Abstract

Many oncogenic mutants of the tumor suppressor p53 are conformationally unstable, including the frequently occurring Y220C mutant. We have previously developed several small-molecule stabilizers of this mutant. One of these molecules, PhiKan083, 1-(9-ethyl-9H-carbazole-3-yl)-N-methylmethanamine, binds to a mutation-induced surface crevice with a KD = 150 μM, thereby increasing the melting temperature of the protein and slowing its rate of aggregation. Incorporation of fluorine atoms into small molecule ligands can substantially improve binding affinity to their protein targets. We have, therefore, harnessed fluorine–protein interactions to improve the affinity of this ligand. Step-wise introduction of fluorines at the carbazole ethyl anchor, which is deeply buried within the binding site in the Y220C–PhiKan083 complex, led to a 5-fold increase in affinity for a 2,2,2-trifluoroethyl anchor (ligand efficiency of 0.3 kcal mol–1 atom–1). High-resolution crystal structures of the Y220C–ligand complexes combined with quantum chemical calculations revealed favorable interactions of the fluorines with protein backbone carbonyl groups (Leu145 and Trp146) and the sulfur of Cys220 at the mutation site. Affinity gains were, however, only achieved upon trifluorination, despite favorable interactions of the mono- and difluorinated anchors with the binding pocket, indicating a trade-off between energetically favorable protein–fluorine interactions and increased desolvation penalties. Taken together, the optimized carbazole scaffold provides a promising starting point for the development of high-affinity ligands to reactivate the tumor suppressor function of the p53 mutant Y220C in cancer cells.
The introduction of fluorine atoms into organic small molecules has become widespread in drug discovery.1−5 Indeed, between 20 and 25% of drugs on the market are estimated to contain at least one fluorine atom.6 Apart from modulating various important compound properties such as logP, metabolic stability, basicity, and bioavailability, fluorine substituents have also been successfully used for improving ligand binding affinities.7−9 This can be achieved via intramolecular stabilization of favorable ligand binding conformations, modulation of polarity or basicity of neighboring functional groups of the ligand, or direct fluorine–protein interactions.1,8 These interactions can be further classified into polar interactions with hydrogen bond donors (e.g., backbone NH, polarized Cα–H, polar side chains, and protein bound water), as observed in the binding of type II statins to the HMGCoA enzyme,10 hydrophobic interactions with lipophilic side chains, and orthogonal multipolar interactions, which can be also described as n → π* interactions,11 with backbone carbonyl groups, amide containing side chains (Asn and Gln), and guanidinium groups (Arg).
A number of case studies on the use of fluorine interactions in rational drug design have been reported. Olsen et al. conducted a systematic fluorine scan with a thrombin inhibitor, revealing favorable interactions with backbone amides and Cα protons that yielded significantly improved binding affinity.12 The introduction of fluorine substituents also significantly increased the potency of kinesin spindle protein inhibitors, abl kinase inhibitors, and peptidic elastase inhibitors.13−16 Vulpetti et al. identified fluorinated fragments binding to trypsin by 19F NMR screening and crystal structure determination and described a general approach to identify fluorophilic hot-spots in proteins using crystal and computational analysis.17 Recently, Pollock et al. investigated the impact of fluorine–protein interactions on the binding affinity of a menin–MLL inhibitor and introduced their computational algorithm FMAP, which aims to facilitate the rational design of fluorine–protein interactions.18
Here, we have harnessed fluorine interactions for the development of mutant p53 rescue drugs. The tumor suppressor p53 plays a key role in regulating cell-cycle arrest, DNA repair, apoptosis, or cellular senescence.19−21 In virtually all human cancers, p53 is inactivated either by mutation or overexpression of negative regulators such as MDM2 or MDMX, which leads to proteasomal degradation of p53.22 The cancer mutation Y220C, which accounts for an estimated 100 000 new cancer cases per year worldwide, significantly destabilizes the p53 DNA-binding domain (DBD) and impairs its function via increased thermal denaturation.21,23 We have previously developed small-molecule stabilizers of p53-Y220C, such as Phikan083, PhiKan5196, and PhiKan7088 (Figure 1), which bind to a mutation-induced surface crevice on the DBD, thereby stabilizing the protein, slowing its unfolding and aggregation, and in some cases restoring tumor suppressor activity in cancer cells harboring the p53-Y220C mutation.24−28 In this study, we aimed at improving the potency of the carbazole-based compound Phikan083 and employed ab initio quantum-chemical calculations to probe potential interaction energy gains upon fluorination of the ethyl anchor. We have synthesized mono-, di-, and tri- fluorinated 9H-fluoroethyl carbazoles; evaluated their binding affinities via differential scanning fluorimetry (DSF) and isothermal titration calorimetry (ITC); and determined their binding mode by X-ray crystallography. We found that trifluorination significantly improved the binding affinity by approximately 5-fold compared with PhiKan083 (1), whereas both monofluoro and difluoro analogues were less potent than the parent compound.
Figure 1.
Chemical structures of the known small-molecule stabilizers of p53-Y220C PhiKan083, PhiKan5196, and PhiKan7088.
Results and Discussion
Quantum Chemical Calculations
In the crystal structure of p53-Y220C in complex with PhiKan083 (PDB: 2VUK), the ethyl moiety of PhiKan083 is in close proximity to the carbonyl groups of Leu145 and Trp146, and the thiol group of Cys220. Given the frequent and well-characterized interactions between organofluorine groups and protein backbone amides, as well as the less frequently observed interactions between fluorine and sulfur atoms,9 we investigated whether gains in binding affinity could be achieved via fluorinated ethyl substituents using DFT-D calculations at the BLYP-D3/def2-SVP level with a truncated model of PhiKan083 bound to the p53-Y220C binding pocket (Figure 2B). Except for the sulfur atom of Cys220, all heavy atoms of the Y220C binding pocket as well as the nitrogen and C-3 atom of the pyrrole ligand model were kept frozen during the calculations.
Figure 2.
Binding mode of the p53-Y220C stabilizer PhiKan083 and fluorinated model systems. (A) Experimentally determined binding mode of PhiKan083 (orange sticks) to the mutation-induced surface crevice of the p53 mutant Y220C (PDB code: 2VUK). (B) Snapshots of DFT-D optimized models of the PhiKan083 N-ethyl group and its fluorinated derivatives (orange sticks) bound to the Y220C surface crevice. For the DFT-D optimizations, truncated models of PhiKan083 (N-ethylpyrrole) and the p53-Y220C pocket (as depicted) were used (only non-hydrogen atoms and polar protons are shown). Interaction energies of each ligand model were compared to the N-ethyl reference interaction energy ΔE to calculate relative interaction energies (ΔΔE = ΔELigand – ΔEN-ethyl). The three distinct orientations of local minima of the 2-fluoroethyl anchor showed different interactions energies, indicating that orientation of the C–F vector toward the backbone carbonyl groups of Leu145 and Trp146 yields the most favorable interaction energy. Pictures were rendered using pymol (www.pymol.org).
