Keywords: nerve regeneration, peripheral nerve injury, sciatic nerve crush, transcriptome sequencing, bioinformatic analysis, canonical pathway, extracellular matrix, matrix metalloproteinase, tissue inhibitors of metalloproteinase, disintegrin, repair, neural regeneration
Abstract
We previously performed transcriptome sequencing and found that genes for matrix metalloproteinases (MMPs), such as MMP7 and 12, seem to be highly upregulated following peripheral nerve injury, and may be involved in nerve repair. In the present study, we systematically determined the expression levels of MMPs and their regulators at 1, 4, 7 and 14 days after sciatic nerve crush injury. The number of differentially expressed genes was elevated at 4 and 7 days after injury, but decreased at 14 days after injury. Among the differentially expressed genes, those most up-regulated showed fold changes of more than 214, while those most down-regulated exhibited fold changes of more than 2−10. Gene sequencing showed that, at all time points after injury, a variety of MMP genes in the “Inhibition of MMPs” pathway were up-regulated, and their inhibitor genes were down-regulated. Expression of key up- and down-regulated genes was verified by quantitative real-time polymerase chain reaction analysis and found to be consistent with transcriptome sequencing. These results suggest that MMP-related genes are strongly involved in the process of peripheral nerve regeneration.
Introduction
Peripheral nerve injury is a common clinical problem that greatly compromises a patient’s quality of life, and much emerging research is focused on the underlying mechanisms. Peripheral nerve injury leads to the damage of axons and surrounding connective tissues, and may ultimately lead to the death of axotomized neurons (Wu and Murashov, 2013). Following injury, the distal stump of the damaged nerve undergoes Wallerian degeneration. Injured axons degenerate and then regenerate, and Schwann cells dedifferentiate, proliferate and migrate to form bands of Büngner (Gaudet et al., 2011; Chen et al., 2015). Meanwhile, Schwann cells actively produce neurotrophic factors and deposit extracellular matrix (ECM) to stimulate axon regeneration and facilitate the directed extension of regenerating axons (Frostick et al., 1998; Gu et al., 2014; Namgung, 2014).
The ECM is a dynamic and complex network of extracellular molecules and provides physical and biochemical support to its surrounding cells. During peripheral nerve regeneration, the ECM secretes biological cues to effect a neuronal cellular response and axonal growth, creates a permissive microenvironment for nerve regeneration, and thus promotes the regeneration of injured nerves (Dubovy, 2004; Chen et al., 2007).
The dynamic remodeling of the ECM is normally regulated by matrix metalloproteinases (MMPs) (Ravanti and Kahari, 2000; Steffensen et al., 2001). MMPs are a large family of zinc-containing endopeptidases and can be grouped into collagenases (MMP1, 8, 13, and 18 in Xenopus), gelatinases (MMP2 and 9), stromelysins (MMP3, 10, and 11), matrilysins (MMP7 and 26), membrane-type MMPs (MMP14, 15, 16, 17, 24, and 25), and enamelysin (MMP20) (Nagase et al., 2006). These proteolytic enzymes are critical in a variety of processes associated with tissue remodeling such as morphogenesis, angiogenesis, cirrhosis, arthritis, metastasis, and tissue repair. MMPs have been implicated in neural development and numerous neurological diseases (Crocker et al., 2004; Agrawal et al., 2008; De Groef et al., 2014). However, whether MMPs are involved in the biological process of peripheral nerve regeneration remains unknown.
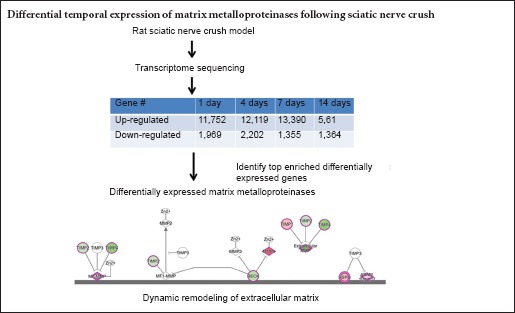
In the study of the mechanisms underlying peripheral nerve regeneration, especially the involvement of MMPs, the rat sciatic nerve crush model is now used to identify the expression patterns of MMP-related genes following peripheral nerve injury. We previously performed transcriptome sequencing to screen out differentially expressed genes in the injured sciatic nerve at different time points, analyzed the top enriched diseases and functions and canonical signaling pathways, and obtained the global transcriptional landscape of rat sciatic nerve crush (Yi et al., 2015).
Since MMPs are critical in tissue remodeling, in the present study, using data from transcriptome sequencing, we analyze the expression levels of MMPs and their regulators.
Materials and Methods
Animals
Thirty adult male Sprague-Dawley rats, weighing 180–220 g, were obtained from the Experimental Animal Center of Nantong University, China (animal license numbers SCXK (Su) 2014-0001 and SYXK (Su) 2012-0031). Animal procedures were performed in accordance with the Institutional Animal Care Guideline of Nantong University and were ethically approved by the Administration Committee of Experimental Animals, Jiangsu Province, China.
