Abstract
Speciation as a process remains a central focus of evolutionary biology, but our understanding of the genomic architecture and prevalence of speciation in the face of gene flow remains incomplete. The Anopheles gambiae species complex of malaria mosquitoes is a radiation of ecologically diverse taxa. This complex is well-suited for testing for evidence of a speciation continuum and genomic barriers to introgression because its members exhibit partially overlapping geographic distributions as well as varying levels of divergence and reproductive isolation. We sequenced 20 genomes from wild A. gambiae s.s., Anopheles coluzzii, Anopheles arabiensis, and compared these with 12 genomes from the “GOUNDRY” subgroup of A. gambiae s.l. Amidst a backdrop of strong reproductive isolation, we find strong evidence for a speciation continuum with introgression of autosomal chromosomal regions among species and subgroups. The X chromosome, however, is strongly differentiated among all taxa, pointing to a disproportionately large effect of X chromosome genes in driving speciation among anophelines. Strikingly, we find that autosomal introgression has occurred from contemporary hybridization between A. gambiae and A. arabiensis despite strong divergence (∼5× higher than autosomal divergence) and isolation on the X chromosome. In addition to the X, we find strong evidence that lowly recombining autosomal regions, especially pericentromeric regions, serve as barriers to introgression secondarily to the X. We show that speciation with gene flow results in genomic mosaicism of divergence and introgression. Such a reticulate gene pool connecting vector taxa across the speciation continuum has important implications for malaria control efforts.
Keywords: Anopheles, speciation, introgression, population genetics, gene flow
Introduction
Speciation is a fundamental evolutionary process generating biodiversity and remains a central focus of evolutionary biology. Following The Modern Synthesis of Evolutionary Biology, speciation was commonly thought to require complete reproductive isolation of nascent taxa, often through geographic isolation (Dobzhansky 1937; Mayr 1942). Under this model, gene pools become separated by geography and diverge in reproductive isolation with no genetic exchange. However, an alternative, more fluid, view of species boundaries and divergence as a continuum may be more appropriate in some cases (reviewed in Harrison and Larson 2014). Both empirical examples and theory support the possibility that the process of speciation can include some gene flow either through intermittent hybridization in the early phases of divergence or through secondary contact (Bolnick and Fitzpatrick 2007; Nachman and Payseur 2012). This model deviates from the geographic isolation model in that it includes intermediate stages where reproductive isolation is incomplete between nascent taxa and hybridization results in genetic exchange between diverging gene pools.
Models of speciation with gene flow posit that speciation in the face of hybridization will result in regions of the genome that are differentiated while others introgress and mix between gene pools (Bazykin 1969; Wu 2001). A number of scenarios could lead to hybridization between taxa including secondary contact following a period of allopatric divergence, divergence in parapatry, sympatric speciation, or other similar models. The geographical details of the divergence process are likely associated with different genetic mechanisms. In the case of sympatric speciation, differentiated regions fail to introgress among taxa presumably due to natural selection against migrant alleles because of local natural selection against those alleles due to ecological misfit, while secondary contact models are more likely to involve selection against alleles that are neutral in the native genomic background but incompatible with the alternative genomic background (i.e., Bateson–Dobzhansky–Muller incompatibilities; Bateson 1909; Dobzhansky 1937; Muller 1940). Of course, these genetic mechanisms need not be mutually exclusive among speciation models. Regardless of the underlying genetic mechanisms, genomic regions in linkage disequilibrium (LD) with the incompatible loci also become differentiated over time, providing suitable genomic context for additional incompatible loci to accumulate, and the proportion of the genome that remains differentiated grows until reproductive isolation is complete. Building on the implied importance of recombination and LD in enabling this process, it has been suggested that genomic regions with restricted meiotic recombination, such as chromosomal inversions and centromeric regions, will facilitate this process and play an important role in speciation in the face of hybridization (Begun et al. 2007; McGaugh and Noor 2012; Mackay et al. 2012). Genomic regions with restricted meiotic recombination in hybrids are thought to be important in the formation and maintenance of species boundaries in the face of gene flow because the hitchhiking process involved in selection against maladaptive introgressed variants will affect larger genomic swaths in lowly recombining regions (Noor et al. 2001; Rieseberg 2001; Navarro and Barton 2003; Butlin 2005; Hoffmann and Rieseberg 2008). However, currently available empirical evidence supporting a role of centromeric regions is confounded by the effects of natural selection on linked sites (Charlesworth 1998; Noor and Bennett 2009; Cruickshank and Hahn 2014).
A number of studies have shown strong differences between sex chromosomes (X or Z) and autosomes in the rates of genetic divergence and introgression and therefore inferred a role for sex chromosomes in speciation among pairs of species in systems ranging from Drosophila to Hominids. Empirical and theoretical evidence also supports the hypothesis that the X chromosome may play an important role in maintaining species boundaries (“large-X effect”; Charlesworth et al. 1987; Coyne and Orr 1989; Garrigan et al. 2012; Sankararaman et al. 2014). Genetic divergence tends to be higher on sex chromosomes relative to autosomes and introgressed regions are underrepresented on the sex chromosomes, presumably owing to reduced recombination rates along sex chromosomes and exposure of recessive incompatible loci in the heterogametic sex (Coyne and Orr 1989; Geraldes et al. 2006; Sankararaman et al. 2014).
Unambiguously demonstrating differential gene flow along the genomes of diverging taxa is challenging for two reasons. First, most pairs of taxa that are most likely to hybridize tend to be very closely related, and may share substantial polymorphism either through inheritance from the ancestral population or due to ongoing genetic exchange, making it difficult to distinguish between the two (Hey 2006). Second, certain population genetic statistics used to measure differentiation are also sensitive to the population genetic effects of background or positive natural selection, thus confounding the signals of differential gene flow and natural selection (Charlesworth 1998; Noor and Bennett 2009). Indeed, many of the well-known empirical examples of genomic regions remaining differentiated in the face of gene flow have been called into question based on the possible confounding effects of selection in generating signals of differentiation (Cruickshank and Hahn 2014). Thus, further work is needed to understand the genomic architecture of speciation in the face of hybridization.
The Anopheles gambiae species complex in sub-Saharan Africa is well-suited for studying the genomic architecture of speciation. Prior to the 1940s, A. gambiae was considered a single biologically variable species, but crossing studies and genetic analysis led to the naming of nine morphologically similar species that vary in their geographic distribution, geographic overlap with other complex members, and ecology (reviewed in Coetzee et al. 2013). It is becoming increasingly appreciated from ecological distinctions and recent discoveries of additional genetic substructure that, even within species, Anopheles species frequently form partially reproductively isolated and differentiated subpopulations (Costantini et al. 2009; Gnémé et al. 2013; Lee et al. 2013). As an example of this dynamic, a new subgroup of A. gambiae s.l. named GOUNDRY was discovered in Burkina Faso that shares larval habitats with other subgroups, but prefers to rest outdoors as adults (Riehle et al. 2011). Although genetic distinctions are clear among most taxa within the species complex, both mixed mating swarms and hybrids have been documented (Marchand 1983; Diabaté et al. 2006), implying an absence of strict geographical isolation and an important role for postzygotic barriers to introgression in some cases.
Among taxa within this species complex, the most substantial effort has been dedicated to understanding the status, history, and genomic consequences of reproductive isolation between two members of the A. gambiae species complex, Anopheles coluzzii (previously the M molecular form of A. gambiae; Coetzee et al. 2013) and A. gambiae (previously the S molecular form of A. gambiae) (Turner et al. 2005; Lawniczak et al. 2010; Neafsey et al. 2010; Weetman et al. 2012). These two taxa are mostly reproductively isolated in the field, although they are compatible in captivity (della Torre et al. 2001; Lawniczak et al. 2010). Internal subdivisions exist even within the molecular forms (Slotman et al. 2006, 2007) and recent evidence indicates an often high level of local A. coluzzii–A. gambiae hybridization (Lee et al. 2013), illustrating the potential for introgression within the A. gambiae group.
