Abstract
Background
Cervical cancer is the second deadliest gynecologic malignancy, characterized by apparently precancerous lesions and cervical intraepithelial neoplasia (CIN), and having a long course from the development of CIN to cervical cancer. Cyclin-dependent kinase inhibitor 2A (CDKN2A) is a well-documented tumor suppressor gene and is commonly methylated in cervical cancer. However, the relationship between methylated CDKN2A and carcinogenesis in cervical cancer is inconsistent, and the diagnostic accuracy of methylated CDKN2A is underinvestigated. In this study, we attempted to quantify the association between CDKN2A methylation and the carcinogenesis of cervical cancer, and its diagnostic power.
Methods
We systematically reviewed four electronic databases and identified 26 studies involving 1,490 cervical cancers, 1,291 CINs, and 964 controls. A pooled odds ratio (OR) with corresponding 95% confidence intervals (95% CI) was calculated to evaluate the association between methylated CDKN2A and the carcinogenesis of cervical cancer. Specificity, sensitivity, the area under the receiver operating characteristic curve, and the diagnostic odds ratio were computed to assess the effect of methylated CDKN2A in the diagnosis of cervical cancer.
Results
Our results indicated an upward trend in the methylation frequency of CDKN2A in the carcinogenesis of cervical cancer (cancer vs control: OR =23.67, 95% CI =15.54–36.06; cancer vs CIN: OR =2.53, 95% CI =1.79–3.5; CIN vs control: OR =9.68, 95% CI =5.82–16.02). The specificity, sensitivity, area under the receiver operating characteristic curve, and diagnostic odds ratio were 0.99 (95% CI: 0.97–0.99), 0.36 (95% CI: 0.28–0.45), 0.93 (95% CI: 0.91–0.95), and 43 (95% CI: 19–98), respectively.
Conclusion
Our findings indicate that abnormal CDKN2A methylation may be strongly correlated with the pathogenesis of cervical cancer. Our results also demonstrate that CDKN2A methylation might serve as an early detector of cervical cancer. These findings require further confirmation.
Keywords: p16, methylation, cervical cancer carcinogenesis
Introduction
Cervical cancer remains the second most common cancer in women worldwide. According to a report by the American Cancer Society, 12,990 new cervical cancers and 4,120 new deaths are projected to occur in the US in 2016,1 although a substantially increasing incidence of cervical cancer has been seen in developing countries, which might be due to the inadequacy of the Pap screening test in these areas.2
The etiology of cervical cancer is related to interactions between host and environmental factors, such as contraceptive use,3 infection with certain types of human papillomavirus (HPV),4 having sex at an early age, and having many sexual partners.5,6 Of all these related components, continuous infection with oncogenic HPV is an indisputable etiologic factor for cervical cancer.7 However, most infections have no clinical syndrome and eventually resolve unaided. In a minority of infected women they may lead to the precancerous lesion of cervical cancer, cervical intraepithelial neoplasia (CIN). Typically, CIN expands slowly and takes several decades to progress into invasive cervical cancer, which is a good model for a multistage disease beginning with low-grade CIN and progressing to high-grade CIN, some of which develop into invasive cancers.8 Most epidemiological studies have demonstrated that low-grade CIN is self-limiting, and that only a small subset with persistent HPV infection progress to high-grade CIN and eventually to invasive cervical cancer,9 thereby attesting that HPV infection alone is not sufficient and that other factors that might accelerate the initiation of cervical cancer are required.10
Cancer is a group of disorders with different biological processes caused by a series of changes in tumor suppressor genes (TSGs),11 including genetic changes and epigenetic alterations. In recent decades, great advances have been made in the identification of epigenetic changes in cancer, especially in characterizing the methylation of deoxyribonucleic acid (DNA).12 Numerous data suggest that cancer is substantially affected by the methylation of multiple TSGs.13,14
Cyclin-dependent kinase inhibitor 2A (CDKN2A) was first reported in early 1994;15 it belongs to a family of cell cycle regulators16 and is widely accepted as a TSG, owing to its ability to inhibit the catalytic activity of CDK4/cyclin D enzymes17 and to block cell cycle progression at the G1/S checkpoint.18 However, the loss of CDKN2A function due mainly to promoter hypermethylation is common in human cancers, including colorectal cancer,19 hepatocellular carcinoma,20 gastric carcinoma,21 and breast cancer.22 The diagnostic accuracy of methylated CDKN2A in discriminating cancer cells from normal tissues has also been investigated. The two requirements for a diagnostic biomarker, specificity and sensitivity, have been investigated in many cancers, demonstrating, for example, a 27% specificity and 70% sensitivity in colorectal cancer, and 100% specificity with 73% sensitivity in the serum of patients with liver cancer.18,23,24
As regards cervical cancer, most studies have demonstrated that the methylation frequency of CDKN2A was significantly higher in cervical cancer than in normal or benign tissue.25,26 However, the changing trend in the methylation frequency of CDKN2A during the carcinogenesis of cervical cancer is contradictory. Some studies have shown that the upward methylation frequency of CDKN2A is observed during the carcinogenesis of cervical cancer,27,28 whereas others have shown inconsistent results.29 Also, the diagnostic accuracy of methylated CDKN2A in the differentiation of cervical cancer is underreported. Therefore, it is essential to combine these data in order to draw reliable conclusions and analyze its diagnostic power.
