Abstract
Reliable assessment of the BRAF mutation status is becoming increasingly important in the clinical management of colorectal carcinomas (CRC). The aim of this study was to investigate the use of a recently developed mutation-specific antibody (VE1, SpringBio, Pleasanton, CA) to detect the BRAF V600E protein in paraffin tissue. We analyzed by immunohistochemistry (IHC) 117 cases that had been evaluated for BRAF mutation using a MALDI-TOF mass-spectrometry based assay. IHC staining was evaluated without the knowledge of the genetic data and was considered positive when there was distinct homogeneous cytoplasmic staining in the tumor cells. The analyzed cases included 4 polyps, 63 primary and 50 metastatic CRC. Forty-five of the 46 (97.8%) cases that were positive by IHC had a BRAF V600E mutation by genetic analysis; the 1 discordant case was notably of signet ring cell type. Similarly, 66 of the 67 (98.5%) cases that were negative by IHC were also negative by genetic analysis. Four cases that showed weak cytoplasmic staining and/or nuclear staining in the tumor cells were considered to be IHC equivocal; by genetic analysis, 2 of the 4 were positive and 2 were negative. The overall sensitivity and specificity of IHC for the detection of a BRAF V600E mutant tumor was 93.7% and 95.6%, respectively. Our results support the use of VE1 IHC for identification of colorectal neoplasms harboring the BRAF V600E mutation. Difficulties in IHC interpretation may arise in a small number of cases, and in those cases molecular testing is required.
Keywords: Colon cancer, BRAF V600E, immunohistochemistry, VE1 antibody
Introduction
Activating mutations of the serine threonine kinase BRAF were first reported in 2002 and are present in a wide range of human tumors including melanoma, colorectal carcinoma (CRC), papillary thyroid carcinoma and ovarian carcinoma1-3. The vast majority of BRAF alterations are characterized by a T1799A transversion which results in a missense substitution of valine at amino acid position 600 by glutamic acid (BRAF V600E)4. This mutation causes constitutive activation of the RAS/RAF/MEK/ERK cell-signaling pathway (also known as the MAP kinase pathway) which regulates cell cycle, growth and survival. Approximately 10% of all CRCs harbor BRAF mutations with the frequency of mutations being lower among cases with stage IV disease5,6.
The determination of the BRAF mutation status in CRC is currently clinically important for several reasons. Detection of the BRAF V600E mutation in tumors showing microsatellite instability (MSI-H) suggests somatic methylation of the MLH1 promoter region instead of a germline mutation when screening for Lynch Syndrome7. In terms of prognosis, several studies have found the presence of a BRAF V600E mutation to predict for poor outcomes8-11. Finally, tumor genotype can help guide targeted therapy as stage IV BRAF mutant CRC may be resistant to epidermal growth factor receptor (EGFR) inhibitors12,13 and may be candidates for combined treatment with BRAF V600E and EGFR inhibitors14.
Currently, evaluation of the BRAF mutational status is based solely on sequencing of the gene using a variety of methods including Sanger sequencing, pyrosequencing and mass spectrometry. Recently, a monoclonal antibody (VE1) has been generated which recognizes the mutant BRAF V600E protein15 using Western blot and immunohistochemistry (IHC) of routinely processed formalin-fixed paraffin embedded tissue (FFPE). Initial published studies have found IHC staining with this antibody to have a high sensitivity and specificity for identifying tumors harboring the V600E mutation in several tumor types15-20. However, experience in colorectal carcinomas is limited, with one group reporting disappointing results16,21-24. In this study we analyzed the IHC application of the VE1 antibody in colorectal neoplasms and compared the results with those obtained from sequencing of the BRAF gene.
Materials and Methods
Case selection
The study was approved by the Institutional Review Board. Cases of CRC and adenomas that were previously tested for the presence of the BRAF V600E mutation were retrieved. The majority of the cases (63%) were tested in a CLIA certified lab, while the remaining were tested for research purposes using the same assay. Cases diagnosed from 1992 until 2011 were included in the study. Both biopsies and resections were analyzed.
