Abstract
Background
Polymalic acid (PMA) is a novel polyester polymer that has been broadly used in the medical and food industries. Its monomer, L-malic acid, is also a potential C4 platform chemical. However, little is known about the mechanism of PMA biosynthesis in the yeast-like fungus, Aureobasidium pullulans. In this study, the effects of different nitrogen concentration on cell growth and PMA biosynthesis were investigated via comparative transcriptomics and proteomics analyses, and a related signaling pathway was also evaluated.
Results
A high final PMA titer of 44.00 ± 3.65 g/L (49.9 ± 4.14 g/L of malic acid after hydrolysis) was achieved in a 5-L fermentor under low nitrogen concentration (2 g/L of NH4NO3), which was 18.3 % higher yield than that obtained under high nitrogen concentration (10 g/L of NH4NO3). Comparative transcriptomics profiling revealed that a set of genes, related to the ribosome, ribosome biogenesis, proteasome, and nitrogen metabolism, were significantly up- or down-regulated under nitrogen sufficient conditions, which could be regulated by the TOR signaling pathway. Fourteen protein spots were identified via proteomics analysis, and were found to be associated with cell division and growth, energy metabolism, and the glycolytic pathway. qRT-PCR further confirmed that the expression levels of key genes involved in the PMA biosynthetic pathway (GLK, CS, FUM, DAT, and MCL) and the TOR signaling pathway (GS, TOR1, Tap42, and Gat1) were upregulated due to nitrogen limitation. Under rapamycin stress, PMA biosynthesis was obviously inhibited in a dose-dependent manner, and the transcription levels of TOR1, MCL, and DAT were also downregulated.
Conclusions
The level of nitrogen could regulate cell growth and PMA biosynthesis. Low concentration of nitrogen was beneficial for PMA biosynthesis, which could upregulate the expression of key genes involved in the PMA biosynthesis pathway. Cell growth and PMA biosynthesis might be mediated by the TOR signaling pathway in response to nitrogen. This study will help us to deeply understand the molecular mechanisms of PMA biosynthesis, and to develop an effective process for the production of PMA and malic acid chemicals.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0547-y) contains supplementary material, which is available to authorized users.
Keywords: Aureobasidium pullulans, Nitrogen availability, Polymalic acid, Proteomics, TOR signaling pathway, Transcriptomics
Background
Polymalic acid (PMA) is a novel polyester polymer composed of L-malic acid as the sole monomer [1, 2]. Due to its good biodegradability, water-solubility, and biocompatibility, it can be used as a new functional material for drug delivery, tissue scaffolds, and food materials, etc. [3, 4]. Its monomer, L-malic acid, which was widely used in the food and pharmaceutical industries, also does not demonstrate toxicity or immunogenicity in the body. In our previous study, a novel process was developed through acid hydrolysis of PMA for the production of L-malic acid [5]. In recent years, PMA and L-malic acid have attracted much attention due to their wide application.
Currently, PMA is mainly produced using the microorganism Aureobasidium pullulans. A. pullulans is a black yeast-like species that is popularly known as secreting melanin. Aureobasidium spp. are ubiquitous species that have evolved an extraordinary tolerance for a broad range of ecological conditions and can be isolated from pollen, leaves, and even antarctic soils, and deep sea water [6]. It is regarded as a polyextremotolerant organism that can survive in hypersaline, acidic and basic, cold and oligotrophic conditions. Due to its inherent universality, A. pullulans can produce a variety of metabolites, such as PMA, pullulan, melanin, amylase, proteinase, xylanase, liamocins, etc. [7, 8].
Culture stress is becoming an efficient strategy for the overproduction of various metabolites by microorganisms. Some genera of filamentous fungi, e.g., Aspergillus and Rhizopus, are known to produce large quantities of malic and fumaric acids and secrete them into the culture broth when cultured under stress conditions [9, 10]. Under nitrogen limitation, these fungi accumulate fumaric and malic acids as the end products of the reductive TCA cycle in the cytosol [11]. In addition, nitrogen limitation has become an important approach for regulating the biosynthesis of some biopolymers, such as polyhydroxyalkanoates (PHA) [12] and poly-3-hydroxybutyrate (PHB) [13]. In our previous study, ammonium nitrate (NH4NO3) was used as the most optimal nitrogen source for the production of PMA [5, 14, 15]; however, cell growth and PMA biosynthesis in response to nitrogen is not yet well understood.
In eukaryotic cells, most of the regulatory events that are induced by nitrogen availability are mediated through “TOR” (target of rapamycin) kinase and the downstream TOR signaling pathway. TOR is a conserved Ser/Thr kinase and plays a crucial role as a global regulator in cell growth and metabolism by sensing and integrating a variety of inputs arising from amino acids, cellular stresses, energy status, and growth factors [16–18]. In Fusarium fujikuroi, Teichert et al. firstly found that TOR kinase was involved in the nitrogen regulation of secondary metabolism. Inhibition of TOR by rapamycin could affect the expression of AreA-controlled secondary metabolite genes involved in the gibberellins (GAs) and bikaverin biosynthesis pathway [19]. Moreover, the TOR pathway was also confirmed to participate in regulating the virulence of Fusarium graminearum [20].
Although PMA has attracted much attention in different fields, the molecular basis of PMA biosynthesis is still unclear. In this study, the effects of different levels of NH4NO3 on cell growth and PMA biosynthesis were investigated on different scales. Comparative transcriptomics and proteomics analyses were used for a global understanding of the nitrogen response. Under rapamycin stress, the outputs of the TOR signaling pathway in A. pullulans were to determine its role in regulating cell growth and PMA biosynthesis.
Methods
Cultures and media
The strain A. pullulans CCTCC M2012223 was isolated by our laboratory and can be obtained from the China Center for Type Culture Collection (Wuhan, China). This strain was maintained on the PDA slant. The seed culture medium contained 60, 2, 0.1, 0.1, 0.1, 0.5 and 20 g/L of glucose, NH4NO3, KH2PO4, MgSO4, ZnSO4, KCl and CaCO3, respectively. The seed culture was grown in a 500-mL shake flask containing 50 mL of liquid medium, and was incubated at 25 °C in a rotary shaker (180 rpm) for 2 days. The fermentation medium contained 90, 0.1, 0.1, 0.1, 0.5 and 30 g/L of glucose, KH2PO4, MgSO4, ZnSO4, KCl and CaCO3, respectively.
