Abstract
Chlamydiaceae are obligate intracellular bacteria that cause a diversity of severe infections among humans and livestock on a global scale. Identification of new species since 1989 and emergence of zoonotic infections, including abortion in women, underscore the need for genome sequencing of multiple strains of each species to advance our knowledge of evolutionary dynamics across Chlamydiaceae. Here, we genome sequenced isolates from avian, lower mammalian and human hosts. Based on core gene phylogeny, five isolates previously classified as Chlamydia abortus were identified as members of Chlamydia psittaci and Chlamydia pecorum. Chlamydia abortus is the most recently emerged species and is a highly monomorphic group that lacks the conserved virulence-associated plasmid. Low-level recombination and evidence for adaptation to the placenta echo evolutionary processes seen in recently emerged, highly virulent niche-restricted pathogens, such as Bacillus anthracis. In contrast, gene flow occurred within C. psittaci and other Chlamydiaceae species. The C. psittaci strain RTH, isolated from a red-tailed hawk (Buteo jamaicensis), is an outlying strain with admixture of C. abortus, C. psittaci, and its own population markers. An average nucleotide identity of less than 94% compared with other Chlamydiaceae species suggests that RTH belongs to a new species intermediary between C. psittaci and C. abortus. Hawks, as scavengers and predators, have extensive opportunities to acquire multiple species in their intestinal tract. This could facilitate transformation and homologous recombination with the potential for new species emergence. Our findings indicate that incubator hosts such as birds-of-prey likely promote Chlamydiaceae evolution resulting in novel pathogenic lineages.
Keywords: Chlamydiaceae, comparative genomics, homologous recombination, molecular clock, population structure, red-tailed hawk
Introduction
Chlamydiaceae are a family of obligate intracellular bacteria with nine species. Until 1992, there were only four recognized species of the genus Chlamydia: Chlamydia trachomatis and Chlamydia psittaci were established in 1968 followed by C. pneumoniae in 1989 and Chlamydia pecorum in 1992 (Fukushi and Hirai 1992). The remaining five species were identified in 1999 (Everett et al. 1999). In 1999, because of shared morphological similarities, Chlamydiaceae species were reclassified into two separate genera, Chlamydia and Chlamydophila (Everett et al. 1999). Despite the differences in chromosome size (∼1.05 Mb) for the Chlamydia species C. trachomatis, C. muridarum and Chlamydia suis compared with the six species of the Chlamydophila lineage (∼1.2 Mb), there was insufficient chromosomal evidence for two separate genera, and they have now been reunited under the genus Chlamydia (Greub 2010; Sachse et al. 2015). In 2014, two new species were accepted: Chlamydia avium infects pigeons and psittacine birds, whereas Chlamydia gallinacea infects chickens, guinea fowl and turkeys (Sachse et al. 2014).
Chlamydia trachomatis exclusively infects humans and remains the leading cause of bacterial sexually transmitted diseases and preventable blindness worldwide (Dean et al. 2013; Shao et al. 2013). Although C. pneumoniae originated from zoonotic transmission, it is primarily a human pathogen responsible for endemic infection as well as epidemic outbreaks of respiratory and ocular disease (Bodetti et al. 2002; Myers et al. 2009; Roulis et al. 2013). Chlamydia psittaci is well recognized for its zoonotic potential, causing psittacosis, an upper respiratory infection that can be life-threatening if it becomes systemic, as well as eye diseases in humans. It also causes disease in wild birds and domesticated animals (Longbottom and Coulter 2003; Harkinezhad et al. 2009; Reinhold et al. 2011). Although Chlamydia abortus is mainly responsible for ovine enzootic abortion, it has emerged as a dangerous pathogen for pregnant women (Longbottom and Coulter 2003). Chlamydia pecorum and C. suis are important livestock pathogens (Mohamad and Rodolakis 2010; Reinhold et al. 2011; Schautteet and Vanrompay 2011), whereas Chlamydia felis, Chlamydia muridarum, and Chlamydia caviae infect cats, rodents, and guinea pigs, respectively.
The ongoing discovery of new Chlamydiaceae species and strains infecting novel animal hosts, such as frogs and koalas, and the recent knowledge of human abortions induced by zoonotic Chlamydia species (Bodetti et al. 2002; Joseph et al. 2011; Somboonna et al. 2011; Sachse et al. 2014) indicate an urgent need to more fully understand Chlamydiaceae evolution. Comparative genomics projects of C. trachomatis using dozens of strains (Harris et al. 2012; Joseph et al. 2012) have revealed much about pathogen population structure, including clade specificity for disease phenotypes and relatively frequent recombination without evidence for gene gain by horizontal gene transfer and little DNA sequence loss by deletion (Joseph et al. 2011, 2012; Joseph and Read 2012).
In contrast, progress in comparative genomics of other Chlamydiaceae has been slow, largely because of technical difficulties in isolating and culturing strains from each species (Read et al. 2013; Bachmann, Polkinghorne, et al. 2014). A study of 20 C. psittaci genomes discovered frequent switching of DNA among strains from different avian and mammalian hosts, and homologous recombination that occurred more often than for C. trachomatis (Read et al. 2013). This study also revealed a clonal epidemic expansion of the species in North America and the recent emergence of the disease psittacosis originating from New World parrots. A comparative study of four C. pecorum genomes identified limited variations between strains, but found differences in the number of pseudogenes (Sait et al. 2014), whereas a study that included these four strains and four additional ones concluded that variation is largely determined by single nucleotide polymorphisms (SNPs) (Bachmann, Fraser, et al. 2014). Another study identified mixed populations of genetically distinct strains of C. pecorum obtained from koalas and sheep (Bachmann et al. 2015). Aside from these studies, only a few genomes are available that cover other Chlamydiaceae species: Two for C. abortus (Thomson et al. 2005); and one each for C. felis (Azuma et al. 2006), C. suis (Donati et al. 2014), C. muridarum (Read et al. 2000), and C. caviae (Read et al. 2003).
Despite the plethora of human and nonhuman diseases that exact a huge economic burden globally, there have been no comparative genomic studies of the different Chlamydiaceae species to date. The fact that similar diseases are caused by the different species and that many species infect a diversity of hosts (supplementary table S1, Supplementary Material online) suggests that genomics, at least in part, could explain host–pathogen interactions and disease. To better understand their interactions and evolutionary processes, we analyzed the genomes of 36 isolates from human, lower mammalian, and avian species.
Materials and Methods
Chlamydiaceae Species/Strains, Clonal Purification, and Generation of Genomic DNA
In this study, we sequenced five C. abortus, three C. pecorum, and two C. psittaci strains (supplementary table S2, Supplementary Material online). In addition, we included the published whole-genome sequencings (WGSs) of 31 other Chlamydiaceae strains, including those from C. pneumoniae, Chlamydia avium, C. felis, C. caviae, C. gallinacea, C. trachomatis, and C. muridarum (supplementary table S2, Supplementary Material online). Although the majority of the strains were of lower mammalian and avian origin, six strains were isolated from humans. Each of the ten new strains was individually propagated in McCoy or HeLa 229 cells, clonally purified, and treated with DNase prior to gDNA purification as previously described (Somboonna et al. 2011; Joseph et al. 2012; Read et al. 2013).
