Abstract
Complex eukaryotic genomes are riddled with repeated sequences whose derivation does not coincide with phylogenetic history and thus is often unknown. Among such sequences, the capacity for transcriptional activity coupled with the adaptive use of reverse transcription can lead to a diverse group of genomic elements across taxa, otherwise known as selfish elements or mobile elements. Short interspersed nuclear elements (SINEs) are nonautonomous mobile elements found in eukaryotic genomes, typically derived from cellular RNAs such as tRNAs, 7SL or 5S rRNA. Here, we identify and characterize a previously unknown SINE derived from the 3′-end of the large ribosomal subunit (LSU or 28S rDNA) and transcribed via RNA polymerase III. This new element, SINE28, is represented in low-copy numbers in the human reference genome assembly, wherein we have identified 27 discrete loci. Phylogenetic analysis indicates these elements have been transpositionally active within primate lineages as recently as 6 MYA while modern humans still carry transcriptionally active copies. Moreover, we have identified SINE28s in all currently available assembled mammalian genome sequences. Phylogenetic comparisons indicate that these elements are frequently rederived from the highly conserved LSU rRNA sequences in a lineage-specific manner. We propose that this element has not been previously recognized as a SINE given its high identity to the canonical LSU, and that SINE28 likely represents one of possibly many unidentified, active transposable elements within mammalian genomes.
Keywords: SINE, RNA polymerase III, LSU, rRNA, 28S
Introduction
Exploitation of recent improvements in both genome sequencing and assembly methodologies has led to increasingly high-quality human genome assemblies since the establishment of the Human Genome Project over a decade ago. Surprisingly, since the first human genome assembly release, studies continue to mount outlining the identification of novel genes as well as an emerging appreciation for the critical role of noncoding regions in shaping genome structure and function. The origin and evolution of noncoding sequences, however, is not always clearly defined by phylogenetic history, in part due to the complex interplay between mobile elements and host genomes.
Short interspersed elements (SINEs) are the most abundant mobile element class in the mammalian genome (Okada 1991). As nonautonomous elements, SINEs require the transposition machinery of another element (typically a long interspersed element [LINE]) to retrotranspose their RNA intermediates (reviewed in Lunyak and Atallah [2011]). These short elements are most often derived from pieces of abundant RNAs found in the cell (reviewed in Weiner [2005]) and have been found to originate from the 7SL gene, like the abundant primate Alus (Ullu and Tschudi 1984; Batzer et al. 1996), tRNA genes (Daniels and Deininger 1985; Okada and Ohshima 1993; Churakov et al. 2005), and 5S rRNAs (Kapitonov and Jurka 2003; Nishihara et al. 2006; Gogolevsky et al. 2009). SINEs typically have a 5′ sequence derived from a progenitor RNA molecule (e.g., 7SL) that carries the RNA polymerase III promoter sequence (Weiner 2005), an intervening sequence, and a 3′ tail that is recognized by the autonomous element through which they retrotranspose (Okada and Ohshima 1993).
While previously considered simply genomic parasites, it is clear that SINEs contribute to genome plasticity and diversity through a variety of means including gene regulation, establishment of chromatin boundaries, recombination hotspots, and gene duplications (reviewed in Lunyak and Atallah [2011]). Moreover, high levels of conservation identified within a subdomain of a new order of ancient SINEs, AmnSINE1, suggests that such retroelements can be exapted for function in vertebrate genomes (Nishihara et al. 2006), although the exact function for these is not yet known. The presence of high-copy numbers of SINEs within eukaryotic genomes offers challenges in genome assembly and annotations, although several sequencing methodologies and computational programs have been developed to meet this challenge (e.g., Natali et al. 2013).
Using traditional genome scans, we have identified a previously unknown, low copy SINE in the human genome derived from the large ribosomal subunit (LSU or 28S); consequently we named this element SINE28. Genome scale analyses reveal that SINE28s show classic signs of transposition and indicate SINE28s have been active within the primate lineage as recently as the human/chimpanzee split (∼6 MYA [Goodman et al. 1998]). We further show that SINE28 is present and conserved in all currently available sequenced mammalian genomes. The high level of conservation observed for SINE28s restricted to mammalian lineages implicates these sequences in an exapted function within the genome.
Materials and Methods
In Silico Analyses
Initial genome screening was performed by aligning the human large-ribosomal subunit consensus sequence (LSU-rRNA_Hsa) as found in Repbase (Jurka et al. 2005) to human assembly hg19 using BLAT (Kent 2002). The RepeatMasker track (Fujita et al. 2011) of the UCSC genome browser (Kent et al. 2002) was used to identify repeats adjacent to or near the BLAT identified LSU sequences (i.e., the SINE28 simple repeat tail). Percent identity between the SINE28 loci and LSU-rRNA_Hsa was determined using BLAST (Altschul et al. 1997). Putative endonuclease (EN) cleavage sites were identified based on a 100% identity to known EN sites and location both within the first 50 bp upstream of the element and immediately after the tail region/3′-target site duplication (TSD). TSDs were identified by capturing areas of 85–100% identity surrounding the LSU-derived portion of candidate SINEs using dotplot in Geneious. Captured regions were subsequently aligned and percent identity calculated using ClustalW. For classification as a TSD, we required the following conditions be met: 1) duplicated region at the 5′-end must be found after predicted EN site, 2) region at the 3′-end must be found immediately after the tail region, 3) predicted duplication annotations cannot contain the annotated EN sites, and 4) if the duplications are not 100% identical, duplications must carry ≥85% identity to each other (as noted in table 1A). Phylogenetic dating of SINE28s was determined using the Comparative Genomics tracks of UCSC (Fujita et al. 2011). SINE28 was identified in other mammalian genomes by aligning the 27 newly identified human SINE28 sequences to genomes found in UCSC using BLAT and referencing the corresponding RepeatMasker tracks. Multiple sequence alignments of mammalian SINE28s were performed using ClustalW (Larkin et al. 2007) and visualized using jalview (Waterhouse et al. 2009). Graphical alignments were performed in Geneious with MUSCLE implementing ClustalW. SINESearch with the SINE28 sequences was performed utilizing default search parameters of 65% sequence identity and 70 nt minimum overlap length (Vassetzky and Kramerov 2013).
Table 1.
