Abstract
In mammals, the cytosine in CG dinucleotides is typically methylated producing 5-methylcytosine (5mC), a chemically less stable form of cytosine that can spontaneously deaminate to thymidine resulting in a T•G mismatched base pair. Unlike other eukaryotes that efficiently repair this mismatched base pair back to C•G, in mammals, 5mCG deamination is mutagenic, sometimes producing TG dinucleotides, explaining the depletion of CG dinucleotides in mammalian genomes. It was suggested that new TG dinucleotides generate genetic diversity that may be critical for evolutionary change. We tested this conjecture by examining the DNA sequence properties of regulatory sequences identified by DNase I hypersensitive sites (DHSs) in human and mouse genomes. We hypothesized that the new TG dinucleotides generate transcription factor binding sites (TFBS) that become tissue-specific DHSs (TS-DHSs). We find that 8-mers containing the CG dinucleotide are enriched in DHSs in both species. However, 8-mers containing a TG and no CG dinucleotide are preferentially enriched in TS-DHSs when compared with 8-mers with neither a TG nor a CG dinucleotide. The most enriched 8-mer with a TG and no CG dinucleotide in tissue-specific regulatory regions in both genomes is the AP-1 motif (TGAC/GTCAN), and we find evidence that TG dinucleotides in the AP-1 motif arose from CG dinucleotides. Additional TS-DHS-enriched TFBS containing the TG/CA dinucleotide are the E-Box motif (GCAGCTGC), the NF-1 motif (GGCA—TGCC), and the GR (glucocorticoid receptor) motif (G-ACA—TGT-C). Our results support the suggestion that cytosine methylation is mutagenic in tetrapods producing TG dinucleotides that create TFBS that drive evolution.
Keywords: tissue specific, TFBS, AP-1, CG methylation, coelacanth, TG dinucleotide
Introduction
The DNA sequence of the genome is a key to the biological form that emerges. We have previously used the abundance of all 65,536 8-mers as a method to describe genomes (Vinson et al. 2011). The abundance of 8-mers in the human genome has a bimodal distribution with all rare 8-mers containing a CG dinucleotide. In contrast, Drosophila has a unimodal distribution of 8-mers (Vinson et al. 2011). In mammalian genomes, the majority of the 20 million cytosines that occur in CG dinucleotides are methylated (5-methylcytosine [5mC]) (Bird 1980; Gardiner-Garden and Frommer 1987). Two to three percent of the genome comprises unmethylated regions (Lister et al. 2009; Chatterjee et al. 2014), which tend to be comprised clusters of CG dinucleotides termed CG islands (CGIs) (Gardiner-Garden and Frommer 1987). 5mC deaminates to thymidine 10–50 times faster than unmethylated cytosine deaminates to uracil (Coulondre et al. 1978; Chen et al. 2014), to create a T•G mismatched base pair. Error-free base excision repair can correct the naturally occurring T•G mismatch to the original C•G base pair and thus CG methylation is not mutagenic (Huff and Zilberman 2014). Although T•G to C•G repair pathways exist in mammals, they are inefficient (Walsh and Xu 2006; Sjolund et al. 2013), resulting in T•G base pairs mutating to T•A base pairs, thus creating a TG dinucleotide. Deamination of 5mC is thought to cause the 4-fold depletion of the CG dinucleotide compared with all other dinucleotides in mammalian genomes (Bird et al. 1995). As expected, the deamination product of 5mCG, TG, is the most abundant dinucleotide in vertebrates, but not other phyla (Gentles and Karlin 2001; Simmen 2008). Both cytosines in the CG dinucleotide are typically methylated and deamination to TG on one strand will give a complementary CA dinucleotide on the other strand. For convenience, we will refer to the TG/CA dinucleotide simply as the TG dinucleotide, knowing that on the second strand it is a CA dinucleotide. Thus, the genome is segregated into islands of genetically stable unmethylated CG dinucleotides surrounded by long stretches of genetically unstable methylated CG dinucleotides (Vinson and Chatterjee 2012).
CG methylation has been implicated in many biological processes, including cell type specificity (Rougier et al. 1998; Takizawa et al. 2001), cellular differentiation (Reik et al. 2001; Laurent et al. 2010), suppression of transposable elements (Akers et al. 2014), X-chromosome inactivation (Bird 2002), genomic imprinting (Reik and Walter 2001), DNA–protein interactions (Rishi et al. 2010), and tumorigenesis (Baylin and Jones 2011; Rodriguez-Paredes and Esteller 2011; Vinson and Chatterjee 2012). A potential additional function of cytosine methylation is to increase the mutation rate (Bird 1980), thereby generating genetic diversity (Leffler et al. 2012). The evolutionary advantage of an increased mutation rate (Giraud et al. 2001) could drive selection for cytosine methylation at nonfunctioning regions of the genome. These mutagenic regions could produce new transcription factor binding sites (TFBS) that become biologically functional. Though Adrian Bird conjectured that 5mC could function as a mutator almost 35 years ago (Bird 1980), at that time it could not be tested at a genomic level. With the advent of genome-wide identification of functional regions of the human and mouse genomes (Stergachis et al. 2014; Vierstra et al. 2014; Yue et al. 2014), we find that TG dinucleotide containing TFBS are enriched in tissue-specific regulatory regions, lending support to the suggestion that one function of cytosine methylation is to be mutagenic, increasing genetic diversity and accelerating evolutionary change.
Materials and Methods
Data Sets
All the DNase I hypersensitive site (DHS) data including human (75 samples, and 125 tissue and cell lines) and mouse (55 samples) DHSs are from the ENCODE Project Consortium (2012) and were obtained from University of California Santa Cruz (UCSC) Genome Bioinformatics website (http://genome.ucsc.edu/) (Rosenbloom et al. 2015). The five DHS groups from human and mouse based on their conservation between species were obtained from Vierstra et al. (2014). The five DHS groups were identified by comparison between human and mouse genomes. Briefly, chain files of pairwise genome alignments between mouse (mm9) and human (hg19) were downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/). Using these chain files, DHSs were mapped between species, and then the DHS groups were identified according to genomic alignment status. These groups included one group of common DHSs shared between human and mouse; two groups of species-specific DHS (mouse-specific or human-specific), which comprise the DHSs that occur only in one species but are conserved in sequence; and two groups of species-unique DHSs (mouse-unique or human-unique), which comprise the DHSs that occur only in one species and are not conserved in the other. The conserved noncoding elements (CNEs) arising in various taxa were obtained from Amemiya et al. (2013).
The sequence assemblies of 25 eukaryotic genomes are as follows: Human (hg19), mouse (mm9), dog (canFam3), elephant (loxAfr3), opossum (momDom5), wallaby (macEug2), minke whale (balAcu1), dolphin (turTru2), chicken (galGal4), lizard (anoCar2), Xenopus tropicalis (xenTro3), coelacanth (latCha1), elephant shark (calMil1), zebrafish (danRer7), Nile tilapia (oreNil2), cod (gadMor1), stickleback (gasAcu1), fugu (fr3), tetrodon (tetNig2), sea urchin (strPur2), Drosophila melanogaster (dm3), honey bee (apiMel3), Caenorhabditis elegans (ce10), Arabidopsis (Tair10), and yeast (sacCer3). The Arabidopsis (Tair10) genome was downloaded from The Arabidopsis Information Resource website (http://www.arabidopsis.org/) and all the other 24 genomes were downloaded from UCSC Genome Bioinformatics website (http://genome.ucsc.edu/) (Rosenbloom et al. 2015).
8-mer Counts and Enrichment Calculations
There are 65,536 (48) possible octameric sequences (8-mers), and of these 256 are palindromic. We added complementary sequences and present the number of occurrences for all 32,640 nonpalindromic 8-mers. For the 256 palindromic 8-mers, each occurrence was counted twice, once for each strand. Thus, the number of possible comparisons can be reduced from 65,536 to 32,896 (32,640 nonpalindromic + 256 palindromic sequences) when both strands of the target sequence are examined. All 32,896 8-mers used in this analysis were automatically generated by a custom-made program.
To determine the enrichment of each 8-mer (continuous (NNNNNNNN), split (NNNNX(1-30)NNNN), glucocorticoid receptor (GR)-like (N-NNN—NNN-N)) in the DHSs, we calculated an enrichment score for each 8-mer (Chatterjee et al. 2014). To avoid sampling bias, we searched for each 8-mer across the whole genome. For each 8-mer M with the length L (for continuous 8-mers such as C/EBP motif (TTGCGCAA), L = 8; for split 8-mers such as GR (G-ACA—TGT-C), L = 13), we denote M (xstart:xend) to record the positions where the motif starts and ends: x1:x1 + L − 1, x2: x2 + L − 1 . . . xN: xN + L − 1, N being the total number of motifs in genome. For each position xi: xi + L − 1, if it overlapped with the examined regions (DHSs), xi = 1, otherwise xi = 0.
For all the DHSs, the observed (OCCobs) and expected (OCCexp) occurrences of the 8-mer are calculated as: and , where N is the total number of that 8-mer in the genome, Lr is the total length of base pairs in the examined regions (DHSs), and Lg is the length of the genome. The enrichment score (E) for 8-mer M is then calculated as follows: , where OCCobs is the observed occurrences, and OCCexp is the expected occurrence of 8-mer M in the examined regions (DHSs).