Fermentation in shake flask and fermentor
In order to evaluate the effect of nitrogen concentrations on cell growth and PMA biosynthesis in different scales, the different levels of NH4NO3 from 0.1 to 10.0 g/L were taken in the initial fermentation medium, respectively. The shake flask fermentation was inoculated with 10 % (v/v) of the above-described seed culture medium and kept at 25 °C with shaking at 220 rpm for 4 day. Batch fermentation kinetics was studied in a 5-L stirred-tank fermentor (Shanghai Baoxing Co. Ltd, China) containing 3 L of the fermentation medium, as well as adding 0.1, 2 or 10 g/L of NH4NO3. The fermentation was inoculated with 300 mL of seed culture grown in a shake flask for 48 h, and operated at 25 °C with agitation and aeration at 400–600 rpm and 1.3 vvm, respectively. All trials were performed in triplicate.
Transcriptomics analysis
As for transcriptomics and proteomics analyses, three independent cell samples under the condition of nitrogen limitation (2 g/L) and nitrogen repletion (10 g/L) were harvested from 5-L stirred-tank fermentor at 36 h, respectively. Total RNA was extracted using Fungal RNA Kit (Omega, USA), and the quality of extracted RNA was measured by NanoDrop 2000 (Thermo scientific, USA). After examination, the magnetic beads with Oligo (dT) are used to isolate mRNA. Mixed with the fragmentation buffer, the mRNA is fragmented into short fragments. Then cDNA is synthesized using the mRNA fragments as templates. Short fragments are purified and resolved with EB buffer for end reparation and single nucleotide A (adenine) addition. After that, short fragments are connected with adapters. The suitable fragments are selected for the PCR amplification as templates. At last, the library was sequenced using Illumina HiSeq™ 2500 equipment. The transcriptome data set of A. pullulans was deposited at NCBI Sequence Read Archive database (SRA—http://www.ncbi.nlm.nih.gov/Traces/sra/) with the BioProject ID PRJNA301913.
In a comparison analysis, two-class unpaired method in the significant analysis of microarray software (SAM, version 3.02) was performed to identify significantly differentially expressed genes between nitrogen limited and sufficient groups, determining with a selection threshold of false discovery rate, FDR <5 % and fold change ≥2. Raw data was log2-transformed and imported.
Protein extraction and two-dimensional (2-D) electrophoresis
Total protein extracts from cells under nitrogen limited and sufficient conditions were prepared via the phenol extraction method. Proteins were separated by two-dimensional gel electrophoresis (2-DE), and the protein spots were visualized by silver staining. For each sample, at least three independent protein extracts and two 2-DE analyses were performed.
2-D gel electrophoresis analysis
In order to analyze the expressed protein patterns, the silver stained gels were scanned using a PowerLook 1100 scanner (UMAX, Taiwan), and protein spots images were analyzed using GE HealthCare software (Amersham Biosciences, Sweden). The protein spots with at least twofold differences in absolute abundance and reproducible changes were considered as the candidate proteins. These protein spots on the corresponding Coomassie-stained gels were excised and digested with trypsin using a Spot Handling Workstation (Amersham Biosciences, Sweden). The digested peptide masses were measured using a MALDI-TOF-TOF mass spectrometer (ABI 4700 system, USA). Data were processed via the Data Explorer software and proteins were identified by searching against a comprehensive non-redundant sequence database using the MASCOT search engine.
Quantitative RT-PCR
The transcription levels of key genes involved in PMA biosynthetic pathway (e.g., GLK, CS, FUM, DAT and MCL) and TOR signaling pathway (e.g., GS, TOR1, Tap42 and Gat1) were tested by quantitative RT-PCR. Total RNA samples from nitrogen limited or nitrogen sufficient fermentation at 36 h were extracted using Fungal RNA Kit (Omega, USA) and reversed to cDNA using reverse transcriptase (Takara, Japan). Primers of different genes were shown in Additional file 1: Table S1. The experiment was repeated three times. The quantitative PCR assay was performed according to Sybr Green method (qPCR Master Mix, ABI, USA) using fluorescence quantitative PCR (ABI, USA).
Rapamycin sensitivity
In order to evaluate the effect of rapamycin on cell growth and PMA biosynthesis, A. pullulans cells were cultured on the plate and shake flask, respectively. In the plate culture, cells were spread on the PDA plate amended with rapamycin from 5 to 50 ng/mL. For the shake flask fermentation, the seed culture (10 %, v/v) was inoculated to 50 mL fermentation media containing various concentrations of rapamycin from 5 to 50 ng/mL. The fermentation cultivation was then operated at 25 °C with shaking at 180 rpm for 4 days. All trials were performed in triplicate. The cell samples at the plate with adding 10 ng/mL of rapamycin were extracted for quantitative PCR assay.
Metabolites analysis
The cell density was determined by the cell dry weight (DCW) method. Prior to the measurement, excess CaCO3 was eliminated from the broth by adding 1 M HCl. The cell suspension was centrifuged at 4000×g and then dried overnight at 105 °C. The concentration of residual sugar was measured using the dinitrosalicylic acid assay method [21]. The concentration of residual NH4+ in the broth was detected by phenol-hypochlorite reaction [22]. For the analysis of intracellular ATP/ADP and NADH/NAD+ ratio, the methods have been introduced in our previous reports [14]. The experiment was repeated three times.
PMA was analyzed by centrifuging the fermentation broth and then adding 1 mL of resulted supernatant to 1 mL of 2 M H2SO4 in an incubator at 85 °C for 8 h. After neutralization of the solution, the hydrolyzed sample was analyzed by HPLC (Agilent 1200, USA), using a Spursil C18-EP organic acid column at 40 °C eluted with 5 mM H2SO4 at a rate of 0.6 mL/min to determine its malic acid content [23].
Results
Investigation of nitrogen availability on PMA biosynthesis
In general, the utilization of a nitrogen source is essential to cell growth, however, the type of nitrogen source affects PMA biosynthesis [15]. In our previous study, NH4NO3 was selected as the best nitrogen source for the production of PMA [5]. Firstly, the effects of different NH4NO3 concentrations on cell growth and PMA biosynthesis were investigated in shake flasks. As shown in Table 1, for NH4NO3 concentrations within the range of 0.1 and 2 g/L, both cell growth and PMA production were gradually increased. The highest PMA titer reached 20.02 ± 2.81 at 2 g/L of NH4NO3, with a corresponding PMA yield (Yp/x) of 0. 96 g/g. However, when the NH4NO3 concentration was >2 g/L in the shake flasks, PMA biosynthesis seemed to be inhibited, as fermentation stopped and there was significant accumulation of residual glucose. At the high level of NH4NO3 (10 g/L), the PMA titer reached 16.57 ± 0.90 g/L, with a relatively low PMA yield (Yp/x) of 0. 69 g/g. It was concluded that the level of nitrogen (NH4NO3) in the media was an important factor to regulate PMA biosynthesis. Therefore, the kinetics of different levels of NH4NO3 (0.1, 2 and 10 g/L) were further investigated in a 5-L stirred-tank fermentor. It is evident from Fig. 1 that cell growth was associated with the level of nitrogen; the highest PMA titer of 44.0 ± 3.65 g/L (49.9 ± 4.14 g/L of malic acid after hydrolysis) was achieved at 2 g/L of NH4NO3 in 96 h, with a corresponding PMA productivity of 0.46 ± 0.04 g/L·h. At the lowest level of NH4NO3 (0.1 g/L), the NH4+ concentration was almost completely consumed at 12 h (only 0.05 mM), which did not support cell growth and seriously hindered PMA biosynthesis. However, at the highest level of NH4NO3 (10 g/L), although the cell growth was faster than that at the lower levels of NH4NO3 (0.1 or 2 g/L), the production of PMA was obviously decreased to 37.2 ± 4.58 g/L in 96 h, which might be attributed to the excess NH4+ concentration in the broth. It was noted that the NH4+ concentration was at a surplus until the end of fermentation (Fig. 1d). In comparison, at 2 g/L of NH4NO3, the NH4+ concentration was almost consumed up at 36 h (0.11 mM). These results indicated that high levels of nitrogen favor cell growth, while on the contrary, low levels of nitrogen are beneficial for PMA biosynthesis.
