Abstract
Automated phenotype identification plays a critical role in cohort selection and bioinformatics data mining. Natural Language Processing (NLP)-informed classification techniques can robustly identify phenotypes in unstructured medical notes. In this paper, we systematically assess the effect of naive, lexically normalized, and semantic feature spaces on classifier performance for obesity, atherosclerotic cardiovascular disease (CAD), hyperlipidemia, hypertension, and diabetes. We train support vector machines (SVMs) using individual feature spaces as well as combinations of these feature spaces on two small training corpora (730 and 790 documents) and a combined (1520 documents) training corpus. We assess the importance of feature spaces and training data size on SVM model performance. We show that inclusion of semantically-informed features does not statistically improve performance for these models. The addition of training data has weak effects of mixed statistical significance across disease classes suggesting larger corpora are not necessary to achieve relatively high performance with these models.
Keywords: phenotyping, classification, natural language processing
Graphical abstract
Introduction
With the proliferation of electronic health records (EHR) in recent years, automated phenotyping for cohort selection has become an area of growing interest for the biomedical informatics community (Pathak, Kho, & Denny, 2013). Despite a wide array of focused work, many challenges still persist for delivering practical phenotyping technologies including high-throughput generalized algorithms that are applicable across different diseases without the need for local- or domain-specific rules (Hripcsak & Albers, 2013). Commonly structured EHR-related information such as ICD-9 codes have been shown to be insufficient for producing state-of-the-art performance (Liao et al., 2010) which is why many phenotyping systems employ semantically-informed Natural Language Processing (NLP) methodologies to unlock the unstructured data contained in clinical narratives (Denny, 2012). Currently, 34 out of 47 published phenotyping algorithms on the eMERGE (McCarty et al., 2011) Phenotype KnowledgBase include NLP components (https://phekb.org/ Accessed: February 13th 2015) .
Problem Definition
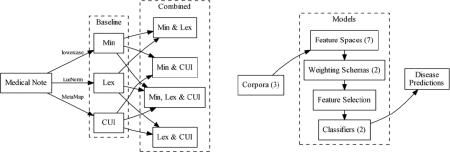
In this paper, we systematically assess the effect of feature spaces, feature weights, and support vector machine (SVM) kernels on model performance on a phenotyping task as the training data is roughly doubled. To do this, we cast the document-level multi-label classification task set out in the 2014 i2b2/UTHealth shared task as a series of five document-level binary classification tasks (one per disease) consistent with the phenotype identification literature. Concretely, given a document from our test set, our goal is to identify the presence (or not) of five different diseases in each patient for each medical record. The five diseases we identify are: obesity, atherosclerotic cardiovascular disease (CAD), hypertension, hyperlipidemia, and diabetes. Using this task as our test bed, we assess the effect of minimally normalized, lexically normalized, and semantically-informed feature spaces on phenotype identification, with various weighting schemes, kernels, and as the training data is doubled. Our primary motivation is not to create the highest performing system possible but to implement reasonable systems and assess the impact of common feature spaces, feature weights, kernels and their combinations on overall system performance. Because of the high cost of annotating data for supervised NLP and machine learning tasks, we also investigate the effect of doubling training data on model performance. To do this, we exploit overlapping annotations from the 2008 i2b2 Obesity Challenge shared task (Uzuner, 2009) and the 2014 i2b2/UTHealth shared task. We evaluate a broad array of models on the 2014 test corpus using the 2008 and 2014 training corpora, and a combined 2008/2014 training corpus.
Related Work
NLP-informed machine learning algorithms have been shown to be successful in identifying patients with rheumatoid arthritis (Carroll, Eyler, & Denny, 2011; Chen et al., 2013), diabetes (Wright, McCoy, Henkin, Kale, & Sittig, 2013), colorectal cancer, and venous thromboembolism (Chen et al., 2013), in risk adjustment for ICU patients (Marafino, Davies, Bardach, Dean, & Dudley, 2014) and for smoking history detection (Khor et al., 2013). In several instances, these methods have been successfully ported across institutions, demonstrating the robustness of the NLP-informed machine learning approach to patient phenotyping (Carroll et al., 2012; Wright et al., 2013). A broad array of tools, techniques and ontologies have been developed for incorporating biomedically-relevant semantic information into machine learning techniques.
One approach for semantically-informed machine learning-based phenotype identification leverages the fixed vocabulary of the Unified Medical Language System's (Bodenreider, 2004; Lindberg, Humphreys, & McCray, 1993) (UMLS) concept unique identifiers (CUIs). The UMLS is a metathesaurus that knits together a wide array of medical vocabularies and provides, among other things, lexical and conceptual crosswalks between constituent terminologies. Many machine-learning approaches to phenotype identification preprocess patient clinical narratives to extract CUIs for use as features. these CUIs are then used, either alone or in concert with other structured EHR information (e.g., ICD-9 codes), for predicting patient membership in a particular phenotype (Bejan, Xia, Vanderwende, Wurfel, & Yetisgen-Yildiz, 2012; Chen et al., 2013; Wei, Tao, Jiang, & Chute, 2010). The process of identifying medical concepts and resolving them to a fixed vocabulary from arbitrary text is not a trivial problem (Bodenreider, 2008). Luckily, several mature tools exist for expediting the process.
MetaMap (formerly MMtx) is a commonly used tool for extracting medical concepts from free-form text and mapping them to the controlled vocabulary of UMLS CUIs (Aronson, 2001). It has a proven track record for high-throughput indexing of medical documents based on semantic content (Aronson & Lang, 2010).
Machine learning techniques to NLP often involve complex pipelines that include normalization, tokenization, sentence breaking, stopping, stemming, word sense disambiguation, part of speech tagging, and information extraction (Manning & Schütze, 1999). Overall system performance can be attributed to many different steps in that pipeline and the effect of a particular implementation choice on system performance is often unclear. From an engineering perspective this can be critically important. Not all steps in the pipeline are equally easy to implement or maintain in a production environment (Sculley et al., 2014). A systematic understanding of the tradeoffs in performance associated with individual implementation decisions can lead to better overall system design and user acceptance (Davis, 1989).
Feature extraction and selection is one area of pipeline design that is critically important to system success. For example, Bejan et al. (2012) used a binary classifier for identifying pneumonia; comparing word n-grams, UMLS concepts and assertion values associated with pneumonia expressions. Using clinical notes from 426 patients, they showed statistical feature selection had a substantial improvement over a baseline system that used the complete set of features. Carrol et al. (2011) saw significant improvement using a SVM over a rule-based system using an expert-defined feature set for phenotype identification of rheumatoid arthritis. Carrol et al. argued that with a curated feature set it should be possible to achieve state--of-theart performance using 50 to 100 annotated documents. Using simple bag-of-words features, Wright et al. (2013) employed SVMs to identify diabetes across different institutions using 2000 progress notes (1000 from each institution) achieving F1 measures of 0.934 and 0.935, respectively. They found that stop word filtering, feature selection, negation extraction, and named entity recognition did not substantially improve performance over a bag of words. Training data size continues to be a key motivating factor in system development (Khor et al., 2013). Annotation of medical data can be difficult and costly especially if private health care information must first be identified and removed. As a result, a wide and inconsistent array of training data sizes are reported in the literature and it is not always clear how increasing or decreasing training data size will affect reported model performance.
In general, literature has shown that machine learning algorithms with simple feature spaces and relatively straightforward applications of biomedical NLP tools for semantic feature extraction can perform well on particular phenotypes, and on records sourced from different hospitals. It is unclear how overall performance of these methods is affected by training corpus size, feature spaces, feature weighting schema, and SVM kernel choices. This paper addresses this gap by systematically comparing the effect of a number of well-established feature spaces and feature weighting schemes on classifier performance with various kernels as training data is roughly doubled.
Background
One of the purposes of the 2014 i2b2/UTHealth shared task was to create a NLP challenge that represented a culmination of the previous shared tasks (Stubbs, Kotfila, Hua, & Uzuner, 2015). Like previous i2b2 shared tasks, the 2014 challenge includes smoking history, identification of obesity and a selection of its comorbidities, medication identification, and temporal classification of medical events. By design, certain aspects of the 2014 i2b2/UTHealth shared task contain highly similar annotations with previous i2b2 shared tasks. For instance, the 2008 i2b2 Obesity Challenge includes annotations for diseases that overlap with the 2014 i2b2/UTHealth annotations.
In 2008, 30 teams participated in the Obesity Challenge which required teams to develop systems for identifying presence or absence in a patient of obesity and fifteen comorbidities based on information from unstructured narratives of medical discharge summaries. The Obesity Challenge task was defined by two experts who studied 50 pilot discharge summaries from the Partners HealthCare Research Patient Data Repository. The experts identified fifteen frequently-occurring comorbidities including CAD, diabetes mellitus, hypercholesterolemia, and hypertension. Obesity Challenge systems were required to make textual and intuitive judgments for each document and each disease. Textual judgements were based on direct references to the diseases in the discharge summary. Intuitive judgements were based on some amount of reasoning on the part of the expert annotators. Textual judgements for each disease fell into four classes: present, absent, questionable, or unmentioned. Intuitive judgements fell into three classes: present, absent, and questionable. For example, the statement “the patient weighs 230 lbs and is 5 ft 2 inches” would lead to a textual judgment of ‘unmentioned’ for obesity and an intuitive judgement of ‘present’ (i.e., obesity is not directly mentioned but can be inferred from the height and weight measurements). Intuitive judgements were primarily intended for the interpretation of textual judgements that fell into the unmentioned category.