Meta-analysis is able to combine data from various studies and help establish relationships across studies30 and can therefore predict a relatively reliable result through quantitative assessment. Therefore, we carried out a meta-analysis to improve our understanding of the role of methylated CDKN2A in the carcinogenesis of cervical cancer. The diagnostic accuracy of methylated CDKN2A in the discrimination of cervical cancer was also analyzed.
Materials and methods
Literature search
Four electronic databases, PubMed, Web of Science, Embase, and China National Knowledge Infrastructure, were searched for relevant studies until January 19, 2016 using the following keywords: (“CDKN2A” or “cyclin-dependent kinase inhibitor 2A” or “p16”) and (“methylation” or “DNA methylation” or “promoter methylation”) and (“cervical cancer” or “cervical carcinoma” or “cancer of uterine cervix”).
Selection criteria
A study could be included if it met the following criteria: 1) it should be a case–control study; 2) it should analyze the methylation status of CDKN2A during the progression of cervical cancer; 3) it should test the methylation level of CDKN2A in human tissues; and 4) it should represent the available methylation data. No studies in cell lines or in animals have been included in the current study. In the current study, the CIN group consists of patients with CIN1, CIN2, and CIN3; normal individuals or patients with benign lesions were included in the control group.
Data extraction
Four authors (MY, HZ, JY, and CCZ) retrieved all the eligible studies and extracted the relevant data independently. The following information was extracted: the first author’s name, the year of publication, the ethnic origins of study subjects, the number of participants, and the frequency of CDKN2A methylation.
Statistical analysis
The strength of association is represented as an overall odds ratio (OR) with corresponding 95% confidence interval (CI). The heterogeneity of all eligible studies was quantified using the I2 statistic and χ2 tests, with corresponding P-value.31 When there was heterogeneity in the meta-analysis a Dersimonian–Laird model (D+L) was applied to calculate a pooled OR (I2>50%, χ2 test with P<0.05), otherwise, a Mantel–Haenszel (M–H) model was applied for the meta-analysis.32 The source of heterogeneity was explored by meta-regression. If the source was uncertain, a sensitivity analysis was performed to assess the stability of the results by omitting a single study in the meta-analysis iteration to determine the effect of the individual data on the overall pooled OR. The stability of our results was also tested by switching the two models, D+L and M–H. Publication bias was quantitatively estimated with Begg’s linear regression test. If a possible publication bias existed, the meta-trim method33 and failsafe number (Nfs) were used to reestimate the effect.34 Diagnostic meta-analyses were also performed. The pooled sensitivity, specificity, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR), and their corresponding 95% CIs were calculated. Summary receiver operating characteristic curves with the areas under the receiver operating characteristic curve (AUC) were then generated. Data were analyzed mainly by the STATA-12.0 software (Stata Corporation, College Station, TX, USA). The Nfs was created using the Meta package in R (version 3.22, http://www.r-project.org/). All P-values are two sides and a P-value <0.05 was deemed statistically significant.
Results
Study characteristics
A total of 293 studies were identified using the search strategy described above, 267 of which were excluded after careful filtration. Of these, 116 studies were duplicates, 93 were without methylation data, 30 were abstracts or reviews, and 28 were irrelevant. Finally, 26 studies (ten published in English and 16 in Chinese) were included in the meta-analysis. The basic characteristics of all the included studies are shown in Table 1, and the selection process is illustrated in Figure 1.
Table 1.
The basic characteristics of all included studies
| Author | Year | Country | Method | Cervical cancer patients (n) | CIN patients (n) | Control individuals (n) |
|---|---|---|---|---|---|---|
| Blanco-Luquin et al51 | 2015 | Spain | MSP | 67 | 95 | 13 |
| Dong et al10 | 2001 | Korea | MSP | 53 | NA | 24 |
| Narayan et al52 | 2003 | Germany/Colombia | MSP | 82 | NA | 8 |
| Lea et al53 | 2004 | USA | MSP | 60 | 30 | 78 |
| Kang et al54 | 2005 | Korea | MSP | 82 | NA | 17 |
| Lin et al55 | 2005 | Korea | MSP | 67 | 30 | 20 |
| Kim et al56 | 2005 | Korea | MSP | 41 | 30 | 11 |
| Kim et al57 | 2010 | Korea | MSP | 69 | 99 | 41 |
| Banzai et al29 | 2014 | Japan | MSP | 53 | 22 | 24 |
| Carestiato et al27 | 2013 | Brazil | MSP | 29 | 84 | 28 |
| Si et al58 | 2012 | China | MSP | 60 | 98 | 40 |
| Xu et al59 | 2007 | China | MSP | 40 | 80 | 20 |
| Yao et al60 | 2012 | China | MSP | 25 | 90 | 10 |
| Ren et al61 | 2007 | China | MSP | 36 | 76 | 71 |
| Li et al62 | 2015 | China | MSP | 100 | NA | 90 |
| Liu et al63 | 2012 | China | MSP | 45 | NA | 10 |
| Guan et al64 | 2008 | China | MSP | 60 | NA | 48 |
| Wu et al65 | 2013 | China | MSP | 64 | 110 | 80 |
| Ji et al66 | 2005 | China | MSP | 60 | NA | 70 |
| Chen et al67 | 2008 | China | MSP | 40 | NA | 20 |
| Wang68 | 2011 | China | MSP | 40 | NA | 10 |
| Hao69 | 2011 | China | MSP | 80 | 105 | 53 |
| Li70 | 2012 | China | MSP | 100 | 142 | 108 |
| Lin et al71 | 2007 | China | MSP | 40 | 80 | 20 |
| Jiao72 | 2012 | China | MSP | 30 | 90 | 30 |
| Lin et al73 | 2007 | Korea | MSP | 67 | 30 | 20 |
Abbreviations: CIN, cervical intraepithelial neoplasia; NA, not available; MSP, methylation-specific PCR.