Table 1.
Effects of different levels of NH4NO3 on cell growth and PMA biosynthesis in shake flasks
| NH4NO3 (g/L) | Residual sugar (g/L) | Cell biomass (g/L) | PMA (g/L) | Productivity (g/L h) | Yield (YP/X, g/g) | Yield (YP/S, g/g) |
|---|---|---|---|---|---|---|
| 0.1 | 56.67 ± 0.50 | 6.23 ± 0.71 | 6.17 ± 0.29 | 0.06 ± 0.003 | 1.00 ± 0.137 | 0.18 ± 0.011 |
| 0.5 | 40.77 ± 0.12 | 13.09 ± 1.09 | 10.86 ± 1.91 | 0.09 ± 0.042 | 0.85 ± 0.246 | 0.25 ± 0.081 |
| 1.0 | 24.57 ± 0.31 | 17.17 ± 0.77 | 14.01 ± 0.04 | 0.15 ± 0.000 | 0.82 ± 0.037 | 0.21 ± 0.000 |
| 2.0 | 16.95 ± 1.91 | 21.31 ± 3.81 | 20.02 ± 2.81 | 0.21 ± 0.029 | 0.96 ± 0.242 | 0.24 ± 0.032 |
| 4.0 | 16.70 ± 0.35 | 23.55 ± 0.85 | 17.31 ± 0.24 | 0.18 ± 0.003 | 0.74 ± 0.024 | 0.24 ± 0.002 |
| 10.0 | 14.52 ± 0.58 | 24.20 ± 1.96 | 16.57 ± 0.90 | 0.17 ± 0.009 | 0.69 ± 0.081 | 0.22 ± 0.013 |
All the values are the means and standard deviations of three independent experiments. Yield (Yp/x): the ratio of PMA to cell biomass concentration (g/g); Yield (Yp/s): the ratio of PMA to consumed sugar concentration (g/g)
Fig. 1.
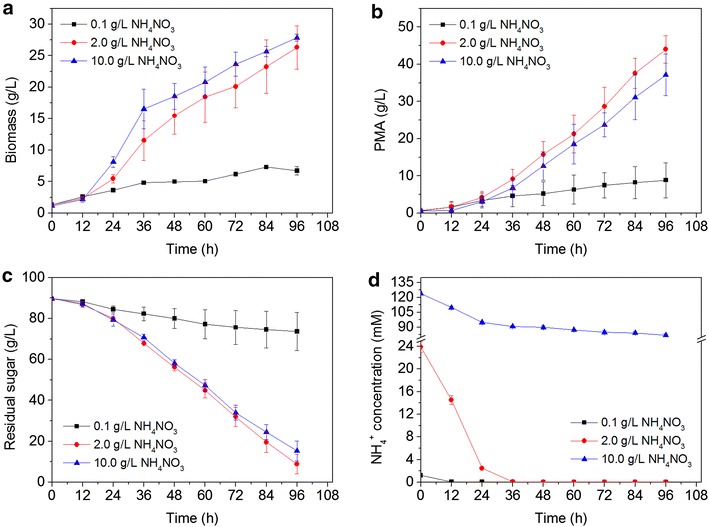
Time course of cell biomass (a), PMA production (b), residual sugar (c) and NH4 + concentration (d) under the different levels of NH4NO3 in a 5-L stirred-tank fermentor. Data are given as the average of triplicate experiments
Figure 2 showed intracellular variation in the ratios of NADH/NAD+ and ATP/ADP under different levels of NH4NO3. Compared to 0.1 g/L of NH4NO3, the ratio of ATP/ADP and NADH/NAD+ at 2 and 10 g/L of NH4NO3 remained at relatively high levels, respectively, which might be helpful for high cell growth and metabolism. However, the ratio of NADH/NAD+ at 10 g/L of NH4NO3 was obviously higher than that at 2 g/L of NH4NO3 during the whole fermentation process, which should generate more ATP necessary for cell metabolism through oxidative phosphorylation. However, the ratio of ATP/ADP at 10 g/L of NH4NO3 was lower than that at 2 g/L of NH4NO3. These results also revealed that the level of nitrogen might be involved in regulating the cofactor regeneration and energy supply during the fermentation process.
Fig. 2.
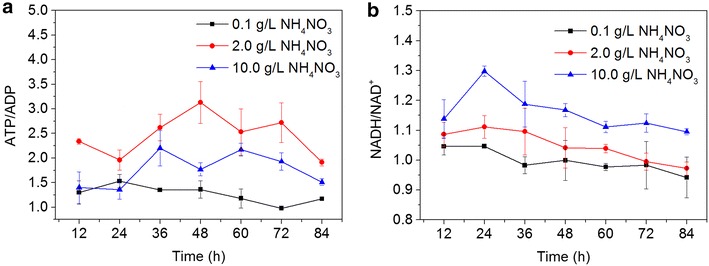
The ratios of ATP/ADP and NADH/NAD+ under the different levels of NH4NO3. a ATP/ADP; b NADH/NAD+. Data are given as the average of triplicate experiments
Transcriptomics and proteomics analysis
In order to further understand cell growth and PMA biosynthesis in response to nitrogen levels, the global transcriptional profiles under nitrogen repletion (10 g/L of NH4NO3) and nitrogen limitation (2 g/L of NH4NO3) were examined at 36 h. In this time node, the NH4+ concentration under nitrogen limitation (2 g/L) was consumed completely, whereas it was sufficient under nitrogen repletion (10 g/L), as previously described. As shown in Additional file 2: Fig. S1, a total of 3706 differentially expressed genes (DEGs), including 1885 upregulated and 1821 downregulated genes, were identified under nitrogen sufficient condition. The enrichment analysis of the KEGG pathway showed that these differentially expressed genes were mainly focused on the ribosome, ribosome biogenesis, proteasome, and nitrogen metabolism (p < 0.05) (Additional file 3: Table S2; Fig. 3), which was associated with cell growth and proliferation that can be mediated by the TOR signaling pathway. Furthermore, these differentially expressed genes were categorized into three major functional groups, including those for biological processes, cellular components, and molecular function via gene ontology (GO) analysis (data no shown). In the biological process group, these significantly expressed genes were mostly involved in metabolic, cellular, and biological regulation processes. In the cellular component group, most of the differentially expressed genes were linked to macromolecular complexes, membranes and organelles, and their molecular functions were primarily focused on catalytic, binding, and transport activity. These results indicated that cell growth and metabolism could be significantly regulated by the level of nitrogen.