In 2014, 27 teams participated in the i2b2/UTHealth shared task which required teams to develop systems for identifying diseases, medications, family history of CAD, and smoking status across a temporally-ordered series of unstructured medical notes for individual patients. Unlike the 2008 Obesity Challenge, disease annotations in the 2014 shared task marked only positive (e.g., present) instances of each disease. Neither directly-negated textual evidence nor inferred absence of a disease was marked. In addition to the presence of a disease, a temporal and indicator component were included with all 2014 disease annotations. The time component had acceptable values of “before document creation time” (before DCT), “during document creation time” (during DCT) and “after document creation time” (after DCT). It was possible to have multiple disease tags with separate time components in each document. The indicator component included values such as ‘mention’ and ‘event’ as well as values that were specific to each disease (e.g., ‘high bp.’ for hypertension, or ‘high chol.’ for hyperlipidemia). For a detailed overview of the 2014 challenge, its construction, and its participants see Stubbs et al. (2015).
The task tackled in this paper is a subset of the 2014 task that overlaps with the 2008 task. Namely, we focus on the component of the 2014 task that would be a key component of any phenotyping system: disease identification. Accordingly, we leverage the disease annotations in the 2014 shared task corpus but make no attempt to classify the time or indicator components. We utilize the 2008 data in support of our explorations of effects of data size in addressing this task on the 2014 data.
Data
The data used in this study originated from two separate i2b2 shared tasks: The 2008 Obesity Challenge training data, and the 2014 i2b2/UTHealth shared task corpus. Models were trained on the 2008 training set, on the 2014 training set, and on a combined 2008/2014 training set. Models were evaluated on the 2014 i2b2/UTHealth test set. The 2014 training corpus contains 790 documents and the 2008 training corpus contains 730 documents. Both 2014 and 2008 documents were drawn from Partners HealthCare Research Patient Data Repository. 2008 data consists solely of discharge summaries, whereas the 2014 data is a diverse combination of discharge summaries, admission notes, and emergency department visit notes. Documents for the 2008 challenge were chosen for patients who were overweight or diabetic (Uzuner, 2009). For the 2008 challenge each discharge summary corresponded to a single patient. The 2014 challenge corpus was designed to track the progression of heart disease in diabetic patients, and it included three to five notes per patient representing the longitudinal medical record and the history of the patient. Roughly a third of the patients in this set had CAD as a precondition, a third of patients were diagnosed with CAD over the course of the patient history contained in this data set, and a third were not diagnosed with CAD by the final discharge summary available for that patient in this data. Documents for both the 2008 and 2014 training corpora contain unstructured and quasi-structured free form text. While the text includes sections, such as lists of medications and tables of lab results, none of these document characteristics are explicitly annotated or guaranteed to be present.
The 2014 data includes five disease classes, all of which also appear in the 2008 data. Unlike the 2008 data, the 2014 annotation guidelines do not include an explicit notion of textual and intuitive document-level classification of diseases. In order to make the 2008 and 2014 data consistent in their treatment of textual and intuitive judgments, we adopted a naive approach to combining them. We mapped both sets of document-level classifications to binary outcome variables for the intersection of the diseases in both corpora. Conceptually, the outcome variable is 1 if a disease is present in the patient (based on the contents of the document) and zero otherwise. Mapping the 2008 data to a binary outcome variable meant considering both the textual and intuitive judgements of the 2008 annotators. We collapsed these judgements into our binary outcome variable by using the following rules: if a judgment is textually present and intuitively present, the outcome variable is 1. If a judgment is textually questionable and intuitively absent, the outcome is 0. If a judgment is textually questionable and intuitively present, the outcome variable is 1. Finally, if a judgment is textually absent and intuitively absent, the outcome variable is 0. Fourteen (14) annotations that included questionable textual or intuitive judgements were dropped. These rules are also outlined in Table 1.
Table 1.
Mapping of textual & intuitive judgments to document-level binary outcome variable.
| Textual | Intuitive | Outcome | Support (# of annotations) |
|---|---|---|---|
| Present | Present | 1 | 1,897 |
| Absent | Absent | 0 | 51 |
| Unmentioned | Present | 1 | 73 |
| Unmentioned | Absent | 0 | 1,208 |
| Present | Absent | Removed | 0 |
| Absent | Present | Removed | 0 |
| Questionable | Present | Removed | 1 |
| Questionable | Absent | Removed | 1 |
| Questionable | Questionable | Removed | 8 |
| Unmentioned | Questionable | Removed | 4 |
In the 2014 corpus, it is possible to have multiple annotations with the same disease if there are either multiple indications for that disease or multiple time components. For the 2014 corpus, our outcome variable took on the value of 1 for a disease if that disease classification was present regardless of the indicator or time components of the annotation. Multiple disease annotations were mapped to a 1 for that disease for that document. Absence of a disease tag for a document meant our outcome variable took the value of zero for that disease in that document. The indicator components in the 2014 annotations provide a potentially more sophisticated mapping between the 2008 and 2014 data. In certain cases, indicators in the 2014 annotations such as “waist circumference” for obesity or “A1C” for hyperlipidemia could be considered similar to positive ‘intuitive’ judgments in the 2008 data. Empirical validation of the relationship between the two data sets at this level was beyond the resources of the authors (e.g., re-annotating 2008 data following the 2014 guidelines). Because of this we did not explore the opportunities to map these datasets at that level; instead favoring a more simplified mapping between 2008 and 2014 annotations as described above.
Table 2 provides descriptive statistics comparing the 2008, 2014, and combined 2008/2014 training data. It includes token counts for minimally normalized tokens, lexically normalized tokens, and UMLS CUI tokens (see Methods for definitions). The support section details the number of annotations of each disease in each of the three training corpora. The overall distribution of disease annotations is relatively consistent between the 2008 and 2014 training corpora with the exception of the obese class which is present in 18.8% of the 2014 corpus, 39% of the 2008 corpus, and 28.5% of the combined 2008/2014 corpus. This is consistent with the intentional bias of the 2008 i2b2 Obesity Challenge toward patients who have obesity.
Table 2.
Descriptive parameters for the 2008 Obesity Challenge training corpus, the 2014 i2b2/UTHealth training and test corpora, and the combined 2008/2014 training corpus.
| 2008 Training Data | 2014 Training Data | Combined 2008/2014 Data | 2014 Test Data | |
|---|---|---|---|---|
| Number of Documents | 730 | 790 | 1520 | 514 |
| Unique: | ||||
| Minimally Normalized Tokens | 45,213 | 46,461 | 77,037 | 34,946 |
| Lexically Normalized Tokens | 23,502 | 18,880 | 33,417 | 15,119 |
| UMLS CUI Tokens* | 17,690 | 18,432 | 24,532 | 14,612 |
| Support: | ||||
| Hyperlipidemia | 314 (43.0%) | 352 (44.6 %) | 666 (43.8%) | 249 (48.4%) |
| Obese | 285 (39.0%) | 149 (18.8%) | 434 (28.5%) | 87 (16.9%) |
| Diabetes | 472 (64.7%) | 536 (67.8%) | 1008 (66.3%) | 364 (70.8%) |
| Hypertension | 511 (70.0%) | 602 (76.2%) | 1113 (73.2%) | 393 (76.4%) |
| CAD | 389 (53.3%) | 344 (43.5 %) | 733 (48.2%) | 226 (43.9%) |
Note: Only Sign or Symptom” (sosy) and “Disease or Syndrome” (dsyn) CUI were included due to the number of candidate CUIs produced. More details are available in the methods section.
Methods
To address the five document-level binary disease classification tasks, we utilize SVMs.SVM classifiers have seen wide use in the phenotyping literature (Marafino et al., 2014; Shivade et al., 2014; Wright et al., 2013).
We investigated three baseline feature spaces (1) minimally normalized token features (referred to as “Min”), (2) lexically normalized token features (referred to as “Lex”), and (3) UMLS CUI token features (referred to as “CUI”). Each of the three baseline feature spaces were then combined to create Min & Lex, Min & CUI, Lex & CUI, and Min, Lex & CUI combined feature spaces for a total of seven (7) feature spaces. Documents were represented as vectors in these feature spaces and individual document features were tested with two weighting schema, (1) documents of scaled count features (See: Weighting Schema section) and (2) documents with term-frequency inverse document frequency weighted (tf-idf) features. Each of the seven feature spaces (three baseline, four combined) were separately weighted with the two weighting schema (count, and tf-idf) producing 14 total feature-space, weight combinations. These feature spaces and weighting schema were selected because of their wide representation in the literature on phenotyping algorithms and more generally in statistical modeling of language (Manning & Schütze, 1999).
Feature Spaces
Minimally normalized tokens were generated by processing the raw text of the documents to remove numbers and non-alphabetical characters. All text was converted to lowercase. A list of 127 common stop words were removed and tokens were split based on sequences of one or more contiguous white space characters. Lexically normalized tokens were generated by passing unformatted document text directly to the LuiNorm tool provided by the Lexical Systems Group of the UMLS (McCray, Srinivasan, & Browne, 1994). This mapped all Unicode symbols to ASCII, split ligatures, stripped diacritics, removed genitives and parenthetic plural forms, replaced punctuation with spaces, removed stop words, lowercased all words, uninflected each word, and mapped it to a randomly chosen canonical form.