Genome Sequencing
Genomes were sequenced using GS-FLX (454 Life Sequencing Inc., Branford, CT). Libraries for sequencing were prepared from 1 to 5 µg of genomic DNA. De novo assembly was performed using Newbler (Knight J, personal communication) and default parameters. Further processing of the genomes was performed as previously described (Joseph et al. 2012). The contigs from C. abortus, C. pecorum and C. psittaci were aligned against the previously sequenced C. abortus S26/3, C. pecorum E58 and C. psittaci 6BC genomes, respectively, using abacus (http://abacas.sourceforge.net/Manual.html, last accessed April, 2015) to create concatenated, ordered “pseudocontigs.” These pseudocontigs were considered as the bacterial chromosome for each strain and annotated using the Prokka bacterial genome annotation pipeline (Seemann 2014). For comparative analysis, all published genomes used in this study were also re-annotated using the same pipeline. Pairwise comparison of genomes was conducted by calculating average nucleotide identity (ANI) as described previously (Richter and Rosselló-Móra 2009) using JSpecies (www.imedea.uib.es/jspecies, last accessed June, 2015) with default parameters. The raw genome data generated for this study are deposited in the SRA database under the following study accession numbers: SRP060457 (EP1), SRP060463 (EP6), SRP060476 (EBA), SRP060475 (AC1), SRP060473 (EAE-LX), SRP060471 (B577), SRP060468 (24-26), SRP060466 (OSP), SRP060465 (JP394), and SRP060464 (757).
Whole-Genome Alignment and Identification of Core Genes
Three whole-genome MAUVE (Darling et al. 2004, 2010) progressive alignments were computed. The first consisted of 36 genomes from C. psittaci, C. abortus, C. pecorum, C. pneumoniae, C. caviae, C. felis, C. avium and C. gallinacea, which was used for ClonalFrame (Didelot et al. 2010), fineSTRUCTURE (Yahara et al. 2013), BAPS (Tang et al. 2009) and phylogenetic analyses (see below). In the second alignment, we added four C. trachomatis genomes from each of the four C. trachomatis clades and a single C. muridarum genome (total of 41 strains). The third alignment consisted of 32 genomes sans C. caviae, C. felis, C. avium, and C. gallinacea for a separate ClonalFrame analysis. The core Locally Collinear Block (LCB) from the second MAUVE alignment was extracted and concatenated to form a super alignment for use in phylogenetic analyses described below.
The complete predicted proteome from all 36 annotated/reannotated genomes was searched against itself using BLASTP with an e-value cutoff of 1e-05. The best BLASTP scores were utilized for identifying orthologous sequences using the OrthoMCL algorithm as described (Li et al. 2003); the pan-matrix containing all information regarding orthologous genes was generated. This pan-matrix was imported into the R-package called micropan (Snipen and Liland 2015) to generate the visualizations describing the Chlamydiaceae pan genome. Core genes are defined as the protein-coding gene clusters that are shared by all Chlamydiaceae strains. Unique genes found in only one strain were also identified. MUSCLE (Edgar 2004) was used with default settings to align core genes; each protein alignment was filtered by GBLOCKS (Castresana 2000) to remove gaps and highly divergent regions.
Phylogenetic Reconstruction
The core LCBs extracted from the second MAUVE alignment of 41 strains (see above) were concatenated for phylogenetic analysis. We also concatenated alignments of all core protein families and reconstructed the phylogeny. The maximum-likelihood (ML)-based phylogenetic reconstruction was implemented using RAxML (Stamatakis et al. 2012). For whole-genome alignment and core protein sequence alignment, the branch lengths/evolutionary distances were estimated, respectively, using the GTR (general time reversible) nucleotide substitution model and Jones–Taylor–Thornton (JTT) amino acid substitution model of rate heterogeneity with four discrete rate categories. To evaluate statistical support, a majority rule-consensus tree of 100 bootstrap replicates was computed. We also used the core of the first and third MAUVE alignments (36 and 32 genomes, respectively) to reconstruct phylogenies using the coalescent-based ClonalFrame method (Didelot et al. 2010) (see below). For the phylogeny of the 17 plasmids, we performed OrthoMCL clustering of all genes, identifying five complete genes found in all 17 plasmids that were concatenated and aligned using progressiveMAUVE. Three sets of ClonalFrame analysis were performed (see below). For the ML tree, we removed the recombinant regions identified by ClonalFrame and performed RAxML using the same parameters as for the chromosome (above).
Analysis of Homologous Recombination
ClonalFrame (version 1.2) (Didelot et al. 2010) was run for 40,000 iterations on the whole-genome core nucleotide alignment identified in the first (36 genomes) and third (32 genomes) MAUVE alignments; the initial half was discarded as Markov chain Monte Carlo (MCMC) burn-in. Three independent and parallel runs of ClonalFrame were performed. Their reconstructed phylogenies and recombination events across the three runs had high congruence. Additionally, for each reconstructed branch substitution event introduced by either mutation or recombination, the number of mutation and recombination events were computed. The relative effect of recombination and mutation on genetic change (r/m) and the relative rate of mutation and recombination (ρ/θ) were estimated as described (Joseph et al. 2012; Read et al. 2013).
Molecular Clock Analysis
Bayesian analysis of evolutionary rates and divergence times was performed based on the MAUVE alignment using BEAST v1.8.2 with the HKY (Hasegawa–Kishino–Yano) substitution model and tip dates defined as year of isolation. We implemented two molecular clock models: Strict molecular clock with a constant size coalescent model, and the relaxed molecular clock model along with the Bayesian skyline demographic model. Three independent runs of BEAST for both models, each with 300 million MCMC iterations, sampling every 10,000 iterations with the first 10% discarded as burn-in, were performed. These three runs were combined to provide robust estimates of posterior parameter distributions (calculated using Tracer v1.5).
ClonalFrame also estimates the clonal genealogy as well as the number of mutations that occurred on each branch of the tree. Combining this information with isolation date for each genome (supplementary table S2, Supplementary Material online) allowed the simultaneous estimation of mutation rate per year and dates of existence of nodes in the ClonalFrame tree. This inference was performed assuming the Kingman coalescent with the temporally offset leaves model (Drummond et al. 2002) for the tree and a uniform prior for mutation rate.
Population Structure Analysis
To elucidate the possible population structure of the 36 Chlamydophila lineage strains, the ChromoPainter algorithm was applied to the genome-wide haplotype data using the linkage model. A recombination map file was created by specifying a uniform recombination rate per-site per-generation using a Perl script called makeuniformrecfile.pl (http://www.paintmychromosomes.com). The output from ChromoPainter is a coancestry matrix that summarized the blockwise homology between the 36 genomes. The fineSTRUCTURE algorithm (Yahara et al. 2013) used the coancestry matrix generated by ChromoPainter to perform model-based clustering using a Bayesian MCMC approach to explore the population structure. FineSTRUCTURE was run for 400,000 iterations; the first 200,000 iterations were discarded as MCMC burn-in. The thinning interval was specified at 100.
To gain further insights into the population structure of these species, we used the BAPS software (Corander and Marttinen 2006) to identify genetically differentiated groups and determine the amount of admixture among these groups. We ran the BAPS clustering model with hierarchical manner to find substructures inside the main clusters (Cheng et al. 2013). To find optimal clustering, we ran five independent iterations with the prior upper bound of the number of clusters set to 35. The clustering was performed with four levels in the hierarchy. The first level gave 10 clusters, whereas the fourth yielded 20 clusters. We also conducted an admixture analysis based on the 20 clusters, considering the minimum number of individuals for a population as one and, other than that, we employed the same values that were used by Castillo-Ramirez et al. (2012).
Attribution of Origins to the Recombination Events
For each branch of the tree reconstructed by ClonalFrame, we defined recombined fragments as genomic intervals with a posterior probability of recombination above 0.50 at every site, reaching 0.95 in at least one site, and a length of at least 100 bp (Didelot et al. 2011). Each such recombined fragment was searched for using BLAST (Basic Local Alignment Search Tool) against the whole database containing all “finished” genomes and plasmid sequences of Chlamydiaceae species minus strains of the clade affected by the import. The hits with the highest normalized BLASTN score along with a percent identity of at least 95% were kept. If these hits were with strains belonging to the same species, the origin of the event was attributed to ancestral nodes and called “ambiguous,” meaning imported from an unknown external source.
