SUMMARY
Objective
In three U.S. state public health laboratories (PHLs) using a fourth-generation immunoassay (IA), an HIV-1/HIV-2 differentiation antibody IA and a nucleic acid test (NAT), we characterized the yield and time to reporting of acute infections, and cost per positive specimen.
Methods
Routine HIV testing data were collected from July 1, 2012-June 30, 2013 for Massachusetts and Maryland PHLs, and from November 27, 2012-June 30, 2013 for Michigan PHL. Massachusetts and Michigan used fourth-generation and differentiation IAs with NAT conducted by a referral laboratory. In Maryland, fourth-generation IA repeatedly reactive specimens were followed by a Western blot (WB), and those with negative or indeterminate results were tested with a differentiation IA and HIV-1 NAT, and if positive by NAT, confirmed by a different HIV-1 NAT. Specimens from WB-positive persons at risk for HIV-2 were tested with a differentiation IA and, if positive, with an HIV-2 WB and/or differential HIV-1/HIV-2 proviral DNA polymerase chain reaction.
Results
Among 7,914 specimens from Massachusetts PHL, 6,069 from Michigan PHL, and 36,266 from Maryland PHL, 0.10%, 0.02% and 0.05% acute infections were identified, respectively. Massachusetts and Maryland PHLs each had 1 HIV-2 positive specimen. The median time from specimen receipt to laboratory reporting of results for acute infections at Massachusetts, Michigan and Maryland PHLs was 8, 11, and 7 days respectively. The laboratory cost per HIV positive specimen was $336 (Massachusetts), $263 (Michigan) and $210 (Maryland).
Conclusions
Acute and established infections were found by PHLs using fourth-generation IA in conjunction with antibody tests and NAT. Time to reporting of acute HIV test results to clients was suboptimal, and needs to be streamlined to expedite treatment and interrupt transmission.
Keywords: Fourth-generation immunoassay, Acute infections, Cost, HIV testing algorithms, Time to reporting
INTRODUCTION
In 2014, the Centers for Disease Control and Prevention (CDC) and the Association of Public Health Laboratories (APHL) recommended an HIV diagnostic testing algorithm [1]: fourth-generation immunoassay (IA) followed, when repeatedly reactive, by an HIV-1/HIV-2 differentiation antibody IA. When that test is negative or indeterminate, an HIV-1 nucleic acid test (NAT) is conducted. Positive NAT results indicate acute infection. Relative to the CDC algorithm from 1989 [2] which used an antibody-based supplemental test (i.e., Western blot or immunofluorescence assay), the recommended algorithm detects HIV-1 infection earlier, and distinguishes HIV-1 from HIV-2 infections [1, 3–7]. With the availability of fourth-generation IAs that detect p24 antigen and HIV-1 and HIV-2 antibodies [8] approximately 80% of acute HIV infections can be detected that would otherwise only be detectable by NAT [9–11]. The commercial availability of fourth-generation HIV-1/2 assays makes simultaneous screening for both acute and established HIV infections feasible for most clinical laboratories in the United States. Fourth-generation IAs perform with high specificity [7, 10, 12–14]; as a result, they are unlikely to have false-positive results when used in the recommended algorithm.
Assessing the performance of the CDC/APHL HIV diagnostic testing algorithm in U.S. state public health laboratories (PHLs) is critical to ensuring its successful implementation in the United States. Little information on turn-around time has been published from laboratories using the algorithm. Timely reporting of HIV test results to clients in the acute stage of HIV infection is important to decrease transmission potential by initiating antiretroviral therapy early [15]. Additionally, there are limited data on the cost of the recommended algorithm, which will impact its future uptake in U.S. state PHLs [16]. Therefore, in three U.S. PHLs, we characterized the yield of acute infections, time to reporting of acute and antibody-positive HIV-1 infections, and cost per HIV positive specimen.
METHODS
Setting
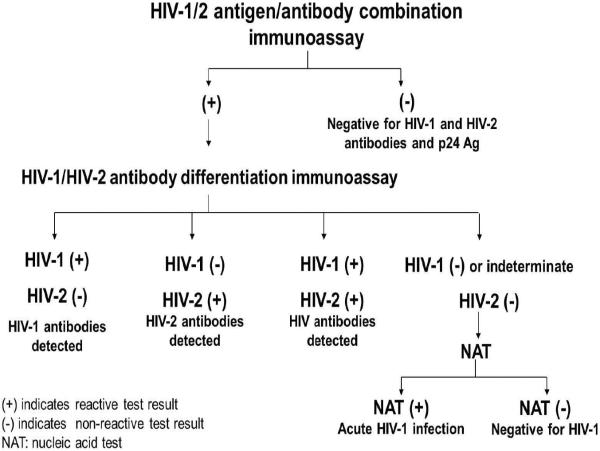
Routine HIV testing data were collected from July 1, 2012- June 30, 2013 for Massachusetts and Maryland PHLs, and from November 27, 2012- June 30, 2013 for Michigan PHL. Maryland and Massachusetts PHLs reported data from clinical testing sites (e.g., sexually transmitted infections clinics, correctional facility clinics, community health centers, and inpatient hospitals) and nonclinical testing sites (e.g., HIV testing sites, schools, and shelters), and Michigan PHL reports data from clinical settings. Massachusetts and Michigan used the CDC/APHL-recommended HIV diagnostic algorithm (Figure 1) with the following tests: GS HIV Combo Ag/Ab IA (Bio-Rad Laboratories, Redmond, WA) (GS Combo) [17], Multispot HIV-1/HIV-2 rapid test (Bio-Rad Laboratories, Redmond, WA) (Multispot) [18], and HIV-1 NAT (APTIMA HIV-1 RNA Qualitative Assay [Gen-Probe Inc., San Diego, CA]) [19]. The APTIMA HIV-1 RNA Qualitative Assay is the only NAT test approved for HIV diagnostic testing. Massachusetts PHL sent specimens needing HIV-1 NAT to the Rhode Island Department of Health, State Health Laboratories, and Michigan PHL sent specimens to Florida Bureau of Public Health Laboratories, Jacksonville, FL as part of a CDC/APHL HIV-1 NAT demonstration project [20].