Finally, each document in the corpus was converted to a set of UMLS CUIs using MetaMap (Aronson & Lang, 2010). MetaMap's performance is significantly reduced if it tries to parse entire unstructured documents and so the Stanford CoreNLP (Finkel, Grenager, & Manning, 2005) sentence breaker was used to pre-process each document into sentences. Raw unnormalized sentences were then fed into MetaMap and candidate CUIs were collected. If a word or phrase in the text produced multiple candidate CUIs, each candidate was included in the final feature space. CUIs were then used as tokens in count and tf-idf weighted feature spaces. Current versions of MetaMap perform built-in negation extraction following Chapman's (2001) NegEx algorithm. Negated CUIs were treated as separate features from their nonnegated counterparts. CUIs were filtered to only include the “Sign or Symptom” (sosy) and “Disease or Syndrome” (dsyn) semantic types due to computing constraints and the large volume of CUIs that are created by MetaMap's default options. MetaMap was run using the 2013AB release of the UMLS USAbase strict and base data sets.
Weighting Schema
Count vector representations of documents were created by converting the training and test document collections to token-document matrices. Tf-idf weighted vector representations of documents were generated by transforming count feature spaces following traditional information retrieval methods (Manning, Raghavan, & Schütze, 2008). Several tf-idf weighting variants were tested including natural and logarithmic term frequency weighting in combination with L1 and L2 term vector normalization. The best overall tf-idf weighting scheme was determined to be logarithmic term frequency weighting using L2 term vector normalization and was applied uniformly to each tf-idf weighted feature space. Count vector feature weights were centered by removing the mean of the features’ weights and scaling the weights to unit variances. This was done because SVMs are not scale invariant, and certain SVM kernels are sensitive to large variations in feature values (Joachims, 1998; Manning, Raghavan, & Schütze, 2008). This is not an issue for tf-idf weighted features because the weighting process accounts for issues like document length. Our count vectors however, are susceptible to variation in feature values and so as a matter of best practice we center and scale them to unit variance.
Feature Selection
The K best features were selected using a univariate parametric filter based on a one way ANOVA F-test (Heiman, 2001, p. 349; Saeys, Inza, & Larrañaga, 2007). One way ANOVA F-test was selected because after count scaling and tf-idf weighting our features become continuous variables. In this context Chi-squared tests are no longer meaningful. The parameter K (i.e., number of features) was chosen from a fixed set of values and jointly optimized along with classifier parameters (see next paragraph). Finally, during pre-processing CUI were limited to “Sign or Symptom (sosy)” and “Disease or Syndrome (dsyn)” semantic types. While the primary motivation for doing this was practical (i.e., computing constraints and the volume of CUI produced from a clinical narrative), it also tacitly participates in the feature selection process.
Classifiers
Classification was performed with two different SVM kernels: a linear kernel and a Gaussian radial basis function (RBF) kernel. The linear kernel SVM was implemented using LibLinear (Fan, Chang, Hsieh, Wang, & Lin, 2008). The RBF kernel SVM was implemented using LibSVM (Chang & Lin, 2011). The penalty parameter of the SVM error term and, in the case of the RBF kernel SVM, the kernel coefficient, were optimized using a brute force grid search across an exponential parameter space. Parameters for models trained on the 2008, 2014, and combined 2008/2014 corpora were validated and selected independently, based on the maximum average F1 measure in five-fold cross-validation. Final parameter values are reported in the supplemental materials.
Linear and RBF kernel SVMs were trained on each individual feature space using the 2008, 2014 and the combined 2008/2014 training corpora. Taking the combination of three separate training datasets, two different SVM kernels and 14 weighted feature spaces gives us 84 total models for evaluation per disease or 420 models total across the five diseases. The final 420 models were evaluated using precision, recall and F1 measure, following the same method outlined in Stubbs et al. (2015). These metrics were calculated against the gold standard i2b2/UTHealth 2014 test set which was transformed using the same process as the 2014 training set to generate binary outcome variables for presence or absence of each disease in each document. F1 for each model is reported in the Results section. Values for precision and recall can be found in the supplemental materials.
Statistical Significance Testing
Statistical significance was determined between all pairs of classifiers, weighting schemes, and feature spaces within a particular disease. Significance was determined using approximate randomization following Chinchor (1992) and Noreen (1989). Approximate randomization involves generating N pseudo-systems and comparing the difference in performance between these pseudo-systems with the difference in performance of the actual systems. Given two systems for comparison such as system A and system B, pseudo-system A’ and pseudo-system B’ are generated by randomly swapping approximately 50% of system A's document level predictions with the corresponding predictions made by system B. The performance of A’ is then compared to B’. We keep track of the number of times (n) the difference in pseudo-system performance is greater than the difference in actual system performance. If (n + 1) / (N + 1) is greater than a cutoff alpha, then we can consider the difference in systems performance to be explained by chance. Intuitively, if system A and system B are not different, then swapping 50% of their system output should have little impact on system A or system B's performance. If system A's performance is significantly better than system B's performance then swapping 50% of their system output should, on average, make system A worse and system B better. By comparing the actual system difference, to the average pseudo-system difference we can get a sense of whether or not the difference in performance between the two systems is significant. For this study we used an N = 9,999 and report cut-off values of alpha at 0.1 and alpha at 0.05. These values are consistent with with both the current and previous i2b2 NLP challenges (Stubbs et al., 2015; Uzuner, Luo, & Szolovits, 2007).
Robustness Checks
Once the best performing models in each disease class were identified, robustness checks were run to ensure the task was sufficiently complex to warrant statistical classifiers of this nature. The top performing model for each disease was re-trained on the corpus that provided the best performance, using the same baseline or combined feature space, without weighting (i.e., binary presence or absence of minimally normalized, lexically normalized and/or CUI tokens) and then evaluated on the test set. To determine if rule-based approaches were sufficient to achieve comparable performance, decision tree classifiers with a maximum tree depth of three were evaluated on the corpus and binary feature space of the top performing model for each disease. In the case of both unweighted SVMs and decision tree classifiers, performance was statistically significantly worse than the weighted SVM for all diseases.
Results
The following tables contain F1 measure results for all models on the 2014 i2b2/UTHealth test set with document-level binary outcome variables for diseases. There is a separate table for each of the five diseases: obesity, CAD, hypertension, hyperlipidemia, and diabetes. Each row represents the combination of an SVM classifier labeled by its kernel (RBF, Linear), the vector weighting scheme (tf-idf, count) and the feature space (Min, Lex, CUI, Min & Lex, Min & CUI, Lex & CUI, Min, Lex & CUI). F1 measures for models trained on the 2008, 2014, and combined 2008/2014 data are reported in the ‘2008’, ‘2014’ and ‘Combined 2008/2014’ columns, respectively. The percentage change in F1 measure for each model, between each training data, is also reported in the “% Change 2008, 2014”, “% Change 2008, Comb.”, and “% Change 2014, Comb” columns, respectively. These should be read “Percentage change from A to B”. The statistical significance of these changes is marked with an asterisk (“*”) for differences at alpha = 0.1 and two asterisks (“**”) for differences at alpha = 0.05.
The highest performing model for each disease has been emphasized. The table is sorted on the column that contains the highest performing model in descending order. Models with F1 measures that are statistically significantly different from the highest performing system at alpha = 0.1 are marked with a double dagger (“††”) and systems that are statistically significantly different from the highest performing system at alpha = 0.05 are marked with a single dagger (“†”). To ease visual identification of the group of models that are not statistically different from the highest performing model at alpha = 0.1, we have marked them with a dark grey background. We refer to this group as the top performing group. Models that are not statistically different at alpha 0.05 have been marked with a light grey background. Tables for models’ precision and recall can be found in the supplemental materials.
Obesity
The highest performing model for obesity was a SVM with an RBF kernel trained on a tfidf weighted feature space of combined minimally normalized, lexically normalized, and CUI tokens with an F1 measure of 0.945. Out of 84 total models for obesity, the highest performing model was statistically not different (alpha = 0.05) in F1 measure to 56 other models ranging in F1 measure from 0.945 to 0.898. 35.71% (20) models in this group were trained on the 2008 training corpus, 25.00% (14) were trained on the 2014 training corpus and %39.29 (22) were trained on the combined 2008/2014 training corpus. 48.21% (27) models were trained on tf-idf weighted vectors while 51.79% (29) of models used scaled count vectors. 39.29% (22) models used linear SVM while 60.71% (34) used RBF kernel SVM. 48.21% (27) models included a CUI feature space. A total of 50 features was selected by the top performing system. Several of the key features were: C0028754 (“Obesity”), obese, obesity, morbid, apnea, C0028756 (“Obesity, Morbid”), sleep, C0520679 (“Sleep Apnea, Obstructive”), and morbidly. A complete list of F1 measures broken down by SVM kernel, weighting scheme, and feature space, compared across training corpora, can be found in Table 3.
Table 3.
