABC transporter G family member 11 | FH150205 | 4.22 |
| 6 | NtPMIa1g122574e1_st | ABC transporter B family member 21 | FI074394 | 3.74 |
| 7 | NtPMIa1g163746e1_st | ABC transporter C family member 10 | FH377610 | 3.69 |
| 8 | NtPMIa1g143376e1_st | ABC transporter C family member 3/4 | FH290450 | 3.55 |
| 9 | NtPMIa1g3211e1_st | Calcium-transporting ATPase | FI052025 | 3.45 |
| 10 | NtPMIa1g61677e3_st | ABC transporter C family member 15 | FH564659 | 3.34 |
| 11 | NtPMIa1g11150e1_st | MRP-like ABC transporter | FH120467 | 3.29 |
| 12 | NtPMIa1g168421e1_st | Sulphate transporter 3 (low affinity) | FH533814 | 3.15 |
| 13 | NtPMIa1g129050e1_st | ABC transporter B family member 25/27 | FH918583 | 3.07 |
| 14 | NtPMIa1g9139e1_st | Anion transporter 2 | ET929814 | 3.02 |
| 15 | NtPMIa1g79753e1_st | Cation-chloride cotransporter 1 | FH091636 | −3.42 |
| Stress proteins | ||||
| 16 | NtPMIa1g123767e1_st | DNA-binding protein SMUBP-2 | FI077868 | 4.26 |
| 17 | NtPMIa1g56604e1_s_st | NAC domain protein NAC5 | ET045577 | 4.09 |
| 18 | NtPMIa1g81083e1_st | REF/SRPP-like protein/or Stress-related protein | FG187326 | 3.66 |
| 19 | NtPMIa1g44375e1_st | Calcium-binding protein/Calmodulin-like protein 4 | EH617111 | 3.44 |
| 20 | NtPMIa1g141096e1_st | Nuclear transport factor 2 (NTF2) family protein | FH231101 | 3.19 |
| 21 | NtPMIa1g49504e2_st | Heat shock 70 kDa protein | ET710960 | 3.17 |
| 22 | NtPMIa1g82819e3_st | ras-related protein Rab7-like/Small GTP-binding protein | ET802996 | 3.17 |
| 23 | NtPMIa1g71923e1_s_st | G-box-binding factor 1-like | ET047835 | 3.16 |
| Stress responsive genes/channels | ||||
| 24 | NtPMIa1g21515e1_s_st | PR-5 Gene | FH357120 | 6.91 |
| 25 | NtPMIa1g138004e1_s_st | NtPOX1 (peroxidase) | FG146731 | 6.29 |
| 26 | NtPMIa1g144683e1_st | str246C gene | FH548471 | 5.88 |
| 27 | NtPMIa1g42792e1_st | Osmotin-like (OLP1) | FH990179 | 4.89 |
| 28 | NtPMIa1g42154e2_st | Peroxidase | FH663398 | 4.79 |
| 29 | NtPMIa1g29557e1_st | Sodium/hydrogen exchanger 8-like | ET764618 | 4.53 |
| 30 | NtPMIa1g201506e1_st | Superoxide dismutase | EH619039 | 4.44 |
| 31 | NtPMIa1g193487e1_st | Glutathione S-Transferase | ET691311 | 4.38 |
| 32 | NtPMIa1g46700e1_st | Sodium/hydrogen exchanger | FH050971 | 4.04 |
| 33 | NtPMIa1g4232e2_s_st | Aquaporin TIP3-2 (tonoplast intrinsic protein) | FH043168 | 3.83 |
| 34 | NtPMIa1g35785e3_st | Potassium channel AKT1-like | ET966045 | 3.70 |
| 35 | NtPMIa1g46700e3_s_st | Na+/H+ antiporter/or sodium/hydrogen exchanger | FI072621 | 3.55 |
| 36 | NtPMIa1g47884e3_st | Calcium channel (two pore) protein 1 | FH134749 | 3.46 |
| Transcription factors | ||||
| 37 | NtPMIa1g31907e2_s_st | Transcription factor (WRKY) 31 | FG161188 | 5.64 |
| 38 | NtPMIa1g80200e5_st | Transcription factor TGA1/Basic leucine zipper protein | FG197996 | 4.60 |
| 39 | NtPMIa1g214476e1_st | Transcription factor (WRKY) 6 | ET043797 | 4.17 |
| 40 | NtPMIa1g51042e1_st | Transcription factor ILR3-like | ET854451 | 3.94 |
| 41 | NtPMIa1g50790e2_st | Transcription factor TCP3 | ET046638 | 3.88 |
| 42 | NtPMIa1g43220e3_st | Transcription factor (WRKY) 21 | FH187095 | 3.72 |
| 43 | NtPMIa1g155166e1_st | Transcription factor LUX | ET694713 | 3.62 |
| 44 | NtPMIa1g50790e1_st | Transcription factor TCP4-like | ET980639 | 3.57 |