Figure 1.
HIV diagnostic testing algorithm of Massachusetts and Michigan Public Health Laboratories.
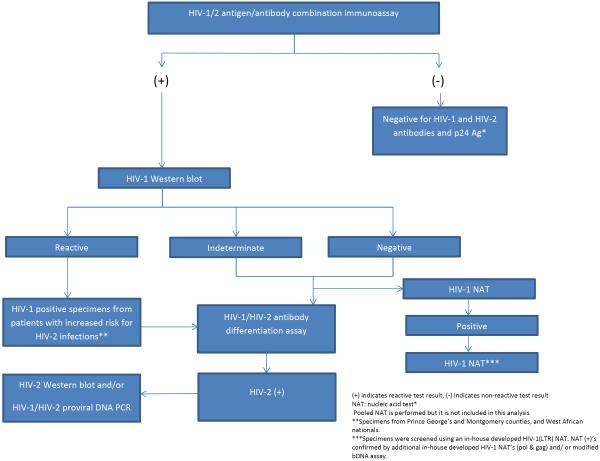
The Maryland PHL used a modified algorithm (Figure 2). ARCHITECT HIV Ag/Ab Combo (Abbott Diagnostics, Abbot Park, IL) (ARCHITECT) [21] was followed, when repeatedly reactive, by GS HIV-1 Western blot (Bio-Rad Laboratories, Redmond, WA) (WB) [22].
Figure 2.
HIV diagnostic testing algorithm of Maryland Public Health Laboratory.
Specimens with negative or indeterminate WB results were tested with Multispot and in-house developed and validated real-time polymerase chain reaction (PCR)-based HIV-1 NATs [23–26]. If the initial NAT targeting the LTR gene was positive, two additional HIV-1 NATs targeting pol and gag gene targets were performed. The presence and relative concentration of HIV-1 RNA in the HIV-1 NAT-positive sera were then confirmed using an off-label validated method of a commercially available HIV-1 viral load assay (VERSANT HIV-1 RNA 3.0 Assay [bDNA] [U.S. IVD] Siemens Health Care Tarrytown, NY). Specimens with positive HIV-1 WB results at increased risk for HIV-2 were tested with Multispot. HIV-2 positive specimens by Multispot were tested with a differential HIV-1/HIV-2 proviral DNA PCR [27, 28] and/or an HIV-2 WB (SIV Western blot Assay [ZeptoMetrix Corporation, Buffalo, NY]) [29].
HIV testing outcomes
For each PHL, we determined the total number of non-reactive and repeatedly reactive specimens using the fourth-generation IA. Among repeatedly reactive specimens, we classified testing outcomes as confirmed HIV-1 antibody positive, acute infection, IA false positive, or HIV-2 positive. Specimens were classified as confirmed HIV-1 antibody positive if the WB or Multispot was HIV-1 positive. They were labeled as acute infections if the fourth-generation IA was repeatedly reactive, the WB or Multispot was negative or indeterminate, and the NAT was reactive. They were labeled as IA false positive if the specimen was fourth-generation IA repeatedly reactive, the WB or Multispot was negative or indeterminate, and NAT was not reactive. The specimen was labeled as HIV-2 positive based on Multispot results in Massachusetts and Michigan PHLs and based on Multispot, HIV-2 WB or differential HIV-1/HIV-2 proviral DNA conventional PCR results in Maryland. We also calculated the specificity of the fourth-generation IA in each PHL.