Obesity: Comparison of F1 measure for obesity models between 2008, 2014 and combined 2008/2014 broken down by SVM kernel, weighting scheme, and feature space.
| Kernel | Weight | Feature Space | 2008 | 2014 | Combined 2008/2014 | % Change 2008, 2014 | % Change 2008, Comb. | % Change 2014, Comb. |
|---|---|---|---|---|---|---|---|---|
| RBF | tf-idf | Min, Lex & CUI | 0.930 | 0.945 | 0.945 | 1.65 | 1.65 | 0.00 |
| RBF | tf-idf | Min | 0.935 | 0.939 | 0.945 | 0.48 | 1.10 | 0.62 |
| RBF | tf-idf | Lex | 0.935 | 0.933 | 0.945 | −0.16 | 1.10 | 1.26 |
| RBF | Count | Lex | 0.940 | 0.945 | 0.945 | 0.55 | 0.55 | 0.00 |
| RBF | tf-idf | Min & Lex | 0.940 | 0.945 | 0.940 | 0.55 | 0.00 | −0.55 |
| RBF | Count | Min, Lex & CUI | 0.915†† | 0.945 | 0.940 | 3.30** | 2.73* | −0.55 |
| RBF | Count | Min & Lex | 0.891†† | 0.945 | 0.940 | 6.04** | 5.46** | −0.55 |
| RBF | Count | Min & CUI | 0.925 | 0.945 | 0.940 | 2.20 | 1.64 | −0.55 |
| RBF | Count | Min | 0.940 | 0.921 | 0.940 | −1.97 | 0.00 | 2.01 |
| RBF | Count | Lex & CUI | 0.935 | 0.940 | 0.940 | 0.55 | 0.55 | 0.00 |
| Linear | tf-idf | Min & CUI | 0.934 | 0.871†† | 0.940 | −6.73** | 0.62 | 7.89** |
| RBF | tf-idf | Lex & CUI | 0.940 | 0.945 | 0.939 | 0.55 | −0.07 | −0.62 |
| RBF | tf-idf | Min & CUI | 0.915†† | 0.907†† | 0.939 | −0.87 | 2.66 | 3.56* |
| Linear | Count | Min & CUI | 0.923 | 0.892†† | 0.933 | −3.41 | 1.11 | 4.68 |
| Linear | Count | Min | 0.923 | 0.840†† | 0.933 | −9.05** | 1.11 | 11.18** |
| Linear | Count | Lex | 0.928 | 0.912† | 0.933 | −1.71 | 0.56 | 2.31 |
| Linear | Count | Min, Lex & CUI | 0.923 | 0.898† | 0.928 | −2.69 | 0.55 | 3.34 |
| RBF | tf-idf | CUI | 0.928 | 0.886†† | 0.928 | −4.49** | 0.00 | 4.70** |
| Linear | tf-idf | Lex | 0.929 | 0.913†† | 0.926 | −1.69 | −0.35 | 1.36 |
| Linear | Count | Min & Lex | 0.909†† | 0.940 | 0.922 | 3.39** | 1.44 | −1.88 |
| Linear | tf-idf | Min | 0.940 | 0.906†† | 0.921 | −3.62* | −1.97 | 1.71 |
| Linear | tf-idf | Min, Lex & CUI | 0.899†† | 0.906†† | 0.920† | 0.78 | 2.40 | 1.61 |
| Linear | tf-idf | Min & Lex | 0.940 | 0.913†† | 0.905†† | −2.83 | −3.71** | −0.91 |
| Linear | Count | Lex & CUI | 0.945 | 0.892†† | 0.903†† | −5.66** | −4.47** | 1.27 |
| Linear | tf-idf | Lex & CUI | 0.939 | 0.878†† | 0.894†† | −6.51** | −4.80** | 1.83 |
| Linear | tf-idf | CUI | 0.867†† | 0.846†† | 0.821†† | −2.42 | −5.26* | ††2.92 |
| RBF | Count | CUI | 0.820†† | 0.385†† | 0.755†† | −53.11** | −7.96 | 96.29** |
| Linear | Count | CUI | 0.693†† | 0.316†† | 0.370†† | −54.44** | −46.57** | 17.28 |
percent change significant at alpha = 0.05
percent change significant at alpha = 0.1
statistically significantly different from highest performing model (emphasized above) at alpha = 0.05
statistically significantly different from highest performing model (emphasized above) at alpha =0.1
Note: For visual reference, models which are not statistically significantly different from the emphasized model at alpha = 0.1 are shaded in dark grey. Models which are not statistically significantly different from the emphasized model at alpha = 0.05 are shaded in light grey.
While 56 models were not statistically significantly different than the top performing model, 12 models were statistically indistinguishable from the top performing model (i.e., these 12 models had the same F1 measure of 0.945). These 12 models are primarily RBF kernel SVMs though one linear kernel is present; they are evenly split between count and tf-idf weighting schemes. Feature spaces among these 12 models range from combined Min, Lex & CUI to simple minimally normalized (Min) suggesting that semantically-informed features are not necessary to identify the presence of obesity in the majority of cases for these medical records. An analysis of errors in model performance of these 12 models shows that 10 documents were consistently misclassified: nine false positives and one false negative. The nine false positives were due to either annotator error or use of modifying adjectives such as ‘minimally’ or ‘slightly.’ However, inclusion of phrase-based features may improve performance for obesity. For example, ‘sleep apnea’ which was encoded as ‘sleep’ and ‘apnea’ lead to some obesity misclassifications for documents that included ‘sleep’ but not in the context of ‘apnea.’
CAD
The highest performing model for CAD was an SVM with an RBF kernel trained on a tf-idf weighted feature space of Min. tokens with a F1 measure of 0.891. Out of 84 total models for CAD, the highest performing model was not statistically different (alpha = 0.05) in F1 measure from 12 other models ranging in F1 measure from 0.883 to 0.868. 50.00% (6) were trained on the 2014 training data and 50.00% (6) were trained on the combined 2008/2014 training data. 100.00% (12) models were trained on tf-idf weighted vectors. 25.00% (3) models used linear SVM while 75.00% (9) used RBF kernel SVM. 41.67% (5) models included a CUI feature space. A total of 50 features were selected by the top performing models. Several of the key features were: coronary, cad, artery, lad, disease, cabg, rca, cardiac, catheterization, aspirin, stent, plavix, circumflex, cath, and angina. A complete list of F1 measures broken down by SVM kernel, weighting scheme, and feature space, compared across training corpora, can be found in Table 4.
Table 4.
CAD: Comparison of F1 measure for CAD models between 2008, 2014 and combined 2008/2014 broken down by SVM kernel, weighting scheme, and feature space.
| Kernel | Weight | Feature Space | 2008 | 2014 | Combined 2008/2014 | % Change 2008, 2014 | % Change 2008, Comb. | % Change 2014, Comb. |
|---|---|---|---|---|---|---|---|---|
| RBF | tf-idf | Min | 0.841†† | 0.896 | 0.878 | 6.54** | 4.37** | −2.03 |
| RBF | tf-idf | Min, Lex & CUI | 0.827†† | 0.891 | 0.872 | 7.73** | 5.47** | −2.10 |
| RBF | tf-idf | Min & Lex | 0.829†† | 0.883 | 0.871† | 6.62** | 5.12** | −1.41 |
| Linear | tf-idf | Min & Lex | 0.805†† | 0.882 | 0.843†† | 9.62** | 4.77** | −4.42** |
| Linear | tf-idf | Lex | 0.815†† | 0.881 | 0.844†† | 8.09** | 3.47** | −4.28** |
| RBF | tf-idf | Lex | 0.748†† | 0.875† | 0.876 | 17.01** | 17.09** | 0.07 |
| Linear | tf-idf | Min, Lex & CUI | 0.793†† | 0.875 | 0.846†† | 10.34** | 6.72** | −3.28* |
| Linear | tf-idf | Min | 0.809†† | 0.872†† | 0.841†† | 7.87** | 3.96** | −3.63** |
| Linear | tf-idf | Min & CUI | 0.788†† | 0.868†† | 0.861†† | 10.15** | 9.30** | −0.77 |
| Linear | tf-idf | Lex & CUI | 0.780†† | 0.863†† | 0.851†† | 10.60** | 9.06** | −1.39 |
| RBF | tf-idf | Min & CUI | 0.820†† | 0.863†† | 0.868† | 5.16** | 5.77** | 0.58 |
| RBF | Count | Min | 0.727†† | 0.861†† | 0.830†† | 18.40** | 14.10** | −3.63 |
| RBF | Count | Lex | 0.792†† | 0.858†† | 0.833†† | 8.37** | 5.14** | −2.97* |
| RBF | Count | Lex & CUI | 0.765†† | 0.857†† | 0.837†† | 12.00** | 9.41** | −2.31 |
| RBF | tf-idf | Lex & CUI | 0.782†† | 0.855†† | 0.881 | 9.29** | 12.68** | 3.10 |
| Linear | Count | Lex & CUI | 0.737†† | 0.851†† | 0.791†† | 15.51** | 7.33** | −7.09** |
| RBF | Count | Min & CUI | 0.696†† | 0.851†† | 0.830†† | 22.23** | 19.16** | −2.51 |
| RBF | Count | Min & Lex | 0.670†† | 0.844†† | 0.843†† | 25.87** | 25.73** | −0.11 |
| Linear | Count | Lex | 0.698†† | 0.841†† | 0.814†† | 20.47** | 16.66** | −3.16 |
| Linear | Count | Min, Lex & CUI | 0.698†† | 0.840†† | 0.767†† | 20.32** | 9.86** | −8.69** |
| Linear | Count | Min & Lex | 0.674†† | 0.835†† | 0.799†† | 23.89** | 18.51** | −4.34* |
| RBF | Count | Min, Lex & CUI | 0.714†† | 0.832†† | 0.842†† | 16.57** | 17.89** | 1.13 |
| Linear | Count | Min | 0.682†† | 0.825†† | 0.795†† | 21.05** | 16.62** | −3.66 |
| Linear | Count | Min & CUI | 0.669†† | 0.824†† | 0.797†† | 23.30** | 19.20** | −3.32 |
| RBF | tf-idf | CUI | 0.608†† | 0.689†† | 0.732†† | 13.29** | 20.43** | 6.30* |
| Linear | tf-idf | CUI | 0.610†† | 0.674†† | 0.654†† | 10.65** | 7.31* | −3.01 |
| RBF | Count | CUI | 0.631†† | 0.616†† | 0.616†† | −2.41 | −2.46 | −0.04 |
| Linear | Count | CUI | 0.542†† | 0.538†† | 0.548†† | −0.65 | 1.23 | 1.89 |
percent change significant at alpha = 0.05
percent change significant at alpha = 0.1
statistically significantly different from highest performing model (emphasized above) at alpha = 0.05
statistically significantly different from highest performing model (emphasized above) at alpha =0.1
Note: For visual reference, models which are not statistically significantly different from the emphasized model at alpha = 0.1 are shaded in dark grey. Models which are not statistically significantly different from the emphasized model at alpha = 0.05 are shaded in light grey
Out of the five disease classes CAD models had the worst overall performance. The top 13 models failed to correctly predict the presence of CAD in 70 unique documents with an average of six false negatives per model. CAD is consistently misclassified by 10 or more of the top 13 models in 22 different documents. One trend emerges in these 22 documents which were frequently misclassified for CAD – CAD was not the primary complaint or reason for admission. These models consistently misclassify CAD as absent for tags in the 2014 test set that are marked with a time component of before DCT and an indicator of ‘event’ or ‘symptom.’ Among the 22 frequently misclassified documents, this usually takes place in the patient history and includes a single feature for the entire document (e.g., “Patient history: CAD” where CAD is the only mention in the document). In these cases, there appears to be insufficient evidence for the NLP system to identify the patient as positive for CAD. Among false positives for CAD, section location (e.g., presence of CAD in family history section) and negation issues play a role. Inclusion of negated CUI concepts does not appear to sufficiently outweigh the presence of minimally normalized or lexically normalized features.