Immunohistochemistry
Four micron thick sections were cut from FFPE tumor blocks. IHC for mutant BRAF V600E was performed on a BenchMark XT automated immunostainer (Ventana Medical Systems, Inc., Tuscon, Ariz). Sections were incubated with the anti-BRAF V600E antibody (clone VE1, SpringBio, Pleasanton, CA) diluted 1:50 for 48 minutes at room temperature. Antigen retrieval was performed by heating at 100°C for 32 minutes, and primary antibody incubation was carried out at 37°C for 32 minutes. Antigen detection was performed using the OptiView DAB Detection kit (Ventana Medical Systems, Inc.). Metastatic melanoma with a documented BRAF V600E mutation was used as a positive control. The immunostained slides were evaluated independently by two pathologists who were blinded to the molecular data and to each other's IHC assessment. Cases were scored as positive when there was diffuse (>80% of tumor cells) staining above any background staining. The intensity of cytoplasmic tumor cell staining in the positive cases was scored as weak, moderate and strong. Cases were considered as equivocal if 1. there was nuclear staining in the tumor cells in addition to cytoplasmic staining or 2. there was disagreement between the two pathologists.
Mutation analysis
For DNA preparation, FFPE tissue or, where available, frozen tissue was used. Serial sectioning of tissue was performed and the presence and extent of tumor was verified using hematoxylin and eosin stain (H&E). Macrodissection was performed if required to ensure >50% tumor content. Genomic DNA was extracted from 10 unstained sections 5 micron thick using the DNeasy Tissue kit (Qiagen) following the manufacturer's protocol. Mutations were detected using the iPLEX assay (Sequenom, Inc.), which is based on a single-base primer extension assay. Briefly, multiplexed PCR and extension primers were designed with the Sequenom Assay Designer v3.1 software for a panel of known mutations in the BRAF gene. After PCR and extension reactions, the resulting extension products were analyzed using a matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF) mass spectrometer. Samples were tested in duplicate and all automated system mutation calls were confirmed by manual review of the spectra. Cases that showed discordant results between molecular results and IHC were also subsequently analyzed by Sanger sequencing of BRAF exon 15.
Results
The total number of cases analyzed in the study was 117. These included 4 polyps, 63 primary CRC and 51 metastatic CRC. Among the polyps one was a sessile serrated polyp (SSP), while 3 were tubulovillous adenomas (TVAs). The metastatic sites included liver (n=39, 76.6% of all metastatic CRC), lung (n=4, 7.8%), lymph nodes (n=4, 7.8%), and soft tissue (n=4, 7.8%). The majority of the specimens tested were resections (n=108, 92.3%) and 9 (7.7%) were biopsies. Fifty (42.7%) cases had been found to be positive for the BRAF V600E mutation upon initial genotyping, and 67 (57.3%) were wild type for this mutation. Among the latter, 2 (1.8% of all tested cases) had a BRAF D594G mutation.
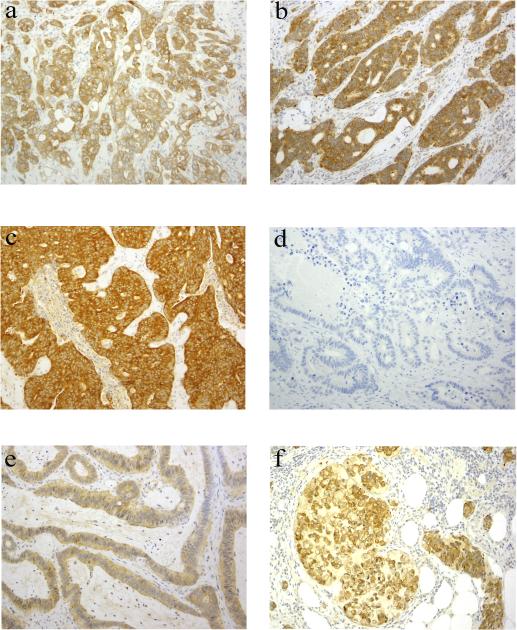
Immunohistochemical staining with the VE1 antibody showed positive staining in 46 (39.3%) cases including the SSP. The vast majority of the cases (44/46, 95.7%) had diffuse homogeneous staining; in two cases there were focal areas (<20%) with no or very weak staining. The staining intensity was scored as weak, moderate or strong in 5 (10.9%), 19 (41.3%) and 22 (47.8%) cases, respectively (Figures 1A-C). Sixty seven (57%) cases were negative for staining in the tumor cells, including the 2 cases that showed a BRAF D594G mutation (Figure 1D).