Time to reporting of HIV test results
In each PHL, we determined the time from specimen collection to receipt in the laboratory, and from receipt in the laboratory to reporting of test results for HIV-1/2 negative specimens as well as for acute and HIV-1 antibody positive specimens. For Massachusetts and Maryland, we calculated the time from reporting of positive test results by the laboratory to receipt of those results by clients separately for acute and HIV-1 antibody positive specimens, and the total time from specimen collection to receipt of positive test results by clients. For Maryland PHL, data on the receipt of test results by clients were only available for HIV tests conducted as part of an HIV Counseling Testing and Referral Program, and the delivery of results was based on direct report by the testing agency. The acute results are usually delivered by local health department disease intervention specialists who conduct field investigations of all acute diagnoses. For Massachusetts PHL, result and date provided are entered by client code on service forms under HIV testing information, and any negative values for time to reporting of HIV test results to clients were set to missing during the analysis.
Cost per HIV positive infection
In each PHL, we determined the total laboratory testing and labor costs per HIV-positive specimen. We collected the cost incurred for each test used in their algorithm. For each test, we collected the test kit brand, cost and size, frequency and number of controls per run, and typical run size. We calculated an adjusted cost per test by adding total kit cost and the cost of extra controls (if applicable), and dividing by the number of tests not used as controls. The total testing costs were obtained by summing the product of the adjusted costs per test and the number of specimens tested with that test. The total labor costs were summed for each test. Labor costs per test were calculated from the product of the labor hours per specimen, the total number of specimens tested, and the labor cost per hour. This was summed for all tests in the algorithm. The labor hours for each test were calculated either by being directly observed at the laboratory or from estimates from literature [16], and was divided by the number of specimens in a run. Labor hours included test processing times, time to prepare for testing the samples as well as time to report the results. Labor hours of the ARCHITECT at Maryland PHL were estimated to be 2.2 hours, and labor hours of the semi-automated GS Combo at Michigan and manual GS Combo at Massachusetts PHLs were estimated on average to be 1.4 and 3.9 contact hours respectively. The WB was estimated to take 3 contact hours [16] and the Multispot was estimated to take 0.5 contact hours. The labor cost per hour was estimated from the Bureau of Labor Statistics' estimate for the 2013 mean salary (including benefits) per hour of a laboratory technologist in each PHL's state [30]. The cost per positive in each of the PHL was calculated by summing laboratory testing and labor costs and dividing by the number of HIV-1 and 2 positive specimens. The cost per specimen in each PHL was calculated by summing laboratory testing and labor costs and dividing by the number of specimens tested. We did not include start-up costs or fixed costs such as space and overhead or non-laboratory variable costs associated with HIV testing such as the cost of specimen collection/phlebotomy and disclosure of HIV test results. All costs are reported in 2013 US dollars.
The project was approved by the Institutional Review Board of each State PHL. The CDC approved the project as post-marketing surveillance because routine de-identified data was submitted to the CDC by the contractor, John Snow Inc.
RESULTS
HIV testing outcomes
Among 7,914 specimens tested from Massachusetts PHL, 6,069 from Michigan PHL and 36,266 from Maryland PHL, 246 (3.11%), 163 (2.68%), and 983 (2.71%) HIV-1 antibody-confirmed infections, and 8 (0.10%), 1 (0.02%) and 19 (0.05%) acute infections were identified, respectively (Table 1). For Massachusetts and Maryland PHL, 1 specimen each was identified as HIV-2 positive. There were 7 (0.09%), 5 (0.08%) and 29 (0.08%) specimens identified as false positive in Massachusetts, Michigan, and Maryland PHL respectively. The specificity of the fourth-generation IAs used in each of the three PHLs was 99.9%.
Table 1.
Results of HIV testing in three U.S. State Public Health Laboratories.
| Massachusetts State Public Health Laboratory, Boston MA | Michigan State Public Health Laboratory, Lansing MI | Maryland State Public Health Laboratory, Baltimore MD | |
|---|---|---|---|