Thirty-Seven Loci Identified in the Human Genome (hg19) Containing a Fragment of 28s rDNA (LSU)
| Chromosome | Coordinates | Size (bp) | Tail Sequence | Strand | LSU Start | LSU End | % ID to LSU | TSD Length | TSD Sequence | EN Sites | Embedded | Adjacent | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. SINE28 sequences | |||||||||||||||||||||||
| 1p36.11 | chr1:25483509–25483621 | 113 | AT_rich | − | 4921 | 5034 | 91.15 | 12 | TTGTATTT-TAA /TTGCATTTCTAA | TTAGAA | /L1ME3E | ||||||||||||
| 1p34.2 | chr1:42779299–42779471 | 172 | A_rich/T_rich | + | 4819 | 4991 | 83.33 | 11 | TTACTAGACAT | ATAGAA | |||||||||||||
| 1p22.1 | chr1:92972728–92972802 | 74 | A_rich | − | 4960 | 5034 | 90.41 | 8 | TTATGTTT | TAAAAA | |||||||||||||
| 1q23.1 | chr1:157139813–157139887 | 74 | A_rich | + | 4960 | 5034 | 91.78 | 9 | AGACATAAT | TGAGAA | |||||||||||||
| 1q24.2 | chr1:169878692–169878920 | 175 | AT_rich | − | 4848 | 5023 | 81.58 | 11 | ACCTGGCAGAA | TTAAAA | MER5B/ | ||||||||||||
| 2p16.1 | chr2:59830731–59830959 | 220 | A_rich | − | 4814 | 5034 | 83.91 | 5 | GAGTG | TTAAAA | |||||||||||||
| 2q24.1 | chr2:159440392-159440797 | 437 | A_rich | - | 4595 | 5032 | 78.08 | 14 | TGCCAG-GGTATTC / TGCCAGCGGAATTC | TTGGAA | |||||||||||||
| 2q37.1 | chr2:232110068-232110184 | 118 | (TTAGGG)n | + | 4909 | 5027 | 85.14 | 28 | GTCACGCGGGTGTGTTCGTTTGCAGGGA/ GTTACACAAATATGTTAGCTAGAAAGTA | CTAAGA | |||||||||||||
| 3q23 | chr3:142384659-142384747 | 89 | (T)n | - | 4945 | 5034 | 90.91 | 15 | TGATGGAACAATTTT | TTTAAT | |||||||||||||
| 3q26.33 | chr3:179497344-179497506 | 128 | (T)n | + | 4906 | 5034 | 89.84 | 9 | GAAAGGGAG/GAAAGGCAG | AAAGAG | AluSc8/ | ||||||||||||
| 4p16.1 | chr4:7584187-7584364 | 177 | AT-rich | + | 4857 | 5034 | 92.66 | 18 | CAATTAAAGGATATAAAA/ CAATTAAAGGATAAAACA | TTAACA | |||||||||||||
| 4q21.3 | chr4:86968242–86968396 | 155 | T-rich | − | 4879 | 5034 | 83.33 | 14 | {LG}TAGAATAGTACTCT/TAAAATAGTACTCT | TCTAAA | |||||||||||||
| 4q26 | chr4:120158525–120158690 | 166 | AT_rich | + | 4867 | 5033 | 91.52 | 13 | AGAATCAGCCTGT/AGAGTCAGCCTGT | TTATGT | Tigger1 | ||||||||||||
| 8q11.21 | chr8:49375957–49376033 | 76 | A-rich | − | 4958 | 5034 | 93.33 | 16 | G–CTGGCTAAGGTTG/ GGTCAAGCCAAGATGG | TCAAAG | |||||||||||||
| 8q12.1 | chr8:56821654–56822036 | 369 | AT-rich | − | 4613 | 4982 | 69.67 | 12 | ACATTTTCTTTA | AAATGT | /L1PB4 | ||||||||||||
| 8q22.1 | chr8:98577734–98577938 | 187 | (TAAA)n | + | 4847 | 5034 | 82.57 | 6 | GGGACA | ACAGAA | |||||||||||||
| 9q21.13 | chr9:79186649–79186950 | 335 | A-rich | + | 4699 | 5034 | 85.37 | 13 | AATAATAATGAGA/AATAATAATAATA | AAGAAA | |||||||||||||
| 9q33.3 | chr9:126813738–126813813 | 75 | A-rich | − | 4959 | 5034 | 93.33 | 14 | AAATCATCATCTC/GAATCACCATCTCA | CTAAAA | |||||||||||||
| 10q21.3 | chr10:68805211–68805496 | 309 | (CAA)n/AT-rich | – | 4725 | 5034 | 87.5 | ND | TTTAAA | /MIRc | |||||||||||||
| 10q25.3 | chr10:115461374−115464039 | 101 | T-rich | – | 4918 | 5019 | 90.32 | 8 | AAAGGTGA/AAAGTTTA | ACAGAA | Tigger1 | ||||||||||||
| 12q24.32 | chr12:127650453–127650987 | 424 | (AAAAAT)n | − | 4610 | 5034 | 93.84 | 16 | AAAAA-ATCAGATCCC/ AAAACCATCAGATCCC | TTAAAA | |||||||||||||
| 17q25.2 | chr17:75158032–75158389 | 417 | (AAAAAT)n | − | 4617 | 5034 | 72.58 | 25 | TCAGCCATGGGGATCTGATGGTTTT/ TCAGCCAATGGGATCTGATT-TTTT | AAAAGT | |||||||||||||
| 18q11.2 | chr18:21191674–21191827 | 130 | A-rich | + | 4904 | 5034 | 87.42 | 9 | CCATTGATA | GAAAAC | |||||||||||||
| 18q23 | chr18:77772847–77773065 | 215 | A-rich | + | 4818 | 5033 | 93.12 | 19 | AAATTTTTTTTTTTTTTTT | CCATTA | /AluY | ||||||||||||
| 22q11.22 | chr22:22210671–22210856 | 211 | A_rich | + | 4823 | 5034 | 69.75 | 6 | CAGATA | ACAAAA | |||||||||||||
| Xq25 | chrX:121106711–121106776 | 65 | A_rich | + | 4963 | 5028 | 95.38 | ND | ATGAAA | L1MA8 | |||||||||||||
| Xq27 | chrX:139858393–139858607 | 215 | ND | + | 4821 | 5035 | 84.7 | ND | AAACAT | L1P3 | |||||||||||||
| B. Segmental duplications of LSU fragment/Alu clusters (sd28alu) | |||||||||||||||||||||||
| 4q21.1 | chr4:76807176–76807697 | 522 | AT-rich | + | 4511 | 5033 | 82.8 | ND | AluSq/ | ||||||||||||||
| 8q11.1 | chr8:46951571–46952097 | 534 | ND | − | 4501 | 5035 | 78.32 | ND | /AluSq2 | ||||||||||||||
| 12p11.1 | chr12:34372315–34372728 | 515 | ND | − | 4513 | 5028 | 94.05 | ND | /AluSq2 | ||||||||||||||
| 12q12 | chr12:38531458–38531930 | 539 | ND | + | 4495 | 5034 | 82.55 | ND | AluSx1/ | ||||||||||||||
| 19q13.31 | chr19:43911666–43912167 | 555 | ND | + | 4480 | 5035 | 79.21 | 7 | CTTCGCA | AluSq/ | |||||||||||||
| 21q11.2 | chr21:15456973–15457500 | 527 | ND | − | 4501 | 5028 | 81.8 | ND | /AluSq2 | ||||||||||||||
| Yq11.221 | chrY:19691914–19692439 | 527 | ND | − | 4501 | 5028 | 79.19 | ND | /AluSx1 | ||||||||||||||
| Yq11.222 | chrY:20487334–20487859 | 527 | ND | + | 4501 | 5028 | 79.19 | ND | AluSx1/ | ||||||||||||||
| C. 3′-LSU fragments of larger size than predicted for a SINE | |||||||||||||||||||||||
| 2q21.2 | chr2:133036251–133039236 | 3,232 | Acro1 | − | 1736 | 4968 | 83.38 | ND | |||||||||||||||
| 19q13.2 | chr19:42069990–42070694 | 771 | (TTTA)n | − | 4253 | 5024 | 78.73 | ND | |||||||||||||||
Sequences given in bold indicate 100% identity across annotated TSD sequence. Sequences given in italics indicate 85–99% identity across TSD sequence, both 5′- and 3′-TSDs are included (separated by slash) with gaps in alignment between them indicated by a hyphen. EN sites: endonuclease recognition sequence. ND, not detected.
Identification of SINE28 loci that overlapped with genes, mRNAs, transcription, and other features were made by position-based queries of hg19 with subsequent visual examination of the annotation tracks within the UCSC Browser Window and analysis of UCSC Table Browser spreadsheet outputs.
To establish whether SINE28s were ancestral or derived SINE sequences within each species interrogated, LSU rDNA sequences and 50 putative SINE28s from representative mammalian lineages were made into a BLAST index. Because very few full-length mammalian LSU (28S) rDNA sequences are available within the ribosomal database SILVA (Quast et al. 2012), we screened the NCBI nonredundant nucleotide database for full-length LSU rDNA sequences using BLAST. Only full-length sequences were included in the compiled index; as such, with the exception of human, no great ape rDNAs were included in this screen. The same 50 SINE28s were then aligned against this SINE28/LSU index using BLAST. A SINE28 was considered ancestral if it had higher sequence identity to a SINE28 from a different species than to the LSU rDNA sequences from its resident genome. Alternatively, if the SINE28 element had higher sequence similarity to the LSU rDNA within the same species, we considered these SINE28s recently derived.