Figure 1.
Immunohistochemical staining of colorectal carcinomas with the BRAF V600E antibody VE1. Figures A-C illustrate positive staining in 3 colorectal carcinomas that harbor a BRAF V600E mutation. Note the varied staining intensity in the 3 cases (A, weak; B, moderate; C, strong). D shows negative staining in a case of colorectal carcinoma without a BRAF V600E mutation. E illustrates equivocal staining in a case of colorectal carcinoma without a BRAF V600E mutation. Note the faint cytoplasmic positivity along with weak nuclear staining in tumor cells. F shows positive staining in a signet ring cell carcinoma without a BRAF V600E mutation.
In the majority of the cases, the determination of the staining being positive or negative and the scoring of the staining intensity were readily achieved, as there was often no interference of background staining. Four (3.4%) cases were scored as equivocal. Three of these 4 cases showed both weak cytoplasmic and nuclear staining in the tumor cells (Figure 1E). The nuclear staining tended to be more pronounced in tumor cells closer to tissue edges. The fourth case showed weak cytoplasmic and focal nuclear staining in the tumor cells in addition to some weak background staining and was scored as negative by one pathologist, while positive by the second pathologist. Repeat IHC staining in the four cases deemed equivocal yielded identical results.
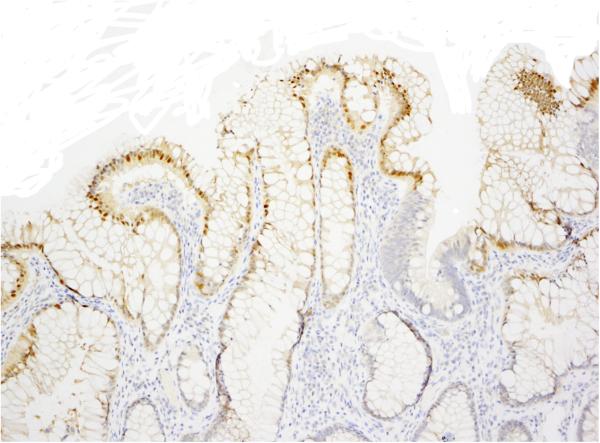
We observed nonspecific nuclear staining consistently in the normal, superficial colonic epithelium (Figure 2) as noted by other investigators21,24. Although considered nonspecific, this staining did serve as an internal positive control. In one case that was found to be entirely negative initially, the absence of the nuclear staining in the normal mucosa raised suspicion for a technical problem; repeat IHC yielded positive staining in the tumor cells (in addition to nuclear staining in the normal, superficial mucosa). Nonspecific staining was also observed in the cilia of benign bronchial glands in one of two CRC metastases to lung that were included in our study, similar to prior observations21.
Figure 2.
An immunohistochemical stain with the BRAF V600E antibody VE1 showing nonspecific nuclear staining in non-neoplastic superficial colonic epithelium.
Overall, after our initial examination there were four cases showing a discrepancy between genotype and IHC in addition to the four cases that were considered equivocal by IHC. These eight cases were tested again for the presence of the BRAF V600E mutation by Sanger sequencing. In two of these cases the result differed from that of the original genotyping, specifically a V600E mutation was detected in cases that were previously classified in a research setting as BRAF wild type. Both these cases showed positive staining by IHC.