Hypertension
The highest performing model for hypertension was an SVM with a linear kernel trained on a count vector feature space of combined Min, Lex & CUI tokens with a F1 measure of 0.957. Out of 84 total models for hypertension, the highest performing model was not statistically different (alpha = 0.05) in F1 measure from 36 other models ranging in F1 measure from 0.956 to 0.944. 19.44% (7) models in this group were trained on the 2008 training data, 30.56% (11) were trained on the 2014 training data and 50.00% (18) were trained on the combined 2008/2014 training data. 50.00% (18) models were trained on tf-idf weighted vectors while 50.00% (18) of models used scaled count vectors. 38.89% (14) models used linear SVM while 61.11% (22) used RBF kernel SVM. 47.22% (17) models included a CUI feature space. A total of 10 features were selected by the top performing models. Several of the key features were: hypertension, htn, bp, and hyperlipidemia. A complete list of F1 measures broken down by SVM kernel, weighting scheme, and feature space, compared across training corpora, can be found in Table 5.
Table 5.
Hypertension: Comparison of F1 measure for hypertension models between 2008, 2014, and combined 2008/2014 broken down by SVM kernel, weighting scheme, and feature space.
| Kernel | Weight | Feature Space | 2008 | 2014 | Combined 2008/2014 | % Change 2008, 2014 | % Change 2008, Comb. | % Change 2014, Comb. |
|---|---|---|---|---|---|---|---|---|
| RBF | Count | Min, Lex & CUI | 0.957 | 0.949 | 0.953 | −0.84 | −0.39 | 0.45 |
| RBF | Count | Min & Lex | 0.953 | 0.949 | 0.953 | −0.45 | 0.00 | 0.45 |
| RBF | tf-idf | Min & Lex | 0.952 | 0.950 | 0.949 | −0.18 | −0.29 | −0.11 |
| RBF | tf-idf | Min, Lex & CUI | 0.952 | 0.939†† | 0.945†† | −1.32** | −0.66 | 0.67 |
| RBF | tf-idf | Min | 0.951 | 0.944†† | 0.952 | −0.69 | 0.10 | 0.80 |
| RBF | tf-idf | Min & CUI | 0.949 | 0.941†† | 0.939†† | −0.84 | −1.06 | −0.22 |
| RBF | Count | Lex & CUI | 0.949†† | 0.950† | 0.953 | 0.04 | 0.42 | 0.38 |
| Linear | tf-idf | Min, Lex & CUI | 0.948† | 0.930†† | 0.956 | −1.99** | 0.78 | 2.82** |
| RBF | Count | Lex | 0.948†† | 0.950† | 0.953 | 0.19 | 0.56 | 0.38 |
| RBF | Count | Min | 0.948†† | 0.947†† | 0.953 | −0.04 | 0.59* | 0.64 |
| RBF | tf-idf | Lex & CUI | 0.947†† | 0.946† | 0.956 | −0.08 | 0.96** | 1.04 |
| Linear | tf-idf | Lex & CUI | 0.946†† | 0.942†† | 0.955 | −0.40 | 0.93 | 1.33** |
| Linear | tf-idf | Min | 0.944†† | 0.941†† | 0.952 | −0.35 | 0.89 | 1.24** |
| Linear | Count | Min & Lex | 0.943†† | 0.955 | 0.950† | 1.23** | 0.66 | −0.57 |
| Linear | Count | Lex & CUI | 0.941†† | 0.944†† | 0.948†† | 0.30 | 0.78 | 0.47 |
| RBF | tf-idf | CUI | 0.938†† | 0.905†† | 0.933†† | −3.51** | −0.53 | 3.08** |
| Linear | Count | Lex | 0.938†† | 0.944†† | 0.946†† | 0.64 | 0.85 | 0.21 |
| Linear | Count | Min, Lex & CUI | 0.937†† | 0.955 | 0.950† | 1.87** | 1.29* | −0.57 |
| Linear | tf-idf | Min & Lex | 0.934†† | 0.948 | 0.951 | 1.52* | 1.81** | 0.29 |
| Linear | Count | Min | 0.934†† | 0.941†† | 0.950† | 0.83 | 1.70** | 0.86 |
| Linear | Count | Min & CUI | 0.931†† | 0.941†† | 0.950† | 1.12 | 1.99** | 0.86 |
| RBF | tf-idf | Lex | 0.929†† | 0.944† | 0.937†† | 1.61** | 0.80 | −0.80 |
| RBF | Count | CUI | 0.928†† | 0.867†† | 0.929†† | −6.66** | 0.02 | 7.16** |
| Linear | tf-idf | CUI | 0.924†† | 0.922†† | 0.929†† | −0.26 | 0.55 | 0.82 |
| RBF | Count | Min & CUI | 0.923†† | 0.947†† | 0.953 | 2.56** | 3.21** | 0.64 |
| Linear | tf-idf | Lex | 0.921†† | 0.946† | 0.941†† | 2.63** | 2.12** | −0.50 |
| Linear | tf-idf | Min & CUI | 0.906†† | 0.941†† | 0.952 | 3.77** | 5.01** | 1.20* |
| Linear | Count | CUI | 0.866†† | 0.861†† | 0.869†† | −0.51 | 0.40 | 0.92* |
percent change significant at alpha = 0.05
percent change significant at alpha = 0.1
statistically significantly different from highest performing model (emphasized above) at alpha = 0.05
statistically significantly different from highest performing model (emphasized above) at alpha =0.1
Note: For visual reference, models which are not statistically significantly different from the emphasized model at alpha = 0.1 are shaded in dark grey. Models which are not statistically significantly different from the emphasized model at alpha = 0.05 are shaded in light grey
False negative predictions of hypertension appear to be systematically due to failure to contextualize high blood pressure measurements. For hypertension, of the 16 documents that 34 or more of the top performing group failed to positively classify, 15 had time components of ‘during DCT’ and indicators of ‘high bp.’ Manual review of these documents indicated that few, if any, features selected for hypertension were present in these documents. To accurately classify these documents, models would have to identify blood pressure measurements in the text. In some contexts this could be done using regular expressions (e.g., presence of the string “BP: 110/70”) but in other contexts, results such as blood pressure are presented inline in semi-structured tab delimited tables. In these situations text marking a test such as blood pressure may be quite far from the test value (i.e., a column value “BP” in such a table may be several lines away from its value “110/70”). Determining blood pressure (as well as other lab results) in these cases would be significantly more complex. Presence of hyperlipidemia without a mention of hypertension appears to be the leading reason for false positives among the top performing group.
Hyperlipidemia
The highest performing model for hyperlipidemia was an SVM with an RBF kernel trained on a tf-idf weighted feature space of combined minimally normalized and CUI tokens with a F1 measure of 0.914. Out of 84 total models for hyperlipidemia, the highest performing model was not statistically different (alpha = 0.05) in F1 measure from 16 other models ranging in F1 measure from 0.904 to 0.887. 31.25% (5) models in this group were trained on the 2008 training data, 37.50% (6) were trained on the 2014 training data, and 31.25% (5) were trained on the combined 2008/2014 training data. 56.25% (9) models were trained on tf-idf weighted vectors while 43.75% (7) of models used scaled count vectors. 37.50% (6) models used linear SVM while 62.50% (10) used RBF kernel SVM. 81.25% (13) models included a CUI feature space. A total of 50 features were selected by the top performing models. Several of the key features were: hyperlipidemia, C0020473 (“Hyperlipidemia”), hypercholesterolemia, C0020443 (“Hypercholesterolemia”), lipitor, cholesterol, C0020538 (“Hypertensive disease”), lad, cad, asa, htn, rca, plavix, ldl, and atorvastatin. A complete list of F1 measures broken down by SVM kernel, weighting scheme, and feature space, compared across training corpora, can be found in Table 6.
Table 6.