Calculation of AP-1 and E-Box Motif Enrichment in Human DHSs
We performed the same strategy described above to calculate the AP-1 and E-Box motif enrichment in human DHSs from 125 cells. Briefly, for AP-1 motif (A/GTGAC/GTCA), we first searched all the AP-1 motifs in human genome (hg19). For each AP-1 MAp-1 with length 8, we denote MAp-1 (xstart:xend) to record the positions where the motif starts and ends: x1:x1 + 7, x2: x2 + 7 . . . xN: xN + 7, N being the total number of AP-1 motifs in genome. For each position xi: xi + 7, we assigned a score of 1 (xi = 1) if it overlapped with a DHS in examined regions (DHSs), otherwise xi = 0. For all DHSs in a given cell type/sample, the observed (OCCobs) and expected (OCCexp) occurrences of AP-1 motifs are calculated as: and , where N is the total number of AP-1 motifs in the genome, Lr is the total length of base pairs in the DHSs, and Lg is the length of the human genome. The enrichment score (E) for MAp-1 is then calculated as follows: , where OCCobs is the observed occurrences, and OCCexp is the expected occurrence of MAp-1 in the DHSs. We calculated the enrichments of E-Box motif (GCAGCTGC) in the same way.
Evolutionary Analysis of AP-1 Motif
To determine the evolutionary history of the AP-1 motif in the human genome, a custom-made program was used to scan the human genome sequence to identify all the AP-1 motifs (TGAC/GTCA). We extended the motif to the 11-mer with the canonical AP-1 motif in the center (NNTGAC/GTCANN). All the pairwise alignments for all other nine species using human as reference genome (human hg19, mouse mm9, dog canFam3, elephant loxAfr3, opossum monDom5, chicken galGal3, lizard anoCar2, Xenopus tropicalis xenTro3, coelacanth latCha1, and stickleback gasAcu1) were downloaded from UCSC Genome Browser (http://genome.ucsc.edu/). Using these genomic alignments, we searched for the most recent common ancestor (MRCA) sequences of homologous instances 11-mers of AP-1 motif in all the other nine genomes, and extracted these sequences in each species. To remove the noise of false alignments, we compared every homologous sequence to canonical AP-1 motifs in human (TGAC/GTCA), and removed sequences with deletions, insertions, or with more than two variations. Thus, only homologous sequences with no more than two variations in each species were used for further analysis.
Cloning and Expression of Mouse AP-1 Members
The DNA binding domain (DBD) of Mouse c-JUN as defined in Pfam cloned into pETGEXCT (C-terminal GST) vector (Sharrocks 1994; Mann et al. 2013). The DBD of c-FOS was cloned into NdeI and HindIII sites in pT5 expression plasmid.
Double Stranding and Methylation of Microarray
The single-stranded oligonucleotide microarrays were double-stranded by primer extension as described by Badis et al. (2009). The primer extension reaction consisted of 1.17 µM HPLC (High-Performance Liquid Chromatography) purified common HK primer (5′-GAGCGGATAACAATTTCACACAGG-3′), 40 µM of dATP, dTTP, dCTP, and dGTP (GE Healthcare), 1.6 µM of Cy3 dCTP (GE Healthcare), 40 Units of Thermo Sequenase DNA polymerase, and 90 µl of 10 × reaction buffer in a total volume of 900 µl. The reaction mixture, microarray, stainless steel hybridization chamber, and single chamber gasket coverslip (Agilent) were assembled according to the manufacturer’s instructions and incubated for 2 h (85 °C for 10 min, 75 °C for 10 min, 65 °C for 10 min, and 60 °C for 90 min). The hybridization chamber was disassembled in a glass staining dish in 500 ml phosphate-buffered saline (PBS)/0.01% Triton X-100 at 37 °C. The microarray was transferred to a fresh staining dish, washed for 10 min in PBS/0.01% Triton X-100 at 37 °C, washed once more for 3 min in PBS at 20 °C, and the arrays dried by dipping in 500 ml PBS and slowly removing the array. The double-stranded arrays were scanned at 570 nm to quantify the amount of incorporated Cy3-conjugated dCTP. Methylation of the double-stranded arrays was performed with 10 μl of CG methyltransferase enzyme M.SssI (20 units/μl) (NEB), 1μl of S-adenosylmethionine, and 15 μl of 10 × NEB buffer 2. Reaction volume was adjusted to 150 μl with water containing 0.005% Triton X-100 and incubated at 37 °C for 3 h. The arrays were stripped for 3 h with Protease (Sigma) in 10% sodium dodecyl sulfate and 10 mM ethylenediaminetetraacetic acid, followed by washing once in 0.5% Tween, once with 0.01% Triton X-100 and once with 1 × PBS.
Protein-Binding Reactions
The protein-binding reactions were carried out as described by Badis et al. (2009). Briefly, the double-stranded microarrays were blocked with 4% nonfat dried milk (Sigma) for 1 h. Microarrays were then washed once with PBS with 0.1% (v/v) Tween-20 for 5 min and once with PBS with 0.01% Triton X-100 for 2 min. Twenty-five microliters of IVT (In Vitro Transcription and Translation) reactions were added to make a total volume of 150 μl protein-binding reaction containing PBS with 2% (w/v) milk, 51.3 ng/μl salmon testes DNA (Sigma), and 0.2 μg/μl bovine serum albumin (NEB), and incubated for 1 h at 20 °C. Preincubated protein-binding mixtures were applied to individual chambers of HK arrays and incubated for 1 h at 20 °C. Microarrays were in a Coplin jar once with 0.5% (v/v) Tween-20 in PBS for 3 min, once with 0.01% Triton X-100 in PBS for 2 min, and then finally washed with PBS for 1 min. Alexa Fluor 647-conjugated GST antibody (Invitrogen) was applied to each chamber and incubated for 1 h at 20 °C. Finally, microarrays were washed thrice with PBS with 0.05% (v/v) Tween-20 for 3 min each, and once in PBS for 2 min. Every protein in this study was assayed in duplicate, once on each of our two separate microarray designs described above.
Image Quantification and Analysis of Microarray Data
Protein-bound microarrays were scanned to detect Alexa Fluor 647-conjugated anti-GST at 640 nm (Red Channel). Microarray images were analyzed using ImaGene (BioDiscovery Inc.), bad spots were manually flagged, and the extracted data were used for further analysis. The Z-score was calculated to estimate the relative binding affinities of proteins to each 8-mer/7-mer as previously described by Mann et al. (2013).
Data Access
Protein-binding microarray data used in this study are available at the NIH public ftp site (ftp://helix.nih.gov/pcf/chuck/Array/). The data are also in the process of submission to the NCBI (National Center for Biotechnology Information) Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo).
Results
The Coelacanth Genome Has a Bimodal 8-mer Distribution
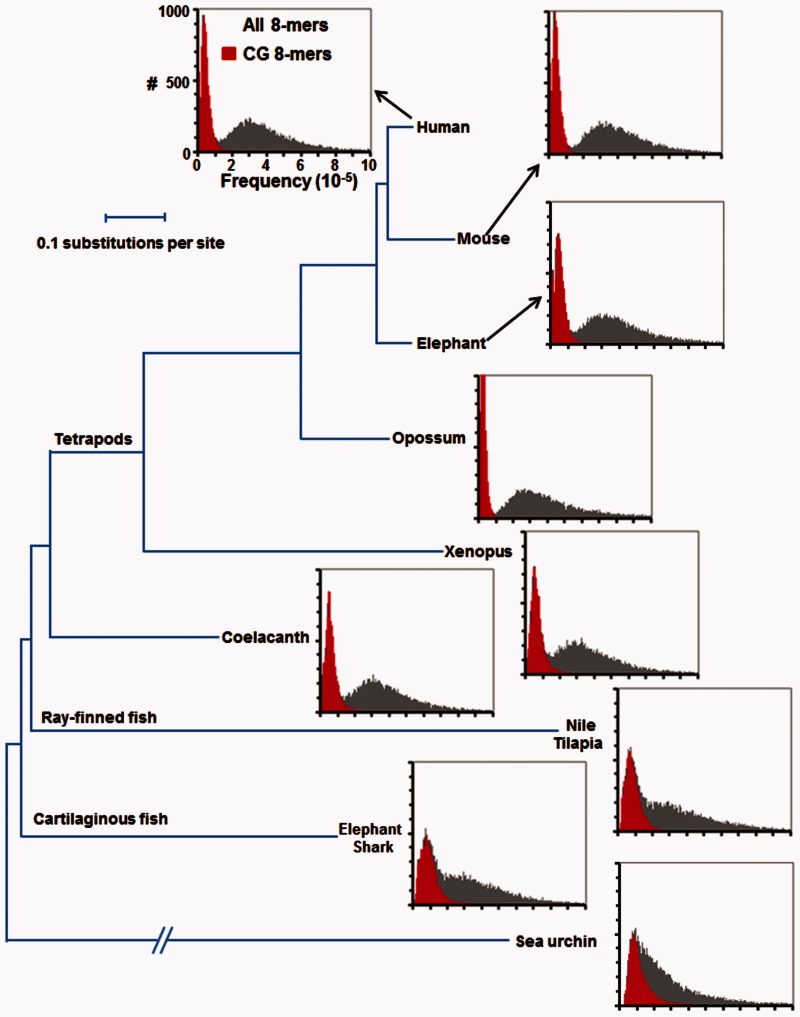
The distribution of 8-mer abundance in the genome is different between Drosophila and humans. In Drosophila, the distribution is unimodal, and in humans it is bimodal (Vinson et al. 2011). We extended our previous analyses to examine when in metazoan evolution a bimodal distribution of 8-mers arises. We examined 8-mers for two reasons: 1) This is the size of many metazoan TFBS (Weirauch et al. 2014), and 2) each 8-mer is abundant enough in the genome to produce strong statistical conclusions. In Drosophila, 8-mers containing CG dinucleotides are slightly less abundant than others potentially reflecting that cytosine is more mutagenic than the other bases (Hwang and Green 2004). A similar trend is observed in the model organisms C. elegans and yeast (supplementary fig. S1, Supplementary Material online). Among the deuterostomes, the sea urchin also has a modest decrease in CGs (fig. 1). The teleost (bony fish), which have 5mC and CGIs (Han and Zhao 2008), do not have a bimodal distribution of 8-mers suggesting that CG methylation in these genomes is not mutagenic and the T•G mismatches are repaired to C•G base pair. The bimodal distribution of 8-mers is initially observed in the coelacanth (Amemiya et al. 2013) which also contains 5mC (Makapedua et al. 2011) and has fewer CGs than expected (Iwasaki et al. 2014) suggesting that at this time the efficient repair of T•G mismatch to a C•G base pair was lost (fig. 1 and supplementary fig. S1, Supplementary Material online). All descendants of the coelacanth, the lineage that crawled out of the water onto the land, also have a bimodal distribution of 8-mer abundance, with all and only rare 8-mers containing a CG dinucleotide. The two marsupial genomes have the most dramatic CG dinucleotide depletion. A closer examination of the abundance of human 8-mers identifies a trimodal distribution, 8-mers with two or more CG dinucleotides are rarer (∼5,000 occurrences) compared with 8-mers with a CG dinucleotide (∼15,000 occurrences). Non-CG 8-mers with a TG dinucleotide have a higher median occurrence (109,000) compared with 8-mers with no TG or CG (96,700) consistent with the conversion of 5mCG to TG (supplementary table S1, Supplementary Material online).