| Results of fourth-generation immunoassay screening | Bio-Rad GS HIV Antigen/Antibody Combo | Bio-Rad GS HIV Antigen/Antibody Combo | Abbott Architect HIV Antigen/Antibody Combo |
|
| |||
| Prospective time period | July 1, 2012-June 30, 2013 | November 27, 2012-June 30, 2013 | July 1, 2012-June 30, 2013 |
|
| |||
| Immunoassay test results | |||
| Total | 7914 | 6069 | 36266 |
| Non-reactive | 7652 (96.69%) | 5898 (97.18%) | 35232 (97.15%) |
| Repeatedly reactive | 262 | 171 | 1034 |
|
| |||
| Results of supplemental testing in specimens with repeatedly reactive results | |||
|
| |||
| Confirmed HIV-1 antibody positivea | 246 (3.11%) | 163 (2.68%) | 983 (2.71%) |
|
|
|||
| Acute infectionb | 8 (0.10%) | 1 (0.02%) | 19 (0.05%) |
|
|
|||
| IA false positivec | 7 (0.09%) | 5 (0.08%) | 29 (0.08%) |
|
|
|||
| HIV-2 positived | 1 (0.01%) | 0 (0.00%) | 1 (0.003%) |
|
|
|||
| Other | 0 (0.00%) | 2 (0.03%)e | 2 (0.005%)f |
|
| |||
| Specificity of the fourth-generation immunoassay | 99.9% | 99.9% | 99.9% |
HIV-1 positive by Western blot (WB) or supplemental HIV-1/HIV-2 antibody test
4th generation immunoassay (IA) reactive, WB or HIV-1/HIV-2 differentiation IA negative or indeterminate, reactive nucleic acid test (NAT)
4th generation IA reactive, WB or HIV-1/HIV-2 differentiation IA negative or indeterminate, NAT not reactive
Positive for HIV-2 by HIV-1/HIV-2 differentiation IA or HIV-2 WB or differential HIV-1/HIV-2 proviral DNA conventional polymerase chain reaction
IA reactive, HIV-1/HIV-2 differentiation IA undifferentiated, WB positive, NAT not reactive (n=1); IA reactive, HIV-1/HIV-2 differentiation IA undifferentiated, NAT not reactive (n=1)
IA reactive, WB indeterminate, HIV-1/HIV-2 differentiation IA positive, NAT not reactive (n=1); IA reactive, WB indeterminate, HIV-1/HIV-2 differentiation IA negative, NAT not reactive (n=1)
Time to reporting of HIV test results
The median time from specimen receipt to laboratory reporting of test results for HIV-1 antibody-confirmed infections was 3, 2 and 3 days, and for acute infections was 8, 11 and 7 days for Massachusetts, Michigan, and Maryland PHLs respectively (Table 2). In Massachusetts and Maryland PHLs, the median time from specimen collection to receipt of test results by client for HIV-1 antibody-confirmed infections was 8 and 14 days, and for acute infections were 10 and 13 days respectively.
Table 2.
Time to reporting of HIV test results in three U.S. State Public Health Laboratories.
| Time interval | Massachusetts State Public Health Laboratory, Boston MA | Michigan State Public Health Laboratory, Lansing MI | Maryland State Public Health Laboratory, Baltimore MD | |||
|---|---|---|---|---|---|---|
|
| ||||||
| n/Na | Median days (range) | n/Na | Median days (range) | n/Na | Median days (range) | |
| Specimen collection to specimen receipt in laboratory | 7914/7914 | 1 (0–10) | 6068/6069 | 3 (0–32) | 36261/36266 | 1 (0–15) |
|
| ||||||
| Receipt of specimen in laboratory to reporting of test results for HIV-1/2 negative specimens | 7652/7652 | 2 (0–14) | 5888/5898 | 1 (0–18) | 35229/35232 | 1 (0–15) |
|
| ||||||
| Receipt of specimen to laboratory reporting of test results for confirmed HIV-1 antibody positive specimens | 246/246 | 3 (0–8) | 163/163 | 2 (0–5) | 981/983 | 3 (0–12) |
|
| ||||||
| Receipt of specimen to laboratory reporting of test results for acute HIV-1 positive specimens | 8/8 | 8 (5–12) | 11b | 19/19 | 7 (3–12) | |
|
| ||||||
| Reporting of test results to receipt of test results by client | ||||||
|
| ||||||
| For acutes | 5/8c | 3 (0–27) | - | 3/19e | 5 (0–5) | |
|
| ||||||
| For confirmed HIV-1 antibody positive | 158/246d | 5 (0–52) | - | 136/983f | 9 (0–128) | |
|
| ||||||
| Total time from specimen collection to receipt of test results by client | ||||||
|
| ||||||
| For acutes | 7/8c | 10 (6–39) | - | 3/19e | 13 (9–13) | |
| For confirmed HIV-1 antibody positive | 169/246d | 8 (0–54) | - | 136/983f | 14 (3–134) | |
There was missing data for timing of results for some specimens.
Only one acute infection was detected in Michigan State Public Health Laboratory.
7 (87.5%) specimens have documentation that the results were delivered to the clients.
169 (68.7%) specimens have documentation that the results were delivered to the clients.
Data on the provision of results to clients were only available for 3 of the 19 specimens (15.8%).
Data on the provision of results to clients were only available for 166 of 983 specimens (16.9%).
Cost per HIV positive infection
The unadjusted and adjusted testing costs, labor costs and total costs by test in each participating PHL are shown in Table 3. Overall, the cost per HIV-positive specimen was $336 in Massachusetts PHL, $263 in Michigan PHL and $210 in Maryland PHL. The cost per specimen tested was $10 in Massachusetts PHL, $7 in Michigan PHL and $5 in Maryland PHL.
Table 3.