Establishing rat models of sciatic nerve crush
The rats were randomized equally into five groups: 1, 4, 7, and 14 days after sciatic nerve crush and the uninjured rats (control group). The animals were anesthetized intraperitoneally with a mixture of 85 mg/kg trichloroacetaldehyde monohydrate, 42 mg/kg magnesium sulfate, and 17 mg/kg sodium pentobarbital, and underwent left sciatic nerve crush surgery as described previously (Yi et al., 2015). In brief, a skin incision was made on the lateral aspect of the mid-thigh of the left hindlimb, and the sciatic nerve was crushed with forceps three times with a force of 54 N (10 seconds each time). The muscle and skin were then sutured. Rats were sacrificed by cervical dislocation at the allocated time after surgery.
RNA extraction and transcriptome sequencing
Sciatic nerve segments (5 mm in length) were harvested from the crush site and total RNA was extracted using TRIzol (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The quality of purified RNA samples was evaluated using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). RNA concentration was measured using a NanoDrop ND-1000 spectrophotometer (Infinigen Biotechnology Inc., City of Industry, CA, USA).
Transcriptome sequencing was performed as described previously (Yi et al., 2015). Briefly, mRNA was isolated from total RNA using magnetic beads with Oligo(dT) and then cut into short fragments. These mRNA fragments were used as templates to synthesize cDNA. cDNA fragments of suitable sizes were selected, purified, and amplified. The amplified library was sequenced using Illumina HiSeq™ 2000 to produce raw reads, which were subjected to quality control to obtain clean reads. Filtered clean reads were then aligned to the reference genome using SOAPaligner/SOAP2. The alignment data were then used to detect gene expression and perform downstream bioinformatic analysis.
Bioinformatic analysis
Transcriptome sequencing data were normalized using the reads per kilobase transcriptome per million mapped reads method (Mortazavi et al., 2008) and then used for independent comparisons. The fold change of the target gene was calculated by comparing its expression at a specific time point following sciatic nerve crush to the control group. mRNAs with a false discovery rate ≤ 0.001 and a fold change ≥ 2 (log2 ratio ≥ 1) were identified as differentially expressed genes. Online Ingenuity pathway analysis software (Ingenuity Systems, Redwood City, CA, USA) was used to investigate the differentially expressed genes at 1, 4, 7, and 14 days after sciatic nerve crush. The canonical signaling pathway “Inhibition of MMPs” was analyzed according to the Ingenuity Pathways Knowledge Base. The observation index is the expression of the target gene at each time point, relative to (divided by) its expression in the control group.
Quantitative real-time polymerase chain reaction (qRT-PCR)
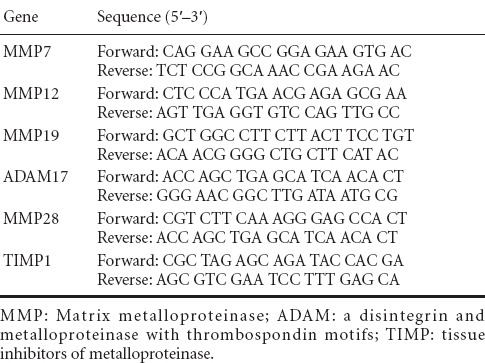
RNA samples were reverse-transcribed to cDNA using the Prime-Script reagent Kit (TaKaRa, Dalian, China). qRT-PCR was performed using SYBR Green Premix Ex Taq (TaKaRa) on a StepOne Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) in triplicate for each sample. The thermocycler program was as follows: 5 minutes at 95°C; 40 cycles of 30 seconds at 95°C, 45 seconds at Tanneal, and 30 seconds at 72°C; and 5 minutes at 72°C. The expression level of each mRNA was calculated as described previously (Yi et al., 2015). Relative expression level was calculated using the comparative 2−ΔΔCt method with GAPDH as the reference gene. Primer sequences are listed in Table 1. Expression level of the target gene relative to GAPDH was calculated by dividing the Ct value of the target gene by that of GAPDH at each time point.
Table 1.
Primer pairs for quantitative real-time polymerase chain reaction
Statistical analysis
Statistical analysis was performed as described previously (Yi et al., 2015). Statistical analysis was performed using SPSS 15.0 software (SPSS, Chicago, IL, USA). In brief, experimental outcomes were expressed as the mean ± SEM for parametric data. Groups were compared using one-way analysis of variance and the least significant difference test. P < 0.05 was considered statistically significant.
Results
Top differentially expressed genes following sciatic nerve crush
Transcriptome sequencing demonstrated that a very large number of genes showed differential expression patterns at 1, 4, 7, and 14 days after sciatic nerve crush compared with control group (Yi et al., 2015). The number of differentially expressed genes increased slightly at 4 and 7 days, but dropped significantly at 14 days. Furthermore, most of the differentially expressed genes were up-regulated, suggesting that many genes were activated in response to sciatic nerve crush (Figure 1).
Figure 1.