RNA Polymerase Inhibitor Treatments
HeLa cells (1 × 107) were seeded in a T75 flask 24-h prior to treatment with RNA polymerase inhibitor reagents. Cells were treated with RNA polymerase III inhibitor, (Calbiochem 557403 EMD Millipore) at a final concentration of 30 uM in culture medium and were harvested or fixed at 4, 8, and 12, and 24-h post-treatment. Cells treated with RNA polymerase II inhibitor, alpha amanitin (Sigma A2263), at a final concentration of 0.5 µg/ml, were harvested or fixed at 48-h post-treatment. A 24-h treatment with CX-5461 (Selleckchem), an RNA polymerase I inhibitor, at a final concentration of 2 uM was used to selectively inhibit rRNA synthesis. Mock treatment was performed in inhibitor resuspension buffer. A final concentration of 0.2 mM 5-ethynyl uridine was added to cells at 50% confluency and subsequently harvested 24-h posttreatment. Total nascent RNA transcripts were captured using the Click-iT Nascent RNA Capture (Invitrogen) assay per manufacturer’s instructions.
Quantitative Real Time-Polymerase Chain Reaction
Small RNA (<200 nt) and total RNA fractions were extracted from cells using the mirVana miRNA Isolation Kit per manufacturer’s instructions (Ambion AM1560). RNAs (>200 bp) were treated with Ambion TURBO DNA-free prior to cDNA synthesis with the qScript cDNA Synthesis Kit Quanta Biosciences. cDNA was prepared from RNAs (<200 bp) using the NCode VILO miRNA cDNA Synthesis Kit (Invitrogen). The Universal NCode primer was used as the reverse primer for all small RNA quantitative polymerase chain reaction (qPCR) analyses. Quantitative reverse transcription PCR (qRT-PCR) of treated and untreated cells was performed using Biorad Sybr Green Supermix on a Biorad iCycler with primers for the LSU (5′-end and middle segment), SINE28, 18S, U1, U6, and beta actin. RT-PCR conditions were as follows: initial denaturation at 94 °C for 3 min, 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s × 35, 95 C for 1 min followed by a melt curve generation. Forward and reverse primer sequences, respectively, from 5′ to 3′, are as follows:
SINE 28: (CCTTGTGTCGAGGGCTGACTT, GTTCGTGTGGAACCTGGCGCTAAAC);
5′-LSU (CGCGACCTCAGATCAGACGTGG, GGGCTCTTCCCTGTTCACTCGC);
SINE28 mid (CAGGGGAATCCGACTGTTTA, CGCGCTTCATTGAATTTCTT);
beta actin (ACAGAGCCTCGCCTTTGC, CACGATGGAGGGGAAGACG);
18S (GTTCGTTCGCTCGCTCGT, AACGACACGCCCTTCTTTCT);
U1 (TACTTACCTGGCAGGGGAGATAC, GGACGCAGTCCCCCACTAC);
U6 (CCATGATCACGAAGGTGGTTT, ATGCAGTCGAGTTTCCCACAT).
PCR efficiency was computed using the equation E = 10(−1/slope) (Pfaffl 2001). Data were first normalized to the appropriate reference RNAs (18S, beta actin, U1, or U6), those that were known to be unaffected by the RNA polymerase inhibitor treatment used in the experiment. Relative expression values were subsequently calculated (Pfaffl 2001) for the control gene (known to be affected by the inhibitor) and the target gene (SINE28) for both sample types (mock-treated cells and cells treated with RNA polymerase inhibitors). Standard deviation and confidence value statistics were evaluated for both the normalization and relative expression calculations.
Results
Unknown SINE Loci within the Human Genome
We identified 37 loci within the reference human genome (hg19) RepeatMasker track (Fujita et al. 2011) of the UCSC genome browser (Kent et al. 2002) containing a fragment of the 3′-end of the 28S rDNA (LSU), with most fragments ranging from 65 bp to 555 bp in length (table 1). A large number of these loci (26) were immediately adjacent to an A-rich, other simple or low complex repeat (table 1A), implicating retrotransposition following aberrant polyadenylation in their origin (Ostertag and Kazazian 2001).
Premature termination of transcripts is known to trigger an aberrant transcript surveillance pathway that proceeds with polyadenylation of such transcripts followed by exosome degradation (reviewed in Wu and Brewer [2012]). Such polyadenylated transcripts are typically transient, but a population of abundant transcripts may be available for reverse transcription and insertion into genomic DNA via enzymes contained within mobile elements, such as LINEs (Ostertag and Kazazian 2001). An LSU rRNA derived fragment resulting from premature termination of the 28 S rRNA would be predicted to be heavily biased to the 5′-end of the LSU sequence and contain poly-A tracts at their 3′-end (Shcherbik et al. 2010). To determine if aberrant polyadenylation of incomplete LSU rRNA transcripts resulted in the genomic loci derived with an immediate 3′ simple repeat tail, rather than our observed bias toward loci derived from the 3′-end of the LSU, we screened the human genome (hg19) with the Repbase consensus human LSU sequence, LSU-rRNA_Hsa. A total of 109 fragments of the LSU were found randomly distributed throughout the human reference genome (supplementary table S1, Supplementary Material online and fig. 1A). This screen validated the original 37 (33.9% of the 109 total fragments identified) we isolated with identity to the 3′ portion of LSU (terminal ∼500 nt); moreover, this group of 3′-end fragments was the most frequent single fragment type found in our search (fig. 1B and supplementary table S1, Supplementary Material online). Further, a total of 33 of the 109 LSU fragments were adjacent to a simple or low complex repeat, with 26 of these 33 (70.3%) “tailed fragments” derived from the 3′ portion of the LSU, thus overlapping with our original set of 37 individual 3′-LSU loci (supplementary table S1, Supplementary Material online and fig. 1A).
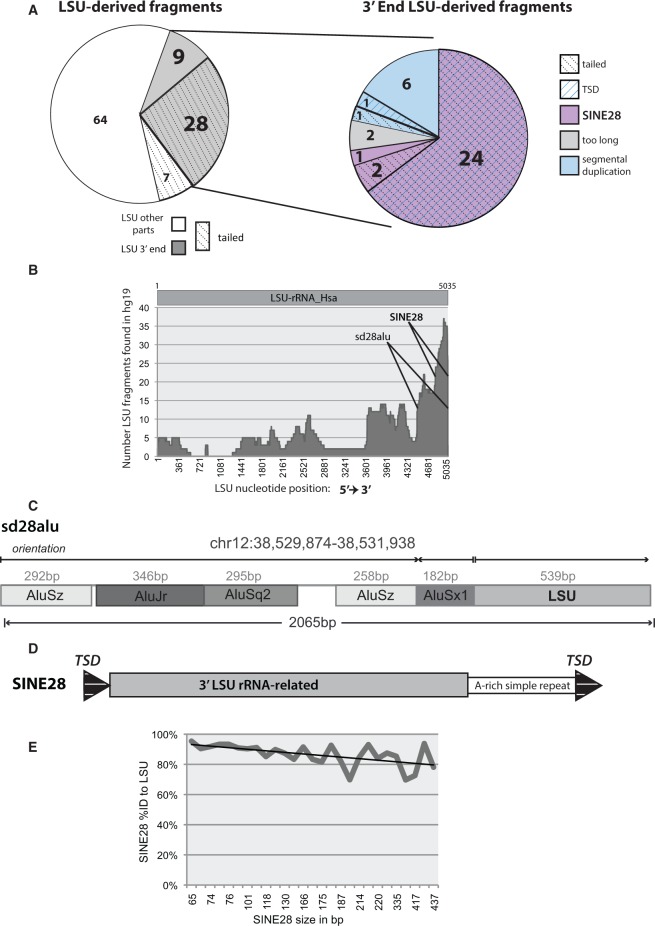
Fig. 1.—
Novel SINE derived from the 3′-end of the LSU rRNA identified in the human genome. (A) (Left) Total distribution of LSU-derived fragments defined by portion (3′-end or other) and whether they contain a tail. (Right) Classification of SINE28s among all 3′-LSU fragments by feature, including: presence of a tail and/or TSD, appropriate length, and evidence for segmental duplication. (B) Nucleotide by nucleotide count of the number of LSU fragments (y axis) found in the human genome corresponding to a specific LSU nucleotide position (x axis) as aligned to the LSU sequence to hg19 via BLAT (Kent 2002). (C) Structural features of the larger 3′-LSU fragments that originate from segmental duplications (sd) and are part of a cluster of repeats that includes several Alu elements (sd28alu) found on chr12. (D) Structural features of SINE28 including the LSU RNA “head,” A-rich simple repeat tail, and TSD. (E) The percent identity of SINE28s (y axis) compared with LSU-rRNA_Hsa consensus sequence is inversely related to element length (as indicated by the trend line; x axis).