Distances between the backbone amides of Leu145 and Trp146 and the nearest C–F groups were between 3.0 and 3.3 Å in the optimized structures, which is in good agreement with typical CF···C=O distances (3.0–3.7 Å) for orthogonal multipolar interactions.16 C–F···S and C–F···HS distances ranged from 2.8 to 3.6 Å, which is also in good agreement with experimentally observed distances for fluorine–sulfur contacts (2.8–3.4 Å) in protein structures.9 Our DFT-D3 calculations indicated that the relative interaction energies (ΔΔE) for all fluorinated ethyl groups improved by at least −2 kcal/mol compared with the N-ethyl reference energy ΔE (ΔE = Ecomplex – (Ereceptor + Eligand); Figure 2B). The energetically most favorable conformation (ΔΔE = −6.5 kcal/mol) of the 2-fluoroethyl group was orientation 1 (Figure 2B), in which the C–F vector points toward the backbone amides of Leu145 and Trp146, predicting two potential orthogonal multipolar interactions between the fluorine atom and both carbonyl groups. Orientations 2 and 3 of the 2-fluoroethyl group, where the fluorines were oriented toward the sulfhydryl group of Cys220, were energetically less favorable, with respective ΔΔE values of −2.2 kcal/mol and −3.7 kcal/mol. The relative interaction energy of conformation 1 of the difluoro ethyl moiety (ΔΔE = −6.9 kcal/mol) was similar to the most favored 2-fluoroethyl conformation (see Figure S1 for difluoro ethyl conformations 2 and 3 and their DFT-D energies), whereas the trifluoro-substituted ethyl anchor was energetically less favorable with a ΔΔE value of −4.2 kcal/mol (Figure 2B). However, the calculated DFT-D3 energies only yield an estimate of the ligand–protein interaction at the chosen computational level in a model system of small size and neglect other contributions to the free energy of binding such as entropic changes and desolvation penalties.
Compound Synthesis
We devised PhiKan083 (1) analogues 2–4 (Figure 3) with a mono-, di-, or trifluoroethyl anchor, in addition to their counterparts 5 and 6 bearing a dimethylamine instead of a monomethylamine group. We envisaged that, having different substitution patterns (e.g., secondary vs tertiary amine) at the level of the pendant, the solvent exposed amino group would provide additional structure–activity relationships and inform on the influence of the amino side chain on the overall potency and binding mode of 1 and fluorinated analogues.
Figure 3.
Structures of compounds 1–6.
Compounds 1 and 2 were obtained from commercial sources (see Materials and Methods). The synthesis of 3, 4, 5, and 6 was straightforward, and is described in Scheme 1. Attempts at alkylation of intermediate 9 were unsuccessful. Alkylation of 9H-carbazole-3-carbaldehyde (9)29 with the appropriate alkyl halide or tosylate was achieved in the presence of Cs2CO3 in DMF, and provided intermediates 7 and 8. Low yields were due to a competing fluorine elimination pathway which has been previously reported by Suehiro et al.(30) Further introduction of the amine side chain by reductive amination afforded 3, 4, 5, and 6. Two reductive amination procedures were attempted, titanium(IV) isopropoxide-sodium borohydride and sodium triacetoxyborohydride, and yields in both cases were comparable.31,32 Indeed, the titanium-based method was used for the reductive amination of 7 and 8, but we anticipate that sodium triacetoxyborohydride would be equally effective.
Scheme 1. Synthesis and Overall Yield of Compounds 3, 4, 5 and 6 Using the Two-Step Synthesis Described.
Not applicable, precursor was bought from TCI UK.
Biophysical Evaluation
We tested PhiKan083 (1) and analogues 2–6 for their stabilization of the p53-Y220C DNA-binding domain (DBD) using differential scanning fluorimetry (DSF; Table 1). The N-2,2,2-trifluoroethyl substituted carbazoles 4 and 6 increased stability of the p53-Y220C DBD by 1.2 K at a compound concentration of 125 μM, showing a clear improvement over their N-ethyl substituted counterparts PhiKan083 (1) and 5 (ΔTm = 0.8 K and ΔTm = 0.6 K, respectively). In contrast, the difluorinated (2) and monofluorinated (3) carbazoles were significantly worse than the parent compound in terms of protein stabilization, with respective thermal shifts of 0.3 and 0.4 K. Dissociation constants (KD) were determined by isothermal titration calorimetry (ITC; Table 1). The substitution pattern of the solvent-exposed amine had a minor effect on affinity, with the additional methyl group in 5 resulting in a 30% drop in affinity. The trifluoro-substituted carbazoles 4 and 6 were the most potent compounds and showed ITC KD values of 28 and 37 μM, respectively (Figure 4), which corresponds to a 5-fold increase in affinity compared to the nonfluorinated parent compounds (1 and 5). Despite prediction of improved interaction energies by DFT-D calculations, the mono- and difluorinated analogues, 2 and 3, failed to display improved affinities. They had KD values of 101 μM and 138 μM, respectively (Figure S2).
Table 1. Thermal Shift Data and KD Values.
| compound | DSF ΔTm at 125 μM | ITC KD (μM) |
|---|---|---|
| 1 | 0.8 | 124a |
| 2 | 0.4 | 138 |
| 3 | 0.3 | 101 |
| 4 | 1.2 | 28 |
| 5 | 0.6 | 169 |
| 6 | 1.2 | 37 |
Data taken from Boeckler et al.(24)
Figure 4.
Binding of compounds 4 (A) and 6 (B) to p53-Y220C as characterized by isothermal titration calorimetry (ITC).