Hyperlipidemia: Comparison of F1 measure for hyperlipidemia models between 2008, 2014 and combined 2008/2014 broken down by SVM kernel, weighting scheme, and feature space.
| Kernel | Weight | Feature Space | 2008 | 2014 | Combined 2008/2014 | % Change 2008, 2014 | % Change 2008, Comb. | % Change 2014, Comb. |
|---|---|---|---|---|---|---|---|---|
| RBF | tf-idf | Min & CUI | 0.807†† | 0.914 | 0.894 | 13.20** | 10.71** | −2.19 |
| Linear | tf-idf | Min & CUI | 0.879†† | 0.900 | 0.887†† | 2.36 | 0.91 | −1.42 |
| RBF | Count | Min & CUI | 0.895 | 0.894 | 0.859†† | −0.17 | −4.02** | −3.85** |
| RBF | tf-idf | Min, Lex & CUI | 0.859†† | 0.892† | 0.888† | 3.82** | 3.39** | −0.42 |
| RBF | tf-idf | Lex & CUI | 0.879†† | 0.892† | 0.891† | 1.47 | 1.42 | −0.04 |
| Linear | tf-idf | Min | 0.781†† | 0.889† | 0.880†† | 13.90** | 12.68** | −1.07 |
| Linear | tf-idf | Min, Lex & CUI | 0.869†† | 0.888 | 0.904 | 2.14 | 3.94** | 1.77 |
| Linear | tf-idf | Lex & CUI | 0.856†† | 0.887 | 0.886†† | 3.58* | 3.46** | −0.12 |
| RBF | tf-idf | CUI | 0.855†† | 0.884 | 0.874†† | 3.31** | 2.25* | −1.03 |
| RBF | tf-idf | Min | 0.795†† | 0.883 | 0.874†† | 11.00** | 9.98** | −0.92 |
| Linear | tf-idf | Lex | 0.881†† | 0.877 | 0.873†† | −0.48 | −0.85 | −0.37 |
| Linear | tf-idf | CUI | 0.845†† | 0.876 | 0.862†† | 3.62** | 2.04 | −1.53* |
| RBF | tf-idf | Min & Lex | 0.866†† | 0.874 | 0.876†† | 0.95 | 1.16 | 0.21 |
| RBF | Count | Lex & CUI | 0.882†† | 0.873 | 0.867†† | −0.99 | −1.64 | −0.65 |
| Linear | Count | Min & CUI | 0.878†† | 0.873 | 0.877†† | −0.66 | −0.14 | 0.52 |
| RBF | tf-idf | Lex | 0.843†† | 0.873 | 0.880†† | 3.51* | 4.37** | 0.83 |
| RBF | Count | Lex | 0.848†† | 0.872 | 0.859†† | 2.83 | 1.26 | −1.52 |
| Linear | tf-idf | Min & Lex | 0.848†† | 0.870 | 0.884†† | 2.58 | 4.26** | 1.64 |
| RBF | Count | Min | 0.890 | 0.869 | 0.856†† | −2.34 | −3.74** | −1.44 |
| RBF | Count | Min, Lex & CUI | 0.890 | 0.869 | 0.872†† | −2.34 | −1.95 | 0.40 |
| RBF | Count | Min & Lex | 0.874†† | 0.868 | 0.860†† | −0.69 | −1.62 | −0.93 |
| Linear | Count | Lex & CUI | 0.887† | 0.868 | 0.892 | −2.14 | 0.52 | 2.72 |
| Linear | Count | Min & Lex | 0.891 | 0.865 | 0.868†† | −2.87 | −2.51 | 0.37 |
| Linear | Count | Min | 0.868†† | 0.865 | 0.868†† | −0.45 | −0.07 | 0.38 |
| Linear | Count | Min, Lex & CUI | 0.875†† | 0.863 | 0.871†† | −1.39 | −0.38 | 1.03 |
| Linear | Count | Lex | 0.873†† | 0.840 | 0.861†† | −3.81** | −1.40 | 2.51* |
| RBF | Count | CUI | 0.843†† | 0.639 | 0.642†† | −24.26** | −23.84** | 0.56 |
| Linear | Count | CUI | 0.840†† | 0.559 | 0.609†† | −33.53** | −27.52** | 9.04** |
percent change significant at alpha = 0.05
percent change significant at alpha = 0.1
statistically significantly different from highest performing model (emphasized above) at alpha = 0.05
statistically significantly different from highest performing model (emphasized above) at alpha =0.1
Note: For visual reference, models which are not statistically significantly different from the emphasized model at alpha = 0.1 are shaded in dark grey. Models which are not statistically significantly different from the emphasized model at alpha = 0.05 are shaded in light grey
Five documents were consistently marked with a false positive and 15 with a false negative by 13 or more models. This made systematic error detection for hyperlipidemia difficult. Like blood pressure, failure to capture the values of cholesterol tests led to an increase in the number of false positives. Additionally, use of single statements like ‘elevated lipids’ appear to be the only supporting evidence in many of the false negatives. While use of bigram or phrase based extraction methods may be more successful at identifying these features, it will not resolve overall lack of supporting features within the document.
Diabetes
The highest performing model for diabetes was an SVM with an RBF kernel trained on a tf-idf weighted feature space of combined minimally-normalized and CUI tokens with a F1 measure of 0.964. Out of 84 total models for diabetes, the highest performing model was not statistically different (alpha = 0.05) in F1 measure from 18 other models ranging in F1 measure from 0.963 to 0.953. 33.33% (6) were trained on the 2014 training data and 66.67 (12) were trained on the combined 2008/2014 training data. No models in of this group were trained on the 2008 training data. 83.33% (15) models were trained on tf-idf weighted vectors while 16.67% (3) of models used scaled count vectors. 22.22% (4) models used linear SVM while 77.78% (14) used RBF kernel SVM. 55.56% (10) models included a CUI feature space. A total of 50 features was selected by the top performing model. Several of the key features were: diabetes, C0011849 (“Diabetes Mellitus”), insulin, mellitus, C0011847 (“Diabetes”), dm, units, C0011860 (“Diabetes Mellitus, Non-Insulin-Dependent”), nph, type, metformin, diabetic, glyburide, dependent and scale. A complete list of F1 measures broken down by SVM kernel, weighting scheme, and feature space, compared across training corpora, can be found in Table 7.
Table 7.
Diabetes: Comparison of F1 measure for diabetes models between 2008, 2014 and combined 2008/2014 broken down by SVM kernel, weighting scheme, and feature space.
| Kernel | Weight | Feature Space | 2008 | 2014 | Combined 2008/2014 | % Change 2008, 2014 | % Change 2008, Comb. | % Change 2014, Comb. |
|---|---|---|---|---|---|---|---|---|
| RBF | tf-idf | Min & CUI | 0.932†† | 0.957 | 0.964 | 2.63** | 3.48** | 0.83 |
| RBF | tf-idf | Min, Lex & CUI | 0.946†† | 0.959 | 0.963 | 1.30 | 1.78** | 0.47 |
| RBF | tf-idf | Lex & CUI | 0.926†† | 0.958 | 0.959 | 3.46** | 3.56** | 0.10 |
| RBF | tf-idf | Min & Lex | 0.939†† | 0.956 | 0.959 | 1.81** | 2.09** | 0.27 |
| Linear | tf-idf | Min, Lex & CUI | 0.939†† | 0.941†† | 0.958 | 0.19 | 2.08** | 1.88** |
| Linear | tf-idf | Min & CUI | 0.934†† | 0.945†† | 0.956† | 1.22 | 2.41** | 1.18 |
| Linear | tf-idf | Lex | 0.907†† | 0.946†† | 0.956 | 4.23** | 5.32** | 1.05 |
| Linear | tf-idf | Lex & CUI | 0.910†† | 0.942†† | 0.955 | 3.46** | 4.95** | 1.44 |
| RBF | tf-idf | Lex | 0.927†† | 0.958 | 0.954† | 3.27** | 2.91** | −0.35 |
| RBF | Count | Min | 0.870†† | 0.949†† | 0.954 | 9.11** | 9.72** | 0.55 |
| RBF | tf-idf | Min | 0.909†† | 0.956† | 0.953†† | 5.15** | 4.86** | −0.27 |
| RBF | Count | Min & Lex | 0.880†† | 0.946†† | 0.953 | 7.43** | 8.25** | 0.77 |
| RBF | Count | Min, Lex & CUI | 0.880†† | 0.942†† | 0.953 | 7.08** | 8.37** | 1.20 |
| Linear | tf-idf | Min | 0.927†† | 0.952†† | 0.949†† | 2.63** | 2.33** | −0.29 |
| Linear | tf-idf | Min & Lex | 0.911†† | 0.949†† | 0.948†† | 4.13** | 4.08** | −0.05 |
| RBF | Count | Lex & CUI | 0.877†† | 0.950†† | 0.948†† | 8.39** | 8.12** | −0.24 |
| RBF | Count | Lex | 0.911†† | 0.950†† | 0.948†† | 4.27** | 4.02** | −0.24 |
| Linear | Count | Min & CUI | 0.842†† | 0.943†† | 0.946†† | 11.99** | 12.26** | 0.24 |
| Linear | Count | Lex & CUI | 0.916†† | 0.946†† | 0.945†† | 3.28** | 3.09** | −0.19 |
| Linear | Count | Lex | 0.830†† | 0.946†† | 0.943†† | 13.99** | 13.64** | −0.31 |
| Linear | Count | Min | 0.830†† | 0.943†† | 0.943†† | 13.72** | 13.68** | −0.03 |
| RBF | Count | Min & CUI | 0.870†† | 0.949†† | 0.943†† | 9.06** | 8.36** | −0.65 |
| Linear | Count | Min, Lex & CUI | 0.796†† | 0.944†† | 0.942†† | 18.53** | 18.27** | −0.22 |
| Linear | Count | Min & Lex | 0.850†† | 0.944†† | 0.942†† | 11.06** | 10.82** | −0.22 |
| RBF | tf-idf | CUI | 0.869†† | 0.866†† | 0.883†† | −0.29 | 1.64 | 1.94 |
| Linear | tf-idf | CUI | 0.864†† | 0.844†† | 0.877†† | −2.32 | 1.50 | 3.91** |
| RBF | Count | CUI | 0.882†† | 0.789†† | 0.812†† | −10.52** | −7.88** | 2.96 |
| Linear | Count | CUI | 0.811†† | 0.799†† | 0.804†† | −1.47 | −0.95 | 0.53 |
* percent change significant at alpha = 0.05
percent change significant at alpha = 0.1
statistically significantly different from highest performing model (emphasized above) at alpha = 0.05
statistically significantly different from highest performing model (emphasized above) at alpha =0.1
Note: For visual reference, models which are not statistically significantly different from the emphasized model at alpha = 0.1 are shaded in dark grey. Models which are not statistically significantly different from the emphasized model at alpha = 0.05 are shaded in light grey
Of the 18 top performing models for diabetes, eight documents were consistently falsely marked positive by 16 or more models. False positives were generally due to use of qualifying adjectives such as ‘borderline’ or inclusion in sections not related to the patient (e.g., family history). In several cases, uses of sentence constructions like “explained that obesity predisposes patients to develop diabetes” caused problems for these models. Among false negatives, seven were shared by 16 or more models which failed to parse glucose readings. At least two documents were misclassified because of the infrequently used token “DMII” for diabetes mellitus 2.