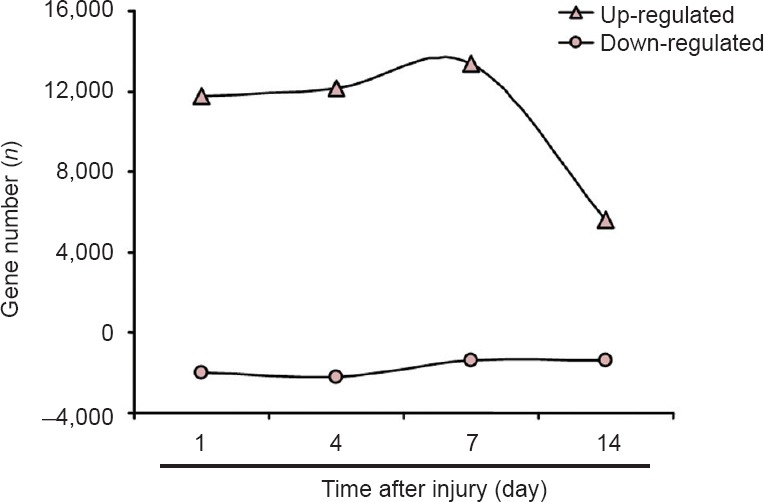
Number of differentially expressed genes after sciatic nerve crush.
Number of up- and down-regulated genes at each time point examined following sciatic nerve crush.
Among these differentially expressed genes, the most up-regulated genes showed fold changes of more than 214 (log2 ratio > 14), whereas those most down-regulated showed fold changes of more than 2−10 (log2 ratio < −10). The top 10 up- and down-regulated mRNAs are listed in Table 2.
Table 2.
Top 20 differentially expressed mRNAs
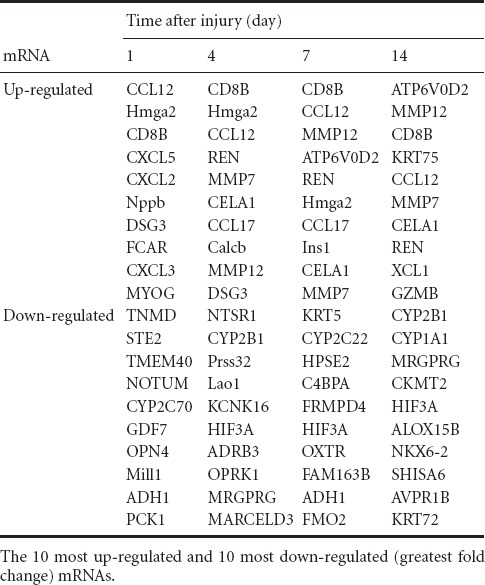
Among the top up-regulated mRNAs, several proteinase-coding genes, including MMP7 and 12, and chymotrypsin-like elastase family member 1 (CELA1), attracted our attention. Transcriptome sequencing data revealed that MMP7 and 12 were not only highly up-regulated at certain time points, but were continuously highly expressed during peripheral nerve regeneration. Compared with control group, MMP7 was up-regulated to 25.27 fold 1 day after injury, 211.81 fold at 4 days, 210.32 fold at 7 days, and 211.43 fold at 14 days. Expression levels of MMP12 were also markedly increased at all time points following nerve crush, reaching 26.34 fold at 1 day, 210.98 fold at 4 days, 211.56 fold at 7 days, and 213.18 fold at 14 days (Figure 2, Table 3). These transcriptome sequencing outcomes suggested that MMPs may play central roles during peripheral nerve regeneration.
Figure 2.
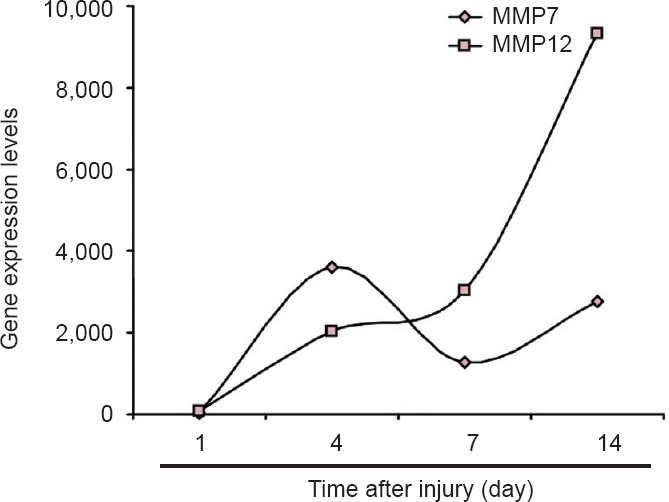
Upregulation of MMP7 and MMP12 mRNA expression following sciatic nerve injury.
Relative amounts of MMP7 and MMP12 mRNA at 1, 4, 7, and 14 days after sciatic nerve crush. MMP: Matrix metalloproteinase.
Table 3.