Unadjusted and adjusted reagent costs and total laboratory costs by test, and cost per specimen and per HIV-positive specimen in three U.S. State Public Health Laboratories.
| Site/Type of Test | Test costs per specimen $ (U.S) | Total specimens | Total tests cost (U.S. $) | Labor cost per specimen (U.S. $)¥ | Total labor cost (U.S. $)††† | Total cost (tests and labor cost) (U.S. $) | Total cost of HIV testing algorithm (tests and labor cost) (U.S. $) | Total cost per specimen (HIV-1 and HIV-2) (U.S. $) | Total positive specimens in HIV testing algorithm | Total cost per positive (HIV-1 and HIV-2) (U.S. $) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cost per test | Adjusted cost per test§ | ||||||||||
| Massachusetts State Public Health Laboratory, Boston MA | |||||||||||
| GS HIV Combo Ag/Ab EIA | 2.78 | 3.44 | 8438 | 29025.80†† | 3.36 | 28376.63 | 57402.44 | ||||
| Multispot HIV-1/HIV-2 Rapid Test | 26.78 | 80.34 | 262 | 21049.08 | 24.44 | 6403.07 | 27452.15 | ||||
| APTIMA HIV-1 RNA Qualitative Nucleic Acid Test (NAT) | 40.00 | 54.55 | 15 | 818.18 | 7.52 | 112.78 | 930.96 | ||||
| 85785.55 | 10.16 | 255 | 336.41 | ||||||||
|
| |||||||||||
| Michigan State Public Health Laboratory, Lansing MI † | |||||||||||
| GS HIV Combo Ag/Ab EIA | 3.50 | 4.46 | 6411 | 28609.09†† | 1.15 | 7388.11 | 35997.20 | ||||
| Multispot HIV-1/HIV-2 Rapid Test | 25.70 | 35.98 | 170 | 6116.60 | 2.99 | 509.05 | 6625.65 | ||||
| APTIMA HIV-1 RNA Qualitative Nucleic Acid Test (NAT) | 8 | 720.00 | - | 0 | 720.00 | ||||||
| 43342.85 | 6.76 | 165 | 262.68 | ||||||||
|
| |||||||||||
| Maryland State Public Health Laboratory, Baltimore MD | |||||||||||
| Architect HIV Ag/Ab Combo | 3.00 | 3.09 | 38334 | 118287.77†† | 0.55 | 21223.98 | 139511.75 | ||||
| GS HIV-1 Western blot | 30.33 | 37.91 | 1034 | 39195.06 | 6.70 | 6931.42 | 46126.48 | ||||
| Multispot HIV-1/HIV-2 Rapid Test | 23.52 | 33.60 | 574 | 19286.40 | 2.55 | 1462.17 | 20748.57 | ||||
| SIV Western blot Assay (HIV-2 WB) Differential | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ||||
| HIV-1/HIV-2 proviral DNA PCR | 22.00 | 1 | 22.00 | 74.63 | 74.48 | 96.48 | |||||
| LTR real time PCR (HIV-1 NAT) | 20.00 | 51 | 1020.00 | 15.90 | 811.19 | 1831.19 | |||||
| Real time PCR gag (HIV-1 NAT) | 6.00 | 19 | 114.00 | 9.98 | 189.62 | 303.62 | |||||
| Real time PCR pol (HIV-1 NAT) | 6.00 | 19 | 114.00 | 9.98 | 189.62 | 303.62 | |||||
| bDNA (HIV-1 NAT) | 72.00 | 19 | 1368.00 | 1.12 | 21.33 | 1389.33 | |||||
| 210311.04 | 5.49 | 1003 | 209.68 | ||||||||
Total kit cost plus cost of controls not provided in kit divided by number of tests not used as controls.
The product of labor hours per specimen and labor costs per hour.
In MI PHL, the estimated cost reimbursed to Florida PHL as part of a CDC/Association of Public Health Laboratories (APHL) NAT demonstration project was $90.
The numbers account for specimens that were run in duplicate when the initial test was reactive.
Labor cost per hour was determined by Bureau of Labor Statistics' estimate for the 2013 mean salary (including benefits). Labor hours per test were calculated either by being directly observed at the laboratory or from the estimates from literature.
-All costs are reported in 2013 US dollars
DISCUSSION
In the current study, acute infections were detected in all laboratories, highlighting the utility of using fourth-generation assays in a variety of settings in the United States. Unlike the previous algorithm, which only detected established HIV-1 infection, the current algorithm works well in high-risk individuals [4], and detects acute [3, 7] and established HIV-1 infections [5]. It also correctly identifies HIV-2 infections that would be missed or misclassified as HIV-1 by WB [6, 31]. The current study provides further evidence that using the recommended algorithm in U.S. PHLs can help in diagnosing infections early, although fourth-generation IAs detect approximately 80% of acute HIV infections [9–11].
Our study showed that the median time from specimen receipt to laboratory reporting of test results for acute infections was at least 7 days in the three PHLs, and for Massachusetts and Maryland PHL the median time from specimen collection to receipt of test results by clients for acute infections was 10 and 13 days respectively. However, documentation of the receipt of test results was not available for many infected individuals. Individuals in the acute stage of HIV infection are usually highly viremic, resulting in more virus shedding at mucosal sites, and are therefore more likely to transmit HIV infection through bodily secretions [32]. Data from previous studies suggest that the rate of sexual transmission during acute infection is up to 26 times as high as that during established HIV-1 infection [33]. Similarly, it has been estimated that the proportion of total HIV transmissions attributable to early infections ranges between 6% and 49.4% [34–36]. Massachusetts Department of Public Health reported that the recommended algorithm reduced the time to reporting of HIV test results for antibody-positive specimens by 1 week compared with their previous algorithm [37]. Nonetheless, time to reporting of acute HIV test results to clients was suboptimal, and needs to be streamlined for timely interruption of transmission. Further, in order to use the recommended algorithm to its full advantage to detect early infections and reduce transmissions, PHLs must be prepared to conduct NAT with rapid turn-around. More affordable NAT options, such as diagnostic point of care NATs, are needed in the U.S. to rapidly identify acute infections. Public health systems must be in place to conduct timely linkage to medical care and partner services [38].