Discussion
Overall performance across each disease class is relatively good despite the lack of complex domain specific feature engineering. F1 measure of the top performing group for diabetes ranges between 0.963 and 0.953, for hypertension between 0.956 and 0.944, for obesity between 0.945 and 0.898, for hyperlipidemia between 0.904 and 0.887, and for CAD between 0.891 and 0.868. At least for diabetes these numbers are consistent with Wright et al. (2013).
In our experiments, doubling the size of the training data did not in general provide statistically significant results. In the models where it was found to provide a statistically significant improvement, the magnitude of that improvement was usually small. Each top performing group of classifiers for each disease included classifiers that were trained on only the 2014 training corpus suggesting that, for these models, addition of the 2008 data did not provide a statistically significant improvement over training with just the 2014 corpus. This begs the question, for each disease, at what point is there sufficient training data to achieve comparable performance to the top performing models trained on an entire corpus? Preliminary analysis of the top performing models’ learning curves suggests that this threshold varies with each disease. The top performing classifiers for obesity, diabetes and CAD appear to level off in performance between 200 and 300 training examples. On the other hand, performance of top performing models for hypertension and hyperlipidemia appear to level off between 500 and 600 examples. It is tempting to infer from this preliminary finding that binary identification of hypertension and hyperlipidemia is a “harder” task than obesity, diabetes and CAD. There are however, confounding factors related to corpus construction that provide an alternate explanation. For example, compared to other diseases, a disproportionate number of the top performing models for obesity were trained exclusively on the 2008 Obesity Challenge data. For CAD and diabetes, which were the focus of the 2014 challenge, all models trained on the 2008 data performed statistically worse (alpha = 0.1) than the highest performing CAD and diabetes models. This suggests that the initial conditions under which a corpus is designed can affect model performance and should be considered when comparing techniques across different corpora. More systematic analysis of classifier learning curves across the 2008, 2014 and combined 2008 and 2014 corpora is needed to better quantify the relationship between corpus construction and classifier performance. This is an area we intend to investigate in future work.
The highest performing models for obesity, hypertension, hyperlipidemia, and diabetes included combined feature spaces of either minimally normalized, lexically normalized, and CUI tokens or minimally normalized and CUI tokens. Despite this, none of these systems was statistically significantly different at alpha = 0.1 than models that included individual non-semantically-informed feature spaces. Often these single feature space models were minimally normalized or lexically normalized and did not rely on the added semantic information of UMLS CUIs. CUI only features spaces are not included in any of the top performing groups with the exception of obesity. This leads us to the conclusion that semantic features as operationalized by UMLS CUI are not a statistically significant determining factor in our models. The CUI only baseline feature space fails to produce top performing models in the context of our data and the common weighting schemes and classifiers investigated in this research.
In both false negative and false positive errors, understanding lab values played a consistent role. While some documents include unstructured lab results in the body of the text, validated, structured EHR components are not included with the i2b2 corpora. While identifying lab values in clinical narratives was not a primary concern of this research, we expect that the addition of structured lab records in conjunction with rule-based feature extraction would likely resolve such lab-related misclassifications and improve overall model performance.To assess this, we considered leveraging a priori knowledge of annotated lab value textual spans from the 2014 annotations. Unfortunately the 2014 annotations guidelines only require annotators to mark the first instance of a lab value and only if that lab value suggests a positive presence of a disease. For example, mention of “BP: 140/90” in a summary would lead to a positive annotation for hypertension – but only if no other mention of hypertension appeared earlier in the document. A mention of “BP: 110/70” would not lead to any annotation. Using the 2014 annotations as a proxy for structured lab values ‘as-is’ would provide a biased perspective on the impact of structured electronic health care record information on modeling false negative errors related to understanding lab values. Without additional annotations including positive and negative indicating values of lab results, it will be difficult to empirically determine the effect of structured lab values on our models.
In certain cases we observed false negative and false positive errors due to lack of proper phrase identification. In the case of obesity for instance we observed several false positives that we attribute to the presence of the feature ‘sleep’ but absence of the feature ‘apnea.’ This suggests that perhaps bi-gram or other phrase based approaches could improve overall performance.
Systematic testing of bi-gram features across all 420 models in conjunction with parameter tuning was not computationally feasible because of time constraints. We did however assess the impact of bi-gram features on the top performing models. In no case did bi-grams produce better overall performance as measured by F1. In some cases (Obesity and CAD) models with bi-gram features were found to not be statistically significantly different (alpha = 0.1) than the top performing model in terms of F1. In general it appears addition of bi-grams caused the feature space to become overly noisy. While features such as ‘diabetes mellitus’ are selected, other less meaningful bi-grams are included such as ‘dependent diabetes’, ‘history diabetes’ and ‘diabetes hypertension’ which appear to primarily rely on the presence of the unigram ‘diabetes’ for selection.
Despite inclusion of negated CUI features, negation was a consistent source of false positive errors. We believe this is related to unintended effects of combining feature spaces. As an example, in combined Min & CUI feature spaces phrases such as “negative for diabetes” included the negated CUI feature “nC0011847” but also the positive minimally normalized feature “diabetes.” In combined Min, Lex & CUI feature spaces this problem is intensified. Given the fragment “negative for premature coronary artery disease.” a combined Min, Lex & CUI feature space will include features ‘coronary’ and ‘artery’ from the Min feature space, as well as ‘coronary and ‘artery’ from the Lex feature space. Even if MetaMap correctly classifies this as a negated mention of CAD, the four features from the Min and Lex feature spaces outweigh the negated CUI feature and fragment would be misclassified as positive for CAD. Even in contexts where negation is not an issue, our feature selection strategy leads to undesirable repetition of features in combined feature spaces. This repetition can prevent important, but less common features such as abbreviations from appearing in our final feature space (e.g., ‘DMII’ for diabetes). As a result, while the top performing models frequently rely on combined feature spaces that include CUIs, the inclusion of semantically-informed features such as UMLS CUIs does not always improve overall performance. This research establishes the importance of minimally and lexically normalized features on model performance. However, to resolve the types of errors we observe in these models, the way in which semantic features are combined with minimally and lexically normalized feature spaces must be carefully considered. Univariate feature selection only considers individual features’ relationship to the outcome variable. Because each feature space is generated from the same underlying document, there are often several identical features included in the combined feature spaces (e.g., ‘obese’ from the minimally normalized feature space and ‘obese’ from the lexically normalized feature space). Simple combinations of orthographically equal features across minimally normalized and lexically normalized feature spaces were investigated to determine the effect of this repetition on model performance. While this marginally changed the order of some models when ranked by F1, its effect on overall performance was negligible (i.e., top performing models remained the same and no model achieved a higher F1 measure). More sophisticated rule-based strategies for collapsing these features could be employed as in Khor et al. (2013) but this begins to introduce domain specific knowledge that may or may not generalize well to different phenotypes or across documents from different institutions. An alternate approach would be collapsing highly co-linear features using dimensionality reduction techniques or pooling (Clemen & Winkler, 2007) using an aggregation function. Once collapsed, use of a more sophisticated feature selection method such as SVM L1 based recursive feature elimination (Guyon & Elisseeff, 2003; Guyon, Weston, Barnhill, & Vapnik, 2002) may improve overall performance.This is an area we intend to investigate in future work.
In general we found the counts of CUI features such as C0028754 (“Obesity”), and Min or Lex features such as “obesity” across documents to be highly similar. This leads us to believe there is a close relationship between the direct textual evidence for these diseases and the CUI that MetaMap generates. CUI features are particularly useful for disambiguating word meaning in the context of a sentence (e.g. “C0010453: Anthropological Culture”, and “C0430400: Laboratory Culture”). The highly co-linear nature of CUI features and their Min/Lex counterparts across our corpora suggests that identification of these phenotypes does not require significant semantic disambiguation. This provides some explanation for why we do not find CUI feature spaces to be statistically significant determining factor in our models. Importantly, a more systematic analysis of the textual spans of CUI tokens and Min & Lex features would be needed to empirically validate this claim..