Eight of the 37 identified 3′-LSU fragments are 535 bp (±20 bp) in length, originate from the exact same 3′-region of the LSU and map to 4q21.1, 8q11.1, 12p11.1, 12q12,19q13.31, 21q11.2, Yq11.221, and Yq11.222 (table 1). Of these 535 bp 3′-LSU fragments, only the LSU fragment at 4q21.1 could be classified as a tailed fragment (i.e., containing a simple repeat tail), but all eight fragments are immediately adjacent to an Alu element and in fact are part of a complex of Alus with a total cluster length of approximately 2,000 bp (fig. 1C). The Alus found within each 2 kb cluster are of the same Alu subfamily and configuration, indicating each Alu cluster and associated 3′-LSU may be a composite transposable element (TE) or derived from large-scale duplication events. Moreover, each cluster is characterized by locus-specific nucleotide insertions and deletions among the clusters, indicating independent accumulation of mutations since their derivation. To determine whether these clusters were bona fide composite elements derived from retrotransposition or the same configuration at distant loci derived simply by segmental duplication, we mapped unique, nonrepeat sequence from 10 kb away in both the 3′- and 5′-direction from one of the 535 bp SINE28/ Alu clusters to hg19 using BLAT (Kent 2002). Six of the eight cluster-containing loci were identified in this search, suggesting at least these six clusters (8q11.1, 12p11.1, 12q12, 21q11.2, Yq11.221, and Yq11.222) originated from interchromosomal segmental duplications of the same progenitor locus. The Alu-adjacent 3′-LSU fragments at 4q21.1 and 19q13.31 were not identified by this segmental duplication search, varied significantly from the first six in sequence and structure, and thus did not appear to be part of the same series of segmental duplication events. However, seven of the eight loci, including both 4q21 and 19q13 were adjacent to regions containing high levels of segmental duplications within the UCSC Genome Browser (Fujita et al. 2011), indicating they likely are part of the complex segmental duplication events that characterize these two loci. Thus, each of these eight loci are derived by segmental duplication events (fig. 1A); we have named these segmental duplication LSU fragment/Alu clusters sd28alu (table 1B and fig. 1C).
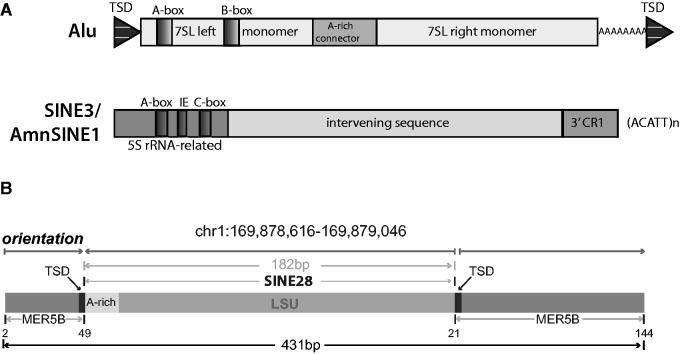
Considering the overall length of the 29 remaining 3′-LSU derived fragments identified in hg19 (65–437 bp, with a mean length of 194 bp and median at 171 bp) and given that 25 of these 29 fragments contained a 3′-simple repeat tail (table 1), we hypothesized that these tailed 3′-LSU fragments may represent a novel SINE, heretofore referred to as SINE28 (fig. 1D). Each SINE28 has varying degrees of identity to LSU-rRNA_Hsa (mean = 86.39%, SD = 7.20%, median = 87.50%) with an expected general trend of longer elements having lower overall identity (table 1A and fig. 1E and supplementary fig. S1, Supplementary Material online). This trend, however, does have some exceptions; for example, SINE28 at 12q24.32 is the second longest SINE28 we identified, yet it maintains 94% identity to the consensus LSU (table 1A). The overall average fragment size and the presence of a 3′ A-rich simple repeat tail is similar to that of Alu elements, the prolific primate SINE (Okada 1991; Batzer et al. 1996) that carries a 7SL RNA head (Ullu and Tschudi 1984) and a LINE1 recognized A-rich tail (Dewannieux et al. 2003; Dewannieux and Heidmann 2005; fig. 2A). Moreover, the presence of an LSU rRNA-derived region comprising the putative SINE28 is analogous to the zebra fish SINE3 and the mammalian AmnSINE1 elements, both of which carry a 5S rRNA derived “head” (Kapitonov and Jurka 2003; Gogolevsky et al. 2009; fig. 2A).
Fig. 2.—
SINE28 shows features of retrotransposition. (A) Schematic of (top) primate Alus are derived from 7SL RNA with a LINE1 recognized A-rich tail (adapted from Mills et al. [2007]) and (bottom) SINE3/AmnSINE1 in vertebrates with a 5S rRNA head and CR1 recognized tail (adapted from Kapitonov and Jurka [2003]). (B) An example of a SINE28 embedded in an otherwise intact TE in the human genome (MER5B).
To further explore whether these SINE28s may be retroelements of the same class as Alus, we tested for signatures of retrotransposition by LINE1-encoded proteins (Dewannieux et al. 2003; Dewannieux and Heidmann 2005). A byproduct of LINE1 retrotransposition is the formation of small direct repeats, called TSDs, bounding the element (Ostertag and Kazazian 2001). Although the absence of recognizable TSDs does not definitively eliminate a candidate from SINE classification because sequence identity between two duplicated target sites will be lower among elements from older insertion events, largely due to genetic drift, the presence of TSDs is a strong indicator of recent retrotransposition. TSDs of varying lengths (5–28)bp bounding 24 of the 29 SINE28 elements (table 1A and C) were identified through visual examination of adjacent genomic DNA for each fragment. The list of 29 putative SINE28 loci was narrowed to 27 because the tailed 3′-LSU fragments at 2q21.2 and 19q13.2 were longer in length (3,232 and 771 bp, respectively) (table 1A and fig. 1A) than canonical SINEs (typically ≤500 bp) and lacked detectable TSDs, thus reducing confidence in classifying these as SINE28s (table 1C). In addition to TSDs, specific signatures of targeted EN cleavage by LINE1-mediated retrotransposition have been identified for mammalian SINEs such as Alus (Feng et al. 1996; Jurka 1997; Cost and Boeke 1998). The most frequently observed hexamers associated with primary nicking sites for Alus and ID elements (Jurka 1997) were used as a database to query sequences surrounding the TSD (or LSU-homologous region for elements lacking TSDs). An EN hexamer was identified for each of the 27 SINE28s (table 1A), further supporting the hypothesis that LINE1 machinery is involved in the insertion of the LSU-fragments into the genome.