| 45 | NtPMIa1g53553e2_x_st | Transcription factor Bhlh041 | FH670186 | 3.53 |
| 46 | NtPMIa1g32403e1_st | Transcription factor (WRKY) 43 | ET049847 | 3.40 |
| 47 | NtPMIa1g99480e1_s_st | Transcription factor (GATA) 25 | FH991549 | 3.17 |
| 48 | NtPMIa1g80077e2_st | transcription factor JUNGBRUNNEN 1-like | FI039736 | 3.03 |
| 49 | NtPMIa1g178470e1_s_st | Transcription factor family-PLATZ- protein | ET042575 | −3.15 |
| 50 | NtPMIa1g93305e1_st | Ethylene-responsive transcription factor | FH965889 | −3.17 |
| Uncharacterised | ||||
| 51 | NtPMIa1g184611e1_s_st | Uncharacterised | EH617968 | 6.21 |
| 52 | NtPMIa1g112984e1_s_st | Uncharacterised | ET991956 | 5.60 |
| 53 | NtPMIa1g59636e1_st | Uncharacterised | FH622649 | 5.58 |
| 54 | NtPMIa1g127287e1_st | Uncharacterised | FH333853 | −5.69 |
| 55 | NtPMIa1g93939e1_st | Uncharacterised | FH331817 | −3.38 |
| 56 | NtPMIa1g125877e1_x_st | Uncharacterised | FI084463 | −3.29 |
| Zinc finger protein | ||||
| 57 | NtPMIa1g1891e1_s_st | Zinc induced facilitator like | FI027112 | 5.19 |
| 58 | NtPMIa1g97020e1_st | Zinc metalloprotease (ATP-dependent) | FH980189 | 3.78 |
| Transcripts significantly differentially expressed under osmotic (300 mM mannitol) stress | ||||
| Transporters | ||||
| 1 | NtPMIa1g179293e1_st | ABC transporter G family member 35 | FH628071 | 4.01 |
| 2 | NtPMIa1g200291e1_st | ABC transporter B family member 21 | FH968229 | 3.35 |
| 3 | NtPMIa1g118531e1_st | ABC transporter A family member 11 | ET925380 | 2.88 |
| 4 | NtPMIa1g19057e2_st | ABC transporter G family member 3 | FH326473 | 2.85 |
| 5 | NtPMIa1g173628e1_s_st | Sucrose transport protein SUC3 | ET942104 | 2.71 |
| 6 | NtPMIa1g42126e1_s_st | ABC transporter B family member 25 | FH321728 | 2.55 |
| 7 | NtPMIa1g47623e1_st | Sulfate transporter | FH062478 | −3.64 |
| Stress responsive proteins/genes/channels | ||||
| 8 | NtPMIa1g136207e1_st | Protein kinase (receptor-like) | FH084028 | 4.92 |
| 9 | NtPMIa1g78543e3_st | Aquaporin-4 | FH137367 | 2.76 |
| 10 | NtPMIa1g57744e1_st | Anionic peroxidase gene/Peroxidase | FH308715 | 2.68 |
| 11 | NtPMIa1g82649e2_st | Plasma membrane H+ ATPase | FH091813 | 2.43 |
| 12 | NtPMIa1g190637e1_x_st | Mitogen-activated protein kinase kinase kinase NPK1 | FH176408 | 2.41 |
| 13 | NtPMIa1g17460e1_s_st | Heat shock protein (class II) | FH127057 | −3.54 |
| 14 | NtPMIa1g40121e1_st | Cytochrome P450 | FG196194 | −3.09 |
| 15 | NtPMIa1g201049e1_st | Heat shock protein 101 | ET915891 | −2.98 |
| Transcription factors | ||||
| 16 | NtPMIa1g2222e1_st | Transcription factor (WRKY) 41 | FI020184 | 2.87 |
| 17 | NtPMIa1g82333e1_st | Transcription factor bHLH55 | FH551099 | 2.38 |
| 18 | NtPMIa1g122488e1_st | Transcription factor (heat stress) B-2a | ET044925 | −2.52 |
| 19 | NtPMIa1g123702e2_st | Transcription factor (WRKY) 33 | FI077699 | −2.33 |
| Uncharacterised | ||||
| 20 | NtPMIa1g140270e1_st | Uncharacterised | FG168624 | 3.49 |
| 21 | NtPMIa1g82079e2_st | Uncharacterised | ET983581 | 3.35 |
| 22 | NtPMIa1g38269e1_st | Uncharacterised | ET920361 | −3.26 |
| 23 | NtPMIa1g180425e1_s_st | Uncharacterised GPI-anchored protein | FH467984 | −2.96 |
No sign indicates up-regulation, whereas “−” sign shows down-regulation. Fold-expression is significant at ANOVA p < 0.05.