Substitution Rate (dN/dS) Calculations
Nonsynonymous (dN) and synonymous (dS) substitutions and dN/dS ratios were calculated, and protein alignments along with corresponding nucleotide sequences were converted to codon alignments using PAL2NAL (Suyama et al. 2006) as described (Joseph et al. 2012).
Results
The C. abortus Species Is a Recent Clonal Expansion within the Chlamydophila Lineage
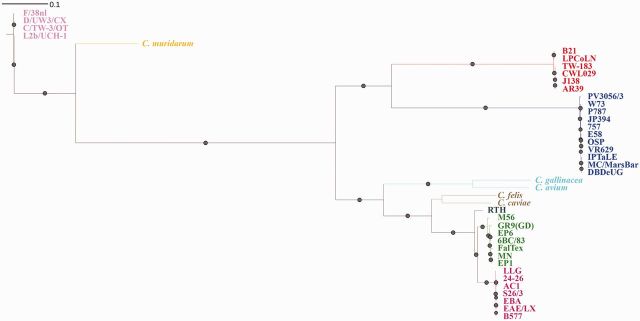
We applied 454 technology for WGS of ten previously unsequenced Chlamydiaceae strains of human and lower mammalian origin, originally typed by serology and disease etiology as C. abortus (supplementary table S2, Supplementary Material online). The ML tree created from an alignment of 36 Chlamydophila genomes (10 from this study + 26) plus 4 C. trachomatis and 1 C. muridarum (fig. 1) determined that two strains needed to be reclassified as C. psittaci and three as C. pecorum. Four of the true C. abortus strains (B577, EBA, AC1, and EAE/LX) caused abortion in sheep and cows, and one (24-26) caused pneumonia in sheep. The three newly sequenced C. pecorum strains (OSP, JP394, and 757) were isolated from sheep abortion specimens whereas the two new C. psittaci strains, EP6 and EPI, caused abortion in a human and pneumonia in a sheep, respectively. None of the C. abortus strains contained the typical 7.5-kb Chlamydiaceae plasmid, whereas all C. psittaci and C. pecorum strains used in this study had the plasmid. However, none of the previously published C. pecorum genomes reported the presence of a plasmid. In the plasmid phylogeny (supplementary fig. S1, Supplementary Material online), each species is separated into monophyletic clades. Surprisingly, the plasmids of C. pneumoniae and C. pecorum shared a more recent ancestor with the Chlamydia species C. trachomatis and C. muridarum than with other Chlamydophila species.
Fig. 1.—
Whole-genome phylogeny of Chlamydia species and strains. The tree was constructed using ML approach utilizing RAxML based on the whole-genome alignment (see Materials and Methods). Chlamydia pneumoniae strains are highlighted in red, C. pecorum strains in blue, C. psittaci strains in green, C. abortus strains in magenta, and C. trachomatis in pink.
Including this study, there are now seven C. abortus genomes in the public domain. These genomes form a monomorphic group showing very little genetic variation (maximum ANI between two strains was 99.5%) (supplementary table S3, Supplementary Material online) and very similar gene content (95% of genes are found in all seven strains). Interspecific genome comparisons show that the most variable portion of Chlamydia chromosomes for gene content is typically around the replication termination region termed the “plasticity zone” (PZ), although its boundaries are not consistently defined (Read et al. 2000; Thomson et al. 2005; Voigt et al. 2012; Sait et al. 2014). The ten strains sequenced in this study had PZ gene content similar to other members of their species (Voigt et al. 2012; Read et al. 2013; Sait et al. 2014).
To investigate further the variation in gene content within the Chlamydophila lineage we created a genome set using the 10 genomes sequenced here plus 26 other published genomes consisting of C. psittaci, C. pneumoniae, C. pecorum, C. avium, C. gallinacea, C. felis, and C. caviae strains (supplementary table S4, Supplementary Material online). The matrix contained 1,449 unique gene clusters/families (supplementary fig. S2, Supplementary Material online). To describe the pan genome, we fitted a binomial mixture model on the pan matrix using the binomixEstimate function on micropan R package; the optimum number of components to characterize the gene clusters was estimated to be 6 (supplementary fig. S2a, Supplementary Material online). Principal component analysis performed using the pan matrix clustered the genomes into groups that were highly congruent to the inferred whole-genome phylogenetic clades (supplementary fig. S2b, Supplementary Material online). Supplementary figure S2c andd, Supplementary Material online, shows the distribution of gene families across the clade and the rarefaction of the e pan-genome, respectively. We identified 668 core genes present among all 36 Chlamydiaceae strains, which represented 48.19–74.80% of the total number of genes in each genome. This is similar to earlier estimates (Collingro et al. 2011), illustrating the parsimony of conserved function within Chlamydiaceae.
Seven genes, present in similarity-based clusters, were only found in C. abortus. Six were short hypothetical genes in the PZ. The other was the rapidly evolving IncA gene, which encodes the inclusion membrane protein A (IncA) protein (Rockey et al. 1995). Chlamydia pecorum had 25 unique gene clusters with 21 hypothetical protein-coding genes and genes coding for aminopeptidase 2, phospholipase D precursor, polymorphic membrane protein 13 (Pmp 13), and IncA. Chlamydia psittaci had only one unique gene cluster encoding a hypothetical protein shared by all C. psittaci strains. All genes of known function found in the five C. abortus, three C. pecorum, and two C. psittaci genomes sequenced in this study have previously been identified in their species. In C. pneumoniae, 80 species-specific protein-coding gene families were identified, of which the majority were hypothetical genes. We also identified unique protein-coding genes (genes that did not cluster with any other genes) for each of the 36 strains.
Comparative analysis of 17 plasmid sequences of all the available Chlamydiaceae species revealed the presence of five core genes in their plasmids, which were replicative DNA helicase, site-specific tyrosine recombinase (XerC, phage integrase family protein), sporulation initiation inhibitor protein (soj), and two hypothetical protein-coding genes. The gene coding for proteins P-6/P-7 was not present in C. avium and C. psittaci M56 strains.
To investigate the type and strength of selection across the 36 strains, we calculated the dN and dS substitutions for each of the 36 combinations of pairwise core genes and assessed the dN/dS ratio of each strain (Rocha et al. 2006; Hershberg and Petrov 2010) (supplementary fig. S4, Supplementary Material online). The median dN/dS ratios given the accumulation of dS substitution for C. abortus strains were higher compared with all other species, a result also suggested by Voigt et al. (2012).