Crystal Structures
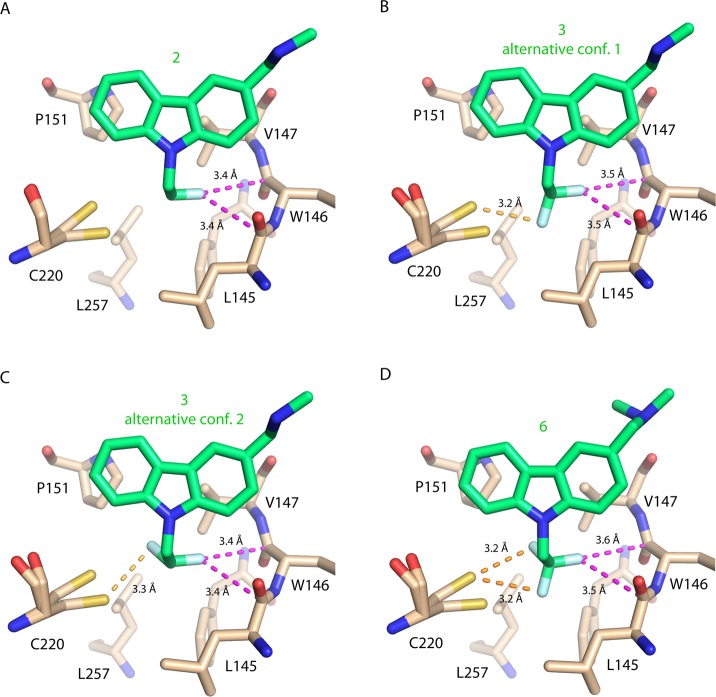
We determined high-resolution crystal structures of the p53-Y220C DBD in complex with 2, 3, and 6 (Table 2) in order to establish the binding modes of mono-, di-, and trifluorinated Phikan083 derivatives. All structures were determined in the same space group as for the parent compound PhiKan083 and contained two molecules in the asymmetric unit (chains A and B). There was excellent electron density for the ligand in chain B in all cases. For the ligand in chain A, there was only partial occupancy for compounds 2 and 3, consistent with the Y220C-PhiKan083 complex structure.24 Numbers given in the following therefore refer to chain B, unless stated otherwise. The binding mode of the carbazoles was almost identical to that of PhiKan083 (1). Small differences in the orientation of the central carbazole scaffold can be attributed to the different methylation state of the amine moiety by comparison with other structures of Y220C ligand complexes (Joerger AC unpublished data).
Table 2. X-Ray Data Collection and Refinement Statistics of Y220C-Carbazole Complexes.
| compound | 2 | 3 | 6 |
|---|---|---|---|
| data collection | |||
| space group | P212121 | P212121 | P212121 |
| a (Å) | 65.03 | 64.05 | 65.16 |
| b (Å) | 71.14 | 71.14 | 71.45 |
| c (Å) | 105.14 | 104.17 | 105.23 |
| molecules/AU | 2 | 2 | 2 |
| resolution (Å)a | 29.6–1.38 | 29.6–1.35 | 29.6–1.48 |
| (1.45–1.38) | (1.42–1.35) | (1.56–1.48) | |
| unique reflections | 99,536 | 107,270 | 82,444 |
| completeness (%)a | 98.9 (98.6) | 99.7 (99.9) | 99.9 (100.0) |
| multiplicitya | 4.8 (4.8) | 4.9 (4.9) | 5.4 (5.5) |
| Rmerge (%)a | 3.9 (50.6) | 4.5 (54.9) | 5.3 (48.7) |
| mean I/σ(I)a | 19.2 (3.4) | 16.2 (3.5) | 16.5 (3.7) |
| Wilson B value (Å2) | 13.4 | 11.8 | 13.0 |
| refinement | |||
| Rwork (%)b | 14.6 | 14.7 | 14.4 |
| Rfree (%)b | 17.0 | 16.9 | 17.1 |
| no. of atoms | |||
| proteinc | 3187 | 3166 | 3186 |
| zinc | 2 | 2 | 2 |
| water | 460 | 430 | 466 |
| ligands | 48 | 50 | 44 |
| RMSD bonds (Å) | 0.005 | 0.005 | 0.005 |
| RMSD angles (deg) | 0.08 | 0.8 | 0.8 |
| mean B (Å2) | 23.4 | 21.2 | 21.3 |
| PDB ID | 5G4M | 5G4N | 5G4O |
Values in parentheses are for the highest resolution shell.
Rwork and Rfree = ∑∥Fobs| – |Fcalc∥/∑|Fobs|, where Rfree was calculated with 5% of the reflections chosen at random and not used in the refinement.
Number includes alternative conformations.
Upon monofluorination, the fluorine atom of 2 points toward the carbonyl groups of Leu145 and Trp146 with a F···C=O interaction distance of 3.4 Å (Figure 5A). The C–F vector and the planes of the backbone carbonyl group of Leu145 and Trp146 show a nearly orthogonal arrangement, with respective O=C···F angles of 97.5° and 80.1°, which is in good agreement with O=C···F angles found in the PDB and CSD for orthogonal multipolar interactions.9,16 This preferred orientation of the C–F vector in the crystal structure is also in good agreement with our initial DFT-D predictions (Figure 5A). Similar to the Y220C-Phikan083 complex, the side chain of Cys220, in immediate vicinity of the ethyl anchor, adopts two alternative, albeit very similar conformations.
Figure 5.
X-ray structures of p53-Y220C with bound fluoro-derivatives of PhiKan083. Multipolar fluorine interactions are highlighted with magenta broken lines and fluorine–sulfur contacts with yellow broken lines. (A) Y220C in complex with monofluorinated compound 2. (B,C) Alternative conformations of the difluorinated compound 3 in chain B of the Y220C mutant. In chain A with only partial occupancy of 3, only one of the two side-chain conformations of Cys220 was observed, with a preferential orientation of 3 as highlighted in panel B. (D) Y220C in complex with 6 (chain B). Interactions with the main conformation of Cys220 are highlighted. The minor conformation of Cys220 is observed in chain B only.
For the N-2-difluoroethyl anchor of 3, we observed two alternative conformations in the p53-Y220C binding pocket (Figure 5B and C). In both conformations, one fluorine atom interacts with the carbonyl groups of Leu145 and Trp146 in an almost identical fashion as observed for compound 2. The second fluorine interacts with the thiol group of Cys220, pointing either toward Pro151 (Figure 5C) or toward Leu145 at the bottom of the binding pocket (Figure 5B), which is essentially the result of a 120° rotation around the ethyl anchor C–C bond. Interestingly, only one of the two Cys conformations was observed in chain A, coinciding with a preferential orientation of the difluoroethyl anchor in the orientation highlighted in Figure 5B, whereby the fluorine forms a weak hydrogen bond with the SH group of Cys220. In both orientations, the fluorine atom interacts with the thiol group at a distance of about 3.2 Å, which is more or less the sum of the van der Waals radii of sulfur and fluorine (rF = 1.47 Å; rS = 1.80 Å).1,33
In the structure of the most potent compound 6, the CF3 group aligns well with the different fluorine positions observed for monofluorinated 2 and difluorinated 3 (Figure 5D); i.e., it interacts with the backbone carbonyl groups of Leu145 and Trp146 as well as with the thiol group of Cys220. Analysis of C–S···F angles in the complexes with di- and trifluorinated compounds suggests that fluorine interacts with Cys220 via weak hydrogen bonding with the polarized proton of the thiol function and via sulfur σ-hole bonding34−36 at an angle close to 180°.