Table 1 summarizes the results after repeat IHC staining and molecular testing of selected cases. The overall sensitivity of VE1 IHC for detection of a BRAF V600E mutant tumor was 93.7%, while the specificity was 95.6%. Concordant results were observed in 111 (94.9%) cases. The 6 cases that were either equivocal by IHC or showed discrepant results between IHC and genotype included 1 biopsy and 5 resection specimens from either primary invasive CRC (n=3), liver metastasis (n=−2) or peritoneal metastasis (n=1). There was no apparent correlation with the age of the block or whether the DNA was obtained from fresh frozen tissue or FFPE tissue.
Table 1.
Comparison of immunohistochemistry and molecular testing for detection of the BRAF V600E mutant protein in colorectal carcinomas
| Positive Genotype | Negative Genotype | Total # of cases | |
|---|---|---|---|
| IHC Positive | 45 | 1 | 46 |
| IHC Negative | 1 | 66 | 67 |
| IHC Equivocal | 2 | 2 | 4 |
| Total # of cases | 48 | 69 | 117 |
IHC, immunohistochemistry.
Forty five of 46 cases (97.8%) that were found to be positive by IHC were also positive by genetic analysis. A discrepant result was observed in one case of a signet ring cell carcinoma arising in the right colon of a 73 year old woman where there was above background cytoplasmic staining (Figure 1F), but genotyping failed to detect a BRAF mutation. Sixty six of the 67 (98.5%) cases that were interpreted as negative for the V600E mutation by IHC were also negative by DNA testing. One case of a microsatellite stable CRC with a positive genotype showed weak IHC staining in the tumor cells that was not readily distinguishable from background staining and, therefore, was considered negative.
Discussion
In our series of 117 colorectal neoplasms we found a high sensitivity (93.7%) and specificity (95.6%) of VE1 IHC for detection of the BRAF V600E mutant protein. Moreover, agreement between the two pathologists was very good as there was a different interpretation of the IHC results in only one case. Our findings are in agreement with other studies and lend support to the use of IHC in the evaluation of CRC for the BRAF V600E mutation while also highlighting potential pitfalls and limitations of IHC.
The majority of prior studies, both in CRC and in other tumors such as melanoma and hairy cell leukemia, have reported highly concordant results between genetic testing and IHC16,17,21,23-27. In their series of CRC, Affolter et al and Sinicorpe et al reported 100% concordance, while three other studies reported isolated discrepant cases 8,16,21,23,24. In contrast to these results, Adackapara et al in their study of 52 CRC found only a 71% sensitivity and 74% specificity with VE1 for the BRAF V600E and concluded that the VE1 mAb is not sufficiently sensitive to serve as an effective screening tool22. The different results obtained in the latter study may be due in part to variable IHC staining methodology as the investigators used a manual technique with overnight antibody incubation. Moreover, Adackapara et al interpreted as positive any perceptible cytoplasmic staining even a blush that was only evident at 400 X magnification, but did not comment on any background staining. In our series, we observed a few cases with very weak staining in tumor cells in addition to background non-specific staining in smooth muscle and/or mucin and these cases were scored as negative.
Cytoplasmic staining in tumor cells was almost always diffuse in all types of tumor samples, including polyps and primary and metastatic CRCs. In a few cases where focal loss of staining was observed, the focal loss seemed attributable to differences in tissue preservation and fixation. Our findings are in agreement with prior studies on CRC and are consistent with the hypothesis that BRAF mutations are an early event in colorectal carcinogenesis. Homogeneous cytoplasmic staining has also been observed in most other tumor types examined17,18,27, with the exception of melanoma where there are suggestions of mutational heterogeneity within primary and metastatic tumors25,28.
We encountered difficulty in the IHC interpretation in two scenarios. First, in three cases we observed nuclear staining in tumor cells in addition to the cytoplasmic staining. This was different from the nuclear staining in normal colonic epithelium that appears to be a consistent finding among studies8,16,21-23. Nuclear staining in tumor cells was reportedly common in the study by Adackapara et al, but was not reported in the other studies. The etiology of the nuclear staining in tumor cells is unclear, but may be due to technical issues such as tissue preservation and/or fixation as the nuclear staining in our three cases was more pronounced in the tumor cells that were closer to tissue edges. Interestingly, the cases showing this type of staining in our study were from mucinous (n=2) or signet ring cell (n=1) carcinomas. Toon et al encountered one signet ring cell carcinoma where BRAF V600E IHC yielded a false negative result and suggested that interpretation of IHC results might be more difficult in cases with mucinous histology24.