Figure 1.
The flow diagram of the stepwise selection from relevant studies.
Abbreviation: CNKI, China National Knowledge Infrastructure.
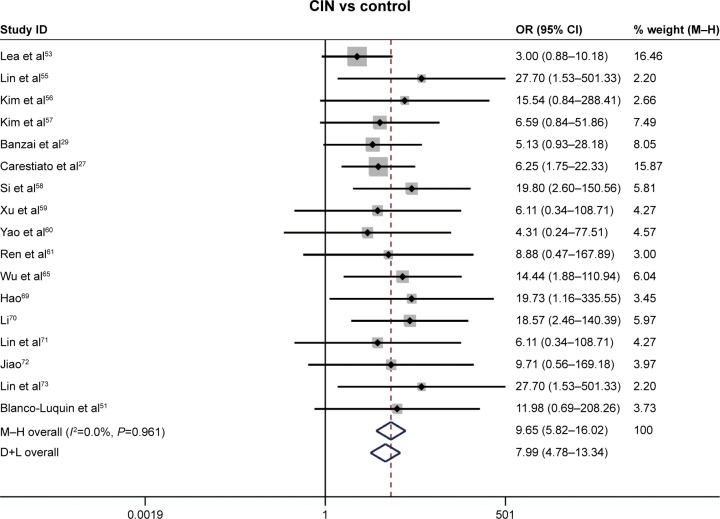
Comparison of the methylation frequency of CDKN2A between cervical cancer and controls
Our study was quantitatively synthesized across 26 studies including 1,490 cervical cancers and 964 controls. For the absence of heterogeneity (I2=0%, χ2=24.21, P=0.51), a D+L model was applied to calculate the association of methylated CDKN2A with cervical cancer. Our results indicated that the frequency of methylated CDKN2A in cervical cancers was significantly higher than in controls (OR =23.67, 95% CI =15.54–36.06, Figure 2). Similar results could be observed by switching to the M–H model to recalculate the pooled OR (Figure 2), and a sensitivity analysis was performed (Table 2), suggesting the stability and credibility of our results. However, the Begg’s test implied the presence of publication bias (P=0.02, Figure 3). To adjust for this, a trim-and-fill method was implemented (Figure 4). After filling eight missing studies the pooled OR was similar to our previous results, indicating the stability of our results. Furthermore, we applied an Nfs to assess the efficacy of the meta-analysis (Nfs0.05=1,744, Nfs0.01=859), which indicated that our results were robust.
Figure 2.
Forest plot of CDKN2A methylation status during the carcinogenesis of cervical cancer.
Note: Weights are from random effects analysis.
Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; CI, confidence interval; CIN, cervical intraepithelial neoplasia; OR, odds ratio.
Table 2.
Sensitivity analysis of pooled OR for CDKN2A methylation between cervical cancer and control
| Study excluded | Year | Estimated OR (95% CI) |
|---|---|---|
| Blanco-Luquin et al51 | 2015 | 23.57 (15.40–36.07) |
| Dong et al10 | 2001 | 23.73 (15.51–36.31) |
| Narayan et al52 | 2003 | 24.77 (16.19–37.89) |
| Lea et al53 | 2004 | 25.31 (15.92–40.22) |
| Kang et al54 | 2005 | 24.26 (15.86–37.12) |
| Lin et al55 | 2005 | 23.12 (15.11–35.39) |
| Kim et al56 | 2005 | 23.45 (15.32–35.89) |
| Kim et al57 | 2010 | 24.86 (16.16–38.26) |
| Banzai et al29 | 2014 | 26.95 (17.32–41.93) |
| Carestiato et al27 | 2013 | 22.56 (14.69–34.65) |
| Si et al58 | 2012 | 24.18 (15.72–37.19) |
| Xu et al59 | 2007 | 23.58 (15.41–36.08) |
| Yao et al60 | 2012 | 23.83 (15.57–36.48) |
| Ren et al61 | 2007 | 23.47 (15.34–35.91) |
| Li et al62 | 2015 | 21.11 (13.39–33.26) |
| Liu et al63 | 2012 | 24.09 (15.74–36.87) |
| Guan et al64 | 2008 | 23.31 (15.23–35.67) |
| Wu et al65 | 2013 | 22.71 (14.77–34.92) |
| Ji et al66 | 2005 | 23.32 (15.24–35.69) |
| Chen et al67 | 2008 | 23.95 (15.65–36.66) |
| Wang68 | 2011 | 24.20 (15.81–37.04) |
| Hao69 | 2011 | 22.98 (15.02–35.17) |
| Li70 | 2012 | 22.39 (14.55–34.44) |
| Lin et al71 | 2007 | 23.58 (15.41–36.08) |
| Jiao72 | 2012 | 23.52 (15.37–35.99) |
| Lin et al73 | 2007 | 23.13 (15.11–35.39) |
Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; CI, confidence interval; OR, odds ratio.
Figure 3.
Funnel plot of publication biases on the relationships between abnormal CDKN2A promoter methylation and the pathogenesis of cervical cancer.
Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; CIN, cervical intraepithelial neoplasia; SE, standard error; OR, odds ratio.
Figure 4.
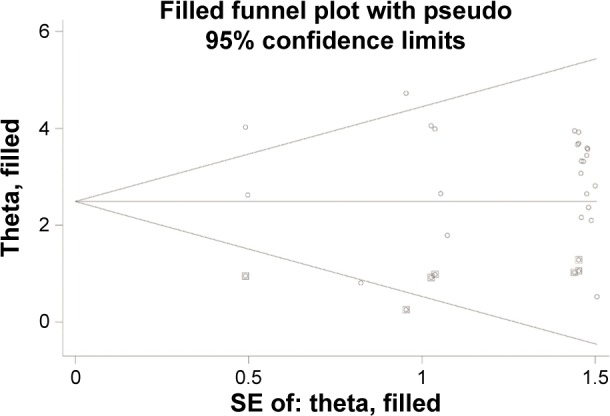
Begg’s funnel plot of publication bias test after trim-and-fill method.
Abbreviation: SE, standard error.
Comparison of the methylation frequency of CDKN2A between cervical cancer and CIN
Our analysis covered 17 studies involving 928 cervical cancers and 1,291 CINs. For the presence of heterogeneity (I2=61.5%, χ2=41.6, P<0.001), the M–H model was used. Our results showed that a higher frequency of methylated CDKN2A was observed in cervical cancers than in CINs (OR =2.53, 95% CI =1.79–3.5, Figure 2). To confirm the existence of heterogeneity among all relevant studies, a meta-regression was performed which showed that no single factor was responsible for the heterogeneity (Table 3). The pooled OR was not significantly changed by switching to the D+L model (Figure 2). The sensitivity analysis results further implied the stability and reliability of our results (Table 4). The Begg’s test for publication bias was not statistically significant (P=0.23, Figure 3).
Table 3.
Mixed effects of meta-regression analysis to identify heterogeneity source
| Heterogeneity sources | Coefficient | t | P-value | 95% CI
|
|
|---|---|---|---|---|---|
| Lower | Upper | ||||
| Publication year | 0.043 | 0.65 | 0.521 | −0.093 | 0.178 |
| Case size | 0.005 | 0.38 | 0.704 | −0.02 | 0.03 |
| Ethnicity | −0.002 | 0 | 0.998 | −1.359 | 1.356 |
Abbreviation: CI, confidence interval.
Table 4.
Sensitivity analysis of pooled OR for CDKN2A methylation between cervical cancer and CIN
| Study excluded | Year | Estimated OR (95% CI) |
|---|---|---|
| Lea et al53 | 2004 | 2.41 (1.96–2.95) |
| Lin et al55 | 2005 | 2.51 (2.04–3.09) |
| Kim et al56 | 2005 | 2.48 (2.02–3.05) |
| Kim et al57 | 2010 | 2.61 (2.12–3.22) |
| Banzai et al29 | 2014 | 2.62 (2.14–3.22) |
| Carestiato et al27 | 2013 | 2.30 (1.88–2.83) |
| Si et al58 | 2012 | 2.79 (2.26–3.45) |
| Xu et al59 | 2007 | 2.39 (1.95–2.95) |
| Yao et al60 | 2012 | 2.43 (1.97–2.98) |
| Ren et al61 | 2007 | 2.44 (1.99–2.99) |
| Wu et al65 | 2013 | 2.39 (1.93–2.95) |
| Hao69 | 2011 | 2.45 (1.99–3.03) |
| Li70 | 2012 | 2.41 (1.94–2.98) |
| Lin71 | 2007 | 2.39 (1.95–2.95) |
| Jiao72 | 2012 | 2.44 (1.99–3.01) |
| Lin et al73 | 2007 | 2.51 (2.04–3.09) |
| Blanco-Luquin et al51 | 2015 | 2.49 (2.01–3.07) |
Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; CI, confidence interval; CIN, cervical intraepithelial neoplasia; OR, odds ratio.
Comparison of the methylation frequency of CDKN2A between CINs and controls
The association between methylated CDKN2A and CIN was analyzed in 17 studies including 1,291 CINs and 667 controls. The pooled OR was computed by a D+L model, as no heterogeneity was observed (I2=0.0%, χ2=7.54, P=0.96). Our results demonstrated that the methylation frequency of CDKN2A was significantly elevated in CIN relative to the controls (OR =9.68, 95% CI =5.82–16.02, Figure 2) and the pooled OR was not significantly transformed by the M–H model (Figure 2). The sensitivity analysis confirmed the stability and credibility of our results (Table 5). No publication bias was observed by Begg’s test (P=0.39, Figure 3).
Table 5.