Discussion
Salinity is an emerging threat to agriculture and it has a detrimental effect on plant growth by imposing two simultaneous stresses; toxic salt ions and water stress. Although several abiotic stress-responsive genes have been characterised, research has focused on identifying novel gene functions for deciphering the salt stress tolerance mechanism and identifying candidate genes for developing resistant crops8,33. To explore potential novel genes for developing salinity tolerance in crops, here, we identify a salt- and drought-responsive gene, SbSDR1, from Salicornia and functionally characterised it in tobacco for the first time. The studied gene can be used to improve the salinity and drought tolerance of the agricultural crop.
No significant similarities were found between the nucleotide and deduced amino acid sequences of SbSDR1 and other genes/proteins in a NCBI-BLAST sequence homology search; therefore, the gene is considered to be novel. The single copy of the intron-less SbSDR1 gene (Figure S2) is differentially expressed under different environmental stresses (Figure S3). The expression of SbSDR1 was elevated under salt (two-fold at 6 h), drought (3.3-fold at 24 h), heat (four-fold at 12 h) and cold (two-fold at 24 h) stress. Similarly, abiotic genes, such as SbpAPX, SbGSTU, SbMT-2 and SbUSP from S. brachiata were differentially expressed under different stress conditions5,20,22,28.
Sub-cellular localisation experiments confirmed that SbSDR1 is a nuclear protein (Fig. 1A) and DNA/protein interactions revealed the binding specificity of SbSDR1 to genomic DNA (Fig. 1B). In silico analyses also predicted that the SbSDR1 protein is nuclear-localised and potentially regulates metabolic process and gene expression. The SbSDR1 protein does not have a fixed conformation, because of the high content of disorder-promoting amino-acid residues (A, E, G, K, P, Q and S) and therefore, is considered to belong to the family of intrinsically disordered proteins (IDPs). Extensive post-translational modifications of IDPs have been reported and SbSDR1 has 14 phosphorylation sites, which potentially facilitate protein–protein interactions34. Plant dehydrins, which are also IDPs, interact with ROS to protect cellular proteins from oxidative damage under stress conditions35. As an IDP, SbSDR1 is protected during stress conditions and might transcriptionally regulate genes26. The SbSDR1 protein also possesses structural flexibility and thus, can easily modulate biological functions by regulating metabolic processes and gene expression. The ability of SbSDR1 to bind genomic DNA of Salicornia and tobacco confirmed that it functions as a transcription factor. The in vitro binding of SbSDR1 enables it to regulate a large number of stress-responsive genes by binding to their regulatory regions. In silico analysis, subcellular localisation and the binding of SbSDR1 to the genomic DNA of Salicornia and tobacco genomic DNA suggest that SbSDR1 might functions as a transcription factor. A novel salt-induced gene, LcSAIN1, cloned from Sheepgrass (Leymus chinensis), conferred salt stress tolerance by activating the expression of transcription factors and functional genes in transgenic plants under salt stress33. Similarly, SbASR-1 protects the plant from salinity and drought stress and functions as a transcription factor in transgenic groundnut26.
Out of sixteen transgenic tobacco lines, three lines – L2, L6 and L10 – showed a high level of GUS expression (Figs 1D and S5) and, therefore, were selected to understand further the role of SbSDR1 in abiotic stress tolerance. A single gene integration, a high expression of the transgene and a high seed germination percentage (Figs 1E, 2A and S6) were observed in the selected lines under stress conditions. Furthermore, the transgenic lines showed enhanced plant growth, including fresh weight, dry weight and shoot and root length under different stress conditions, compared to the control plants (Figs 2 and S7). The enhanced plant growth in the transgenic lines demonstrates that overexpression of SbSDR1 reduces the adverse effects of stress. The degree of injuries caused by stress in a plant are commonly studied by assessing the physiological status, by measuring EL, the MSI, RWC, lipid peroxidation (MDA content), and the concentration of proline and total soluble sugars12,13. Notably, the ectopic expression of SbSDR1 in tobacco promotes salt and osmotic tolerance by modulating the physiology of plants, including RWC, EL, MSI, proline, total sugar and MDA (Figs 3 and 4). A significant increase in RWC, MSI, proline and total soluble sugars alleviates stress injury, and thus, stress-induced damage is reduced in transgenic lines compared to WT, whereas lipid peroxidation and EL, which are common stress markers, decreased in transgenic lines. Proline is considered to be an osmolyte, a ROS scavenger and also a molecular chaperone that protects the cell from stress-induced damage12. Cells undergo metabolic rearrangements and activate regulatory networks in response to different environmental stresses, including salinity and drought12,13.