Estimating the cost per positive of the HIV diagnostic algorithm is challenging because of differences in laboratory HIV testing practices and volumes, and the prevalence of established and acute HIV infections [16]. Maryland PHL likely had the lowest cost per positive as compared to Massachusetts and Michigan PHLs because of the high number of confirmed HIV-1 antibody tests. However, when estimating the cost per specimen, the cost was still lowest for Maryland followed by Michigan and Massachusetts probably because of the lower adjusted cost for the screening test. High-volume laboratories are more likely to access volume pricing of the screening IAs than medium-volume laboratories [16]. In terms of the supplemental tests, Massachusetts ran controls each time the Multispot was run, which increased the cost but provided results to clients in a faster time frame for action at that site. However, Maryland and Michigan PHLs batched Multispot, which decreased their cost per HIV-positive specimen, but increased turn-around time. PHLs should assess whether excess controls are being used and develop an Individualized Quality Control Plan [39] for Multispot that follows the manufacturer's package insert. As in the Maryland PHL, a previous study has shown that for HIV-antibody positive specimens, laboratory costs in medium- (<50,000 IAs/year) and high-volume (≥50,000 IAs/year) laboratories for the recommended algorithm were generally less than those for the previous algorithm [16]. NAT is expensive, but only has to be used after a reactive fourth-generation IA, and negative or indeterminate antibody differentiation test to distinguish specimens with acute infection from false-positive IAs. Maryland PHL used validated in-house HIV-1 NATs and HIV-1/HIV-2 conventional PCR assays, which were less expensive to run than commercial assays. PHLs without NAT that wish to use the recommended algorithm may need to partner with other PHLs or clinical sites with access to a NAT.
Our study is subject to a few limitations. Firstly, comparison across sites regarding the cost and time to reporting of test results may not be reliable because of variations in the HIV diagnostic algorithms used by the participating PHLs. Secondly, the analysis is based on specimens and not individuals; there may be instances where more than one specimen belongs to the same individual. Thirdly, although Maryland PHL used a different HIV testing algorithm than the recommended algorithm, they used fourth-generation IA, an HIV-1/HIV-2 differentiation antibody IA, and a HIV-1 NAT that helped identifying acute infections. Lastly, the information on time from specimen receipt to receipt of test results by clients is missing for a large number of test results, therefore the results should be interpreted with caution.
To conclude, fourth-generation IA, coupled with antibody tests and NAT detected acute infections in PHLs that may have been missed if WB was the only confirmatory test in use. Algorithm costs were driven by the screening fourth-generation IA in the PHLs; reduction in their cost may help reduce the overall laboratory cost of diagnosing HIV infections. The identification of acute infection in PHLs can be coupled with public health intervention in order to interrupt disease transmission.
ACKNOWLEDGMENTS
We thank:
Centers for Disease Control and Prevention: Mr. Emeka Oraka for data analysis support, and Kristina Bowles for acting as Contracting Officer Representative (COR) for the project.
John Snow Inc.: Jeanne Day and Amy Flynn
Supporting personnel at Massachusetts Department of Public Health: Tracy Stiles, Arthur Kazianis, Paul Borne, Corrie Noonan, Betsey John, Maura Miminos, and Monica Morrison.
Supporting personnel at Michigan Department Health and Human Services: Bruce A. Robeson, and Robert R. White
Supporting personnel at Maryland Department of Health and Mental Hygiene: Michele Rand, Hope Cassidy-Stewart, and Steven Montgomery for compiling and reviewing the data and staffs of the Retrovirology and Viral Disease Assessment Laboratories for their technical contributions
FUNDING The project was funded by the U.S. Centers for Disease Control and Prevention (Task Order #200-2009-30955-0003).
Footnotes
Conflict of interest statement: No financial disclosures were reported by the authors of this paper.
Publisher's Disclaimer: The findings and conclusions in this article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services.
REFERENCES
- 1.Centers for Disease Control and Prevention and Association of Public Health Laboratories Laboratory Testing for the Diagnosis of HIV Infection: Updated Recommendations. 2014 [cited 2015 February 9]; Available from: http://stacks.cdc.gov/view/cdc/23447.