Firm conclusions regarding the degree of reproductive isolation and the age of these two taxa have been controversial. This is due in part to the fact that studies of A. coluzzii–A. gambiae were based on single nucleotide polymorphism (SNP) panels that preclude measurement of absolute sequence divergence, low-resolution sequencing data sets, and sequence data sets from small samples of laboratory mosquito colonies. In addition, they relied on statistical approaches that are incapable of distinguishing between multiple confounding population genetic processes. In particular, these analyses relied on measures of relative divergence, which are not robust to variation in recombination rates and natural selection across the genome and do not explicitly distinguish between lineage sorting of ancestral polymorphism and introgression (Charlesworth 1998; Noor and Bennett 2009; Hahn et al. 2012; Cruickshank and Hahn 2014). Relatively few studies have addressed divergence among other members of the species complex, and the same analytical concerns apply here as well (Wang-Sattler et al. 2007; Neafsey et al. 2010; O’Loughlin et al. 2014). A recent study used a phylogenetic approach to test for introgression among members of the A. gambiae species complex and found evidence for substantial introgression among many members of the complex, also arguing that the X chromosome has played an important role in speciation (Fontaine et al. 2015). However, additional questions remain about the nature of species boundaries and the genomic architecture of barriers to introgression in this system. In particular, it remains unclear whether signals of introgression reflect only historical hybridization or also signify the result of contemporary hybridization. In addition, whether certain autosomal genomic regions may also serve as barriers to introgression among a background of extensive autosomal introgression remains to be established. Understanding the nature of species boundaries in this system may reveal principles underlying the process of speciation in the absence of geographic isolation, but it also has relevance for public health because this complex includes several major vectors of malaria, which continues to place a devastating burden on local human populations (World Health Organization 2013).
In this study, we analyzed a panel of 32 newly generated and previously available genomes from wild-caught females from the A. gambiae species complex, representing multiple points along the speciation continuum within this complex. We conducted detailed population genetic analysis of these data to address questions regarding introgression among taxa of varying levels of divergence as well as questions regarding the genomic architecture of barriers to introgression.
Materials and Methods
Details regarding mosquito samples, DNA sequencing, next-generation sequencing bioinformatics, and standard population genetic analyses can be found as supplementary material, Supplementary Material online.
Introgression Analysis and D Statistics
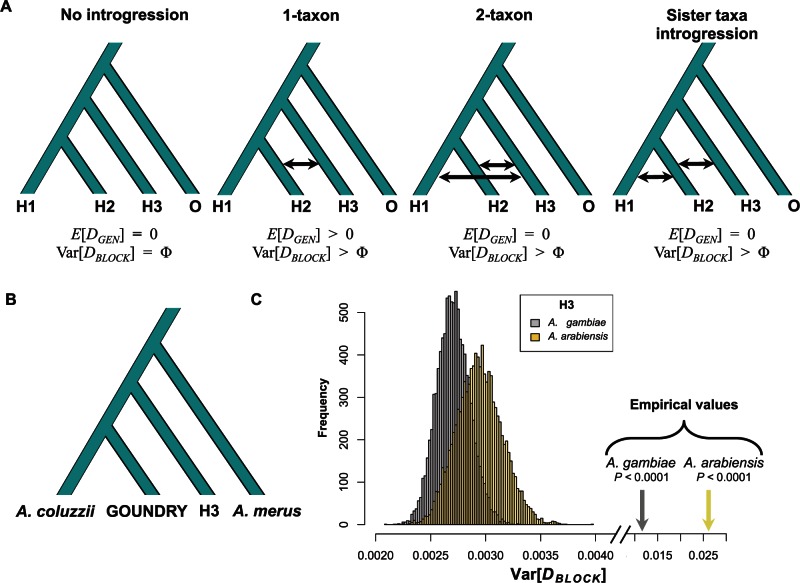
To explicitly differentiate between introgression and ancestral lineage sorting, we used a modification of the ABBA-BABA test (Green et al. 2010; Durand et al. 2011). This test uses the D statistic to compare the distribution of alleles on the four taxon tree ((H1,H2),H3),O), where H1 and H2 are sister taxa and H3 and O are the outgroups. Under the null hypothesis of a perfect tree structure and no gene flow, the number of derived mutations that are shared only between the genomes of H2 and H3 (ABBA) is expected to equal the number of those that are shared only between H1 and H3 (BABA). D is then calculated as the standardized difference between the numbers of ABBAs and the number of BABAs with an expectation that D is zero under the null hypothesis (Green et al. 2010). Significant excess sharing of derived alleles between H3 and either H1 or H2 will result in a nonzero D and provides evidence of introgression. In our case, such tests are not appropriate because the alternative to no introgression is not only asymmetric introgression between an outgroup and one of the ingroups, but also potentially approximately equal amounts of introgression between the outgroup and each of the two ingroups (fig. 1). For that reason we use a modified test in which we instead consider the length distribution of fragments of shared ancestry. Because haplotypes are broken down by recombination over successive generations, the length distribution of shared haplotypes among populations is informative regarding the time since the most recent introgression event (Pool and Nielsen 2009; Gravel 2012). After t generations, the mean length of a shared haplotype is approximately , where r is the recombination rate and m is the proportion of introgressed individuals in the population (Gravel 2012). Because t will be smaller for introgressed haplotypes than for shared ancestral haplotypes, the mean length of introgressed haplotypes will be longer resulting in longer clusters of excess ABBAs or BABAs. We used patterns of LD to approximate this expectation and deviations from it by calculating variance in the D statistic among genomic blocks, Var[DBLOCK], to detect physical clusters of correlated genomic segments consistent with an excess of long-shared haplotypes, and compared it with its null distribution generated by permuting fragments among blocks in order to manually break up possible correlations. If the observed value of Var[DBLOCK] is significantly larger than that predicted from permuted fragments of size, which should be >10× the average length of the decay of LD, we conclude that the genomes harbor introgressed haplotypes.
Fig. 1.—
Excess variance in DBLOCK indicates recent introgression. (A) Four-taxon trees used in ABBA-BABA tests with three alternative introgression models. The expected genome-wide value of the D statistic (E[DGEN]) is presented below in addition to the expected variance among D statistics calculated in genomic blocks (Var[DBLOCK]). Var[DBLOCK] under the “No introgression” model is unknown and indicated here by Φ for comparison in other models. The “2-taxon” and “Sister taxa introgression” models may result in E[DGEN] of 0 but are expected to have increased Var[DBLOCK] relative to the No introgression model providing a test for introgression. (B) The four-taxon test tree used for analysis. (C) The distributions of Var[DBLOCK] calculated from 104 permuted genomes (see Methods) for test trees with Anopheles gambiae (grey) and Anopheles arabiensis from Burkina Faso (yellow) as the H3 are presented. The true Var[DBLOCK] values from each empirical data set, presented on a broken x-axis for comparison, are greater than all permuted genomes in each case consistent with the presence of introgressed haplotypes in these genomes.
We divided the genome into blocks of 500 informative sites (i.e., ABBAs and BABAs). These genomic blocks were then divided into 100 segments of 5 informative sites. We chose this block size because this number of sites corresponded to a physical size of ∼100 × LLD, where LLD is the physical distance at which point LD decays to background levels. As a result, we could then divide the genomic blocks into 100 segments that would be larger in physical size than LLD. Because introgressed haplotypes will be highly correlated over exceptionally high physical distances relative to ancestral haplotypes, we expect that the size of introgressed haplotypes would exceed LLD while ancestral haplotypes would not. For the tests involving A. gambiae and Anopheles arabiensis as the H3 taxon, the mean genomic block length was ∼250 and ∼350 kb, respectively. Because LLD is approximately 200 bp in this system (supplementary fig. S1, Supplementary Material online), and segments within the blocks were approximately 2.5 or 3.5 kb in size, segments exceeded LLD by a factor of more than 10×. These analyses were conducted using pseudohaploidized genomes from the individuals from each population/species with the highest short-read coverage. In general, sites involved in calculating D are not entirely independent due to LD, so permutation and jackknife analyses are necessary to properly test for significance. We have used these corrected tests for significance in the following ways.
For the first test, we calculated the D statistic for each genomic block (hereafter DBLOCK) and the variance among DBLOCK, hereafter Var[DBLOCK], to test for an excess in variance among genomic blocks that would be consistent with the presence of correlated genomic segments (haplotypes) with shared derived mutations. Under the null hypothesis of no introgression, Var[DBLOCK] among true genomic blocks derives largely from relatively small ancestral haplotypes such that random permutation of segments among blocks will not affect the variance among blocks. If the genome contains introgressed haplotypes that are larger than the segments, Var[DBLOCK] will be larger when these haplotypes are intact in the empirical data and smaller after random permutation that dissolves correlations among segments. We calculated Var[DBLOCK] among genomic blocks in the empirical data. Then, for each comparison, we permuted the internal ∼2.5 kb segments among genomic blocks and recalculated Var[DBLOCK] for each permuted genome. Then Var[DBLOCK] from the empirical data was compared to the distribution of Var[DBLOCK] from permuted genomes to ask whether variance among genomic blocks is higher in the empirical data where true correlations remain intact relative to the permuted genomes where variance in D will be driven largely by segregating ancestral haplotypes.