Another genomic signature observed for some actively transposing elements is that their insertion site lies embedded within another transposable element (Hughes and Coffin 2001). Four of the 27 putative SINE28s (1q24.2, 4q26, 10q25.3, and Xq25) are embedded within another transposable element (table 1 and fig. 2B). For example, chromosome 1q24.2 SINE28 is bounded by TSDs (table 1) and is found within a MER5B element (fig. 2B), disrupting the open reading frame of this transposable element. While none of these descriptive features, small overall length, presence of identifiable TSDs, carrying a simple repeat tail, and residing within another mobile element, on their own define a genomic SINE, the presence of these in combination adds considerable support to the hypothesis these 27 SINE28 loci (fig. 1A and D) are previously unknown SINEs within the human genome.
SINE28s Are Transcriptionally Active and RNA Polymerase III Dependent
All currently classified SINEs are derived from cellular RNAs originating from RNA polymerase III (Pol III) transcripts (e.g., tRNAs [Daniels and Deininger 1985; Okada and Ohshima 1993], 7SL RNA [Ullu and Tschudi 1984], or 5S rRNA [Kapitonov and Jurka 2003; Gogolevsky et al. 2009]). Transcription of these mobile elements is required for their subsequent reverse transcription and reintegration into the host genome. The 28S ribosomal subunit, from which the putative SINE28 is derived, is transcribed by RNA polymerase I (Srivastava and Schlessinger 1991). We reasoned that SINE28 would, like other SINE classes, carry the capacity to promote RNA polymerase III transcription.
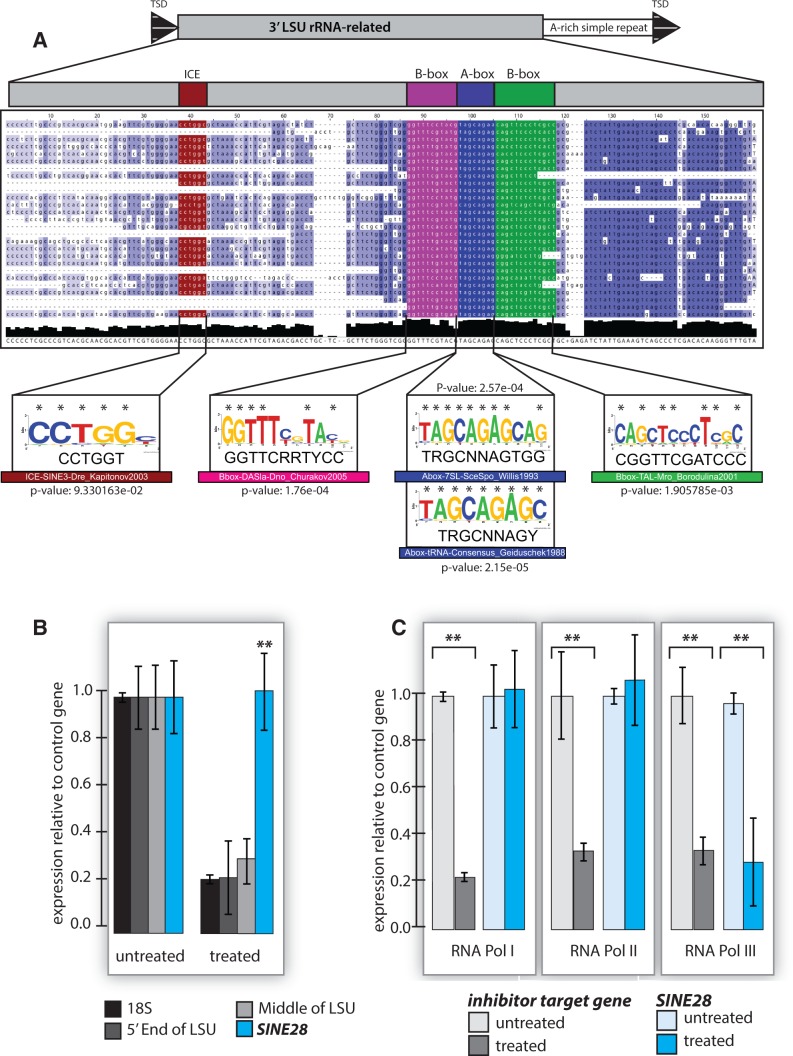
Three types of RNA polymerase III promoters have been described, two of which are distinguished from other polymerases as they are internal to the transcription unit itself (reviewed in Schramm and Hernandez [2002] and Dieci et al. [2007]). Type 1 promoters are internal promoters found in 5S rDNA and contain an internal control region consisting of an A-box, an intervening sequence (IE) and a C-box (Paule 2000). The other internal RNA polymerase III promoter (Type 2), characteristic of tRNAs and 7SL RNAs, has an A-box separated by some intervening sequence from a B-box (Paule 2000). Type 3 RNA polymerase III promoters have external regulatory elements, distal sequence element, and proximal sequence element (Paule 2000).
A multiple sequence alignment of all identified human SINE28s (table 1 A) compared with previously annotated RNA Pol III promoter elements (supplementary table S2, Supplementary Material online) showed a statistically significant conservation of nucleotides within the predicted motifs of IE, A-box and B-box (fig. 3A), indicating a potential for active transcription via RNA polymerase III. However, the orientation of these putative promoter elements differs from those of most SINE classes, rendering an assumption for RNA polymerase III transcription circumstantial. To confirm RNA polymerase III recognition, we assessed expression levels of SINE28 in cells treated with RNA polymerase inhibitors for RNA polymerase I, II, or III. Given that rRNA transcripts may be stable despite the inhibition of new transcription, our assay targeted only nascent SINE28 transcripts using an ethylene uridine ribonucleotide analog (Click-iT technology) and primers that distinguish SINE28 from the canonical 28S LSU. SINE28 transcripts as measured by quantitative RT-PCR are not affected by inhibition of RNA polymerase inhibitors for either RNA polymerase I or RNA polymerase II; however, SINE28 transcripts are significantly reduced in cells treated with RNA polymerase III inhibitor (fig. 3B and C).
Fig. 3.—
SINE28 is RNA polymerase III dependent. (A) A multiple sequence alignment of all identified human SINE28s (table 1A) with putative RNA polymerase III promoter elements shown (supplementary table S2, Supplementary Material online). Below, DNA logos of predicted motifs with RNA polymerase III; conserved nucleotides are indicated by an asterisk. The degree of conservation among SINE28s is depicted at the bottom (black). (B) qRT-PCR of SINE28 (blue), the 18S (black), 5′-end of the 28S LSU (dark gray) and a portion in the middle of the 28S LSU (light gray) normalized to U1, pre and post RNA polymerase I inhibition (**P < 0.001). (C) qRT-PCR of SINE28 (blue) compared with a positive control target gene (gray) pre- and postspecific RNA polymerase inhibition (**P < 0.001). All data are normalized to an invariant transcript (control); confidence intervals (95%) are indicated. From left: RNA polymerase I inhibition—positive target 18SrRNA and control β-actin; RNA polymerase II inhibition—positive target β-actin and control 18SrRNA; RNA polymerase III inhibition—positive target U6 and control U1.
As independent validation that SINE28s are transcriptionally competent, transcripts from specific SINE28 loci in human were identified via in silico analyses. Sequence reads that distinguish SINE28 from canonical 28S rRNAs were identified for 27 SINE28 loci within one or more publicly available transcriptome databases within annotation tracks of the Groups “mRNA and expressed sequence tag” and “expression” following position-based queries within UCSC Genome Browser. Transcription of SINE28 (as assayed by RNA-seq on nine cell lines from ENCODE) was also evident within the “regulation” annotation tracks (data not shown).