Conclusion
In this paper, we systematically compared classifier performance on phenotype identification. Performance was compared across a wide array of weighted feature spaces including minimally normalized, lexically normalized, and UMLS CUI tokens. Combinations of these features were also evaluated and groups of top performing systems were identified. These groups ranged in performance and complexity. We found in many cases that simple feature spaces performed as well as combinations of feature spaces. Among the top performing groups in each disease, term frequency inverse document frequency weighting was found more often than count based weighting schemes, but was not found to be a statistically significant factor in determining model performance. SVM with RBF kernels generally outperformed linear kernels but all diseases included models with linear kernels that could not be found to be statistically significantly different than the top performing model for that disease. Finally, we tested classifier performance on the phenotype identification task using two small (730, & 790) training sets and a larger (1520) combined training set. In general, we found that doubling your data is not always necessary to get good performance with these feature spaces and models, for these phenotypes.
Highlights.
Systematic assessment of common machine learning methods on document phenotype classification
Five diseases studied: Obesity, CAD, Hyperlipidemia, Hypertension, and Diabetes
Statistical testing of model performance using approximate randomization techniques
As expected, semantic features (UMLS - CUI) present in most top performing models
Unexpectedly, many models without semantic features not statistically significantly different from top performing model in each disease.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proceedings / AMIA ... Annual Symposium. AMIA Symposium. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010;17(3):229–236. doi: 10.1136/jamia.2009.002733. http://doi.org/10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejan CA, Xia F, Vanderwende L, Wurfel MM, Yetisgen-Yildiz M. Pneumonia identification using statistical feature selection. Journal of the American Medical Informatics Association : JAMIA. 2012;19(5):817–823. doi: 10.1136/amiajnl-2011-000752. http://doi.org/10.1136/amiajnl-2011-000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32(SI):D267–D270. doi: 10.1093/nar/gkh061. http://doi.org/10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearbook of Medical Informatics. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- Carroll RJ, Eyler AE, Denny JC. Naïve Electronic Health Record Phenotype Identification for Rheumatoid Arthritis. AMIA Annual Symposium Proceedings. 2011;2011:189–196. [PMC free article] [PubMed] [Google Scholar]
- Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, Denny JC. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. Journal of the American Medical Informatics Association : JAMIA. 2012;19(e1):e162–169. doi: 10.1136/amiajnl-2011-000583. http://doi.org/10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C-C, Lin C-J. LIBSVM: A Library for Support Vector Machines. ACM Transactions on Intelligent Systems and Technology. 2011;2(3):1–27. 27. http://doi.org/10.1145/1961189.1961199. [Google Scholar]
- Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A simple algorithm for identifying negated findings and diseases in discharge summaries. Journal of Biomedical Informatics. 2001;34(5):301–310. doi: 10.1006/jbin.2001.1029. http://doi.org/10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- Chen Y, Carroll RJ, Hinz ERM, Shah A, Eyler AE, Denny JC, Xu H. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e2):e253–e259. doi: 10.1136/amiajnl-2013-001945. http://doi.org/10.1136/amiajnl-2013-001945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchor N. Proceedings of the 4th conference on Message understanding. Association for Computational Linguistics; 1992. The statistical significance of the MUC-4 results. pp. 30–50. [Google Scholar]
- Clemen RT, Winkler RL. Aggregating probability distributions. Advances in Decision Analysis: From Foundations to Applications. 2007:154–176. [Google Scholar]
- Davis FD. Perceived usefulness, perceived ease of use, and user acceptance of information technology. MIS Quarterly. 1989:319–340. [Google Scholar]
- Denny JC. Chapter 13: Mining Electronic Health Records in the Genomics Era. PLoS Computational Biology. 2012;8(12) doi: 10.1371/journal.pcbi.1002823. http://doi.org/10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J. LIBLINEAR: A Library for Large Linear Classification. Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]
- Finkel JR, Grenager T, Manning C. Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics. Association for Computational Linguistics; 2005. Incorporating non-local information into information extraction systems by gibbs sampling. pp. 363–370. [Google Scholar]
- Guyon I, Elisseeff A. An introduction to variable and feature selection. The Journal of Machine Learning Research. 2003;3:1157–1182. [Google Scholar]
- Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Machine Learning. 2002;46(1-3):389–422. [Google Scholar]
- Heiman GW. Understanding Research Methods and Statistics: An Integrated Introduction for Psychology. Houghton Mifflin. 2001 Retrieved from http://books.google.com/books?id=r2UNAAAACAAJ.
- Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. Journal of the American Medical Informatics Association. 2013;20(1):117–121. doi: 10.1136/amiajnl-2012-001145. http://doi.org/10.1136/amiajnl-2012-001145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims T. Text categorization with support vector machines: Learning with many relevant features. Springer; 1998. [Google Scholar]
- Khor R, Yip W-K, Bressel M, Rose W, Duchesne G, Foroudi F. Practical implementation of an existing smoking detection pipeline and reduced support vector machine training corpus requirements. Journal of the American Medical Informatics Association : JAMIA. 2013;21(1):27–30. doi: 10.1136/amiajnl-2013-002090. http://doi.org/10.1136/amiajnl-2013-002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao KP, Cai T, Gainer V, Goryachev S, Zeng-Treitler Q, Raychaudhuri S, Plenge RM. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care & Research. 2010;62(8):1120–1127. doi: 10.1002/acr.20184. http://doi.org/10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg D, Humphreys B, McCray A. The Unified Medical Language System. Methods of Information in Medicine. 1993;32(4):281–291. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning CD, Raghavan P, Schütze H. Introduction to information retrieval. Vol. 1. Cambridge University Pess; Cambridge: 2008. [Google Scholar]
- Manning CD, Schütze H. Foundations of Statistical Natural Language Processing. MIT Press; Cambridge, MA, USA: 1999. [Google Scholar]
- Marafino BJ, Davies JM, Bardach NS, Dean ML, Dudley RA. N-gram support vector machines for scalable procedure and diagnosis classification, with applications to clinical free text data from the intensive care unit. Journal of the American Medical Informatics Association. 2014;21(5):871–875. doi: 10.1136/amiajnl-2014-002694. http://doi.org/10.1136/amiajnl-2014-002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, Wolf WA. The eMERGE Network: A consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Medical Genomics. 2011;4:13–13. doi: 10.1186/1755-8794-4-13. http://doi.org/10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proceedings / the ... Annual Symposium on Computer Application [sic] in Medical Care. Symposium on Computer Applications in Medical Care. 1994:235–9. [PMC free article] [PubMed] [Google Scholar]
- Noreen EW. Computer-intensive methods for testing hypotheses: an introduction. Wiley; New York: 1989. [Google Scholar]
- Pathak J, Kho AN, Denny JC. Electronic health records-driven phenotyping: challenges, recent advances, and perspectives. Journal of the American Medical Informatics Association : JAMIA. 2013;20(e2):e206–e211. doi: 10.1136/amiajnl-2013-002428. http://doi.org/10.1136/amiajnl-2013-002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeys Y, Inza I, Larrañaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23(19):2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Sculley D, Holt G, Golovin D, Davydov E, Phillips T, Ebner D, Young M. Machine Learning: The High Interest Credit Card of Technical Debt. SE4ML: Software Engineering for Machine Learning (NIPS 2014 Workshop) 2014.
- Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, Lai AM. A review of approaches to identifying patient phenotype cohorts using electronic health records. Journal of the American Medical Informatics Association. 2014;21(2):221–230. doi: 10.1136/amiajnl-2013-001935. http://doi.org/10.1136/amiajnl-2013-001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs A, Kotfila C, Hua X, Uzuner O. Practical applications for NLP in Clinical Research: the 2014 i2b2/UTHealth shared tasks. Journal of Biomedical Informatics. 2015 doi: 10.1016/j.jbi.2015.10.007. (i2b2 NLP Proceedings 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs A, Uzuner O. Annotating Risk Factors for Heart Disease in Clinical Narratives for Diabetic Patients. Journal of Biomedical Informatics. 2015 doi: 10.1016/j.jbi.2015.05.009. (i2b2 NLP Proceedings 2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner Ö. Recognizing Obesity and Comorbidities in Sparse Data. Journal of the American Medical Informatics Association. 2009;16(4):561–570. doi: 10.1197/jamia.M3115. http://doi.org/http://dx.doi.org/10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner O, Luo Y, Szolovits P. Evaluating the state-of-the-art in automatic de-identification. Journal of the American Medical Informatics Association. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. http://doi.org/10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W-Q, Tao C, Jiang G, Chute CG. A High Throughput Semantic Concept Frequency Based Approach for Patient Identification: A Case Study Using Type 2 Diabetes Mellitus Clinical Notes. AMIA Annual Symposium Proceedings. 2010;2010:857–861. [PMC free article] [PubMed] [Google Scholar]
- Wright A, McCoy AB, Henkin S, Kale A, Sittig DF. Use of a support vector machine for categorizing free-text notes: assessment of accuracy across two institutions. Journal of the American Medical Informatics Association. 2013;20(5):887–890. doi: 10.1136/amiajnl-2012-001576. http://doi.org/10.1136/amiajnl-2012-001576. [DOI] [PMC free article] [PubMed] [Google Scholar]