For the second test, we established confidence intervals for estimates of the DBLOCK statistic in order to identify individual genomic blocks with significant evidence for introgression. To do so, we conducted block jackknife analyses within each genomic block (Green et al. 2010) by dropping each genomic segment within a given block in turn and recalculating DBLOCK. We calculated 95% confidence intervals for each genomic block using variance estimated from this jackknife procedure. These confidence intervals are presented as ribbons in figure 2.
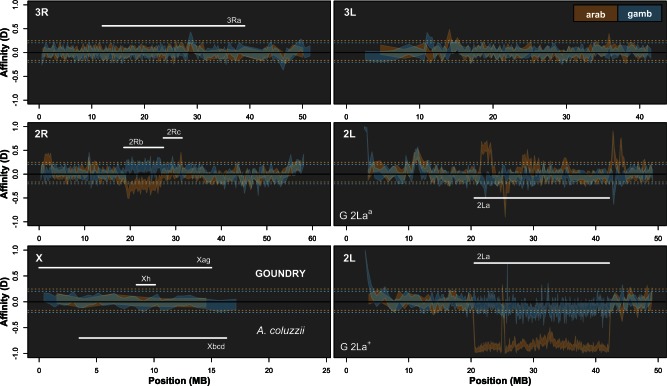
Fig. 2.—
Significant autosomal introgression between pairs of Anopheles species and subspecies. ABBA-BABA statistics were calculated in nonoverlapping windows of 500 informative sites using Anopheles merus as the outgroup. Blue ribbon indicates 95% confidence region for introgression between Anopheles gambiae (2Laa/+) and GOUNDRY (positive D; 2Laa/a and 2La+/+; 3R+; Xag) and Anopheles coluzzii (negative D; 2Laa/a; 3R+; Xag). Orange ribbon indicates 95% confidence region for introgression between Anopheles arabiensis (2Laa/a; 3Ra; Xbcd) and GOUNDRY (positive D) and A. coluzzii (negative D). Horizontal dotted lines (orange = A. arabiensis; blue = A. gambiae) indicate genome-wide significance level after correction for multiple testing. Positions of relevant chromosomal inversions are indicated with horizontal white lines. A full list of genes within significant windows is given in supplementary table S2, Supplementary Material online.
For the third test, we established genome-wide thresholds corrected for multiple testing in order to identify genomic blocks exceeding these thresholds consistent with recent introgression. We conducted the permutation of segments within blocks procedure as above, but for each permuted genome, we calculated DBLOCK and retained the maximum and minimum values of DBLOCK. To determine whether any individual true genomic blocks showed evidence of significant excess sharing of derived alleles, we established 95% critical thresholds (table 1) from this permutation procedure and compared the value of DBLOCK among true blocks. These genome-wide critical thresholds are presented as dashed lines in figure 2.
Table 1.
Modified Block-based ABBA-BABA Test of Introgression
| H1a | H2a | H3a | Upper 95%b | Lower 95%b | Proportion of the genome in sig Anopheles coluzzii Windowsc | Proportion of the genome in sig GOUNDRY Windowsc |
|---|---|---|---|---|---|---|
| A. coluzzii | GOUNDRY | Anopheles gambiae | 0.204 | −0.204 | 0.0114 | 0.0325 |
| A. coluzzii | GOUNDRY | Anopheles arabiensis (Burkina Faso) | 0.248 | −0.164 | 0.0364 | 0.0354 |
| A. coluzzii | GOUNDRY | A. arabiensis (Tanzania) | 0.256 | −0.156 | 0.0323 | 0.0312 |
Note.—Genome-wide 95% thresholds and the proportions of the genome in significant windows are presented for three comparisons.
aTaxonomic assignment in the ABBA-BABA test tree ((H1,H2),H3),O).
bBoundaries of the genome-wide 95% threshold region estimated using block jackknife estimates of standard error within genomic regions after permutation (n = 104 replicates).
cFor each comparison, windows exceeding the 95% thresholds were identified, and the sum of their length was compared with the total length of the A. gambiae PEST reference.
To determine whether introgression has been very recent between A. arabiensis and either A. coluzzii or GOUNDRY, we compared the proportion of the genome in windows with significant D values between sympatric A. arabiensis from Burkina Faso and allopatric A. arabiensis from Tanzania (Marsden et al. 2014). Because the standard assumption of introgression with only one of the two sister taxa holds for this test, we calculated the standard error of D for each comparison using the block jackknife approach and used a Z-test to assess significance (Green et al. 2010; Durand et al. 2011).
Comparing Genetic Divergence among Genomic Regions
To test hypotheses related to the role of recombination in determining the genomic architecture of reproductive isolation in this system, we divided the genome into regions based on expected levels of recombination in hypothetical hybrids. A fine-scale genetic map is not yet available for Anopheles mosquitoes, but it has been shown in Drosophila that recombination rates approach zero within several megabases on each side of the centromere and also near the telomeres (Fiston-Lavier et al. 2010; Chan et al. 2012). Although patterns of LD are also affected by processes other than local meiotic recombination rates, estimated recombination rates should give a rough approximation of expected LD across the genome. In fact, patterns of LD have been used to define genetic maps in some vertebrates and correspond approximately to genetic maps based on experimental crosses or pedigrees (McVean et al. 2004; Auton et al. 2013). We measured background LD (see Supplementary Material online for details) in our A. coluzzii and A. arabiensis samples, taking average r2 values within 10 kb physical windows across the genome. We found that LD was relatively constant across the genome except for large increases near the autosomal centromeres and smaller increases near the telomeres (supplementary fig. S2, Supplementary Material online). Based on this pattern and the assumption that recombination rates in Anopheles correspond approximately to the Drosophila genetic map, we defined several broad recombinational categories for analysis. We first defined the “Pericentromeric–Telomeric” regions of the autosomes to be all windows within 10 MB on either side of the centromere or within 1 MB from the telomere. It should be noted that we assumed that the starting and ending coordinates of the A. gambiae PEST (Pink Eye STandard) reference chromosomal sequences were reliable indicators for distance from centromeres and telomeres. Unless a chromosomal inversion was present, all remaining regions on the autosomes were assigned to the “Freely Recombining” category. For the comparison between A. gambiae and Anopheles merus, we assigned all windows inside the 2Rop chromosomal inversion complex to the “Autosomal-Inversion” category. We used the outer coordinates for 2Ro and 2Rp breakpoint regions estimated by Kamali et al. (2012).
The X chromosome was categorized for each comparison, according to species-specific conditions. We did not define a general Pericentromeric–Telomeric category for two reasons: 1) We did not observe an increase in LD in the euchromatic regions near centromeres and telomeres (supplementary fig. S2, Supplementary Material online) similar to increases observed on the autosomes. 2) In Drosophila (Fiston-Lavier et al. 2010; Chan et al. 2012), the pericentromeric reduction in recombination affects a relatively small region on the X relative to the autosomes, and we have excluded a large heterochromatic region around the centromere that likely encompasses the affected region in Anopheles. For the comparison between A. gambiae and A. merus, and the comparison between A. coluzzii and A. gambiae, the entire euchromatic region on the X was considered Freely Recombining because no inversions differentiate these groups on the X. For the comparison between A. gambiae and A. arabiensis, we assigned the entire euchromatic region of the X as “X-Inversion,” because these species are differentiated across nearly 75% of the entire chromosome and introgression rates have been estimated to be 0 in laboratory crosses (Slotman et al. 2005). For the comparison between GOUNDRY and both A. coluzzii and A. gambiae, the entire euchromatic X was categorized as Freely Recombining except for the region spanning 8.47–10.1 MB, which was categorized as X-Inversion. As described in Supplementary Material online, we were not able to identify inversion breakpoints for the GOUNDRY inversion, but these coordinates correspond to the outer boundaries of the region with reduced nucleotide diversity (fig. 3).