Discussion
Incorporation of fluorine atoms into the N-ethyl anchor of the p53-Y220C stabilizer PhiKan083 (1) yielded two compounds, 4 and 6, with substantially increased p53-Y220C stabilization (1.2 K at 125 μM) and Y220C binding affinity. Compared with the parent compounds PhiKan083 (1) and 5, the ITC KD values improved by a factor of 5 and the free energy of binding by approximately −0.9 kcal/mol (at 293 K). Typically, C–F···C=O orthogonal multipolar interactions with optimal geometry raise the binding free enthalpy by about −0.2 to −0.3 kcal/mol,37 which suggests that the observed C–F···C=O interactions with the carbonyl groups of Leu145 and Trp146 are the main factor for the potency increase of the trifluorinated PhiKan083 derivatives. The DFT-D calculations are also consistent with the binding modes 2 and 3 observed in our crystal structures, as the preferred orientation of fluorine toward the backbone carbonyl groups of Leu145 and Trp146 in these structures indicates that interactions of the protein carbonyl groups with fluorine atoms contribute more strongly to the affinity increase than interactions with the thiol group of Cys220 or apolar protons of hydrophobic side chains. The preferred interaction with the protein backbone then seems to direct the orientation of additional fluorine atoms in the binding pocket, interacting with the thiol group of Cys220 via either a hydrogen bond or a planar sulfur σ-hole interaction.
Although DFT-D calculations predicted improved interaction energies for monofluorinated and difluorinated PhiKan083 derivatives, this was not reflected in the experimentally determined dissociation constants and stability data for compounds 2 and 3, which were close to that of the parent compound PhiKan083. These discrepancies between theoretical and experimental data are likely due to different desolvation penalties associated with each fluorinated group. Fluorine can act as a weak hydrogen-bond acceptor in a 2,2-difluoromethyl group and even more strongly in a 2-fluoromethyl group, although hydrogen-bond strength was found to be significantly weaker than for the conventional hydrogen-bond acceptor acetophenone.38 For example, difluoroalkyl groups have been used as thiol surrogates in drug discovery because of their similar steric properties and the acidity of the terminal hydrogen resulting from the high polarization of the C–H bond by the geminal fluorine atoms.39 To estimate desolvation penalties of fluorinated N-ethyl anchors, we calculated solvation energies for N-ethylcarbazole, N-2-fluoroethylcarbazole, N-2,2-difluoroethylcarbazole, and N-2,2,2-trifluoroethylcarbazole with Jaguar using the SM8 water model at the BLYP-D3/6-31G** level of theory (Table 3 and Figure S3).40 Both mono- and difluorinated carbazoles showed solvation energies that were by −1.0 and −2.8 kcal/mol larger than the solvation energy of N-ethylcarbazole. The solvation energy of the trifluorinated carbazole was identical to N-ethylcarbazole (−3.1 kcal/mol). Hence, the higher polarity of the 2-fluoroethyl and 2,2-difluoroethyl moieties in the PhiKan083 derivatives 2 and 3 and the associated desolvation penalties are most likely the key factors counteracting and quenching favorable fluorine–protein interactions in these compounds.
Table 3. Calculated Solvation Energies for DFT-D3 Optimized Carbazoles.
| carbazole anchora | solvation energy (kcal/mol)b |
|---|---|
| N-ethyl | –3.1 |
| N-2-fluoroethyl | –5.9 |
| N-2,2-difluoroethyl | –4.1 |
| N-2,2,2-trifluoroethyl | –3.1 |
Substitution of 9H-carbazole. Structures of DFT-D3 optimized compounds are shown in Figure S3.
Solvation energies were calculated from single point calculations of the global minimum conformation of each compound in Jaguar at the BLYP-D3/6-31G** level using the SM8 water model.
In conclusion, targeting Cys220 and the backbone carbonyl groups of Leu145 and Trp146 via fluorine bonding helped to further optimize the carbazole scaffold. N-3,3,3-Trifluoroethyl substituted carbazoles 4 and 6 exhibit a high ligand efficiency (LE = 0.3 kcal mol–1 atom–1), although binding is still relatively weak for these fragment-like molecules. Nevertheless, they represent a promising starting point for the development of high affinity small-molecule stabilizers of the oncogenic Y220C mutant.
Materials and Methods
Protein Expression and Purification
Stabilized p53-Y220C DBD (residues 94–312) was expressed and purified as previously described.26
Differential Scanning Fluorimetry (DSF)
DSF was performed as described.26 Briefly, melting temperatures (Tm) were determined using 8 μM protein (stabilized p53-Y220C DBD) and 10 × SYPRO orange (Life Technologies) in a 25 mM KPi (pH 7.2), 150 mM NaCl, and 1 mM TCEP assay buffer at a final DMSO concentration of 5% (v/v). ΔTm values were calculated as ΔTm= Tm (protein + compound) – Tm (protein)). All samples were measured in triplicate.
Isothermal Titration Calorimetry (ITC)
ITC experiments were conducted as described.26 Samples for the cell unit contained 50 μM protein in a 25 mM KPi (pH 7.2), 150 mM NaCl, 1 mM TCEP, and 5% (v/v) DMSO assay buffer. The syringe contained 2–5 mM compound in the same assay buffer.
Quantum Chemical Calculations
QM calculations were conducted with TURBOMOLE 6.4, applying a BLYP(RI)-D3-def2-SVP (DFT-D3) level of theory. A truncated model of the p53 Y220C binding pocket with an N-ethyl-pyrrole ligand (truncated PhiKan083 model), comprising 140 atoms, was derived from the crystal structure (PDB code: 2VUK). For geometry optimization, all heavy atoms of the binding pocket, except the sulfur atom of Cys220, and the nitrogen atom plus one carbon atom of the ligand pyrrole ring were frozen (see Supporting Information). Interaction energies were calculated as ΔE = Ecomplex – (Ereceptor + Eligand). Global minimum conformers, which were used as input structures for solvation energy calculations, were determined by DFT-D3 calculations at the BLYP(RI)-D3-def2-TZVP level of theory with TURBOMOLE 6.4. For N-2-fluoroethylcarbazole and N-2,2-difluoroethylcarbazole, we rotated the C–C axis of the ethyl moiety stepwise by 60° to obtain six different conformers that were used as input structures for the DFT-D3 calculations. Solvation energies were calculated with Jaguar 8.9 (Schrödinger, USA, NY) as single point calculations using the SM8 water model at the BLYP-D3/6-31G** level of theory.