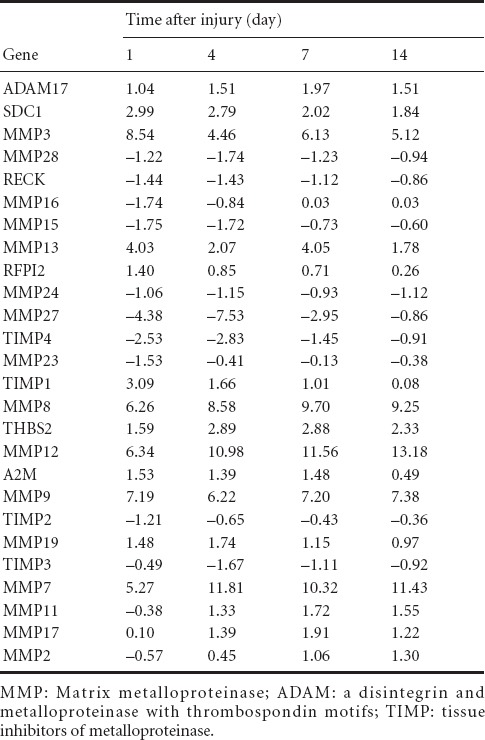
Expression of genes (log2 ratio) involved in “Inhibition of MMPs” pathway at 1, 4, 7, and 14 days following sciatic nerve crush
“Inhibition of MMPs” pathway analysis
To elucidate the involvement of MMPs in the peripheral nerve regeneration process, we examined the canonical pathway associated with MMP-related genes. The canonical “Inhibition of MMPs” pathway describes the cellular movement and organismal growth and development controlled by MMPs and their regulators. We therefore chose to study this specific canonical pathway in detail.
MMPs can be regulated by gene transcription, activation of the latent enzyme, and inactivation by specific inhibitors such as tissue inhibitors of metalloproteinases (TIMPs). Differentially expressed or activated MMPs affect proteolytic degradation or activation at the cell surface and in the ECM, influence the components as well as dynamics of the ECM, modulate cell-cell and cell-ECM interactions, and thus regulate cellular growth, organismal development, wound repair, and tissue remodeling (Daley et al., 2008; Bosse, 2012). In addition to MMPs, the degradation of ECM is also regulated by a disintegrin and metalloproteinase with thrombospondin motifs (ADAM) family member proteins, including ADAM10, 12, and 17 (Lu et al., 2011).
Analysis of the top enriched canonical pathways according to the Ingenuity Pathways Knowledge Base suggested that the “Inhibition of MMPs” pathway still ranked highly following sciatic nerve crush. It had P values of 10−5.13, 10−4.61, 10−6.91, and 10−3.04 at 1, 4, 7, and 14 days post-injury, and affected 21, 20, 20, and 12 differentially expressed genes involved in the pathway. As the schematic network of the “Inhibition of MMPs” pathway shows (Figures 3, 4), mRNA expression of MMPs and ADAMs was altered at different time points following nerve crush, thus affecting cell-cell and cell-ECM interactions.
Figure 3.
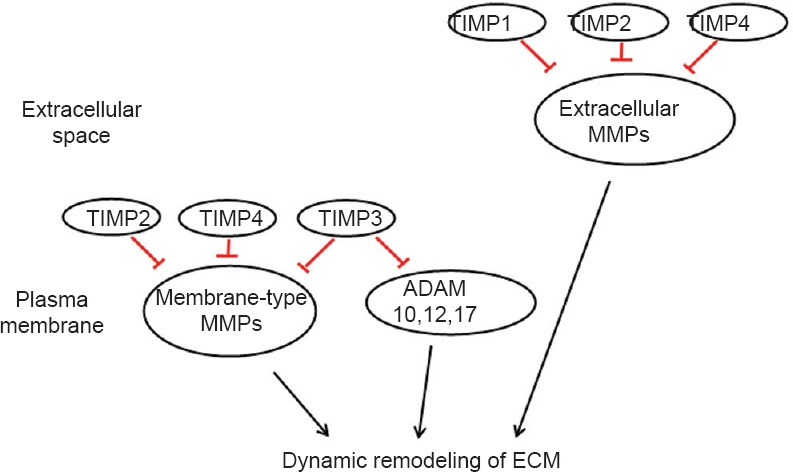
Schematic of “Inhibition of MMPs” pathway.
TIMPs inhibit membrane-type MMPs, extracellular MMPs, and ADAMs, and thus mediate the dynamic remodeling of ECM. TIMP: Tissue inhibitors of metalloproteinase; MMPs: matrix metalloproteinases; ADAM: a disintegrin and metalloproteinase with thrombospondin motifs; ECM: extracellular matrix.
Figure 4.