Fig. 3.
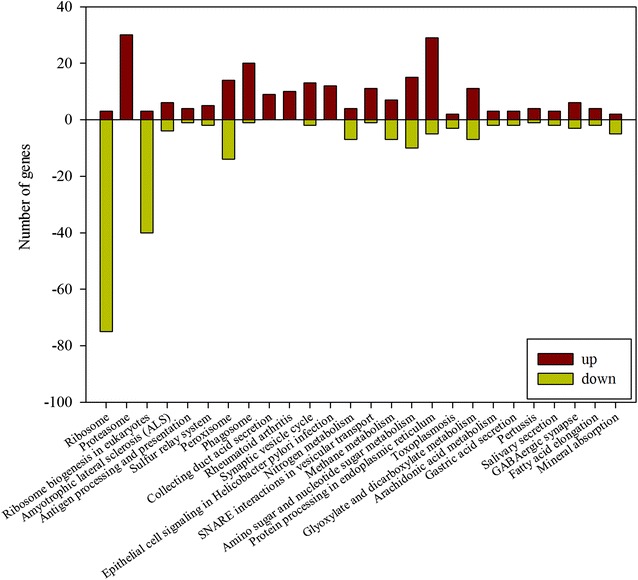
Major function categories of differentially expressed genes under nitrogen-sufficient conditions
Figure 4 shows representational gel maps of the proteins extracted from A. pullulans cells at 36 h under nitrogen limitation (2 g/L of NH4NO3) and nitrogen repletion (10 g/L of NH4NO3). Based on the comparison and analysis of the gel images, a total of 27 protein spots from the whole cell protein gels were excised and subjected to MALDI-TOF/TOF analysis. Fourteen protein spots were successfully identified, and were matched to known proteins from the fungal species and other microorganisms. Among them, nine proteins were upregulated under nitrogen repletion and five proteins were upregulated under nitrogen limitation, as shown in Table 2. Under nitrogen sufficient condition, the upregulated proteins were involved in cell division and growth (14, 60S ribosomal protein L11, histone H2B, and meiotic expression upregulated protein 14), energy metabolism (ADP/ATP carrier protein-like protein, and adenylate kinase), the glycolytic pathway (fructose-bisphosphate aldolase, and pyruvate decarboxylase), etc. The 60S ribosomal protein L11 and histone H2B take part in nucleic acid replication and cell division. Fructose-bisphosphate aldolase (FBA) catalyzes the reversible reaction between fructose-1, 6-diphosphate, and dihydroxyacetone phosphate (DHAP), plus 3-phosphate glyceraldehyde (G-3-P) [24], which is one of the key enzymes in the glycolytic pathway and affects the rate of glycolysis. It was worth noting that the ADP/ATP carrier protein plays a key role in the maintenance of energetic fluxes in eukaryotic cells, which is an integral protein of the mitochondrial inner membrane that exchanges cytoplasmic ADP for mitochondrial ATP under the conditions of oxidative phosphorylation [25]. All these upregulated proteins revealed that the high level of nitrogen could affect glycolysis and aerobic metabolism, which would promote A. pullulans cells to grow faster.
Fig. 4.
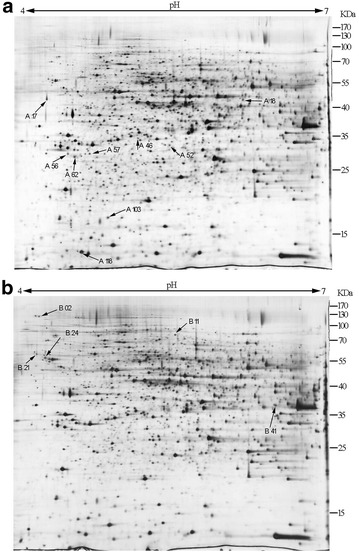
Comparative 2-DE gel analyses of cellular proteins of A. pullulans cultivated under nitrogen-sufficient (a) and nitrogen-limited (b) conditions. Spots marked on the maps with arrows and numbers represent the 14 identified proteins analyzed with MALDI-TOF/TOF
Table 2.