Phylogenetic Analysis Places the C. psittaci Red-Tailed Hawk Strain As a Potential New Species within the Chlamydiaceae Family
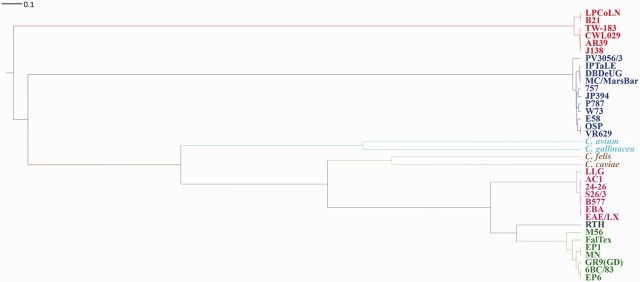
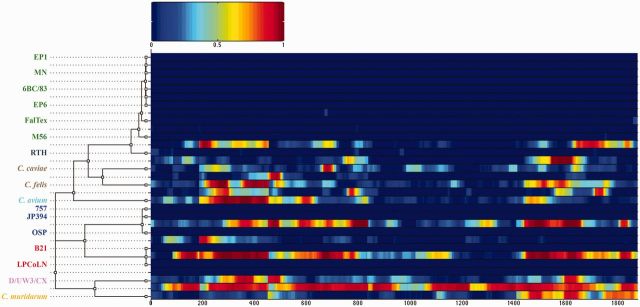
We inferred a phylogeny for the Chlamydophila clade, which accounted for the effects of recombination by using ClonalFrame (Didelot and Falush 2007), where branch lengths were estimated in coalescent units of time (figs. 2 and 3). We used a whole-genome alignment of the 36 genomes consisting of 549 homologous regions (LCBs) identified by MAUVE software (Darling et al. 2004). These regions represented 676,097 bp of the 1.2 Mb average genome size. Predicted recombination events occurred across the entire portion of the conserved core alignment, as was seen in C. trachomatis and C. psittaci alignments (Harris et al. 2012; Joseph and Read 2012; Read et al. 2013). ClonalFrame provided two output statistics of the recombination rate in the Chlamydophila lineage: ρ/θ represents the frequency of occurrence of recombination relative to mutation, whereas r/m shows the importance of recombination on genetic diversification relative to mutation (supplementary table S4, Supplementary Material online). In C. abortus, the recombination rate was significantly lower than the other species, whereas its closest relative C. psittaci had the highest rate in the genus. For plasmids, ClonalFrame analysis on the core alignment of 17 Chlamydiaceae strains revealed significant evidence of recombination (fig. 4) except for all C. psittaci strains including the red-tailed hawk (RTH) strain. The estimated r/m ratio for the plasmids was 1.18 (95% CI; 0.37–3.144) and 0.076 (95% CI; 0.015–0.186) for ρ/θ.
Fig. 2.—
Clonal genealogy inferred by ClonalFrame. Whole-genome alignment data for the 36 Chlamydiaceae genomes in supplementary table S2, Supplementary Material online, except for the sole representative of the C. felis, C. caviae, C. avium, and C. gallinacea species, were used for analysis. The branch lengths are shown in coalescent units of time.
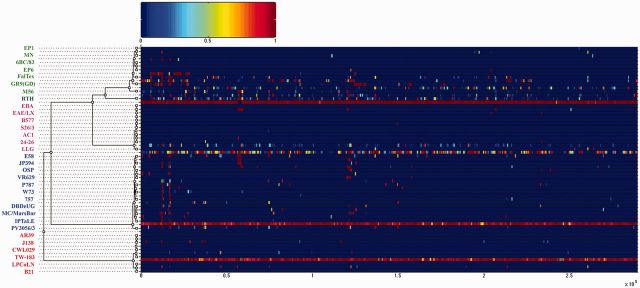
Fig. 3.—
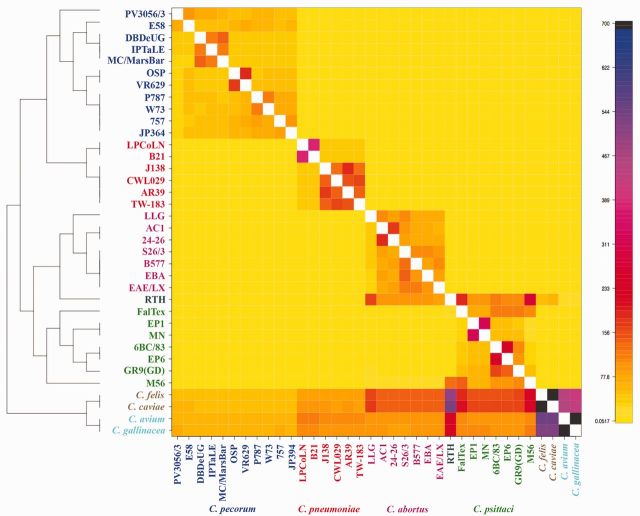
Results of the ClonalFrame analysis on an alignment of 32 chlamydial genomes. Inferred clonal genealogy is shown on the left. Each branch of the tree corresponds to a row of the heat map, which is horizontally aligned according to the core MAUVE whole-genome alignment. Each row shows the posterior probability of recombination estimated by ClonalFrame on the corresponding branch (y axis) and along positions of the alignment (x axis; ×105 bp). Chlamydia psittaci, C. abortus, C. pecorum and C. pneumoniae strains are in green, magenta, blue and red, respectively. The C. psittaci RTH is shown in dark blue.
Fig. 4.—
Results of the ClonalFrame analysis on an alignment of 17 Chlamydiaceae plasmids. The inferred clonal genealogy is shown on the left. Each branch of the tree corresponds to a row of the heat map, which is horizontally aligned according to the core MAUVE whole-genome alignment. Each row of the heat map shows the posterior probability of recombination estimated by ClonalFrame on the corresponding branch (y axis) and along the positions of the alignment (x axis). C. psittaci, C. pecorum, and C. pneumoniae species are in green, magenta, blue and red, respectively.
In the recombination-corrected ClonalFrame phylogeny (figs. 2 and 3), all species clustered monophyletically into clades except the C. psittaci RTH strain (figs. 1 and 2). The two recently reported species, C. avium and C. gallinacea, formed a distinct clade, as did C. caviae and C. felis. In the ML tree, RTH was an outgroup of both C. abortus and the main C. psittaci group. However, in the ClonalFrame tree, RTH was in the C. psittaci clade, as previously reported (Read et al. 2013). The difference in ML phylogeny was likely due to extensive recombination with C. abortus found in RTH (see below). It is notable that the RTH had an ANI less than 94% with other C. psittaci or C. abortus members (supplementary table S3, Supplementary Material online), which is below the threshold for assignment of the strain to a new species.
There was a reasonable correlation (r = 0.4) with date of sample and root-to-tip distance on the ClonalFrame phylogeny, which allowed a molecular clock rate to be estimated (supplementary fig. S3, Supplementary Material online). Based on ClonalFrame analysis that differentiates between recombination and mutation events separately, the average mutation rate in Chlamydiaceae was found to be high (1.42e-5 per site per year with a 95% credibility interval of 1.41 e-5–1.43 e-5). BEAST-estimated mutation rates were even higher: 2.4778 e-4 (95% HPD interval 3.374 e-11–1.4353 e-7) for the relaxed skyline model and 1.8576 e-4 (95% HPD interval 3.363 e-12, 1.3408 e-7) for the strict molecular clock with a constant size coalescent model, in line with our previous estimates of 1.683 e-4 for BEAST and 1.74 e-5 for ClonalFrame (Read et al. 2013). With this fast rate, the molecular clock based on ClonalFrame placed the root of the Chlamydophila lineage at only 5570 BC. The split at the common ancestor of C. abortus and C. psittaci to form the two new species was predicted at AD 1–500, the split between RTH and C. psittaci at AD 719, the modern C. psittaci lineage seems to have emerged in the past 200 years (AD 1678 as we noted previously [Read et al. 2013]), and the split between C. psittaci and C. abortus at AD 1881 (supplementary fig. S3, Supplementary Material online). In the absence of historical data or specimens, the dates were based solely on mutation rates derived from recent genome sequences and should therefore be treated with caution.
Population Structure of the Chlamydophila Lineage
As ClonalFrame revealed that recombination had been a major factor in Chlamydiaceae evolution, we investigated how genetic variation is apportioned across species using BAPS (fig. 5) and fineSTRUCTURE software (fig. 6). Both methods establish genetically differentiated groups and infer possible admixture occurring among those groups. We used BAPS to implement a hierarchical Bayesian model-clustering based on the core SNP alignment, which produced 20 clusters at the finest hierarchical level (fig. 5 and supplementary table S5, Supplementary Material online). fineSTRUCTURE was a two-stage analysis consisting of in silico chromosome painting that created a coancestry matrix containing the number of blocks copied from each donor strain to each recipient strain. The fineSTRUCTURE algorithm enabled use of this coancestry matrix to conduct model-based linkage clustering and assign individual strains to 18 populations (fig. 6).