Weak cytoplasmic staining in the tumor cells was the source of false negatives in 2 cases that were positive for the BRAF V600E mutation by molecular testing. In these cases, in addition to the weak staining in the tumor cells there was background staining that resulted in one or both pathologists interpreting the tumor as negative for the presence of the BRAF mutant protein. Weak staining intensity in a minority of cases with a confirmed BRAF V600E genotype has been reported for many tumor types. Capper et al used a pan-BRAF antibody in parallel with VE1 IHC and suggested that weak staining may be the result of reduced BRAF expression. Regardless of its etiology, a weak staining reaction can lead to wrong conclusions, especially when it is accompanied by some background staining. In contrast to the findings of Adackapara et al., it was not a frequent occurrence in our study.
We had one discrepant case that was positive by IHC analysis while negative by molecular testing. This was a right sided signet ring cell carcinoma in an older female patient that was genotyped both by Sequenom mass-spectrometry performed in a CLIA-certified laboratory as well as Sanger sequencing. Of interest, expression of the mismatch repair proteins MLH1, MSH2, MSH6 and PMS2 was retained in the tumor cells, arguing against an MSI-H tumor. In their study, Thiel et al reported one case that was positive by IHC while negative by molecular testing, but they were able to detect a mutation upon repeat sequencing after macrodissection to enrich for tumor content23. Our case had an adequate tumor content, therefore it is unlikely that a mutation was not detected due to a small number of tumor cells. Capper et al also encountered a case in the right colon of a 92-year old woman with a negative genotype that was positive by IHC; in their case repeat sequencing even after LASER-assisted microdissection failed to detect any V600E mutant alleles16. The reason for the positive IHC staining in these isolated CRC cases remains unclear, but it has been hypothesized that it might be due to mutations in proteins that share similar epitopes, e.g. ARAF or CRAF.
Availability of IHC for BRAF V600E testing in archival tissue specimens can facilitate the screening of CRCs. IHC offers the advantage of a faster and easier to perform assay compared to molecular testing and as such can allow more laboratories to test for the BRAF V600E mutation. IHC can also provide an alternative method for mutation detection in cases where there might not be adequate tissue for molecular testing, e.g. in biopsies that may have few tumor cells. It should be noted, however, that most studies to date have focused on resection specimens. A recent study looking at IHC staining on tissue microarrays observed an increased number of false negatives due to heterogeneous staining24. In our series, we did not observe any trend towards more discrepant or ambiguous results in biopsies, however, a larger number of biopsies needs to be examined.
In conclusion, we observed a high sensitivity and specificity of IHC staining with the VE1 antibody for detection of BRAF V600E mutant CRC cases. Our data suggest that it can be successfully used in the diagnostic setting, as the majority of cases can be diagnosed as negative or positive. There are, however, limitations and in cases where ambiguous IHC staining is observed additional genetic analysis is required to determine the mutational status of the BRAF gene.
Footnotes
Disclosure/conflict of interest: The authors declare no conflict of interest
This study was presented in part at the 102nd annual meeting of the United States and Canadian Academy of Pathology in Baltimore, MD, in March 2013.