SINE28s Have Been Transpositionally Active in the Primate Lineage
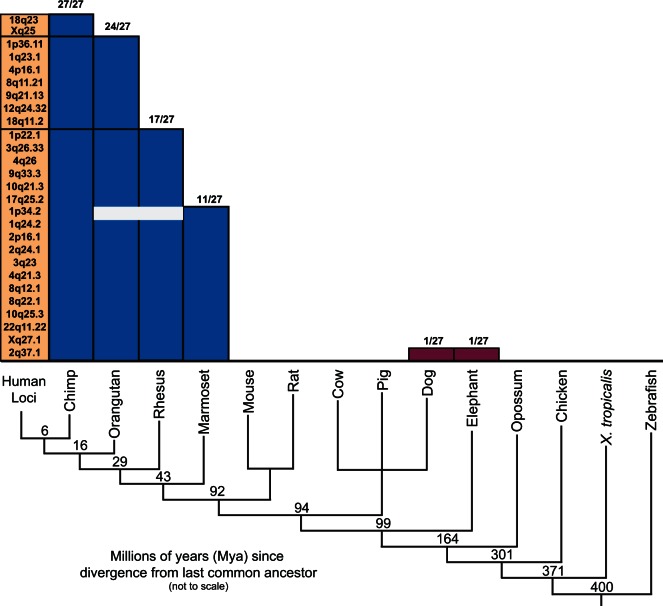
To investigate whether the SINE28s found in the human genome are evolutionarily ancient or recent insertions, we examined the Comparative Genomics annotation tracks in the UCSC genome browser (Fujita et al. 2011) for conservation of insertions across primate genomes. All of the 27 SINE28s identified in hg19 are present at orthologous chromosome locations in chimpanzee (fig. 4), indicating they have been derived by descent from the chimp/human common ancestor. The SINE28s at 18q23 and Xq25 are not found in orthologous locations in orangutan, indicating activity sometime between 16 and 6 Mya, prior to the human/chimp split (Goodman et al. 1998). The SINE28 at 18q23 carries signatures of recent transposition, including TSDs that retain 100% identity. The SINE28 at Xq25, however, lacks a detectable TSD, lowering the likelihood it is a recently inserted element. Moreover, this SINE is embedded within a LINE (L1MA8) that is not found at this orthologous region of the orangutan X chromosome. Thus, it is likely this element traveled with its resident LINE rather than inserted within the LINE at this location. Seven SINE28 loci were found to be restricted to human/chimp/orangutan and six SINE28 loci were found within human/chimp/orangutan/rhesus, indicating activity after a last common ancestor was shared among these groups, 29 and 43 Ma, respectively.
Fig. 4.—
Phylogenetic dating of SINE28. Left are listed all 27 SINE28 loci in the human genome (hg19). Blue bars indicate the presence of SINE28 at the orthologous locus of the species indicated at the bottom. White boxes indicate an element missing at an orthologous locus. Purple are “outlier” orthologous SINE28s found in the dog and elephant genomes. Cladogram is not to scale; numbers on cladogram represent millions of years since a last shared common ancestor (Hedges et al. 2006).
Confirmation of our phylogenetic dating of SINE28s was accomplished by comparing other elements in each orthologous region for presence or absence in other genomes (supplementary table S3, Supplementary Material online). For instance, the SINE28 at 10q21.3 is not found in marmoset but is found in chimp, orangutan, and rhesus macaque (fig. 4 and supplementary fig. S2, Supplementary Material online). In the region immediately surrounding this SINE28, other repeats verify the correct assembly of the orthologous regions within these other genomes. Slightly upstream of the SINE28 at 10q21.3 is a MIR element, a mammal-specific SINE element (Veyrunes et al. 2006) that is found only in mammalian conservation tracks. In addition, in the immediate vicinity of the SINE 28 at 10q21.3 is an L1PA16, a primate-specific LINE1 (Veyrunes et al. 2006), and as such only found in the primate conservation tracks.
The SINE28 at 1p34.2 (fig. 4) is found at orthologous loci in both chimp and marmoset, but not within orangutan or rhesus macaque. Such phylogenetically incongruent orthologs could represent independent insertional events at orthologous locations in each lineage that this specific SINE28 is found within, including human, chimp, and marmoset. Conversely, such phylogenetic incongruence could be the result of independent loss at this specific locus in both orangutan and rhesus macaque. However, the observation of phylogenetically incongruent orthologs of the SINE28 at 1p34.2 is more likely a reflection of incomplete assemblies for the orangutan and rhesus macaque genomes rather than independent insertion or loss events across multiple lineages. Interestingly, this phylogenetic dating shows that the segmentally duplicated sd28alu loci have been recently derived as well. None of the sd28alu loci (table 1) are found in orthologous regions in species more divergent than rhesus macaque, and two are found restricted to humans and great apes (4q21.1 and 19q13.31), indicating derivation within the last 29 Myr (fig. 4). Notably, all of the human SINE28s had corresponding positional orthologs restricted to primate lineages, with the exception of the SINE28 at 2q37.1. The 2q37.1 SINE28 is found at orthologous loci in both the dog and elephant comparative genome tracks.
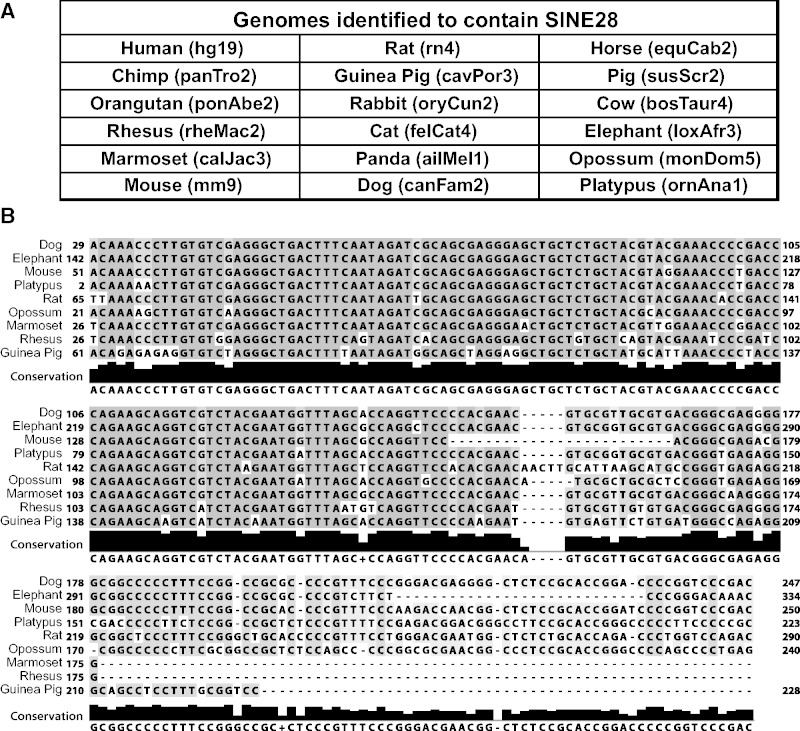
SINE28s Are Present in All Mammals
All SINE28 loci annotated in hg19 herein appear restricted to the primate lineage with the exception of the SINE28 locus at 2q37.1. The inclusion of the 2q37.1 SINE28 within the assembly of both dog and elephant prompted a closer investigation to determine if these are in fact orthologs, assembly errors or perhaps human contamination (Longo et al. 2011) in the respective assemblies. The scaffold_55 of elephant (loxAfr3), indicated in the UCSC comparative genomic track as orthologous to the human the 2q37.1 SINE28 region, did not have any identifiable (using BLAT and RepeatMasker) SINE28 sequences, suggesting the apparent ortholog in elephant was the result of a computational error in the elephant/human comparison or in the assembly of this elephant contig. The human 2q37.1 SINE28 locus is orthologous to dog chromosome 25, where a SINE28 can be identified. However, due to limited sequence availability, we were not able to confirm that the dog chromosome 25 SINE28 locus was a completely orthologous locus to human 2q37.1 rather than a recent insertion event. Human contamination was eliminated as a likely confounding factor as there is no identifiable, unique human sequence within this dog chromosome. Thus, the presence of SINE28 in a species outside of the primate lineage prompted a search for SINE28s beyond the human orthologous SINE28 loci.