Fig. 1.—
8-mer distribution in deuterostome genomes. Histogram showing the abundance of 32,896 continuous 8-mers in nine deuterostome genomes: Human, mouse, elephant, opossum, Xenopus, coelacanth, Nile tilapia, elephant shark, and sea urchin. The x axis displays the normalized frequency of each 8-mer in the genome (8-mer occurrence per 100 kb), and the y axis indicates the number of 8-mers with that frequency in the genome. 8-mers containing a CG dinucleotide are in red. The phylogenetic tree of nine deuterostome genomes is modified from (Amemiya et al. 2013) and is based on multiple sequence alignments of 251 genes.
Sequence Properties of CNEs
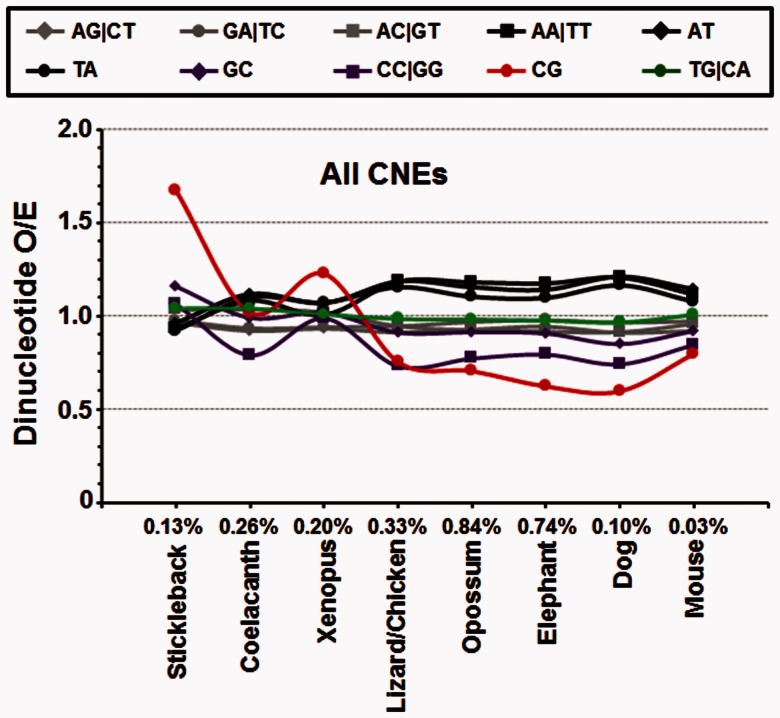
We next examined whether 8-mers containing a TG dinucleotide are enriched in putative regulatory regions that evolved throughout the tetrapod lineage. We obtained CNEs based on sequence alignments of nine genomes (stickleback, coelacanth, Xenopus, lizard/chicken, opossum, elephant, dog, mouse, and human), producing eight groups of CNEs (Amemiya et al. 2013) representing 2.63% of the human genome (fig. 2 and supplementary table S2, Supplementary Material online). We divided all 8-mers into three groups: 1) 8-mers with at least one CG dinucleotide (+CG ± TG), 2) 8-mers with at least one TG and no CG dinucleotides (+TG − CG), and 3) 8-mers with neither a TG nor a CG dinucleotide (−CG − TG), which serves as a negative control as any observed signal cannot be attributed to the conversion of 5mCG to TG. Although a small subset of all three groups of 8-mers are enriched in all CNEs (supplementary fig. S2, Supplementary Material online), +CG ± TG containing 8-mers are enriched in older CNEs arising in the stickleback, coelacanth, and Xenopus (supplementary fig. S3, Supplementary Material online). Newer CNEs show only modest enrichment for 8-mers in all three groups.
Fig. 2.—
Sequence properties of CNEs. Dinucleotide enrichment in eight groups of CNEs from stickleback to mouse. The specific CNEs for each species are determined by comparing the genome sequences between the human and other species. The percentage value for each species indicates the percentage of specific CNEs to the same species in human genome.
We next focused our attention to general properties (dinucleotide enrichment) across all groups of CNEs. The more ancient CNEs, conserved back to the stickleback, are enriched 1.7-fold for the CG dinucleotide (fig. 2). CNEs conserved since Xenopus also show enrichment for CG dinucleotides. More recent CNEs from the lizard to the present are all depleted for the CG dinucleotide. These results indicate that CG dinucleotides are enriched in ancient CNEs and depleted in more recent CNEs perhaps reflecting the emergence of CGIs in the older lineages. In contrast, TG dinucleotides are neither enriched nor depleted in these eight groups of CNEs, indicating that the new TG dinucleotides by themselves have not gained regulatory function.
Abundance of 8-mers in Tissue-Specific DHSs and Housekeeping DHSs
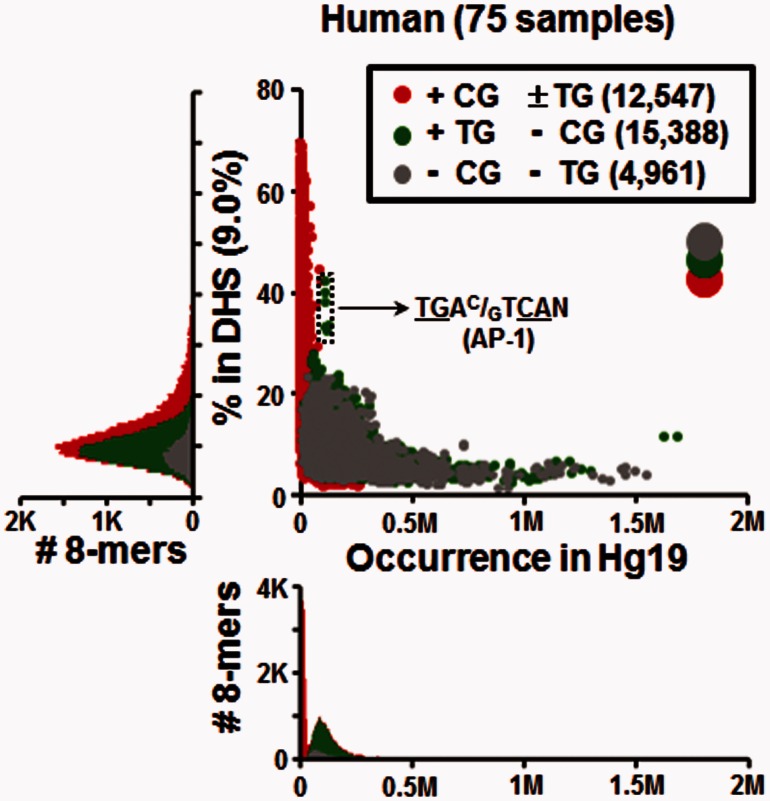
We next determined the DNA sequence properties of experimentally identified DHSs (Vierstra et al. 2014) which would represent both highly conserved and newer regulatory regions that would not be in CNEs. We examined 8-mer enrichment in DHSs found in human (9.0% of genome) and mouse (10.2% of genome) genomes (fig. 3, supplementary figs. S4 and Supplementary Data, Supplementary Material online). Figure 3 presents the abundance of 8-mers in the genome versus their occurrence in all human DHSs. Some 8-mers containing + CG ± TG dinucleotides are highly enriched in DHSs, with up to 70% of occurrences in the genome occurring within DHSs. Among the most enriched + CG ± TG 8-mers are those that contain two or three CG dinucleotides (supplementary fig. S6A and Supplementary Data, Supplementary Material online) (Chatterjee et al. 2012). +TG − CG 8-mers are more abundant than −CG −TG 8-mers across the genome, but overall, both groups show similar abundance within DHSs, except for a subset of +TG –CG 8-mers being highly enriched in DHSs, with approximately 40% of their genome occurrences found within a DHS (fig. 3 and supplementary fig. S4, Supplementary Material online). These 8-mers are all variants of the AP-1 motif, TGAC/GTCAN, a pseudopalindrome with two TG/CA dinucleotides (Lee et al. 1987).