X-Ray Crystallography
Crystals of the p53-Y220C DBD were grown at 18 °C as described.41 Crystals were soaked for either 90 min (6) or 4 h (2 and 3) in a 20 mM solution of compound in cyro buffer (19% polyethylene glycol 4000, 20% glycerol, 10 mM sodium phosphate, pH 7.2, 100 mM Hepes, pH 7.2, 150 mM NaCl) and flash frozen in liquid nitrogen. X-ray data sets were collected at 100 K at beamlines I03 and I04 of the Diamond Light Source, Oxford, UK. Indexing of the data sets was performed using XDS42 and scaling using the program SCALA43 within the CCP4 software suite.44 The structures of the Y220C-carbazole complexes were then solved using the program PHENIX:45 initial rigid body refinement was done using PDB entry 2J1X as a starting model. The models were further refined using iterative cycles of manual model building in COOT46 and refinement with PHENIX. While there was clear electron density for the ligands in chain B, there was only weak ligand density in chain A of the Y220C complex with 2 and 3, indicating partial occupancy. The same observation had been made for the parent compound PhiKan083.24 Accordingly, alternative states of the binding pocket were refined: bound ligand and water network of the ligand-free structure. Data collection and refinement statistics are given in Table 2. Structural figures were prepared using PyMOL (www.pymol.org). The coordinates and structure factors of the complexes are deposited in the Protein Data Bank (PDB ID: 5G4M, 5G4N, 5G4O).
General Procedures
All reactions were carried out in air unless otherwise stated, using commercial grade starting materials, solvents, and reagents. The progress of all reactions was monitored by thin layer chromatography (TLC) using commercially available glass silica gel plates (60 Å, F254). The mobile phase was generally a solvent mixture, and the visualization was undertaken using UV light. Microwave reactions were conducted in a CEM Discovery microwave reactor. All NMR spectra were measured on a Varian NMR 500 spectrometer at either 500 MHz (1H) or 126 MHz (13C). Chemical shifts are quoted in parts per million (ppm; δ relative to a residual solvent peak for 1H and 13C). Chromatographic purifications were undertaken using an ISCO purification unit, Combi Flash RF 75 PSI, using Biotage silica gel columns. LC-MS purity analyses were undertaken using a 5 μm C18 110 Å column. Percentage purities were performed using a 30 min method in water/acetonitrile with 0.1% formic acid (5 min at 5%, 5–95% over 20 min, 5 min at 95%) with the UV set to 254 nm. All high-resolution mass spectrometry was carried out at the EPSRC UK National Mass Spectrometry Facility (NMSF), Swansea University, using a Thermo Scientific LTQ Orbitrap XL spectrometer.
Materials
1-(9-(2-Fluoroethyl)-9H-carbazol-3-yl)-N-methylmethanamine (2) was purchased from Enamine; its purity was assessed by LC-MS and found to be higher than 95% (LC-MS purity > 95% (UV), ret. time = 10.83 min). 9-Ethyl-9H-carbazole-3-carbaldehyde was purchased from TCI UK (>98% purity by manufacturer). 1-Iodo-2,2-difluoroethane and 2,2,2-trifluoroethyl p-toluenesulfonate were purchased from fluorochem. 9H-Carbazole-3-carbaldehyde was synthesized according to a known procedure.29 Final compounds that were tested had an LC purity of >95%.
Representative Procedure for the Synthesis of Intermediates 7 and 8
9-(2,2-Difluoroethyl)9H-carbazole-3-carbaldehyde (7)
To a suspension of 9H-carbazole-3-carbaldehyde (196 mg, 1 mmol) and Cs2CO3 (652 mg, 2 mmol) in anhydrous DMF (5 mL) was added 1-iodo-2,2-difluoroethane (172 mg, 2 mmol). The resulting suspension was stirred under microwave irradiation at 150 °C for 30 min. The suspension was cooled and diluted with H2O (20 mL). Crude product was extracted with ethyl acetate (EtOAc; 3 × 20 mL). The organic extracts were washed with brine (3 × 20 mL) and dried over anhydrous MgSO4. The suspension was filtered and was concentrated to dryness in vacuo. Crude product was purified by chromatography on silica gel using dichloromethane (DCM) as an eluent to yield product as a pale yellow solid, yield 44% (115 mg, 0.44 mmol). 1H NMR (500 MHz, DMSO-d6): δ 10.06 (s, 1H), 8.73 (s, 1H), 8.33–8.17 (m, 1H), 7.99 (d, J = 8.4 Hz, 1H), 7.81 (d, J = 8.4 Hz, 1H), 7.76–7.66 (m, 1H), 7.60–7.47 (m, 1H), 7.36–7.24 (m, 1H), 6.50 (tt, 2JFH = 54.5 Hz, 3JHH = 3.2 Hz, 1H), 4.97 (td, 3JFH = 16.1 Hz, 3JHH = 3.2 Hz, 2H). 13C NMR (126 MHz, DMSO-d6): δ 192.3, 144.6, 141.7, 129.4, 127.3, 127.2, 124.0, 123.0, 122.8, 121.1, 121.0, 115.1 (t, 1JFC = 241.9 Hz), 110.9, 110.8, 45.0 (t, 2JFC = 24.5 Hz). HRMS-ESI (m/z) found: 260.0884. Calcd: 260.0881 for [C15H11F2NO+H]+.
9-(2,2,2-Trifluoroethyl)-9H-carbazole-3-carbaldehyde (8)
Using 2,2,2-trifluoroethyl p-toluenesulfonate as an electrophile, the reaction was carried out on a 0.48 mmol scale. Pale yellow solid, yield 50% (68 mg, 0.24 mmol). 1H NMR (500 MHz, DMSO-d6): δ 10.07 (s, 1H), 8.75 (s, 1H), 8.28 (d, J = 8.3 Hz, 1H), 8.02 (d, J = 8.5 Hz, 1H), 7.90 (d, J = 8.5 Hz, 1H), 7.80 (d, J = 8.3 Hz, 1H), 7.61–7.49 (m, 1H), 7.40–7.25 (m, 1H), 5.51 (q, 3JFH = 9.3 Hz, 2H).13C NMR (126 MHz, DMSO-d6): δ 192.3, 144.2, 141.3, 129.8, 127.5, 127.4, 125.4 (q, 1JFC = 281.8 Hz), 124.0 123.3, 122.9, 121.5, 121.2, 110.9, 110.8, 44.1 (q, 2JFC = 33.5 Hz). HRMS-ESI (m/z) found: 278.0789. Calcd: 278.0787 for [C15H10F3NO+H]+.