References
- 1.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Cohen Y, Xing M, Mambo E, et al. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 3.Singer G, Oldt R, 3rd, Cohen Y, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–486. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 4.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–358. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaughn CP, Zobell SD, Furtado LV, et al. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer. 2011;50:307–312. doi: 10.1002/gcc.20854. [DOI] [PubMed] [Google Scholar]
- 6.Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajagopalan H, Bardelli A, Lengauer C, et al. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature. 2002;418:934. doi: 10.1038/418934a. [DOI] [PubMed] [Google Scholar]
- 8.Sinicrope FA, Foster NR, Yoon HH, et al. Association of obesity with DNA mismatch repair status and clinical outcome in patients with stage II or III colon carcinoma participating in NCCTG and NSABP adjuvant chemotherapy trials. J Clin Oncol. 2012;30:406–412. doi: 10.1200/JCO.2011.39.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Souglakos J, Philips J, Wang R, et al. Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer. 2009;101:465–472. doi: 10.1038/sj.bjc.6605164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Cutsem E, Kohne CH, Lang I, et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011–2019. doi: 10.1200/JCO.2010.33.5091. [DOI] [PubMed] [Google Scholar]
- 11.Samowitz WS, Sweeney C, Herrick J, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65:6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 12.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 13.Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 14.Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 15.Capper D, Preusser M, Habel A, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathol. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 16.Capper D, Voigt A, Bozukova G, et al. BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. Int J Cancer. 2013 doi: 10.1002/ijc.28183. [DOI] [PubMed] [Google Scholar]
- 17.Koperek O, Kornauth C, Capper D, et al. Immunohistochemical detection of the BRAF V600E-mutated protein in papillary thyroid carcinoma. Am J Surg Pathol. 2012;36:844–850. doi: 10.1097/PAS.0b013e318246b527. [DOI] [PubMed] [Google Scholar]
- 18.Preusser M, Capper D, Berghoff AS, et al. Expression of BRAF V600E mutant protein in epithelial ovarian tumors. Appl Immunohistochem Mol Morphol. 2013;21:159–164. doi: 10.1097/PAI.0b013e31825d7402. [DOI] [PubMed] [Google Scholar]
- 19.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 20.Capper D, Berghoff AS, Magerle M, et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathol. 2012;123:223–233. doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 21.Affolter K, Samowitz W, Tripp S, et al. BRAF V600E mutation detection by immunohistochemistry in colorectal carcinoma. Genes Chromosomes Cancer. 2013 doi: 10.1002/gcc.22070. [DOI] [PubMed] [Google Scholar]
- 22.Adackapara CA, Sholl LM, Barletta JA, et al. Immunohistochemistry using the BRAF V600E mutation-specific monoclonal antibody VE1 is not a useful surrogate for genotyping in colorectal adenocarcinoma. Histopathology. 2013;63:187–193. doi: 10.1111/his.12154. [DOI] [PubMed] [Google Scholar]
- 23.Thiel A, Heinonen M, Kantonen J, et al. BRAF mutation in sporadic colorectal cancer and Lynch syndrome. Virchows Arch. 2013 doi: 10.1007/s00428-013-1470-9. [DOI] [PubMed] [Google Scholar]
- 24.Toon CW, Walsh MD, Chou A, et al. BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol. 2013;37:1592–1602. doi: 10.1097/PAS.0b013e31828f233d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busam KJ, Hedvat C, Pulitzer M, et al. Immunohistochemical analysis of BRAF(V600E) expression of primary and metastatic melanoma and comparison with mutation status and melanocyte differentiation antigens of metastatic lesions. Am J Surg Pathol. 2013;37:413–420. doi: 10.1097/PAS.0b013e318271249e. [DOI] [PubMed] [Google Scholar]
- 26.Long GV, Wilmott JS, Capper D, et al. Immunohistochemistry is highly sensitive and specific for the detection of V600E BRAF mutation in melanoma. Am J Surg Pathol. 2013;37:61–65. doi: 10.1097/PAS.0b013e31826485c0. [DOI] [PubMed] [Google Scholar]
- 27.Andrulis M, Penzel R, Weichert W, et al. Application of a BRAF V600E mutation-specific antibody for the diagnosis of hairy cell leukemia. Am J Surg Pathol. 2012;36:1796–1800. doi: 10.1097/PAS.0b013e3182549b50. [DOI] [PubMed] [Google Scholar]
- 28.Yancovitz M, Litterman A, Yoon J, et al. Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS One. 2012;7:e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]