It is well established that plants often accumulate ROS under abiotic stress conditions, which leads to oxidative stress9. A reduction in the total chlorophyll content, which is a marker of cellular stress was observed in stress conditions, due to ROS generation in the chloroplasts36. Transgenic plants showed a significantly higher chlorophyll content and reduced leaf senescence and H2O2 content under oxidative stress (Figs 3 and 4). In this study, it was observed that SbSDR1 inhibits ROS accumulation under salinity and oxidative stress (Fig. 4). Abiotic stress also leads to the generation of superoxide radicals10,11. The results confirmed that control plants (WT) accumulate more MDA, O2− and H2O2 than transgenic plants and thus, confirm the role of SbSR1 in contributing to oxidative stress tolerance by regulating ROS scavenging activities. Other abiotic stress-responsive genes, such as SbpAPX1, SbNHX1, SbUSP and SbASR-1 from S. brachiata, contribute towards ROS homeostasis under salt and osmotic stress condition5,6,25,26.
Abiotic stress tolerance, including salt and drought tolerance, depends on the growth stage and is regulated at different developmental stages. Because the stress tolerance at one stage of plant growth does not correlate with tolerance at other stages5. Therefore, the performance of SbSDR1-overexpressing tobacco lines was assessed at different stages and in field conditions (Figs 5 and 6). The growth of WT plants was severely affected in comparison to that of transgenic tobacco lines, which were unaffected. Tobacco plants that overexpressed SbSDR1 were significantly taller and healthier than WT plants, which showed retarded growth, wilting and chlorosis symptoms. Typical flowering and seed-setting were observed in transgenic lines but not in WT. In fact, the SbSDR1 overexpressing plants performed better in the field and completed their life cycle in time under abiotic stress conditions, whereas the WT plants did not (Figs 5, 6 and S8). This study demonstrates that the ectopic expression of SbSDR1 confers salt and osmotic tolerance at different developmental stages of the plant in pots and also in the field. Many studies have demonstrated that different salt-responsive genes, such as SbNHX1, SbpAPX1 and SbASR-1, improved the tolerance of crops such as jatropha, castor and peanut to oxidative stress24,25,26,37. The SbpAPX gene confers salinity and drought tolerance to tobacco plants at different developmental stages5. Similarly, the overexpression of the novel gene(s) LcSAIN1, MsZEP and the TF, Zmhdz10 in plants leads to the improved tolerance to oxidative stress that is caused by drought and salt stresses16,17,33.
Quantitative real-time PCR confirmed that the SbSDR1 gene is involved in the transcriptional regulation of host stress-responsive genes and transcription factors, such as NtAP2, NtPLC, NtLEA, NtP5CS and NtERF8 under stress conditions (Fig. 7A). Microarray analysis demonstrated the differential expression of several stress-responsive genes and transcription factors in SbSDR1 transgenic plants compared to in WT plants under salt and osmotic stress (Table 1 and Fig. 7D). In this study, genes that were differentially expressed in transgenic plants under stress conditions were normalised using transcripts of WT plants treated with the same stress, to rule out the possibility that genes in WT tobacco plants are differentially expressed due to stress. Microarray analysis revealed the upregulation of genes encoding transporters, stress-related proteins, signalling components and transcription factors, as well as uncharacterised proteins. Salinity and drought stresses have a strong impact on gene expression, and many genes and stress-associated transcription factors associated with cellular metabolism are differentially expressed, to modulate the physiology of the plant to combat stress-induced injuries7,12,13. Previous microarray studies demonstrated that the overexpression of genes such as OsACA6 and OsCPK4 leads to the up-regulation of stress responsive genes/proteins, oxidative-burst proteins and transcription factors in transgenic rice38,39. The functional categorisation and distribution of differentially expressed genes in transgenic SbSDR1 tobacco plants revealed that the majority of these genes encode proteins/enzymes involved in the maintenance of cellular metabolism39. Microarray results also suggest that GmDREB1 activates the expression of many soybean-specific stress-responsive genes under different abiotic stress conditions40. Similarly, the overexpression of DPB3–1 enhanced heat-stress tolerance via unknown transcription factors without growth retardation and yield reduction in rice41. In conclusion, the up-regulation of transcription factors, salt-responsive channels, transporters and enzymes/proteins involved in cellular metabolism in transgenic plants confirms that SbSDR1 functions as a molecular switch and thus, confers abiotic stress tolerance to crops (Figure S12).
Conclusion
In this study, a novel gene, SbSDR1, was functionally characterised. The overexpression of SbSDR1 resulted in enhanced tolerance against salinity and drought stress. Morphological, biochemical, physiological and molecular evidence confirmed that transgenic plants are more tolerant to salinity and drought stress than wild type. Furthermore, the results provide direct evidence that SbSDR1 contributes to tolerance to salinity and drought stress without yield loss by regulating stress-responsive genes. The functional characterisation of a novel abiotic stress-responsive gene provides new insights into the bio-physiological responses of plants at the molecular level to salt and drought stresses. The results further support the potential of SbSDR1 as a candidate gene for the genetic engineering of crop plants to enhance tolerance to salt and drought stress.