Fig. 3.—
Abundance of 8-mers in the human genome versus their occurrence in DHSs. For each continuous 8-mer, the occurrence in human genome is plotted on the horizontal axis versus the percentage in the regulatory regions represented 9.0% of the genome (DHSs) plotted on the vertical-axis. The histogram below the scatterplot shows the distribution of abundance of all 8-mers in human, and the histogram on the left shows the distribution of the percentage of 8-mers within DHSs. 8-mers are divided into three groups based on the presence of the CG and TG dinucleotides as indicated in the legend. The relatively big three circles colored in gray, green, and red shown in the upper right indicate layer orders of these three groups of 8-mers.
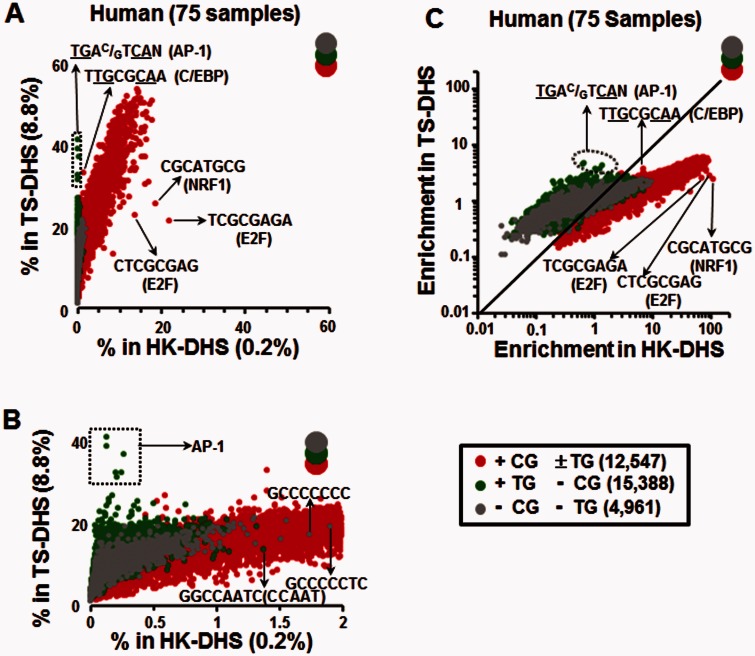
As the human DHS data set contained 75 samples derived from different tissue types and cell lines (supplementary table S3, Supplementary Material online) we reasoned that DHSs occurring in all samples are likely to represent housekeeping DHSs (HK-DHS), regulating older more established components of cellular life. In contrast, those DHSs that occur in a subset of samples are likely tissue-specific DHSs (TS-DHS), representing newer regulatory regions (i.e., related to cell differentiation). We thus divided the DHSs into two groups: 8,376 DHSs, representing 0.2% of the genome, that occur in all 75 human samples and are termed HK-DHS with 83% (6,965) in CGIs. The remaining 1,149,570 DHSs, which do not occur in all samples, comprise 8.8% of the genome, and are termed TS-DHS (Chatterjee et al. 2012). We compared 8-mers in the HK-DHSs and the TS-DHSs in two ways: 1) The fraction of total occurrences in the human genome that occur in HK-DHSs and TS-DHSs (fig. 4A and B), and 2) their enrichment, normalizing their occurrences within each type of DHS according to their abundance across the genome (fig. 4C).
Fig. 4.—
TG dinucleotide containing 8-mers are enriched in TS-DHSs in human. (A) Comparison of 8-mer occurrences within TS- and HK-DHSs. For each continuous 8-mer, the percentage of occurrences within HK-DHSs common to all 75 human samples that represent 0.2% of the genome is plotted on the horizontal axis versus the percentage in TS-DHS representing 8.8% of the genome is plotted on the vertical axis. The 8-mers are divided into three groups based on the presence of the CG and TG dinucleotide as in figure 3. (B) Same as in (A) but with the x axis zoomed in. (C) Similar as in (A) but using a normalized enrichment score (see Materials and Methods).
Figure 4 A presents the fraction of 8-mers in the human genome found in HK-DHSs versus TS-DHSs. +CG ± TG 8-mers are the most enriched class in both HK-DHSs and TS-DHSs. Over 20% of all occurrences of the 8-mer TCGCGAGA representing the E2F motif are in the 0.2% of the genome that are HK-DHSs. Noteworthy among the enriched 8-mers with two CG dinucleotides are the binding sites for NRF-1 and E2F, thus linking these TFBS with housekeeping functions (Evans and Scarpulla 1990; Muller and Helin 2000) (fig. 4A). −CG −TG 8-mers show modest enrichment in both HK-DHSs and TS-DHSs. A small number are enriched in HK-DHS and are C+G rich (fig. 4A and B), possibly reflecting the C + G richness of regulatory regions (Tillo et al. 2010) or contain poly dC stretches, reflecting potential binding sites for SP-1 and KLF transcription factors (TFs) (Lania et al. 1997; Suske 1999; Kaczynski et al. 2003). Additional −CG −TG 8-mers enriched in HK-DHS include those containing the CCAAT motif as previously described (Rozenberg et al. 2008) (fig. 4A and B).
A subset of 8-mers in the +TG −CG group are more enriched in TS-DHSs relative to −CG −TG 8-mers (fig. 4A and B). Specifically, the top 20 8-mers in TS-DHS are the consensus AP-1 motif (TGAC/GTCAN), single nucleotide polymorphisms of the AP-1 consensus, and the E-box motif (GCAGCTGC) (supplementary table S4, Supplementary Material online). Presenting the data as an enrichment (fig. 4C) also highlights the enrichment of +CG ± TG TFBS in HK-DHS (chi-square test, P < 10−324 for E2F and NRF1; supplementary table S5, Supplementary Material online), and the enrichment of the AP-1 motif in TS-DHSs (chi-square test, P < 10−324) relative to HK-DHSs. In general, all three classes of 8-mers show a bias toward being more enriched in HK-DHSs as compared with TS-DHSs perhaps indicating that the HK-DHSs have more regulatory information than TS-DHSs.
We see similar results when the DHSs are divided based on their overlap with CNEs. DHSs in CNEs (DHS + CNE) would be conserved across evolution, whereas the DHSs not in CNEs (DHS − CNE) would be newer regulatory regions. Again, +CG ± TG 8-mers are more enriched in DHSs with CNEs, whereas +TG −CG 8-mers are more enriched in DHSs without CNEs (DHS−CNE) with AP-1 motifs being the most enriched (supplementary fig. S7, Supplementary Material online).
A more fine grained analysis that examined 2,233,542 DHSs from 125 human tissues and cell lines (supplementary fig. S8, Supplementary Material online) representing 18% of the genome (Vierstra et al. 2014) in which TS-DHSs were separated into five groups based on their frequency of occurrence in all samples showed similar results (supplementary fig. S9, Supplementary Material online). Again, certain +TG −CG 8-mers (AP-1 and E-box motifs) are enriched in TS-DHSs, particularly in DHS shared between 6 and 60 cell lines (supplementary fig. S9, Supplementary Material online).
An identical analysis using mouse DHS data from 55 tissues and cell lines (Vierstra et al. 2014) yielded similar results, with the AP-1 and E-box motifs being the most enriched +TG −CG 8-mer in TS-DHS (supplementary fig. S10, Supplementary Material online).
Although our remaining analyses focus on TG-dinucleotide containing 8-mers, specifically the AP-1 TFBS, we also find evidence that the enrichment of TG-dinucleotide containing TFBS within TS-DHSs holds for longer sequences. These included discontinuous (gapped) 8-mers separated by 1- to 30-bp gaps (NNNN-(1-30)NNNN, where “-”s indicate a gap), and 8-mers of the form N-NNN—NNN-N, which are bound by hormone binding proteins (He et al. 2013). The longer TG-dinucleotide containing motifs that are enriched in TS-DHSs in both human and mouse genomes include those for nuclear factor 1 (NF1, TGGC—–GCCA) (de Vries et al. 1987; Blomquist et al. 1999; Whittle et al. 2009), and the canonical GR motif (G-ACA—TGT-C) (Yamamoto 1985; Beato 1989) (supplementary figs. S11 and Supplementary Data, Supplementary Material online).