Fig. 5.—
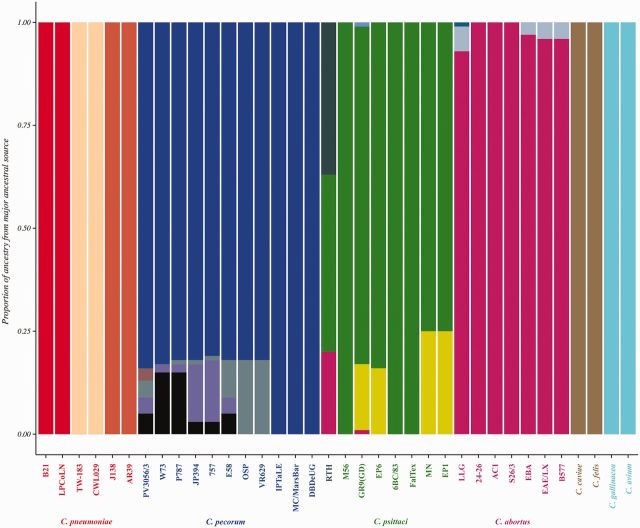
Admixture analysis of Chlamydiaceae. Mixture and admixture analyses were conducted using BAPS. Each color represents one of the genetically differentiated groups, and each vertical colored bar corresponds to one isolate. When a vertical bar shows two or more colors, each color corresponds to one of the groups, showing evidence for admixture; the proportion of every color in the bar reflects the proportion of the isolate coming from the group represented by that particular color. For example, the ancestral source of strain GR9(GD) consists predominately of C. psittaci (green) but also contains sources from C. abortus (magenta) and from two different external sources (yellow and pale blue).
Fig. 6.—
ChromoPainter co-ancestry matrix with population structure assignment based on fineSTRUCTURE analysis. The color of each cell of the matrix indicates the expected number of DNA chunks copied from a donor genome (x axis) to a recipient genome (y axis).
As expected, both methods mapped populations to distinct clades of the phylogenetic tree (figs. 1 and 2). fineSTRUCTURE grouped C. pneumoniae strains into two populations: The LPCoLN and B21 strains isolated from lower mammals; and the remaining four strains isolated from humans. However, BAPS clustered C. pneumoniae strains into three groups: TW-183 and CWL029 in one group; J138 and AR39 into a second group; and B21 and LPCoLN into a third group (fig. 5). The 11 C. pecorum strains were segregated into 5, 5, and 6 groups by phylogeny, fineSTRUCTURE, and BAPS analysis, respectively. The three koala-infecting C. pecorum strains, MC/MarsBar, DBDeUG and IPTaLE, were grouped together in one well-defined clade by all methods. Chlamydia pecorum sheep strains W73, P787, 757, and JP396 that clustered together by fineSTRUCTURE were regrouped into two distinct clusters at the fourth finest hierarchical level by BAPS. The patterns of admixture for those strains inferred by BAPS support this partition (see below; fig. 5 and supplementary table S5, Supplementary Material online). The C. pecorum bovine strains E58 and PV3056/3 were grouped into distinct single strain clusters by both fineSTRUCTURE and BAPS. E58 did not group with the clade formed by sheep strains VR629 and OSP in the ML phylogeny but grouped with JP396 by ClonalFrame phylogenetic analysis. Even though the phylogenetic methods and fineSTRUCTURE appeared to have three distinct clades for C. abortus with the LLG strain as an outgroup, BAPS inferred the presence of four clusters, placing the B577 strain as a distinct cluster (figs. 5 and 6; supplementary table S5, Supplementary Material online).
The new C. psittaci EP1 strain grouped with the MN strain forming a distinct clade by all three methods. In a previous analysis (Read et al. 2013), the MN strain did not form a distinct clade. The other novel C. psittaci strain, EP6, isolated from a human grouped with the 6BC/83 strain by phylogeny, fineSTRUCTURE and BAPS hierarchical analysis (supplementary table S5, Supplementary Material online), indicating that this strain is also part of the recent clonal expansion of C. psittaci in the United States. Interestingly, the C. psittaci RTH strain was assigned into a single cluster by both population structure methods.
The newly identified species C. avium and C. gallinacea grouped together forming a single clade by ML-based phylogenetic inference and by fineSTRUCTURE, indicating that they may be part of a similar population despite the ANI score (supplementary table S3, Supplementary Material online) that puts them into different species groups (fig. 6). Similarly, C. caviae and C. felis formed a single clade (fig. 6).
Signatures of Admixture Suggest a Genetic and/or Ecological Barriers between Species Groups
Based on the fineSTRUCTURE coancestry matrix visualized as a heatmap (fig. 6), there appeared to be many events of genetic exchange happening among the different subgroups within and across each of the species. The color of each cell of the matrix indicated the expected number of genetic markers imported from the donor (x axis) to a recipient genome (y axis). Chlamydia psittaci RTH strain received DNA sequences from all C. abortus and C. psittaci strains, primarily C. abortus LLG, C. psittaci FalTex, and C. psittaci M56 genomes. Admixtures of DNA exchange were previously observed by STRUCTURE analysis (Read et al. 2013) between the RTH strain and C. psittaci strains M56, FalTex, and MN. The subgroup of C. felis and C. caviae received DNA from all C. abortus and C. psittaci strains along with smaller amounts from C. pneumoniae and C. pecorum strains. This trend was also observed in the C. avium and C. gallinacea subgroup, but to a lower extent; the RTH strain was the most significant donor to that subgroup (fig. 6).
The fineSTRUCTURE algorithm assumed that all genetic exchanges occurred within the sequenced strains, whereas ClonalFrame allowed recombination events from “outside” the sample of genomes under consideration (although they do not model the source). Figure 5 shows the proportion of ancestry from all major ancestral sources in separate colors for each species. There were no signs of admixture signals in the three distinct ancestral populations of C. pneumoniae. Except for the koala-infecting C. pecorum strains, which had a 100% proportion of ancestry from a single ancestral source of C. pecorum (shown in blue), admixture signals from four other unknown/external ancestral populations were estimated (colors other than blue). For C. psittaci strains, evident admixture signals were estimated from at least two other unknown ancestral populations (gray and yellow) along with exchanges from C. abortus ancestral population (magenta). The C. psittaci RTH strain had evidence of admixture signals from three ancestral populations; 43% from C. psittaci (green), 20% from C. abortus (magenta) sources, and 37% from an unknown ancestral population (gray). Chlamydia abortus strains also showed events of admixtures from two other unknown ancestral populations, but at very low proportions. In the BAPS model, the two novel species, C. gallinacea and C. avium did not show any evidence of admixtures; both were estimated to have 100% ancestry from a single ancestral source. Similarly, C. caviae and C. felis did not have evidence of exchange from other Chlamydiaceae species (fig. 5).
We assigned the origin of each recombination event identified by ClonalFrame by postprocessing the output (see Materials and Methods) (supplementary table S6, Supplementary Material online). Figure 7 summarizes the flux of recombination among the three major Chlamydiaceae species, C. abortus, C. psittaci and C. pecorum, that showed significant amounts of recombination. We also estimated the number and origin of recombination events for the C. psittaci RTH strain (fig. 7).