Methods
Cloning, bioinformatics and transcript profiling
The Sal-C-53.e1 gene clone (EB484704), showing no significant homology/identity with existing database (NCBI/EMBL) and exhibited high expression under salt and drought stress, therefore, selected for the study32. Primers were designed from partial EST gene sequence of Sal-C-53.e1 gene clone (EB484704), a gene of unknown function (designated as Salicornia brachiata salt & drought responsive 1, SbSDR1) was made full length using RACE (rapid amplification of cDNA ends), cloned and sequenced (Table S1). The SbSDR1 gene sequences were analysed for homology and conserved motifs. Amino acid sequences, deduced from nucleotide sequences, were characterised in silico using Expert Protein Analysis System42 (ExPASy). For expression analysis, one-month-old S. brachiata seedlings were transferred to a hydroponic system (½ MS with 8/16 h dark/light cycle at 25 °C) for 15 days. Acclimatised plants were subjected to different abiotic stresses such as salinity (250 mM NaCl), desiccation, heat (45 °C) and cold (4 °C). Total RNA was isolated from each treated plants (and control i.e. unstressed plants), cDNA was prepared, and transcript analysis was performed using quantitative real-time (qRT-PCR) PCR (Table S1). The relative fold expression was calculated using the CT method; gene β-tubulin was used as an internal reference and compared with control plants43.
Genome organisation and subcellular localisation
Plant (S. brachiata) genomic DNA was extracted, quantified using ND-1000 spectrophotometer and qualitatively analysed by agarose gel electrophoresis. The SbSDR1 gene was amplified using genomic DNA as template with a gene-specific primer pair (Table S1), cloned and sequenced. The copy number of SbSDR1 gene was determined using Southern blot analysis6,26. For subcellular localisation, a translational fusion cassette of SbSDR1 along with RFP (red fluorescent protein) was generated using the gateway technology25,26. Expression cassette (RFP:SbSDR1) and control vector (pSITE-4CA:RFP) were transferred to onion epidermal cells using microprojectile bombardment (PDS-1000/He biolistic system, Biorad, USA) and transient expression of RFP was observed using an epifluorescence microscope (Axio Imager, Carl Zeiss AG, Germany).
Heterologous expression and DNA binding assay
The coding sequence of the SbSDR1 gene was cloned into the pET28a expression vector (Table S1), transformed into E. coli BL21 (DE3) cells and recombinant protein expression was induced26. The SbSDR1 protein was purified, evaluated on SDS-PAGE (12%) and used for analysing DNA binding property by the electrophoretic mobility shift assay (EMSA). Genomic DNA, extracted from Salicornia and tobacco plants were digested (BamH1), denatured at 94 °C for 5 min followed by cooling at 4 °C for 5 min and incubated with SbSDR1 protein at 25 °C for 16 h. The SbSDR1 protein and DNA/protein complexes were electrophoresed on non-denaturing polyacrylamide gel (6% native PAGE) at 50 V for three hours at 4 °C. The gel was silver stained, and electrophoretic mobility shift was studied.
Genetic transformation of tobacco plants and molecular analysis
The SbSDR1 CDS was amplified (Table S1), cloned in pCAMBIA2301 through an intermediate pRT101 plant expression vector and transformed into tobacco (Nicotiana tabacum cv. Petit Havana) plants by Agrobacterium tumefacians (strain LBA4404) mediated leaf disc method44. Putative transgenic tobacco plants were regenerated, and transgenic lines (T0 and T1) were screened by growing on kanamycin (50 mgL−1). The presence of transgenes was confirmed by PCR amplification (of uidA and SbSDR1 genes), and integration (transgene events) was determined by Southern blot (Table S1). Transgenic lines (T1) were further screened for histochemical β-glucuronidase activity45 and lines showing high GUS activities were selected for further analysis. Overexpression of SbSDR1 gene was analysed in selected transgenic lines by semiquantitative reverse transcriptase PCR (Rt-PCR), and the actin gene was used as an internal reference (Table S1).
Abiotic stress treatments and evaluation of transgenic lines
All transgenic lines were maintained under controlled containment facility. Percent seed germination of transgenic plants was calculated under abiotic stress (salinity, 200 mM NaCl and osmotic, 300 mM mannitol) and compared with non-transgenic plants (wild type, WT) and unstressed (control) condition46. Seeds were germinated on MS containing kanamycin (50 mgL−1); germinated T1 transgenics and WT (germinated on only MS) seedlings were grown on MS supplemented with NaCl (200 mM) or mannitol (300 mM) for 21 days. Plant morphology, growth parameters, including shoot (SL) & root (RL) length, and fresh (Fw) & dry (Dw) weight were studied and compared with controls (WT and control condition; plants grown on MS only were taken as a control condition).