Representative Procedure for the Synthesis of Compounds 3, 4, 5, and 6
1-(9-(2,2-Difluoroethyl)-9H-carbazol-3-yl)-N-methylmethanamine (3)
To a solution of 7 (75 mg, 0.29 mmol) in anhydrous EtOH/DCM (10 mL) was added methylamine hydrochloride (39 mg, 0.58 mmol), triethylamine (61 μL, 0.44 mmol), and titanium(IV) isopropoxide (172 μL, 0.58 mmol). The resulting solution was stirred at RT for 18 h before the addition of sodium borohydride (22 mg, 0.58 mmol). The solution was stirred at RT for 8 h before pouring into 2 M aqueous ammonia (25 mL). The suspension was filtered through Celite, and to the filtrate was added H2O. Crude product was extracted with DCM, dried over anhydrous K2CO3. The suspension was filtered, and the filtrate was concentrated in vacuo to yield crude product that was purified by chromatography on silica gel using DCM/MeOH 9:1 as an eluent to yield the product as a pale yellow oil, yield 58% (46 mg, 0.16 mmol). 1H NMR (500 MHz, acetonitrile-d3): δ 8.19–8.13 (m, 1H), 8.10 (s, 1H), 7.62–7.56 (m, 1H), 7.56–7.45 (m, 3H), 7.32–7.25 (m, 1H), 6.31 (tt, 2JFH = 55.0 Hz, 3JHH = 3.2 Hz, 1H), 4.76 (td, 3JFH = 15.6 Hz, 3JHH = 3.2 Hz, 2H), 3.89 (s, 2H), 2.43 (s, 3H) (NH missing due to D-H exchange). 13C NMR (126 MHz, acetonitrile-d3): δ 141.1, 139.9, 132.5, 126.6, 125.9, 122.9, 122.8, 120.1, 119.7, 119.6, 114.8 (t, 1JFC = 241.9 Hz), 109.3, 109.0, 55.7, 44.9 (t, 2JFC = 25.6 Hz), 35.1. LC-MS purity = 96% (UV), ret. time = 11.01 min. HRMS-ESI (m/z) found: 275.1353. Calcd: 275.1354 for [C16H16F2N2+H]+.
1-(9-(2,2,2-Trifluoroethyl)-9H-carbazol-3-yl)-N-methylmethanamine (4)
The reaction was carried out on a 0.31 mmol scale. The product was isolated as a pale yellow solid, yield 40% (37 mg, 0.12 mmol). 1H NMR (500 MHz, acetonitrile-d3): δ 8.20–8.14 (m, 1H), 8.12 (s, 1H), 7.64–7.58 (m, 1H), 7.58–7.47 (m, 3H), 7.36–7.28 (m, 1H), 5.07 (q, 3JFH = 9.1 Hz, 2H), 3.89 (s, 2H), 2.43 (s, 3H) (NH missing due to D-H exchange). 13C NMR (126 MHz, DMSO-d6) 140.9, 140.2, 128.3, 127.7, 126.6, 126.1 (q, 1JFC = 281.2 Hz), 122.9 122.8, 121.3, 120.6, 120.5, 110.4, 110.1, 53.9, 44.0 (q, 2JFC = 33.4 Hz), 34.2. LC-MS purity = 97% (UV), ret. time = 11.55 min. HRMS-ESI (m/z) found: 293.1259. Calculated: 293.1260 for [C16H15F3N2+H]+.
1-(9-Ethyl-9H-carbazol-3-yl)-N,N-dimethylmethanamine (5)
The reaction was carried out on a 0.44 mmol scale. The product was isolated as a white solid, yield 82% (91 mg, 0.36 mmol). 1H NMR (500 MHz, chloroform-d): δ 8.14–8.10 (m, 1H), 8.06 (s, 1H), 7.51–7.34 (m, 4H), 7.26–7.20 (m, 1H), 4.37 (q, J = 7.2 Hz, 2H), 3.64 (s, 2H), 2.33 (s, 5H), 1.45 (t, J = 7.2 Hz, 3H). 13C NMR (126 MHz, chloroform-d): δ 140.28, 139.3, 129.3, 127.1, 125.5, 122.9 (2C – quaternary), 121.0, 120.4, 118.7, 108.4, 108.1, 64.7, 45.3 (2C), 37.5, 13.8. LC-MS purity 97% (UV), ret. time = 11.36 min. HRMS-ESI (m/z) found: 208.1120. Calculated: 208.1121 for [C15H14N]+ (loss of NMe2).
1-(9-(2,2,2-Trifluoroethyl)-9H-carbazol-3-yl)-N,N-dimethylmethanamine (6)
The reaction is carried out on a 0.10 mmol scale. The product was isolated as a white solid, yield 56% (17 mg, 0.05 mmol). 1H NMR (500 MHz, chloroform-d): δ 8.04–8.00 (m, 1H), 7.99 (s, 1H), 7.45–7.39 (m, 2H), 7.37–7.29 (m, 2H), 7.27–7.20 (m, 1H), 4.73 (q, 3JFH = 8.7 Hz, 2H), 3.62 (s, 2H), 2.28 (s, 6H). 13C NMR (126 MHz, chloroform-d): δ 140.9, 140.1, 129.8, 127.8, 126.4, 124.3 (q, 1JFC = 281.5 Hz), 123.6, 123.4, 121.3, 120.6, 120.4, 108.7, 108.5, 45.3 (q, 2JFC = 35.7 Hz), 44.9 (2C). LC-MS purity >99% (UV), ret. time = 11.68 min. HRMS-ESI (m/z) found: 307.1419, Calcd: 307.1417 for [C16H17F3N2+H]+.
1-(9-Carbazol-3-yl)-N,N-dimethylmethanamine (9)
To a solution of 9H-carbazol-3-carbaldehyde (45 mg, 0.26 mmol) in anhydrous THF (5 mL) was added dimethylamine (130 μL, 2.0 M solution in THF, 0.26 mmol) and acetic acid (12 μL, 0.26 mmol). Sodium triacetoxyborohydride (80 mg, 0.39 mmol) was added, and the resulting solution was stirred for 18 h. Solvent was removed under reduced pressure, and to the residue was added DCM and H2O. The solution was filtered through a hydrophobic frit, and the filtrate was concentrated in vacuo to yield crude product, which was purified by chromatography on silica gel (using DCM/MeOH 9/1 as an eluent) to yield product as a pale yellow solid, yield 88% (51 mg, 0.22 mmol). 1H NMR (500 MHz, chloroform-d): δ 8.59 (s, 1H), 8.09–8.03 (m, 1H), 8.02 (s, 1H), 7.46–7.38 (m, 3H), 7.38–7.33 (m, 1H), 7.25–7.18 (m, 1H), 3.77 (s, 2H), 2.40 (s, 6H).13C NMR (126 MHz, chloroform-d): δ 139.9, 139.2, 127.5, 126.8, 125.9, 123.3, 123.0, 121.4, 120.3, 119.4, 110.7, 110.6, 63.9, 44.2 (2C). HRMS-ESI (m/z) found: 180.0805. Calculated: 180.0808 for [C13H10N]+ (loss of NMe2).