Screening the UCSC genomes with both LSU consensus sequence and SINE28 sequences identified SINE28s in all mammalian species examined (fig. 5A and supplementary fig. S3, Supplementary Material online). In species encompassing all mammalian infraclasses (Eutheria, Metatheria, and Prototheria), we found the 3′-LSU fragment adjacent to a simple or low-complex repeat (supplementary fig. S3, Supplementary Material online). Like the primate SINE28s, many of the mammalian elements examined are surrounded by TSDs (59%). Interestingly, these elements were highly conserved across the mammalian order, although they were notably not present at orthologous loci across all of these lineages (e.g., fig. 5B).
Fig. 5.—
SINE28 is present and conserved in mammals. (A) Table of all genomes with identifiable SINE28s. (B) A multiple sequence alignment of SINE28 showing a high degree of similarity (dark gray) even across very distant mammalian species. The degree of conservation is depicted at the bottom (black).
The high degree of identity for SINE28s among divergent mammalian lineages could either be the result of stabilizing selection of an ancestor element (with no assumption on its presumed mobility) or the derivation of new SINEs from nascent LSU sequences in a lineage-specific fashion. We reasoned that if the broad range of SINE28s were derived from a conserved element(s) in a common mammalian ancestor, the SINE28s observed within a single mammalian species would be more similar to other SINE28s in divergent, but related taxa, than to the ribosomal sequences within that same lineage. If, however, these SINE28s are rederived from LSU RNAs in each lineage independently, we would expect SINE28s to be more similar to the host LSU RNA sequences than to SINE28s from any other species. To test for either scenario, we first identified annotated LSU sequences by screening the NCBI nucleotide database with the most conserved 100 bp of the multispecies SINE28 alignment (fig. 5B). LSUs identified in this screen were combined with 50 mammalian SINE28s spanning all lineages (as in fig. 4A) and converted into a reference index (supplementary table S4, Supplementary Material online). The same 50 SINE28s were aligned to this index to identify the closest relative for each element by sequence identity through BLAST (Altschul et al. 1997). If a SINE28 was more similar to an interspecies SINE28 and/or interspecies LSU RNA than to an intraspecies SINE28, then we considered that element to have originated from an ancestral SINE28, thus representing a sequence shared by descent. If, on the other hand, a SINE28 carried higher sequence identity to intraspecies LSU RNA than to any other sequence, we considered it to be a newly derived element within that lineage.
For many species, the percent identities of alignments were equivalent between interspecies SINE28s and intraspecies LSU RNA, preventing delineation of identity by descent or rederivation of new elements from the LSU RNA progenitor. For some species, including rabbit and rat, it is clear that SINE28s have been derived both by descent and recent derivation from host rRNAs. In each of these lineages, we found example SINE28s that have high similarity to the host LSU RNA as well as elements that were very different from the host LSU RNA and nearly identical to other SINE28s found in a different species (supplementary table S4, Supplementary Material online). Our phylogenetic dating (fig. 4) indicates that the most recent human insertion occurred prior to the chimp/human split (∼6 Mya). In other species (e.g., cow), all of the examined elements are most similar to interspecies SINE28s or native LSU RNAs, indicating they were all of ancestral origin. Interestingly, all of the SINE28s we examined in mouse appeared to be recently derived from mouse LSU RNA, perhaps a reflection of the extensive genomic evolution the Mus lineage has experienced (Veyrunes et al. 2006; Mlynarski et al. 2010).
Discussion
Herein we have described a previously unidentified SINE element derived from the 3′-end of the 28S rRNA (LSU), have named this element SINE28, and have verified that this element is present in all mammalian genomes examined, encompassing all mammalian infraclasses. The canonical SINE28 carries an A-rich repeat tail and is found flanked by TSDs, like the primate Alus, implicating LINE1s as the likely mechanism of transposition ([Feng et al. 1996; Cost and Boeke 1998] and reviewed in Kramerov and Vassetzky [2005]). Interestingly, while fragments derived from portions spanning the entire length of the LSU can be found at low incidence within the human genome (Munro et al. 1986; Wang et al. 1997), only the 3′ most portion (SINE28) has characteristic features of a transposable element. Nascent transcripts for SINE28 are the result of RNA polymerase III activity rather than the RNA polymerase I activity of its parent rRNA sequence (fig. 3). Furthermore, unlike the RNA polymerase II transcribed R superfamily of non-LTR elements which specifically insert into 28S rRNA genes and encode for ribozymes (thus permitting self-cleavage and processing of the 5′-end of the element out of the 28S cotranscript; Eickbush and Eickbush 2012), SINE28 elements are reliant upon RNA polymerase III for transcription (fig. 3) and are clearly rRNA derived. Finally, while recent data shows that posttranscriptional polyadenylation of abundant 28S fragments occurs in human cells, the addition of these homopolymeric or heteropolymeric poly(A) tails occurs on fragments prematurely truncated and thus completely lacking the 3′ portion of the LSU found within SINE28 (Slomovic et al. 2006).
SINE28s are not reported in either RepBase (Jurka et al. 2005) or SINEBase (Vassetzky and Kramerov 2013) and are not annotated in the human genome (likely because automated genome annotation methods identify them simply as LSU fragments). Thus given that these SINE28 loci are relatively short (<700 bp), nonautonomous (transcribed by the cellular RNA polymerase III from an internal promoter), are likely reliant on LINE1s for reverse transcription, and are consistent with the typical head, body, and tail structure of SINES, we have identified a previously unknown class of SINEs in the human genome.
According to our phylogenetic analysis, this SINE has been recently active within the primate lineage with definable transposition events as recent as the human and chimpanzee divergence (∼6 Mya). Curiously, we identify SINE28s in all mammalian genomes, yet the human elements cataloged herein are also found in orthologous regions within primates, indicating they may no longer be transpositionally active in humans, although they may still undergo transcription as evidenced by sequencing data in public archives for the ENCODE consortium. We could infer two possibilities from this observation: 1) that there is a tendency for SINE28s to be “lost” from the genome over evolutionary time, or 2) that active copies reside in regions of the genome that are not fully annotated, such as centromeres, pericentromeres, and telomeres. In support of this, a recent study of the tammar wallaby centromere-specific contigs within the genome assembly (Renfree et al. 2011) identified SINE28 copies within active centromere regions (Lindsay et al. 2012). Moreover, these SINE28 copies were associated with transcriptional activity as well as centromere-specific histone occupancy (Lindsay et al. 2012), indicating they may be critical players among the elements that demarcate centromere function and/or stability (Hall et al. 2012; Carone et al. 2013).
The high level of sequence identity among SINE28s, even across distantly related species, is particularly interesting. A survey of AmnSINE1 sequences lends support to the proposal that a high level of sequence identity among these SINEs across multiple lineages is a signature of evolutionary constraint and possible exaptation for function (Nishihara et al. 2006). While the sequence similarity observed across SINE28 elements indicate these SINEs are continuously being rederived from the intact LSU sequence in many lineages simultaneously, our comparisons among mammalian SINE28s, their endogenous rRNA counterparts and interspecies SINE28s indicates that elements are also maintained in some lineages from a common ancestor yet experience lower levels of overall nucleotide substitutions (supplementary data S1, Supplementary Material online). Given the high conservation for ribosomal sequences across all eukaryotes, it is intriguing that a ribosomally derived SINE, SINE28, is only found within mammals. This may reflect a divergent composition of other retrotransposons found in nonmammalian genomes since SINE transposition requires the protein machinery of another element (i.e., LINE1s).