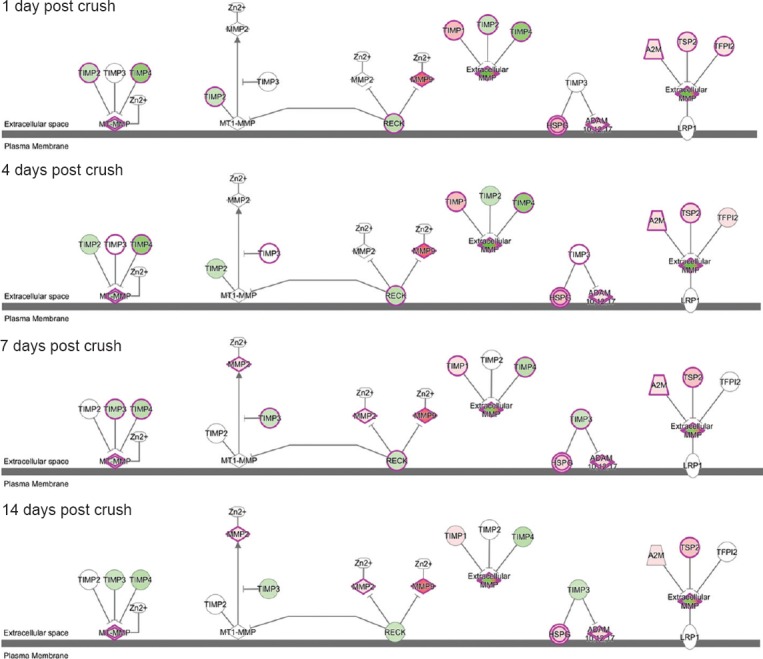
Schematic of “Inhibition of MMPs” signaling pathway at 1, 4, 7, and 14 days after nerve crush injury.
Differentially expressed genes are shown in red (up-regulated) and green (downregulated). Stronger colors represent higher fold changes. MMP: Matrix metalloproteinase; ADAM: a disintegrin and metalloproteinase with thrombospondin motifs; TIMP: tissue inhibitors of metalloproteinase.
A global genome heatmap with hierarchical cluster analysis was also performed to illustrate the expression of those genes involved in the “Inhibition of MMPs” pathway (Figure 5). Many MMPs, including MMP2, 3, 7, 8, 9, 11, 12, 13, 17, and 19, were up-regulated following peripheral nerve injury, while some, such as MMP15, 16, 24, 23B, 27, and 28 were down-regulated. The expressions of the inhibitors of MMPs followed the same pattern, in that that some inhibitors, such as TIMP2, 3, and 4, were down-regulated, whereas inhibitor TIMP1 was up-regulated following peripheral nerve injury. A full list of differentially expressed genes in the “Inhibition of MMPs” pathway is provided in Table 3.
Figure 5.
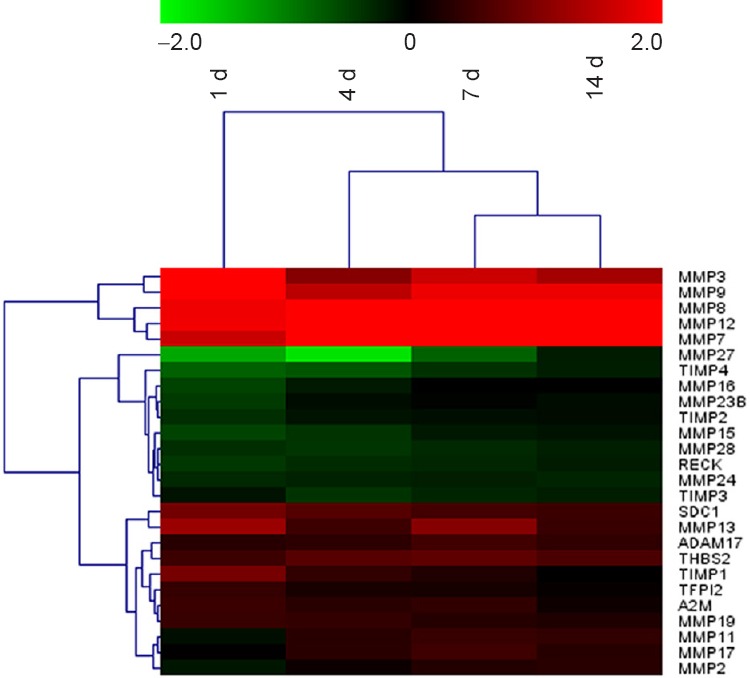
Heatmap and hierarchical clustering of genes involved in “Inhibition of MMPs” pathway.
Gene expression levels are indicated by the color bar above the heatmap (red, up-regulated; green, down-regulated) compared to day 0 (control). MMP: Matrix metalloproteinase; ADAM: a disintegrin and metalloproteinase with thrombospondin motifs; TIMP: tissue inhibitors of metalloproteinase; d: day(s).
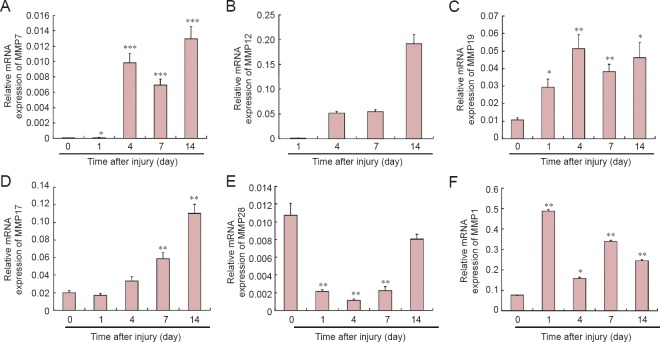
Validation of the expression levels of MMP-related mRNAs
To validate the reliability of outcomes from transcriptome sequencing, the expression patterns of several key molecules involved in the “Inhibition of MMPs” pathway were selected for qRT-PCR examination. Consistent with sequencing data, outcomes from qRT-PCR showed that, compared with control group, MMP7 was notably up-regulated at 1 day after nerve crush, and further up-regulated at 4, 7, and 14 days (Figure 6A). Elevated expression of MMP12 at 4, 7, and 14 days post-injury was also observed (Figure 6B). No comparison of MMP12 expression to the control was made due to non-efficient amplification of MMP12 in the uninjured samples (Ct value was not defined). This is in accordance with the long reads per kilobase transcriptome per million mapped reads of MMP12 in the uninjured samples in transcriptome sequencing. Expression of MMP19 and ADAM-17 was also measured; consistent with the sequencing data, expression of both was significantly elevated (Figure 6C, D).