Acknowledgments
This work was funded by Worldwide Cancer Research Grant 14-1002 “Rescuing thermally unstable p53 mutants with small molecule stabilisers; new targeted cancer therapies” and ERC Advanced Grant 268506. R.N.J. is funded by the University of Sussex (Ph.D. studentship). We thank the staff at Diamond beamlines I03 and I04 for technical assistance during data collection. Access was supported in part by the EU FP7 infrastructure grant BIOSTRUCT-X (contract no. 283570).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.6b00315.
Additional DFT-D data and coordinates used for calculations; ITC data for 2, 3, and 5; global minimum conformers of fluorinated carbazole analogues and their calculated solvation energies; scans of NMRs; LC purity (PDF)
Author Contributions
# M.R.B and R.N.J. contributed equally to this work
The authors declare no competing financial interest.
Supplementary Material
References
- Muller K.; Faeh C.; Diederich F. (2007) Fluorine in pharmaceuticals: looking beyond intuition. Science 317, 1881–1886. 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- Wang J.; Sanchez-Rosello M.; Acena J. L.; del Pozo C.; Sorochinsky A. E.; Fustero S.; Soloshonok V. A.; Liu H. (2014) Fluorine in pharmaceutical industry: fluorine-containing drugs introduced to the market in the last decade (2001–2011). Chem. Rev. 114, 2432–2506. 10.1021/cr4002879. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Wang J.; Gu Z.; Wang S.; Zhu W.; Acena J. L.; Soloshonok V. A.; Izawa K.; Liu H. (2016) Next Generation of Fluorine-Containing Pharmaceuticals, Compounds Currently in Phase II-III Clinical Trials of Major Pharmaceutical Companies: New Structural Trends and Therapeutic Areas. Chem. Rev. 116, 422–518. 10.1021/acs.chemrev.5b00392. [DOI] [PubMed] [Google Scholar]
- Hagmann W. K. (2008) The many roles for fluorine in medicinal chemistry. J. Med. Chem. 51, 4359–4369. 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]
- Kirk K. L. (2006) Fluorine in medicinal chemistry: Recent therapeutic applications of fluorinated small molecules. J. Fluorine Chem. 127, 1013–1029. 10.1016/j.jfluchem.2006.06.007. [DOI] [Google Scholar]
- Vulpetti A.; Dalvit C. (2012) Fluorine local environment: from screening to drug design. Drug Discovery Today 17, 890–897. 10.1016/j.drudis.2012.03.014. [DOI] [PubMed] [Google Scholar]
- Gerebtzoff G.; Li-Blatter X.; Fischer H.; Frentzel A.; Seelig A. (2004) Halogenation of drugs enhances membrane binding and permeation. ChemBioChem 5, 676–684. 10.1002/cbic.200400017. [DOI] [PubMed] [Google Scholar]
- Bohm H. J.; Banner D.; Bendels S.; Kansy M.; Kuhn B.; Muller K.; Obst-Sander U.; Stahl M. (2004) Fluorine in medicinal chemistry. ChemBioChem 5, 637–643. 10.1002/cbic.200301023. [DOI] [PubMed] [Google Scholar]
- Zhou P.; Zou J.; Tian F.; Shang Z. (2009) Fluorine bonding--how does it work in protein-ligand interactions?. J. Chem. Inf. Model. 49, 2344–2355. 10.1021/ci9002393. [DOI] [PubMed] [Google Scholar]
- Istvan E. S.; Deisenhofer J. (2001) Structural mechanism for statin inhibition of HMG-CoA reductase. Science 292, 1160–1164. 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- Kamer K. J.; Choudhary A.; Raines R. T. (2013) Intimate interactions with carbonyl groups: dipole-dipole or n-- > pi*?. J. Org. Chem. 78, 2099–2103. 10.1021/jo302265k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. A.; Banner D. W.; Seiler P.; Obst Sander U.; D’Arcy A.; Stihle M.; Muller K.; Diederich F. (2003) A fluorine scan of thrombin inhibitors to map the fluorophilicity/fluorophobicity of an enzyme active site: evidence for C-F···C=O interactions. Angew. Chem., Int. Ed. 42, 2507–2511. 10.1002/anie.200351268. [DOI] [PubMed] [Google Scholar]
- Cox C. D.; Breslin M. J.; Mariano B. J.; Coleman P. J.; Buser C. A.; Walsh E. S.; Hamilton K.; Huber H. E.; Kohl N. E.; Torrent M.; Yan Y.; Kuo L. C.; Hartman G. D. (2005) Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP. Bioorg. Med. Chem. Lett. 15, 2041–2045. 10.1016/j.bmcl.2005.02.055. [DOI] [PubMed] [Google Scholar]
- Cowan-Jacob S. W.; Fendrich G.; Floersheimer A.; Furet P.; Liebetanz J.; Rummel G.; Rheinberger P.; Centeleghe M.; Fabbro D.; Manley P. W. (2007) Structural biology contributions to the discovery of drugs to treat chronic myelogenous leukaemia. Acta Crystallogr., Sect. D: Biol. Crystallogr. 63, 80–93. 10.1107/S0907444906047287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. L.; Sieker L. C.; Bieth J.; Dimicoli J. L. (1982) Crystallographic study of the binding of a trifluoroacetyl dipeptide anilide inhibitor with elastase. J. Mol. Biol. 162, 645–658. 10.1016/0022-2836(82)90393-X. [DOI] [PubMed] [Google Scholar]
- Bissantz C.; Kuhn B.; Stahl M. (2010) A medicinal chemist’s guide to molecular interactions. J. Med. Chem. 53, 5061–5084. 10.1021/jm100112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulpetti A.; Schiering N.; Dalvit C. (2010) Combined use of computational chemistry, NMR screening, and X-ray crystallography for identification and characterization of fluorophilic protein environments. Proteins: Struct., Funct., Genet. 78, 3281–3291. 10.1002/prot.22836. [DOI] [PubMed] [Google Scholar]