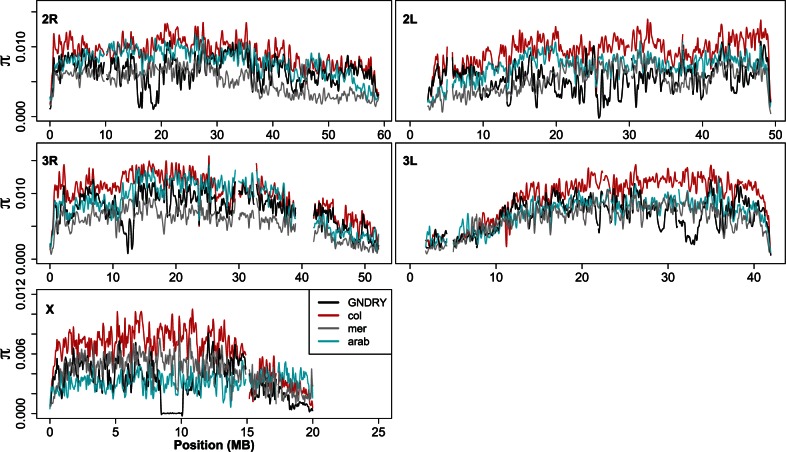
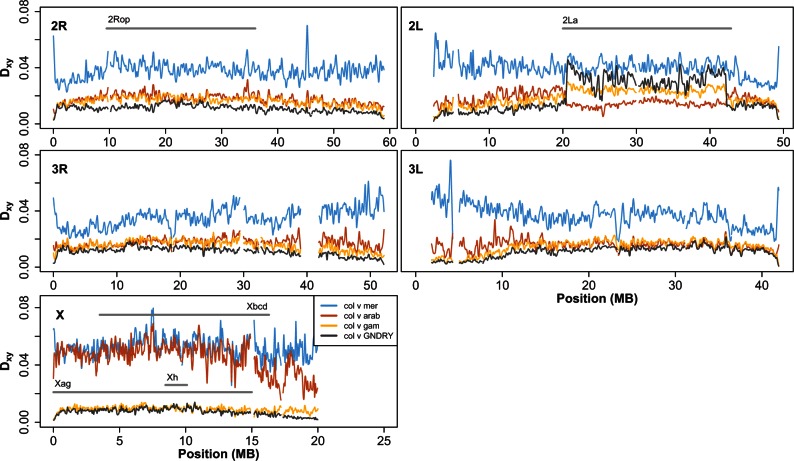
Fig. 3.—
Chromosomal distributions of nucleotide diversity (π) at intergenic sites (LOESS-smoothed with span of 1% using 10 kb nonoverlapping windows). Low complexity and heterochromatic regions were excluded. col = Anopheles coluzzii; GNDRY = GOUNDRY (2La+/+ individual excluded in 2 L estimate); mer = Anopheles merus; arab = Anopheles arabiensis.
To identify regions that are barriers to introgression in the A. gambiae species complex, we compared genetic divergence among the four genomic categories using the following logic. Because genome-wide variation in mutation rate and the effects of linked selection could also lead to genomic variation in divergence among species even in the absence of introgression, we jointly analyzed absolute genetic divergence (Dxy) and nucleotide diversity (π) in 10 kb nonoverlapping windows and asked whether differences in genetic divergence among genomic regions are observed that cannot be explained by differences in nucleotide diversity (where diversity approximates the effects of linked selection or variation in mutation rate). Dxy is an estimate of 2μt + 4Neμ, where Ne is the effective size of the ancestral population, μ is the mutation rate, and t is the divergence time. π provides an estimate of 4Neμ, where Ne is the effective size of the current population. By jointly considering these, we can make comparisons among genomic regions that differ substantially in 4Neμ such that differences in Dxy largely reflect differences in 2μt. We note that this analysis assumes that estimates of 4Neμ from current populations (or in most cases, average of the estimates from the two subgroups) are reliable estimates of 4Neμ in the ancestral population, and we believe this to be a reasonable assumption given the recency of radiation of the A. gambiae species complex. Under scenarios with gene flow among diverging subgroups or species, t will be smaller in regions that have introgressed, so regional barriers to gene flow are expected to have especially large values of t, and therefore values of Dxy that are larger than regions with similar 4Neμ but more introgression. In contrast, under a model of divergence in allopatry with no introgression, t is approximately equal among genomic regions such that divergence should be determined largely by 4Neμ, and thus correlate well with nucleotide diversity, even if elevated. We made comparisons among genomic regions using this framework and assuming that most freely recombining autosomal regions will have been introgressed at some point in the history of divergence because high rates of recombination inhibit associations between barriers to introgression (e.g., hybrid sterility factors) and surrounding chromosomal regions. Therefore, we compared other genomic regions to freely recombining autosomal regions with respect to both divergence and nucleotide diversity to ask whether these other regions harbor excess divergence consistent with less introgression in these regions. Comparisons of distributions of genetic divergence and nucleotide diversity were made using the M–W test in R (R Development Core Team 2011).
For this analysis, we used only 10 kb windows containing at least 600 sequenced sites with data for the taxa involved. We chose to use a focal species comparison to minimize the number of comparisons for concise presentation, but we obtain similar results in other comparisons (e.g., GOUNDRY vs. A. arabiensis) because most variation in divergence depends on variance in coalescence in the ancestral population. We note that estimates of nucleotide diversity in GOUNDRY are not reliable estimates of the ancestral population because GOUNDRY is partially inbred and has the large swept region on the X (Crawford JE, et al. submitted), so we used estimates of nucleotide diversity from A. coluzzii only for the comparison with GOUNDRY.
Results
Genome Sequencing and Population Genetic Analysis
We have completely sequenced the genomes of 20 field-captured female Anopheles mosquitoes from Burkina Faso and Guinea using the Illumina HiSeq2000 platform. We sequenced A. coluzzii (n = 10), A. gambiae (n = 1), and A. arabiensis (n = 9) and compared these sequences with a panel of 12 A. gambiae GOUNDRY genomes. Most individuals were sequenced to an average read depth of 9.79 × (range 5.94–20.03 per individual), while one individual each from GOUNDRY, A. coluzzii, and A. gambiae was sequenced to at least 16.44 × (supplementary table S1, Supplementary Material online). We also used publicly available genome sequences from A. merus (n = 6) as an outgroup and an A. arabiensis genome from Tanzania for a geographically distinct comparison (Marsden et al. 2014). We conducted population genetic analysis of aligned short-read data using genotype likelihoods and genotype calls calculated using the probabilistic inference framework ANGSD (Korneliussen et al. 2014).
Broad-scale genetic relationships among the Anopheles included in this analysis were confirmed in a companion analysis (Crawford JE, et al. submitted). Briefly, a neighbor-joining tree based on genome-wide genetic distance shows that GOUNDRY is more closely related to A. coluzzii than to A. gambiae, and these three taxa are more closely related to A. arabiensis than to A. merus. This topology is consistent with other phylogenetic estimates for this species complex (Kamali et al. 2012; Fontaine et al. 2015).
Recent Autosomal Introgression
An important first step toward understanding the nature of species boundaries in A. gambiae species complex is to determine whether introgression has occurred among diverging Anopheles taxa and whether introgression is restricted to closely related taxa or occurs among more distant taxa. We tested for historical and contemporaneous introgression along the speciation continuum using a variance-based modification of the D statistic (Var[DBLOCK]) that allows for introgression among multiple taxa in a four-taxon tree (Methods; fig. 1) by comparing the variance in D among genomic blocks with variance expected under the null hypothesis of shared ancestry due to lineage sorting. We applied this test using two four-taxon trees and find evidence for recent introgression among species and subgroups of the A. gambiae species complex. We calculated the D statistic using a four-taxon tree with A. coluzzii and GOUNDRY as the sister taxa (H1 and H2, respectively), A. merus as the outgroup (O), and either A. gambiae or A. arabiensis as the test group, H3. After comparing the Var[DBLOCK] in the empirical data with Var[DBLOCK] from 104 randomly permutated genomes, we find significant evidence for introgression between the A. coluzzii–GOUNDRY clade and A. gambiae (P < 0.0001, fig. 1). Using A. coluzzii and GOUNDRY as the ingroups again, but with A. arabiensis as the test taxon (H3), we also find that the Var[DBLOCK] is significantly larger than all 104 permuted genomes (P < 0.0001, fig. 1). In fact, Var[DBLOCK] is larger than the largest value of the 104 permuted genomes by a factor of 3.7 in the A. gambiae test (Var[DBLOCK] = 0.0122) and by a factor of 6.7 in the A. arabiensis test (Var[DBLOCK] = 0.0268) (fig. 1). These results indicate that the observation of shared polymorphism between these taxa can be attributed in part to introgression.