Identification of differentially expressed protein spots from A. pullulans cells under different levels of nitrogen via proteomics analysis with 2D-GE and MALDI-TOF/TOF
| Spot no. | Protein name | Putative function | pI | Mascot score | Coverage (%) | Mass (Da) | Fold change | E value |
|---|---|---|---|---|---|---|---|---|
| A17 | Meiotic expression up regulated protein 14 | Cell differentiation | 5.44 | 70 | 18 | 39,244 | 4.93 | 0.0013 |
| A18 | S-adenosylmethionine synthase | Amino acid metabolism | 5.75 | 128 | 12 | 43,115 | 1,000,000a | 1.9e−009 |
| A46 | Pyruvate decarboxylase | Glycolytic pathway | 5.57 | 163 | 21 | 64,181 | 2.97 | 6e−013 |
| A52 | Fructose-bisphosphate aldolase, class II | Carbohydrate metabolism | 5.41 | 282 | 22 | 39,637 | 2.24 | 7.5e−025 |
| A56 | ADP/ATP carrier protein-like protein | Energy metabolism | 9.81 | 64 | 30 | 33,892 | 2.32 | 0.0045 |
| A57 | Adenylate kinase | Energy metabolism | 8.84 | 87 | 17 | 30,980 | 8.36 | 2.6e−005 |
| A62 | ADP/ATP carrier protein-like protein | Energy metabolism | 9.81 | 243 | 30 | 33,892 | 2.14 | 6e−021 |
| A103 | 60S ribosomal protein L11 | Ribosome | 10.08 | 81 | 18 | 21,863 | 3.10 | 9.5e−005 |
| A118 | Histone H2B | Nucleosome | 10.16 | 159 | 37 | 15,047 | 2.25 | 1.5e−012 |
| B02 | Lysophospholipase Plb2 | Lipid metabolism | 4.31 | 44 | 2 | 71,790 | 1,000,000 | 0.00022 |
| B11 | NADH-quinone oxido-reductase | Electron transport | 5.71 | 245 | 24 | 82,010 | 1.68 | 3.8e−021 |
| B21 | Casein kinase II subunit beta | Cell differentiation | 4.40 | 122 | 7 | 38,463 | 2.01 | 7.5e−009 |
| B24 | Nucleosome assembly protein-like protein | Cell differentiation | 4.26 | 299 | 25 | 45,383 | 1.97 | 1.5e−026 |
| B41 | Glyceraldehyde-3-phosphate dehydrogenase | Glycolytic pathway | 6.25 | 260 | 21 | 36,436 | 2.40 | 1.2e−022 |
A17–A118 upregulated proteins under nitrogen repletion; B02–B41 upregulated proteins under nitrogen limitation
aThis value means infinite because the protein in the control is not be detected
In comparison, under the nitrogen-limited condition, the upregulated proteins were involved in cell division (casein kinase II subunit beta and the nucleosome assembly protein), the glycolytic pathway (glyceraldehyde-3-phosphate dehydrogenase, GAPDH), and the electron transport chain (NADH-quinone oxidoreductase). GAPDH is an important enzyme that can drive carbon flux to form pyruvate in the glycolytic pathway [26]. Moreover, the expression levels of key genes involved in the PMA biosynthetic pathway were further confirmed by the qRT-PCR method. It is evident in Figs. 5 and 6 that the expression levels of glucokinase (GLK), citrate synthase (CS), and fumarase (FUM) involved in glycolysis and the TCA cycle were upregulated by 25.93, 2.42 and 2.33-fold, respectively, under nitrogen limitation. Mitochondrial dicarboxylate transporter (DAT) was responsible for the transport of malic acid from the mitochondrion to the cytoplasm. In our previous study, the expression level of DAT was upregulated by the inducement of Tween-80, which was beneficial for PMA biosynthesis [14]. In addition, malate-CoA ligase (MCL) was responsible for PMA polymerization from malic acid [27]. As shown in Fig. 5, the expression levels of the genes (DAT and MCL) were upregulated by 3.09 and 3.25-fold, respectively. These results indicated that carbon flux from glycolysis and the TCA cycle was strengthened to produce more malic acid, and the upregulation of DAT and MCL was responsible for transferring more malic acid from mitochondrion and polymerize them to produce PMA. Furthermore, under nitrogen limitation, the expression levels of the genes (GS, TOR1, Tap42 and Gat1) involved in the TOR signaling pathway were upregulated by 7.49, 3.33, 3.36 and 2.83-fold, respectively. This result further revealed that the TOR pathway, via the Tap42-PP2A branch, was activated and might positively take part in regulating PMA biosynthesis.
Fig. 5.
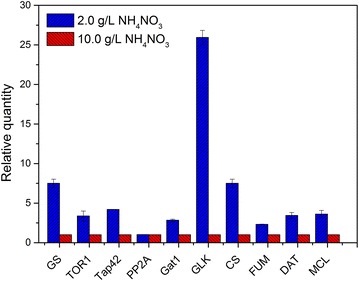
Transcription levels of key genes in the PMA and TOR pathways under the different levels of nitrogen. Data are given as the average of triplicate experiments
Fig. 6.
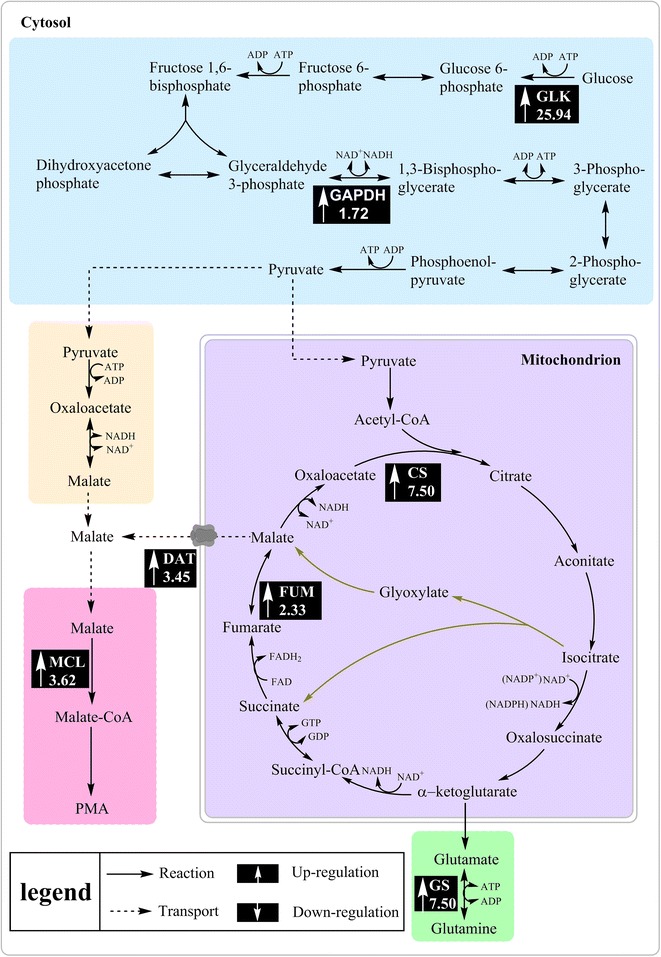
PMA biosynthesis metabolic network and the transcriptional changes of the genes encoding the enzymes catalyzing those steps. Up arrows represent the upregulated genes under nitrogen limitation (2 g/L of NH4NO3)
Rapamycin treatment on cell growth and PMA biosynthesis
As previously described, the TOR pathway responds to nutrients and growth factors to orchestrate cell growth and proliferation in eukaryotic cells. Usually, TOR can be inhibited by rapamycin, an immunosuppressant drug. Therefore, we further investigated the role of the TOR signaling pathway on cell growth and PMA biosynthesis under the treatment of rapamycin. As shown in Fig. 7a, radial growth of A. pullulans was severely inhibited on a PDA plate when the rapamycin concentration was >5 ng/mL. Correspondingly, the production of PMA in shake flasks decreased gradually (Fig. 7b); after adding 50 ng/mL of rapamycin, the PMA titer was decreased by 21.3 % compared to that in the control. Moreover, the corresponding PMA yield (Yp/x) was also decreased to 0.92 g/g (the control as 1.08 g/g), which showed that PMA biosynthesis was inhibited by rapamycin with a dose-dependent manner. These results suggested that the TOR pathway indeed participated in regulating PMA biosynthesis.
Fig. 7.