Fig. 7.—
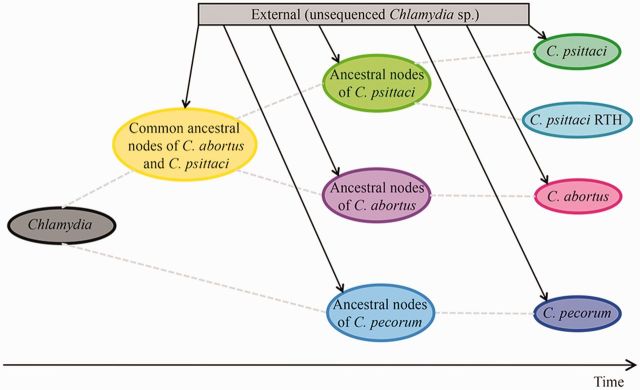
Recombination flux between the Chlamydiaceae species. Based on ClonalFrame analysis, the flux of recombination among the three major Chlamydiaceae species, C. abortus, C. psittaci and C. pecorum, is shown on an axis of time. Dashed lines indicate vertical inheritance of the Chlamydiaceae genomes. Solid lines indicate heritance from an external source.
Recombination events occurred on the ancestral nodes and are still present in each of the species. For C. pecorum, the origins for all 18 recombination events were attributed to an unknown “external” source (probably as yet undiscovered Chlamydiceae species), which corroborated with the results from BAPS. There were 21 recombination events where the origin was from an unknown external source that occurred at the common ancestral nodes of C. abortus and C. psittaci. All 21 recombination events were also detected in the C. psittaci RTH genome. There were 14 recombination events in C. psittaci whose origin of recombination was assigned to C. abortus. These data suggested that DNA exchanges were occurring between C. abortus, C. psittaci, and the RTH strains. For the plasmids, there were 18 recombination events identified. Four recombination events that occurred on the ancestral node of C. trachomatis and C. muridarum were traced back to import from the C. pneumoniae plasmid, suggesting that the event of acquisition of plasmids in C. trachomatis and C. muridarum might not be an ancient evolutionary event but a recent one, and most probably acquired from C. pneumoniae. We were not able to assign the origins of imports for the rest of the plasmid recombination events.
Discussion
One of the main aims of this work was to present a detailed analysis of the population genomics of Chlamydiaceae species isolated from avian, mammalian and human hosts and, in particular, for those in the Chlamydophila lineage. Taking into consideration that species designation for this lineage is often based on clinical symptoms (e.g., C. abortus is found in abortion) rather than molecular analysis, the fact that five of the ten novel genomes sequenced in this work were originally typed as C. abortus but were in fact either C. psittaci or C. pecorum indicates that we will remain ignorant of the true diversity of Chlamydiaceae without the use of appropriate methodology. The discovery of genetically admixed strains illustrates the importance of whole-genome as opposed to partial gene analysis. Indeed, it is likely that many of the strains not yet sequenced are mistyped.
Here, we extend our knowledge of C. abortus by showing it to be a highly monomorphic group with very similar gene content. Chlamydia abortus is the most recently emerged species and closely related to the more diverse C. psittaci, not only in terms of genetics but also host association and disease pathology. The high dN/dS ratios for C. abortus have previously been reported in other rapidly evolving pathogens such as Clostridium difficile (He et al. 2010) and could be caused by ongoing adaptation of the C. abortus population to a new pathogenic lifestyle (Shapiro et al. 2009). Furthermore, the low-level of recombination relative to other species and evidence for adaptation to the placenta in C. abortus echo evolutionary processes seen in other recently emerged, highly virulent pathogens with restricted ecological niches, such as Bacillus anthracis and Mycobacterium tuberculosis (Liu et al. 2006; Didelot and Maiden 2010; Zwick et al. 2012).
We showed that gene-flow occurred within the species and between C. psittaci and C. abortus. However, there was no evidence here of gene-flow between or among C. felis, C. caviae, C. avium, C. gallinacea, C. pecorum, and C. pneumoniae. Homologous recombination among Chlamydiaceae species has emerged as a central feature from comparative genomics studies in the past few years (Gomes et al. 2007; Jeffrey et al. 2010; Harris et al. 2012; Joseph et al. 2012; Read et al. 2013). The exact mechanism for DNA transfer between Chlamydiaceae infected cells is unknown, but laboratory studies have shown that recombination can occur after host cell coinfection (DeMars and Weinfurter 2008; Suchland et al. 2009; Srinivasan et al. 2012). There is no absolute requirement for transducing bacteriophage. However, that does not rule out a role in some exchanges since a number of bacteriophages have been identified for C. psittaci, C. abortus, C. felis, C. caviae, C. pecorum, and C. pneumoniae (Pawlikowska-Warych et al. 2015).
Not all species are permissive to interspecies recombination. Barriers to recombination have been documented for C. caviae and C. suis; C. caviae and C. trachomatis (Suchland et al. 2009); and C. muridarum and C. psittaci (Millman et al. 2001). Without additional laboratory experiments, we do not know whether the lack of recombination discovered in the present study between C. psittaci (except for the RTH strain) and C. abortus, and C. pecorum and C. pneumoniae is primarily the result of an ecological or a genetic barrier. In the latter case, the reduced efficiency of mismatch repair following homologous recombination between divergent nucleotide sequences may be a major factor (Matic et al. 1995).
With the proviso that the r/m statistic for recombination frequency has several well-known ascertainment biases (Shapiro 2014), it may still be significant that the highest rates of exchange are found in C. psittaci strains. Chlamydia psittaci has an extremely broad avian host range encompassing 460 species in 30 orders including 9 domestic species (e.g., turkey, duck, pigeon, quail, chicken, goose, peafowl) and is most commonly detected in the orders Psittaciformes, followed by Lariformes (gulls), Alciformes (alks), Sphenisciformes (penguins), and Anseriformes (ducks and geese) (Kaleta and Taday 2003). It is also noteworthy that the most highly genetically admixed C. psittaci strain, RTH, was discovered in a wild bird-of-prey. Similar to the RTH strain, other Chlamydiaceae species have been isolated from a number of other predatory birds (Kaleta and Taday 2003; Van Loock et al. 2003; Schettler et al. 2003; Blomqvist et al. 2012), some of which do not fit easily within either the C. psittaci or C. abortus species, based on limited genetic data. 16S rRNA and ompA gene analysis of four samples revealed one novel Chlamydia species as well as two novel C. psittaci strains in peregrine falcons and white-tailed sea eagles in Sweden (Blomqvist et al. 2012). Other strains that are seemingly intermediate between C. psittaci and C. abortus include the R54 strain from the subantartic brown skua (Herrmann et al. 2000), the 84/2334 strain isolated from a yellow-crowned amazon in Germany (Vanrompay et al. 1997), the parakeet strains Prk/Daruma (Fukushi and Hirai 1988) and strain VS225 (Van Loock et al. 2003) isolated in Japan, and in 1991 in Laredo, TX, respectively. It would be informative to see whether isolates from these and other birds-of-prey harbor diverse strains with evidence of recent recombination between species.
Theoretically, infection by different Chlamydiaceae species either simultaneously or sequentially would facilitate transformation and homologous recombination (Gomes et al. 2007). The red-tailed hawk (Buteo jamaicensis), from which the RTH strain was isolated, is one of the most common and widely distributed raptors in North America, living in various habitats ranging from forestland to agricultural and even urban landscapes (Fitch et al. 1946; Stout et al. 2006). This variety of nesting and hunting grounds also requires great flexibility in their diet. Red-tailed hawks prey on small mammals (squirrels, mice, and rabbits), birds, such as pigeons and pheasants (Gates 1972), as well as reptiles including snakes and lizards (Fitch et al. 1946). They also feed on carrion (Marti and Kochert 1995) and, if living near agricultural landscapes, may seize dead chickens that were spread on the fields with manure (Orians and Kuhlman 1956). Because a variety of Chlamydiaceae species infect the intestinal tract of many avian and lower mammalian species, infected feces are ubiquitous in these environments and available for ingestion along with carrion or other animals. These dietary habits are important because horizontal gene transfer of entire metabolic pathways, fitness and virulence factors, as well as antibiotic resistance genes, has been reported in the gut ecosystem, particularly among closely related bacterial species (Stecher et al. 2013). These findings lend support to our hypothesis that the RTH and other raptors serve as a nexus for Chlamydiaceae evolution.