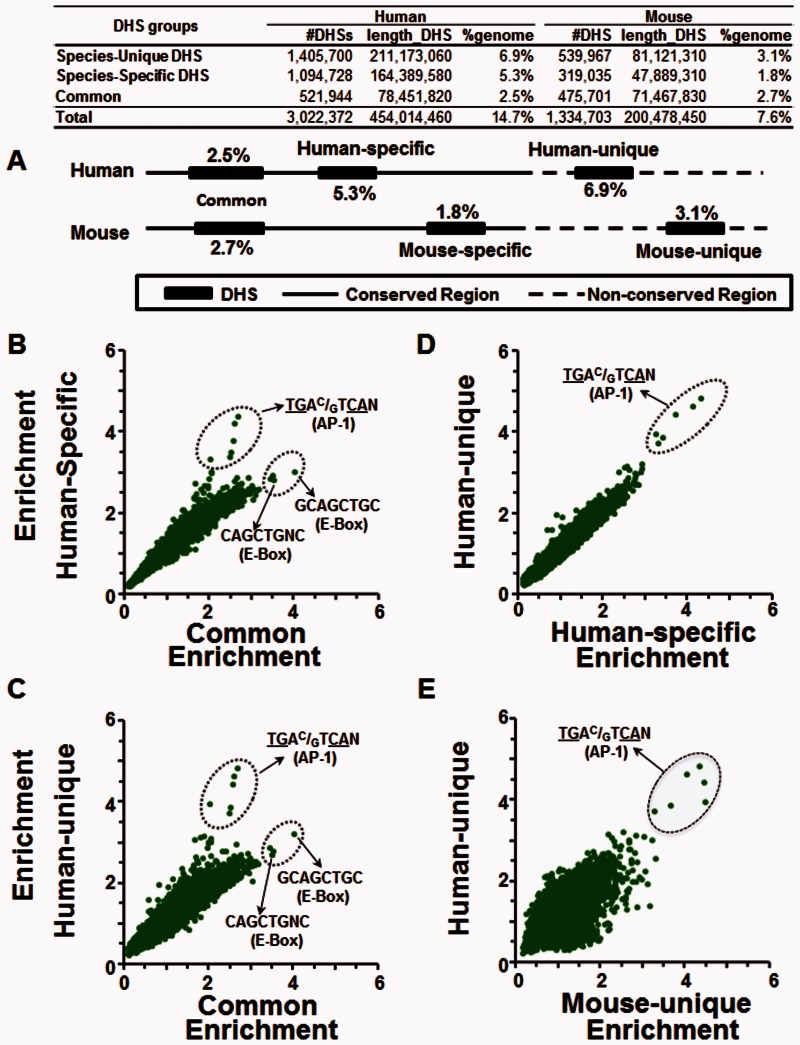
AP-1 Motifs Are Enriched in Mouse- and Human-Specific DHSs
We compared the enrichment of +TG −CG 8-mers in five groups of DHSs from human and mouse tissues defined based on their conservation between the two species, allowing us to further differentiate the DHSs based on their evolutionary origin (Vierstra et al. 2014) (fig. 5A). These groups cover between 1.8% of the genome for the least frequently occurring group and 6.9% of the genome for the most frequently occurring group (fig. 5A). One group contains DHSs shared between the two species (common). Each species also contains two DHS groups that are specific to that species. Two groups comprise DHSs that occur in only one species even though the DNA sequence is conserved (species-specific). The other two groups in each species comprise DHSs that occur in one species and contain a DNA sequence that is not conserved between species (species-unique) (fig. 5A and table 1). The most enriched +TG −CG 8-mer in the four species-specific groups of DHS is the AP-1 motif (fig. 5B–E and supplementary figs. S13–S15, Supplementary Material online). The E-Box motif (GCAGCTGC) is the most enriched +TG −CG containing 8-mer in common DHSs (fig. 5B and C), perhaps suggesting an association with more conserved regulatory functions.
Fig. 5.—
AP-1 motifs are most enriched in the human-unique TS-DHSs, whereas E-Box motifs are enriched in shared DHSs. (A) Schematic of the five types of DHSs were identified by comparing human and mouse DHSs: DHSs common between the two species (Common), and two groups of DHSs for each species, species-specific DHSs where the DNA is either conserved (human-specific and mouse-specific) or not conserved between the two species (human-unique and mouse-unique). Enrichment of +TG-CG 8-mers in TS-DHSs in (B) common versus human-specific, (C) common versus human-unique, (D) human-specific versus human-unique, and (E) mouse-unique versus human-unique.
Table 1.
Human and Mouse DHSs with AP-1 Motifs
| Human DHSs |
Mouse DHSs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % in Genome | #DHS with AP-1 Motif | %DHS with AP-1 Motif | With AP-1 motif in mouse |
% in Genome | #DHS with AP-1 Motif | %DHS with AP-1 Motif | With AP-1 Motif in Human |
|||
| N | % | N | % | |||||||
| Species-specific DHSs | 5.3 | 59,126 | 5.4 | 5,552 | 9.4 | 1.8 | 20,606 | 6.5 | 1,533 | 7.4 |
| Common DHSs | 2.5 | 27,302 | 5.2 | 10,207 | 37.4 | 2.7 | 28,318 | 6.0 | 10,022 | 35.4 |
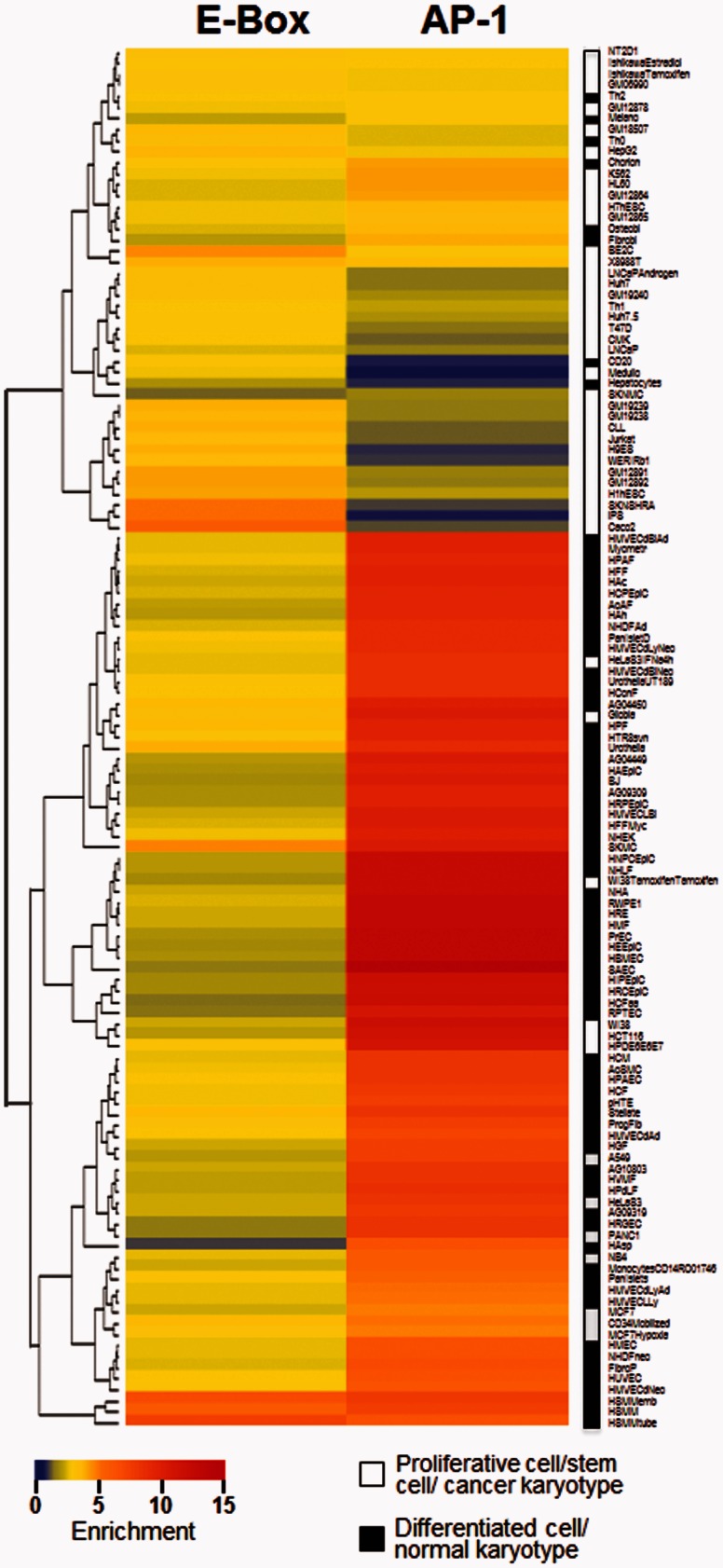
Consistent with this idea, when we examine the enrichment of the conserved E-Box and tissue-specific AP-1 motifs in DHSs of different cell types, we find that the E-box motif (GCAGCTGC) is enriched in growing cells including pluripotent stem cells (iPSC) and cancer cells (fig. 6 and supplementary table S6A, Supplementary Material online). In contrast, the enrichment of the AP-1 motif in DHSs is most prominent in differentiated tissues including astrocytes and epithelial cells and is depleted in embryonic cell, such as iPSC (fig. 6 and supplementary table S6B, Supplementary Material online). That is, E-box motifs tend to be found in regulatory regions involved in cell proliferation and growth, whereas the AP-1 motifs tend to be found in regulatory regions in differentiated tissues, suggesting a link between the conservation and enrichment of AP-1 and E-box motifs with their regulatory functions and phenotypes.
Fig. 6.—
Heat-map of enrichment of AP-1 and E-Box motifs in DHS derived from 125 human samples. Hierarchical clustering of E-box or AP-1 enrichment in DHS derived from 125 samples. Samples were classified according to their karyotype (cancer vs. normal) and cell type (differentiated vs. proliferative) as indicated by the white/black legend.
We next evaluated whether the AP-1 motif positively correlates with formation of new DHSs by examining the species-specific DHSs groups. There are 59,126 human-specific DHSs with an AP-1 motif, but only 9.4% of these have an AP-1 motif in mouse, linking the presence of an AP-1 motif and the formation of DHSs (table 1). In contrast, 37% of the 27,302 common human DHSs that have an AP-1 motif also have an AP-1 motif in the mouse (table 1). The common DHSs with AP-1 sites in human that lack an AP-1 motif in mouse have high intrinsic nucleosome sequence preference (INOS, Intrinsic Nucleosome Occupancy Score) (Tillo and Hughes 2009) (supplementary fig. S16C, Supplementary Material online), consistent with G+C rich sequences being associated with regulatory function (Tillo et al. 2010; Bird 2011) and may also contain other TFBS that produce the DHSs. Similar results were obtained for the 20,606 mouse-specific DHSs with an AP-1 motif (table 1).