- Pollock J.; Borkin D.; Lund G.; Purohit T.; Dyguda-Kazimierowicz E.; Grembecka J.; Cierpicki T. (2015) Rational Design of Orthogonal Multipolar Interactions with Fluorine in Protein-Ligand Complexes. J. Med. Chem. 58, 7465–7474. 10.1021/acs.jmedchem.5b00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. P. (1992) Cancer. p53, guardian of the genome. Nature 358, 15–16. 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Vousden K. H.; Prives C. (2009) Blinded by the Light: The Growing Complexity of p53. Cell 137, 413–431. 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Joerger A. C.; Ang H. C.; Fersht A. R. (2006) Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl. Acad. Sci. U. S. A. 103, 15056–15061. 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger A. C.; Fersht A. R. (2010) The tumor suppressor p53: from structures to drug discovery. Cold Spring Harbor Perspect. Biol. 2, a000919. 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger A. C.; Bauer M. R.; Wilcken R.; Baud M. G.; Harbrecht H.; Exner T. E.; Boeckler F. M.; Spencer J.; Fersht A. R. (2015) Exploiting Transient Protein States for the Design of Small-Molecule Stabilizers of Mutant p53. Structure 23, 2246–2255. 10.1016/j.str.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckler F. M.; Joerger A. C.; Jaggi G.; Rutherford T. J.; Veprintsev D. B.; Fersht A. R. (2008) Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl. Acad. Sci. U. S. A. 105, 10360–10365. 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Wilcken R.; Joerger A. C.; Chuckowree I. S.; Amin J.; Spencer J.; Fersht A. R. (2013) Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 41, 6034–6044. 10.1093/nar/gkt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcken R.; Liu X. R.; Zimmermann M. O.; Rutherford T. J.; Fersht A. R.; Joerger A. C.; Boeckler F. M. (2012) Halogen-Enriched Fragment Libraries as Leads for Drug Rescue of Mutant p53. J. Am. Chem. Soc. 134, 6810–6818. 10.1021/ja301056a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcken R.; Wang G.; Boeckler F. M.; Fersht A. R. (2012) Kinetic mechanism of p53 oncogenic mutant aggregation and its inhibition. Proc. Natl. Acad. Sci. U. S. A. 109, 13584–13589. 10.1073/pnas.1211550109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Fersht A. R. (2012) First-order rate-determining aggregation mechanism of p53 and its implications. Proc. Natl. Acad. Sci. U. S. A. 109, 13590–13595. 10.1073/pnas.1211557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Mihalic J.; Fan P.; Liang L.; Lindstrom M.; Wong S.; Ye Q.; Fu Y.; Jaen J.; Chen J. L.; Dai K.; Li L. (2012) Discovery and characterization of a potent and selective antagonist of melanin-concentrating hormone receptor 2. Bioorg. Med. Chem. Lett. 22, 363–366. 10.1016/j.bmcl.2011.10.125. [DOI] [PubMed] [Google Scholar]
- Suehiro M.; Yang G.; Torchon G.; Ackerstaff E.; Humm J.; Koutcher J.; Ouerfelli O. (2011) Radiosynthesis of the tumor hypoxia marker [18F]TFMISO via O-[18F]trifluoroethylation reveals a striking difference between trifluoroethyl tosylate and iodide in regiochemical reactivity toward oxygen nucleophiles. Bioorg. Med. Chem. 19, 2287–2297. 10.1016/j.bmc.2011.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Magid A. F.; Carson K. G.; Harris B. D.; Maryanoff C. A.; Shah R. D. (1996) Reductive Amination of Aldehydes and Ketones with Sodium Triacetoxyborohydride. Studies on Direct and Indirect Reductive Amination Procedures(1). J. Org. Chem. 61, 3849–3862. 10.1021/jo960057x. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S. (1995) Reductive Alkylations of Dimethylamine Using Titanium(Iv) Isopropoxide and Sodium-Borohydride - an Efficient, Safe, and Convenient Method for the Synthesis of N,N-Dimethylated Tertiary-Amines. J. Org. Chem. 60, 4928–4929. 10.1021/jo00120a044. [DOI] [Google Scholar]
- Beno B. R.; Yeung K. S.; Bartberger M. D.; Pennington L. D.; Meanwell N. A. (2015) A Survey of the Role of Noncovalent Sulfur Interactions in Drug Design. J. Med. Chem. 58, 4383–4438. 10.1021/jm501853m. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Gong Z.; Li J.; Lu T. (2015) Intermolecular Sulfur···Oxygen Interactions: Theoretical and Statistical Investigations. J. Chem. Inf. Model. 55, 2138–2153. 10.1021/acs.jcim.5b00177. [DOI] [PubMed] [Google Scholar]
- Murray J. S.; Lane P.; Politzer P. (2008) Simultaneous alpha-Hole and Hydrogen Bonding by Sulfur- and Selenium-Containing Heterocycles. Int. J. Quantum Chem. 108, 2770–2781. 10.1002/qua.21753. [DOI] [Google Scholar]
- Lange A.; Gunther M.; Buttner F. M.; Zimmermann M. O.; Heidrich J.; Hennig S.; Zahn S.; Schall C.; Sievers-Engler A.; Ansideri F.; Koch P.; Laemmerhofer M.; Stehle T.; Laufer S. A.; Boeckler F. M. (2015) Targeting the Gatekeeper MET146 of C-Jun N-Terminal Kinase 3 Induces a Bivalent Halogen/Chalcogen Bond. J. Am. Chem. Soc. 137, 14640–14652. 10.1021/jacs.5b07090. [DOI] [PubMed] [Google Scholar]
- Paulini R.; Muller K.; Diederich F. (2005) Orthogonal multipolar interactions in structural chemistry and biology. Angew. Chem., Int. Ed. 44, 1788–1805. 10.1002/anie.200462213. [DOI] [PubMed] [Google Scholar]
- Dalvit C.; Invernizzi C.; Vulpetti A. (2014) Fluorine as a Hydrogen-Bond Acceptor: Experimental Evidence and Computational Calculations. Chem. - Eur. J. 20, 11058–11068. 10.1002/chem.201402858. [DOI] [PubMed] [Google Scholar]
- Narjes F.; Koehler K. F.; Koch U.; Gerlach B.; Colarusso S.; Steinkuhler C.; Brunetti M.; Altamura S.; De Francesco R.; Matassa V. G. (2002) A designed P1 cysteine mimetic for covalent and non-covalent inhibitors of HCV NS3 protease. Bioorg. Med. Chem. Lett. 12, 701–704. 10.1016/S0960-894X(01)00842-3. [DOI] [PubMed] [Google Scholar]
- Chamberlin A. C.; Cramer C. J.; Truhlar D. G. (2008) Performance of SM8 on a test to predict small-molecule solvation free energies. J. Phys. Chem. B 112, 8651–8655. 10.1021/jp8028038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger A. C.; Ang H. C.; Fersht A. R. (2006) Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl. Acad. Sci. U. S. A. 103, 15056–15061. 10.1073/pnas.0607286103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (2010) Xds. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 62, 72–82. 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.