Sensitivity analysis of pooled OR for CDKN2A methylation between CIN and control
| Study excluded | Year | Estimated OR (95% CI) |
|---|---|---|
| Lea et al53 | 2004 | 10.96 (6.24–19.25) |
| Lin et al55 | 2005 | 9.25 (5.52–15.48) |
| Kim et al56 | 2005 | 9.49 (5.68–15.88) |
| Kim et al57 | 2010 | 9.90 (5.87–16.69) |
| Banzai et al29 | 2014 | 10.04 (5.91–17.08) |
| Carestiato et al27 | 2013 | 10.29 (5.92–17.89) |
| Si et al58 | 2012 | 9.02 (5.34–15.24) |
| Xu et al59 | 2007 | 9.81 (5.87–16.40) |
| Yao et al60 | 2012 | 9.91 (5.93–6.56) |
| Ren et al61 | 2007 | 9.67 (5.78–16.18) |
| Wu et al65 | 2013 | 9.34 (5.54–15.77) |
| Hao69 | 2011 | 9.29 (5.55–15.55) |
| Li70 | 2012 | 9.09 (5.39–15.34) |
| Lin71 | 2007 | 9.81 (5.87–16.40) |
| Jiao72 | 2012 | 9.65 (5.77–16.13) |
| Lin et al73 | 2007 | 9.25 (5.52–15.48) |
| Blanco-Luquin et al51 | 2015 | 9.56 (5.72–15.99) |
Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; CI, confidence interval; CIN, cervical intraepithelial neoplasia; OR, odds ratio.
Diagnostic accuracy of methylated CDKN2A in distinguishing cervical cancer from controls
The diagnostic accuracy of methylated CDKN2A in the detection of cervical cancer was analyzed from 26 studies involving 1,490 cervical cancers and 964 controls. The summary specificity and sensitivity of methylated CDKN2A for distinguishing cervical cancer from controls were 0.99 (95% CI: 0.97–0.99) and 0.36 (95% CI: 0.28–0.45), respectively (Figure 5). The summary receiver operating characteristic curves based on the specificity and sensitivity is shown in Figure 6, and the AUC for methylated CDKN2A-diagnosed cervical cancer was 0.93 (95% CI: 0.91–0.95). The summary diagnostic OR was 43 (95% CI: 19–98). The PLR and NLR were 27.9 (95% CI: 12.5–62.2) and 0.64 (95% CI: 0.57–0.73), respectively. As indicated by the value of PLR, cervical cancer patients have a nearly 28 times higher chance of having methylated CDKN2A than normal controls. As indicated by the value of NLR, noncancer controls have a 1.5-fold greater chance (the reciprocal of the value of NLR) of having unmethylated CDKN2A than patients with cervical cancer. The Fagan plot analyses based on the PLR and NLR demonstrated that the probability of a patient being diagnosed with cervical cancer was respectively 90%, 97%, and 99% following a positive methylated CDKN2A result, whereas the pretest probability of being diagnosed with cervical cancer was 25%, 50%, and 75%, respectively. However, the probability of an exclusion diagnosis of cervical cancer was 82%, 51%, and 34% following a negative methylated CDKN2A result, namely unmethylation of CDKN2A. The Fagan plots are shown in Figure 7.
Figure 5.
Forest sensitivity and specificity of methylated CDKN2A for differentiation of pancreatic mass.
Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; CI, confidence interval; df, degrees of freedom.
Figure 6.

SROC plot with best-fitting asymmetric curve of methylated CDKN2A for cervical cancer diagnosis.
Abbreviations: AUC, area under the receiver operating characteristic curve; SROC, summary of receiver operating characteristic; SENS, sensitivity; SPEC, specificity.
Figure 7.
Fagan plot analysis to evaluate the clinical utility of methylated CDKN2A for identification of cervical cancer.
Note: With a pretest probability of cervical cancer of 25% (A), 50% (B), and (C) 75% the post-test probability of cervical cancer.
Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; LR, likelihood ratio.
Discussion
CDKN2A has been proved to bear a striking resemblance to classic TSGs such as p53, and to be an important negative regulator of cell growth and proliferation.35 It is largely reported that a critical mechanism for silencing CDKN2A is hypermethylation of its regulatory region.36 Abnormal methylation of the CDKN2A gene is a common event in many types of human cancer, as well as in cervical cancer.27,37–40 However, conclusions regarding the role of methylated CDKN2A during the carcinogenesis of cervical cancer are inconsistent. Thus, in order to address inconsistent conclusions and to provide a better understanding of the relationship between the aberrant methylation of CDKN2A and the progression of cervical cancer, we performed a comprehensively quantitatively synthesized analysis across all relevant studies. Our results show that the abnormal methylation of CDKN2A is significantly higher in cervical cancer than in control tissues (including both normal individual tissues and benign tissues), as well as in precancerous lesions (including CIN1–CIN3), and that the frequency of CDKN2A methylation is higher in CINs than in control tissues. Our results suggest that abnormal CDKN2A methylation might be involved in the initiation and progression of cervical cancer. 5-Azacytidine was the first hypomethylating agent to be approved by the US Food and Drug Administration for the treatment of myelodysplastic syndrome,41 and subsequent studies further confirmed its anticancer effectiveness.42,43 Advanced studies have also demonstrated that the methylation status of cell cycle regulators is associated with the sensitivity of chemotherapeutic agents in breast cancer.44 From this perspective, hypomethylation of methylated CDKN2A using a hypomethylating agent might be a whole new approach to cervical cancer therapy.