Evolution of AP-1 and E-Box Motifs
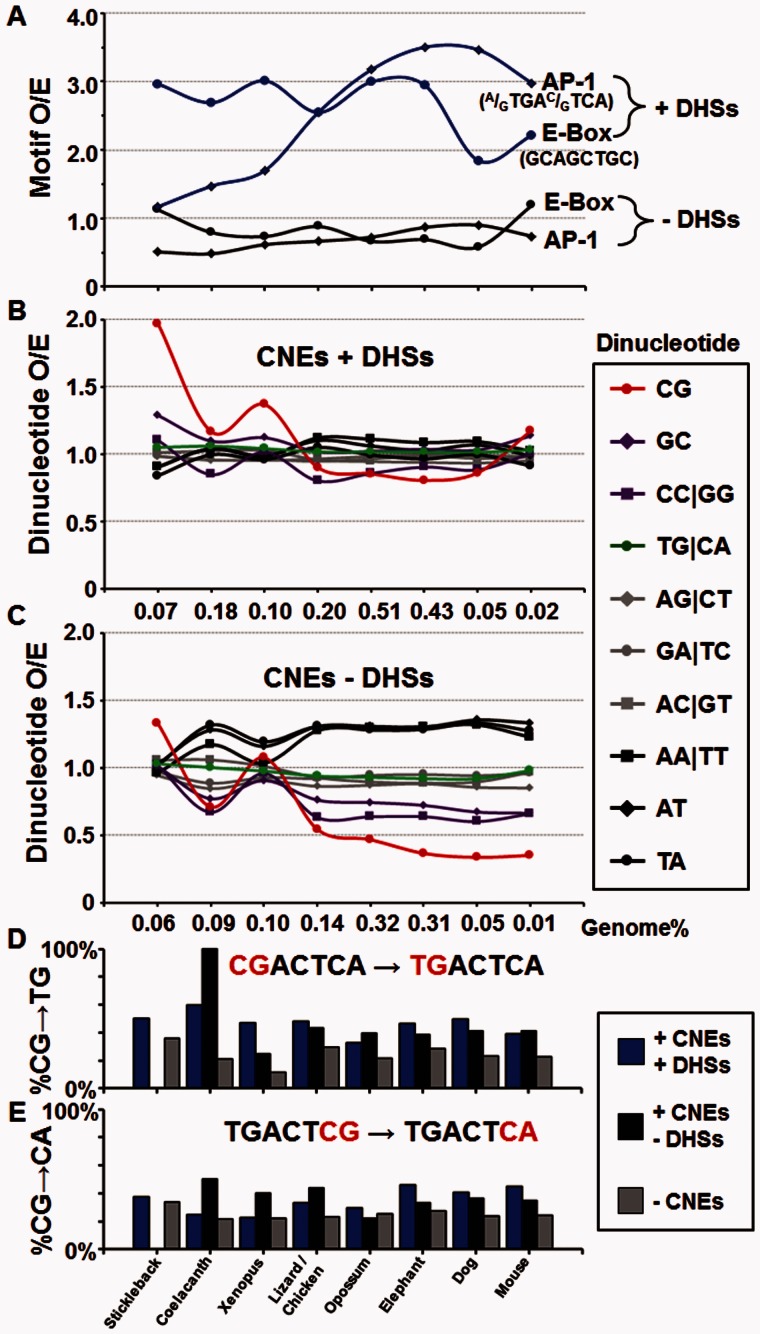
We next reexamined the CNEs focusing on the enrichment and evolution of functional AP-1 and E-box motifs. CNEs were divided into two groups based on their overlap with human DHSs resulting in two similarly sized groups (371,061 CNEs+DHS and 368,536 CNEs−DHS; supplementary table S2, Supplementary Material online, and fig. 7). The E-box motif is enriched in both ancient and recent CNEs in DHSs (fig. 7A), having 3-fold enrichment in the three more ancient groups of CNEs (stickleback, coelacanth, and Xenopus), coinciding with the enrichment of the CG dinucleotides in CNEs (fig. 2). In contrast, the AP-1 motif becomes highly (4-fold) enriched in more recent CNEs in DHSs, starting with the chicken, coinciding with the lineages in which the CG dinucleotide becomes depleted (fig. 2). The E-Box or AP-1 motifs are not enriched in CNE that are not in DHSs suggesting that these CNEs do not recruit TFs (fig. 7A).
Fig. 7.—
Enrichment of dinucleotides and AP-1 and E-Box motifs in CNES ± DHSs. (A) The enrichment for the AP-1 (A/GTGAC/GTCA) and E-Box (GCAGCTGC) motifs in eight groups of CNEs from stickleback to mouse is presented for CNEs in TS-DHSs and CNEs not in DHSs. Dinucleotide enrichment in eight groups of CNEs from stickleback to mouse for (B) CNEs in DHS and (C) CNEs not in DHSs. The percentage of T being C at different positions in human AP-1 motifs in three groups of regions based on conservation of sequence and accessibility to DNase I digestion (+DHS+CNEs, blue; +DHS−CNEs, black; −CNEs, gray) for (D) position 1 CG→TG and (E) position 7 CG→CA.
Examination of general properties (dinucleotide abundance) of CNEs within DHSs shows that the CG dinucleotide is enriched in the three more ancient groups of CNEs (stickleback, coelacanth, and Xenopus) at the same time the E-box motif is enriched. There is a notable increase in CG dinucleotides in human CNE + DHSs only shared with the mouse, possibly reflecting the higher number of CGIs in the human genome relative to mouse (Antequera and Bird 1993; Han et al. 2008). In the more recent CNEs, the CG dinucleotide becomes depleted, coinciding with the increased enrichment of the AP-1 motif (fig. 7A and B). The remaining dinucleotides including the TG dinucleotide show little change in abundance. CNEs not in DHSs show more variability in dinucleotide enrichment, with AA/TT, TA, and AT dinucleotides becoming enriched and CG, GC, and CC/GG dinucleotides being depleted in newer CNEs (fig. 7C).
New AP-1 Motifs Previously Contained a CG Dinucleotide
We examined whether AP-1 motifs are derived from sequences containing a CG dinucleotide by looking at the evolutionary history of the 460,228 AP-1 motifs (TGAC/GTCA) in the human genome. Of all genomes examined (see Materials and Methods), the dog and elephant comparisons show strong evidence that AP-1 motifs in human previously contained a CG dinucleotide (fig. 7D and E, table 2, and supplementary tables S7 and Supplementary Data, Supplementary Material online). For example, nearly half of the human AP-1 motifs can be identified in elephant, with 110,317 containing a single nucleotide difference at most from the consensus motif, with 52,812 being identical (table 2 and supplementary table S7, Supplementary Material online). The 110,317 single nucleotide differences of AP-1 motifs were placed into three groups based on their presence in CNEs and DHSs (CNE + DHS, CNE − DHS, and AP-1 not in CNE; fig. 7D and E, table 2, and supplementary tables S7 and Supplementary Data, Supplementary Material online). In the case of AP-1 sites in DHSs, over 95% of each base is conserved (supplementary table S8G, Supplementary Material online). For the AP-1 sites in human that are not conserved in elephant, 46.6% contain a cytosine in the elephant sequence at the position of the first thymidine of the motif (fig. 7D, table 2, and supplementary table S8G, Supplementary Material online). This percentage is lower in AP-1 sites in nonregulatory regions (AP-1 not in CNE), where 28.7% contain a C at this position (fig. 7D, table 2, and supplementary table S8G, Supplementary Material online), suggesting that selection is not acting to preserve newly created AP-1 sites as occurs for AP-1 motifs in regulatory regions (fig. 7D and E and supplementary table S8A–I, Supplementary Material online).
Table 2.
The MRCA Sequence in Elephant to Variations of 11mers with TGACTCA in Human
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Consensus | N | N | T | G | A | C | T | C | A | N | N |
| AP1 + CNEs + DHSs (11,741) | |||||||||||
| A | 30.5% | 32.3% | 25.4% | 44.9% | — | 49.2% | 20.4% | 24.6% | — | 18.1% | 29.2% |
| C | 17.3% | 26.3% | 46.6% | 17.5% | 20.5% | — | 60.8% | — | 28.7% | 22.1% | 22.2% |
| G | 22.8% | 24.9% | 28.0% | — | 68.0% | — | 18.8% | 21.4% | 46.3% | 28.8% | 18.6% |
| T | 29.5% | 16.5% | — | 37.5% | 11.5% | 50.8% | — | 54.0% | 25.1% | 31.0% | 30.0% |
| AP1 + CNEs − DHSs (2,966) | |||||||||||
| A | 33.7% | 29.3% | 32.1% | 51.1% | — | 48.8% | 32.5% | 20.3% | — | 28.3% | 32.4% |
| C | 17.3% | 23.8% | 38.5% | 24.4% | 19.4% | — | 58.1% | — | 29.9% | 15.4% | 16.4% |
| G | 16.1% | 16.5% | 29.4% | — | 66.5% | — | 9.4% | 26.7% | 33.3% | 26.6% | 17.3% |
| T | 32.9% | 30.4% | — | 24.4% | 14.1% | 51.2% | — | 52.9% | 36.8% | 29.6% | 33.9% |
| AP1 − CNEs (95,610) | |||||||||||
| A | 27.9% | 25.9% | 37.3% | 52.8% | 0.0% | 49.7% | 23.4% | 21.4% | — | 24.1% | 31.3% |
| C | 20.5% | 29.2% | 28.7% | 20.6% | 21.8% | — | 70.5% | — | 33.9% | 20.2% | 20.9% |
| G | 20.1% | 22.0% | 34.0% | — | 63.2% | — | 6.1% | 24.1% | 27.7% | 31.0% | 19.8% |
| T | 31.5% | 22.8% | — | 26.5% | 14.9% | 50.3% | — | 54.5% | 38.4% | 24.8% | 28.0% |
Note.—The MRCA sequences to variations of 11mers with TGACTCA in human are divided into three groups: 1) In CNEs and in DHSs, 2) in CNEs and no DHSs, and 3) no CNEs for elephant. For the first and last two “N”, the percentage indicates the base component at each position. For the middle seven positions of AP-1 motif, the “—” indicates the consensus base, whereas the percentage of each base indicates the variation component at each position. For example, in position 3, the variation component is A→T, C→T, and G→T. The bold percentage highlights the C→T and G→A variations may be arose from CG dinucleotide.