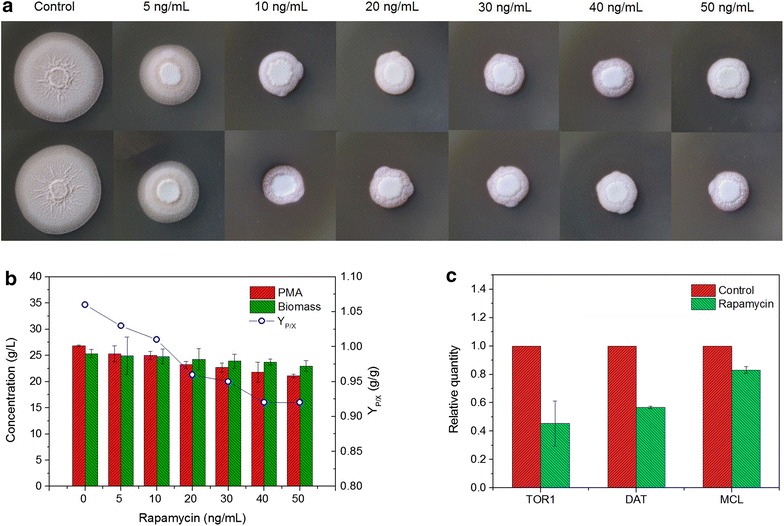
Effect of rapamycin treatment on cell growth and PMA biosynthesis. a Growth on PDA plate; b Shake flask fermentation; c Transcription levels of genes in the PMA and TOR pathways. Yield (Yp/x): the ratio of PMA to cell biomass concentration (g/g). Data are given as the average of triplicate experiments
As previously described, the production of PMA under nitrogen repletion was lower than that under nitrogen limitation. The inhibitory effect of rapamycin on PMA biosynthesis was similar to that under nitrogen repletion. The transcription levels of key genes involved in the PMA and TOR pathways after adding 10 ng/mL of rapamycin are shown in Fig. 7c. As the target of rapamycin, the expression level of TOR1 was obviously downregulated. The transcription levels of MCL and DAT were downregulated by 0.83 and 0.56-fold, which might be the direct evidence to explain the decrease of the PMA titer.
Discussion
Culture stress, especially the level of nitrogen, is becoming a useful strategy for the overproduction of organic acids and biopolymers. In this study, the effect of nitrogen availability on PMA biosynthesis was investigated. While a high level of nitrogen favored cell growth, it inhibited the production of PMA. Under nitrogen limitation (2 g/L of NH4NO3), a final PMA titer of 44.0 ± 3.65 g/L (49.9 ± 4.14 g/L of malic acid after hydrolysis) was achieved at 96 h in a 5-L fermentor, which was increased by 18.3 % compared to that under nitrogen repletion (10 g/L of NH4NO3). It was noted that, during the whole fermentation process, the ratio of ATP/ADP under nitrogen limitation was also higher than that under nitrogen repletion. According to the biosynthetic pathway of PMA proposed by Willibald et al. [28], adequate energy supply was necessary to polymerize the monomer, L-malic acid, to form PMA. Therefore, more ATP produced under nitrogen limitation was preferable to match the energy demand for the polymerization of PMA.
Transcriptomics analysis resulted in some genes involved in ribosome, ribosome biogenesis, peroxisome, and nitrogen metabolism that were expressed significantly at different levels of nitrogen. After the analysis of proteomics was integrated, it was clear that the levels of nitrogen regulate some proteins involved in cell growth and differentiation, glycolysis and energy metabolism. Under nitrogen repletion, cell growth was obviously faster than that under nitrogen limitation, which was associated with some upregulated proteins such as fructose-bisphosphate aldolase (FBA). Sheng et al. found that (NH4)2SO4 could regulate the fructose-bisphosphate aldolase that led to carbon flux toward the glycolytic pathway for the growth of biomass in A. pullulans [29], which was consistent with our experimental data. In comparison, under the nitrogen-limited condition, the upregulated proteins were mainly involved in glycolysis and energy metabolism, which meant producing more ATP and substrates for the synthesis of PMA.
As noted, the reversible oxidative phosphorylation of G-3-P to 1, 3-bisphosphoglycerate (1, 3-BPG) is mediated by GAPDH using NAD+ as a cofactor [26]. GAPDH is known as having higher relative activity compared to other enzymes in the glycolytic pathway, and is regarded as a switching control of glycolysis [30]. NADH-quinone oxidoreductase (NDH-1) is the first enzyme in the respiratory chain in mitochondria. It catalyzes oxidation of NADH in the mitochondrial matrix and reduction of quinone in the membrane, and is coupled to proton translocation through the inner mitochondrial membrane [31]. Under nitrogen-limited conditions, the enhanced activity of GAPDH and NDH-1 would strengthen the glycolytic pathway and oxidize more NADH, driving carbon flux toward pyruvate and subsequent malic acid for PMA biosynthesis. In addition, as shown in Figs. 5 and 6, the transcriptional levels of key genes involved in the PMA biosynthetic pathway (e.g., GLK, FUM, CS, DAT, and MCL) were upregulated under nitrogen limitation. It was worth noting that the expression level of glucokinase (GLK) was upregulated by 25.93-fold, which would extremely promote the rate of glycolysis from glucose.
Moreover, the low level of nitrogen upregulated the transcriptional levels of genes involved in the TOR signaling pathway, as shown in Fig. 5. Glutamine, catalyzed exclusively by glutamine synthetase (GS), is an upstream regulator of the TOR pathway. GS plays an important role not only in providing glutamine, but also as a key regulator in the nitrogen regulatory network in yeast and filamentous fungi [32]. Glutamine starvation affects a subset of TOR-controlled transcription factors including GLN3, RTG1, and RTG3 [33]. Among the downstream effectors of TOR kinase, Tap42-PP2A is the most relevant effector for stress response. Nitrogen starvation and TOR kinase inactivation result in Tap42p dephosphorylation and subsequent dissociation of the Tap42-PP2A and Tap42-PP2A-like phosphatase complex [34, 35], thereby regulating several transcription factors (including Gat1, Gln3, Gaf1, etc.) to drive nitrogen catabolism [36, 37]. Compared to nitrogen repletion, the expression levels of genes (GS, TOR1, Tap42, and Gat1) involved in the TOR pathway were upregulated, indicating that the TOR signaling pathway, via Tap42-PP2A branch, was activated and positively regulated PMA biosynthesis. Furthermore, A. pullulans cell growth was obviously inhibited by the rapamycin treatment, accompanied with a dose-depended decrease in the PMA titer. The expression levels of genes (TOR1, DAT, and MCL) were also downregulated after the treatment of rapamycin. These results further revealed that the TOR pathway indeed participated in regulating cell growth and PMA biosynthesis. In Fusarium fujikuroi, Teichert et al. found that TOR kinase is involved in the nitrogen regulation of secondary metabolism [19]. Under nitrogen starvation, protein phosphatase 2A (PP2A)-branch signaling in the downstream TOR pathway can be activated [36].