Contemporary Introgression between Anopheles gambiae and Anopheles arabiensis
Although the above analysis is sensitive to contemporary introgression, we used an additional approach comparing signals of introgression with sympatric and allopatric populations to explicitly ask whether introgression has occurred via contemporary hybridization. If introgression has occurred recently, we expect stronger affinity among sympatric populations relative to allopatric populations (Nosil et al. 2003; Grant et al. 2005; Noor and Bennett 2009). We tested whether introgression between A. coluzzii and A. arabiensis has been recent with sympatric A. arabiensis versus allopatric A. arabiensis from Tanzania. In this case, we used the standard ABBA-BABA test because we are explicitly testing a simple “1-taxon” model (fig. 1). We tested for introgression using a four-taxon tree of high-coverage individuals with the two A. arabiensis individuals as the ingroups (H1 and H2) and A. coluzzii as the test group (H3) and found a significant excess of shared derived mutations between A. coluzzii and sympatric A. arabiensis relative to allopatric A. arabiensis from Tanzania (D = −0.0542, Block Jackknife Z-score = −13.1533, P = 1.63 × 10−39; table 1). Similarly, we used GOUNDRY as H3 and found evidence for significant introgression between sympatric A. arabiensis and GOUNDRY (D = −0.0441, Block Jackknife Z-score = −11.7559, P = 6.58 × 10−32; table 1). In line with recent evidence of contemporary hybridization between A. gambiae and A. arabiensis in Uganda (Weetman et al. 2014), our results provide strong evidence that introgression continues to occur via contemporary hybridization in Burkina Faso that has the potential to impact the evolution of both ecologically and epidemiologically relevant traits.
Introgressed Genes
Introgressed haplotypes may contain genes with potentially important phenotypic effects, and a local reduction in the concentration of introgressed haplotypes along the genome can be indicative of barriers to introgression, so we partitioned the signal of introgression across the genome to identify introgressed chromosomal segments. To do so, we compared each empirical DBLOCK value with genome-wide significance thresholds established by analyzing the distribution of the most extreme DBLOCK values from each of the 104 permuted genomes (Methods). Because each DBLOCK statistic is polarized, we can identify windows that show significant introgression between each of the sister taxa (i.e., A. coluzzii and GOUNDRY) and H3 (fig. 2). After conservatively correcting for multiple testing by comparing the empirical values with a distribution of genome-wide extreme values from permutations, we find significant evidence of introgression between A. coluzzii and both A. gambiae and A. arabiensis, and the proportions of the genome that are represented by significant windows are 1.1% and 3.6% for A. gambiae and A. arabiensis, respectively. These introgressed chromosomal blocks include 97 annotated protein-coding sequences introgressed between A. coluzzii and A. gambiae and 543 introgressed between A. coluzzii and A. arabiensis (supplementay table S2, Supplementary Material online). Interestingly, we find strong evidence for introgression of the pericentromeric region on chromosome 2L between GOUNDRY and A. gambiae, which contrasts starkly with previous suggestions that this region may be a barrier to introgression and important for speciation between these taxa (Turner et al. 2005; Neafsey et al. 2010; Lawniczak et al. 2010). A recent study based on SNP genotype data identified a signal of introgression between A. coluzzii and A. gambiae in this genomic region and attributed the high frequency of the introgressed haplotype to the sharing of an adaptive insecticide resistance allele at the kdr locus (Clarkson et al. 2014).
One window that shows an exceptionally strong introgression signal between A. coluzzii and A. arabiensis harbors the GABA receptor gene, also known as the “resistance to dieldrin” locus because of the role of this receptor in conferring resistance to the insecticide dieldrin and related insecticides (Ffrench-Constant et al. 1993). Although any resistance phenotype conferred by introgressed alleles is unknown at this point, our finding that the Rdl locus has been introgressed between A. coluzzii and A. arabiensis would be contradictory to previous reports that these species have acquired resistance at this locus through independent but convergent mutations (Du et al. 2005). Moreover, introgression of an allele with adaptive value in the face of insecticide pressure further supports the occurrence of contemporary hybridization between these taxa.
We used the same approach to identify genomic blocks shared between GOUNDRY and two H3 taxa, A. gambiae and A. arabiensis. We find that windows representing 3.2% of the GOUNDRY genome and 369 protein-coding sequences share a significant excess of derived mutations with A. gambiae (table 1). In addition, we find that 3.5% of the GOUNDRY genome harboring 499 protein-coding sequences shares a significant excess of derived mutations with A. arabiensis. In line with results above showing evidence of more introgression with sympatric A. arabiensis relative to allopatric A. arabiensis, we find that the windows with significant evidence of introgression with sympatric (Burkina Faso) A. arabiensis cover slightly more of the genome than windows with significant evidence of introgression with allopatric (Tanzania) A. arabiensis (table 1). Although only relatively small percentages of these individual genomes show significant evidence of recent introgression, hundreds of protein-coding genes have been shared in each case such that homogenization of these genomic regions may have large effects on phenotypic evolution, exhibited most prominently by the sharing of presumably functional mutations at an insecticide locus in the A. coluzzii comparisons.
Although identifying introgressed regions provides insight into the homogenizing effects of hybridization, identifying genomic regions depauperate of recent introgression can provide evidence for genomic barriers to introgression. Despite evidence for considerable introgression on the autosomes, we find no evidence of recent introgression of X chromosome sequence among any subgroups or species, which suggests a disproportionately large role for the X in speciation among these taxa.
Excess Genetic Divergence on the X Chromosome
We hypothesized that long-term differences in introgression along the genome will be reflected in patterns of genetic divergence, because genetic divergence will be partially determined in part by selection for and against introgressed material. This pattern has been notoriously difficult to demonstrate, in part because measurable differences in divergence are slow to develop (Cruickshank and Hahn 2014). As such, we will focus largely on comparisons between more divergent species pairs (A. coluzzii vs. A. arabiensis and A. coluzzii vs. A. merus) where differences are most easily identified. To test for genomic barriers among taxa in the A. gambiae species complex, we asked whether sequence divergence (Dxy) between taxa differed among genomic regions as expected under a model of differential rates of introgression. However, Dxy is an estimator of 2μt + 4Nμ, where μ is the mutation rate, t is the number of generations since the species split, and N is the effective population size of the ancestral population, but 4Nμ can vary among genomic regions for reasons unrelated to differential introgression (i.e., variable mutation rate or effects of natural selection on linked sites; fig. 4). Therefore, we jointly analyzed nucleotide diversity (π) as a proxy for 4Nμ among genomic regions to avoid confounding intrapopulation effects with differential introgression (see Methods).
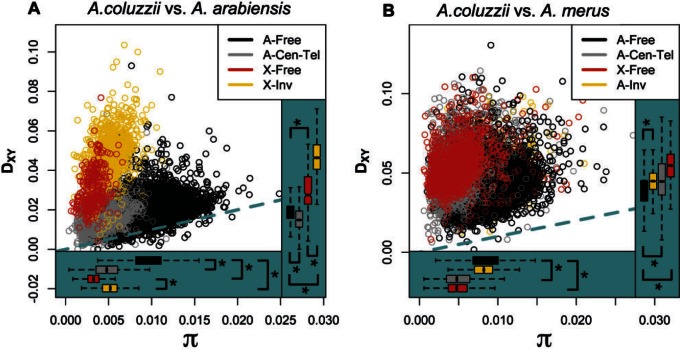
Fig. 4.—
Patterns of genetic divergence (Dxy) between populations as a function of nucleotide diversity (π) reveal differential gene flow during speciation. Genomic regions defined by expected rates of recombination in hybrids (see Methods) differ in their distributions of nucleotide diversity and genetic divergence, but not always in the same direction, suggesting that gene flow has been restricted on the X and lowly recombining regions in some cases. (A) Anopheles coluzzii versus Anopheles arabiensis; (B) A. coluzzii versus Anopheles merus. Panel legends indicate colors corresponding to genomic location of each 10 kb window where “Free” indicates freely recombining regions, “Cen-Tel” indicates centromeric and telomeric autosomal regions, and “Inv” indicates chromosomal inversions. “A-” and “X-” indicate autosome or X chromosome. Dashed blue-green line indicates perfect correlation. Asterisks indicate M–W tests with P < 3.92 × 10−5 for comparisons indicated with brackets. Note that the y-axis scale differs among panels.