DNA Binding Affinity of c-Jun|c-Fos Heterodimers to Motifs with CG, 5mCG, and TG Dinucleotides
Previously, it was shown that methylation of the CG dinucleotide in the sequence CGAGTCA increased binding of the c-Jun|c-Fos heterodimer (Gustems et al. 2014). To test whether deamination of 5mCG to TG creates a TFBS that is better bound by the c-Jun|c-Fos heterodimer, we used a protein-binding microarray in which the CG dinucleotides were enzymatically methylated using Msss1 (Mann et al. 2013) (table 3), and examined heterodimer binding to three sequences CGAGTCA, 5mCGAGTCA, and TGAGTCA. We find that the deamination product, TGAC/GTCA, is better bound by the c-Jun|c-Fos heterodimer compared with 5mCGAGTCA, which in turn is better bound than CGAGTCA, demonstrating that deamination creates a better AP-1 motif (table 3).
Table 3.
cJUN-GST + cFOS Heterodimers Binding the AP-1 Motifs (Z-Score)
| Sequence | Replicate 1 | Replicate 2 |
|---|---|---|
| TGAC/GTCA | 18.15 | 14.45 |
| mCGAGTCA | 6.78 | 5.63 |
| mCGACTCA | 6.21 | 5.27 |
| CGAGTCA | 1.47 | 1.50 |
| CGACTCA | 1.36 | 1.41 |
Discussion
Deamination of 5mC as a Mutator System
Genetic mutator systems have been described (Degnen and Cox 1974; Goodman et al. 1993) including systems that develop resistance to antibiotics (Chopra et al. 2003) and pesticides (Travis JM and Travis ER 2002) demonstrating the importance of an increased mutation rate in particular selective situations. These mutator systems often compromise DNA repair pathways. We have explored the suggestion that one function of cytosine methylation in CG dinucleotides is to increase the mutation rate resulting in greater genetic diversity (Bird 1980). 5mC deamination is not inherently mutagenic as the T•G mismatch base pair can be repaired back to the original C•G base pair as happens in algal genomes, which contain CG methylation but no depletion of CG dinucleotides (Huff and Zilberman 2014). In vertebrates, the deamination of 5mC produces the T•G mismatch base pair that sometimes results in the formation of a T•A base pair producing a TG dinucleotide. This has produced a dramatic depletion in the CG dinucleotide causing a bimodal distribution of 8-mer abundance, which initially occurs in the coelacanth (fig. 1). We speculate that mutagenic CG methylation is a trait that has been selected for because it generates genetic diversity (Leffler et al. 2012). To test this idea, we examined whether TS-DHSs, which are more recently evolved than HK-DHSs, are enriched in sequences containing the TG dinucleotide, the deamination product of 5mC in CG dinucleotides. We find that the most enriched TG-containing sequences in TS-DHS are TFBS (AP-1 and E-box). In the case of the AP-1 motif, this enrichment is more pronounced in the more recent DHSs. Additionally, we have presented evidence that the AP-1 motif previously contained CG dinucleotides, suggesting that they may be molecular fossils of 5mCG deamination. Thus, deamination of 5mCG to TG dinucleotides increases genetic diversity, forming TFBS that create new regulatory regions and the emergence of novel phenotypes.
Sequence Properties of Ancient and Recent CNEs
In our examination of CNEs from eight lineages among the vertebrates (Amemiya et al. 2013), we observe enrichment of CG dinucleotides and C + G rich CNEs in the stickleback, coelacanth, and Xenopus, suggesting that the appearance of CGIs occurs within these lineages. The origins of CGIs have been discussed (Han et al. 2008), but the mechanisms driving their emergence remain unclear. Stickleback, coelacanth, and Xenopus lineages also contain C + G poor CNEs, though they are not particularly enriched for either TG containing 8-mers or 8-mers with neither TG nor CG. The enrichment of TG containing 8-mers in CNEs that overlap with DHSs only becomes evident among mammals, and only when these TG containing 8-mers represent TFBS, suggesting that selection is acting upon these sequences because of their regulatory function. It is worth pointing out that, although some proportion of TG dinucleotides derived from mutation of 5mCG dinucleotide may obtain regulatory function, most CG-derived TGs do not gain function and remain in the genome. Nevertheless, the accelerated creation and destruction (Mann et al. 2013) of TFBS by 5mCG deamination could explain the rapid change in TF localization observed in mammalian systems that is not observed in Drosophila (Schmidt et al. 2010; Villar et al. 2014). It will be interesting to place TFBS into groups based on the presence of CG and/or TG dinucleotides and determine whether those without these dinucleotides are evolving as quickly as in mammalian genomes. CNEs not in DHS do not have any enrichment of TG-containing TFBS, and have different patterns of dinucleotide enrichment compared with CNEs + DHS, suggesting that these sequences are not involved in recruiting TFs. It would be interesting to examine these CNEs in more detail to determine why they are conserved across evolution.
AP-1 Motif in Ancient and Recent DHSs
The most enriched TG dinucleotide containing 8-mers in TS-DHSs are motifs for TFs from the B-ZIP or B-HLH families, two of the largest classes of TFs in vertebrates (Weirauch and Hughes 2011). These TF families function as dimers, with dimerization important for specificity and fine-tuning of regulatory control (Vinson et al. 1989; Klemm et al. 1998; Amoutzias et al. 2008). In addition, these 8-mers are palindromic, unlike older CG-containing sequences enriched in HK-DHSs. Palindromic sequences are biologically attractive, as they produce two copies of the identical sequence and therefore a 2-fold increase in local DNA sequence concentration, which is exploited by dimeric TFs.
Our analyses show that AP-1 motifs (Lee et al. 1987) are enriched in TS-DHS, particularly the more recent DHSs that are human- and mouse-specific. However, the tissue specificity of AP-1 motif enrichment is general and not limited to a particular tissue, which is consistent with the widespread expression of Jun and Fos and their roles as facilitators (Ravasi et al. 2010). To achieve this kind of tissue specificity, the combination of tissue-specific TFs and facilitators is required. For example, GR binding to cisregulatory elements is dependent on AP1 activity in the murine mammary epithelial cell line (Biddie et al. 2011). Moreover, these functional AP-1 sites in TS-DHS occur in regions that are also predicted to be well-bound by nucleosomes. Pairs of TFs can cooperatively bind to DNA directly (Chatterjee and Vinson 2012; Martinez and Rao 2012) or indirectly by competition with nucleosomes, providing increased specificity and control of gene regulation (Polach and Widom 1996; He et al. 2013). We suggest that this indirect competition mechanism is operating in recently evolved mammalian regulatory sequences.
The AP-1 motif is often bound by heterodimers containing an Fos and a Jun family member. Fos and Jun proteins are conserved from Drosophila (Fassler et al. 2002) and have been reduplicated in the vertebrate lineage creating multiple heterodimeric complexes that can bind the AP-1 motif. Classically, AP-1 TFs have been implicated in cell growth (Olive et al. 1996; Eferl et al. 2003; Gerdes et al. 2006), with c-Jun and Fos proteins viewed as protooncogenes (Bohmann et al. 1987). However, these TFs do not show up as candidates in cancer screens or GWAS studies and our analysis indicates that their binding sites are enriched in DHS from normal cells and tissues and not in embryonic and cancer cells, suggesting a role in differentiation (fig. 6). We propose that the newer AP-1 motifs are involved in cellular differentiation (Andreucci et al. 2002). We also show here that the c-Jun|c-Fos heterodimer preferentially binds mCGACTCA better than unmethylated CGACTCA (Gustems et al. 2014) but binds the best to the deamination product TGACTCA which creates a canonical AP-1 motif. This is in contrast to the C/EBP motif (TTGCGCAA), which contains a central CG dinucleotide and deamination of methylated cytosine (5mCG) decreases C/EBP binding (Mann et al. 2013). The observation that methylated cytosine in the first position of AP-1 motif enhances the binding ability of c-Jun|c-Fos compared with unmethylated cytosine may indicate that methylated cytosines can mimic thymine in DNA-protein interactions (Dickerson et al. 2009). It will be interesting to evaluate how deamination changes binding of additional heterodimers of Jun and Fos family members revealing which heterodimers preferentially bind the deamination product.
In conclusion, we have presented evidence that one consequence of methylation of cytosine in CG dinucleotides is to increase the mutation rate, producing new TG dinucleotides that can create TFBS that function in a tissue-specific manner to drive evolution. These TFBS are important for the emergence of new regulatory regions and ultimately novel phenotypes.