The overall 5-year survival rate of cervical cancer is 68%, whereas if patients are diagnosed when the cervical cancer is localized, the 5-year survival rate is 92%.1 This implies that early diagnosis is of practical significance in the evaluation of survival rate. Advanced research has reported that DNA methylation is considered a vigorous tool for the diagnosis of cancers such as lung cancer,45 and that methylation of CDKN2A is a diagnostic biomarker for many human cancers, including oral cancer and lung cancer.46,47 However, the diagnostic value of methylated CDKN2A in cervical cancer is not so well investigated. Therefore, we performed diagnostic meta-analyses to evaluate the diagnostic performance of CDKN2A in detecting cervical cancer. The most commonly used terms to estimate diagnostic accuracy are specificity and sensitivity, which in our study were 0.99 and 0.36, respectively. As an independent indicator of prevalence combining the strengths of specificity and sensitivity, the DOR is the ratio of the odds of true positivity to the odds of negative positivity, which ranges from zero to infinity. A higher DOR value represents better diagnostic accuracy.48 In our study the value of DOR was 43, supporting the good performance of methylated CDKN2A in distinguishing cervical cancer tissues from controls. The ROC plot is an index of diagnostic accuracy and an AUC >0.7 is deemed a good risk predictor.49,50 In the current study the AUC is 0.93, suggesting that methylated CDKN2A could be an extremely useful biomarker for the detection of cervical cancer. Further, the clinical performance of CDKN2A methylation was explored using Fagan plots. The Fagan plot analysis results showed that a positive result for methylated CDKN2A could be used to distinguish cervical cancer tissue from normal tissue, with a 90% probability of diagnosing cervical cancer. Also, if the pretest probability was low there was a >80% probability that a diagnosis of cervical cancer could be ruled out following a negative result for methylated CDKN2A, such as unmethylated CDKN2A. Altogether, methylated CDKN2A has good diagnostic power to discriminate cervical cancer from benign or normal tissues.
However, some limitations of this study should be taken into consideration. First, studies on CDKN2A methylation in cervical cancer with statistical significance were more likely to be published, and unpublished studies were not included in our study. Second, relevant studies in other languages were also not included. Third, a significant heterogeneity was observed; therefore, our conclusions should be interpreted with caution.
In summary, CDKN2A methylation has been shown to be involved in the carcinogenesis of cervical cancer and is regarded as a useful biomarker for the identification of cervical cancer. Future large-scale studies, especially regarding the accurate evaluation of CDKN2A methylation, are required to verify our conclusions.
Acknowledgments
The research was supported by the grants from the Zhejiang Provincial Natural Science Foundation of China (LY16H160005), Project of Scientific Innovation Team of Ningbo (2015B11050), and the Ningbo Natural Science Foundation (2014A610235). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author contributions
JL, CCZ, HJZ, MY: Conceived and designed the experiments. JL, CCZ, TLB, TJG: Performed the experiments. JL CCZ: Analyzed the data. CCZ: Contributed analysis tools. JL, CCZ, MY: Wrote the manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Society AC. Cancer Facts and Figures 2016. Atlanta, GA: American Cancer Society; 2016. [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Moreno V, Bosch FX, Munoz N, et al. Effect of oral contraceptives on risk of cervical cancer in women with human papillomavirus infection: the IARC multicentric case–control study. Lancet. 2002;359(9312):1085–1092. doi: 10.1016/S0140-6736(02)08150-3. [DOI] [PubMed] [Google Scholar]
- 4.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2(5):342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 5.Edelstein ZR, Madeleine MM, Hughes JP, et al. Age of diagnosis of squamous cell cervical carcinoma and early sexual experience. Cancer Epidemiol Biomarkers Prev. 2009;18(4):1070–1076. doi: 10.1158/1055-9965.EPI-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang ZF, Parkin DM, Yu SZ, Esteve J, Yang XZ. Risk factors for cancer of the cervix in a rural Chinese population. Int J Cancer. 1989;43(5):762–767. doi: 10.1002/ijc.2910430503. [DOI] [PubMed] [Google Scholar]
- 7.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Anderson MC. The pathology of cervical cancer. Clin Obstet Gynaecol. 1985;12(1):87–119. [PubMed] [Google Scholar]
- 9.Kataja V, Syrjanen S, Mantyjarvi R, Yliskoski M, Saarikoski S, Syrjanen K. Prognostic factors in cervical human papillomavirus infections. Sex Transm Dis. 1992;19(3):154–160. doi: 10.1097/00007435-199205000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Dong SM, Kim HS, Rha SH, Sidransky D. Promoter hypermethylation of multiple genes in carcinoma of the uterine cervix. Clin Cancer Res. 2001;7(7):1982–1986. [PubMed] [Google Scholar]
- 11.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 12.Das PM, Singal R. DNA methylation and cancer. J Clin Oncol. 2004;22(22):4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 13.Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56(4):722–727. [PubMed] [Google Scholar]