Hybrid male sterility maps to the X chromosome in A. gambiae–A. arabiensis crosses, and two large X-linked chromosomal inversion complexes (Xag and Xbcd) suppress recombination on the X in hybrids of these species, so we hypothesized that less frequent introgression may lead to exceptionally high sequence divergence on the X relative to other genomic regions. In support of this hypothesis, we found that nucleotide diversity on the X is significantly lower than on the autosomes (Mann–Whitney [hereafter M–W] P < 2.2 × 10−16), but genetic divergence is significantly higher on the X (M–W P < 2.2 × 10−16; fig. 3). Moreover, genetic divergence is significantly higher in the region harboring inversions (M–W P < 2.2 × 10−16) than in the surrounding chromosome. Nucleotide diversity, however, is also higher in the inverted region relative to the centromere-proximal region (M–W P < 2.2 × 10−16), where nucleotide diversity is especially low, presumably due to the effects of linked selection on neutral genetic variation. Although we cannot formally rule out an elevated mutation rate inside the inverted region, there is no reason to expect the inverted region to be more mutable. This excess genetic divergence on the X is consistent with our results in the ABBA-BABA analysis above showing that the X chromosome lacks evidence of recent introgression among these taxa. In general, these observations are in line with previous analyses of genetic differentiation among A. arabiensis and A. gambiae, as well as with laboratory backcrossing experiments (Slotman et al. 2005; Neafsey et al. 2010; O’Loughlin et al. 2014), indicating that introgression is particularly inhibited on the X chromosome, and that the X chromosome plays a disproportionately large role in driving speciation.
The X chromosomes of A. coluzzii and A. gambiae are collinear with that of A. merus, so recombination is not expected to be suppressed in contemporary or historical hybrids among these groups. To test whether sequence divergence is exceptionally high on the X relative to the autosomes in these comparisons despite the lack of inverted regions, we compared genetic divergence and nucleotide diversity among genomic regions as described above. We find that genetic divergence between A. gambiaes.l. (A. coluzzii used as proxy for ancestral population here) and A. merus does not scale with nucleotide diversity (fig. 3). Nucleotide diversity is lower on the X than on the autosomes (M–W P < 2.2 × 10−16), but genetic divergence is significantly higher on the X (M–W P < 2.2 × 10−16). This is strong evidence that the autosome continued to be homogenized by introgression after the X chromosome had become a barrier to introgression among these taxa.
Interestingly, despite the relatively recent split time (<0.5 Ne) between A. coluzzii and A. gambiae (Cruickshank and Hahn 2014; Fontaine et al. 2015), inspection of chromosomal distributions of divergence between A. coluzzii and A. gambiae reveals a slight elevation in genetic divergence in a chromosomal region where nucleotide diversity is low relative to the rest of the chromosome (∼16–20 MB; figs. 4 and 5). Indeed, this particular region coincides with a genomic region recently shown to play a role in assortative mating between these taxa (Aboagye-Antwi et al. 2015), further supporting its role in speciation, especially the early stages. Such signals of excess divergence on the X provide further evidence for a large role of the X in driving speciation in Anopheles and suggests that inversions may facilitate speciation but are not required.
Fig. 5.—
Patterns of divergence among subgroups of Anopheles gambiae s.l. follow similar curves (LOESS-smoothed with a span of 1% using 10 kb nonoverlapping windows), although differing slightly in magnitude. One exception to this pattern is an increase in the X chromosome pericentromeric region (∼15–20 Mb) in the Anopheles coluzzii versus A. gambiae comparison and inside the 2La inversion where these populations differ in karyotype (G-2La+/+, A. coluzzii–2Laa/a, A. gambiae–2Laa/+). Divergence between A. coluzzii and both Anopheles arabiensis and Anopheles merus is increased on the X chromosome, especially inside the inverted Xag and Xbcd region (A. coluzzii vs. A. arabiensis) and in pericentromeric regions (A. coluzzii vs. A. merus). Grey bars indicate locations of differentially fixed chromosomal inversions as well as the 2La inversion and the large sweep on the GOUNDRY X (Xh). Low complexity and heterochromatic regions were excluded.
Autosomal Barriers to Introgression
Evidence for a large role of the X chromosome is strong, but it remains unclear which, if any, autosomal regions may serve as barriers to introgression. We divided the autosome into three classes based on expected levels of meiotic recombination (freely recombining, chromosomal inversions, pericentromeric and telomeric) and asked whether the regions where recombination may be restricted (i.e., inversions and pericentromeric/telomeric) harbor excess divergence consistent with barriers to introgression. In the comparison between A. coluzzii and A. merus, we find that pericentromeric and telomeric regions are significantly more diverged as a class than the freely recombining autosome (M–W P < 2.2 × 10−16), but nucleotide diversity is significantly lower in these regions (M–W P < 2.2 × 10−16). In the comparison between A. coluzzii and A. arabiensis, we generally find remarkably little evidence for elevated divergence across the autosome considering the level of divergence on the X, indicating a long history of introgression and few autosomal barriers. However, inspection of the genomic distribution of nucleotide diversity and divergence (figs. 4 and 5) reveals that the difference between nucleotide divergence and intraspecies diversity is especially high in the pericentromeric region of chromosome 3. Nucleotide diversity is reduced in this region, presumably resulting in part from the effects of intrapopulation positive and negative selection on linked sites. However, the relative increase in Dxy, which is not affected by positive selection, suggests that migrant alleles have been selected against in this region such that this region harbors both fewer shared polymorphisms as well as more private mutations relative to the nearby freely recombining regions.
A nopheles merus is fixed for a large private chromosomal inversion complex on chromosome 2 (2Rop), while other members of the complex carry the alternative forms of the overlapping 2Ro and 2Rp inversions. We compared genetic divergence in this region with freely recombining autosomal regions and find that the inverted region is significantly more diverged than surrounding genomic regions (M–W P < 2.2 × 10−16; fig. 3). Within-population nucleotide diversity is also slightly higher within the inverted region, but only without correcting for multiple testing (M–W uncorrected P = 0.0203; fig. 3), implying that inherently higher levels of diversity in this region could explain increased divergence in this region. These results suggest that, while this inversion may serve as a more recent barrier to introgression, 2Rop was not likely a primary barrier to introgression among these species, perhaps because it originated subsequent to the evolutionary periods with the highest rates of introgression between A. gambiaes.l. and A. merus.
Discussion
The permeability of species boundaries among species and subgroups of Anopheles has been controversial (Turner et al. 2005; Lawniczak et al. 2010; Neafsey et al. 2010; Turner and Hahn 2010; Cruickshank and Hahn 2014). The original observation of centromeric “islands of divergence” between the M and S (now A. coluzzii and A. gambiae, respectively) molecular forms led to the conclusion that gene flow among these species was extensive and ongoing, but these islands remained differentiated for reasons presumably related to their role in speciation among these taxa (Turner et al. 2005). Subsequent higher resolution studies observed especially high levels of differentiation in the islands of divergence against a background of differentiation across most of the genome, leading to the conclusion that reproductive isolation among these taxa is more advanced than previously thought (Lawniczak et al. 2010; Neafsey et al. 2010). However, these studies, like others claiming evidence of differential introgression along the genome, did not determine the relative roles of genetic drift, natural selection on linked sites, and introgression in determining the observed levels of differentiation (Cruickshank and Hahn 2014). A recent phylogenomic analysis made a strong case for extensive introgression among taxa within the A. gambiae species complex (Fontaine et al. 2015). In line with that conclusion, we use an alternative approach to show that chromosomal segments have introgressed among species at varying points on a speciation continuum (fig. 2). From these and previous results, a consensus is beginning to emerge that, rather than a radiation of discrete well-defined species, the A. gambiae species complex exists as a reticulate network of gene pools with offshoot populations and ancestral species remaining connected by periodic hybridization of differential efficiency across the genome. Such a model conflicts with traditional notions of species boundaries and raises questions about phenotypic evolution within a network of ecologically specialized gene pools.
Although the proportion of the individual genomes tested here that we identify with statistical evidence for recent introgression is relatively small (∼3%), the effect on the autosome is large over evolutionary time. The relationship between A. gambiae and A. arabiensis provides an extreme example of variation in the permeability of species boundaries, with evidence of autosomal introgression over the evolutionary history of these two species, including the putative sharing of an insecticide resistance allele in the last 60 years. In stark contrast to this pattern, the X chromosomes of these species remain highly diverged relative to the autosomes, implying that introgression has been ineffective on the X and that the X chromosome is the primary driver of speciation. A recent independent comparison between the A. arabiensis and A. gambiae genomes supports our conclusion that introgression is a major factor in the evolutionary history of the A. gambiae species complex (Fontaine et al. 2015).