Author Contributions
X.H. conducted the majority of data analysis. D.T., C.D., G.J.R., P.C.F. conducted data analysis. J.V. conducted identification of DHSs. S.K.S. conducted the experiments of microarray. X.H. and C.V. conceived and designed the study. C.V. oversaw the study. X.H., D.T., J.V., J.S., P.C.F. and C.V. contributed to writing of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Health (NIH), Center for Cancer Research, National Cancer Institute. The authors thank Dr Alison P. Lee for providing the annotations for conserved noncoding elements (CNEs) in tetrapods and Dr Sanjit Mukherjee for helpful discussion. They also thank the high-performance computational capabilities of the Helix & Biowulf Systems at the NIH, Bethesda, MD (http://helix.nih.gov).
Literature Cited
- Akers SN, et al. 2014. LINE1 and Alu repetitive element DNA methylation in tumors and white blood cells from epithelial ovarian cancer patients. Gynecol Oncol. 132:462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, et al. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoutzias GD, Robertson DL, Van de Peer Y, Oliver SG. 2008. Choose your partners: dimerization in eukaryotic transcription factors. Trends Biochem Sci. 33:220–229. [DOI] [PubMed] [Google Scholar]
- Andreucci JJ, et al. 2002. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J Biol Chem. 277:16426–16432. [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A. 1993. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 90:11995–11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, et al. 2009. Diversity and complexity in DNA recognition by transcription factors. Science 324:1720–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. 2011. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 11:726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beato M. 1989. Gene regulation by steroid hormones. Cell 56:335–344. [DOI] [PubMed] [Google Scholar]
- Biddie SC, et al. 2011. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 43:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6–21. [DOI] [PubMed] [Google Scholar]
- Bird A. 2011. The dinucleotide CG as a genomic signalling module. J Mol Biol. 409:47–53. [DOI] [PubMed] [Google Scholar]
- Bird A, et al. 1995. Studies of DNA methylation in animals. J Cell Sci Suppl. 19:37–39. [DOI] [PubMed] [Google Scholar]
- Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8:1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomquist P, Belikov S, Wrange O. 1999. Increased nuclear factor 1 binding to its nucleosomal site mediated by sequence-dependent DNA structure. Nucleic Acids Res. 27:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann D, et al. 1987. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science 238:1386–1392. [DOI] [PubMed] [Google Scholar]
- Chatterjee R, et al. 2012. Overlapping ETS and CRE Motifs ((G/C)CGGAAGTGACGTCA) preferentially bound by GABPalpha and CREB proteins. G3 (Bethesda) 2:1243–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee R, et al. 2014. High-resolution genome-wide DNA methylation maps of mouse primary female dermal fibroblasts and keratinocytes. Epigenetics Chromatin. 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee R, Vinson C. 2012. CpG methylation recruits sequence specific transcription factors essential for tissue specific gene expression. Biochim Biophys Acta. 1819:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Miller BF, Furano AV. 2014. Repair of naturally occurring mismatches can induce mutations in flanking DNA. Elife 3:e02001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I, O’Neill AJ, Miller K. 2003. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat. 6:137–145. [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274:775–780. [DOI] [PubMed] [Google Scholar]
- de Vries E, van Driel W, van den Heuvel SJ, van der Vliet PC. 1987. Contactpoint analysis of the HeLa nuclear factor I recognition site reveals symmetrical binding at one side of the DNA helix. EMBO J. 6:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnen GE, Cox EC. 1974. Conditional mutator gene in Escherichia coli: isolation, mapping, and effector studies. J Bacteriol. 117:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson SJ, et al. 2009. Methylation-dependent binding of the Epstein-Barr virus BZLF1 protein to viral promoters. PLoS Pathog. 5:e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, et al. 2003. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell 112:181–192. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Scarpulla RC. 1990. NRF-1: a trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 4:1023–1034. [DOI] [PubMed] [Google Scholar]
- Fassler J, et al. 2002. B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res. 12:1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. 1987. CpG islands in vertebrate genomes. J Mol Biol. 196:261–282. [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Karlin S. 2001. Genome-scale compositional comparisons in eukaryotes. Genome Res. 11:540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes MJ, et al. 2006. Activator protein-1 activity regulates epithelial tumor cell identity. Cancer Res. 66:7578–7588. [DOI] [PubMed] [Google Scholar]
- Giraud A, et al. 2001. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science 291:2606–2608. [DOI] [PubMed] [Google Scholar]
- Goodman MF, Creighton S, Bloom LB, Petruska J. 1993. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 28:83–126. [DOI] [PubMed] [Google Scholar]
- Gustems M, et al. 2014. c-Jun/c-Fos heterodimers regulate cellular genes via a newly identified class of methylated DNA sequence motifs. Nucleic Acids Res. 42:3059–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Su B, Li WH, Zhao Z. 2008. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 9:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Zhao Z. 2008. Comparative analysis of CpG islands in four fish genomes. Comp Funct Genomics. 2008:565631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, et al. 2013. Contribution of nucleosome binding preferences and co-occurring DNA sequences to transcription factor binding. BMC Genomics 14:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff JT, Zilberman D. 2014. Dnmt1-independent CG methylation contributes to nucleosome positioning in diverse eukaryotes. Cell 156:1286–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DG, Green P. 2004. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc Natl Acad Sci U S A. 101:13994–14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki Y, et al. 2014. Evolutionary changes in vertebrate genome signatures with special focus on coelacanth. DNA Res. 21:459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm JD, Schreiber SL, Crabtree GR. 1998. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 16:569–592. [DOI] [PubMed] [Google Scholar]
- Lania L, Majello B, De Luca P. 1997. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 29:1313–1323. [DOI] [PubMed] [Google Scholar]
- Laurent L, et al. 2010. Dynamic changes in the human methylome during differentiation. Genome Res. 20:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Mitchell P, Tjian R. 1987. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell 49:741–752. [DOI] [PubMed] [Google Scholar]
- Leffler EM, et al. 2012. Revisiting an old riddle: what determines genetic diversity levels within species?. PLoS Biol.:10e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makapedua DM, et al. 2011. Genome size, GC percentage and 5mC level in the Indonesian coelacanth Latimeria menadoensis. Mar Genomics. 4:167–172. [DOI] [PubMed] [Google Scholar]
- Mann IK, et al. 2013. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res. 23:988–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez GJ, Rao A. 2012. Immunology. Cooperative transcription factor complexes in control. Science 338:891–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H, Helin K. 2000. The E2F transcription factors: key regulators of cell proliferation. Biochim Biophys Acta. 1470:M1–M12. [DOI] [PubMed] [Google Scholar]
- Olive M, Williams SC, Dezan C, Johnson PF, Vinson C. 1996. Design of a C/EBP-specific, dominant-negative bZIP protein with both inhibitory and gain-of-function properties. J Biol Chem. 271:2040–2047. [DOI] [PubMed] [Google Scholar]
- Polach KJ, Widom J. 1996. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 258:800–812. [DOI] [PubMed] [Google Scholar]
- Ravasi T, et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089–1093. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. 2001. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2:21–32. [DOI] [PubMed] [Google Scholar]
- Rishi V, et al. 2010. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 107:20311–20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M. 2011. Cancer epigenetics reaches mainstream oncology. Nat Med. 17:330–339. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, et al. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res. 43:D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougier N, et al. 1998. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 12:2108–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenberg JM, et al. 2008. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics 9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328:1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD. 1994. A T7 expression vector for producing N- and C-terminal fusion proteins with glutathione S-transferase. Gene 138:105–108. [DOI] [PubMed] [Google Scholar]
- Simmen MW. 2008. Genome-scale relationships between cytosine methylation and dinucleotide abundances in animals. Genomics 92:33–40. [DOI] [PubMed] [Google Scholar]
- Sjolund AB, Senejani AG, Sweasy JB. 2013. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat Res. 743–744:12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, et al. 2014. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature 515:365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G. 1999. The Sp-family of transcription factors. Gene 238:291–300. [DOI] [PubMed] [Google Scholar]
- Takizawa T, et al. 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 1:749–758. [DOI] [PubMed] [Google Scholar]
- Tillo D, Hughes TR. 2009. G + C content dominates intrinsic nucleosome occupancy. BMC Bioinformatics 10:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillo D, et al. 2010. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One 5:e9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis JM, Travis ER. 2002. Mutator dynamics in fluctuating environments. Proc Biol Sci. 269:591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra J, et al. 2014. Mouse regulatory DNA landscapes reveal global principles of cis-regulatory evolution. Science 346:1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Flicek P, Odom DT. 2014. Evolution of transcription factor binding in metazoans—mechanisms and functional implications. Nat Rev Genet. 15:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Chatterjee R. 2012. CG methylation. Epigenomics 4:655–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson C, Chatterjee R, Fitzgerald P. 2011. Transcription factor binding sites and other features in human and Drosophila proximal promoters. Subcell Biochem. 52:205–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson CR, Sigler PB, McKnight SL. 1989. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 246:911–916. [DOI] [PubMed] [Google Scholar]
- Walsh CP, Xu GL. 2006. Cytosine methylation and DNA repair. Curr Top Microbiol Immunol. 301:283–315. [DOI] [PubMed] [Google Scholar]
- Weirauch MT, et al. 2014. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Hughes TR. 2011. A catalogue of eukaryotic transcription factor types, their evolutionary origin, and species distribution. Subcell Biochem. 52:25–73. [DOI] [PubMed] [Google Scholar]
- Whittle CM, Lazakovitch E, Gronostajski RM, Lieb JD. 2009. DNA-binding specificity and in vivo targets of Caenorhabditis elegans nuclear factor I. Proc Natl Acad Sci U S A. 106:12049–12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto KR. 1985. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet. 19:209–252. [DOI] [PubMed] [Google Scholar]
- Yue F, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515:355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.