Plants (transgenic and WT) grown in control condition for 21 days were further transferred to hydroponics (½ MS) and plastic cups (containing garden soil) and grown for 30 days. Salinity (200 mM NaCl) or osmotic (300 mM mannitol) stress treatments were given for 24 hours to 30 days hydroponically grown plants. Physio-biochemical analyses, including electrolyte leakage, membrane stability index, relative water, proline, total soluble sugar, lipid peroxidation (MDA content), H2O2 content were performed for T1 transgenic lines under different abiotic stress treatments and compared with controls47,48,49,50,51,52,53. Similarly, generation of H2O2 and superoxide (O2−) was visualised by in vivo localisation studies54. Healthy leaves of 30 days grown (in hydroponics) plants (transgenic and WT) of a control condition were taken; 8 mm discs were punched out, subjected to stress condition (salinity, 200 mM NaCl or osmotic, 300 mM mannitol) and leaf senescence study was performed along with estimation of chlorophyll contents21,23,55,56.
Plants (transgenic and WT) transferred to plastic cups (containing garden soil) were grown for 30 days under green-house containment facility. For control condition, plants were irrigated (every alternate day) with ½ MS only, whereas irrigation with ½ MS containing 200 mM NaCl and no irrigation were considered salinity and drought stress treatment, respectively5. After 15 days of stress, plants were observed morphologically and compared with controls, whereas recovery study was performed for plants under drought stress by re-irrigating with tap water.
Plants (transgenic and WT) grown in plastic cups (for 30 days in green house under control condition) were shifted to controlled field condition and allow to grow further for 30 days. After that, salinity and drought stress were employed (as above) for 15 days and physiology including, net photosynthesis rate and stomatal conductance were measured by portable photosynthesis system (LI6400XT, LI-COR Biosciences, USA) and compared with controls.
Expression profiling and microarray analysis
Abiotic stress (salinity, 200 mM NaCl or osmotic, 300 mM mannitol) was given to 30 days hydroponically grown plants (transgenic and WT) for 24 hours. Plants were harvested, total RNA was extracted for cDNA synthesis, and relative fold expression of selected transcription factors (NtTFs) was studied by qRT-PCR. Extracted RNA of samples (WT and a selected transgenic line of control and treated with NaCl or mannitol) was converted to first strand cDNA. After that proceed to second strand cDNA synthesis, cRNA amplification, single stranded cDNA synthesis. Finally, fragmentation and terminal labelling were performed by following whole transcript (WT) expression arrays user manual (Affymetrix, USA). Labelled cDNAs were hybridised with tobacco whole transcript expression gene chip (containing total 272410 gene probes), washed and stained using fluidics module (GeneChip Fluidics Station 450, Affymetrix, USA) as per user instruction. Hybridised chips were scanned (Scanner 3000 7G, Affymetrix, USA), and scanned images were processed and analysed using expression console and transcriptome analysis console (Affymetrix, USA). Microarray analysis was performed in duplicate (n=2) and genes exhibiting significant fold expression (ANOVA p-value < 0.05) were considered for the study.
Statistical analysis
Data from five replicates (for each set of the experiment), each containing fifteen plants were presented as mean ± SE and subjected to analysis of variance (ANOVA) to determine the significance of difference amongst the means of WT and transgenic plants of every treatment set. Data exhibited p < 0.05 was considered significantly different and designated by similar letters. All dataset was also analysed individually and in combination with principal component analysis (PCA), and respective heat maps were generated.
Additional Information
How to cite this article: Singh, V. K. et al. A novel transcription factor-like gene SbSDR1 acts as a molecular switch and confers salt and osmotic endurance to transgenic tobacco. Sci. Rep. 6, 31686; doi: 10.1038/srep31686 (2016).
Supplementary Material
Acknowledgments
CSIR-CSMCRI Communication No. PRIS-033/2016. This study was supported by the Council of Scientific and Industrial Research (CSIR; www.csir.res.in), Government of India, New Delhi [BSC0109-SIMPLE]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Dr. Narendra Singh Yadav and Dr. Pradeep K Agarwal are duly acknowledged for 5’ RACE of the gene. Authors are grateful to Arabidopsis Biological Resource Centre, Ohio State University, USA for providing the pSITE-4CA vector.
Footnotes
Author Contributions Conceived and designed the experiments: A.M. and B.J. Performed the experiments: V.K.S. and I.H. Analysed the data: V.K.S. and A.M. Wrote the paper: A.M.
References
- FAO. The State of the World’s Land and Water Resources for Food and Agriculture (SOLAW)-Managing Systems at Risk http://www.fao.org/nr/solaw/solaw-home/en/ (2011).