Figure 6.
Quantitative real-time polymerase chain reaction analysis of MMP-related genes in the sciatic nerve at 0, 1, 4, 7, and 14 days following injury.
The relative expression of each mRNA was calculated using comparative Ct with GAPDH as the reference gene. *P < 0.05, **P < 0.01, ***P < 0.0001, vs. control group (uninjured rats). Data are summarized from 3–4 paired experiments and values are shown as the mean ± SEM (one-way analysis of variance and the least significant difference test). GAPDH: Glyceraldehyde-3-phosphate dehydrogenase; MMP: matrix metalloproteinase; ADAM: a disintegrin and metalloproteinase with thrombospondin motifs; TIMP: tissue inhibitors of metalloproteinase.
In addition to those genes revealed by sequencing data to be upregulated, we also measured the amount of MMP28, a proteolytic peptidase, that was down-regulated according to sequencing data. qRT-PCR results showed that MMP28 expression decreased following nerve crush, reaching a valley at 4 days, and increasing slightly thereafter, but not reaching the expression level in the control group (Figure 5E). This down-regulation of MMP28 might be correlated with increased TIMP1 expression (Figure 6F).
Discussion
In the present study, we examined the molecular changes of genes involved in the “Inhibition of MMPs” canonical pathway in the sciatic nerve at 1, 4, 7, and 14 days following sciatic nerve crush. Deep sequencing revealed that a number of genes were up-regulated after nerve crush injury. Of these, cytokines and chemokines were two important classes, including chemokines (C-C motif) ligand 12 (CCL12) and ligand 17 (CCL17); chemokines (C-X-C motif) ligand 5 (CXCL5), ligand 2 (CXCL2), and ligand 3 (CXCL3); and chemokine (C motif) ligand 1 (XCL1). Other up-regulated mRNAs were related to the immune response and include the CD8b molecule (CD8B), Fc fragment of IgA receptor (FCAR), and granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) (GZMB). These observations were consistent with the current understanding of the process of peripheral nerve regeneration that Schwann cells secrete cytokines and chemokines to promote the clearance of axonal and myelin debris and to stimulate axonal regrowth (Bosse, 2012; Dubovy et al., 2013). These observations were also in accordance with our previous finding that inflammatory and immune response-related functions are the top enriched biological functions following peripheral nerve regeneration (Yi et al., 2015).
In addition to the above genes, mRNA levels of MMPs, ADAMs, and other genes related to ECM degradation were also differentially expressed after peripheral nerve crush injury. These genes might modulate the component and dynamics of ECM, influence the intracellular interactions and the interactions between cell and ECM, and thus affect the process of peripheral nerve regeneration. Accordingly, these genes became the focus of the present study.
MMP-related genes influence cell differentiation, migration, proliferation, and survival, and thus influence a variety of tissues and organs, including the nervous system (George and Dwivedi, 2004; Kim and Joh, 2012; Apte and Parks, 2015). The essential roles of MMPs following ischemia/reperfusion have been studied extensively (Dejonckheere et al., 2011; Palladini et al., 2015). Following cerebral ischemia, MMPs are activated and subsequently degrade the ECM and disrupt the blood-brain barrier, leading to hemorrhagic damage, infiltration of inflammatory cells at the infarct site, and neuronal death (Candelario-Jalil et al., 2009; Jin et al., 2010). It has been demonstrated that elevated intranuclear MMP2 and 9 disrupt tight junctions, decrease the activation of poly (ADP-ribose) polymerase, degrade nuclear DNA repair proteins, and mediate neuronal apoptosis (Yang et al., 2007; Hill et al., 2012). It has also been shown that MMP3 knockout mice with bilateral carotid artery occlusion have fewer microglial cells and less neuronal death compared with wild-type mice, suggesting that MMP-3 inhibitors can be used to delay inflammation and reduce hippocampal neuronal death following ischemia (Walker and Rosenberg, 2009). Besides their effects on neuronal viability, MMPs also affect the hydrolysis and activation of growth factors and neurotrophic factors. MMP2 promotes the conversion of active nerve growth factor from its precursor, and thus exhibits beneficial effects during neuronal growth (Saygili et al., 2011). MMP3 and 7 mediate the release and activation of brain derived neurotrophic factor, and thus promote axonal growth (Hwang et al., 2005). These studies suggested that the specific roles of MMPs in the nerve regrowth are complicated and may be bidirectional.