Despite the homogenizing effects of extensive introgression among these taxa, distinct genetic clades can be identified implying that some genomic regions have remained differentiated and are incompatible with introgressing haplotypes. Over evolutionary time scales, we expect that natural selection will play a large role relative to drift in determining the composition of ancestry along the genome, resulting in a mosaic of introgressed segments along the genome, as has been observed for Neanderthal ancestry in modern humans (Sankararaman et al. 2014). To test for heterogeneity in ancestry that may reflect the action of natural selection and therefore barriers to introgression, we measured genetic sequence divergence among taxa and showed that long-term differential introgression has indeed shaped patterns of divergence among members of this species complex (figs. 3 and 5). By placing genomic patterns of divergence observed in our data into a phylogenetic context, a model of speciation and its genomic architecture emerges. In conjunction with known ecological differences between the species profiled here (Lehmann and Diabate 2008; Coetzee et al. 2013), our data point to a four-step speciation model in these mosquitoes in which 1) a small founder population expands into and adapts to an available ecological niche in the face of ongoing gene flow, 2) barriers to gene flow establish on the X chromosome, sometimes facilitated by suppressed recombination related to chromosomal inversions, 3) lowly recombining autosomal regions secondarily become restricted from gene flow, and 4) either pre- or postzygotic processes reduce effective gene flow entirely, and freely recombining autosomal regions become reproductively isolated and accumulate genetic divergence. Implicit to this model is that different regions become barriers to introgression in stages while introgression continues to homogenize the remaining genomic regions as in standard speciation with gene flow models (Wu 2001).
We show that the X chromosome is the most diverged and harbors the least evidence for introgression, suggesting that it is likely a barrier to introgression in multiple Anopheles species (figs. 2 and 3). In some cases, chromosomal inversions seem to play an important role. The observation that the X chromosome plays a disproportionately large effect in driving speciation (large-X) is in line with studies from Anopheles as well as many other organisms ranging from Drosophila to mammals (Coyne and Orr 1989; Geraldes et al. 2008; Garrigan et al. 2012; Sankararaman et al. 2014), but a unifying explanation for this pattern has yet to emerge. Importantly, a recent study of mating behavior showed that assortative mating between A. coluzzii and A. gambiae is controlled by the pericentromeric region on the X chromosome, providing a functional role for this region in speciation (Aboagye-Antwi et al. 2015). From an evolutionary genetic perspective, however, multiple hypotheses have been posited to explain the underlying evolutionary mechanisms underlying this pattern including the “faster-X” hypothesis, sex ratio meiotic drive, and misregulation of X-linked genes in males (reviewed in Presgraves 2008). The faster-X hypothesis posits that X-linked loci adapt faster than autosomal loci because X-linked recessive mutations are exposed to selection in males that have only a single X chromosome, resulting in faster accumulation of hybrid sterility factors (Charlesworth et al. 1987; Coyne and Orr 1989). Another popular hypothesis involves sex ratio meiotic drive where species-specific sex ratio distorter suppressors are disrupted in hybrids causing hybrid sterility (Frank 1991; Hurst and Pomiankowski 1991). A third hypothesis to explain this pattern is that gene expression dosage compensation of X-linked genes is misregulated in hybrids, causing sterility in hybrids (Lifschytz and Lindsley 1972). Although data have accumulated in Drosophila allowing more detailed speculation about the mechanisms underlying the large-X effect (Presgraves 2008), more data are needed to fully understand this pattern in Anopheles.
We show that pericentromeric regions also harbor especially high levels of divergence among more distantly related species pairs. This pattern is most apparent in the comparison between A. gambiae (A. coluzzii) and A. merus (figs. 3 and 5) and provides strong evidence that both A. arabiensis as well as A. merus diverged from A. gambiae while continuing to hybridize at a nonnegligible rate in at least the early stages of speciation. Although we could not test for recent introgression among A. gambiae and A. merus using the ABBA-BABA tests, the pattern of excess divergence in some genomic regions is in contrast to what we would expect under a divergence in allopatry model and is more consistent with historical introgression in freely recombining autosomal regions among A. gambiae and A. merus, likely in the evolutionary period following the initial species split. In the case of A. arabiensis, we show that contemporary hybridization with A. gambiae continues to homogenize all autosomal regions except the pericentromeric region of 3L despite strong barriers to genetic introgression across the X chromosome. Similar patterns of elevated divergence in lowly recombining pericentromeric and telomeric regions have also been observed in comparisons between Drosophila species (Begun et al. 2007; Langley et al. 2012; Mackay et al. 2012; Garrigan et al. 2014), but our results are the first demonstration of this pattern of excess sequence divergence in Anopheles. It is important to note that our results, especially the observation of excess divergence between A. gambiae and A. merus in low-diversity pericentromeric regions, are robust to the issues confounding previous observations of high differentiation in these regions, because our results derive from absolute measures of divergence instead of relative divergence measures that are highly sensitive to other population genetic processes including natural selection (Charlesworth 1998; Noor and Bennett 2009; Cruickshank and Hahn 2014). Importantly, we conservatively excluded heterochromatic centromeric regions, so the signals we identify reach well into euchromatic autosomal regions that are more robust to bioinformatic artifacts that plague analyses of centromeric regions. These results provide a clear empirical example of the important role of lowly recombining regions as barriers to introgression among hybridizing species.
The ABBA-BABA test has become a preferred method for detecting introgression, but there are several caveats and concerns relating to both the standard test as well as our modified DBLOCK test. First, it is possible that the signals of introgression we have detected are not from the species used in the test, but in fact we have detected introgression from an unsampled, or “ghost,” species. Durand et al. (2011) showed that such introgression can affect the results of ABBA-BABA tests. The presence of ghost Anopheles species hybridizing with the species sampled here is certainly a possibility and could impact some of our results. However, the possibility of introgression from “ghost taxa” does not change our conclusion that introgression continues among Anopheles species, shaping patterns of divergence regardless of exactly which subgroup is the donor. Second, results from our divergence-based analysis suggest that A. merus introgressed with an ancestral population of the A. gambiae species complex, potentially compromising its use as an outgroup in the ABBA-BABA test. Although such historical introgression could contribute marginally to the false positive rate in our inference, it is not likely to change our conclusion of introgression among taxa because our analysis is focused on long-shared haplotypes that are not likely to be affected by such old introgression. And third, a recent analysis showing that a similar block-wise test of introgression lacked power to identify introgressed haplotypes (Martin et al. 2014) raises questions about the robustness of our DBLOCK analysis. However, there are two important differences between our approach and the one evaluated by Martin et al. (2014). First, these authors implemented the test based on constant-sized physical windows of the genome, but we used constant-sized blocks of informative sites that varied in their physical size. This is an important distinction because our approach controls for the amount of information in each window, while the number of ancestry informative sites is bound to vary greatly among physical windows in the approach of Martin et al., which is likely to impact the sensitivity of this approach. The second difference between the approaches is that our approach explicitly controlled for LD in the data. We chose block sizes of 500 informative sites because this resulted in average physical window sizes of ∼ 250–350 kb (see Methods), which allowed each block to be divided into 100 segments larger in size than the expected distance that LD decays to background levels in this system. As a result, we believe that our approach is a robust approach for identifying introgressed genomic regions and is not likely to suffer from the same concerns raised by Martin et al. (2014).
In sum, our results suggest that species and subgroups in the A. gambiae species complex comprise a diffuse and interconnected gene pool that may confer access to beneficial genetic variants from a broad geographic and environmental range. Such genetic affinity has important implications for malaria control. On one hand, transgenes may spread more easily among subgroups and species of malaria vectors, which could reduce the effort needed to reach and manipulate all populations involved in disease transmission. On the other hand, our analysis suggests that certain genomic regions are less likely to cross species boundaries, especially the X chromosome, providing ideal settings for locating transgenes that are not intended for broad and general distribution across the species complex. In both cases, our results underscore the complexities involved in vector control on a continental scale.