- Qadir M. et al. Economics of salt‐induced land degradation and restoration. Nat. Resour. Forum 38, 282–295 (2014). [Google Scholar]
- Munns R. & Gilliham M. Salinity tolerance of crops–what is the cost? New Phytol. 208, 668–673 (2015). [DOI] [PubMed] [Google Scholar]
- Panta S. et al. Halophyte agriculture: Success stories. Environ. Exp. Bot. 107, 71–83 (2014). [Google Scholar]
- Singh N., Mishra A. & Jha B. Over-expression of the peroxisomal ascorbate paroxidase (SbpAPX) gene cloned from halophyte Salicornia brachiata confers salt and drought stress tolerance in transgenic tobacco. Mar. Biotechnol. 16, 321–332 (2014). [DOI] [PubMed] [Google Scholar]
- Udawat P., Jha R. K., Sinha D., Mishra A. & Jha B. Overexpression of a cytosolic abiotic stress responsive universal stress protein (SbUSP) mitigates salt and osmotic stress in transgenic tobacco plants. Front. Plant Sci. 7, 518 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H. & Probst N. Tolerance to drought and salt stress in plants: unravelling the signaling networks. Front. Plant Sci. 5, 151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himabindu Y. et al. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 124, 39–63 (2016). [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M. & Van Breusegem F. Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 (2004). [DOI] [PubMed] [Google Scholar]
- Gill S. S. & Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Bioch. 48, 909–930 (2010). [DOI] [PubMed] [Google Scholar]
- Demidchik V. Mechanisms of oxidative stress in plants: from classical chemistry to cell biology. Environ. Exp. Bot. 109, 212–228 (2015). [Google Scholar]
- Krasensky J. & Jonak C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinlein U. et al. Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani M. A. et al. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 133, 1755–1767 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A. et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149 (2008). [DOI] [PubMed] [Google Scholar]
- Zhang Z. et al. MsZEP, a novel zeaxanthin epoxidase gene from alfalfa (Medicago sativa), confers drought and salt tolerance in transgenic tobacco. Plant Cell rep. 35, pp. 439–453 (2016). [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. A novel maize homeodomain–leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and arabidopsis. Plant Cell Physiol. 55, 1142–1156 (2014). [DOI] [PubMed] [Google Scholar]
- Wang F. et al. A novel Cys2/His2 zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta 243, 783–797 (2016). [DOI] [PubMed] [Google Scholar]
- Tiwari V., Chaturvedi A. K., Mishra A. & Jha B. The transcriptional regulatory mechanism of the peroxisomal ascorbate peroxidase (pAPX) gene cloned from an extreme halophyte, Salicornia brachiata. Plant Cell Physiol. 55, 1774–1471 (2014). [DOI] [PubMed] [Google Scholar]
- Jha B., Sharma A. & Mishra A. Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol. Biol. Rep. 38, 4823–4832 (2011). [DOI] [PubMed] [Google Scholar]
- Joshi M., Jha A., Mishra A. & Jha B. Developing transgenic Jatropha using the SbNHX1 gene from an extreme halophyte for cultivation in saline wasteland. PLoS One. 8, e71136 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A. K., Mishra A., Tiwari V. & Jha B. Cloning and transcript analyses of type 2 metallothionein gene (SbMT-2) from extreme halophyte Salicornia brachiata and its heterologous expression in E. coli. Gene 499, 280–287 (2012). [DOI] [PubMed] [Google Scholar]
- Chaturvedi A. K., Patel M. K., Mishra A., Tiwari V. & Jha B. The SbMT-2 gene from a halophyte confers abiotic stress tolerance and modulates ROS scavenging in transgenic tobacco. PLoS One 9, e111379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Mishra A. & Jha B. Ectopic over-expression of peroxisomal ascorbate paroxidase (SbpAPX) gene confers salt tolerance in transgenic peanut (Arachis hypogea). Gene 547, 119–125 (2014). [DOI] [PubMed] [Google Scholar]
- Patel M., Joshi M., Mishra A. & Jha B. Ectopic expression of SbNHX1 gene in transgenic castor (Ricinus communis L.) enhances salt stress by modulating physiological process. Plant Cell Tiss. Org. 122, 477–490 (2015). [Google Scholar]
- Tiwari V., Chaturvedi A. K., Mishra A. & Jha B. Introgression of the SbASR-1 gene cloned from a halophyte Salicornia brachiata enhances salinity and drought endurance in transgenic groundnut (Arachis hypogea) and act as a transcription factor. PLoS One 10, e0131567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V., Patel M. K., Chaturvedi A. K., Mishra A. & Jha B. Functional characterization of the tau class glutathione-S-transferases gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic stress. PLoS One 11, e0148494 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udawat P., Mishra A. & Jha B. Heterologous expression of an uncharacterized universal stress protein gene (SbUSP) from the extreme halophyte, Salicornia brachiata, which confers salt and osmotic tolerance to E. coli. Gene 536, 163–170 (2014). [DOI] [PubMed] [Google Scholar]