Although emerging evidence demonstrates the complex roles of MMPs in the nervous system, the involvement of MMP-related genes in the process of peripheral nerve regeneration has not been fully understood. A previous study showed that, following sciatic nerve injury, MMP12 mRNA was markedly increased in both the proximal and distal segments of the injured nerve during Wallerian degeneration, providing preliminary evidence that MMPs are important for nerve regeneration (Jiang et al., 2014). In the present study, for the first time, we systematically measured the mRNA expression of MMP-related genes and analyzed the “Inhibition of MMPs” signaling pathway following peripheral nerve injury. Our data suggest that numerous MMP-related genes show temporal differential expression following sciatic nerve crush. Most of these differentially expressed MMPs were up-regulated, suggesting that elevated MMPs may accelerate the degradation and remodeling of ECM, promote cellular movement and tissue morphology, and build a suitable extracellular environment for peripheral nerve regeneration. Conversely, some MMPs, such as MMP28, were down-regulated after nerve crush, indicating that MMPs have diverse biological functions. Further research will be performed to determine the regulation of each MMP on neuronal survival, neurite outgrowth, and phenotype modulation of Schwann cells.
Collectively, in the present study, using transcriptome sequencing and bioinformatic analysis, we have gained a better understanding of the changes of MMP-related genes following rat sciatic nerve crush. These results may advance our comprehension of the essential roles of MMPs underlying peripheral nerve regeneration, and contribute to the identification of potential targets for treating neurological pathology.
Footnotes
Funding: This study was supported by the Natural Science Foundation of Jiangsu Province of China, No. BK20150409; the Natural Science Foundation of Jiangsu Higher Education Institutions of China, No. 15KJB180013; the Scientific Research Foundation of Nantong University in China, No. 14R29; the Natural Science Foundation of Nantong City in China, No. MS12015043; the Priority Academic Program Development of Jiangsu Higher Education Institutions in China.
Conflicts of interest: None declared.
Plagiarism check: This paper was screened twice using Cross-Check to verify originality before publication.
Peer review: This paper was double-blinded and stringently reviewed by international expert reviewers.
Copyedited by Slone-Murphy J, Wysong S, Wang J, Qiu Y, Li CH, Song LP, Zhao M
References
- 1.Agrawal SM, Lau L, Yong VW. MMPs in the central nervous system: where the good guys go bad. Semin Cell Dev Biol. 2008;19:42–51. doi: 10.1016/j.semcdb.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Apte SS, Parks WC. Metalloproteinases: a parade of functions in matrix biology and an outlook for the future. Matrix Biol. 2015;44-46:1–6. doi: 10.1016/j.matbio.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Bosse F. Extrinsic cellular and molecular mediators of peripheral axonal regeneration. Cell Tissue Res. 2012;349:5–14. doi: 10.1007/s00441-012-1389-5. [DOI] [PubMed] [Google Scholar]
- 4.Candelario-Jalil E, Yang Y, Rosenberg GA. Diverse roles of matrix metalloproteinases and tissue inhibitors of metalloproteinases in neuroinflammation and cerebral ischemia. Neuroscience. 2009;158:983–994. doi: 10.1016/j.neuroscience.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen P, Piao X, Bonaldo P. Role of macrophages in Wallerian degeneration and axonal regeneration after peripheral nerve injury. Acta Neuropathol. 2015;130:605–618. doi: 10.1007/s00401-015-1482-4. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZL, Yu WM, Strickland S. Peripheral regeneration. Annu Rev Neurosci. 2007;30:209–233. doi: 10.1146/annurev.neuro.30.051606.094337. [DOI] [PubMed] [Google Scholar]
- 7.Crocker SJ, Pagenstecher A, Campbell IL. The TIMPs tango with MMPs and more in the central nervous system. J Neurosci Res. 2004;75:1–11. doi: 10.1002/jnr.10836. [DOI] [PubMed] [Google Scholar]
- 8.Daley WP, Peters SB, Larsen M. Extracellular matrix dynamics in development and regenerative medicine. J Cell Sci. 2008;121:255–264. doi: 10.1242/jcs.006064. [DOI] [PubMed] [Google Scholar]
- 9.De Groef L, Van Hove I, Dekeyster E, Stalmans I, Moons L. MMPs in the neuroretina and optic nerve: modulators of glaucoma pathogenesis and repair? Invest Ophthalmol Vis Sci. 2014;55:1953–1964. doi: 10.1167/iovs.13-13630. [DOI] [PubMed] [Google Scholar]
- 10.Dejonckheere E, Vandenbroucke RE, Libert C. Matrix metalloproteinases as drug targets in ischemia/reperfusion injury. Drug Discov Today. 2011;16:762–778. doi: 10.1016/j.drudis.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Dubovy P. Schwann cells and endoneurial extracellular matrix molecules as potential cues for sorting of regenerated axons: a review. Anat Sci Int. 2004;79:198–208. doi: 10.1111/j.1447-073x.2004.00090.x. [DOI] [PubMed] [Google Scholar]