Among the above upregulated genes, Gat1 belongs to a conserved family of zinc-finger- containing transcriptional regulators known as GATA-factors, which can activate the transcription of nitrogen catabolite repression (NCR)-sensitive genes when preferred nitrogen sources are absent or limited [38]. It was found that AreA, the ortholog of Gat1, could regulate secondary metabolite biosynthesis in different species. In F. graminearum, AreA could regulate the production of the mycotoxins deoxynivalenol (DON) and zearalenone [39]. Disruption of AreA in Acremonium chrysogenum could reduce cephalosporin production [40]. In comparison, how it acts on sensitive genes in the PMA biosynthetic pathway via Gat1 or the other TOR-controlled transcription factors were not fully understood.
Under nitrogen limitation, our assumption might be confirmed that more α-ketoglutarate was produced by an activated TCA cycle, which resulted in the production of more GS; then, TOR1 was activated by GS and resulted in the phosphorylation of Tap42. In this case, PP2A was separated from Tap42 and then was inhibited, which will promote the migration of Gat1 into nucleus from the cytoplasm and positively regulate the expression of key genes in the PMA biosynthetic pathway.
Conclusions
In this study, the level of nitrogen was found to regulate cell growth and PMA biosynthesis. The low level of nitrogen was beneficial for PMA biosynthesis, which could upregulate the expression levels of key genes involved in the PMA biosynthesis pathway. Cell growth and PMA biosynthesis in response to nitrogen might be mediated by the TOR signaling pathway. This study will help us to deeply understand the molecular mechanisms of PMA biosynthesis regulated by the different levels of nitrogen, and to develop an effective process for the production of PMA and malic acid chemicals.
Authors’ contributions
YW participated in the fermentation and helped to draft the manuscript. XS performed the analysis of transcriptome. YZ performed the analysis of proteomics. BW participated in the design of the study. XZ participated in the design of the study, and drafted the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors thank Dr. Wenjun Zhang for providing the transcriptomics analysis helps.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the NCBI Sequence Read Archive database (SRA—http://www.ncbi.nlm.nih.gov/Traces/sra/) with the BioProject ID PRJNA301913.
Funding
This study was supported in part by grants from the National Natural Science Foundation of China (Grant No. 31571816), National High Technology Research and Development Program of China (863 Program) (2015AA021005 and 2014AA021205), Chongqing Social and People’s Livelihood Guarantee Special Program (cstc2016shmszx80075), Chongqing Intellectual Property Special Foundation Program (CQIPO2015224), and Visiting Scholar Foundation of Key Laboratory of Biorheological Science and Technology (Chongqing University), Ministry of Education, China (CQKLBST-2014-006).
Additional files
10.1186/s12934-016-0547-y Primers for the gene transcription level analysis.
10.1186/s12934-016-0547-y Totals of differentially expressed genes (DEGs) under nitrogen-sufficient conditions.
10.1186/s12934-016-0547-y Numbers of genes in mainly enriched pathways under nitrogen-sufficient conditions.
Contributor Information
Yongkang Wang, Email: 1121236203@qq.com.
Xiaodan Song, Email: 1012448871@qq.com.
Yongjun Zhang, Email: yjzhang@swu.edu.cn.
Bochu Wang, Email: wangbc2000@126.com.
Xiang Zou, Email: zhx1030@swu.edu.cn.
References
- 1.Portilla-Arias JA, Garcia-Alvarez M, de Ilarduya AM, Holler E, Munoz-Guerra S. Thermal decomposition of fungal poly(beta, L-malic acid) and poly(beta, L-malate) Biomacromolecules. 2006;7:3283–3290. doi: 10.1021/bm060710i. [DOI] [PubMed] [Google Scholar]
- 2.Portilla-Arias JA, Garcia-Alvarez M, de Ilarduya AM, Holler E, Galbis JA, Munoz-Guerra S. Synthesis, degradability, and drug releasing properties of methyl esters of fungal poly(beta, L-malic acid) Macromol Biosci. 2008;8:540–550. doi: 10.1002/mabi.200700248. [DOI] [PubMed] [Google Scholar]
- 3.Portilla-Arias J, Patil R, Hu J, Ding H, Black KL, Garcia-Alvarez M, Munoz-Guerra S, Ljubimova JY, Holler E. Nanoconjugate platforms development based in poly(beta,L-malic acid) methyl esters for tumor drug delivery. J Nanotechnol. 2010;2010:825363. doi: 10.1155/2010/825363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiang N, Yang W, Li L, Dong P, Zhu J, Wang T, Zeng C, Quan D. Synthesis of pendent carboxyl-containing poly(epsilon-caprolactone-co-beta-malic acid)-block-poly(L-lactid e) copolymers for fabrication of nano-fibrous scaffolds. Polymer. 2012;53:4993–5001. doi: 10.1016/j.polymer.2012.09.010. [DOI] [Google Scholar]
- 5.Zou X, Zhou Y, Yang S-T. Production of polymalic acid and malic acid by Aureobasidium pullulans fermentation and acid hydrolysis. Biotechnol Bioeng. 2013;110:2105–2113. doi: 10.1002/bit.24876. [DOI] [PubMed] [Google Scholar]
- 6.Gostincar C, Ohm RA, Kogej T, Sonjak S, Turk M, Zajc J, Zalar P, Grube M, Sun H, Han J, et al. Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genomics. 2014;15:549. doi: 10.1186/1471-2164-15-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi ZM, Wang F, Chi Z, Yue LX, Liu GL, Zhang T. Bioproducts from Aureobasidium pullulans, a biotechnologically important yeast. Appl Microbiol Biotechnol. 2009;82:793–804. doi: 10.1007/s00253-009-1882-2. [DOI] [PubMed] [Google Scholar]
- 8.Leathers TD, Price NPJ, Bischoff KM, Manitchotpisit P, Skory CD. Production of novel types of antibacterial liamocins by diverse strains of Aureobasidium pullulans grown on different culture media. Biotechnol Lett. 2015;37:2075–2081. doi: 10.1007/s10529-015-1892-3. [DOI] [PubMed] [Google Scholar]
- 9.Knuf C, Nookaew I, Brown SH, McCulloch M, Berry A, Nielsen J. Investigation of malic acid production under nitrogen staration conditions. Appl Environ Microbiol. 2013;79:6050–6058. doi: 10.1128/AEM.01445-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding Y, Li S, Dou C, Yu Y, Huang H. Production of fumaric acid by Rhizopus oryzae: role of carbon-nitrogen ratio. Appl Biochem Biotechnol. 2011;164:1461–1467. doi: 10.1007/s12010-011-9226-y. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg I, Rokem JS, Pines O. Organic acids: old metabolites, new themes. J Chem Technol Biotechnol. 2006;81:1601–1611. doi: 10.1002/jctb.1590. [DOI] [Google Scholar]