- Jha B., Singh N. P. & Mishra A. Proteome profiling of seed storage proteins reveals the nutritional potential of Salicornia brachiata Roxb., an extreme halophyte. J. Agr. Food Chem. 60, 4320–4326 (2012). [DOI] [PubMed] [Google Scholar]
- Mishra A., Joshi M. & Jha B. Oligosaccharide mass profiling of nutritionally important Salicornia brachiata, an extreme halophyte. Carbohyd. Polym. 92, 1942–1945 (2013). [DOI] [PubMed] [Google Scholar]
- Mishra A., Patel M. K. & Jha B. Non-targeted metabolomics and scavenging activity of reactive oxygen species reveal the potential of Salicornia brachiata as a functional food. J. Funct. Foods 13, 21–31 (2015). [Google Scholar]
- Jha B. et al. Identification of salt-induced genes from Salicornia brachiata, an extreme halophyte through expressed sequence tags analysis. Genes Genet. Syst. 84, 111–120 (2009). [DOI] [PubMed] [Google Scholar]
- Li X. et al. LcSAIN1, a novel salt-induced gene from sheepgrass, confers salt stress tolerance in transgenic Arabidopsis and rice. Plant Cell Physiol. 54, 1172–1185 (2013). [DOI] [PubMed] [Google Scholar]
- Uversky V. N. & Dunker A. K. Understanding protein non-folding. Biochim. Biophys. Acta 1804, 1231–1264 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanin M. et al. Plant dehydrins and stress tolerance. Plant Signal. Behav. 6, 1503–1509 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels D. & Sunkar R. Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58 (2005). [Google Scholar]
- Joshi M., Mishra A. & Jha B. NaCl plays a key role for in vitro micropropagation of Salicornia brachiata, an extreme halophyte. Ind. Crop. Prod. 35, 313–316 (2012). [Google Scholar]
- Huda K. M. et al. OsACA6, a P‐type IIB Ca2+ ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing the expression of stress‐responsive genes. Plant J. 76, 997–1015 (2013). [DOI] [PubMed] [Google Scholar]
- Campo S. et al. Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidokoro S. et al. Soybean DREB1/CBF‐type transcription factors function in heat and drought as well as cold stress‐responsive gene expression. Plant J. 81, 505–518 (2015). [DOI] [PubMed] [Google Scholar]
- Sato H. et al. The Arabidopsis transcriptional regulator DPB3‐1 enhances heat stress tolerance without growth retardation in rice. Plant Biotechnol. J. 10.1111/pbi.12535 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artimo P. et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 40, W597–W603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analyses of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C (T) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Horsch R. B. et al. A simple and general method for transferring genes into plants. Science 227, 1229–1231 (1985). [DOI] [PubMed] [Google Scholar]
- Jefferson R. A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405 (1987). [Google Scholar]
- Pandey S., Patel M. K., Mishra A. & Jha B. Physio-biochemical composition and untargeted metabolomics of cumin (Cuminum cyminum L.) make it promising functional food and help in mitigating salinity stress. PLoS One 10, e0144469 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutts S., Kinet J. M. & Bouharmont J. NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann. Bot. 78, 389–398 (1996). [Google Scholar]
- Sairam R. K. Effect of homobrassinolide application on metabolism and grain yield under irrigated and moisture stress conditions of two wheat varieties. J. Plant Growth Regul. 14, 173–181 (1994). [Google Scholar]
- Barrs H. D. & Weatherle P. E. A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust. J. Biol. Sci. 15, 413–428 (1962). [Google Scholar]
- Bates L. S., Waldern R. & Teare I. D. Rapid determination of free proline for water stress studies. Plant Soil 39, 205–207 (1973). [Google Scholar]
- Irigoyen J. J., Einerich D. W. & Sánchez‐Díaz M. Water stress induced changes in concentrations of proline and total soluble sugars in nodulated alfalfa (Medicago sativd) plants. Physiol. Plantarum 84, 55–60 (1992). [Google Scholar]
- Hodgson R. A. & Raison J. K. Lipid peroxidation and superoxide dismutase activity in relation to photoinhibition induced by chilling in moderate light. Planta 185, 215–219 (1991). [DOI] [PubMed] [Google Scholar]
- Mukherjee S. P. & Choudhuri M. A. Implications of water stress-induced changes in the leaves of indigenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol. Plantarum 58, 166–170 (1983). [Google Scholar]
- Shi J. et al. Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol. 30, 914–922 (2010). [DOI] [PubMed] [Google Scholar]
- Arnon D. I. Copper enzymes in isolated chloroplasts polyphenol oxidase in Beta vulgaris. Plant Physiol. 24, 1–15 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S., Patel M. K., Mishra A. & Jha B. In planta transformed cumin (Cuminum cyminum L.) plants, overexpressing the SbNHX1 gene showed enhanced salt endurance. PloS one. 11, e0159349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.