- 12.Dubovy P, Jancalek R, Kubek T. Role of inflammation and cytokines in peripheral nerve regeneration. Int Rev Neurobiol. 2013;108:173–206. doi: 10.1016/B978-0-12-410499-0.00007-1. [DOI] [PubMed] [Google Scholar]
- 13.Frostick SP, Yin Q, Kemp GJ. Schwann cells, neurotrophic factors, and peripheral nerve regeneration. Microsurgery. 1998;18:397–405. doi: 10.1002/(sici)1098-2752(1998)18:7<397::aid-micr2>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 14.Gaudet AD, Popovich PG, Ramer MS. Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J Neuroinflammation. 2011;8:110. doi: 10.1186/1742-2094-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George SJ, Dwivedi A. MMPs, cadherins, and cell proliferation. Trends Cardiovasc Med. 2004;14:100–105. doi: 10.1016/j.tcm.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 16.Gu X, Ding F, Williams DF. Neural tissue engineering options for peripheral nerve regeneration. Biomaterials. 2014;35:6143–6156. doi: 10.1016/j.biomaterials.2014.04.064. [DOI] [PubMed] [Google Scholar]
- 17.Hill JW, Poddar R, Thompson JF, Rosenberg GA, Yang Y. Intranuclear matrix metalloproteinases promote DNA damage and apoptosis induced by oxygen-glucose deprivation in neurons. Neuroscience. 2012;220:277–290. doi: 10.1016/j.neuroscience.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang JJ, Park MH, Choi SY, Koh JY. Activation of the Trk signaling pathway by extracellular zinc. Role of metalloproteinases. J Biol Chem. 2005;280:11995–12001. doi: 10.1074/jbc.M403172200. [DOI] [PubMed] [Google Scholar]
- 19.Jiang N, Li H, Sun Y, Yin D, Zhao Q, Cui S, Yao D. Differential gene expression in proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration. Neural Regen Res. 2014;9:1186–1194. doi: 10.4103/1673-5374.135325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin R, Yang G, Li G. Molecular insights and therapeutic targets for blood-brain barrier disruption in ischemic stroke: critical role of matrix metalloproteinases and tissue-type plasminogen activator. Neurobiol Dis. 2010;38:376–385. doi: 10.1016/j.nbd.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim YS, Joh TH. Matrix metalloproteinases, new insights into the understanding of neurodegenerative disorders. Biomol Ther (Seoul) 2012;20:133–143. doi: 10.4062/biomolther.2012.20.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 3. 2011 doi: 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 24.Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res. 2006;69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Namgung U. The role of Schwann cell-axon interaction in peripheral nerve regeneration. Cells Tissues Organs. 2014;200:6–12. doi: 10.1159/000370324. [DOI] [PubMed] [Google Scholar]
- 26.Palladini G, Ferrigno A, Richelmi P, Perlini S, Vairetti M. Role of matrix metalloproteinases in cholestasis and hepatic ischemia/reperfusion injury: A review. World J Gastroenterol. 2015;21:12114–12124. doi: 10.3748/wjg.v21.i42.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravanti L, Kahari VM. Matrix metalloproteinases in wound repair (review) Int J Mol Med. 2000;6:391–407. [PubMed] [Google Scholar]
- 28.Saygili E, Schauerte P, Pekassa M, Saygili E, Rackauskas G, Schwinger RH, Weis J, Weber C, Marx N, Rana OR. Sympathetic neurons express and secrete MMP-2 and MT1-MMP to control nerve sprouting via pro-NGF conversion. Cell Mol Neurobiol. 2011;31:17–25. doi: 10.1007/s10571-010-9548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steffensen B, Hakkinen L, Larjava H. Proteolytic events of wound-healing-coordinated interactions among matrix metalloproteinases (MMPs), integrins, and extracellular matrix molecules. Crit Rev Oral Biol Med. 2001;12:373–398. doi: 10.1177/10454411010120050201. [DOI] [PubMed] [Google Scholar]
- 30.Walker EJ, Rosenberg GA. TIMP-3 and MMP-3 contribute to delayed inflammation and hippocampal neuronal death following global ischemia. Exp Neurol. 2009;216:122–131. doi: 10.1016/j.expneurol.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu D, Murashov AK. Molecular mechanisms of peripheral nerve regeneration: emerging roles of microRNAs. Front Physiol. 2013;4:55. doi: 10.3389/fphys.2013.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, Estrada EY, Thompson JF, Liu W, Rosenberg GA. Matrix metalloproteinase-mediated disruption of tight junction proteins in cerebral vessels is reversed by synthetic matrix metalloproteinase inhibitor in focal ischemia in rat. J Cereb Blood Flow Metab. 2007;27:697–709. doi: 10.1038/sj.jcbfm.9600375. [DOI] [PubMed] [Google Scholar]
- 33.Yi S, Zhang H, Gong L, Wu J, Zha G, Zhou S, Gu X, Yu B. Deep sequencing and bioinformatic analysis of lesioned sciatic nerves after crush injury. PLoS One. 2015;10:e0143491. doi: 10.1371/journal.pone.0143491. [DOI] [PMC free article] [PubMed] [Google Scholar]