- 12.Basak B, Ince O, Artan N, Yagci N, Ince BK. Effect of nitrogen limitation on enrichment of activated sludge for PHA production. Bioproc Biosyst Eng. 2011;34:1007–1016. doi: 10.1007/s00449-011-0551-x. [DOI] [PubMed] [Google Scholar]
- 13.Sun J, Peng X, Van Impe J, Vanderleyden J. The ntrB and ntrC genes are involved in the regulation of poly-3-hydroxybutyrate biosynthesis by ammonia in Azospirillum brasilense Sp7. Appl Environ Microbiol. 2000;66:113–117. doi: 10.1128/AEM.66.1.113-117.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu G, Wang Y, Ji Y, Zou X. The effect of Tween 80 on the polymalic acid and pullulan production by Aureobasidium pullulans CCTCC M2012223. World J Microbiol Biotechnol. 2015;31:219–226. doi: 10.1007/s11274-014-1779-9. [DOI] [PubMed] [Google Scholar]
- 15.Zan Z, Zou X. Efficient production of polymalic acid from raw sweet potato hydrolysate with immobilized cells of Aureobasidium pullulans CCTCC M2012223 in aerobic fibrous bed bioreactor. J Chem Technol Biotechnol. 2013;88:1822–1827. doi: 10.1002/jctb.4033. [DOI] [Google Scholar]
- 16.Kim SG, Buel GR, Blenis J. Nutrient regulation of the mTOR complex 1 signaling pathway. Mol Cells. 2013;35:463–473. doi: 10.1007/s10059-013-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 18.Rohde J, Heitman J, Cardenas ME. The TOR kinases link nutrient sensing to cell growth. J Biol Chem. 2001;276:9583–9586. doi: 10.1074/jbc.R000034200. [DOI] [PubMed] [Google Scholar]
- 19.Teichert S, Wottawa M, Schoenig B, Tudzynski B. Role of the Fusarium fujikuroi TOR kinase in nitrogen regulation and secondary metabolism. Eukaryot Cell. 2006;5:1807–1819. doi: 10.1128/EC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu F, Gu Q, Yun Y, Yin Y, Xu J-R, Shim W-B, Ma Z. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014;203:219–232. doi: 10.1111/nph.12776. [DOI] [PubMed] [Google Scholar]
- 21.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 22.Weatherburn M. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. doi: 10.1021/ac60252a045. [DOI] [Google Scholar]
- 23.Zou X, Tu G, Zan Z. Cofactor and CO2 donor regulation involved in reductive routes for polymalic acid production by Aureobasidium pullulans CCTCC M2012223. Bioproc Biosyst Eng. 2014;37:2131–2136. doi: 10.1007/s00449-014-1182-9. [DOI] [PubMed] [Google Scholar]
- 24.Thomson GJ, Howlett GJ, Ashcroft AE, Berry A. The dhnA gene of Escherichia coli encodes a class I fructose bisphosphate aldolase. Biochem J. 1998;331:437–445. doi: 10.1042/bj3310437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lousa CD, Trezeguet W, Dianoux AC, Brandolin G, Lauquin GJM. The human mitochondrial ADP/ATP carriers: kinetic properties and biogenesis of wild-type and mutant proteins in the yeast Saccharomyces cerevisiae. Biochemistry. 2002;41:14412–14420. doi: 10.1021/bi0261490. [DOI] [PubMed] [Google Scholar]
- 26.Nelson K, Whittam TS, Selander RK. Nucleotide polymorphism and evolution in the glyceraldehyde-3-phosphate dehydrogenase gene in natural-populations of Salmonella and Escherichia coli. PNAS. 1991;88:6667–6671. doi: 10.1073/pnas.88.15.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Zhou F, Tu G, Zou X. Gene cloning, expression and characterization of malate-CoA ligase in the polymerization pathway of polymalic acid from Aureobasidium pullulans. Acta Microbiol Sin. 2014;54:919–925. [PubMed] [Google Scholar]
- 28.Willibald B, Bildl W, Lee BS, Holler E. Is beta-poly(L-malate) synthesis catalysed by a combination of beta-L-malyl-AMP-ligase and beta-poly(L-malate) polymerase? Eur J Biochem. 1999;265:1085–1090. doi: 10.1046/j.1432-1327.1999.00834.x. [DOI] [PubMed] [Google Scholar]
- 29.Sheng L, Zhu G, Tong Q. Comparative proteomic analysis of Aureobasidium pullulans in the presence of high and low levels of nitrogen source. J Agric Food Chem. 2014;62:10529–10534. doi: 10.1021/jf503390f. [DOI] [PubMed] [Google Scholar]
- 30.Cho H-S, Seo SW, Kim YM, Jung GY, Park JM. Engineering glyceraldehyde-3-phosphate dehydrogenase for switching control of glycolysis in Escherichia coli. Biotechnol Bioeng. 2012;109:2612–2619. doi: 10.1002/bit.24532. [DOI] [PubMed] [Google Scholar]
- 31.Brandt U. Energy converting NADH: quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 32.Tudzynski B. Nitrogen regulation of fungal secondary metabolism in fungi. Front Microbiol. 2014;5:1–15. doi: 10.3389/fmicb.2014.00656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crespo JL, Powers T, Fowler B, Hall MN. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. PNAS. 2002;99:6784–6789. doi: 10.1073/pnas.102687599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang Y, Broach JR. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 1999;18:2782–2792. doi: 10.1093/emboj/18.10.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot Cell. 2004;3:1261–1271. doi: 10.1128/EC.3.5.1261-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hallett JEH, Luo X, Capaldi AP. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics. 2014;198:773–786. doi: 10.1534/genetics.114.168369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laor D, Cohen A, Kupiec M, Weisman R. TORC1 regulates developmental responses to nitrogen stress via regulation of the GATA transcription factor Gaf1. MBIO. 2015;6:e00959. doi: 10.1128/mBio.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tate JJ, Georis I, Dubois E, Cooper TG. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J Biol Chem. 2010;285:17880–17895. doi: 10.1074/jbc.M109.085712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giese H, Sondergaard TE, Sorensen JL. The AreA transcription factor in Fusarium graminearum regulates the use of some nonpreferred nitrogen sources and secondary metabolite production. Fungal Biol. 2013;117:814–821. doi: 10.1016/j.funbio.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Pan Y, Liu G. Disruption of the nitrogen regulatory gene AcareA in Acremonium chrysogenum leads to reduction of cephalosporin production and repression of nitrogen metabolism. Fungal Genet Biol. 2013;61:69–79. doi: 10.1016/j.fgb.2013.10.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset supporting the conclusions of this article is available in the NCBI Sequence Read Archive database (SRA—http://www.ncbi.nlm.nih.gov/Traces/sra/) with the BioProject ID PRJNA301913.