- 2.Centers for Disease Control and Prevention Interpretation and use of the western blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. MMWR Morb Mortal Wkly Rep. 1989;38:1–7. [PubMed] [Google Scholar]
- 3.Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol. 2011;52(Suppl 1):S17–22. doi: 10.1016/j.jcv.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Delaney KP, Heffelfinger JD, Wesolowski LG, Owen SM, Meyer WA, 3rd, Kennedy S, et al. Performance of an alternative laboratory-based algorithm for HIV diagnosis in a high-risk population. J Clin Virol. 2011;52(Suppl 1):S5–10. doi: 10.1016/j.jcv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Wesolowski LG, Delaney KP, Hart C, Dawson C, Owen SM, Candal D, et al. Performance of an alternative laboratory-based algorithm for diagnosis of HIV infection utilizing a third generation immunoassay, a rapid HIV-1/HIV-2 differentiation test and a DNA or RNA-based nucleic acid amplification test in persons with established HIV-1 infection and blood donors. J Clin Virol. 2011;52(Suppl 1):S45–9. doi: 10.1016/j.jcv.2011.09.026. [DOI] [PubMed] [Google Scholar]
- 6.Nasrullah M, Ethridge SF, Delaney KP, Wesolowski LG, Granade TC, Schwendemann J, et al. Comparison of alternative interpretive criteria for the HIV-1 Western blot and results of the Multispot HIV-1/HIV-2 Rapid Test for classifying HIV-1 and HIV-2 infections. J Clin Virol. 2011;52(Suppl 1):S23–7. doi: 10.1016/j.jcv.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 7.Nasrullah M, Wesolowski LG, Meyer WA, 3rd, Owen SM, Masciotra S, Vorwald C, et al. Performance of a fourth-generation HIV screening assay and an alternative HIV diagnostic testing algorithm. AIDS. 2013;27:731–7. doi: 10.1097/QAD.0b013e32835bc535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ly TD, Ebel A, Faucher V, Fihman V, Laperche S. Could the new HIV combined p24 antigen and antibody assays replace p24 antigen specific assays? J Virol Methods. 2007;143:86–94. doi: 10.1016/j.jviromet.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Pandori MW, Hackett J, Jr., Louie B, Vallari A, Dowling T, Liska S, et al. Assessment of the ability of a fourth-generation immunoassay for human immunodeficiency virus (HIV) antibody and p24 antigen to detect both acute and recent HIV infections in a high-risk setting. J Clin Microbiol. 2009;47:2639–42. doi: 10.1128/JCM.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chavez P, Wesolowski L, Patel P, Delaney K, Owen SM. Evaluation of the performance of the Abbott ARCHITECT HIV Ag/Ab Combo Assay. J Clin Virol. 2011;52(Suppl 1):S51–5. doi: 10.1016/j.jcv.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Bentsen C, McLaughlin L, Mitchell E, Ferrera C, Liska S, Myers R, et al. Performance evaluation of the Bio-Rad Laboratories GS HIV Combo Ag/Ab EIA, a 4th generation HIV assay for the simultaneous detection of HIV p24 antigen and antibodies to HIV-1 (groups M and O) and HIV-2 in human serum or plasma. J Clin Virol. 2011;52(Suppl 1):S57–61. doi: 10.1016/j.jcv.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 12.Bennett B, Neumann D, Fordan S, Villaraza R, Crowe S, Gillis L. Performance of the new HIV-1/2 diagnostic algorithm in Florida's public health testing population: a review of the first five months of utilization. J Clin Virol. 2013;58(Suppl 1):e29–33. doi: 10.1016/j.jcv.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 13.Lee K, Park HD, Kang ES. Reduction of the HIV seroconversion window period and false positive rate by using ADVIA Centaur HIV antigen/antibody combo assay. Ann Lab Med. 2013;33:420–5. doi: 10.3343/alm.2013.33.6.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muhlbacher A, Schennach H, van Helden J, Hebell T, Pantaleo G, Burgisser P, et al. Performance evaluation of a new fourth-generation HIV combination antigen-antibody assay. Med Microbiol Immunol. 2013;202:77–86. doi: 10.1007/s00430-012-0250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365:493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchinson AB, Ethridge SF, Wesolowski LG, Shrestha RK, Pentella M, Bennett B, et al. Costs and outcomes of laboratory diagnostic algorithms for the detection of HIV. J Clin Virol. 2013;58(Suppl 1):e2–7. doi: 10.1016/j.jcv.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 17.Bio-Rad Laboratories . Product insert, GS HIV Combo Ag/Ab EIA. Bio-Rad; Redmond, WA, USA: 2011. [Google Scholar]
- 18.Bio-Rad Laboratories . Product insert, Multispot HIV-1/HIV-2 Rapid Test. Bio-Rad; Redmond, WA, USA: 2004. [Google Scholar]
- 19.Gen-Probe Incorporated . Product insert, APTIMA HIV-1 RNA Qualitative assay. Gen-Probe Incorporated; San Diego, CA, USA: 2006. [Google Scholar]
- 20.Wesolowski LG, Wroblewski K, Bennett SB, Parker MM, Hagan C, Ethridge SF, et al. Nucleic acid testing by public health referral laboratories for publichealth laboratories using the U.S. HIV diagnostic testing algorithm. J Clin Virol. 2015 doi: 10.1016/j.jcv.2015.01.017. 10.1016/j.jcv.2015.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbott Laboratories . Product insert, Architect HIV Ag/Ab Combo. Abbott Laboratories; Abbot Park, IL, USA: 2010. [Google Scholar]
- 22.Bio-Rad Laboratories . Product insert, Human Immunodeficiency Virus Type1, Genetic Systems HIV-1 Western Blot. Bio-Rad; Redmond, WA, USA: 2004. [Google Scholar]
- 23.Kozaczynska K, Cornelissen M, Reiss P, Zorgdrager F, van der Kuyl AC. HIV-1 sequence evolution in vivo after superinfection with three viral strains. Retrovirology. 2007;4:59. doi: 10.1186/1742-4690-4-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers RA. HIV NAAT testing of HIV antibody negative samples. 2005 [cited 2015 March 31]; Available from: http://www.hivtestingconference.org/hivtesting2005/Conference-Abstracts.htm.