Based on the plasmid phylogeny, each species separated into monophyletic clades (supplementary fig. S1, Supplementary Material online). Surprisingly, the plasmids of C. pneumoniae and C. pecorum were ancestral to C. trachomatis and C. muridarum, whereas these four species shared a more recent ancestor than the other Chlamydia species. The unexpected similarity of plasmid genes in the C. trachomatis and C. muridarum lineage to C. pneumoniae and C. pecorum suggest a potential ancestral transfer event. Chlamydia trachomatis and C. pneumoniae are both human pathogens that infect the respiratory tract (Webley et al. 2009), providing an opportunity for genetic exchange. Mice could potentially acquire C. pecorum with its plasmid from feces that are ubiquitous in a farmyard setting. This would create the potential opportunity for DNA/plasmid exchange with C. muridarum in the intestinal tract.
This work also raises issues in terms of how we define Chlamydia species given the Chlamydiaceae population structure. Chlamydia avium and C. gallinacea have recently been adopted as new species based on ANI scores of at most 94% with the closest other genome (Sachse et al. 2014). The cutoff for strain assignment to the same species has been suggested to be approximately 95–96% ANI (Richter and Rosselló-Móra 2009). However, population structure results place them in the same genetic group as another existing species (figs. 5 and 6). Chlamydia avium and C. gallinacea (and C. caviae/C. felis) could be divergent clades of genetic congruent species (fig. 2) or have undergone sympatric speciation with ongoing frequent genetic exchanges.
Finally, the C. psittaci RTH strain has an ANI relationship that suggests that it is a separate species from both C. psittaci and C. abortus and may be a representative of a new Chlamydia species altogether. Although there is evidence of DNA exchange, the RTH strain still has unique population markers and possibly a unique ecology that would qualify it to be treated as a distinct species. The molecular clock data would support this contention as the RTH strain is ancestral to C. abortus. As with many other stories concerning Chlamydiaceae, the answers are likely to come from further genomic sequencing and bioinformatics efforts.
Conclusion
The family Chlamydiaceae are bacteria that cause mild to severe diseases in humans, livestock and wild animals, such as koalas, frogs, and birds-of-prey. The estimated economic burden of these diseases is over $10 billion annually. Given their zoonotic potential for ocular, respiratory, and systemic diseases as well as abortion, we examined the evolutionary dynamics of Chlamydia species using genomes from 36 isolates of diverse origins. Analyses revealed that C. abortus is the most recently emerged species with significantly lower recombination compared with all other species and lacks the virulence-associated plasmid. Through gene-flow analysis, the RTH isolate was found to have extensive genomic admixture from chlamydial and external sources, and may be a member of a novel intermediary species linking C. psittaci to C. abortus evolution. Predators such as birds-of-prey may serve as incubator hosts that acquire chlamydiae and other pathogens in their guts resulting in species emergence by recombination.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Art Andersen for providing his collection of Chlamydiaceae species and strains to the Dean Lab without which this research would not have been possible. They thank Manu Sharma for excellent technical assistance. This work was supported in part by the Public Health Service grant from the National Institute of Health (R01 AI098843 to D.D. and T.D.R.); and the National Science Foundation (NIH) (2009-65109-05760 to D.D.) and an Early Postdoctoral Mobility Fellowship grant from the Swiss National Science Foundation (SNSF) (P2ZHP3_158590 to H.M.).
Literature Cited
- Azuma Y, et al. 2006. Genome sequence of the cat pathogen, Chlamydophila felis. DNA Res. 13:15–23. [DOI] [PubMed] [Google Scholar]
- Bachmann NL, et al. 2015. Culture-independent genome sequencing of clinical samples reveals an unexpected heterogeneity of infections by Chlamydia pecorum. J Clin Microbiol. 53:1573–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann NL, Fraser TA, et al. 2014. Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genomics 15:667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann NL, Polkinghorne A, Timms P. 2014. Chlamydia genomics: providing novel insights into chlamydial biology. Trends Microbiol. 22:464–472. [DOI] [PubMed] [Google Scholar]
- Blomqvist M, et al. 2012. Chlamydia psittaci in birds of prey, Sweden. Infect Ecol Epidemiol. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodetti TJ, et al. 2002. Molecular evidence to support the expansion of the host range of Chlamydophila pneumoniae to include reptiles as well as humans, horses, koalas and amphibians. Syst Appl Microbiol. 25:146–152. [DOI] [PubMed] [Google Scholar]
- Castillo-Ramírez S, et al. 2012. Phylogeographic variation in recombination rates within a global clone of methicillin-resistant Staphylococcus aureus .Genome Biol. 13:R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cheng L, Connor TR, Sirén J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol. 30:1224–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingro A, et al. 2011. Unity in variety—the pan-genome of the Chlamydiae. Mol Biol Evol. 28:3253–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J, Marttinen P. 2006. Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol. 15:2833–2843. [DOI] [PubMed] [Google Scholar]
- Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14:1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean D, Rothschild J, Ruettger A, Kandel RP, Sachse K. 2013. Zoonotic Chlamydiaceae species associated with trachoma, Nepal. Emerg Infect Dis. 19:1948–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMars R, Weinfurter J. 2008. Interstrain gene transfer in Chlamydia trachomatis in vitro: mechanism and significance. J Bacteriol. 190:1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, et al. 2011. Recombination and population structure in Salmonella enterica. PLoS Genet. 7:e1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Lawson D, Darling A, Falush D. 2010. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics 186:1435–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Maiden MCJ. 2010. Impact of recombination on bacterial evolution. Trends Microbiol. 18:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati M, et al. 2014. Genome sequence of Chlamydia suis MD56, isolated from the conjunctiva of a weaned piglet. Genome Announc. 2:e00425–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Nicholls GK, Rodrigo AG, Solomon W. 2002. Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data. Genetics 161:1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett KD, Bush RM, Andersen AA. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 49(Pt 2):415–440. [DOI] [PubMed] [Google Scholar]
- Fitch HS, Swenson F, Tillotson DF. 1946. Behavior and food habits of the red-tailed hawk. Condor 48:205–237. [Google Scholar]
- Fukushi H, Hirai K. 1988. Immunochemical diversity of the major outer membrane protein of avian and mammalian Chlamydia psittaci. J Clin Microbiol. 26:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi H, Hirai K. 1992. Proposal of Chlamydia pecorum sp. nov. for Chlamydia strains derived from ruminants. Int J Syst Bacteriol. 42:306–308. [DOI] [PubMed] [Google Scholar]
- Gates JM. 1972. Red-tailed hawk populations and ecology in east-central Wisconsin. Wilson Bull. 84:421–433. [Google Scholar]
- Gomes JP, et al. 2007. Evolution of Chlamydia trachomatis diversity occurs by widespread interstrain recombination involving hotspots. Genome Res. 17:50–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greub G. 2010. International Committee on Systematics of Prokaryotes Subcommittee on the taxonomy of the Chlamydiae Minutes of the inaugural closed meeting, 21 March 2009, Little Rock, AR, USA. Int J Syst Evol Microbiol. 60:2691–2693. [DOI] [PubMed] [Google Scholar]
- Harkinezhad T, et al. 2009. Prevalence of Chlamydophila psittaci infections in a human population in contact with domestic and companion birds. J Med Microbiol. 58:1207–1212. [DOI] [PubMed] [Google Scholar]