- 14.Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163(3):1101–1107. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378(2):F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 16.Nakashima R, Fujita M, Enomoto T, et al. Alteration of p16 and p15 genes in human uterine tumours. Br J Cancer. 1999;80(3–4):458–467. doi: 10.1038/sj.bjc.6690379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 18.Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264(1):42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 19.Maeda K, Kawakami K, Ishida Y, Ishiguro K, Omura K, Watanabe G. Hypermethylation of the CDKN2A gene in colorectal cancer is associated with shorter survival. Oncol Rep. 2003;10(4):935–938. [PubMed] [Google Scholar]
- 20.Csepregi A, Ebert MP, Rocken C, et al. Promoter methylation of CDKN2A and lack of p16 expression characterize patients with hepatocellular carcinoma. BMC Cancer. 2010;10:317. doi: 10.1186/1471-2407-10-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lima EM, Leal MF, Burbano RR, et al. Methylation status of ANAPC1, CDKN2A and TP53 promoter genes in individuals with gastric cancer. Braz J Med Biol Res. 2008;41(6):539–543. doi: 10.1590/s0100-879x2008000600017. [DOI] [PubMed] [Google Scholar]
- 22.Guan RJ, Fu Y, Holt PR, Pardee AB. Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology. 1999;116(5):1063–1071. doi: 10.1016/s0016-5085(99)70009-0. [DOI] [PubMed] [Google Scholar]
- 23.Wong IH, Lo YM, Zhang J, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59(1):71–73. [PubMed] [Google Scholar]
- 24.Zou HZ, Yu BM, Wang ZW, et al. Detection of aberrant p16 methylation in the serum of colorectal cancer patients. Clin Cancer Res. 2002;8(1):188–191. [PubMed] [Google Scholar]
- 25.Jeong DH, Youm MY, Kim YN, et al. Promoter methylation of p16, DAPK, CDH1, and TIMP-3 genes in cervical cancer: correlation with clinicopathologic characteristics. Int J Gynecol Cancer. 2006;16(3):1234–1240. doi: 10.1111/j.1525-1438.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 26.Jha AK, Nikbakht M, Jain V, Capalash N, Kaur J. p16(INK4a) and p15(INK4b) gene promoter methylation in cervical cancer patients. Oncol Lett. 2012;3(6):1331–1335. doi: 10.3892/ol.2012.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carestiato FN, Afonso LA, Moyses N, Almeida Filho GL, Velarde LG, Cavalcanti SM. An upward trend in DNA p16ink4a methylation pattern and high risk HPV infection according to the severity of the cervical lesion. Rev Inst Med Trop Sao Paulo. 2013;55(5):329–334. doi: 10.1590/S0036-46652013000500006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang LW, Pan HS, Lin YH, Seow KM, Chen HJ, Hwang JL. P16 methylation is an early event in cervical carcinogenesis. Int J Gynecol Cancer. 2011;21(3):452–456. doi: 10.1097/IGC.0b013e31821091ea. [DOI] [PubMed] [Google Scholar]
- 29.Banzai C, Nishino K, Quan J, et al. Promoter methylation of DAPK1, FHIT, MGMT, and CDKN2A genes in cervical carcinoma. Int J Clin Oncol. 2014;19(1):127–132. doi: 10.1007/s10147-013-0530-0. [DOI] [PubMed] [Google Scholar]
- 30.Higgins JP, Green S. Cochrane Handbook for Systematic Reviews of Interventions. Vol. 5. Wiley Online Library; 2008. [Google Scholar]
- 31.Coory MD. Comment on: Heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2010;39(3):932. doi: 10.1093/ije/dyp157. author reply 933. [DOI] [PubMed] [Google Scholar]
- 32.Jackson D. The power of the standard test for the presence of heterogeneity in meta-analysis. Stat Med. 2006;25(15):2688–2699. doi: 10.1002/sim.2481. [DOI] [PubMed] [Google Scholar]
- 33.Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341x.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- 34.Orwin RG. A fail-safe N for effect size in meta-analysis. J Educ Stat. 1983;8(2):157. [Google Scholar]
- 35.Foulkes WD, Flanders TY, Pollock PM, Hayward NK. The CDKN2A (p16) gene and human cancer. Mol Med. 1997;3(1):5–20. [PMC free article] [PubMed] [Google Scholar]
- 36.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21(2):163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 37.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4(9):707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 38.Florl AR, Franke KH, Niederacher D, Gerharz CD, Seifert HH, Schulz WA. DNA methylation and the mechanisms of CDKN2A inactivation in transitional cell carcinoma of the urinary bladder. Lab Invest. 2000;80(10):1513–1522. doi: 10.1038/labinvest.3780161. [DOI] [PubMed] [Google Scholar]
- 39.Linder J. Immunohistochemistry in surgical pathology. The case of the undifferentiated malignant neoplasm. Clin Lab Med. 1990;10(1):59–76. [PubMed] [Google Scholar]
- 40.Wu CL, Roz L, McKown S, et al. DNA studies underestimate the major role of CDKN2A inactivation in oral and oropharyngeal squamous cell carcinomas. Genes Chromosomes Cancer. 1999;25(1):16–25. [PubMed] [Google Scholar]
- 41.Issa J, Kantarjian HA. Azacitidine. Nat Rev Drug Discov. 2005;(Suppl):S6–S7. doi: 10.1038/nrd1726. [DOI] [PubMed] [Google Scholar]
- 42.Christman JK. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21(35):5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 43.Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13(6):1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 44.Klajic J, Busato F, Edvardsen H, et al. DNA methylation status of key cell-cycle regulators such as CDKNA2/p16 and CCNA1 correlates with treatment response to doxorubicin and 5-fluorouracil in locally advanced breast tumors. Clin Cancer Res. 2014;20(24):6357–6366. doi: 10.1158/1078-0432.CCR-14-0297. [DOI] [PubMed] [Google Scholar]