- 25.Drosten C, Seifried E, Roth WK. TaqMan 5'-nuclease human immunodeficiency virus type 1 PCR assay with phage-packaged competitive internal control for high-throughput blood donor screening. J Clin Microbiol. 2001;39:4302–8. doi: 10.1128/JCM.39.12.4302-4308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo W, Yang H, Rathbun K, Pau CP, Ou CY. Detection of human immunodeficiency virus type 1 DNA in dried blood spots by a duplex real-time PCR assay. J Clin Microbiol. 2005;43:1851–7. doi: 10.1128/JCM.43.4.1851-1857.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allan JS, Short M, Taylor ME, Su S, Hirsch VM, Johnson PR, et al. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J Virol. 1991;65:2816–28. doi: 10.1128/jvi.65.6.2816-2828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao F, Yue L, Robertson DL, Hill SC, Hui H, Biggar RJ, et al. Genetic diversity of human immunodeficiency virus type 2: evidence for distinct sequence subtypes with differences in virus biology. J Virol. 1994;68:7433–47. doi: 10.1128/jvi.68.11.7433-7447.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ZeptoMetrix Corporation . Product insert, SIV Western Blot Assay. ZeptoMetrix Corporation; Buffalo, NY: [cited 2015 November 14]; Available from: http://www.zeptometrix.com/docs/PI0801500.pdf?1456512898. [Google Scholar]
- 30.Bureau of Labor Statistics. Occupational employment statistics. United States Department of Labor; 2014. [cited 2014 November 14]; Available from: http://www.bls.gov/oes/tables.htm. [Google Scholar]
- 31.Torian LV, Forgione LA, Punsalang AE, Pirillo RE, Oleszko WR. Comparison of Multispot EIA with Western blot for confirmatory serodiagnosis of HIV. J Clin Virol. 2011;52(Suppl 1):S41–4. doi: 10.1016/j.jcv.2011.09.017. [DOI] [PubMed] [Google Scholar]
- 32.Pilcher CD, Eron JJ, Jr., Galvin S, Gay C, Cohen MS. Acute HIV revisited: new opportunities for treatment and prevention. J Clin Invest. 2004;113:937–45. doi: 10.1172/JCI21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hollingsworth TD, Anderson RM, Fraser C. HIV-1 transmission, by stage of infection. J Infect Dis. 2008;198:687–93. doi: 10.1086/590501. [DOI] [PubMed] [Google Scholar]
- 34.Pinkerton SD. How many sexually-acquired HIV infections in the USA are due to acute-phase HIV transmission? AIDS. 2007;21:1625–9. doi: 10.1097/QAD.0b013e32826fb6a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiridou M, Geskus R, de Wit J, Coutinho R, Kretzschmar M. Primary HIV infection as source of HIV transmission within steady and casual partnerships among homosexual men. AIDS. 2004;18:1311–20. doi: 10.1097/00002030-200406180-00010. [DOI] [PubMed] [Google Scholar]
- 36.Brenner BG, Roger M, Routy JP, Moisi D, Ntemgwa M, Matte C, et al. High rates of forward transmission events after acute/early HIV-1 infection. J Infect Dis. 2007;195:951–9. doi: 10.1086/512088. [DOI] [PubMed] [Google Scholar]
- 37.Goodhue T, Kazianis A, Werner BG, Stiles T, Callis BP, Dawn Fukuda H, et al. 4th generation HIV screening in Massachusetts: a partnership between laboratory and program. J Clin Virol. 2013;58(Suppl 1):e13–8. doi: 10.1016/j.jcv.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Kelly JA, Morin SF, Remien RH, Steward WT, Higgins JA, Seal DW, et al. Lessons learned about behavioral science and acute/early HIV infection. The NIMH Multisite Acute HIV Infection Study: V. AIDS Behav. 2009;13:1068–74. doi: 10.1007/s10461-009-9579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Center for Medicare and Medicaid Services Individualized Quality Control Plan (IQCP) 2015 [cited 2015 August 17]; Available from: https://www.cms.gov/Regulations-and-Guidance/Legislation/CLIA/Individualized_Quality_Control_Plan_IQCP.html.