Supplementary Material
Acknowledgments
We thank Matteo Fumagalli, Filipe Vieira, and Tyler Linderoth for assistance with next-generation sequence data analyses and ANGSD. We thank members of the Nielsen group for helpful discussions on various aspects of this work. We also thank Russ Corbett-Detig, Wynn Meyer, and three anonymous reviewers for helpful comments on an earlier version of this manuscript. We are thankful for the use of the Extreme Science and Engineering Discovery Environment, which is supported by National Science Foundation grant number OCI-1053575. This work was supported by National Institutes of Health grant AI062995. This work was also supported by a Cornell Center for Comparative and Population Genomics Graduate Fellowship and the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number F32GM103258 (J.E.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Literature Cited
- Aboagye-Antwi F, et al. 2015. Experimental swap of Anopheles gambiae’s assortative mating preferences demonstrates key role of X-chromosome divergence island in incipient sympatric speciation. PLoS Genet. 11:e1005141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A, et al. 2013. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet. 9:e1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W. 1909. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and modern science. Cambridge: Cambridge University Press; p. 85–101. [Google Scholar]
- Bazykin AD. 1969. Hypothetical mechanism of speciation. Evolution 23:685–687. [DOI] [PubMed] [Google Scholar]
- Begun DJ, et al. 2007. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 5:e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick DI, Fitzpatrick BM. 2007. Sympatric speciation: models and empirical evidence. Annu Rev Ecol Evol Syst. 38:459–487. [Google Scholar]
- Butlin RK. 2005. Recombination and speciation. Mol Ecol. 14:2621–2635. [DOI] [PubMed] [Google Scholar]
- Chan AH, Jenkins PA, Song YS. 2012. Genome-wide fine-scale recombination rate variation in Drosophila melanogaster. PLoS Genet. 8:e1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. 1998. Measures of divergence between populations and the effect of forces that reduce variability. Mol Biol Evol. 15:538–543. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. 1987. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 130:113–146. [Google Scholar]
- Clarkson CS, et al. 2014. Adaptive introgression between Anopheles sibling species eliminates a major genomic island but not reproductive isolation. Nat Commun. 5:4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee M, et al. 2013. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 3619:246–274. [PubMed] [Google Scholar]
- Costantini C, et al. 2009. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 1989. Patterns of speciation in Drosophila. Evolution 43:362–381. [DOI] [PubMed] [Google Scholar]
- Cruickshank TE, Hahn MW. 2014. Reanalysis suggests that genomic islands of speciation are due to reduced diversity, not reduced gene flow. Mol Ecol. 23:3133–3157. [DOI] [PubMed] [Google Scholar]
- della Torre A, et al. 2001. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 10:9–18. [DOI] [PubMed] [Google Scholar]
- Diabaté A, et al. 2006. Mixed swarms of the molecular M and S forms of Anopheles gambiae (Diptera: Culicidae) in sympatric area from Burkina Faso. J Med Entomol. 43:480–483. [DOI] [PubMed] [Google Scholar]
- Dobzhansky TG. 1937. Genetics and the origin of species. New York: Columbia University Press. [Google Scholar]
- Du W, et al. 2005. Independent mutations in the Rdl locus confer dieldrin resistance to Anopheles gambiae and An. arabiensis. Insect Mol Biol. 14:179–183. [DOI] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ffrench-Constant RH, Rocheleau TA, Steichen JC, Chalmers AE. 1993. A point mutation in a Drosophila GABA receptor confers insecticide resistance. Nature 363:449–451. [DOI] [PubMed] [Google Scholar]
- Fiston-Lavier AS, Singh ND, Lipatov M, Petrov DA. 2010. Drosophila melanogaster recombination rate calculator. Gene 463:18–20. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, et al. 2015. Mosquito genomics. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Science 347:1258524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SA. 1991. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45:262–267. [DOI] [PubMed] [Google Scholar]
- Garrigan D, Kingan SB, Geneva AJ, Vedanayagam JP, Presgraves DC. 2014. Genome diversity and divergence in Drosophila mauritiana: multiple signatures of faster X evolution. Genome Biol Evol. 6:2444–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrigan D, et al. 2012. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 22:1499–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Ferrand N, Nachman MW. 2006. Contrasting patterns of introgression at X-linked loci across the hybrid zone between subspecies of the European rabbit (Oryctolagus cuniculus). Genetics 173:919–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, et al. 2008. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Mol Ecol. 17:5349–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnémé A, et al. 2013. Equivalent susceptibility of Anopheles gambiae M and S molecular forms and Anopheles arabiensis to Plasmodium falciparum infection in Burkina Faso. Malar J. 12:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PR, Grant BR, Petren K. 2005. Hybridization in the recent past. Am Nat. 166:56–67. [DOI] [PubMed] [Google Scholar]
- Gravel S. 2012. Population genetics models of local ancestry. Genetics 191:607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, White BJ, Muir CD, Besansky NJ. 2012. No evidence for biased co-transmission of speciation islands in Anopheles gambiae. Philos Trans R Soc Lond B Biol Sci. 367:374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison RG, Larson EL. 2014. Hybridization, introgression, and the nature of species boundaries. J Hered. 105(Suppl. 1):795–809. [DOI] [PubMed] [Google Scholar]
- Hey J. 2006. Recent advances in assessing gene flow between diverging populations and species. Curr Opin Genet Dev. 16:592–596. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 39:21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pomiankowski A. 1991. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics 128:841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamali M, Xia A, Tu Z, Sharakhov IV. 2012. A new chromosomal phylogeny supports the repeated origin of vectorial capacity in malaria mosquitoes of the Anopheles gambiae complex. PLoS Pathog. 8:e1002960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneliussen T, Albrechtsen A, Nielsen R. 2014. ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192:533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawniczak MKN, et al. 2010. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 330:512–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, et al. 2013. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc Natl Acad Sci U S A. 110:19854–19859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Diabate A. 2008. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 8:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifschytz E, Lindsley DL. 1972. The role of X-chromosome inactivation during spermatogenesis (Drosophila-allocycly-chromosome evolution-male sterility-dosage compensation). Proc Natl Acad Sci U S A. 69:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand RP. 1983. Field observations on swarming and mating in Anopheles Gambiae mosquitoes in Tanzania. Neth J Zool. 34:367–387. [Google Scholar]
- Marsden CD, et al. 2014. Diversity, differentiation, and linkage disequilibrium: prospects for association mapping in the malaria vector Anopheles arabiensis. G3 (Bethesda) 4:121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SH, Davey JW, Jiggins CD. 2014. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol Biol Evol. 32:244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. 1942. Systematics and the origin of species, from the viewpoint of a zoologist. New York: Columbia University Press. [Google Scholar]
- McGaugh SE, Noor MAF. 2012. Genomic impacts of chromosomal inversions in parapatric Drosophila species. Philos Trans R Soc Lond B Biol Sci. 367:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GAT, et al. 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304:581–584. [DOI] [PubMed] [Google Scholar]
- Muller HJ. 1940. Bearing of the Drosophila work on systematics. In: Huxley JS, editor. The new systematics. Oxford: Clarendon Press; p. 185–268. [Google Scholar]
- Nachman MW, Payseur BA. 2012. Recombination rate variation and speciation: theoretical predictions and empirical results from rabbits and mice. Philos Trans R Soc Lond B Biol Sci. 367:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Barton NH. 2003. Accumulating postzygotic isolation genes in parapatry: a new twist on chromosomal speciation. Evol Int J Org Evol. 57:447–459. [DOI] [PubMed] [Google Scholar]
- Neafsey DE, et al. 2010. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330:514–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J. 2001. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 98:12084–12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noor MAF, Bennett SM. 2009. Islands of speciation or mirages in the desert? Examining the role of restricted recombination in maintaining species. Heredity 103:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ, Sandoval CP. 2003. Reproductive isolation driven by the combined effects of ecological adaptation and reinforcement. Proc Biol Sci. 270:1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin SM, et al. 2014. Genomic analyses of three malaria vectors reveals extensive shared polymorphism but contrasting population histories. Mol Biol Evol. 31:889–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. 2009. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics 181:711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. [Google Scholar]
- Riehle MM, et al. 2011. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science 331:596–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. 2001. Chromosomal rearrangements and speciation. Trends Ecol Evol. 16:351–358. [DOI] [PubMed] [Google Scholar]
- Sankararaman S, et al. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, della Torre A, Calzetta M, Powell JR. 2005. Differential introgression of chromsomal regions between Anopheles gambiae and An. arabiensis . Am J Trop Med Hyg. 73:326–335. [PubMed] [Google Scholar]
- Slotman MA, et al. 2006. Genetic differentiation between the BAMAKO and SAVANNA chromosomal forms of Anopheles gambiae as indicated by amplified fragment length polymorphism analysis. Am J Trop Med Hyg. 74:641–648. [PubMed] [Google Scholar]
- Slotman MA, et al. 2007. Evidence for subdivision within the M molecular form of Anopheles gambiae. Mol Ecol. 16:639–649. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW. 2010. Genomic islands of speciation or genomic islands and speciation? Mol Ecol. 19:848–850. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. 2005. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 3:e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Sattler R, et al. 2007. Mosaic genome architecture of the Anopheles gambiae species complex. PLoS One. 2:e1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, Wilding CS, Steen K, Pinto J, Donnelly MJ. 2012. Gene flow-dependent genomic divergence between Anopheles gambiae M and S forms. Mol Biol Evol. 29:279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weetman D, et al. 2014. Contemporary gene flow between wild An. gambiae s.s. and An. arabiensis. Parasit Vectors. 7:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. 2013. World malaria report. Geneva (Switzerland): World Health Organization. [Google Scholar]
- Wu CI. 2001. The genic view of the process of speciation. J Evol Biol. 14:851–865 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.