The identification of novel transposable elements in the human genome is of note, considering how extensively analyzed and annotated the human reference genome has become. What remains to be determined, however, is not whether novel, and active, transposable elements can be found within the human genome, but rather, why only the 3′-end of the LSU rRNA is captured as a transposable element. Known pathways for RNA decay processing in eukaryotes cannot explain the observation of SINE28s derived specifically from only a 3′ portion of the LSU and carrying a homopolymeric tail interrupted by a short spacer sequence. For example, other fragments from the 5′ portion of the LSU rRNA have been shown to carry poly-A tails but are rapidly shuttled to the exosome for degradation from the 3′-end (Slomovic et al. 2006) as part of the 3′-5′ exosome-mediated decay process. Another form of aberrant mRNA transcript processing proceeds from the 5′-end of transcripts following decapping and could thus conceivably produce a fragment of only the 3′ portion of a given transcript (reviewed in Eulalio et al. [2007]). However, this process is often coupled with deadenylation, reducing the likelihood of identifying 5′-truncated rRNAs carrying a tail as observed for SINE28s.
The identification of RNA polymerase III conserved target sequences within the 3′-region of the LSU and within its derived SINE28s may provide a clue to their origin from aberrant rRNA processing. These conserved RNA polymerase III motifs may serve a secondary function as a docking site for an RNA binding protein that protects SINE RNAs, as has been observed for Alus (Chang et al. 1994). If rRNA processing and fragmentation resulted in polyadenylation of 5′-truncated fragments that would typically proceed through exosome degradation, perhaps a small number of transcripts containing only a 3′-end of the sequence escape this fate if blocked by inappropriate recognition of these motifs by a SINE RNA binding protein.
Although the mechanism for initial derivation of SINE28s from rRNAs is highly speculative, the fact that these elements have been rederived in many independent lineages suggests a dynamic process that is active and maintained, albeit at a low frequency. Developing a better understanding of the targeted sequence preference in the derivation of SINE28s may elucidate a more general mechanism underlying the origin of SINEs as well as the genomic consequences of such retrotransposition events.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Thanks to Craig Obergfell and the Center for Applied Genetics and Technology for data analysis and management support. M.S.L. and R.J.O. are supported by the NSF 1449974 and M.J.O. is supported by the NIH 1R01NS057607.
Literature Cited
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, et al. Standardized nomenclature for Alu repeats. J Mol Evol. 1996;42:3–6. doi: 10.1007/BF00163204. [DOI] [PubMed] [Google Scholar]
- Carone DM, et al. Hypermorphic expression of centromeric retroelement-encoded small RNAs impairs CENP-A loading. Chromosome Res. 2013;1:49–62. doi: 10.1007/s10577-013-9337-0. [DOI] [PubMed] [Google Scholar]
- Chang DY, et al. A human Alu RNA-binding protein whose expression is associated with accumulation of small cytoplasmic Alu RNA. Mol Cell Biol. 1994;14:3949–3959. doi: 10.1128/mcb.14.6.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Smit AFA, Brosius J, Schmitz J. A novel abundant family of retroposed elements (DAS-SINEs) in the nine-banded armadillo (Dasypus novemcinctus) Mol Biol Evol. 2005;22:886–893. doi: 10.1093/molbev/msi071. [DOI] [PubMed] [Google Scholar]
- Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. 1998;37:18081–18093. doi: 10.1021/bi981858s. [DOI] [PubMed] [Google Scholar]
- Daniels GR, Deininger PL. Repeat sequence families derived from mammalian tRNA genes. Nature. 1985;317:819–822. doi: 10.1038/317819a0. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Heidmann T. LINEs, SINEs and processed pseudogenes: parasitic strategies for genome modeling. Cytogenet Genome Res. 2005;110:35–48. doi: 10.1159/000084936. [DOI] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Eickbush DG, Eickbush TH. R2 and R2/R1 hybrid non-autonomous retrotransposons derived by internal deletions of full-length elements. Mob DNA. 2012;3:10. doi: 10.1186/1759-8753-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogolevsky KP, Vassetzky NS, Kramerov DA. 5S rRNA-derived and tRNA-derived SINEs in fruit bats. Genomics. 2009;93:494–500. doi: 10.1016/j.ygeno.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Goodman M, et al. Toward a phylogenetic classification of Primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- Hall LE, Mitchell SE, O'Neill RJ. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res. 2012;20:535–546. doi: 10.1007/s10577-012-9297-9. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM. Evidence for genomic rearrangements mediated by human endogenous retroviruses during primate evolution. Nat Genet. 2001;29:487–489. doi: 10.1038/ng775. [DOI] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol Biol Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS. Short retroposons in eukaryotic genomes. Int Rev Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lindsay J, et al. Unique small RNA signatures uncovered in the tammar wallaby genome. BMC Genomics. 2012;13:559. doi: 10.1186/1471-2164-13-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo MS, O'Neill MJ, O'Neill RJ. Abundant human DNA contamination identified in non-primate genome databases. PLoS One. 2011;6:e16410. doi: 10.1371/journal.pone.0016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Atallah M. Genomic relationship between SINE retrotransposons, Pol III-Pol II transcription, and chromatin organization: the journey from junk to jewel. Biochem Cell Biol. 2011;89:495–504. doi: 10.1139/o11-046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Mlynarski EE, Obergfell CJ, O'Neill MJ, O'Neill RJ. Divergent patterns of breakpoint reuse in Muroid rodents. Mamm Genome. 2010;21:77–87. doi: 10.1007/s00335-009-9242-1. [DOI] [PubMed] [Google Scholar]
- Munro J, Burdon RH, Leader DP. Characterization of a human orphon 28 S ribosomal DNA. Gene. 1986;48:65–70. doi: 10.1016/0378-1119(86)90352-5. [DOI] [PubMed] [Google Scholar]
- Natali L, et al. The repetitive component of the sunflower genome as shown by different procedures for assembling next generation sequencing reads. BMC Genomics. 2013;14:686. doi: 10.1186/1471-2164-14-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Smit AFA, Okada N. Functional noncoding sequences derived from SINEs in the mammalian genome. Genome Res. 2006;16:864–874. doi: 10.1101/gr.5255506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N. SINEs. Curr Opin Genet Dev. 1991;1:498–504. doi: 10.1016/s0959-437x(05)80198-4. [DOI] [PubMed] [Google Scholar]
- Okada N, Ohshima K. A model for the mechanism of initial generation of short interspersed elements (SINEs) J Mol Evol. 1993;37:167–170. doi: 10.1007/BF02407352. [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Paule MR. Survey and summary transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfree MB, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12:R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation of ribosomal RNA in human cells. Nucleic Acids Res. 2006;34:2966–2975. doi: 10.1093/nar/gkl357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schlessinger D. Structure and organization of ribosomal DNA. Biochimie. 1991;73:631–638. doi: 10.1016/0300-9084(91)90042-y. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Vassetzky NS, Kramerov DA. SINEBase: a database and tool for SINE analysis. Nucleic Acids Res. 2013;41:D83–D89. doi: 10.1093/nar/gks1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veyrunes F, et al. Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse. Proc Biol Sci. 2006;273:2925–2934. doi: 10.1098/rspb.2006.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Pirtle IL, Pirtle RM. A human 28S ribosomal RNA retropseudogene. Gene. 1997;196:105–111. doi: 10.1016/s0378-1119(97)00214-x. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. SINEs and LINEs: troublemakers, saboteurs, benefactors, ancestors. In: Gesteland R, Cech T, Atkins J, editors. The RNA World. Cold Spring Harbor, NY: CSHL press; 2005. pp. 507–533. [Google Scholar]
- Wu X, Brewer G. The regulation of mRNA stability in mammalian cells: 2.0. Gene. 2012;500:10–21. doi: 10.1016/j.gene.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.