- Harris SR, et al. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 44:413–419, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, et al. 2010. Evolutionary dynamics of Clostridium difficile over short and long time scales. Proc Natl Acad Sci U S A. 107:7527–7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann B, Rahman R, Bergström S, Bonnedahl J, Olsen B. 2000. Chlamydophila abortus in a Brown skua (Catharacta antarctica lonnbergi) from a subantarctic island. Appl Environ Microbiol. 66:3654–3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. 2010. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 6:e1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, et al. 2010. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun. 78:2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Didelot X, Gandhi K, Dean D, Read TD. 2011. Interplay of recombination and selection in the genomes of Chlamydia trachomatis. Biol Direct. 6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, et al. 2012. Population genomics of Chlamydia trachomatis: insights on drift, selection, recombination, and population structure. Mol Biol Evol. 29:3933–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Read TD. 2012. Genome-wide recombination in Chlamydia trachomatis. Nat Genet. 44:364–366. [DOI] [PubMed] [Google Scholar]
- Kaleta EF, Taday EMA. 2003. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 32:435–461. [DOI] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gutacker MM, Musser JM, Fu Y-X. 2006. Evidence for recombination in Mycobacterium tuberculosis. J Bacteriol. 188:8169–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longbottom D, Coulter LJ. 2003. Animal chlamydioses and zoonotic implications. J Comp Pathol. 128:217–244. [DOI] [PubMed] [Google Scholar]
- Marti CD, Kochert MN. 1995. Are red-tailed hawks and great horned owls diurnal-nocturnal dietary counterparts? Wilson Bull.. 107:615–628. [Google Scholar]
- Matic I, Rayssiguier C, Radman M. 1995. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell 80:507–515. [DOI] [PubMed] [Google Scholar]
- Millman KL, Tavare S, Dean D. 2001. Recombination in the ompA Gene but not the omcB gene of Chlamydia contributes to serovar-specific differences in tissue tropism, immune surveillance, and persistence of the organism. J Bacteriol. 183:5997–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamad KY, Rodolakis A. 2010. Recent advances in the understanding of Chlamydophila pecorum infections, sixteen years after it was named as the fourth species of the Chlamydiaceae family. Vet Res. 41:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers GSA, et al. 2009. Evidence that human Chlamydia pneumoniae was zoonotically acquired. J Bacteriol. 191:7225–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orians G, Kuhlman F. 1956. Red-tailed hawk and horned owl populations in Wisconsin. Condor 58:371–385. [Google Scholar]
- Pawlikowska-Warych M, Śliwa-Dominiak J, Deptuła W. 2015. Chlamydial plasmids and bacteriophages. Acta Biochim Pol. 62:1–6. [DOI] [PubMed] [Google Scholar]
- Read TD, et al. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, et al. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read TD, et al. 2013. Comparative analysis of Chlamydia psittaci genomes reveals the recent emergence of a pathogenic lineage with a broad host range. MBio 4:e00604–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhold P, Sachse K, Kaltenboeck B. 2011. Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens? Vet J.. 189:257–267. [DOI] [PubMed] [Google Scholar]
- Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 106:19126–19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, et al. 2006. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 239:226–235. [DOI] [PubMed] [Google Scholar]
- Rockey DD, Heinzen RA, Hackstadt T. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol. 15:617–626. [DOI] [PubMed] [Google Scholar]
- Roulis E, Polkinghorne A, Timms P. 2013. Chlamydia pneumoniae: modern insights into an ancient pathogen. Trends Microbiol. 21:120–128. [DOI] [PubMed] [Google Scholar]
- Sachse K, et al. 2014. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst Appl Microbiol. 37:79–88. [DOI] [PubMed] [Google Scholar]
- Sachse K, et al. 2015. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst Appl Microbiol. 38:99–103. [DOI] [PubMed] [Google Scholar]
- Sait M, et al. 2014. Genome sequencing and comparative analysis of three Chlamydia pecorum strains associated with different pathogenic outcomes. BMC Genomics 15:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schautteet K, Vanrompay D. 2011. Chlamydiaceae infections in pig. Vet Res. 42:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schettler E, et al. 2003. Newcastle disease virus and Chlamydia psittaci in free-living raptors from eastern Germany. J Wildl Dis. 39:57–63. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- Shao R, Hu J, Billig H. 2013. Toward understanding Chlamydia infection-induced infertility caused by dysfunctional oviducts. J Infect Dis. 208:707–709. [DOI] [PubMed] [Google Scholar]
- Shapiro BJ. 2014. Signatures of natural selection and ecological differentiation in microbial genomes. Adv Exp Med Biol. 781:339–359. [DOI] [PubMed] [Google Scholar]
- Shapiro BJ, David LA, Friedman J, Alm EJ. 2009. Looking for Darwin’s footprints in the microbial world. Trends Microbiol. 17:196–204. [DOI] [PubMed] [Google Scholar]
- Snipen L, Liland KH. 2015. micropanmicropan: an R-package for microbial pan-genomics. BMC Bioinformatics 16:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somboonna N, et al. 2011. Hypervirulent Chlamydia trachomatis clinical strain is a recombinant between lymphogranuloma venereum (L(2)) and D lineages. MBio 2:e00045–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan T, et al. 2012. In vitro recombinants of antibiotic-resistant Chlamydia trachomatis strains have statistically more breakpoints than clinical recombinants for the same sequenced loci and exhibit selection at unexpected loci. J Bacteriol. 194:617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A, et al. 2012. RAxML-Light: a tool for computing terabyte phylogenies. Bioinformatics 28:2064–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Maier L, Hardt W-D. 2013. “Blooming” in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 11:277–284. [DOI] [PubMed] [Google Scholar]
- Stout WE, Temple SA, Cary JR. 2006. Landscape features of red-tailed hawk nesting habitat in an urban/suburban environment. J Raptor Res. 40:181–192. [Google Scholar]
- Suchland RJ, Sandoz KM, Jeffrey BM, Stamm WE, Rockey DD. 2009. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother. 53:4604–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34:W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Hanage WP, Fraser C, Corander J. 2009. Identifying currents in the gene pool for bacterial populations using an integrative approach. PLoS Comput Biol. 5:e1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson NR, et al. 2005. The Chlamydophila abortus genome sequence reveals an array of variable proteins that contribute to interspecies variation. Genome Res. 15:629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loock M, et al. 2003. Missing links in the divergence of Chlamydophila abortus from Chlamydophila psittaci. Int J Syst Evol Microbiol. 53:761–770. [DOI] [PubMed] [Google Scholar]
- Vanrompay D, Butaye P, Sayada C, Ducatelle R, Haesebrouck F. 1997. Characterization of avian Chlamydia psittaci strains using omp1 restriction mapping and serovar-specific monoclonal antibodies. Res Microbiol. 148:327–333. [DOI] [PubMed] [Google Scholar]
- Voigt A, Schöfl G, Saluz HP. 2012. The Chlamydia psittaci genome: a comparative analysis of intracellular pathogens. PLoS One 7:e35097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webley WC, et al. 2009. Occurrence of Chlamydia trachomatis and Chlamydia pneumoniae in paediatric respiratory infections. Eur Respir J. 33:360–367. [DOI] [PubMed] [Google Scholar]
- Yahara K, et al. 2013. Chromosome painting in silico in a bacterial species reveals fine population structure. Mol Biol Evol. 30:1454–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick ME, et al. 2012. Genomic characterization of the Bacillus cereus sensu lato species: backdrop to the evolution of Bacillus anthracis. Genome Res. 22:1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.