- 45.Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21(35):5450–5461. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- 46.Belinsky SA, Nikula KJ, Palmisano WA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95(20):11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Cao J, Lu Z, Liu H, Deng D. A 115-bp MethyLight assay for detection of p16 (CDKN2A) methylation as a diagnostic biomarker in human tissues. BMC Med Genet. 2011;12:67. doi: 10.1186/1471-2350-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol. 2003;56(11):1129–1135. doi: 10.1016/s0895-4356(03)00177-x. [DOI] [PubMed] [Google Scholar]
- 49.Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann Thorac Surg. 2005;79(1):16–20. doi: 10.1016/j.athoracsur.2004.09.040. [DOI] [PubMed] [Google Scholar]
- 50.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39(4):561–577. [PubMed] [Google Scholar]
- 51.Blanco-Luquin I, Guarch R, Ojer A, et al. Differential role of gene hypermethylation in adenocarcinomas, squamous cell carcinomas and cervical intraepithelial lesions of the uterine cervix. Pathol Int. 2015;65(9):476–485. doi: 10.1111/pin.12332. [DOI] [PubMed] [Google Scholar]
- 52.Narayan G, Arias-Pulido H, Koul S, et al. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Mol Cancer. 2003;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lea JS, Coleman R, Kurien A, et al. Aberrant p16 methylation is a biomarker for tobacco exposure in cervical squamous cell carcinogenesis. Am J Obstet Gynecol. 2004;190(3):674–679. doi: 10.1016/j.ajog.2003.09.036. [DOI] [PubMed] [Google Scholar]
- 54.Kang S, Kim JW, Kang GH, et al. Polymorphism in folate- and methionine-metabolizing enzyme and aberrant CpG island hypermethylation in uterine cervical cancer. Gynecol Oncol. 2005;96(1):173–180. doi: 10.1016/j.ygyno.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 55.Lin Z, Gao M, Zhang X, et al. The hypermethylation and protein expression of p16 INK4A and DNA repair gene O6-methylguanine-DNA methyltransferase in various uterine cervical lesions. J Cancer Res Clin Oncol. 2005;131(6):364–370. doi: 10.1007/s00432-004-0657-5. [DOI] [PubMed] [Google Scholar]
- 56.Kim NR, Lin Z, Kim KR, Cho HY, Kim I. Epstein-Barr virus and p16INK4A methylation in squamous cell carcinoma and precancerous lesions of the cervix uteri. J Korean Med Sci. 2005;20(4):636–642. doi: 10.3346/jkms.2005.20.4.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JH, Choi YD, Lee JS, Lee JH, Nam JH, Choi C. Assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Gynecol Oncol. 2010;116(1):99–104. doi: 10.1016/j.ygyno.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 58.Si T-b. Experimental study of the relationship between jointly detecting p16INK4A and HPV in the prognosis of cervical cancer. Gansu Medical Journal. 2013;32(4):241–245. [Google Scholar]
- 59.Xu J. The expression and significance of hypermethylation of p16 and CDH1 in cervical tissue. Progress in Obstetrics and Gynecology. 2007;16(2):127–129. [Google Scholar]
- 60.Yao H-x. Association between p16 gene methylation and cervical diseases. Journal of Tongji University (Medical Science) 2012;33(3):17–19. 36. [Google Scholar]
- 61.Ren L-q, Liu P, Wang Y-x, Luo L-y. Discussion of the Value of Inhibiting Gene Methylic P16 in Early Diagnosis of Cervical Cancer. Journal of Practical Medical Techniques. 2007;14(32):4391–4393. [Google Scholar]
- 62.Li XM, Gu YL, Wang XZ, et al. Relationship between p16, DAPK and ESRβ gene promoter methylation and cervical cancer. Chinese Journal of Diagnostic Pathology. 2015;22(2):78–81. [Google Scholar]
- 63.Liu J. Detection of p16 gene promoter methylation in cervical carcinoma patients. Journal of Modern Oncology. 2012;20(7):1430–1433. [Google Scholar]
- 64.Guan C-j, Mu K-h. Detection of p16 Promoter Hypermethylation in Cervical Carcinoma. Applied Journal of General Practice. 2008;6(1):3–5. [Google Scholar]
- 65.Wu TT, Wang J-t, Ding L, et al. Folate deficiency and aberrant methylation of p16 CpG islands on cervical cancer and its precancerous lesions. Chin J Dis Control Prev. 2013;(01):9–12. [Google Scholar]
- 66.Ji RY, Li Y, Hu S-J, Li P. Methylation and Aberrant Expression of the p16 Gene in Cervical Carcinoma. Hereditas (Beijing) 2005;27(1):39–43. [PubMed] [Google Scholar]
- 67.Chen W. The clinical significance of p16 methylation in cervical cancer. Chinese Journal of Obstetrics and Gynecology. 2008;43(5):375–376. [Google Scholar]
- 68.Wang L. Association between HPV and P16 methylation in cervical cancer [master] Sanxia University; 2011. [Google Scholar]
- 69.Hao J. Research of p16 gene methylation, expression of p16 protein and DNMT1 in cervical lesions [master] Sanxi Medicine University; 2011. [Google Scholar]
- 70.Li Y. Interaction between abnormal methylation and expression of FHIT, p16 and HPV16 infection in cervical carcinogenesis [master] Sanxi Medicine University; 2012. [Google Scholar]
- 71.Lin X, Xu J, Wang H. The expression and significant of hypermethylation of p16 and RASSF1A genes in cervical carcinoma tissues. Tumor. 2007;(9):727–729. [Google Scholar]
- 72.Jiao X. Correlation of the expression of P16INK4A, P16 gene methylation and the infection of HPV in cervical carcinoma [master] Sanxi Medicine University; 2012. [Google Scholar]
- 73.Lin ZH, Li ZH, Ren XS, Liu SP, Zhao YW. Clinical significance of Dmnt-1 expression and aberrant methylation of P16/MGMT gene promoter in uterine cervical neoplasm. China Journal of Modern Medicine. 2007;17(4):394–397. [Google Scholar]